Abstract
The cellular response to DNA damage is essential for maintaining the integrity of the genome. Recent evidence has identified E2F7 as a key player in DNA damage-dependent transcriptional regulation of cell-cycle genes. However, the contribution of E2F7 to cellular responses upon genotoxic damage is still poorly defined. Here we show that E2F7 represses the expression of genes involved in the maintenance of genomic stability, both throughout the cell cycle and upon induction of DNA lesions that interfere with replication fork progression. Knockdown of E2F7 leads to a reduction in 53BP1 and FANCD2 foci and to fewer chromosomal aberrations following treatment with agents that cause interstrand crosslink (ICL) lesions but not upon ionizing radiation. Accordingly, E2F7-depleted cells exhibit enhanced cell-cycle re-entry and clonogenic survival after exposure to ICL-inducing agents. We further report that expression and functional activity of E2F7 are p53-independent in this context. Using a cell-based assay, we show that E2F7 restricts homologous recombination through the transcriptional repression of RAD51. Finally, we present evidence that downregulation of E2F7 confers an increased resistance to chemotherapy in recombination-deficient cells. Taken together, our results reveal an E2F7-dependent transcriptional program that contributes to the regulation of DNA repair and genomic integrity.
INTRODUCTION
Mammalian E2F transcription factors (E2F1-E2F8) are key components of the Retinoblastoma (RB) pathway that control cell-cycle progression through the activation or repression of target genes. Deregulation of E2F activity has a high impact on health and disease (1). An insight into the specific functions of E2F family members has been provided by the identification of a large set of genes regulated by each individual factor (2). These studies have revealed a key role for RB-dependent classical E2Fs (E2F1-5) in cell-cycle control and DNA damage response (DDR). By contrast, the contribution of RB-independent atypical E2F factors, E2F7-8, to these processes has not been clearly defined.
E2F7, a predominantly transcriptional repressor, is known to be induced in late G1 by E2F1, together with a large array of E2F target genes (3,4). E2F7 binds to promoters of microRNA and protein-coding genes bearing E2F consensus motifs, such as E2F1, CDC6, MCM2 or miR-25 during S-phase, thereby repressing their expression (4,5). These findings have raised the possibility that E2F7 protein may be a key component of a negative feedback loop required to turn off transcription of E2F-driven G1/S target genes, thus allowing progression through the cell cycle. Accordingly, overexpression of E2F7 blocks S-phase entry (4,6,7), whereas acute loss of E2F7 accelerates cell-cycle progression (5).
Involvement of E2F7 in stress responses is supported by various lines of evidence, although the mechanisms by which E2F7 participates in these processes remain unresolved. E2F7 and E2F8 double knockout mouse embryos exhibit widespread apoptosis, suggesting a role for these E2Fs in cell survival (8). Furthermore, depletion of atypical E2Fs has been shown to reduce survival of tumor cells, primary mouse keratinocytes and embryonic fibroblasts after treatment with several DNA damaging compounds, indicating that sensitivity to cytotoxic/genotoxic stimuli is enhanced by loss of E2F7 or by the combined loss of E2F7/8 (8–10). Co-depletion of E2F1 under these circumstances could rescue stress-induced apoptosis (8,11) and accelerate tumorigenesis in a two-stage skin carcinogenesis model (10), implying a key role for E2F1 in E2F7/8-dependent stress responses. Additional mediators of E2F7-dependent resistance to DNA damaging drugs include the sphingosine kinase SPHK1 and its downstream target AKT (12), although the precise role of E2F7 in this pathway remains to be elucidated.
Both transcription-independent and transcription-dependent roles of E2F7 in the response to DNA damage have been suggested. On the one hand, a recruitment of E2F7 to the sites of DNA breaks has been reported, and it has been suggested that E2F7 represses DNA repair process directly on the lesion (13). On the other hand, a p53-dependent E2F7 transactivation has been described after treatment with DNA topoisomerase inhibitors, which leads to repression of a subset of cell-cycle genes, including DHFR, RRM2 and E2F1 (14), suggesting a key transcriptional role for E2F7 in cell-cycle arrest upon DNA damage.
Genes involved in DNA repair have been reported as targets of E2F factors, including E2F7 (4,15), but whether E2F7 modulates responses to DNA damage through regulation of DNA repair gene expression remains to be established. In this work we have investigated the role of E2F7 in the transcriptional regulation of genes involved in DNA repair, and the functional consequences of E2F7-mediated transcriptional program upon genotoxic damage. Our results suggest that E2F7 plays a p53-independent role in the attenuation of DNA repair function through transcriptional repression of target genes that are required for the timely regulation of replication fork-associated DNA damage repair.
MATERIALS AND METHODS
Cell culture and flow cytometry
Human cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum (10% for U2OS and HeLa cells; 20% for CAPAN-1 cells). For cell synchronization in G1/S, exponentially growing U2OS cells were incubated with 4 mM hydroxyurea (HU) for 24 h and subsequently washed and cultured in complete medium. For cell synchronization at mitosis, cell cultures were incubated with nocodazole (50–100 ng/ml) for the last 14 h of culture. To assess cell-cycle distribution, cells were fixed with chilled 70% ethanol, stained with 50 μg/ml propidium iodide (PI) and analyzed by flow cytometry (FACSCalibur, BD). To analyze the percentage of mitotic or γ-H2AX-positive cells, ethanol-fixed cells were stained with an antibody recognizing Histone H3 phosphorylated on Serine 10 (pH3) conjugated with FITC (06-570, Millipore), or an antibody recognizing γ-H2AX protein conjugated with FITC (05-636, Millipore), subsequently incubated with PI and analyzed by flow cytometry. Cell-cycle distribution, mitotic index and γ-H2AX accumulation analyses were performed with Summit 4.3 software. To analyze the percentage of cells replicating DNA, cells were pulse-labeled with 10 μM BrdU for the last 2 h of cell culture, washed in ice-cold phosphate-buffered saline and fixed in ice-cold 70% ethanol. Cells were stained with an antibody recognizing BrdU (M0744, Dako) and analyzed by flow cytometry as described (16).
Generation of U2OS E2F7 knockout cells
E2F7 knockout cells were generated using the CRISPR/Cas9 system. A CRISPR guide RNA (gRNA) targeting the first coding exon of E2F7 was designed using Benchling, and cloned into the BbsI site of pX330 (42230, Addgene). U2OS cells were co-transfected with this plasmid, together with a plasmid containing a gRNA to the zebrafish TIA gene (5′-GGTATGTCGGGAACCTCTCC-3′) and a P2A-puromycin resistance cassette flanked by two TIA target sites. Co-transfection results in excision of the cassette and subsequent sporadic incorporation at the site of the targeted genomic locus as previously described (17). Successful integration of the cassette into the targeted gene disrupts the allele and renders cells resistant to puromycin. After puromycin selection, resistant clones were expanded, screened for cassette integration and indels into the target gene.
Clonogenic survival assays
Cells were treated with cisplatin (CSP) at the indicated concentrations for 24 h. Cells were then washed free of the drug and incubated in fresh medium for 14 days or left untreated. The number of colonies of more than 50 cells in each dish was counted after staining with crystal violet.
Analysis of chromosomal aberrations
Chromosomal aberrations were visualized in chromosome spreads following published protocols, with minor modifications (18). Cells were arrested in metaphase after treating cell cultures with Karyomax Colcemid (Life Technologies) for 12 h at a final concentration of 100 ng/ml. Metaphase-arrested cells were subsequently harvested and fixed in Carnoy solution. An aliquot of the cellular suspension was dropped onto microscopy slides to obtain chromosome spreads, which were stained and mounted with ProLong Gold Antifade with DAPI reagent (Life Technologies). Image acquisition was performed on a Leica DMI 6000B fluorescence microscope.
Transfections and homologous recombination assay
To silence endogenous expression of E2F7, p53, BRCA2 and RAD51, cells were transfected with commercial siRNAs (Life Technologies), at a final concentration of 10 nM (sequences provided in Supplementary Table S1) using Lipofectamine RNAiMAX (Life Technologies) following manufacturer’s recommendation. Plasmid transfection was performed with various amounts of DNA in 6-well culture dishes using XtremeGENE HP transfection reagent (Roche Pharma), following manufacturer’s recommendations. The mixture was incubated for 15 min at room temperature and added dropwise to cell cultures.
