Abstract
Correct localization of the centromeric histone variant CenH3/CENP-A/Cse4 is an important part of faithful chromosome segregation. Mislocalization of CenH3 could affect chromosome segregation, DNA replication and transcription. CENP-A is often overexpressed and mislocalized in cancer genomes, but the underlying mechanisms are not understood. One major regulator of Cse4 deposition is Psh1, an E3 ubiquitin ligase that controls levels of Cse4 to prevent deposition into non-centromeric regions. We present evidence that Chromatin assembly factor-1 (CAF-1), an evolutionarily conserved histone H3/H4 chaperone with subunits shown previously to interact with CenH3 in flies and human cells, regulates Cse4 deposition in budding yeast. yCAF-1 interacts with Cse4 and can assemble Cse4 nucleosomes in vitro. Loss of yCAF-1 dramatically reduces the amount of Cse4 deposited into chromatin genome-wide when Cse4 is overexpressed. The incorporation of Cse4 genome-wide may have multifactorial effects on growth and gene expression. Loss of yCAF-1 can rescue growth defects and some changes in gene expression associated with Cse4 deposition that occur in the absence of Psh1-mediated proteolysis. Incorporation of Cse4 into promoter nucleosomes at transcriptionally active genes depends on yCAF-1. Overall our findings suggest CAF-1 can act as a CenH3 chaperone, regulating levels and incorporation of CenH3 in chromatin.
INTRODUCTION
One of the unique features of eukaryotic centromeres is a specific histone H3 variant, CenH3. CenH3 is known as CENP-A in human and Cse4 in budding yeast. Nucleosomes containing this histone variant serve as the foundation for kinetochore assembly and attachment of chromosomes to spindle microtubules for faithful segregation. Eukaryotes have evolved stringent regulatory mechanisms to promote exclusive centromeric localization of CenH3/Cse4 (1).
Histone chaperones are indispensable for their role in regulation of histone deposition into chromatin. They facilitate proper nucleosome assembly and disassembly (2). In addition, histone chaperones play roles in nuclear import, storage and stability of histones. Many chaperones regulate the canonical histones, creating functional redundancy. Most histone variants have dedicated chaperones (2). CenH3 assembly into centromeric nucleosomes is regulated by the evolutionarily conserved histone chaperone known as Scm3 in budding yeast (3–6), and its ortholog HJURP in humans (7,8).
Regulating the deposition of Cse4 is critical to prevent mislocalization of Cse4 to non-centromeric regions. In addition to being regulated by the Cse4-specific chaperone Scm3, Cse4 levels are controlled by multiple mechanisms, including ubiquitin-mediated proteolysis (9). An E3 ubiquitin ligase Psh1 in conjunction with Casein Kinase 2 (CK2) facilitates ubiquitylation and proteasomal degradation of Cse4 (10–12). In addition, several other factors play roles in ubiquitin-mediated proteolysis of Cse4 (13–18). Additional chromatin regulating proteins, including remodelers, have been identified that assist in efficient removal of mislocalized Cse4 nucleosomes (19–21).
Chaperones in addition to Scm3 may play a role in the dynamics of Cse4 deposition, especially under conditions of overexpression when the balance of H3 and Cse4 levels is dramatically altered. Elevated expression and misincorporation of CenH3/CENP-A is reported in many human cancers (22–26). In human cells the transcription-coupled histone H3.3 chaperones DAXX/ATRX target CENP-A to ectopic locations (23,26,27). Mislocalization of CenH3 could disrupt chromatin structure and lead to negative consequences for growth by causing ectopic centromere/kinetochore formation, chromosome instability and missegregation (27–30). Mislocalized CenH3 could also affect chromatin based processes like DNA replication and transcription (31). Therefore, it is important to identify and characterize factors that regulate promiscuous CenH3 localization.
We present evidence that the evolutionarily conserved histone H3/H4 chaperone Chromatin Assembly Factor-1 (CAF-1) can function as a Cse4 chaperone in budding yeast. Subunits of CAF-1 had previously been reported to be important for building functional kinetochores (32), for recruitment of CenH3/Cnp1 and Scm3 to centromeres in Schizosaccharomyces pombe (33,34) and for regulating Cse4/H3 exchange kinetics (35). We report that recombinant CAF-1 can assemble Cse4 nucleosomes in vitro and CAF-1 subunits co-immunoprecipitate with Cse4 in vivo. CAF-1 is responsible for the majority of Cse4 incorporated into chromatin when Cse4 is overexpressed. The negative impact of Cse4 misincorporation on growth is likely multifactorial, including disrupted gene expression and other chromatin-based processes. CAF-1 may also promote the assembly of Cse4 nucleosomes at the centromere and facilitate proteolysis of Cse4 by Psh1 under certain conditions. Overall our findings help to establish how the histone H3/H4 chaperone CAF-1 regulates deposition of Cse4.
MATERIALS AND METHODS
Yeast strains
All the yeast strains are listed in Supplementary Table S1.
Whole cell extract Co-IP
Cultures grown to mid-log phase in appropriate media were used to prepare cell extracts in lysis buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol (DTT), 10% glycerol and protease inhibitors). Protein concentration was determined using Bradford assay. Cell extracts were diluted with dilution/wash buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% NP-40). Diluted cell extracts were incubated with the antibody overnight followed by 2 h with protein G dynabeads (Invitrogen-10004D) at 4°C. Some co-IPs was performed using antibody conjugated beads. The beads were washed three times with dilution/wash buffer and proteins were eluted with sodium dodecyl sulphate (SDS) buffer (10 mM Tris pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1% SDS). Immunoprecipitates were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
Recombinant protein expression and purification
Methods for histone expression and purification have been published (36).
GST-Psh1 expression and purification
Plasmid containing GST-Psh1 (pSB1535 is a kind gift from Biggins lab, Fred Hutchinson Cancer Research Center, Seattle, WA, USA) was transformed into BL21-CodonPlus(DE3)-RIL cells. A total of 1 l of a bacterial culture was grown to OD600 of 0.5 at 37°C. Protein expression was induced with isopropyl-β-D-thiogalactoside (IPTG, 0.5 mM) at 25°C, overnight. Cells were harvested, frozen in liq. N2, and stored in −80°C until used. Cells were re-suspended in 15 ml of ice-cold STE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl with protease inhibitors). The cell suspension was incubated with Lysozyme (0.5 mg/ml, Sigma #62971) on ice until the texture changes (∼20–30 min) and added 10 mM DTT and 1% sarkosyl. The cell lysate was sonicated using a digital sonifier (run time: 10 min, pulse ON: 15 s, pulse OFF: 20 s) and centrifuged at 15 000 reads per million (rpm) for 20 min at 4°C. The supernatant was incubated with 500 µl of glutathione sepharose beads (GE #17-0756-01) suspension in the presence of 2% Triton X-100 for 2 h at 4°C. The beads were spun down and washed two times with 10 ml of cold STET (STE with 1% Triton X-100) and two more times with 10 ml of cold STE (without Triton). The beads were transferred into a 1.5 ml spin column (Bio-Rad #732–6204, Micro Bio-Spin column). Psh1-GST was eluted using a gradient of elution buffers containing 1, 10, 20, 30, 40 and 50 mM reduced Glutathione in 50 mM Tris pH8.0. After running a gel, fractions containing GST-Psh1 was combined and the buffer was exchanged with storage buffer (50 mM HEPES, pH 7.9, 100 mM NaCl, 2 mM MgCl2 and 10% glycerol) using 30K MWCO centrifuge filters.
CAF-1 expression and purification
CAF-1 complex expression and purification was performed as described previously (37). Briefly, optimum levels of Cac1, Cac2 and Cac3 viruses were used to infect the SF9 cells for 72h. After infection, cells were collected for the nuclei isolation and subsequent purification. The extraction buffer (15 mM Tris–HCl, pH 7.5, 400 mM NaCl, 10% sucrose, 1 mM EDTA,1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride (PMSF)) was used to resuspend the nuclear pellet. Supernatant with the CAF-1 complex was collected after spinning at 33 000 rpm for 40 min and 15% of ammonium sulfate was added to precipitate the protein. After the clarification at 12 000 rpm for 30 min, the supernatant was adjusted to 65% saturation with ammonium sulfate to precipitate the remaining proteins. After spinning at 12 000rpm for 30 min, the precipitated proteins containing CAF-1 complex, were dissolved in buffer B0 (20 mM HEPES, pH7.5, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.01% Triton X-100). After final clarification, the supernatant was loaded onto a HiTrap SP HP column and proteins were eluted between gradient of B100 to B1000. Fractions with the CAF-1 complex were detected by western and pooled for overnight binding with M2-FLAG agarose beads. Elution buffer (B100+1 mg/ml 3× FLAG) was added to elute the CAF-1 complex. Purified CAF-1 complex was analyzed by silver staining and dialyzed against a storage buffer (20 mM HEPES, pH7.5, 1 mM EDTA, 1 mM DTT, 10% glycerol, 25 mM NaCl).
In vitro ubiquitylation assay
Human E1 (UBE1), E2 (UbcH3-human homolog of budding yeast Ubc3) and HA-ubiquitin from Boston Biochem (E-305, K-980B and U-110, respectively) were used for the assay. Ubiquitylation mixture was prepared by mixing E1 (0.1 μg), E2 (0.05 μg), E3 (GST-Psh1, 0.1μg) and HA-ubiquitin (2.5 μg) in the E3 ligase reaction buffer (Boston Biochem, B-71). Energy regeneration solution-ERS (Boston Biochem, B-10) was used as the energy source. 0.4 μg of Cse4 was added as the substrate. Reactions were performed in the presence or absence of 1 μg of CAF-1. Total reaction volume was 30 μl. Samples were incubated for 1 h at 30°C and diluted with 400 μl of Tris-buffered saline (TBS). A total of 60 μl of 25% slurry of pre-washed poly-ubiquitin affinity resin (Agarose-TUBE-1, Life Sensors, UM401) was added and incubation proceeded overnight at 4°C. The beads were washed three times with TBS. Proteins were eluted with 30 μl of SDS buffer (10 mM Tris pH 7.5, 1 mM EDTA and 1% SDS) and boiled with 10 μl of 4× SDS sample buffer. Ubiquitylated proteins were analyzed by SDS-PAGE and western blotting.
