Abstract
Cell division of rod-shaped bacteria requires the Z ring, a ring of FtsZ filaments associated with the inner-membrane wall. The MinCDE proteins help localize the Z ring to the center of the Escherichia coli cell. MinC, which inhibits Z-ring assembly, is a passenger on MinD. Previous studies have shown that MinC–MinD from E. coli and Aquifex aeolicus assemble in vitro into extended filaments with a 1:1 stoichiometry. However, a recent study has raised questions about the function of the MinC–MinD copolymer in vivo, because its assembly appears to require a high concentration of these two proteins and has a long lag time, and its blockade does not affect in vivo activities. Here, we found that MinC and MinD from Pseudomonas aeruginosa coassemble into filaments with a 1:1 stoichiometry. We also found that the minimal concentration of ∼4 μm required for assembly applies only to MinD because above 4 μm MinD, even very low MinC concentrations sustained coassembly. As previously reported, the MinC–MinD coassembly exhibited a long lag of ∼100 s when initiated by ATP. Premixing MinD with ATP eliminated this lag, suggesting that it may be due to slow MinD dimerization following ATP activation. We also discovered that MinC–MinD copolymers quickly bound FtsZ filaments and formed huge bundles. Our results resolve previous questions about the low concentration of MinC and the lag time, insights that may inform future investigations into the exact role of the MinC–MinD copolymer in vivo.
Keywords: bacteria, cell division, electron microscopy (EM), protein dynamic, cytoskeleton, FtsZ, min system, MinC–MinD copolymer, MinCDE proteins, Z-ring
Introduction
Division of rod-shaped bacteria is effected by the Z ring, a ring of FtsZ filaments that may constrict the inner membrane and serves to dock downstream proteins that remodel the peptidoglycan wall (1–4). The Z ring is localized with remarkable precision, within 2.9% standard deviation from the center of the rod (5). Localization is imposed by two systems: nucleoid exclusion and MinCDE (6–8), although there is evidence for additional factors (9). Our interest here is the Min system, which comprises three proteins that oscillate from one end of the rod to the other, leaving a minimum average concentration at midcell.
The oscillation is set up by a feedback loop of the ATPase MinD and its activator MinE (10–14). ATP-bound MinD favors its dimerization, which exposes its C-terminal amphipathic helices to anchor MinD into the membrane (15, 16). The MinD assembles into a patch at one end of the cell. Its activator MinE stimulates MinD's ATPase, causing dissociation of monomeric MinD from the membrane (11, 17). The released MinD exchanges ADP for ATP and reforms a patch at the opposite end. Self-organized MinDE proteins act as a dynamic reaction–diffusion device leading to their oscillation in vivo (18).
MinC is the protein that inhibits FtsZ, by disassembling FtsZ protofilaments (19), blocking lateral association of protofilaments (20), or both. MinC forms a tight dimer that binds MinD and oscillates with MinD as a passenger. MinC has two domains (21). The N-terminal domain, MinCN, is the primary inhibitor of FtsZ filament assembly (19, 22). The C-terminal domain, MinCC, does three things: it forms a tight homodimer, it binds MinD, and it binds the conserved C-terminal peptide of FtsZ (22). One question for this simple scenario is why the MinC concentration in vivo is so low. It is ∼0.7 μm, which is 6–8 times less than that of MinD and FtsZ (1, 23). How the small amount of MinC regulates Z-ring assembly is unclear.
In the simplest scenario a MinC dimer binds a MinD dimer and is carried as a passenger as the MinD oscillates from one end of the cell to the other; MinD also activates MinC, at least in part by bringing it to the membrane (6, 7). Because its average concentration is highest at the poles, the inhibitory action of MinC blocks Z-ring formation at the poles and favors Z rings at the center. This scenario was complicated by two independent studies, which found that a mixture of MinC and MinD could assemble long filaments (24, 25). Ghosal et al. (24) obtained a crystal structure showing a dimer of MinCC flanked by monomers of MinD, which they used to model an extended filament of alternating dimers of MinC and MinD. The 1:1 stoichiometry of the filaments was confirmed by pelleting assays, which also indicated that a minimal concentration of ∼7 μm was needed for assembly of an equimolar mixture of MinC and MinD. Both groups found that MinE caused the MinCD filaments to disassemble over a period of 10–15 min. They proposed that these copolymers were likely the operational agents in the Min system in vivo.
The proposal that MinCD filaments were the active agents in vivo was challenged by a subsequent study. Park et al. (26) suggested that the copolymers were unlikely to exist in vivo because the previous study (24) showed that the assembly of MinC–MinD copolymers required a high concentration of proteins and had a long lag time. Park et al. (26) created mutants of MinC and MinD that would disrupt the interfaces seen in the crystal structure and showed that heterodimers of the mutants and WT protein were still active in vivo. These heterodimers should block assembly of filaments of MinC–MinD copolymers, suggesting that the filament assembly was not needed for function in vivo.
The previous studies were done with Min proteins from Escherichia coli and Aquifex aeolicus. Here we have extended the study to Min proteins from Pseudomonas aeruginosa. We have confirmed several results of the previous studies and discovered some new features of the coassembly that may be relevant to the Min system in vivo.
Results
MinC–MinD from P. aeruginosa forms copolymers with a 1:1 ratio; assembly requires a minimal concentration of MinD
Recent studies reported that MinC and MinD from E. coli and from A. aeolicus could assemble into copolymers in the presence of ATP (24, 25). In the present study we have confirmed similar copolymers assembled by MinC and MinD from P. aeruginosa. All experiments (except the indicated EDTA) were done in assembly buffer: 50 mm HEPES, pH 7.5, 5 mm MgAc, and 100 mm KAc.
