Abstract
Long non-coding RNAs (lncRNAs) constitute an important layer of chromatin regulation that contributes to various biological processes and diseases. By interacting with chromatin, many lncRNAs can regulate that state of chromatin by recruiting chromatin-modifying complexes and thus control large-scale gene expression programs. However, the available information on interactions between lncRNAs and chromatin is hidden in a large amount of dispersed literature and has not been extensively collected. We established the LnChrom database, a manually curated resource of experimentally validated lncRNA–chromatin interactions. The current release of LnChrom includes 382 743 interactions in human and mouse. We also manually collected detailed metadata for each interaction pair, including those of chromatin modifying factors, epigenetic marks and disease associations. LnChrom provides a user-friendly interface to facilitate browsing, searching and retrieving of lncRNA–chromatin interaction data. Additionally, a large amount of multi-omics data was integrated into LnChrom to aid in characterizing the effects of lncRNA–chromatin interactions on epigenetic modifications and transcriptional expression. We believe that LnChrom is a timely and valuable resource that can greatly motivate mechanistic research into lncRNAs.
Database URL: http://biocc.hrbmu.edu.cn/LnChrom/
Introduction
Long non-coding RNAs (lncRNAs) represent a new class of RNA molecule that has vital roles in various biological processes, such as differentiation, development, cell proliferation and apoptosis (1, 2). LncRNAs carry out diverse functional roles that are generally mediated through intimate interactions with chromatin and are essential for epigenetic gene control and the functional organization of the chromosomes (3). For example, HOTAIR, which is a lncRNA of the HOXC locus, targets genomic DNA and reprograms the chromatin state to promote cancer metastasis (4), and Xist is a well-known lncRNA that contributes to the X inactivation process through the recruitment of polycomb repressive complex 2 (PRC2) to its genomic binding sites (5). Another example is lincRNA-p21, which represses the canonical p53 pathway genes to increase tumor proliferation through interactions with chromatin and the guidance of heterogeneous nuclear ribonucleoprotein K (hnRNP-K) localization (6).
Given the clear potential of lncRNAs to act in the widespread regulation of chromatin modification and gene expression, the identification of chromatin-interacting lncRNAs and their genomic targets is important for dissecting lncRNA function. Many researchers have made efforts to investigate lncRNA–chromatin interactions by combining multiple types of low-throughput experiments. For example, Hu and colleagues (7) employed RNA-immunoprecipitation, lncRNA knockdown, quantitative real-time polymerase chain reaction (qRT-PCR) and chromatin immunoprecipitation (ChIP) to identify the genomic targets that are co-occupied by the lncRNA FAL1 and chromatin repressive complex PRC2. Analogous strategies have been successfully applied to various lncRNAs (e.g. HOTAIR, Braveheart and linc-UBC1) to examine their physical interactions with chromatin and, in turn, to understand their functional roles (8, 9). With advances in next-generation sequencing, several high-throughput techniques, such as chromatin isolation by RNA purification (ChIRP-seq) (10) and capture hybridization analysis of RNA targets (CHART-seq) (11), have been employed to probe the binding sites of various chromatin-interacting lncRNAs on the whole-genome scale. For example, using ChIRP-Seq, a high-resolution map of the genomic sites occupied by the prostate-specific lncRNA PCGEM1 was created. The majority of the genomic targets of PCGEM1 were found to be associated with androgen receptor-dependent gene activation events in prostate cancer cells (12).
The current knowledge of lncRNA–chromatin interactions could aid in the understanding of the functional characteristics of lncRNAs. However, the wealth of information about chromatin regulation by lncRNAs is still scattered and hidden in a large amount of literature. Here, we developed LnChrom, which is a manually curated resource containing information about experimentally validated lncRNA–chromatin interactions. LnChrom provides the first comprehensive map of lncRNA interactions with chromatin. Currently, LnChrom contains 382 743 interactions involving 2390 lncRNAs and 34 345 target genes in human and mouse. LnChrom further offers detailed metadata and allows for the intuitive visualization and analysis of multi-omics data sets for the characterization of lncRNA–chromatin interactions. Therefore, LnChrom has the potential to serve as an up-to-date knowledgebase for the facilitation of the understanding of the regulatory mechanisms of lncRNAs.