SDS-PAGE and western blotting
SDS-PAGE was performed using 4–12% pre-cast gels (Invitrogen). After running the gels, proteins were transferred onto PVDF membrane (Millipore, Immobilon-P) and Western blotting was performed according to standard protocol.
Antibodies/conjugated beads
The antibodies used are as follows: anti-Myc (Bio Legend-626802), anti-FLAG-HRP (Roche-6952, Sigma-A8592), anti-FLAG (Sigma-F3165), anti-FLAG-HRP (Roche-6952), anti-Cse4 (polyclonal rabbit antibody against recombinant Cse4), anti-H2A (Active Motif-39235), anti-Pgk1 (Invitrogen-459250), anti-ubiquitin (Covance-MMS257P), anti-HA (Covance-PRB101P), anti-FLAG beads (Sigma-F2426), secondary antibodies were HRP linked, anti-rabbit IgG (from donkey) and anti-mouse IgG (from sheep) (GE Healthcare, NA934V and NA931V, respectively).
ChIP-qPCR
Cultures were grown to mid-log phase in appropriate medium. If gal induction of Cse4 was needed, galactose was added to a final concentration of 4% and incubated for 6 h. Cells were fixed in 1% formaldehyde for 15 min at room temperature, then quenched with glycine (final concentration 0.125 M) for 5 min. Cells were washed twice with cold TBS, frozen in liq. N2, and stored in −80°C until used. Cells were lysed by bead beating for 1 h at 4°C in FA-Lysis SDS buffer (50 mM Hepes (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.2% SDS, protease inhibitors) and centrifuged to remove cell debris. The lysate was sonicated using a Biorupter for 30 min, 30 s on/off, medium intensity. Protein concentration was determined and samples were normalized before proceeding with the chromatin immunoprecipitation (ChIP). About 20% of total chromatin extract was used for IP and incubated with antibody at 4°C overnight. A total of 25 μl of Protein G Dynabeads (Invitrogen 100-04D) pre-washed in FA-Lysis buffer (50 mM Hepes (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) were added and incubated at 4°C for 2 h. Beads were washed at room temperature with the following sequence of buffers; FA-Lysis buffer, FA-Lysis buffer with 1M NaCl, FA-Lysis buffer with 500 mM NaCl, TEL buffer (0.25M LiCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate), twice with 1×TE. Chromatin fragments were eluted twice using 200 μl of Elution Buffer (1% SDS, 250 mM NaCl, 1×TE) at 65°C with agitation. Two elutions were combined and treated with Proteinase K for 1 h at 55°C and incubated overnight at 65°C. DNA was extracted with phenol–chloroform and precipitated using 100% ethanol. About 2% of the total chromatin extract was processed for the input sample. Cse4-ChIPs were performed in triplicate with two no-antibody controls for each strain. Antibodies used for ChIP experiments are 2 μl of α-Cse4 antibody (polyclonal rabbit antibody against recombinant Cse4) and anti-FLAG (Sigma-F3165).
Quantitative polymerase chain reaction (qPCR) was performed in triplicate for each sample using a Perfecta SYBR Green FastMix, ROX (Quanta Biosciences-95073). CAS4200 robot and ABI 7900HT cycler (ABI SDS 2.4 program) were used for plate setup and qPCR respectively. Cycling parameters for qPCR: Stage-1: 95C° 3 min, 1 cycle; Stage 2: 95°C 15 s → 60°C 1min, 40 cycles; Stage 3: 95°C 15 s → 60°C 15 s → 95°C 15 s, 1 cycle. Total volume of a PCR reaction was 10 μl and 5–25 ng of DNA was used per reaction. Primers used for qPCR: PHO5-1F/R (Forward: 5′-CCCATTTGGGATAAGGGTAAAC-3′; Reverse: 5′-GATGAAGCCATACTAACCTCGA-3′, amplicon-95 bp from promoter of PHO5-1/YBR093C), rDNA 37-1F/R (Forward: 5′-AGTTTGTCCAAATTCTCCGCTC-3′; Reverse: 5′-ACGTCCCTGCCCTTTGTACACA-3′, amplicon-121 bp within rDNA 37-1/RDN37-1), CEN3 F/R (Forward: 5′-AAG TCA CAT GAT GAT ATT TGA T-3′; Reverse: 5′-ATT TCT TTT TTA ACT TTC GGA A-3′, amplicon-125 bp from CEN3). UCSC In-Silico PCR tool and qPCR-melt curves were used to determine the specificity of each primer set. Standard curves were generated including six standards for each primer set (PHO5-1 F/R (slope: −3.91, efficiency:80%, y-intercept:26.82, r2:0.995); rDNA37-1 F/R (slope: −3.76, efficiency:85%, y-intercept:19.36, r2:0.998); CEN3 F/R (slope: −4.27, efficiency:71%, y-intercept:30.66, r2:0.979)). Total DNA to be used in a reaction was determined based on linear dynamic range (LDR), Limit of Detection (LoD) and Cq values of each primer set/standard curve. qPCR data were analyzed manually using mean quantity values to calculate IP/No antibody ratios after normalizing to total input DNA. The standard deviation (error bars) was calculated using IP/No antibody values for ChIPs performed in triplicate. P-values were calculated using a two-sided t-test. Each qPCR experiment was repeated to check reproducibility.
ChIP-seq
ChIP was performed as mentioned in ChIP-qPCR. About 30 and 3.5% of total chromatin extract were used for the IP and the input samples, respectively. IPs were performed at least in duplicate for two biological replicates of each strain. Libraries were prepared using the KAPA HTP Library Prep Kit for Illumina and Bioo Scientific NEXTflex DNA barcodes. The resulting libraries were purified using the Agencourt AMPure XP system (Beckman Coulter) then quantified using a Bioanalyzer (Agilent Technologies) and a Qubit fluorometer (Life Technologies). Post amplification size selection was performed on libraries where there was enough available material to do so using a Pippin Prep (Sage Science). Libraries were re-quantified, normalized, pooled and sequenced on an Illumina NextSeq 500 instrument as Mid Output 75 bp paired read run. Following sequencing, Illumina NextSeq Real Time Analysis version 2.4.6.0 was ran to demultiplex reads and generate FASTQ files.
Paired-end reads were aligned to version sacCer3 of the yeast genome from UC Santa Cruz using bowtie2 with -k 2. Concordant alignments were analyzed in R using Bioconductor to generate coverage and normalize the data in rpm. Locations of enrichment for each protein were identified using MACS2 version 2.1.0.20140616. Metagene plots are constructed by converting coverage of the Cse4 ChIP into rpm per base, averaging the replicates for each condition and then extracting out coverage for the promoters of a given set of genes, defined as −500 to +100 of the transcription start site (TSS), and plotting the average of the gene set at each base pair.
RNA-seq
Overnight cultures of wild-type (WT) and mutant strains grown in duplicate in appropriate raffinose medium were diluted to OD600 of 0.1. After 2 h at 30°C, Cse4 overexpression was induced by adding galactose to a final concentration of 4% and cultures were grown at 30°C for 10 h. Cells were pelleted, frozen in liq. N2 and stored in −80°C until used. The cell pellet was resuspended in 800 µl of cold AE Buffer (50 mM NaOAc (pH 5.2), 10 mM EDTA in RNase Free water). A total of 80 μl of 10% SDS and 800 µl acid phenol (pH 4.3) (Ambion# AM9720) were added and incubated for 10 min at 65°C. The cell suspension was incubated on ice for 5 min and centrifuged for 15 min at 14K rpm, 4°C. The supernatant was transferred to a fresh tube, 800 µl of chloroform was added and centrifuged for 15 min at 14K rpm, 4°C. Chloroform extraction step was repeated one more time and the supernatant was transferred to a fresh tube. Next 10 μg of linear acrylamide, 3M NaOAc, pH 5.2–5.6 (1/10th of total volume) and isopropanol (volume equal to total volume) were added and centrifuged for 15 min at 14K rpm, 4°C. The pellet was washed with 1 ml 70% ethanol and resuspended in ∼100 μl TE buffer (10 mM Tris pH8.0, 0.1 mM EDTA in RNase Free water). The RNA extract was chilled first on ice for 5 min, then 20 s at 65°C. It was mixed by gentle vortexing and chilled on ice. RNA integrity was checked following isolation using an Agilent Technologies 2100 Bioanalyzer. The RNA concentration was measured using Qubit 2.0 Fluorometer. Ribosomal RNA was depleted from 1000 ng total RNA using Ribo-Zero™ Magnetic Gold Kit (Yeast) (Illumina, MRZY1324) per manufacturer’s instructions. Following depletion, Illumina libraries were generated using the TruSeq Stranded RNA LT kit with Ribo-Zero Gold, 48; Set A (Illumina, RS-122-2301) and resulting libraries were checked for quality and concentration using an Agilent 2100 Bioanalyzer and Invitrogen Qubit 2.0 Fluorometer. Libraries were normalized and pooled together with an average base pair size of 279, and sequenced as 50 bp single reads on the HiSeq 2500 (Illumina) using HiSeq Control Software v2.2.58. Following sequencing, Illumina Primary Analysis version RTA 1.18.61 and Secondary Analysis bcl2fastq2 v2.17 were run to demultiplex reads from all libraries and generate FASTQ files.