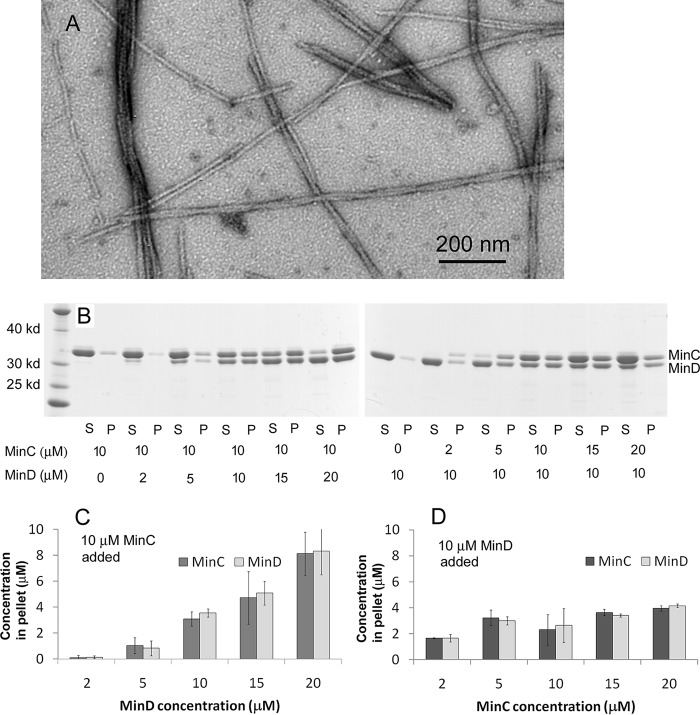
Fig. 1A shows the filamentous polymers observed by negative stain EM. The widths of single copolymeric filaments were 8.5 ± 1.2 nm. Some filaments appeared to be twisted pairs with a pitch ∼170 ± 17 nm, and some further assembled into small bundles. We used sedimentation and SDS-PAGE to analyze the stoichiometry of the copolymers (Fig. 1, B–D). Most copolymers were pelleted following centrifugation at 50,000 rpm. Regardless of the ratio of MinC and MinD in the assembly mixture, the ratio in the pelleted copolymers was always 1:1. Interestingly, the coassembly appeared to require a minimum concentration of ∼4–5 μm MinD, even at high concentrations of MinC. With 10 μm MinC, almost no copolymers were assembled if the MinD concentration was 2 μm (Fig. 1, B, left panel, and C). In contrast, when the MinD concentration was 10 μm, almost all of 2 or 5 μm MinC was in the pellet. The pellet increased only slightly when the MinC concentration increased to 20 μm (Fig. 1, B, right panel, and D). These results confirm the 1:1 stoichiometry and the minimal concentration of ∼7 μm observed by Ghosal et al. (24) for E. coli MinC and MinD. A significant extension in our results is that the minimal concentration applies only to MinD. The lower minimal concentration in our work, 4–5 versus 7 μm, may be a species difference.
Figure 1.
MinC and MinD from P. aeruginosa assembled into copolymers with a 1:1 stoichiometry in the presence of ATP. A, MinC and MinD assembled into straight filaments and bundles observed by EM. Bar is 200 nm. B, SDS-PAGE analysis of cosedimentation in different ratios of MinC and MinD showed that the pellet always contained a 1:1 MinC:MinD, and assembly required a minimum of 4–5 μm MinD. S, supernatant; P, pellet. C and D, the concentrations of MinC and MinD in the pellet were calculated through SDS-PAGE analysis. Each value is the average of four independent analyses.
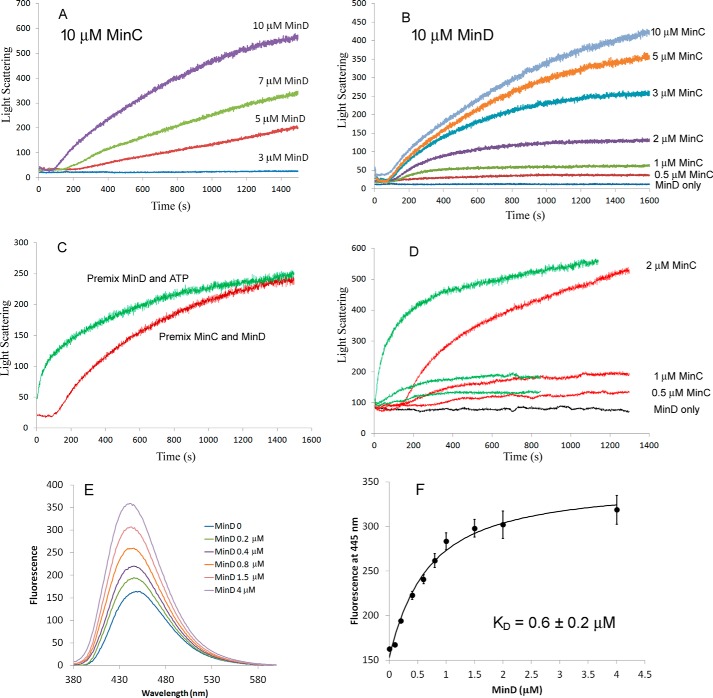
Light scattering was used to follow the kinetics of MinC–MinD copolymer assembly. Fig. 2 (A and B) shows the copolymer assembly kinetics at different concentrations of MinC and MinD. Here we premixed different concentrations of MinC and MinD and tracked the light-scattering signal after adding 1 mm ATP to initiate assembly. The curves displayed a typical cooperative assembly which had several phases: a 100–200 s lag time, followed by a fast-rising phase, and then a slower phase approaching a plateau. The light-scattering measurements confirmed the ∼4–5 μm minimal concentration of MinD needed for coassembly. In 10 μm MinC there was no detectable signal at 3 μm MinD (Fig. 2A). MinC appears to have no minimal concentration, because 10 μm MinD gave a detectable light scattering signal at 2, 1, and even 0.5 μm MinC (Fig. 2, B and D). Also, the lag time was reduced when MinD concentration increased (Fig. 2A) but showed little change when MinC concentration increased (Fig. 2B). With equimolar MinC and MinD, assembly was virtually not detectable for protein concentrations 4 μm or below (data not shown), which is consistent with the previous report using equimolar mixtures of MinC and MinD (24). Our results suggest that the minimal concentration is determined solely by MinD. In the following experiments, we used 10 μm MinD protein and lower concentrations of MinC protein, which is close to the physiological conditions.
Figure 2.