Materials and methods
Extraction of the interaction information
Using the keywords ‘lncRNA’, ‘chromatin’, ‘RNP’, ‘triplex’, ‘duplex’, ‘guide’, ‘scaffold’, ‘ChIRP’, ‘CHART’, ‘RAP’ and ‘ChOP’, we retrieved approximately 8000 pertinent literature from PubMed. We required that the lncRNA–chromatin interactions were strictly verified via a combination of multiple types of low-throughput experiments. We also collected the interactions in which the target genes were not reported as single, specific genes in the original literature. Although the information about these interactions was not specific to exact target genes, we considered this information as supportive for the roles of some lncRNAs and that it should thus be provided to researchers for understanding the mechanisms of lncRNA-mediated chromatin regulation. For example, it has been reported that the lncRNA TUG1 recruits the chromatin remodelling complex PRC2 to silence cell-cycle regulation (13).
Additionally, we also searched the GEO dataset (http://www.ncbi.nlm.nih.gov/geo) with the keywords ‘ChIRP-seq’, ‘CHART-seq’, ‘RAP-seq’, ‘ChOP-seq’ and ‘MARGI’ to collect interaction pairs that have been confirmed by high-throughput experiments. In total, 30 high-quality datasets were found. We processed the raw data of 27 datasets using the standard pipeline (10) and downloaded the processed lncRNA binding site data from the remaining three datasets from the supplementary data of the original articles. These data sets were converted to the same genome assembly (hg19 for human, mm9 for mouse) using UCSC liftOver tools.
Identification of target gene
The genes closest to the lncRNA binding sites were identified as their target genes. Then, the target genes were divided into the following four groups: ‘P-type’, ‘B-type’, ‘F-type’ and ‘C-type’. The ‘P-type’ target genes were defined as the genes whose promoters are occupied by lncRNAs. The ‘B-type’ target genes were defined as the genes whose gene boundaries overlapped with the lncRNA binding sites. The ‘F-type’ target genes were defined as the genes whose flanking regions (±10 kb of the gene boundary) overlapped with the lncRNA binding sites. ‘C-type’ target genes were defined as the closest genes of lncRNAs which were >10 kb away from the lncRNA target regions. Given that a majority of the interactions collected by literature mining provided target genes but lacked the exact genomic coordinates of lncRNAs bound, we introduce ‘E-type’ (evidence based) target genes for the literature-based interactions. Several of these literature have demonstrated that lncRNA may regulate the target genes by binding to the promoter region. Similar to ‘P-type’ target genes, we thus used the target gene promoters as lncRNA target regions for visualization purposes in LnChrom. The software bedtools 2.24.0 (14) was employed to determine the target genes. Genomic location information about the transcription start site (TSS) and gene boundary and chromosome length files were downloaded from the UCSC genome browser. Promoters were defined as −2 kb to +0.5 kb relative to the TSS.
Metadata collection
To further consolidate the curated interaction pairs, we collected metadata for each interaction that included the following 18 items: species, lncRNA name, lncRNA Ensembl ID, target gene name, target gene ID, reference genome, target region, chromatin-modifying (or transcription) factors, associated epigenetic modifications, state of epigenetic modification (enriched or depleted), regulation type (in cis or in trans), regulation direction (positive or negative), function, disease association, cell type, experiment methods, description of supporting evidence and original article.
Integration of multi-omics data sets
Transcriptome data
The raw RNA-seq data for 16 normal tissues were downloaded from the Illumina Human Body Map project (http://www.ebi.ac.uk/gxa/experiments/E-MTAB-513). We mapped the reads against the human genome (hg38) using TopHat 2.0.13 (15) with the default parameters. According to the annotation of GENCODE (v27) (16), we calculated the fragments per kilobase of exon per million mapped reads for each lncRNA.
The gene expression (RNA-seq data, level 3) and lncRNA expression profiles for 13 cancer types were downloaded from The Cancer Genome Atlas (TCGA) data portal (http://cancergenome.nih.gov/) and ‘The Atlas of Noncoding RNAs in Cancer’ (TANRIC)(17), respectively.
The differentially expressed gene (DEG) sets observed after lncRNA knockdown or overexpression that are identified in the Gene Perturbation Atlas (18) and LncRNA2Target (19) were downloaded. The functional consequences of the lncRNA–chromatin interactions that have been validated by high-throughput experiments were investigated using the lncRNA-DEG datasets.