Homologous recombination (HR)-dependent DNA double stranded break (DSB) repair was assessed using the DR-GFP/SceI assay described by M. Jasin’s group (19). For these experiments we used a U2OS cell line that carries a recombination substrate, DR–GFP, inserted in the genome (U2OS DR-GFP cell line). To induce a double-strand break in the HR reporter, U2OS-DR-GFP cells were transfected with I-SceI restriction enzyme expression construct (pCBAI-SceI) (20). HR repair was analyzed using flow cytometry by scoring GFP-positive cells.
RNA expression analyses
Total RNA extraction was performed with TRIzol Reagent (Life Technologies) and purified using the RNeasy Mini kit (Qiagen) following the manufacturer’s recommendations. mRNA was used to build a cDNA library using the reagents provided in the Illumina TruSeq RNA Sample Preparation Kit following the manufacturer’s instructions. The resulting purified cDNA library was sequenced on the Genome Analyzer IIx with SBS TruSeq v5 reagents following manufacturer’s protocols.
Sequencing reads obtained in each condition were mapped to the human reference genome (GRCh37/hg19) with TopHat software tool (21). After running TopHat, the resulting alignment files were supplied to Cufflinks tool to generate a transcriptome assembly for each sample. The reads were subsequently fed to Cuffdiff, which calculates the expression levels of each identified transcript and tests the statistical significance of the expression changes between conditions. This tool assumes that the number of sequencing reads generated from a transcript is directly proportional to the relative abundance of that transcript in the sample. Expression levels were represented by FPKM values (fragments per kilobase per million sequenced reads), which incorporate two normalization steps to ensure that expression levels of different transcripts can be compared across different runs (longer transcripts produce more sequencing fragments than shorter transcripts and different sequencing runs may produce different volumes of sequencing reads). Changes in gene expression between samples were considered significant at a false discovery rate (FDR)-adjusted P-value (q-value) < 0.05.
For individual mRNA expression analysis, RNA was reverse-transcribed into cDNA with the High-Capacity cDNA RT Kit (Life Technologies) and qPCR was performed as described previously (22), following Minimal Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines. Sequences of RT-qPCR primers are listed in Supplementary Table S2.
Bioinformatic tools
For gene set enrichment analysis (GSEA) analyses we tested whether 137 pathways obtained from the Pathway Interaction Database (NCI-Nature) are enriched among E2F7-regulated genes. We considered as statistically enriched those pathways with normalized enrichment scores (NES) higher than 1.5 and FDRs < 10%.
Search for E2F motifs in E2F7-regulated genes was carried out with the MotifLocator tool from TOUCAN program (https://gbiomed.kuleuven.be/english/research/50000622/lcb /tools/toucan) (23). The search was restricted to the proximal promoter region (−1000 and +500 bp relative to the transcription start site). Cutoffs of 0.8, 0.85 and 0.9 were applied, and the ‘Human 1 Kb Proximal 1000 ENSMUSG’ was used as background.
Identification of over-represented transcription factor binding motifs in the regulatory regions of E2F7-regulated genes was performed using the DiRE server (http://DiRE.dcode.org/) (24). For these analyses, the list of up-regulated genes and the list of downregulated genes obtained in the RNA-seq experiments were submitted independently. A list of 5000 human genes randomly selected by DiRE were used as background.
Protein expression and chromatin immunoprecipitation analyses
For western blot analyses, cells were lysed in buffer containing 10 mM NaH2PO4 pH 7.2; 1 mM ethylenediaminetetraacetic acid; 1 mM Ethylene glycol tetraacetic acid (EGTA); 150 mM NaCl; 1% NP-40, and a cocktail of protease and phosphatase inhibitors (Roche). Protein concentrations in supernatants were determined using a commercially available kit (DC Protein Assay from Bio-Rad). A total of 20 μg of protein were loaded per lane, fractionated in 8–10% sodium dodecyl sulphate-polyacrylamide gels and transferred onto nitrocellulose membranes (Bio-Rad). Antibodies against the following proteins were used: E2F7 (sc-32574, Santa Cruz), Cyclin E1 (4129, Cell Signaling), p53 (sc-1312, Santa Cruz), RAD51 (sc-8349, Santa Cruz), pH3 (06-570, Millipore), α-Tubulin (T-9026, Sigma), β-Actin (A5441, Sigma). Immunocomplexes were visualized with horseradish peroxidase-conjugated anti-mouse, anti-goat or anti-rabbit IgG antibodies (Santa Cruz), followed by chemiluminiscence detection (ECL, Amersham) with a ChemiDoc camera (Bio-Rad).
Chromatin immunoprecipitations (ChIPs) and the quantification of immunoprecipitated DNA sequences by qPCR were performed as described previously (25). Sequences of qPCR primers are listed in Supplementary Table S3. Antibodies used for ChIP analysis were: E2F7 (sc-66870, Santa Cruz), and SV40LT (sc-147, Santa Cruz).
Immunoflorescence/high-throughput microscopy (HTM)
For standard immunofluorescence, cells were grown on coverslips in 12-well plates. For FANCD2 staining, cells were fixed for 10 min with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized in PBS containing 0.5% Triton X-100. Primary antibodies against FANCD2 (sc-20022, Santa Cruz) and RAD51 (sc-8349, Santa Cruz) were applied to the coverslips for 2 h at room temperature. After washing twice with PBS-T, samples were incubated with the corresponding diluted fluorescent secondary antibody for 1 h. Finally, samples were stained with DAPI and mounted on a microscopy slide using Prolong Gold antifade (Life Technologies) mounting medium. Image acquisition was performed on a Leica DMI 6000B fluorescence microscope. FANCD2 foci quantification was performed with the Definiens Tissue Phenomics analysis software.
High-throughput microscopy (HTM) was performed with the protocol described above, except that in this case cells were grown in 96-well plates with flattened glass bottom (mCLEAR, Greiner Bio-One) at a density of 7500 cells per well. An antibody against 53BP1 (NB100-304, Novus) was used. As a final step nuclei were stained with a DAPI containing solution and the preparations were kept in PBS. Images were automatically acquired with the Opera High Content Screening platform (Perkin Elmer). Data analysis was performed with the Acapella Imaging and analysis software (Perkin Elmer) as described previously (26). Data were represented with the Prism software (GraphPad Software).
Statistical analysis
Data are presented as mean ± SD. The significance of the difference between two groups was assessed using the Student two-tailed t-test. A P < 0.05 was considered statistically significant.
RESULTS
E2F7-regulated gene expression profiling in the cell cycle
To better define E2F7 function we analyzed global gene expression profiles during the cell cycle after acute depletion of E2F7. To this end, U2OS cells were synchronized in G1/S transition with HU, and subsequently transfected with siRNAs specific for E2F7 (siE2F7) or with non-target control siRNAs (siNT), as previously described (5). RNA was isolated at three time-points following exit from cell-cycle arrest, which represent G1/S transition (0 h), S phase (3 h) and G2/M boundary (12 h) of the cell cycle (Supplementary Figure S1A). Kinetics of CCNE1 protein levels confirmed the cell-cycle phases of the selected time-points, with high levels at 0 h (G1/S) and a stepwise reduction in the following time-points (Supplementary Figure S1B). Furthermore, we showed efficient E2F7 protein depletion upon siE2F7 transfection, with a concomitant increase in CCNE1 levels, in line with an E2F7-dependent regulation of this gene (Supplementary Figure S1B) (7).
RNA samples were harvested from three independent experiments and subsequently pooled. PolyA+ enriched samples from siNT- and siE2F7-transfected cells were used to build cDNA libraries that were sequenced by RNA-seq. Close to 107 high quality reads were obtained per sample. Changes in gene expression between siE2F7-transfected relative to siNT-transfected cells were scored as significant at q-value < 0.05. In all three time-points under study, RNA-seq analyses showed close to 500 genes with altered expression upon E2F7 knockdown in comparison with control cells. The proportion of overexpressed and underexpressed genes was similar (data not shown).