Assembly kinetics measured by light-scattering assay. A, assembly kinetics of 10 μm MinC and different concentration of MinD. B, assembly kinetics of 10 μm MinD and different concentration of MinC. C, comparison of the MinC–MinD copolymer assembly kinetics of premixed MinC and MinD proteins or premixed MinD and ATP. Assembly was initiated by adding ATP or MinC, respectively. MinD was 10 μm, MinC was 3 μm, and ATP was 1 mm. D, comparison of the assembly kinetics of 10 μm MinD and low concentrations of MinC (0.5, 1, or 2 μm). Red curves are premixed MinC and MinD, and green curves are premixed MinD and ATP. ATP was 1 mm. E, the fluorescence recorded from 380 to 600 nm of 0.2 μm mant-ATP after adding different concentration of MinD proteins. F, fluorescence at the 445 nm peak plotted against the concentration of MinD (average and standard deviation of two assays). The curve is fitted using a simple binding model (“Experimental Procedures”) giving a KD of 0.6 μm for the best fit.
As noted above, when assembly was initiated by adding ATP to premixed MinC and MinD, there was a substantial lag (∼100 s; Fig. 2C) before assembly began. We found that the lag was completely eliminated if we premixed MinD and ATP and initiated assembly by adding MinC (Fig. 2, C and D). Fig. 2D shows the assembly kinetics when adding 0.5, 1, or 2 μm MinC into 10 μm MinD premixed with 1 mm ATP (Fig. 2D, green curves), compared with adding 1 mm ATP into premixed 10 μm MinD and MinC (Fig. 2D, red curves). These results suggested that the lag may be due to a slow binding of ATP by MinD or to slow dimerization of MinD-ATP after binding.
To check the binding of ATP by MinD, we used mant-ATP, whose fluorescence increases upon binding to proteins. Fig. 2 (E and F) shows the fluorescence changes of 0.2 μm mant-ATP after adding different concentration of MinD proteins. We found that the binding process was very fast, and the fluorescence increased to a stable peak within seconds (data not shown). The dissociation constant KD = 0.6 ± 0.2 μm was calculated using a one-site binding model, as described under “Experimental procedures.” This suggests that the lag time of MinC–MinD coassembly may be due to the slow dimerization of MinD after it is activated by ATP binding.
Assembly of MinD premixed with ATP, although simplified by elimination of the lag, still shows complex kinetics. Fig. 2D shows a rapid rise up to ∼50 s, followed by a slower rise to ∼1,000 s and perhaps an even slower phase thereafter. We note that light scattering is not a simple measure of polymer mass but is strongly affected by the size of the polymers. Light scattering should therefore be interpreted as a qualitative assay of assembly dominated by bundling.
FtsZ enhanced the formation of the MinC–MinD copolymers
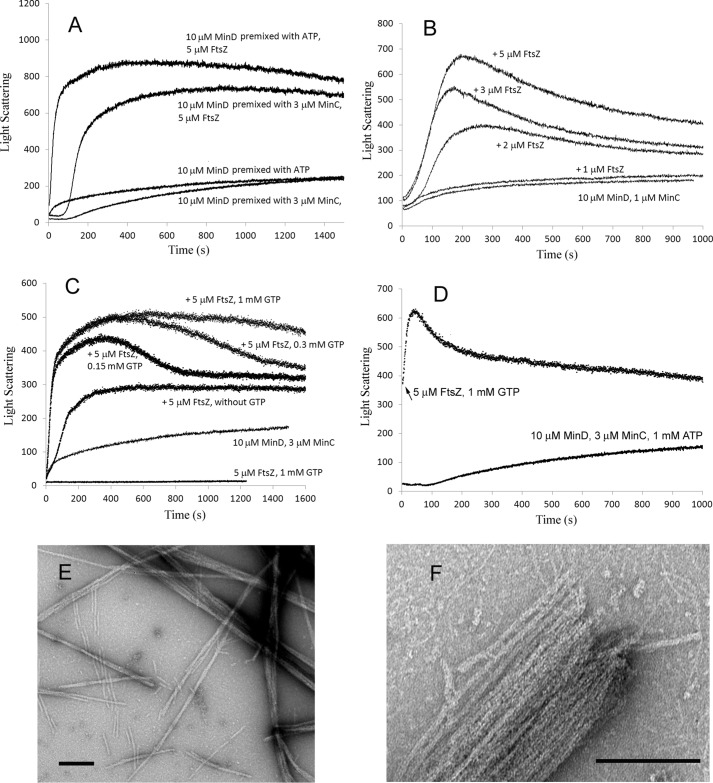
We next investigated the interaction between FtsZ filaments and MinC–MinD copolymers. FtsZ filaments strongly enhanced the final light scattering signal (Fig. 3A). Fig. 3B shows the effect of increasing concentration of FtsZ. The light-scattering signal showed little increase with 1 μm FtsZ, which is close to its critical concentration and therefore lacking assembled FtsZ protofilaments. Higher concentrations of FtsZ accelerated the assembly and enhanced the peak light scattering, suggesting that FtsZ filaments are incorporated into the MinC–MinD copolymers.
Figure 3.
FtsZ enhanced the MinC–MinD copolymer assembly. A, comparison of the assembly kinetics of 10 μm MinD and 3 μm MinC with or without 5 μm FtsZ. The concentration of ATP and GTP was 1 mm. MinD was premixed with MinC or ATP; the premix also contained 5 μm FtsZ (and 1 mm GTP where indicated). Assembly was initiated by adding ATP or MinC. A control of FtsZ-GTP alone is shown in C. Importantly, the single FtsZ filaments have negligible scattering compared with the MinC–MinD bundles. B, assembly kinetics of premixed 10 μm MinD, 1 μm MinC, 1 mm GTP, and different concentrations of FtsZ initiated by adding ATP. C, the assembly kinetics of 10 μm MinD premixed with ATP and initiated by addition of 3 μm MinC plus 5 μm FtsZ preassembled with different concentrations of GTP. D, MinD plus MinC was assembled for 100 s as shown in the bottom curve. Then 5 μm FtsZ, preassembled with 1 mm GTP, was added (arrow for upper curve, whose time 0 corresponds to 1,000 s of the lower curve). E and F, the mixture of MinC, MinD, and FtsZ assembled into large bundles in the presence of ATP and GTP. Bars are 200 nm.
Fig. 3C shows FtsZ-MinC–MinD coassembly at different concentrations of GTP. MinC and MinD could not assemble in the presence of GTP (data not shown). Interestingly, 5 μm FtsZ still enhanced the peak light-scattering signal without GTP. This suggests that oligomers of FtsZ at 5 μm in the presence of Mg-GDP (27) can cross-link MinC–MinD copolymers into larger bundles. In this experiment higher concentrations of GTP maintained a longer plateau of light scattering. This can be attributed to consumption of the GTP by the FtsZ, suggesting that the GTPase and recycling of FtsZ subunits are still occurring in the MinC–MinD–FtsZ bundles.