Reads of 51 bases from an Illumina HiSeq 2500 were aligned to version sacCer3 of the Saccharomyces cerevisiae genome from UC Santa Cruz using tophat2 and the -g 1 alignment parameter. The resulting BAM files were analyzed in R using the GenomicRanges library from Bioconductor to quantify reads on genes and the edgeR library (38) for normalization and calculation of P-values between treatments. P-values were adjusted for multiple hypothesis testing by the method of Benjamini and Hochberg (39).
In vitro chromatin assembly with purified CAF-1 complex
Purified CAF-1 complex with Cse4-containing histone octamer was used for the in vitro nucleosome assembly. Briefly, Topoisomerase I relaxed 200 ng of plasmid was incubated with CAF-1 complex or Nap1 for 2 h at 30°C in the presence of Topoisomerase I with 8.3 mM HEPES pH7.4, 0.5 mM EGTA, 0.65 mM MgCl2, 1.7% glycerol, 0.005% NP-40, 33 mM KCl, 0.33 mM DTT and 0.02 mg/mL BSA. Topoisomers of the plasmids were deproteinized and isolated by standard protocol and resolved in 0.8% agarose gel. Topoisomerase I was a kind gift from S. Venkatesh, Stowers Institute. Two plasmids were used: (i) pG5E4-5S containing five repeats of 5S flanking each side of an E4 core promoter downstream of five Gal4- binding sites (gift from the Workman lab, Stowers Institute) and (ii) pCEN1-10X containing 10 tandem repeats of the centromere 1 sequence.
RESULTS
CAF-1 subunits interact with Cse4
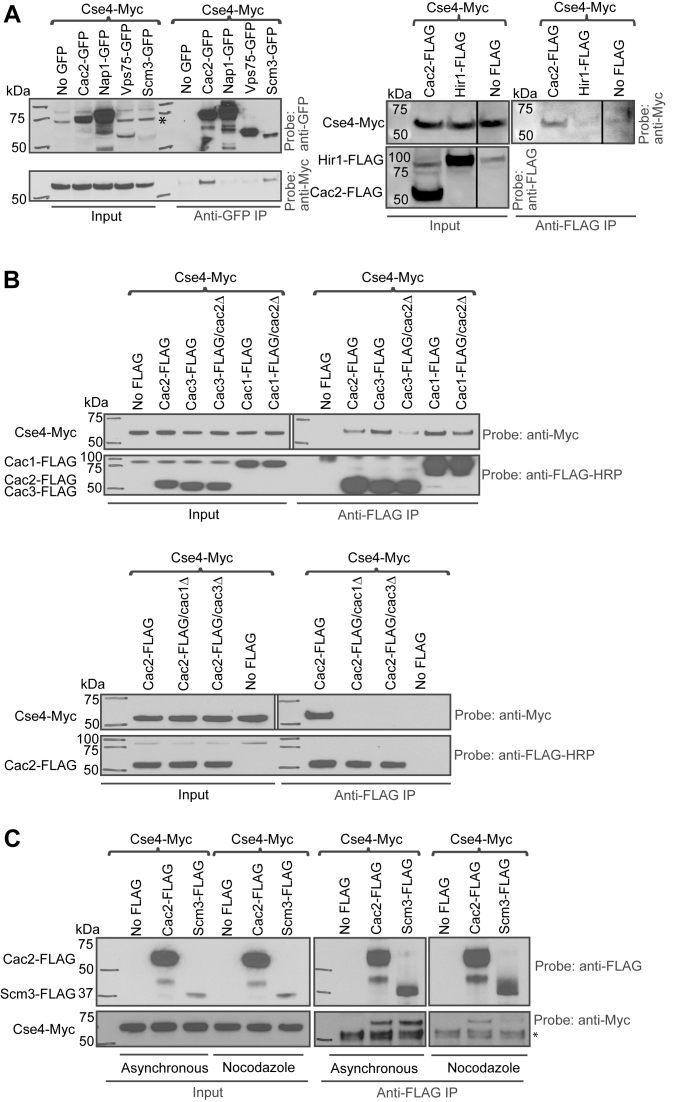
We took a candidate approach to identify potential chaperones for Cse4. Known histone chaperones Vps75, Nap1 and the Cac2 subunit of CAF-1 were GFP tagged. Whole cell extracts were used to perform co-immunoprecipitation (co-IP) with anti-GFP antibody followed by detection of Cse4-Myc in western blots with anti-Myc antibody (Figure 1A, left panel). Scm3, the chaperone that targets Cse4 to the centromere, was used as a positive control. Of these three candidates, only Cac2 pulled down Cse4. Hir1 (Histone regulator 1) is another histone H3/H4 chaperone whose biological role significantly overlaps with CAF-1 (35). Hir1-FLAG was also tested for interaction with Cse4. Cac2-FLAG served as a control. While Hir1-FLAG did not pull down Cse4, suggesting Hir1 does not associate with Cse4 in vivo, Cac2-FLAG pulled down Cse4-Myc, further confirming the interaction between Cac2 and Cse4 (Figure 1A, right panel).
Figure 1.
CAF-1 subunits interact with Cse4. Whole cell extracts were used to carry out co-IPs in (A–C). (A) Left panel: strains carrying GFP-tagged chaperones, Scm3, Vps75, Nap1 and Cac2 subunit of CAF-1 were used in co-IP. Only Cac2-GFP pulled down Cse4. A control IP was performed from a strain lacking the GFP tag. Scm3-GFP IP was the positive control. Right panel: affinity-tagged strains (Cse4-Myc/Cac2-FLAG and Cse4-Myc/Hir1-FLAG) were used for co-IP. A control IP was performed from a strain lacking the FLAG tag. In anti-FLAG co-IP, Hir1-FLAG did not pull down Cse4, only Cac2-FLAG pulled down Cse4-Myc. (B) Cac1 and Cac3 subunits of CAF-1 are required for interaction with Cse4. Upper panel: WT or cac2Δ strains carrying FLAG-tagged Cac1 or Cac3 were used for co-IP. A strain lacking the tag on Cac1 and Cac3 was used as the negative control. In anti-FLAG co-IP, both Cac1 and Cac3 pulled down noticeably more Cse4-Myc. These interactions became weaker with CAC2 deletion. Lower panel: WT, cac1Δ or cac3Δ strains carrying FLAG-tagged Cac2 were used in anti-FLAG co-IP. A strain lacking the tag was used as the control. Cac2/Cse4 interaction was completely abolished in cac1Δ or cac3Δ strains. (C) CAF-1 can interact with Cse4 outside of S phase. Affinity-tagged strains (Cse4-Myc/Cac2-FLAG and Cse4-Myc/Scm3-FLAG) were used in anti-FLAG co-IP using asynchronously grown cells and G2/M-arrested cells. A strain lacking the FLAG tag served as a negative control. Scm3 was used as a positive control. Cac2 interacted with Cse4 in both asynchronously grown cells as well as G2/M-arrested cells. A non-specific band is marked with an asterisk.
CAF-1 is a heterotrimeric protein complex. In human, the three subunits are p150, p60 and p48 (also named RbAp48). The budding yeast subunits are named Cac1 (p90), Cac2 (p60) and Msi1/Cac3 (p50) (40). The Cac3 ortholog RbAp48 (also named p55) in Drosophila co-purified with CenH3/CID and assembled CID nucleosomes in vitro (41,42). The human CAF-1 subunits p48/RbAp48 and p150 also co-purified with CENP-A (7,8). Taken together, these findings suggest the CAF-1 complex or subunits thereof could serve as an evolutionarily conserved CenH3 chaperone.
The Cac2 subunit of CAF-1 is essential for efficient histone H3/H4 binding and nucleosome assembly (43). To identify the subunits of CAF-1 important for physical interaction with Cse4, WT strains carrying epitope tagged Cse4-Myc/Cac1-FLAG, Cse4-Myc/Cac2-FLAG and Cse4-Myc/Cac3-FLAG were used to perform co-immunoprecipitation and western blotting (Figure 1B). All three subunits of CAF-1 pulled down Cse4-Myc. However, Cac1 and Cac3 subunits pulled down noticeably more Cse4-Myc (Figure 1B, upper panel). Furthermore, the interaction between Cse4 and Cac3, and to a lesser extent Cac1, was dependent on CAC2. Cse4 interaction with Cac2 was completely abolished without CAC1 or CAC3 (Figure 1B, lower panel). These observations suggest that Cac1 and Cac3 subunits are required for Cse4-CAF-1 interaction, while loss of Cac2 reduces the interaction. Cse4 may interact with the entire CAF-1 complex in budding yeast.
CAF-1 deposits histone H3/H4 into nucleosomes during DNA synthesis, although a previous report argued that the exchange of Cse4 for H3 mediated by CAF-1 can be replication independent (35). Also, CAF-1 deposits histone H3.1 outside of S phase at repair sites after UV damage in human cells (44). Both results suggest CAF-1 can operate outside of S phase. We investigated whether the Cse4–CAF-1 interaction was cell cycle dependent using co-IP and western blotting. Whole cell extracts were prepared from cells growing asynchronously or arrested in G2/M (nocodazole treated). Scm3 was used as a positive control. Like Scm3, Cac2 interacted with Cse4 in extracts generated from both asynchronously grown cells as well as G2/M-arrested cells, suggesting the interaction between Cse4 and CAF-1 is not confined to S phase (Figure 1C). Together, these experiments suggest that CAF-1 can interact with Cse4 outside of S phase and may therefore be able to assemble and disassemble Cse4 nucleosomes independent of DNA replication.