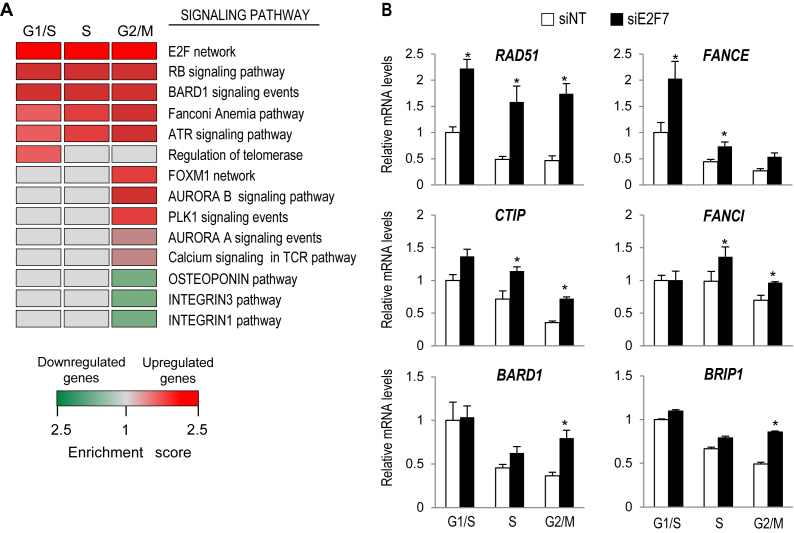
We analyzed the potential pathways regulated by E2F7 by performing GSEA. We considered as significantly enriched the pathways with an NES above 1.5 and an FDR below 10%. In concordance with the role of E2F7 as transcriptional repressor of RB/E2F-regulated cell-cycle genes (4), GSEA analyses showed highest enrichment values for E2F and RB pathways (Figure 1A) among the set of E2F7-repressed genes. This group included many genes previously described as E2F targets: CCND3, CDC6, DHFR and several MCM-s among others.
Figure 1.
E2F7 represses the expression of genes involved in cell cycle and DNA repair pathways. (A) Functional classification of E2F7-regulated genes by GSEA analysis using Pathway Interaction Database. The heat map shows the enriched pathways among the list of genes differentially expressed in cells lacking E2F7. Normalized enrichment ratios obtained in GSEA analyses are represented as colors (red for upregulated genes, green for downregulated genes). Only pathways with FDR < 10% were considered significantly enriched. (B) Validation of RNA-seq results. U2OS cells transfected with NT control or E2F7 siRNAs were synchronized in the cell cycle by HU treatment. RT-qPCR analyses of indicated genes were carried out with RNA samples harvested at 0h (G1/S), 3 h (S phase) and 12 h (G2/M) after HU treatment release. Expression values are normalized to the expression of EIF2C2, used as standard control. Data are represented as fold-change (mean ± SEM) relative to siNT-transfected samples from three independent experiments (*, P < 0.05).
Interestingly, pathways involved in (DDR) and repair, including BARD1, Fanconi anemia (FA) and Ataxia telangiectasia and Rad3-related protein (ATR) pathways were also over-represented among the genes repressed by E2F7 in all cell-cycle phases (Figure 1A). We confirmed E2F7-mediated repression of genes belonging to these functional groups by RT-qPCR analysis (Figure 1B). RNAi-mediated depletion of E2F7 resulted in a significantly increased expression of genes involved in HR-mediated repair of damaged DNA (RAD51, CTIP, BARD1) or in FA pathway (FANCE, FANCI, BRIP1). These results suggest that, in addition to the previously reported regulation of cell-cycle genes, E2F7 might also mediate repression of genes involved in DNA damage response and repair pathways.
E2F7 is recruited to the promoters of DNA damage repair genes
A search for promoter regulatory elements of the differentially expressed genes showed that about 40% of overexpressed genes in E2F7-depleted cells harbor E2F binding sites. In fact, the canonical E2F-binding motif was the most over-represented transcription factor-binding site among the upregulated set of genes in the three time-points analyzed according to DiRE analysis. By contrast, this motif was not over-represented among the set of genes displaying decreased mRNA levels in cells lacking E2F7. This finding supports a role for E2F7 in transcriptional repression through binding to consensus E2F motifs, in agreement with previous data (4,5).
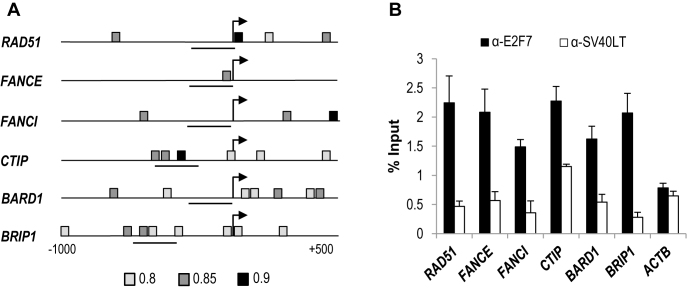
To identify the E2F motifs within E2F7-repressed RNAs we made use of MotifLocator tool provided by TOUCAN program. We focused our search on the genes belonging to BARD1, FA and ATR pathways that showed aberrant expression upon E2F7 attenuation in at least two of the analyzed time-points. Taking into account previous evidence suggesting that E2F factors are predominantly recruited to the proximal promoter of their target genes (4,15,27,28), we limited our search to a region spanning −1000 to +500 bp relative to transcription initiation. Using a threshold level of 0.8 for similarity with the canonical E2F motif recorded in the JASPAR database, we found that all E2F7-repressed genes included in the selected subset harbored at least one canonical E2F motif (Figure 2A and Supplementary Table S4).
Figure 2.
E2F7 occupies the promoter regions of a set of genes involved in the DNA damage response. (A) Schematic representation of human RAD51, FANCE, FANCI, CTIP, BARD1, BRIP1 promoter regions (−1000, +500), indicating the localization of consensus E2F motifs detected by Motiflocator tool from TOUCAN at 0.8 (light gray), 0.85 (gray) and 0.9 (dark gray) threshold levels. Horizontal lines depict the chromatin sequences amplified by qPCR. (B) ChIP-qPCR analyses of E2F7-regulated genes. Cell lysates were harvested 3 h after HU release and used for ChIP assays with an antibody against E2F7. Promoter regions near E2F consensus sites were amplified by qPCR. The promoter of β-Actin (ACTB) was used as a negative control. An unrelated antibody against the SV40 large T antigen (SV40LT) was used as a control for background immunoprecipitation. Data are presented as percentage of input chromatin (representative experiment of three independent experiments where the values are the mean ± SD of triplicate determinations).
We next assessed E2F7 binding activity to the promoters of E2F7-regulated genes by performing ChIP analyses with an anti-E2F7 specific antibody followed by qPCR using promoter-specific primers. Amplification of the β-actin promoter was used as a negative control, since this promoter lacks E2F binding sites but is highly expressed in U2OS cells (5). In addition, as a control for antibody specificity, we used an irrelevant antibody (anti-SV40LT), which has no affinity for chromatin and is unable to immunoprecipitate any of the various E2F target sequences (25). As shown in Figure 2B, ChIP analyses revealed efficient binding of E2F7 to all analyzed promoters, suggesting an important role for E2F7 in the direct transcriptional repression of DNA repair genes.
E2F7 controls cellular responses after genotoxic damage
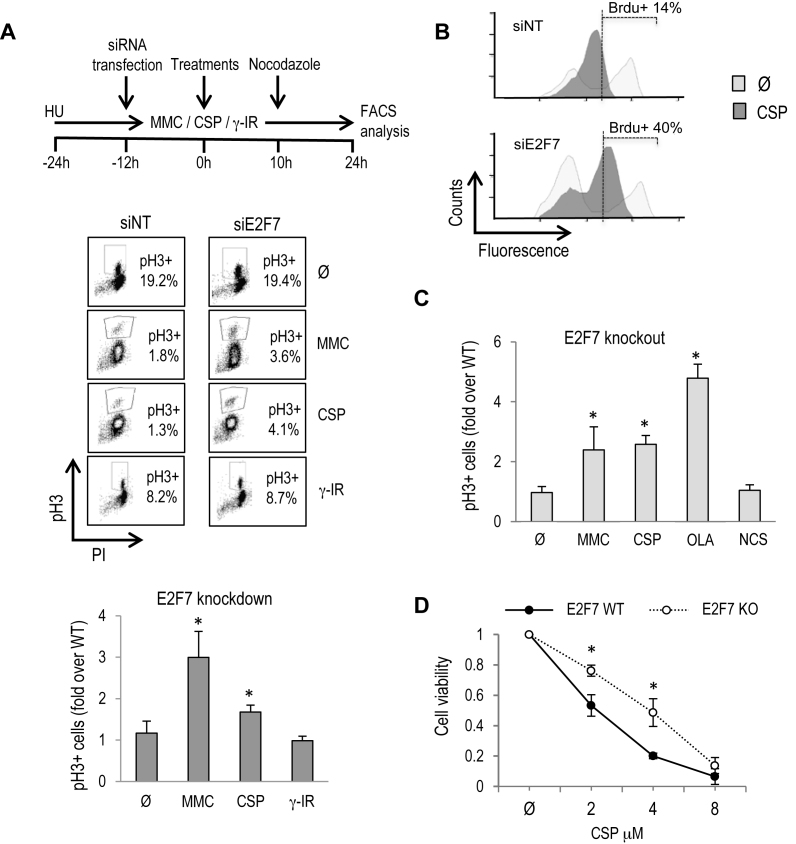
Given the enrichment in DDR and DNA repair genes within the list of E2F7-regulated transcripts, and having validated several of them as direct E2F7 target genes, we hypothesized that E2F7 could control cellular responses following DNA damage. To test this possibility, G1/S-synchronized cells were transfected with siNT or siE2F7, and subsequently cultured under several genotoxic conditions (Figure 3A): mitomycin C (MMC) and CSP are known to generate DNA interstrand crosslinks (ICL), in which FA repair pathway is involved (29), whereas γ-irradiation (IR) or neocarzinostatin (NCS), which mimics DNA damage caused by γ-IR (30) are known to induce DNA double-stranded breaks (31).