TF binding data
The encyclopaedia of DNA elements transcription factor (TF) binding tracks were downloaded from the UCSC genome browser. These data include 161 unique TFs across 91 human cell types (20). The software bedtools was used to identify co-bindings between TFs and lncRNAs.
Mutation data
The mutation profiles detected by whole-genome sequencing technology were downloaded from the International Cancer Genome Consortium (21) and involved 27 cancer types. Genetic mutations within the ±5 kb regions around the lncRNA binding sites were identified using bedtools.
Motif analysis
For binding sites of each lncRNA, we used HOMER (22) to search for overrepresented sequence motifs using the programs default parameter values. The top three most significant motifs from knownResults or homerResults are represented.
Database statistics
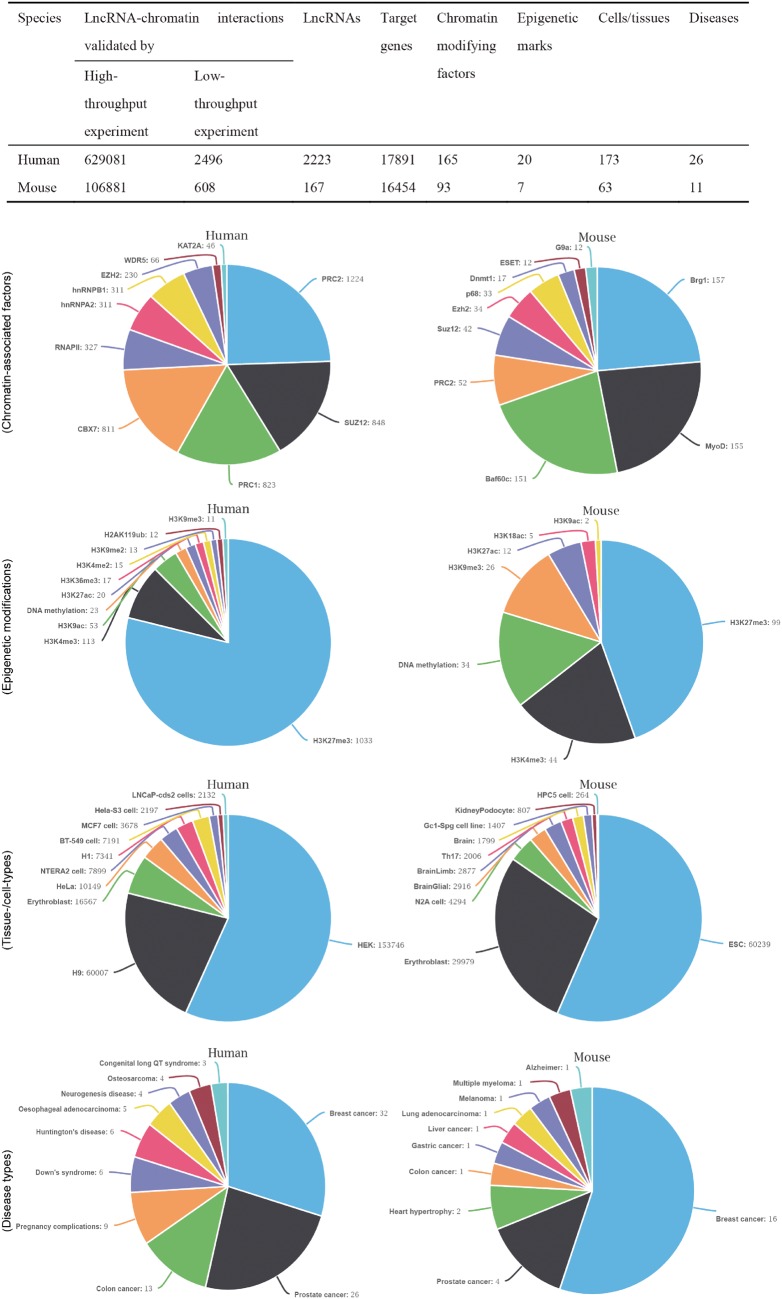
The current release contains 382 743 lncRNA–chromatin interactions in 263 human and mouse cell types/tissues that involve 34 345 target genes and 2390 lncRNAs. The interaction pairs were validated by combined low-throughput or high-throughput experiments. To characterize the curated records of lncRNA-mediated chromatin regulation, we also gathered a series of metadata for each interaction. Figure 1 lists detailed information about the interaction pairs. In total, 258 types of chromatin-modifying factors and 27 types of epigenetic marks are associated with lncRNA-mediated chromatin regulation. The epigenetic modifications associated with the interactions can be grouped into five categories that cover most aspects of epigenetic regulation and include histone methylation, acetylation, phosphorylation, ubiquitination and DNA methylation. Additionally, 37 types of diseases are associated with lncRNA-mediated chromatin regulation.