CAF-1 can assemble Cse4 nucleosomes in vitro
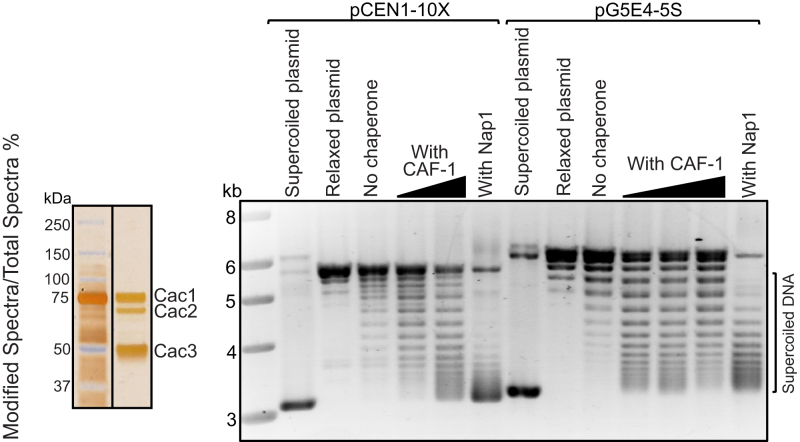
The Cac3 ortholog RbAp48 in Drosophila can assemble CID nucleosomes in vitro (41,42). We tested whether recombinant budding yeast CAF-1 could facilitate the assembly of Cse4 nucleosomes in vitro using a plasmid supercoiling assay (6). In this assay, wrapping of DNA around the histone core particle induces supercoiling in relaxed, closed, plasmid DNA. Following the assembly reaction, DNA is deproteinized and plasmid topoisomers are resolved by agarose gel electrophoresis. We tested nucleosome assembly using a plasmid containing 10 copies of a 5S nucleosome positioning sequence (pG5E4-5S) as well as one containing 10 tandem copies of a yeast centromere 1 (CEN1) repeat unit (pCEN1-10X). Incubation of purified CAF-1 (37) and Cse4 octamers with either pG5E4-5S or pCEN1-10X resulted in the induction of supercoiling in a dose dependent manner (Figure 2), demonstrating that CAF-1 can assemble Cse4 containing nucleosomes on both plasmids. Nap1 was used as a positive control. Although Nap1 does not co-immunoprecipitate with Cse4 in whole cell extracts, the high concentrations of purified proteins used in the assembly assay in vitro, combined with the absence of H3, may allow Nap1 to promote the assembly of Cse4 nucleosomes. These experiments demonstrate that CAF-1 can assemble Cse4 nucleosomes in vitro irrespective of DNA sequence.
Figure 2.
CAF-1 can assemble Cse4 nucleosomes in vitro. Recombinant purified CAF-1 is shown in a silver stained gel (left). Nucleosome assembly activity of CAF-1 was studied using a plasmid supercoiling assay (right). Supercoiled plasmids were purified from Escherichia coli and relaxed by addition of topoisomerase I. Supercoiled plasmid, relaxed plasmids and no chaperone samples were included as controls for each assembly experiment. Chromatin assembly was performed by incubating the relaxed plasmids with increasing amounts of purified CAF-1 and Cse4 octamers. DNA and Cse4 octamer amounts are held constant. Nucleosome assembly was performed using a plasmid (pCEN1-10X) containing 10 tandem copies of a yeast centromere 1 (CEN1) repeat unit (left side), as well as a plasmid (pG5E4-5S) containing 10 copies of a 5S nucleosome positioning sequence (right side). Histone chaperone Nap1 was used as a control. CAF-1 resulted in the induction of supercoiling on both plasmids in a dose dependent manner.
Loss of CAF-1 rescues growth of psh1Δ and cka2Δ strains
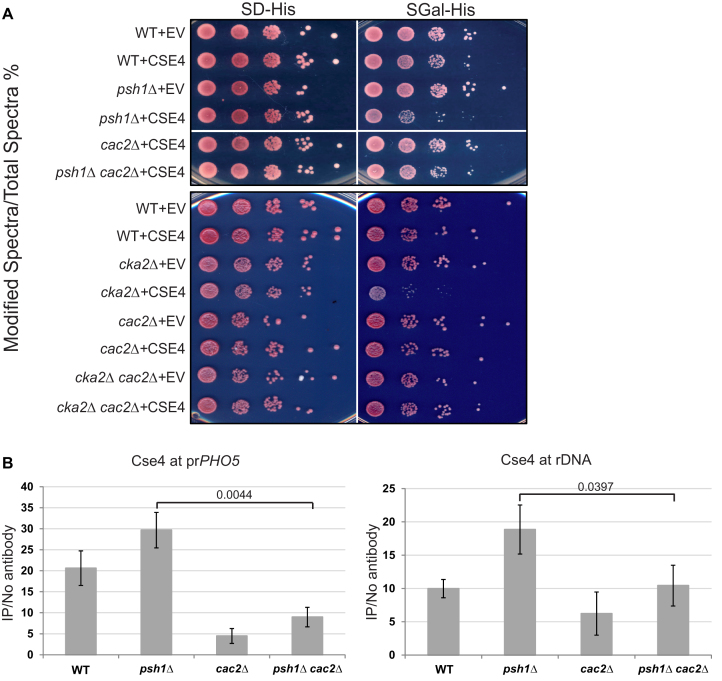
The E3 ubiquitin ligase Psh1, regulated in part by Casein Kinase 2 (CK2), controls Cse4 levels by proteolysis. Deletion of PSH1 or CKA2, one of the two catalytic subunits of CK2, resulted in poor growth when Cse4 was overexpressed and higher levels of Cse4 incorporation at non-centromeric sites (10–12). Deletion of PSH1 with overexpression of Cse4 could potentially affect kinetochore function, and consistently, Psh1 may contribute to plasmid segregation (45), but this does not seem to fully explain the growth defect (11,46). If part of the growth defect is due to disruption of chromatin-based processes from misincorporation of Cse4, and CAF-1 is responsible for targeting Cse4 into ectopic chromosome regions, then deletion of CAF-1 subunits might rescue the slow growth observed upon Cse4 overexpression in psh1Δ and cka2Δ strains. To test this, we generated CAC2 deletion mutants in a psh1Δ or cka2Δ W303a background and performed growth assays (Figure 3A). We observed improved growth with psh1Δ cac2Δ and cka2Δ cac2Δ double mutants, compared to psh1Δ or cka2Δ single mutants (Figure 3A, compare psh1Δ+CSE4 and cka2Δ+CSE4 with psh1Δ cac2Δ+ CSE4 and cka2Δ cac2Δ+CSE4 respectively). Similar rescue of growth was observed with CAC1 and CAC3 deletion (Supplementary Figure S1A). Therefore, deletion of any single subunit of the CAF-1 complex rescues the growth phenotype of psh1Δ or cka2Δ mutants, suggesting that CAF-1 is partly responsible for the poor growth phenotype.
Figure 3.
Loss of CAF-1 rescues growth of psh1Δ and cka2Δ strains and reduces Cse4 incorporation at promoters. (A) Cse4 was overexpressed from the gal promoter on a 2 micron plasmid in WT, psh1Δ, cac2Δ, cka2Δ, psh1Δ cac2Δ and cka2Δ cac2Δ strains in the W303a background. ‘EV’ indicates an empty vector control as a point of comparison. 10-fold serial dilutions of overnight cultures were plated to either SD-His or SGal-His medium. psh1Δ and cka2Δ strains overexpressing Cse4 grow poorly as expected whereas psh1Δ cac2Δ and cka2Δ cac2Δ strains grow similar to WT strains. (B) ChIP was performed for Cse4 using Cse4 overexpressing strains from (A). qPCR was used to detect Cse4 levels for the PHO5 promoter and rDNA. The y-axis indicates arbitrary units representing the enrichment for each sequence from ChIP performed with and without antibody, with respect to the signal for total chromatin for each sample. The error bars represent the standard deviation for ChIP performed in triplicate. P-values were calculated for psh1Δ cac2Δ strain relative to psh1Δ using a two-sided t-test.
Nap1, Vps75, Hir1, Rtt106 and Asf1 are all known histone H3/H4 chaperones (2,47). We tested whether deletion of the genes encoding any of these five chaperones in the psh1Δ W303a background with Cse4 overexpression would rescue growth (Supplementary Figure S1B); none rescued growth. In fact, deletion of HIR1, RTT106 or ASF1 in the psh1Δ strain compromised growth further when Cse4 was overexpressed. Deletion of RTT106 or ASF1 alone also causes a growth phenotype when Cse4 is overexpressed. These three histone H3/H4 chaperones play roles in several chromatin dependent processes including promoter fidelity, heterochromatin silencing and histone gene transcription, and general H3 assembly and disassembly (2,48–56). Disruption of any of these processes could negatively affect growth. In addition, defective H3 nucleosome dynamics may make cells more susceptible to Cse4 misincorporation by CAF-1, causing a synergistic effect that makes the double mutant strains grow more poorly. Another possibility is that these chaperones facilitate removal of mislocalized Cse4 by disassembling Cse4 nucleosomes and/or by recruiting factors that promote Cse4 nucleosome disassembly and proteolysis. If this is true, then the removal of these chaperones will make the psh1Δ strain more vulnerable to Cse4 accumulation in chromosome arms. Supporting this idea, Rtt106 physically interacts with the FACT complex (57) which associates with Psh1 to facilitate efficient degradation of mislocalized Cse4 (19). While additional chaperones may affect H3-Cse4 exchange, the only chaperone whose loss relieves poor growth in the psh1Δ strain is CAF-1, suggesting CAF-1 may be the main chaperone contributing to deleterious deposition of Cse4 in this context.