Figure 3.
E2F7 controls cellular recovery after DNA lesions that interfere with fork progression. (A) As shown in the schematic diagram, U2OS cells were HU-synchronized and transfected with siNT and siE2F7. Subsequently, cells were released into the cell cycle and treated for 24 h with 250 nM MMC, 8 μM CSP and a dose of 2.5 Gy of γ-IR. Nocodazole was present in the culture for the last 14 h of culture. The percentage of mitotic pH3-positive cells is shown in a representative figure obtained by FACS analysis. The graphs represent fold-change over siNT values (mean ± SD) of E2F7-depleted pH3-positive cells from four independent experiments. (B) Asynchronously growing U2OS cells were transfected with siNT and siE2F7 and subsequently treated with 8 μM CSP for 12 h. BrdU was present in the cultures for the last 2 h. Cells were stained with anti-BrdU conjugated with FITC and with propidium iodide. A representative FACS analysis is shown. (C) E2F7 knockout and wild-type cells were treated and analyzed as in (A). NCS (20 ng/ml) and OLA (4 μM) were also analyzed in these cells. The graphs represent fold-change of E2F7-knockout pH3-positive cells over parental wild-type values (mean ± SD) from three independent experiments. (D) Clonogenic survival assays were carried out with E2F7-knockout and parental U2OS cells treated with indicated doses of CSP. For each cell line tested, cell viability of untreated cells was defined as 1. Data represent mean ± SD from two independent experiments. Ø, untreated.
Cell-cycle progression was assessed by recording the accumulation of pH3-positive mitotic cells after nocodazole addition. Only a small fraction of siNT-transfected cells exposed to non-lethal doses of DNA-damaging agents was able to enter mitosis, reflecting the efficient arrest in G2 caused by these genotoxic agents (Figure 3A and Supplementary Figure S2A). Strikingly, knockdown of E2F7 led to a significant increase in the fraction of cells capable of exiting from the G2 arrest imposed by MMC and CSP relative to siNT-transfected cells (Figure 3A). This difference in recovery capacity between siNT and siE2F7-transfected cells was not observed upon ionizing radiation-induced G2 arrest. Results compiled from multiple biological replicates of synchronized as well as asynchronous cells confirmed the significant increase in the percentage of E2F7-silenced mitotic cells after treatment with MMC and CSP (Figure 3A and Supplementary Figure S2B). Similar results were obtained after treatment with Olaparib (OLA), an inhibitor of poly(ADP-ribose) polymerase-1 (PARP1) that induces single-strand break accumulation and reduced fork stability (32) (Supplementary Figure S2B and C).
Given the known effect of these drugs in replication fork progression, we next examined DNA replication rates after knockdown of E2F7 by measuring BrdU incorporation in asynchronously growing cells treated with CSP for 12 h. As expected, DNA synthesis rate was reduced in siNT cells under this treatment. By contrast, the reduction in DNA replication was alleviated in siE2F7 cells (Figure 3B), suggesting that E2F7 inhibits DNA replication when DNA lesions that interfere with fork progression are generated.
To confirm the functional significance of E2F7 in DNA damage responses, a CRISPR/Cas9 mediated E2F7 gene knockout was established in U2OS cells using a gRNA that targets the N-terminal region of E2F7 protein. Efficient E2F7 knockout of a selected cell line was demonstrated by DNA sequencing and Western analysis (Supplementary Figure S3). E2F7 knockout cells were treated with DNA damaging compounds, and pH3-positive mitotic cells were scored. Similarly to our finding with E2F7-knockdown cells, the fraction E2F7 knockout cells that was positive for pH3 was significantly increased relative to parental E2F7 wild-type cells after ICL induction or PARP inhibition (Figure 3C).
We next determined whether the improved cell-cycle progression of E2F7-deficient cells after genotoxic damage impacted their long-term clonogenic survival. To this end, we treated E2F7-knockout cells with CSP for 24 h and cultured them for two additional weeks to allow for colony formation from individual surviving cells. As shown in Figure 3D, the number of colonies that were scored in E2F7-knockout cultures exposed to CSP was significantly higher than in wild-type cultures, in concordance with cell-cycle analyses (Supplementary Figure S4). Altogether, our results suggest that lack of E2F7 confers an increased checkpoint recovery competence upon treatment with compounds that affect replication fork progression (CSP, MMC, OLA).
E2F7 expression and activity in cells exposed to DNA crosslinkers is p53-independent
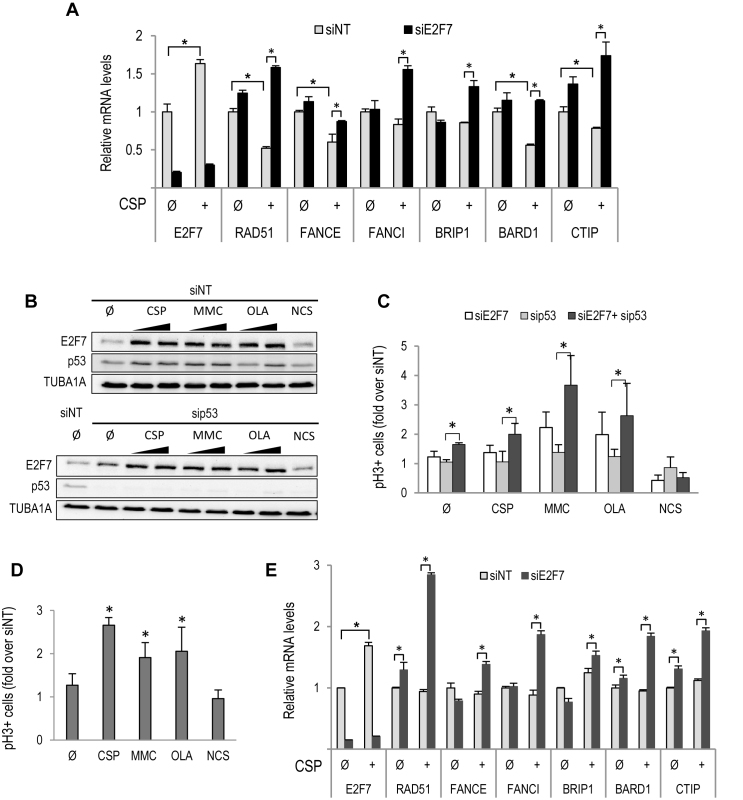
It has been reported that E2F7 expression is induced when DNA lesions are generated upon treatment with selected drugs (11,14). We tested whether E2F7 expression and transcriptional activity are also regulated upon ICL induction. Asynchronously growing U2OS cells transfected with siE2F7 or siNT were treated with CSP or left untreated for 24 h, and gene expression was analyzed at the mRNA level. A significant increase in E2F7 levels was detected upon CSP exposure, which was blocked in siE2F7-transfected cells (Figure 4A). In contrast to E2F7 expression, the mRNA levels of target genes involved in DNA replication and repair identified in our RNA-seq were consistently reduced upon CSP treatment. Importantly, silencing of E2F7 led to a robust overexpression of target genes in CSP treated cells (Figure 4A and Supplementary Figure S5). Similar results were obtained after MMC treatment (data not shown). These findings point to a role for E2F7 in the negative regulation of genes involved in DNA damage responses during cell-cycle progression, but also following ICL induction.
Figure 4.