We next tested the effect of adding FtsZ protofilaments into preassembled MinC–MinD polymers. This caused a rapid increase in turbidity followed by a slow decrease (Fig. 3D). The slow decrease following a peak light scattering may indicate an initial assembly of large bundles followed by rearrangement into smaller ones. This was seen also in Fig. 3B, whereas Fig. 3A showed a more stable plateau. We do not understand the reason for this variability but emphasize again that the light scattering is only a qualitative assay of assembly and is dominated by bundling.
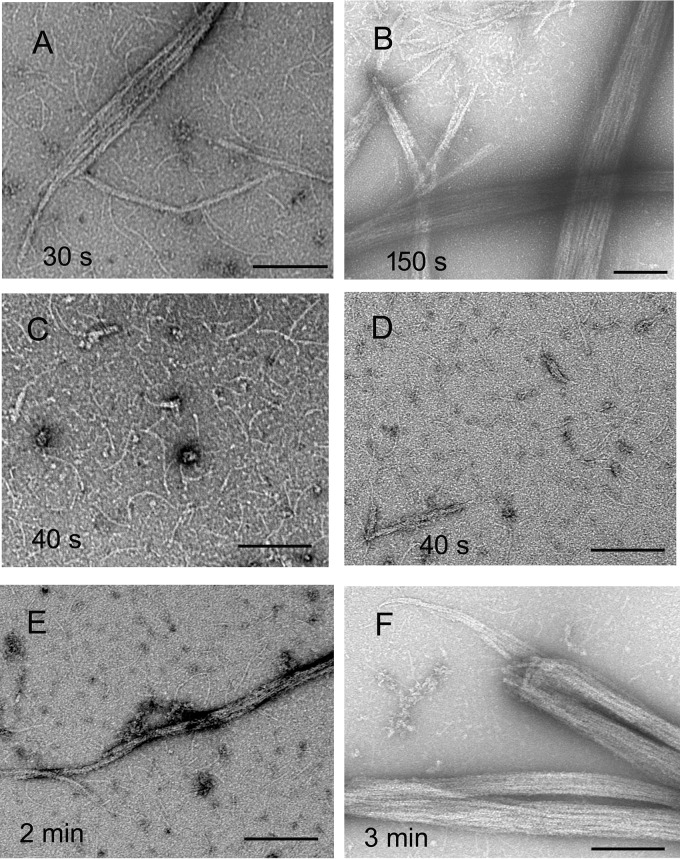
The enhanced light scattering induced by FtsZ suggests that bundling is enhanced by FtsZ. This was confirmed by EM, which showed that MinC, MinD, and FtsZ together assembled into large bundles (Fig. 3, E and F). Some thin filaments likely to be FtsZ are seen outside the large bundles. FtsZ filaments are presumably also inside the bundles but are not resolved. We also imaged the MinC–MinD–FtsZ polymers assembled at different times. Adding MinC into premixed MinD, FtsZ, ATP, and GTP showed a fast assembly without a lag (Fig. 3A). Consistent with this, bundles appeared at 30 s (Fig. 4A), followed by more and much larger bundles after 2.5 min (Fig. 4B). In contrast, when premixed MinC, MinD, and FtsZ proteins were induced by addition of 1 mm ATP and 1 mm GTP, the assembly showed a lag time of ∼100 s (Fig. 3A). EM showed there were only some short filaments assembled at 40 s (Fig. 4, C and D), which resemble single FtsZ protofilaments. After 2 min, some filaments associated into small and medium size bundles (Fig. 4E), and after 3 min, very large bundles appeared (Fig. 4F).
Figure 4.
EM images of MinC–MinD–FtsZ copolymers at different assembly times. A and B, premixed MinD, FtsZ, ATP, and GTP, initiated by addition of MinC, showed bundle formation within 30 s (A) and very large bundles at 150 s (B). C–F, premixed MinC, MinD, and FtsZ induced by the addition of ATP showed only small filaments at 40 s, which is within the lag period (two examples are shown, C and D). Single FtsZ filaments are seen in high contrast in C and in lower contrast in D (this field was chosen to show rare small bundles). Bundles appeared at 2 min (E), and very large bundles were obvious at 3 min (F). The concentration of MinD was 10 μm, MinC was 5 μm, and FtsZ was 5 μm. Both ATP and GTP were 1 mm. Bars are 200 nm.
As described above, we observed that 5 μm FtsZ without GTP enhanced the light scattering signal of MinC–MinD coassembly (Fig. 3C). However, EM of MinC–MinD showed very similar bundles with or without FtsZ (no GTP) in the presence of only ATP (Fig. S1, D and E). A sedimentation assay confirmed that FtsZ without GTP could be copelleted with MinC–MinD copolymers (Fig. S2). The FtsZ monomers may be binding to the outside of the MinC–MinD copolymers, increasing the light scattering without a structural change that is obvious in the EM images. This emphasizes again the qualitative nature of EM.