CAF-1 promotes localization of Cse4 to gene promoters and active subtelomeric regions
We speculated that CAF-1 may promote Cse4 deposition into promoter regions. Deletion of CAC2 partially disrupts Cse4/CAF-1 interaction and, unlike the other subunits, Cac2 does not function outside of the CAF-1 complex. Therefore, we utilized cac2Δ to further analyze CAF-1 regulation of Cse4 deposition. To examine Cse4 levels, we performed ChIP/qPCR at the promoter of PHO5 (YBR093C) and at the rDNA (RDN37-1) region. As previously shown, Cse4 localization to these regions was elevated in a psh1Δ strain compared to a WT strain (Figure 3B) (10). We observed a significant reduction in Cse4 levels at both locations in the psh1Δ cac2Δ strain compared to the psh1Δ strain. Cse4 levels were also significantly reduced in a cac2Δ single deletion strain relative to WT at the PHO5 promoter. These data are consistent with the idea that CAF-1 facilitates localization of Cse4 to promoter regions.
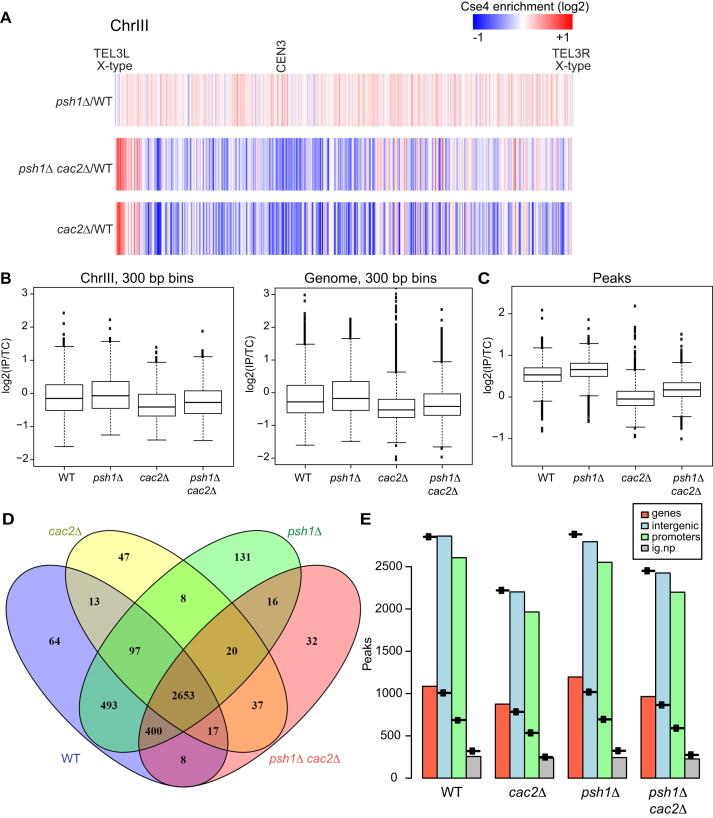
To further analyze the contribution of CAF-1 to global misincorporation of Cse4, we performed ChIP seq for the strains depicted in Figure 3. A heat map of Cse4 levels for chromosome III is shown in Figure 4A. In agreement with previous reports (10,11,31) we observed that deletion of PSH1 with overexpression of Cse4 results in elevated Cse4 levels at many non-centromeric locations (red is enriched and blue is depleted). We observed a significant reduction in global incorporation of Cse4 in both psh1Δ cac2Δ and cac2Δ strains, although there are select regions at which Cse4 is enriched relative to a WT strain. All 16 chromosomes show a similar reduction in global Cse4 incorporation upon loss of CAC2 (Supplementary Figure S2). The pattern of incorporation in the cac2Δ and psh1Δ cac2Δ strains was similar, although there is more Cse4 in the psh1Δ cac2Δ heatmaps (Supplementary Figure S2, middle rows) compared to the cac2Δ mutant (Supplementary Figure S2, bottom rows) which could stem from the higher levels of Cse4 protein when Psh1 is absent. However, the pattern differed substantially from the pattern in the psh1Δ strain, which has higher levels of Cse4 compared to WT (Supplementary Figure S2, top rows). The IP:total chromatin ratio for each mutant was divided by the same ratio for WT for 300 bp bins for Chromosome 3 and plotted as a box plot (Figure 4B). The differences in Cse4 signal were statistically significantly different between WT and each of the mutants, with higher Cse4 levels in the psh1Δ strain and lower Cse4 levels in the cac2Δ and psh1Δ cac2Δ mutant strains. The P-values approach zero when all chromosomes are considered together (Figure 4B). Overall the pattern suggests that CAF-1 is responsible in large part for Cse4 incorporation not only in the psh1Δ strain but also in a WT background.
Figure 4.
CAF-1 promotes localization of Cse4 genome-wide. (A) The heat map shows Cse4 patterns of incorporation for chromosome III. Cse4-ChIP-seq ratio tracks were generated using Cse4 levels from psh1Δ, psh1Δ cac2Δ and cac2Δ strains relative to WT. The grid library in R was used to generate heat maps. Red and blue indicates relative enrichment and depletion of Cse4 signal, respectively. The telomeres and the centromere are indicated. Both telomeres on Chromosome III are X-type. (B) The IP:total chromatin ratio for each mutant was divided by the IP:total chromatin ratio for WT for 300 bp bins across chromosome III, and across the entire genome. For chromosome III the P-value for each mutant compared to WT is <0.005 and for the genome, the P-value for each mutant compared to WT is <2.2e-16. (C) The mean IP:total chromatin signal for peaks of Cse4 is plotted for each of the four genotypes indicated, P < 2.2e-16 for each mutant compared to WT. (D) The overlap of the peaks is shown for each genotype. (E) The number of peaks observed is plotted with a symbol indicating the number expected for each genotype. Genic and intergenic regions are in red and blue, respectively, and intergenic regions are further broken down by promoter (green) and non-promoter (gray).
To further assess Cse4 misincorporation, we used PE MACS to determine peaks of Cse4 in each of the four genetic backgrounds. The mean of the peak signal for each of the four genotypes is plotted (Figure 4C). The average peak signal in the psh1Δ background is higher than WT while the average peak signal without CAC2 is significantly lower than WT, consistent with the idea that yCAF-1 promotes deposition of Cse4. We examined the overlap of peak locations in the different genetic backgrounds and found the most unique peaks in the psh1Δ background, although overall, Cse4 incorporation tends to occur in similar positions in all four strain backgrounds (Figure 4D). To further determine where Cse4 is incorporated, we separated the genome into genic and intergenic regions, and further categorized the intergenic regions by promoter and non-promoter regions. We plotted the number of observed peaks and a symbol that indicates the expected number of peaks for each type of region. The number of expected peaks per region was determined by taking the number of each type of region in the genome, and then calculating the number of peaks expected if the peaks were distributed randomly. This analysis suggests that Cse4 is depleted on genes in all backgrounds and enriched in intergenic regions, and especially at promoter regions (Figure 4E). Taken together our data suggests that Cse4 incorporation may be most predominant at promoter regions but is diminished in the absence of yCAF-1.
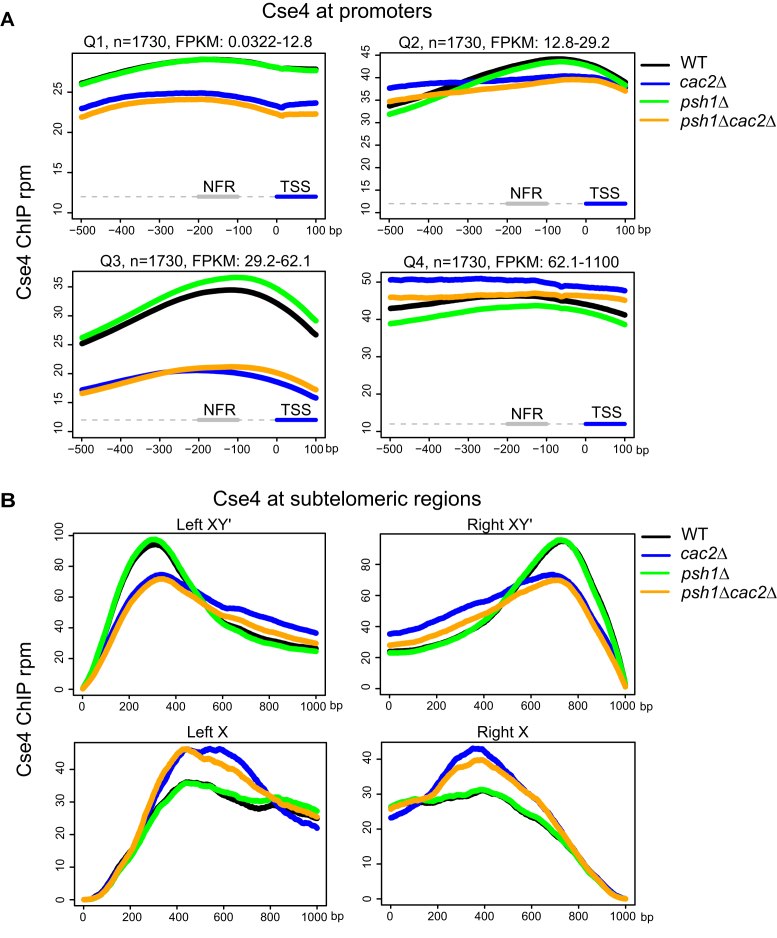
The pattern of incorporation in a psh1Δ strain is associated with poor growth. Ectopic localization of Cse4 at promoters in a psh1Δ strain is associated with decreased gene expression, suggesting misincorporation of Cse4 at promoters might compromise gene expression (31) and growth. We generated metagene plots to examine Cse4 enrichment at promoters of genes binned into quartiles based on the transcription levels in a WT strain overexpressing Cse4, ranked from lowest transcription to highest transcription (Q1–Q4) (Figure 5A). We found evidence that CAF-1 contributes to misincorporation of Cse4 at promoter regions of genes for medium to highly transcribed genes (Q3). In the psh1Δ and WT strains, Cse4 accumulates in the promoter regions of these genes (black and green lines, Figure 5A), with slightly more incorporation in the psh1Δ background. This pattern is resolved when CAC2 is absent. Thus CAF-1 appears to be responsible for Cse4 incorporation into active promoter regions into both the WT and psh1Δ strains.