E2F7 expression and activity are regulated in a p53-independent manner after ICL induction and PARP inhibition. (A) Asynchronously growing U2OS cells were transfected with siNT and siE2F7 and subsequently treated with 8 μM CSP for 24 h. RT-qPCR analyses of indicated genes are shown. Expression values are normalized to the expression of EIF2C2, used as standard control. Data are represented as fold-change (mean ± SEM) relative to siNT-transfected samples from three independent experiments. (B) U2OS cells were transfected with siRNA molecules specific for p53 or with siRNA control, and 24 h later treated with MMC (250 and 500 nM), CSP (4 and 8 μM), OLA (2 and 4 μM) or NCS (40 ng/ml) for an additional 24 h period. E2F7 and p53 protein levels were analyzed by western blots using specific antibodies. (C) U2OS cells were HU-synchronized and transfected with siNT, siE2F7 and/or sip53. Subsequently, cells were treated as in Figure 3B. The graphs represent fold-change over siNT values (mean ± SD) of E2F7- and/or p53-depleted pH3-positive cells from three independent experiments. (D) Asynchronously growing HeLa cells were transfected and treated as in (A). Shown are RT-qPCR analyses of indicated genes. (E) HeLa cells were treated as in Figure 3B, and the percentage of mitotic pH3-positive cells was analyzed by flow cytometry. The graph represents fold-change of E2F7-depleted pH3-positive cells over siNT values (mean ± SD) from three independent experiments. Ø, untreated. (*, P < 0.05)
ICL damage and PARP inhibition also led to a substantial induction of E2F7 at the protein level, concomitant with p53 accumulation (Figure 4B). To determine whether the observed accumulation of E2F7 levels was mediated by p53, as had been reported previously for cells treated with DNA topoisomerase II inhibitors (14), we silenced p53 expression by specific siRNA transfection and examined E2F7 expression upon genotoxic treatment. Remarkably, loss of p53 did not reduce E2F7 levels (Figure 4B). In functional assays, we found that depletion of p53 had no effect on the recovery of U2OS cells exposed to CSP, MMC or OLA, whereas concomitant depletion of p53 and E2F7 led to a significant increase in the number of mitotic cells (Figure 4C), suggesting that E2F7’s role in cell-cycle recovery from ICL damage or PARP inhibition is p53-independent.
To confirm these results we made use of HeLa cells, in which p53 activity is very low due to human papillomavirus-derived E6 protein expression in these cells (33). HeLa cells that were transfected with siRNA molecules specific for E2F7 and subsequently treated with genotoxic compounds accumulated a significantly higher percentage of pH3-positive mitotic cells compared to control cells transfected with non-target siRNAs (Figure 4D). Furthermore, exposure to CSP led to an induction of E2F7 mRNA levels and to an upregulation of target gene expression in E2F7-depleted HeLa cells (Figure 4E). These results suggest a p53-independent role in the regulation of cellular responses by E2F7 after DNA damage by ICLs and PARP inhibition.
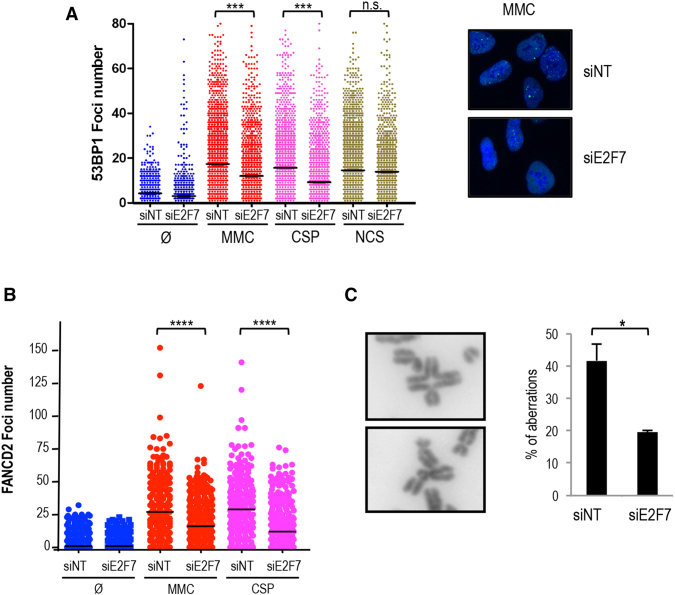
Reduced number of DNA repair foci and chromosome breaks after E2F7 silencing
We next considered the possibility that E2F7 could contribute to the modulation of DNA repair pathways involved in ICL resolution. To test this hypothesis, we examined the accumulation of 53BP1 foci in the nuclei of damaged cells. 53BP1 has been involved in DNA damage signaling and repair, and is well characterized for its ability to localize to DNA lesions in cells exposed to genotoxic agents, including ICL-inducing agents (34). U2OS cells were transfected with E2F7-specific siRNAs and subsequently treated with MMC, CSP or the radiomimetic drug NCS. Twenty-four hours after treatments, cells were fixed and 53BP1 foci were analyzed and quantified by immunofluorescence and high-throughput microscopy (Figure 5A). As expected, treatments with DNA damaging agents resulted in an increased number of foci relative to untreated cells. Interestingly, depletion of E2F7 caused a significant decrease in MMC or CSP-induced 53BP1 foci, but did not alter NCS-derived foci number. Furthermore, E2F7-null cells showed lower levels of γ-H2AX compared to E2F7-competent cells after 24 h of genotoxic treatment, whereas the γ-H2AX levels were comparable at earlier time points in both cell lines (Supplementary Figure S6). These data imply that E2F7 does not affect foci formation, but instead E2F7 plays a role in the negative control of pathways involved specifically in ICL repair.
Figure 5.
E2F7 knockdown results in reduced foci and chromosome break number after ICL induction. (A) siNT and siE2F7 transfected U2OS cells were treated with MMC, CSP and NCS and fixed 24 h later. Cells were stained for 53BP1 with a FITC-conjugated specific antibody. Nuclear DNA was stained with DAPI. The number of 53BP1 foci was scored by HTM. Data are representative of three independent analyses. Horizontal lines indicate mean vales. (B) siNT and siE2F7 transfected U2OS cells were treated with 250nM MMC or with 4 μM CSP. Twenty fours hours later, samples were fixed and stained for FANCD2. The number of FANCD2 foci was scored on fluorescence microscope images. Continuous horizontal lines indicate mean values. Data are representative of three independent analyses. (C) siNT and siE2F7 transfected U2OS cells were treated with 250 nM MMC for 48 h and scored for chromosomal aberrations by analyzing metaphase spreads. Representative images of a radial chromosome and a chromatid break are shown. Chromosomal aberrations are expressed as the average breaks and radial chromosomes found per metaphase (n = 50 cells) in three independent experiments. (*, P < 0.05; ***, P < 0.001), Ø, untreated, n.s. not significant.
It has been shown that FANCD2 recruitment to sites of DNA crosslinks is an essential step for ICL repair (35,36). Thus, we analyzed FANCD2 foci by immunofluorescence, and quantified the number of foci per nucleus in E2F7-depleted cells exposed to MMC or CSP. As expected, MMC and CSP treatments increased the number of FANCD2 foci/nucleus. Importantly, depletion of E2F7 caused a significant decrease in MMC or CSP-induced FANCD2 foci (Figure 5B).
We next assessed the accumulation of chromosomal aberrations, a hallmark of ICL-inducing agents (29,37), visualized in metaphase spreads. Numerous control cells (siNT) displayed radial and broken chromosomes (nearly 0.5 aberrations per metaphase) upon MMC exposure. Silencing of E2F7 provided partial resistance against MMC-induced chromosomal aberrations, with a reduced presence of radial and broken chromosomes (Figure 5C). Thus, E2F7 appears to have a negative role in the repair of chromosomal aberrations resulting from MMC treatment. The reduction in 53BP1 and FANCD2 foci upon MMC treatment shown by cells lacking E2F7 supports this hypothesis.