The effects of MinC alone on the FtsZ filaments
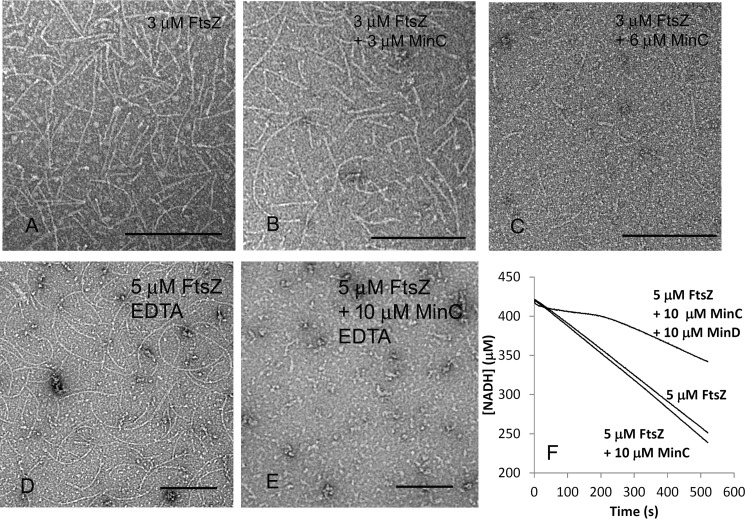
It is generally accepted that MinC is an inhibitor of FtsZ polymerization. We therefore examined the effects of MinC alone on FtsZ assembly. Fig. 5 (A–C) shows EM images of 3 μm FtsZ filaments assembled with or without MinC. With an equimolar amount of MinC (3 μm), the FtsZ protofilaments showed little change from FtsZ alone (Fig. 5, A and B). However, increasing MinC to 6 μm greatly reduced the number and length of FtsZ protofilaments (Fig. 5C). Similar results were obtained when we used 5 μm FtsZ: no effect up to stoichiometric MinC and almost no assembly with a 2-fold excess of MinC (data not shown). We also examined the effects of MinC on FtsZ filaments in EDTA buffer, which blocks the GTPase. Fig. 5 (D and E) shows that a 2-fold excess of MinC virtually eliminated the FtsZ protofilaments, the same as in magnesium. This contrasts with an earlier study of the E. coli proteins, where MinC inhibited FtsZ in magnesium, where GTPase was active, but had no effect when GTPase was blocked by EDTA (20).
Figure 5.
A 2-fold excess of MinC shortens FtsZ filaments. A–C, EM images of 3 μm FtsZ without MinC (A), with 3 μm MinC (B), and 6 μm MinC (C) in 50 mm HEPES, pH 7.5, 100 mm KAc, and 5 mm MgAc. D and E, EM images of 5 μm FtsZ without MinC (D) and with 10 μm MinC (E) in 50 mm MES, pH 6.5, 100 mm KAc, and 1 mm EDTA. F, GTPase activity of 5 μm FtsZ alone, with 10 μm MinC, or 10 μm MinC and MinD. The changing slope of the line for the three-part mixture is probably due to the turbidity of the MinC–MinD bundles. Both ATP and GTP are 1 mm. Bars are 200 nm.
Previous studies have reported that, notwithstanding its inhibition of FtsZ protofilament assembly, MinC had no effect on the GTPase (19, 20). Our results agreed completely: a 2-fold excess of MinC had no effect on the GTPase of 5 μm FtsZ: 19.8 ± 1.5 GTP and 21.1 ± 1.6 GTP per minute in the absence or presence of 10 μm MinC (Fig. 5F and three independent measurements). We also examined whether MinC–MinD copolymers would affect FtsZ GTPase. With the addition of 10 μm MinC and 10 μm MinD, the GTPase activity of 5 μm FtsZ was reduced to 12 ± 2.1 GTP per minute (Fig. 5F). The reduced GTPase activity may be due to reduced subunit exchange of the FtsZ protofilaments incorporated into the large bundles with MinC–MinD filaments. It is clear, however, that FtsZ retained substantial GTPase activity even within these large bundles.
MinE inhibits MinC–MinD copolymers
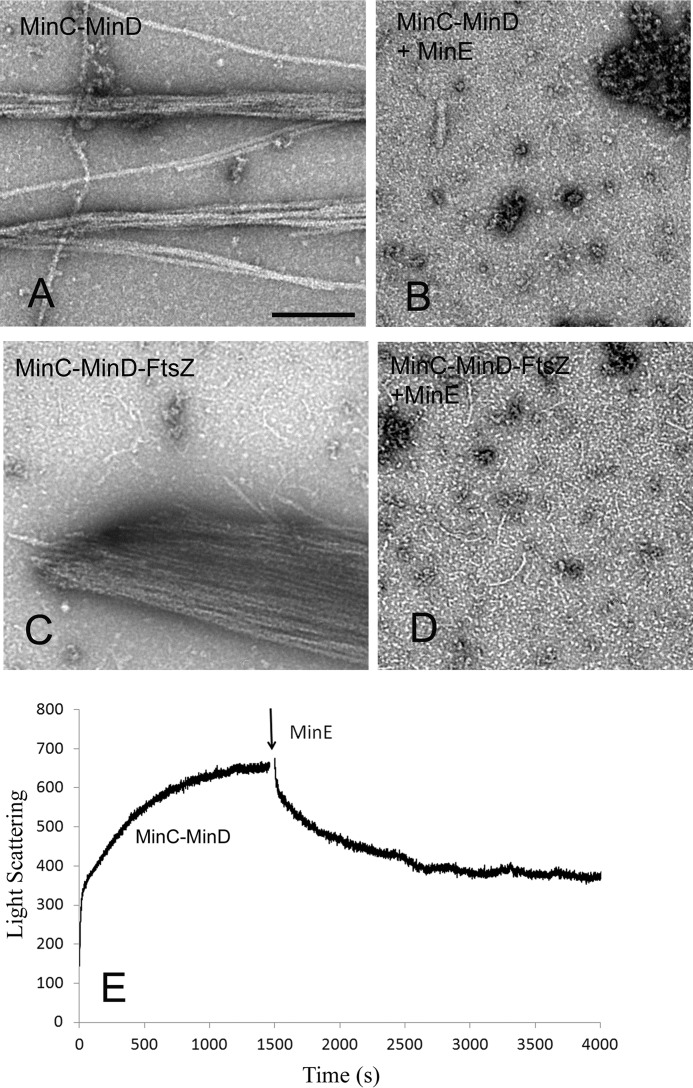
Purified MinE inhibited MinC–MinD copolymer assembly. Fig. 6 compares the EM images of MinC–MinD and MinC–MinD–FtsZ copolymers assembled without and with MinE. 10 μm MinD and 5 μm MinC alone assembled into filament bundles (Fig. 6A). When 15 μm MinE was added before assembly, assembly was almost completely blocked (Fig. 6B). Similar results were observed for MinC–MinD–FtsZ copolymer assembly. The very large bundles of MinC–MinD–FtsZ (Fig. 6C) were completely blocked by 15 μm MinE (Fig. 6D) and only shortened FtsZ filaments were seen.
Figure 6.