Figure 5.
CAF-1 promotes localization of Cse4 to active gene promoters and subtelomeric regions. (A) Metagene plots showing mean Cse4-ChIP coverage at gene promoters. Genes were binned into quartiles based on the transcription levels in a WT strain overexpressing Cse4. Q1, Q2, Q3 and Q4 are gene expression quartiles from lowest to highest expression. A segment of −500 to +100 bp relative to the TSS is shown. The nucleosome free region (NFR, −200 to −100 bp) is indicated. (B) Metagene plots showing mean Cse4-ChIP coverage at X and XY’ type telomeres. A segment of 1000 bp from left or right telomere ends are shown separately. In (A and B), the y-axis is the Cse4 signal in mean rpm and the x-axis is the position in base pairs.
Metagene plots of XY’ subtelomeric regions, which are transcriptionally active due to the presence of an expressed helicase gene, show a similar pattern observed at the gene promoters in Q1–Q3 (Figure 5B). Transcriptionally inactive subtelomeric regions, which contain only X, do not show this pattern. Cse4 has been previously shown to accumulate at nucleosomes that turn over due to transcription (58); we suggest this accumulation may be primarily due to incorporation by CAF1. We do not currently understand why this is not the case for the highly transcriptionally active genes in the Q4 grouping, but we speculate that additional chaperones and nucleosome remodelers may influence the dynamics at these regions.
Gene expression and Cse4 misincorporation
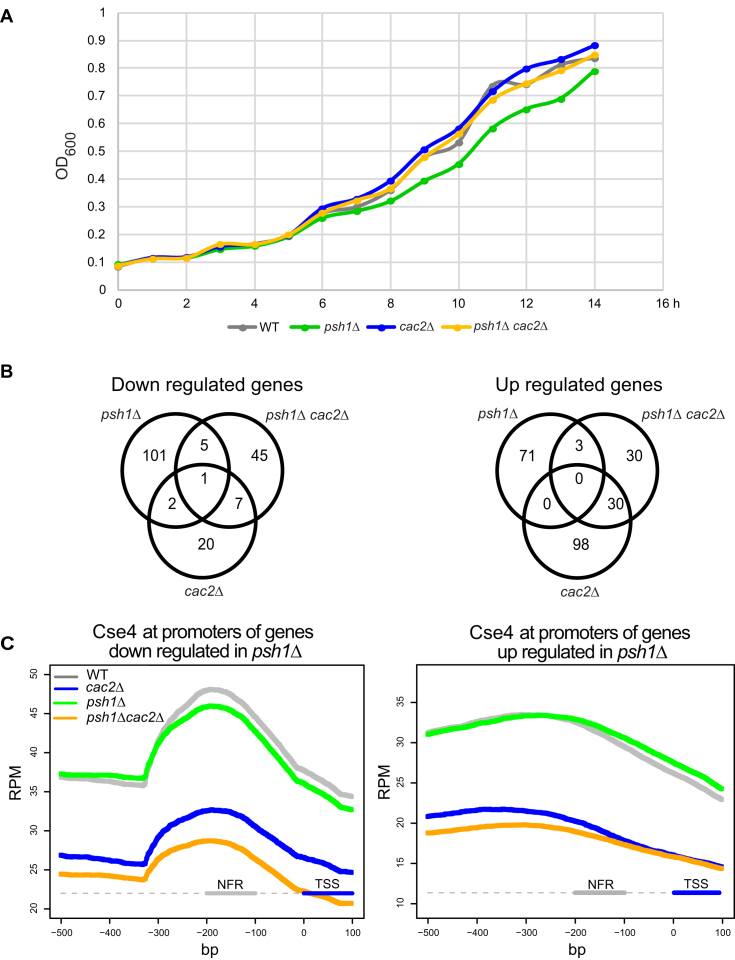
To examine how deposition of Cse4 nucleosomes affects gene expression, we carried out RNA seq experiments. To begin, we determined a timepoint of galactose induction at which the differences in growth between the strains becomes apparent (Figure 6A). We postulated that at this time gene expression could be compromising growth. We collected RNA from two biological replicates from strains with each of the four genotypes after 10 h galactose induction. The expression profiles were compared. While all three mutant genotypes had differentially expressed genes compared to the WT strain, the psh1Δ strain had the most differentially expressed genes (Figure 6B and Supplementary Table S2). The Venn diagram shows that in the psh1Δ cac2Δ double mutant strain, fewer genes were differentially expressed as compared to psh1Δ (121 versus 183 based on adjusted P-value < 0.01 and fold change >1.5). If instead we compare the number of differentially expressed genes based solely on an adjusted P-value < 0.01, we get 571 genes differentially expressed in psh1Δ and 328 genes differentially expressed in the double mutant. Taken together our results are consistent with the idea that gene expression is most disrupted in the psh1Δ background and could be one contributor to the slow growth phenotype, although it is likely that disruption of additional chromatin-based processes also contributes to slow growth.
Figure 6.
CAF-1 is responsible for Cse4 incorporation at promoters of differentially expressed genes. (A) Growth curves show OD600 of WT, psh1Δ, cac2Δ and psh1Δ cac2Δ strains over time after galactose induction of Cse4 overexpression. Overnight cultures grown in raffinose medium were diluted to OD600 of 0.1. After 2 h at 30°C, Cse4 overexpression was induced by adding galactose to a final concentration of 4% (t 0) and cultures were grown for 14 h. OD600 was measured at each hour after galactose addition. (B) Venn diagrams indicate the number of significantly up- and downregulated genes between psh1Δ, cac2Δ and psh1Δ cac2Δ strains relative to WT (adjusted P < 0.01 and fold change > 1.5). (C) Metagene plots show the mean Cse4-ChIP coverage at promoters of genes downregulated or upregulated in the psh1Δ strain relative to WT. A segment of −500 to +100 bp relative to TSS is shown. The nucleosome free region (NFR, −200 to -100 bp) is indicated.
To further understand the gene expression profiles in the different mutant backgrounds, we carried out gene ontology (GO) analysis. Downregulated genes in the psh1Δ strain are enriched for biological processes such as ion transport, glutathione metabolic processes, and aldehyde metabolic processes whereas upregulated genes are enriched for oxidation-reduction and carbohydrate transport (Supplementary Figure S3). Misregulation of these important metabolic processes could contribute to poor growth in the psh1Δ background. Furthermore, this pattern is substantially different from the profile in the double mutant (for a complete list of genes see Supplementary Table S2). Taking the 571 and 328 genes differentially expressed in the psh1Δ background as compared the double mutant, only 71 are in common. The GO terms associated with the 500 genes uniquely differentially expressed in the psh1Δ background include oxidation-reduction, electron transport and purine metabolism whereas the GO terms associated with the differentially expressed genes are completely non-overlapping in the psh1Δ cac2Δ background and include transposition, spore wall biogenesis and reproductive processes (Supplementary Figure S4), but overlap with the GO terms for genes differentially expressed in the cac2Δ strain. Furthermore, GO term analysis for differentially expressed genes specific to psh1Δ and not occurring in the psh1Δ cac2Δ background, reveal similar GO terms obtained for overall differential expression in the psh1Δ background, indicating this is a specific signature of the psh1Δ background that is absent in the double mutant. Therefore, expression of genes critical for metabolic processes and growth are unaffected when CAC2 is deleted, possibly contributing to the restoration of normal growth in the psh1Δ cac2Δ double mutant and cac2Δ background relative to the psh1Δ background.
The gene expression pattern in the psh1Δ background is likely a combination of direct effects due to genome-wide deposition of Cse4 and indirect effects. We speculated that the deposition of Cse4 may compromise the expression of some genes in the psh1Δ background which does not occur when CAF1 function is reduced or absent. We used our ChIP seq data to examine Cse4 levels at the promoters of genes that were downregulated in the psh1Δ background (affected promoters). These promoters had evidence of Cse4 incorporation that was strongly reduced when CAC2 was deleted. The same trend was observed for upregulated genes, but the peak of deposition was not nearly as striking and was much more diffuse (Figure 6C). The trends most resemble the pattern observed for the Q3 genes in Figure 5A and are overall consistent with the idea that CAF1 may be responsible for depositing Cse4 at transcriptionally active promoters. However, it is not clear that Cse4 deposition is inhibiting gene expression since a similar pattern is observed in WT. Many of the effects on gene expression are likely to be indirect. Processes in addition to promoter deposition of Cse4, and accumulation of targets in addition to Cse4, may contribute to poor growth in the psh1Δ background. Although the metagene plots of promoters suggest Cse4 is present at similar levels in WT and psh1Δ strains, in the psh1Δ background we speculate there could be more H3-Cse4 hybrid nucleosomes which are ultrastable (59) and could present an issue for chromatin based processes. Overall the poor growth in the psh1Δ strain is likely to be due to multiple factors, with the effect of Cse4 deposition on gene expression being only one of them.