E2F7 modulates homology-directed DNA repair
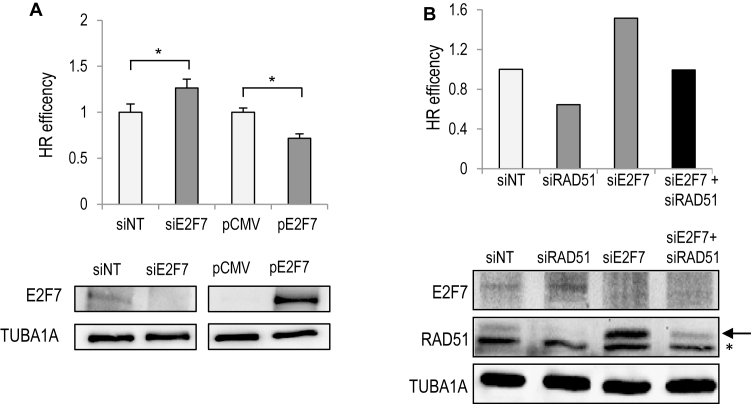
We next investigated the efficiency of HR in cells depleted of E2F7. We used a U2OS cell line with an integrated direct repeat recombination reporter (DR-GFP). With this reporter, homology-directed DNA repair is detected when a DSB introduced into the chromosome by the I-SceI endonuclease is repaired by HR to give rise to GFP-positive cells (20). Knockdown of E2F7 resulted in a significant increase in HR efficiency (Figure 6A). Conversely, overexpression of E2F7 in U2OS-DR-GFP cells to levels that are comparable to those observed after ICL induction led to a significant reduction in HR efficiency, suggesting that E2F7 inhibits HR-mediated repair.
Figure 6.
E2F7 suppresses HR through transcriptional repression of DNA repair genes. (A) U2OS-DR-GFP cells were transfected with siRNAs specific for E2F7 or with a plasmid expressing E2F7 together with an SceI expression vector. GFP-positive cells were analyzed by FACS. Data are shown as a percentage over siNT or over empty pCMV transfection. The values shown represent the mean ± SD of three independent experiments (*, P < 0.05). (B) U2OS-DR-GFP cells were transfected with siRNAs specific for E2F7, RAD51 or with a combination of both and analyzed as in (A). Western blot analysis confirms knockdown of E2F7 and RAD51 (arrow). Shown is a representative experiment of two independent experiments. A non-specific band in RAD51 blot is indicated with an asterisk.
To better define the mechanism underlying E2F7-mediated modulation of DNA repair, we analyzed whether the improved genomic stability conferred by loss of E2F7 could be attributed to increased expression of E2F7 target genes necessary for HR repair. We examined RAD51 recombinase activity, a surrogate marker of HR efficiency and transcriptional target of E2F7 (Figures 1B and 2). In E2F7-depleted U2OS cells, RAD51 foci were significantly increased under both basal and ICL-inducing conditions (Supplementary Figure S7), suggesting that HR may be hyperactive upon loss of E2F7. Using the DR-GFP reporter system, siRNA-mediated RAD51 depletion led to a reduction in HR repair, whereas E2F7 depletion resulted in increased HR rates, as measured by the differences in the percentages of GFP-positive cells detected by flow cytometry (Figure 6B). Western blot analysis of protein extracts derived from E2F7-silenced cells showed overexpression of RAD51 protein levels, in line with the mRNA results described in Figure 1B. We next co-transfected E2F7-specific siRNAs with a concentration of RAD51-specific siRNAs that would attenuate RAD51 expression to the levels found in E2F7 competent cells. Interestingly, the increased HR repair efficiency conferred by loss of E2F7 was abrogated under these conditions, and the percentage of GFP positive cells decreased to the levels found in control cells (Figure 6B and Supplementary Figure S8). These results suggest that E2F7 modulates DNA repair through the transcriptional repression of target genes that play a central role in the resolution of DNA lesions requiring homology-directed repair, such as RAD51. In the absence of E2F7 the HR pathway could become hyperactive and potentially harmful.
Improved genomic stability in HR-deficient cells after E2F7 depletion
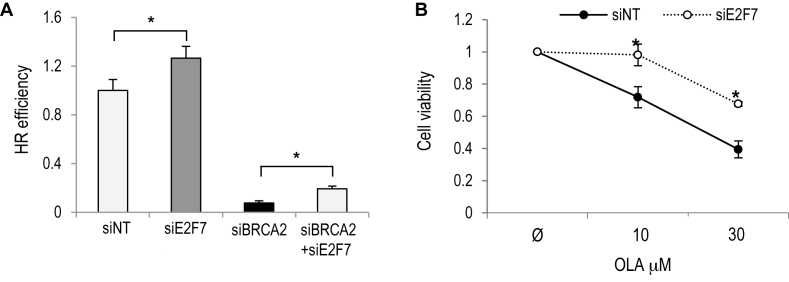
Given that E2F7 displays features of an HR inhibitor, we next tested whether downregulation of E2F7 could suppress genomic instability in cells with an underlying genetic defect in HR. We hypothesized that the increased recombination conferred by E2F7 depletion might promote DNA repair and protect cells from genomic instability and cell death. To test this possibility we used RNA interference to attenuate the expression of BRCA2 in the U2OS DR-GFP cell line. As expected, knockdown of BRCA2 abolished HR repair in this assay. Interestingly, we observed that E2F7 co-depletion could improve HR in cells with reduced BRCA2 activity, therefore ensuring genomic stability (Figure 7A and Supplementary Figure S9). We next made use of CAPAN-1 pancreatic adenocarcinoma cells, which are defective in HR due to a loss-of-function mutation of BRCA2 (38). Treatment of these cells with PARP1 inhibitor OLA compromised cell viability as seen in short-term and long-term clonogenic survival assays (Figure 7B and Supplementary Figure S10), consistent with the finding that cancer cells deficient in HR repair through loss of BRCA2 are hypersensitive to PARP inhibitors (39). Importantly, downregulation of E2F7 expression in CAPAN-1 cells was associated with increased resistance to the PARP1 inhibitor OLA (Figure 7B and Supplementary Figure S10). Thus, E2F7 knockdown confers an increased resistance to chemotherapy in cells carrying defects in genes involved in HR.
Figure 7.
Improved genomic stability and survival upon E2F7 depletion in BRCA2-deficient cells. (A) Direct repeat recombination was measured in U2OS-DR-GFP cells transfected with siRNAs specific for E2F7, BRCA2 or with a combination of both. E2F7 knockdown partly rescued the severe defect caused by BRCA2 depletion. Data are represented as fold-change (mean ± SEM) relative to siNT-transfected samples from two independent experiments. (B) Clonogenic survival assays were carried out with siE2F7 or siNT transfected CAPAN-1 cells treated with indicated doses of OLA. For each siRNA, cell viability of untreated cells was defined as 1. Data represent mean ± SD from two independent experiments. Ø, untreated. (*, P < 0.05).
DISCUSSION
In this work we have investigated the role of the atypical E2F member E2F7 in DNA damage repair by analyzing its contribution to the control of gene expression and to cellular responses upon exposure to genotoxic damage. We find that in addition to controlling the timely expression of genes necessary for G1/S transition and DNA replication in unperturbed conditions, E2F7 is involved in the negative regulation of genes controlling DNA repair pathways. Consequently, E2F7 activity is associated with a suppression of DNA repair reactions.
The expression of genes that are involved in various aspects of the DDR and DNA repair pathways is cell-cycle regulated, showing highest expression in G1/S transition and decreasing thereafter. Here, we show that the downregulation of these genes throughout the cell cycle is E2F7-dependent. Interestingly, all upregulated genes included in the DNA damage repair functional group harbor at least one E2F binding site in their promoters, and although many of those have been previously identified as targets of classical E2F proteins (15,40–44), their regulation by E2F7 has only been demonstrated for some of them (4). Our RNA-seq and ChIP experiments have extended the collection of direct E2F7 target genes involved in DNA repair by demonstrating that E2F7 is recruited to the promoter regions of RAD51, FANCE, FANCI, CTIP, BARD1 and BRIP1, implying their direct transcriptional repression by E2F7. An E2F7-dependent downregulation of replication fork-associated DNA damage repair genes in the cell cycle could help restrict DNA repair activities to the S phase, which might otherwise give rise to unscheduled DNA repair activity and genome instability.Exposure of siE2F7-transfected cells to ICL-inducing DNA damaging agents results in higher levels of DNA replication and mitotic cells compared to siNT-transfected cells, consistent with a role for E2F7 in cell-cycle arrest. Supporting this possibility, we demonstrate that loss of E2F7 confers an increased recovery competence upon treatment with DNA damage-inducing doses of CSP, MMC or OLA, suggesting that E2F7 is a factor that controls cellular recovery during an ongoing DNA damage response. In contrast to our findings, it has been reported that lack of E2F7 sensitizes cells to topoisomerase inhibitors by inducing apoptosis through a mechanism involving E2F1 upregulation (10,11). Several reasons could explain the disparity between our results and those from previous studies. On the one hand, we have used a set of genotoxic agents that are known to differ in their mechanism of DNA damage and in the elicited response from the previously analyzed ones. On the other hand, the drug doses used in our study were non-lethal although sufficient to induce checkpoint arrest in G2, whereas previous studies employed doses sufficiently high to induce apoptosis. Thus, there could be a DNA damage threshold below which cells lacking E2F7 could be involved in repairing the damage, but above which these cells would activate cell death pathways. Systematic analyses using a wide range of doses of a variety of compounds may help resolve these differences.