Figure 1.
The statistics of lncRNA–chromatin interactions. The table shows the basic statistics of lncRNA–chromatin interactions. The pie chart shows the distribution of interactions based on cell/tissue types, disease types, chromatin-associated factors and epigenetic modifications. For each chart, the top 10 groups of interactions are shown.
Web interface
Data browsing, searching and retrieving
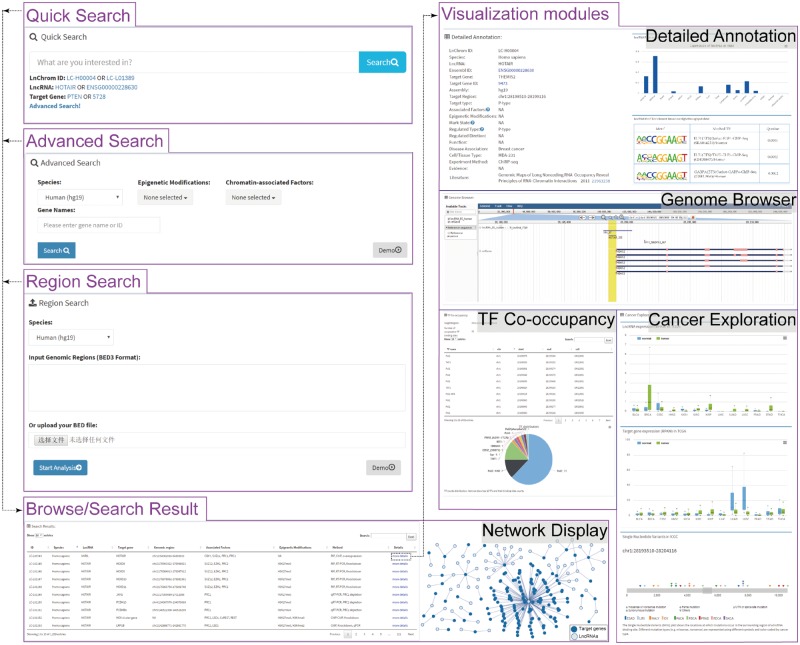
LnChrom provides a user-friendly web interface that allows users to browse, query and download the lncRNA–chromatin interaction data (Figure 2).
Figure 2.
Schematic view of the LnChrom web interface. Users can query the resource through the ‘Quick search’, ‘Advanced search’, ‘Region search’ and ‘Browse’ panels, and the results include a brief table and a network display of the interactions. Users interested in individual interactions can click ‘more details’ within the table to access the visualization modules, which include ‘Detailed Annotation’, ‘Genome Browser’, ‘TF Co-occupancy’ and ‘Cancer Exploration’. In general, these modules were developed by integrating multi-omics data to gain insight into the regulatory mechanisms of lncRNAs. The ‘Detailed Annotation’ module illustrates a set of metadata that was curated from the original literature, lncRNA expression across different tissues from the Human Body Map project and sequence motif discovery of the lncRNA binding sites. ‘Genome Browser’ provides an intuitive illustration of the lncRNA binding sites and gene annotation. ‘TF Co-occupancy’ shows the potential cooperative TFs of lncRNAs in humans based on an integration of the binding sites for 161 TFs from the ENCODE project. The ‘Cancer Exploration’ module provides the expressions of lncRNAs and their target genes from the TCGA project and the genetic mutations around the lncRNA binding sites across cancers from the ICGC project.
A browser view has been made available as an expandable tree, and the user can easily browse all of the interaction pairs in human and mouse by ‘lncRNAs’, ‘chromatin-modifying factors’ or ‘epigenetic marks’. To provide easier access, we added a quick search box on the home page for fast browsing and searching. Alternatively, a user can query the resource through the ‘Advanced search’ and ‘Region search’ panels. The ‘Advanced search’ panel provides options that include species, associated epigenetic modifications, chromatin-modifying protein/complex and lncRNA or target gene name, which allows users to perform customized searches of the interesting contents. The ‘Region search’ panel is used to investigate potential lncRNA-mediated chromatin regulations within the user-defined genomic region. The user is required to select an organism and then type in or upload the intended genomic regions in BED format. The server will return the related interaction pairs after calculation.