CAF-1 may regulate ubiquitylation of Cse4
The Scm3 chaperone protects Cse4 from ubiquitylation by Psh1 in vitro (10). Therefore, we examined the role of CAF-1 in proteolysis of Cse4 by Psh1. We measured ubiquitylation of Cse4 by Psh1 in the presence or absence of CAF-1 using recombinant proteins in vitro (Supplementary Figure S5A). Ubiquitylated proteins were pulled down from reaction mixtures using poly-ubiquitin affinity resin and eluates were probed using western blotting with anti-HA-HRP (to HA-ubiquitin) and anti-Cse4 antibodies. The reaction in the presence of CAF-1 (Supplementary Figure S5A, sample 3) shows more ubiquitylated Cse4 compared to the reaction without CAF-1 (sample 1), suggesting CAF-1 can promote ubiquitylation of free Cse4, opposite to the effect of Scm3. To further explore whether there is a direct interaction between CAF-1 and Psh1 in vivo that could promote their cooperation in the destruction of Cse4, we examined whether they co-immunoprecipitate in whole cell extracts, However, CAF-1 does not co-immunoprecipitate with Psh1 (Supplementary Figure S5B). Instead, we speculate that soluble Cse4 bound to CAF-1 may expose ubiquitylation sites on Cse4, promoting ubiquitylation by Psh1.
CAF-1 and centromeric localization of Cse4
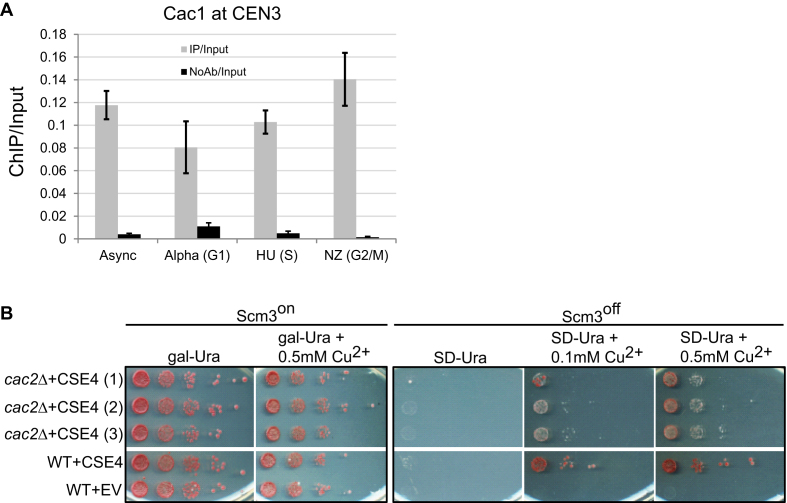
Previous findings suggest that CAF-1 may facilitate centromeric localization of CenH3. The Cac3 orthologs, Mis16 in fission yeast and RbAp46/RbAp48 in human, are required for efficient Cnp1 and CENP-A loading at centromeres, respectively (34,60). Interestingly, HJURP (human ortholog of Scm3) contains five highly conserved tryptophan residues resembling the tryptophan-aspartate (WD40) repeats found in CAF-1 subunits p60 and RbAp48/RbAp46 (8). In budding yeast CAF-1 subunits are recruited to centromeres and CAF-1 and Hir1 are required for proper centromere/kinetochore structure and function (32). We tested whether the localization of CAF-1 at centromeres is cell-cycle dependent. We used asynchronously growing cells and also G1, S and G2/M arrested cells to perform Cac1 ChIP followed by qPCR to detect CEN3 enrichment (Figure 7A). We detected CEN3 enrichment in all four samples, indicating that CAF-1 localizes to centromeres regardless of the phase of the cell cycle.
Figure 7.
When Scm3 is absent and Cse4 levels are high, CAF-1 may help target Cse4 to the centromere. (A) Cac1 localization to centromeres is cell-cycle independent. ChIP was performed for Cac1 using a FLAG-tagged strain using asynchronously growing cells and also G1, S and G2/M arrested cells. qPCR was used to detect Cac1 levels at CEN3. The y-axis indicates arbitrary units representing the Cac1 enrichment at CEN3 from ChIP performed with and without antibody, with respect to the signal for total chromatin for each sample. The error bars represent the standard deviation for ChIP performed in triplicate. (B) Rescue of Scm3off strain by overexpression of Cse4 is less efficient when CAC2 is deleted. Strains containing a single copy of Scm3 under the control of the gal promoter, allowing the expression of Scm3 to be controlled with glucose/galactose, were used in the spotting assay. Cse4 was under the control of a copper-inducible promoter on a plasmid. ‘EV’ indicates empty vector. When Scm3 is expressed (Scm3on), no growth differences are observed in 10-fold serial dilutions. Induction of Cse4 does not efficiently rescue growth of the Scm3off strain when CAC2 is deleted as compared to WT. Three independent isolates of the cac2Δ strain (1, 2 and 3) were tested.
Although Scm3 is the main chaperone responsible for assembly of Cse4 nucleosomes at centromeres in budding yeast, we wondered whether CAF-1 could in principle facilitate the assembly of Cse4 at centromeres. CAF-1 can assemble Cse4 nucleosomes in vitro on a plasmid containing centromere sequence (Figure 2). Although deletion of Scm3 is lethal, Scm3 can be deleted or turned off (Scm3off) if Cse4 is overexpressed, and Cse4 still localizes to the centromere (36), arguing other chaperones can mediate the assembly of Cse4 nucleosomes. To test whether CAF-1 might be able to assemble Cse4 at centromeres, we used the Scm3on/off strain, which can be toggled by galactose, along with copper-inducible Cse4 overexpression. The level of Cse4 can be controlled by the concentration of copper. In this strain background we deleted genes encoding different chaperones, including CAC2 from CAF1. A growth assay was performed to compare growth under Scm3on and Scm3off conditions (Figure 7B). Three independent transformants with CAC2 deletion were examined for growth. When Scm3 is on (Scm3on), there is no difference in growth between WT and cac2Δ strains. However, when CAC2 is deleted and Scm3 is turned off, the rescue of growth by Cse4 overexpression is poor (Figure 7B, compare WT+CSE4 and cac2Δ+CSE4). Deletion of CAC2 from the Scm3off strain does not completely abolish growth when Cse4 is overexpressed, suggesting either a partially functioning CAF-1 complex or some other chaperone might target Cse4 to centromeres under these conditions. These results suggest that when Scm3 is absent and Cse4 levels are high, CAF-1 may be the primary chaperone targeting Cse4 to the centromere. Deletion of the genes encoding chaperones Nap1, Vps75 and Hir1 did not compromise growth in Scm3off conditions (Supplementary Figure S6). Even though deletion of the gene encoding chaperone Rtt106 compromised growth in Scm3off conditions (Supplementary Figure S6) the growth defect is not as severe as loss of CAC2. Furthermore, the overexpression of Cse4 in rtt106Δ cells causes a growth defect (Supplementary Figure S1B), making it difficult to interpret the growth defect under Scm3off conditions since Cse4 is overexpressed. While a previous report suggested that CAF-1 subunits can recruit the chaperone Scm3 to centromeres in S. pombe (33), our result suggests that when Cse4 is present at high levels, CAF-1 can locate to centromeres and incorporate Cse4 independent of Scm3. We speculate that under normal conditions, these two chaperones may both play important roles in the deposition of Cse4 at centromeres.
DISCUSSION
Our data are consistent with CAF-1 being the main H3 chaperone in S. cerevisiae that can interact with and deposit Cse4. CAF-1 can deposit Cse4 at promoter nucleosomes, especially those experiencing medium to high turnover. We examined multiple other chaperones for physical interaction with Cse4 and for growth rescue of psh1Δ strains under Cse4 overexpression conditions and in both respects CAF-1 was unique. Although Cse4 is still misincorporated into chromatin in the absence of CAF-1 when Cse4 is overexpressed, suggesting other chaperones can incorporate Cse4 when Cse4 levels are high, CAF-1 is likely unmatched with respect to genome-wide Cse4 deposition in S. cerevisiae. We speculate that the ability of Cse4/CenH3 to physically interact with subunits of CAF-1 in yeast, flies and human is unlikely to be an evolutionary accident, and that CAF-1 or subunits thereof play an evolutionarily conserved role in Cse4 biology that have been difficult to discern since CAF-1 is also an H3/H4 chaperone.
Misregulation of CENP-A and the factors that regulate CENP-A localization are hallmarks of many human cancers (22). Proper stoichiometric balance between CenH3 and its canonical counterparts is critical for exclusive centromere targeting of CenH3 by chaperones. Overexpressed CenH3 may lead to the unintended deposition of CenH3 by H3 chaperones with deleterious consequences. Chaperones that misincorporate CenH3 into non-centromeric sites are beginning to be identified. In human cancer cells, overexpressed CENP-A occupies transcription factor binding sites and subtelomeric locations, potentially chaperoned in part by the transcriptionally coupled histone variant H3.3 chaperones DAXX and ATRX (23,26,27). We present evidence that the evolutionarily conserved histone H3/H4 chaperone CAF-1 facilitates Cse4 misincorporation at active gene promoters and subtelomeric regions (XY’ type). Under normal cellular conditions, histone H3 is present in large excess over Cse4. Under these conditions, misincorporation of Cse4 occurs at low levels and may not be particularly harmful. However, when Cse4 is overexpressed, CAF-1-mediated assembly of Cse4 may become deleterious. We suggest that Cse4 incorporation may negatively impact multiple chromatin-based processes and ultimately, growth.
CAF-1 is normally considered a replication and repair coupled histone H3 chaperone. However, Cse4/H3 exchange mediated by CAF-1 can be replication independent (35), suggesting CAF-1 may play a role in replication independent nucleosome assembly. Interestingly, the human CAF-1 subunit p60 was one of the overexpressed chaperones in CENP-A overexpressing breast cancer cells (26). Our study indicates that CAF-1 can target Cse4 into promoters and subtelomeric regions in budding yeast; given the evolutionary conservation of both the proteins and the pattern of mislocalization, we suggest human CAF-1 could be partly responsible for mislocalization of CENP-A in cancer cells.