E2F7 expression and cell-cycle target gene repression have been previously linked to p53 after DNA damage by topoisomerase inhibitors (14). Unexpectedly, we found that E2F7 expression and E2F7-modulated cellular recovery upon ICL damage is largely p53-independent. Our results point to a fundamental difference in the DNA damage-mediated regulation of E2F7 expression and function between DNA topoisomerase inhibitors and interstrand crosslink inducers. Further studies should determine whether other p53 family members are involved in E2F7 regulation upon ICL induction or whether a distinct pathway mediates E2F7 regulation in this context. In contrast to DNA crosslinkers, ionizing radiation did not induce E2F7 expression in U2OS cells, which may explain why depletion of E2F7 did not confer increased cellular recovery after radiation.
Remarkably, our data show that E2F7 has additional roles beyond inhibition of DNA replication in the presence of DNA damage. In fact, DNA damage foci are significantly reduced upon MMC and CSP treatments in E2F7-depleted cells. These results point to a uniquely increased DNA repair competence upon treatment with ICL-inducing agents in cells lacking E2F7, leading to an earlier release from the chromatin of γ-H2AX, 53BP1 and FANCD2. Our observation that E2F7 knockdown has a protective effect against chromosomal aberrations induced by MMC treatment supports this hypothesis.
Interestingly, the FA pathway, which is known to be involved in ICL repair (45), is highly enriched among E2F7-repressed genes. ICL-resistant cell lines are known to have elevated gene expression involving the FA/BRCA pathway, including FANCF and RAD51C, which was suggested to be causally related with enhanced removal of ICLs by the resistant cells (46,47). We have found evidence that the expression of at least FANCE, FANCI, BRIP1 (also called FANCJ) or RAD51 (also called FANCR) is directly regulated by E2F7, and that E2F7 depletion results in enhanced expression of these FA genes, particularly during S/G2 phases or after DNA damage, two cellular contexts exhibiting high E2F7 levels (5) (Figure 4B). By contrast, this enhanced expression of E2F7 target genes was not observed in asynchronously growing cells, probably because most of these cells are in G1, a time-point where E2F7 levels are very low (5) and therefore unable to repress target gene expression. It will be interesting to analyze whether a correlation can be found between ICL resistance and E2F7 levels in different cancer cell lines in unperturbed conditions, but also upon exposure to ICL-inducing chemotherapy.
ICL repair is known to involve homology-directed repair machinery and increased HR is associated with resistance to ICL-inducing agents in human tumor cells (48,49). Our results are consistent with a negative role for E2F7 in HR repair activity. Indeed, using a DR-GFP assay to measure the effect of E2F7 in HR, we found that E2F7 negatively affects HR activity. A transcription-independent contribution to DNA repair process for E2F7 and E2F1 has been previously reported, which involves the binding of these E2Fs and recruitment of several factors to damaged DNA sites (13,50–52). We cannot discard the possibility that there is a transcription-independent contribution to E2F7-mediated regulation of ICL lesion repair in our system, which should be important to analyze. However, our data strongly suggest that a major DNA damage response function of E2F7 is through transcription-dependent regulation of DNA repair genes. Several genes involved in HR were found upregulated upon E2F7 depletion, including RAD51, CTIP and BARD1, among others. Most importantly, the results obtained in our E2F7/RAD51 co-depletion experiments suggest that increased HR activity in E2F7 silenced cells is associated with increased levels of RAD51 recombinase, implying a transcriptional role for E2F7 in repair of ICL lesions, through upregulation of target genes involved in homology-directed DNA repair. Thus, the transcriptional landscape regulated by E2F7 could provide an additional level of recombination control in addition to that described for several recombinases (53,54), whereby cells can interfere with HR at different steps in the process.
Increasing recombination in HR-deficient cells might result in protective effects. Our results have revealed an intriguing link between genomic integrity of DNA repair-deficient cells and E2F7. HR-deficient (BRCA2 mutated) cells exhibit increased genomic instability and accumulation of mutations that ultimately disrupt cell-cycle control pathways, leading to cancer. In this scenario, increased HR activity conferred by inactivation of E2F7 might prevent genomic instability in the cells of these patients and protect against cancer onset, as has been proposed for the depletion of the PCNA-binding protein PARI (54). However, dysregulated hyper-recombination has also been associated with increased genomic instability and resistance to genotoxic therapy in some cellular contexts, such as after RAD51 upregulation (55). In fact, the increased survival of BRCA2-deficient tumor cells treated with a PARP inhibitor that we observe after knockdown of E2F7 implies that loss of E2F7 in the context of HR deficiency confers resistance to chemotherapy, a potentially harmful outcome for cancer treatment.
Although further research will be needed to elucidate the molecular mechanisms underlying E2F7-dependent control of genomic stability, our data are consistent with an antioncogenic function for E2F7 whereby E2F7 functions to inhibit or to switch off repair pathways for specific DNA lesions. It has been reported that efficient ICL repair requires negative regulation of the FA pathway. Once repair is completed, the repair factors have to be inactivated to avert inappropriate action and corruption of genetic information (56). Thus, the inability to turn off or reset the FA pathway after the repair of specific DNA damage sites may have deleterious effects on genome integrity. In a similar manner, E2F7 might counter-balance the transcriptional program activated in response to ICL repair to fine-tune the cellular response to DNA lesions and ensure response termination.
DATA AVAILABILITY
RNA-seq datasets generated in this study were deposited in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107216.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Zubiaga and Malumbres laboratory for helpful discussions, Naiara Zorrilla for technical support, José Antonio Rodríguez for critical reading of the manuscript, D. Olmos and J. Surrallés for kindly providing cell lines and R. Medema for providing CRISPR/Cas9 plasmids.