MinE inhibited MinC–MinD copolymer formation. A–D, EM images of copolymers with or without MinE. 10 μm MinD plus 5 μm MinC assembles into copolymers and bundles in the presence of 1 mm ATP without MinE (A), but 15 μm MinE completely blocked the assembly (B). 10 μm MinD, 5 μm MinC, and 5 μm FtsZ assembles into large bundles in the presence of 1 mm ATP and GTP without MinE (C), but the bundles are completely blocked by 15 μm MinE (D). The remaining thin filaments are identified as FtsZ protofilaments. E, light-scattering measurements show that MinE disassembles preformed MinC–MinD copolymers. Bar represents 200 nm.
MinE can also disassemble preassembled MinC–MinD copolymers. Fig. 6E shows assembly of MinD plus ATP, induced by MinC at time zero. At 1,500 s, 15 μm MinE was added, and the light-scattering signal decreased slowly to a new plateau equal to that at the end of the initial sharp rise.
Discussion
In this study we have confirmed that MinC and MinD from P. aeruginosa coassemble into filaments with a 1:1 stoichiometry, very similar to the coassembly of the E. coli and A. aeolicus proteins (24, 25). We have confirmed a number of features of the coassembly, and we also extended the study to discover several new features.
We used MinC and MinD proteins from P. aeruginosa because they show a stable activity that may be better than the E. coli proteins. Both E. coli and P. aeruginosa are γ-proteobacteria. MinD and MinE are well-conserved between E. coli and P. aeruginosa; they share 90 and 80% sequence similarity. MinC is much less conserved, with sequence similarity of 54%. The C-terminal of MinC (MinCC), which mediates the interaction with MinD, is more conserved, with sequence similarity of 68%. Interestingly, FtsZ from E. coli could coassemble with MinC–MinD copolymers from P. aeruginosa to form large bundles (data not shown). MinE from E. coli could also prevent the assembly of the MinC–MinD copolymers from P. aeruginosa (data not shown). These cross-species interactions suggest similar mechanisms for the MinC/D/E systems from E. coli and P. aeruginosa.
We discovered new features of the coassembly of MinC and MinD. MinD dominates two properties of coassembly: critical concentration and lag time. If MinD is above ∼4 μm, it will copolymerize all MinC up to a 1:1 stoichiometry. Ghosal et al. (24) reported that an equimolar mixture of MinC and MinD required a minimal concentration of ∼7 μm before they would assemble in solution. We have extended this result and discovered that only the concentration of MinD is important. In our experiments the minimal concentration of P. aeruginosa MinD was ∼4–5 μm. If MinD was below 4 μm, there was no assembly even with 10 μm MinC. In contrast, 10 μm MinD generated assembly of copolymers at MinC concentrations as low as 0.5 μm. The requirement for a large amount of MinD but a low amount of MinC for the coassembly is consistent with the protein concentrations in vivo. Ribosome profiling estimated that there is ∼5 μm MinD and ∼0.7 μm MinC in an E. coli cell (23). This suggests that MinC–MinD could assemble into copolymers in the cytoplasm, the amount being limited by the concentration of MinC. Notably, Ghosal et al. (24) found that MinC–MinD could assemble at much lower concentration if they attached onto membrane surfaces.
The lag could be eliminated by preincubating the MinD with ATP. Because ATP binding to MinD is rapid (Fig. 2E and data not shown), it is likely that the lag is due to slow formation of MinD dimers after binding ATP. The lag may be relevant to the oscillations in vivo, because it would probably apply to MinD released by the action of MinE.
We found that MinE completely blocked assembly of MinC–MinD copolymers when added in excess to MinD before assembly. MinE also caused depolymerization of copolymers when added after assembly. This disassembly was slow, requiring ∼15 min for completion. This confirms the observations of the two previous studies, where disassembly also occurred over 10–15 min (24, 25). This disassembly is much slower than the ∼50-s oscillation of MinD–MinE in vivo, suggesting that the in vivo MinC–MinD polymers may be much shorter than the filament bundles observed in vitro.
An important consideration for mechanisms is the number of molecules and how they might be dispersed. The ribosome profiling study of Li, Weissman, and co-workers (23) determined that there are 857 copies of MinC, 5,358 of MinD, 3,597 of MinE, and 6,750 of FtsZ in an average E. coli MG1655 cell (see Ref. 1 for a summary of their quantitation of all cell division proteins.) If all 430 MinC dimers were incorporated into MinC–MinD copolymers, the 8-nm repeat (24) would mean a total polymer length of 3,440 nm, enough to encircle the cell one time. If the polymer were divided into 10 shorter filaments, they would be just resolved in the light microscope. However, fluorescence microscopy shows that MinC and MinD are not in discrete, resolvable filaments but are in diffuse patches that cover ∼1/3 of the cell when clustered at one pole (10, 11). A bacterial cell 3,000 nm long by 1,000-nm diameter will have a surface area of 9.4 × 106 nm2. If the 2,700 MinD dimers were spaced equally in a patch of 1/3 of the total surface area, they would have 1,160 nm2/MinD dimer, spacing them on average 35 nm apart. MinC dimers would have 7,255 nm2, averaging 85 nm apart. These spacings are very large compared with the ∼3–5 nm size of the dimers, raising the question of what kind of interactions keep them concentrated in a patch at one end of the cell. The MinC–MinD copolymers may play a role in the organization of the patches, but they would probably have to be very short to produce a diffuse patch rather than resolvable filaments.
MinC is widely recognized as an inhibitor of FtsZ. We found that MinC alone had no effect on FtsZ protofilaments up to a 1:1 stoichiometry, but at 2:1 MinC:FtsZ assembly of filaments visible by EM was blocked. The fact that GTP hydrolysis continued may suggest that the excess MinC has shortened FtsZ filaments to a length difficult to see by EM. FtsZ in the cell is eight times higher in concentration than MinC, yet we found in vitro that MinC disassembled FtsZ only when it was in excess, a seeming contradiction. However, we found that FtsZ filaments are able to bind to MinC–MinD copolymers to form large bundles. This bundling could be mediated by multiple interactions of the conserved C-terminal peptides of FtsZ protofilaments with the MinCCs of MinC–MinD filaments and let MinC shorten FtsZ filaments efficiently in vivo at a very low concentration level. We note, however, that our results contrast with previous studies where MinC from E. coli caused FtsZ disassembly at substoichiometric concentrations (19).