When Cse4 is overexpressed, it may form histone complexes not present under normal conditions. Cse4 may normally exist predominately as a dimer with H4. However, when the Cse4 levels are high, there may be an excess of Cse4 monomers in the soluble pool. This free Cse4 may compete with histone H3 and bind to CAF-1. CAF-1 association with free Cse4 may promote ubiquitylation and proteolysis. In this way CAF-1 may cooperate with Psh1 to regulate proteolysis of Cse4. Analysis of ectopic CENP-A nucleosomes from colorectal cancer cells reveals a subpopulation of structurally distinct hybrid nucleosomes containing CENP-A and H3.3 (23,26). This CENP-A/H3.3 nucleosome forms a highly stable structure compared to CENP-A nucleosomes (59). In budding yeast Cse4 nucleosomes are highly enriched at sites of high-nucleosome turnover genome-wide (58). We speculate CAF-1 may facilitate assembly of heterotypic (Cse4/H3) octasomes at gene promoters and active subtelomeric regions when Cse4 levels are high and proteolytic removal of Cse4 by Psh1 is compromised. These unusual nucleosomes might pose difficulties for chromatin-based processes.
A recent report (31) indicates Cse4 misincorporation can negatively affect gene expression. Our results suggest the majority of differential gene expression in the psh1Δ mutant may not be directly attributed to Cse4 at the promoter nucleosome. Instead, Cse4 deposition may affect multiple processes, including gene expression, that impact growth. We highlight one molecular mechanism by which misincorporation can occur by identifying and characterizing an evolutionarily conserved histone H3 chaperone CAF-1 that can incorporate Cse4 into non-centromeric nucleosomes.
DATA AVAILABILITY
Genomic data been deposited with GEO under accession number GSE98397.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Manjunatha Shivaraju and Priyanka Nandakumar for technical assistance. We thank Karolin Luger (University of Colorado Boulder) for providing baculovirus for CAF-1 purification. We thank Richard Shrock for assistance with manuscript preparation. The authors have no competing financial interests to declare.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at https://www.stowers.org/research/publications/libpb-1279.
FUNDING
National Institutes of Health [R01 GM080477 to J.G.]; Stowers Institute for Medical Research. Funding for open access charge: Stowers Institute for Medical Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Choy J.S., Mishra P.K., Au W.C., Basrai M.A.. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2012; 1819:776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Das C., Tyler J.K., Churchill M.E.. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 2010; 35:476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C.. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007; 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- 4. Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R.E.. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U.S.A. 2007; 04:10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L.. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007; 26:853–865. [DOI] [PubMed] [Google Scholar]
- 6. Shivaraju M., Camahort R., Mattingly M., Gerton J.L.. Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 2011; 286:12016–12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G.. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009; 137:485–497. [DOI] [PubMed] [Google Scholar]
- 8. Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. 3rd, Bassett E.A., Wood S., Black B.E., Cleveland D.W.. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009; 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins K.A., Furuyama S., Biggins S.. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004; 14:1968–1972. [DOI] [PubMed] [Google Scholar]
- 10. Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., Florens L., Workman J.L., Gerton J.L.. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell. 2010; 40:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ranjitkar P., Press M., Yi X., Baker R., MacCoss M.J., Biggins S.. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 2010; 40:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hewawasam G.S., Mattingly M., Venkatesh S., Zhang Y., Florens L., Workman J.L., Gerton J.L.. Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J. Bio.l Chem. 2014; 289:29297–29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Au W.C., Dawson A.R., Rawson D.W., Taylor S.B., Baker R.E., Basrai M.A.. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013; 194:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng H., Bao X., Rao H.. The F-box protein Rcy1 is involved in the degradation of histone H3 variant Cse4 and genome maintenance. J. Biol. Chem. 2016; 291:10372–10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohkuni K., Takahashi Y., Fulp A., Lawrimore J., Au W.C., Pasupala N., Levy-Myers R., Warren J., Strunnikov A., Baker R.E. et al.. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell. 2016; 27:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohkuni K., Abdulle R., Kitagawa K.. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl cis-trans isomerase. Genetics. 2014; 196:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mishra P.K., Guo J., Dittman L.E., Haase J., Yeh E., Bloom K., Basrai M.A.. Pat1 protects centromere-specific histone H3 variant Cse4 from Psh1-mediated ubiquitination. Mol. Biol. Cell. 2015; 26:2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Canzonetta C., Vernarecci S., Iuliani M., Marracino C., Belloni C., Ballario P., Filetici P.. SAGA DUB-Ubp8 deubiquitylates centromeric histone variant Cse4. G3 (Bethesda). 2016; 6:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deyter G.M., Biggins S.. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 2014; 28:1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi E.S., Stralfors A., Catania S., Castillo A.G., Svensson J.P., Pidoux A.L., Ekwall K., Allshire R.C.. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 2012; 8:e1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gkikopoulos T., Singh V., Tsui K., Awad S., Renshaw M.J., Scholfield P., Barton G.J., Nislow C., Tanaka T.U., Owen-Hughes T.. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011; 30:1919–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zink L.M., Hake S.B.. Histone variants: nuclear function and disease. Curr. Opin. Genet. Dev. 2016; 37:82–89. [DOI] [PubMed] [Google Scholar]
- 23. Athwal R.K., Walkiewicz M.P., Baek S., Fu S., Bui M., Camps J., Ried T., Sung M.H., Dalal Y.. CENP—a nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet. Chromatin. 2015; 8:2–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F.. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003; 63:3511–3516. [PubMed] [Google Scholar]
- 25. Li Y., Zhu Z., Zhang S., Yu D., Yu H., Liu L., Cao X., Wang L., Gao H., Zhu M.. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011; 6:e17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G.. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014; 53:631–644. [DOI] [PubMed] [Google Scholar]
- 27. Shrestha R.L., Ahn G.S., Staples M.I., Sathyan K.M., Karpova T.S., Foltz D.R., Basrai M.A.. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 2017; 8:46781–46800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Au W.C., Crisp M.J., DeLuca S.Z., Rando O.J., Basrai M.A.. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008; 179:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amato A., Schillaci T., Lentini L., Di Leonardo A.. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer. 2009; 8:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collins K.A., Camahort R., Seidel C., Gerton J.L., Biggins S.. The overexpression of a Saccharomyces cerevisiae centromeric histone H3 variant mutant protein leads to a defect in kinetochore biorientation. Genetics. 2007; 175:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hildebrand E.M., Biggins S.. Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 2016; 12:e1005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharp J.A., Franco A.A., Osley M.A., Kaufman P.D.. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002; 16:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X. et al.. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009; 33:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M.. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004; 118:715–729. [DOI] [PubMed] [Google Scholar]
- 35. Lopes da Rosa J., Holik J., Green E.M., Rando O.J., Kaufman P.D.. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011; 187:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L.. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009; 35:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winkler D.D., Zhou H., Dar M.A., Zhang Z., Luger K.. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012; 40:10139–10149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995; 57:289–300. [Google Scholar]
- 40. Ramirez-Parra E., Gutierrez C.. The many faces of chromatin assembly factor 1. Trends Plant Sci. 2007; 12:570–576. [DOI] [PubMed] [Google Scholar]
- 41. Furuyama T., Dalal Y., Henikoff S.. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Furuyama T., Henikoff S.. Biotin-tag affinity purification of a centromeric nucleosome assembly complex. Cell Cycle. 2006; 5:1269–1274. [DOI] [PubMed] [Google Scholar]
- 43. Mattiroli F., Gu Y., Balsbaugh J.L., Ahn N.G., Luger K.. The Cac2 subunit is essential for productive histone binding and nucleosome assembly in CAF-1. Sci. Rep. 2017; 7:46274–46285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polo S.E., Roche D., Almouzni G.. New histone incorporation marks sites of UV repair in human cells. Cell. 2006; 127:481–493. [DOI] [PubMed] [Google Scholar]
- 45. Metzger M.B., Scales J.L., Dunklebarger M.F., Weissman A.M.. The ubiquitin ligase (E3) Psh1p Is Required for Proper Segregation of both Centromeric and Two-Micron Plasmids in Saccharomyces cerevisiae. G3 (Bethesda). 2017; 7:3731–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng H., Bao X., Gan X., Luo S., Rao H.. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci. Rep. 2017; 7:8565–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bowman A., Ward R., Wiechens N., Singh V., El-Mkami H., Norman D.G., Owen-Hughes T.. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol. Cell. 2011; 41:398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silva A.C., Xu X., Kim H.S., Fillingham J., Kislinger T., Mennella T.A., Keogh M.C.. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 2012; 287:1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spector M.S., Raff A., DeSilva H., Lee K., Osley M.A.. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 1997; 17:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sutton A., Bucaria J., Osley M.A., Sternglanz R.. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001; 158:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adkins M.W., Howar S.R., Tyler J.K.. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004; 14:657–666. [DOI] [PubMed] [Google Scholar]
- 52. Williams S.K., Truong D., Tyler J.K.. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A.. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007; 27:393–405. [DOI] [PubMed] [Google Scholar]
- 54. Burgess R.J., Zhang Z.. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013; 20:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang S., Zhou H., Katzmann D., Hochstrasser M., Atanasova E., Zhang Z.. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13410–13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang S., Zhou H., Tarara J., Zhang Z.. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007; 26:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang J., Zhang X., Feng J., Leng H., Li S., Xiao J., Liu S., Xu Z., Xu J., Li D. et al.. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016; 14:1128–1141. [DOI] [PubMed] [Google Scholar]
- 58. Krassovsky K., Henikoff J.G., Henikoff S.. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., Kagawa W., Fukagawa T., Almouzni G., Kurumizaka H.. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 2014; 4:7115–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mouysset J., Gilberto S., Meier M.G., Lampert F., Belwal M., Meraldi P., Peter M.. CRL4(RBBP7) is required for efficient CENP-A deposition at centromeres. J. Cell Sci. 2015; 128:1732–1745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data been deposited with GEO under accession number GSE98397.