The initial output of the browsing or searching is presented as a table that contains the basic information about each interaction and is comprised of the interaction ID, species, lncRNA name, target gene name, lncRNA binding site (including ‘chr’, ‘start’ and ‘end’), associated protein/complex, epigenetic modification and method. The results table can be sorted flexibly based on each column. Users can also use the keyword search box to quickly find the interesting interaction pairs from the results table. To further learn about and explore the interaction of interest, users can click ‘more details’ within the table to launch a new page that contains the corresponding metadata and visualizations of the multi-omics data. Additionally, a network-based visualization of the resulting interactions is presented in the browse and search pages. LnChrom also provides a download page from which users can retrieve all of the interaction data.
Visualization modules for exploring lncRNA–chromatin interactions
Recent advances indicate that lncRNAs can target specific genomic sequences and participate in the epigenetic regulation of gene expression (23). Considering the complexity of lncRNA-mediated chromatin regulation, LnChrom provides not only a map of lncRNA–chromatin interactions but also the detailed metadata and analytic results of large scale of multi-omics datasets and thus allows for the elucidation of the possible mechanisms of lncRNA action. Accordingly, four visualization modules, including ‘Detailed Annotation’, ‘Genome Brower’, ‘TF Co-occupancy’ and ‘Cancer Exploration’, were built. Users can explore these modules by clicking ‘more details’ about the interaction of interest in the results table from the browse or search page.
The ‘Detailed Annotation’ module illustrates a set of metadata that was curated from the original literature. Each interaction is accompanied by 18 items that include regulation type, cell/tissue type and experiment method. We further provide additional annotation information on the lncRNAs that includes external links (Ensembl for lncRNAs and NCBI for target genes), lncRNA expressions across 16 normal human tissues, functional characterizations and motif enrichment analyses. Additionally, the ‘Genome Brower’ module was developed to intuitively illustrate the lncRNA binding sites.
A number of studies have indicated that lncRNAs can function through cooperation with TFs to modulate gene expression programs (24–26). ‘TF Co-occupancy’ was built to identify potential cooperative TFs in human. We used the genomic binding profiles of 161 TFs to identify the TFs with binding sites that overlap with lncRNA target regions. A brief table lists all the cooperative TFs and their binding sites, and a pie chart displays the top 10 cooperative TFs.
By interacting with chromatin, lncRNAs play an important role in the control of gene expression and contribute to cancer pathogenesis. The ‘Cancer Exploration’ module provides the expressions of lncRNAs and their target genes in 4351 tumor samples and 556 normal samples of 13 cancers from the TCGA for assessments of functional significance of lncRNA–chromatin interactions. This module also presents the genetic mutations around the lncRNA binding sites across 27 cancer types.
Implementation
The interaction information and analysis results of the multi-omics data sets were stored and queried by MySQL. The web interface was implemented in the JavaScript and Cascading Styling Sheets languages and has been tested on Firefox, Chrome and Safari. The tables are visualized by datatables, the interactive network visualization is based on Cytoscapeweb (27), the single nucleotide variants view is presented by gd3 library (28), the embedded pie charts, box plots and bar graphs are generated by HighCharts. A local installation of the UCSC genome browser hosted on our server is used to visualize the relevant annotations of the lncRNA binding sites on a locus-by-locus basis (29).
Summary and prospective works
This work provides a comprehensive collection of experimentally validated lncRNA–chromatin interactions for the scientific community. LnChrom currently contains 382 743 interaction pairs in 263 cell types/tissues in human and mouse. For each interaction, we further summarized the metadata, including associated proteins/complexes, epigenetic modifications and diseases, from original publications, which is helpful for promoting the study of lncRNAs. LnChrom also provides user-friendly web interfaces for flexibly searching, browsing and accessing the interaction data. Coupled with intuitive visualization and integrative analysis of multi-omics data, LnChrom enables researchers to translate existing interaction information into novel biologic insights.