Ghosal et al. (24) proposed that the MinC–MinD copolymers should bind FtsZ filaments much more strongly than MinC could bind FtsZ monomers, because of cooperativity. When they added MinCD copolymers and GMPCPP-polymerized FtsZ filaments to liposomes and imaged them with electron cryotomography, they found FtsZ filaments covering up the MinCD copolymers that bound to the lipid membranes. Our observation of enhanced bundling of MinC–MinD copolymers by FtsZ filaments is consistent with this. In addition to, or apart from, inhibiting FtsZ filaments, MinC–MinD polymers near the poles might capture FtsZ filaments. The capture would be modified when the MinC–MinD are attached to the membrane, which would probably limit the bundles to filament pairs or ribbons. Parallel copolymers of MinC–MinD and captured FtsZ filaments on the membrane might facilitate disassembly of FtsZ by mechanisms that are not obvious from the large bundles in solution. See Fig. 4K in Ghosal et al. (24) for additional speculation on the structural aspects of MinC–MinD copolymers with FtsZ filaments.
The study of Park et al. (26) showed that overexpression of mutants of MinC or MinD that block copolymer assembly did not block functions in vivo, suggesting that extended filaments of MinC–MinD are not necessary in vivo. Mutants limiting assembly to a dimer of MinC or MinD flanked by one or two dimers of the other retained in vivo activities. Nevertheless the coassembly in vitro has been observed now for three bacterial species, including the very distant A. aeolicus, which suggests that the interactions producing the filaments are biologically relevant. We cannot really reconcile the results of Park et al. (26). One possibility is that the functional copolymers in vivo may be very short filaments, utilizing the interfaces revealed in the Aquifex crystal structure but comprising only a few dimers of MinC and MinD. Short copolymers may also be important to achieve the ∼50-s oscillations seen in vivo versus the 10–15 min required for disassembly of the large filament bundles assembled in vitro.
Experimental procedures
Plasmid construction and protein purification
FtsZ protein from P. aeruginosa was constructed in the plasmid pET15b at the NdeI/BamHI sites and was purified as described previously (28, 29). Briefly, the soluble His6 protein was purified by affinity chromatography on a Talon column (Clutch Lab, Inc.). After incubation with 2 units/ml of thrombin for 2 h at room temperature, protein was further purified by chromatography on a source Q 10/10 column (GE healthcare) with a linear gradient of 50–500 mm KCl in 50 mm Tris, pH 7.9, 1 mm EDTA, and 10% glycerol and was stored at −80 °C.
Expression vectors for P. aeruginosa MinC, MinD, and MinE were constructed in the plasmid pET15b at the NdeI/BamHI sites. Proteins were expressed in BL21 at 37 °C for 4 h. After sonication and centrifugation, the soluble protein was applied to a Talon column and eluted with a gradient containing 10–100 mm imidazole. The buffer was changed to 50 mm HEPES, pH 7.5, 5 mm MgAc, and 100 mm KAc using the Amicon Ultra-15 centrifugal filter (Merck Millipore). This buffer was used for all assembly experiments, except the indicated EDTA buffer. The P. aeruginosa Min proteins used in these experiments retained their His tags, because we found that their removal had little effect on their assembly. The His tag was removed from FtsZ.
Light-scattering measurement
A light-scattering assay was used to measure the kinetics of MinC–MinD assembly as described previously (30). Light scattering was measured using a Shimadzu RF-5301 PC spectrofluorometer, with both excitation and emission at 350 nm. Two different assays were used to measure the kinetics. In the first, MinC and MinD proteins were premixed, and the measurement started immediately after adding ATP. In the second, MinD and ATP were premixed, and the measurement started after adding MinC. Each measurement was repeated two or three times, with consistent results.
Sedimentation assay
Assembly of MinC and MinD was also assayed by sedimentation. MinC and MinD at different concentrations were assembled with 1 mm ATP at room temperature for 30 min and centrifuged at 50,000 rpm for 30 min at 25 °C in a Beckman TLA100 rotor. The supernatant was carefully removed, and the pellet was resuspended in the same volume of buffer. The protein in the pellet and supernatant was assayed by SDS-PAGE. The ratio of supernatant and pellet was measured using ImageJ software, and the protein concentrations were calculated from the percentage of total protein concentration. The measurement was repeated more than four times.
GTPase measurement
The GTPase activity of FtsZ with or without MinC was measured using a regenerative coupled GTPase assay as described previously (28). Our assay mixture included 1 mm phosphoenolpyruvate, 0.8 mm NADH, 20 units/ml pyruvate kinase, and lactate dehydrogenase (Sigma–Aldrich), and 0.5 mm GTP. In this assay, all free GDP in solution is rapidly regenerated into GTP, in a reaction that consumes one NADH per GDP. The NADH concentration was measured by the absorption in a Shimadzu UV-2401PC spectrophotometer, using the extinction coefficient 6,220 m−1 cm−1 at 340 nm. The absorbance showed a linear decrease over time, giving the hydrolysis rate for each FtsZ concentration. These rates were plotted versus FtsZ concentration, and the overall hydrolysis rate in GTP per minute per FtsZ was the slope of the line above the critical concentration. Each assay was repeated two or three times.
Binding of mant-ATP to MinD
We determined the binding affinity of MinD for mant-ATP (Thermo Fisher Scientific) using the enhanced fluorescence enhancement that occurs upon binding. The fluorescence emission of 0.2 μm mant-ATP from 380–600 nm with excitation at 350 nm was recorded for titration with 0–4 μm MinD proteins. The equilibrium dissociation constant KD was calculated using a simple binding model with the following equation,
| (1) |
where F is the measured fluorescence, Fo is the fluorescence of 0.2 μm mant-ATP alone, Fm is the maximum fluorescence if all mant-ATP is bound to MinD, R is the concentration of mant-ATP, and G is the concentration of MinD. Origin software was used to find the value of KD giving the best fit to the data, averaged from two titrations.
Electron microscopy
MinC–MinD copolymers and FtsZ filaments were visualized by negative stain EM. Approximately 10 μl of the assembly mixture was incubated at room temperature for several minutes and then applied to a carbon-coated copper grid. The samples were stained with 2% uranyl acetate for 10 s. The images were obtained with a Philips 420 electron microscope at 49,000× or 82,000× magnification.