Notes
Present Address: Jone Mitxelena, Department of Molecular Mechanisms of Disease, University of Zurich, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from the Spanish Ministry [SAF2012-33551 and SAF2015-67562-R, co-financed by FEDER funds, and SAF2014-57791-REDC], the Basque Government [IT634-13 and KK-2015/89], and the University of the Basque Country UPV/EHU [UFI11/20] to AMZ; and grants from the Spanish Ministry [SAF2015-69920-R], and Worldwide Cancer Research [15-0278] to MM. JM was recipient of a Basque Government fellowship for graduate studies and JVR is recipient of a UPV/EHU fellowship for graduate studies. M.A.F. was supported by a young investigator grant from MINECO [SAF2014-60442-JIN; co-financed by FEDER funds]. Funding for open access charge: Spanish Ministry [SAF2015-67562-R, co-financed by FEDER funds]; Basque Government [IT634-13].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen H.Z., Tsai S.Y., Leone G.. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009; 9:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimova D.K., Dyson N.J.. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005; 24:2810–2826. [DOI] [PubMed] [Google Scholar]
- 3. Bertoli C., Skotheim J.M., de Bruin R.A.. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013; 14:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Westendorp B., Mokry M., Groot Koerkamp M.J., Holstege F.C., Cuppen E., de Bruin A.. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 2012; 40:3511–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitxelena J., Apraiz A., Vallejo-Rodriguez J., Malumbres M., Zubiaga A.M.. E2F7 regulates transcription and maturation of multiple microRNAs to restrain cell proliferation. Nucleic Acids Res. 2016; 44:5557–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Bruin A., Maiti B., Jakoi L., Timmers C., Buerki R., Leone G.. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2003; 278:42041–42049. [DOI] [PubMed] [Google Scholar]
- 7. Di Stefano L., Jensen M.R., Helin K.. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003; 22:6289–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J., Ran C., Li E., Gordon F., Comstock G., Siddiqui H., Cleghorn W., Chen H.Z., Kornacker K., Liu C.G et al.. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell. 2008; 14:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Endo-Munoz L., Dahler A., Teakle N., Rickwood D., Hazar-Rethinam M., Abdul-Jabbar I., Sommerville S., Dickinson I., Kaur P., Paquet-Fifield S et al.. E2F7 can regulate proliferation, differentiation, and apoptotic responses in human keratinocytes: implications for cutaneous squamous cell carcinoma formation. Cancer Res. 2009; 69:1800–1808. [DOI] [PubMed] [Google Scholar]
- 10. Thurlings I., Martinez-Lopez L.M., Westendorp B., Zijp M., Kuiper R., Tooten P., Kent L.N., Leone G., Vos H.J., Burgering B et al.. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene. 2017; 36:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zalmas L.P., Zhao X., Graham A.L., Fisher R., Reilly C., Coutts A.S., La Thangue N.B.. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008; 9:252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hazar-Rethinam M., Cameron S.R., Dahler A.L., Endo-Munoz L.B., Smith L., Rickwood D., Saunders N.A.. Loss of E2F7 expression is an early event in squamous differentiation and causes derepression of the key differentiation activator Sp1. J. Invest. Dermatol. 2011; 131:1077–1084. [DOI] [PubMed] [Google Scholar]
- 13. Zalmas L.P., Coutts A.S., Helleday T., La Thangue N.B.. E2F-7 couples DNA damage-dependent transcription with the DNA repair process. Cell Cycle. 2013; 12:3037–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carvajal L.A., Hamard P.J., Tonnessen C., Manfredi J.J.. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012; 26:1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laresgoiti U., Apraiz A., Olea M., Mitxelena J., Osinalde N., Rodriguez J.A., Fullaondo A., Zubiaga A.M.. E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res. 2013; 41:10185–10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iglesias-Ara A., Zenarruzabeitia O., Fernandez-Rueda J., Sanchez-Tillo E., Field S.J., Celada A., Zubiaga A.M.. Accelerated DNA replication in E2F1- and E2F2-deficient macrophages leads to induction of the DNA damage response and p21(CIP1)-dependent senescence. Oncogene. 2010; 29:5579–5590. [DOI] [PubMed] [Google Scholar]
- 17. Blomen V.A., Majek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A et al.. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015; 350:1092–1096. [DOI] [PubMed] [Google Scholar]
- 18. Remeseiro S., Cuadrado A., Carretero M., Martinez P., Drosopoulos W.C., Canamero M., Schildkraut C.L., Blasco M.A., Losada A.. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012; 31:2076–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richardson C., Moynahan M.E., Jasin M.. Homologous recombination between heterologs during repair of a double-strand break. Suppression of translocations in normal cells. Ann. N. Y. Acad. Sci. 1999; 886:183–186. [DOI] [PubMed] [Google Scholar]
- 21. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osinalde N., Olea M., Mitxelena J., Aloria K., Rodriguez J.A., Fullaondo A., Arizmendi J.M., Zubiaga A.M.. The Nuclear Protein ALY binds to and modulates the activity of transcription factor E2F2. Mol. Cell Proteomics. 2013; 12:1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aerts S., Van Loo P., Thijs G., Mayer H., de Martin R., Moreau Y., De Moor B.. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005; 33:W393–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gotea V., Ovcharenko I.. DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 2008; 36:W133–W139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Infante A., Laresgoiti U., Fernandez-Rueda J., Fullaondo A., Galan J., Diaz-Uriarte R., Malumbres M., Field S.J., Zubiaga A.M.. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008; 7:3915–3927. [DOI] [PubMed] [Google Scholar]
- 26. Lopez-Contreras A.J., Specks J., Barlow J.H., Ambrogio C., Desler C., Vikingsson S., Rodrigo-Perez S., Green H., Rasmussen L.J., Murga M et al.. Increased Rrm2 gene dosage reduces fragile site breakage and prolongs survival of ATR mutant mice. Genes Dev. 2015; 29:690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rabinovich A., Jin V.X., Rabinovich R., Xu X., Farnham P.J.. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008; 18:1763–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu X., Bieda M., Jin V.X., Rabinovich A., Oberley M.J., Green R., Farnham P.J.. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007; 17:1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCabe K.M., Olson S.B., Moses R.E.. DNA interstrand crosslink repair in mammalian cells. J. Cell Physiol. 2009; 220:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang P., Lee J.W., Yu Y., Turner K., Zou Y., Jackson-Cook C.K., Povirk L.F.. Gene rearrangements induced by the DNA double-strand cleaving agent neocarzinostatin: conservative non-homologous reciprocal exchanges in an otherwise stable genome. Nucleic Acids Res. 2002; 30:2639–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeggo P.A., Lobrich M.. DNA double-strand breaks: their cellular and clinical impact. Oncogene. 2007; 26:7717–7719. [DOI] [PubMed] [Google Scholar]
- 32. Pommier Y., O’Connor M.J., de Bono J.. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016; 8:362ps317. [DOI] [PubMed] [Google Scholar]
- 33. Haupt Y., Rowan S., Shaulian E., Vousden K.H., Oren M.. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995; 9:2170–2183. [DOI] [PubMed] [Google Scholar]
- 34. Hicks J.K., Chute C.L., Paulsen M.T., Ragland R.L., Howlett N.G., Gueranger Q., Glover T.W., Canman C.E.. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell. Biol. 2010; 30:1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D’Andrea A.D.. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001; 7:249–262. [DOI] [PubMed] [Google Scholar]
- 36. Liu T., Ghosal G., Yuan J., Chen J., Huang J.. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010; 329:693–696. [DOI] [PubMed] [Google Scholar]
- 37. Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D’Andrea A.D., Bernards R.. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005; 17:331–339. [DOI] [PubMed] [Google Scholar]
- 38. McCabe N., Lord C.J., Tutt A.N., Martin N.M., Smith G.C., Ashworth A.. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol. Ther. 2005; 4:934–936. [DOI] [PubMed] [Google Scholar]
- 39. Lord C.J., Ashworth A.. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J.R.. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001; 21:4684–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D.. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002; 16:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bindra R.S., Glazer P.M.. Basal repression of BRCA1 by multiple E2Fs and pocket proteins at adjacent E2F sites. Cancer Biol. Ther. 2006; 5:1400–1407. [DOI] [PubMed] [Google Scholar]
- 43. Tategu M., Arauchi T., Tanaka R., Nakagawa H., Yoshida K.. Systems biology-based identification of crosstalk between E2F transcription factors and the Fanconi anemia pathway. Gene Regul. Syst. Bio. 2007; 1:1–8. [PMC free article] [PubMed] [Google Scholar]
- 44. Hoskins E.E., Gunawardena R.W., Habash K.B., Wise-Draper T.M., Jansen M., Knudsen E.S., Wells S.I.. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008; 27:4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clauson C., Scharer O.D., Niedernhofer L.. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 2013; 5:a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Q., Van der Sluis P.C., Boulware D., Hazlehurst L.A., Dalton W.S.. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005; 106:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hazlehurst L.A., Enkemann S.A., Beam C.A., Argilagos R.F., Painter J., Shain K.H., Saporta S., Boulware D., Moscinski L., Alsina M et al.. Genotypic and phenotypic comparisons of de novo and acquired melphalan resistance in an isogenic multiple myeloma cell line model. Cancer Res. 2003; 63:7900–7906. [PubMed] [Google Scholar]
- 48. Slupianek A., Schmutte C., Tombline G., Nieborowska-Skorska M., Hoser G., Nowicki M.O., Pierce A.J., Fishel R., Skorski T.. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell. 2001; 8:795–806. [DOI] [PubMed] [Google Scholar]
- 49. Xu Z.Y., Loignon M., Han F.Y., Panasci L., Aloyz R.. Xrcc3 induces cisplatin resistance by stimulation of Rad51-related recombinational repair, S-phase checkpoint activation, and reduced apoptosis. J. Pharmacol. Exp. Ther. 2005; 314:495–505. [DOI] [PubMed] [Google Scholar]
- 50. Chen J., Zhu F., Weaks R.L., Biswas A.K., Guo R., Li Y., Johnson D.G.. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle. 2011; 10:1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo R., Chen J., Mitchell D.L., Johnson D.G.. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011; 39:1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guo R., Chen J., Zhu F., Biswas A.K., Berton T.R., Mitchell D.L., Johnson D.G.. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J. Biol. Chem. 2010; 285:19308–19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barber L.J., Youds J.L., Ward J.D., McIlwraith M.J., O’Neil N.J., Petalcorin M.I., Martin J.S., Collis S.J., Cantor S.B., Auclair M et al.. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008; 135:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moldovan G.L., Dejsuphong D., Petalcorin M.I., Hofmann K., Takeda S., Boulton S.J., D’Andrea A.D.. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 2012; 45:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin R.W., Orelli B.J., Yamazoe M., Minn A.J., Takeda S., Bishop D.K.. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007; 67:9658–9665. [DOI] [PubMed] [Google Scholar]
- 56. Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D’Andrea A.D.. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 2009; 16:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets generated in this study were deposited in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107216.