The current understanding of and research on the interactions between lncRNA and chromatin are rapidly progressing. We will pay close attention to new lncRNA–chromatin interaction data and add this information to the database as it becomes available. As more experimentally validated data become available, we hope to build several supervised predictive models for lncRNA–chromatin interactions in the future. Furthermore, we will continue to extend the amount of storage space and improve the performance of our computer servers for storing and analysing the newly generated data. We expect that the continuous efforts to develop and improve LnChrom will contribute to the understanding of lncRNA-mediated chromatin regulation.
Funding
This work was supported in part by the National Program on Key Basic Research Project [973 Program, Grant Nos. 2014CB910504]; the National High Technology Research and Development Program of China [863 Program, Grant Nos. 2014AA021102]; the National Natural Science Foundation of China [Grant Nos. 61473106, 61573122]; the China Postdoctoral Science Foundation (2016M600260); Wu lien-teh youth science fund project of Harbin medical university [Grant Nos. WLD-QN1407]; Special funds for the construction of higher education in Heilongjiang Province [Grant Nos. UNPYSCT-2016049]; the Heilongjiang Postdoctoral Foundation (LBH-Z16098) and the Creative Research Groups of the National Natural Science Foundation of China (81421063).
Conflict of interest. None declared.
References
- 1. Wapinski O., Chang H.Y. (2011) Long noncoding RNAs and human disease. Trends Cell Biol., 21, 354–361. [DOI] [PubMed] [Google Scholar]
- 2. Xu J., Bai J., Zhang X.. et al. (2017) A comprehensive overview of lncRNA annotation resources. Brief. Bioinformatics, 18, 236–249. [DOI] [PubMed] [Google Scholar]
- 3. Goff L.A., Rinn J.L. (2015) Linking RNA biology to lncRNAs. Genome Res., 25, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta R.A., Shah N., Wang K.C.. et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pontier D.B., Gribnau J. (2011) Xist regulation and function explored. Hum. Genet., 130, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huarte M., Guttman M., Feldser D.. et al. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell, 142, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu X., Feng Y., Zhang D.. et al. (2014) A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell, 26, 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchese F.P., Huarte M. (2014) Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics, 9, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang K.C., Chang H.Y. (2011) Molecular mechanisms of long noncoding RNAs. Mol. Cell, 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu C., Qu K., Zhong F.L.. et al. (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mole. Cell, 44, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simon M.D., Pinter S.F., Fang R.. et al. (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature, 504, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang L., Lin C., Jin C.. et al. (2013) lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature, 500, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khalil A.M., Guttman M., Huarte M.. et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A, 106, 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trapnell C., Roberts A., Goff L.. et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols, 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrow J., Frankish A., Gonzalez J.M.. et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., Han L., Roebuck P.. et al. (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in Cancer. Cancer Res., 75, 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao Y., Gong Y., Lv Y.. et al. (2015) Gene Perturbation Atlas (GPA): a single-gene perturbation repository for characterizing functional mechanisms of coding and non-coding genes. Sci. Rep., 5, 10889.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Q., Wang J., Wu X.. et al. (2015) LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucl. Acids Res., 43, D193–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Euskirchen G.M., Rozowsky J.S., Wei C.L.. et al. (2007) Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res., 17, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hudson (Chairperson T.J., Anderson W., Aretz A.. et al. (2010) International network of cancer genome projects. Nature, 464, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heinz S., Benner C., Spann N.. et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lalevee S., Feil R. (2015) Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics, 7, 877–879. [DOI] [PubMed] [Google Scholar]
- 24. Leveille N., Melo C.A., Rooijers K.. et al. (2015) Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun., 6, 6520.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo M., Jeong M., Sun D.. et al. (2015) Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell, 16, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vance K.W., Ponting C.P. (2014) Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet., 30, 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopes C.T., Franz M., Kazi F.. et al. (2010) Cytoscape Web: an interactive web-based network browser. Bioinformatics, 26, 2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leiserson M.D., Gramazio C.C., Hu J.. et al. (2015) MAGI: visualization and collaborative annotation of genomic aberrations. Nat. Methods, 12, 483–484. [DOI] [PubMed] [Google Scholar]
- 29. Rosenbloom K.R., Armstrong J., Barber G.P.. et al. (2015) The UCSC Genome Browser database: 2015 update. Nucl. Acids Res., 43, D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]