Author contributions
H. H., P. W., L. B., and Y. C. data curation; H. H., P. W., L. B., M. O., and Y. C. investigation; H. H., P. W., L. B., M. O., H. P. E., and Y. C. writing-review and editing; P. W., H. P. E., and Y. C. writing-original draft; M. O. resources; M. O., H. P. E., and Y. C. methodology; H. P. E. and Y. C. supervision; H. P. E. and Y. C. funding acquisition; Y. C. software; Y. C. project administration.
Supplementary Material
Footnotes
This work was supported by First-class University and Academic Programs of Northwest University (to Y. C.) and National Institutes of Health Grant GM066014 (to H. P. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
References
- 1. Erickson H. P., and Osawa M. (2017) FtsZ constriction force: curved protofilaments bending membranes. Subcell. Biochem. 84, 139–160 10.1007/978-3-319-53047-5_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erickson H. P. (2017) How bacterial cell division might cheat turgor pressure: a unified mechanism of septal division in Gram-positive and Gram-negative bacteria. Bioessays 39, 1700045 10.1002/bies.201700045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haeusser D. P., and Margolin W. (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 10.1038/nrmicro.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Du S., and Lutkenhaus J. (2017) Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 10.1111/mmi.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guberman J. M., Fay A., Dworkin J., Wingreen N. S., and Gitai Z. (2008) PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput. Biol. 4, e1000233 10.1371/journal.pcbi.1000233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lutkenhaus J., and Du S. (2017) E. coli cell cycle Machinery. Subcell. Biochem. 84, 27–65 10.1007/978-3-319-53047-5_2 [DOI] [PubMed] [Google Scholar]
- 7. Szwedziak P., and Ghosal D. (2017) FtsZ-ring architecture and its control by MinCD. Subcell. Biochem. 84, 213–244 10.1007/978-3-319-53047-5_7 [DOI] [PubMed] [Google Scholar]
- 8. Schumacher M. A. (2017) Bacterial nucleoid occlusion: multiple mechanisms for preventing chromosome bisection during cell division. Subcell. Biochem. 84, 267–298 10.1007/978-3-319-53047-5_9 [DOI] [PubMed] [Google Scholar]
- 9. Bailey M. W., Bisicchia P., Warren B. T., Sherratt D. J., and Männik J. (2014) Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet. 10, e1004504 10.1371/journal.pgen.1004504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raskin D. M., and de Boer P. A. (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 4971–4976 10.1073/pnas.96.9.4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Z., and Lutkenhaus J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34, 82–90 10.1046/j.1365-2958.1999.01575.x [DOI] [PubMed] [Google Scholar]
- 12. Loose M., Fischer-Friedrich E., Ries J., Kruse K., and Schwille P. (2008) Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792 10.1126/science.1154413 [DOI] [PubMed] [Google Scholar]
- 13. Loose M., Fischer-Friedrich E., Herold C., Kruse K., and Schwille P. (2011) Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat. Struct. Mol. Biol. 18, 577–583 10.1038/nsmb.2037 [DOI] [PubMed] [Google Scholar]
- 14. Vecchiarelli A. G., Li M., Mizuuchi M., Hwang L. C., Seol Y., Neuman K. C., and Mizuuchi K. (2016) Membrane-bound MinDE complex acts as a toggle switch that drives Min oscillation coupled to cytoplasmic depletion of MinD. Proc. Natl. Acad. Sci. U.S.A. 113, E1479–E1488 10.1073/pnas.1600644113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szeto T. H., Rowland S. L., Habrukowich C. L., and King G. F. (2003) The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 278, 40050–40056 10.1074/jbc.M306876200 [DOI] [PubMed] [Google Scholar]
- 16. Hu Z., and Lutkenhaus J. (2003) A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47, 345–355 10.1046/j.1365-2958.2003.03321.x [DOI] [PubMed] [Google Scholar]
- 17. Park K. T., Wu W., Battaile K. P., Lovell S., Holyoak T., and Lutkenhaus J. (2011) The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell 146, 396–407 10.1016/j.cell.2011.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kretschmer S., and Schwille P. (2016) Pattern formation on membranes and its role in bacterial cell division. Curr. Opin. Cell Biol. 38, 52–59 10.1016/j.ceb.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 19. Hu Z., Mukherjee A., Pichoff S., and Lutkenhaus J. (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. U.S.A. 96, 14819–14824 10.1073/pnas.96.26.14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dajkovic A., Lan G., Sun S. X., Wirtz D., and Lutkenhaus J. (2008) MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18, 235–244 10.1016/j.cub.2008.01.042 [DOI] [PubMed] [Google Scholar]
- 21. Cordell S. C., Anderson R. E., and Löwe J. (2001) Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20, 2454–2461 10.1093/emboj/20.10.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Z., and Lutkenhaus J. (2000) Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol 182, 3965–3971 10.1128/JB.182.14.3965-3971.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li G. W., Burkhardt D., Gross C., and Weissman J. S. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosal D., Trambaiolo D., Amos L. A., and Löwe J. (2014) MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat. Commun. 5, 5341 10.1038/ncomms6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conti J., Viola M. G., and Camberg J. L. (2015) The bacterial cell division regulators MinD and MinC form polymers in the presence of nucleotide. FEBS Lett. 589, 201–206 10.1016/j.febslet.2014.11.047 [DOI] [PubMed] [Google Scholar]
- 26. Park K. T., Du S., and Lutkenhaus J. (2015) MinC/MinD copolymers are not required for Min function. Mol. Microbiol. 98, 895–909 10.1111/mmi.13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rivas G., López A., Mingorance J., Ferrándiz M. J., Zorrilla S., Minton A. P., Vicente M., and Andreu J. M. (2000) Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer: the primary steps for FtsZ assembly. J. Biol. Chem. 275, 11740–11749 10.1074/jbc.275.16.11740 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Milam S. L., and Erickson H. P. (2012) SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51, 3100–3109 10.1021/bi201669d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Y., Huang H., Osawa M., and Erickson H. P. (2017) ZipA and FtsA* stabilize FtsZ-GDP miniring structures. Sci. Rep. 7, 3650 10.1038/s41598-017-03983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y., Porter K., Osawa M., Augustus A. M., Milam S. L., Joshi C., Osteryoung K. W., and Erickson H. P. (2017) The chloroplast tubulin homologs FtsZA and FtsZB from the red alga Galdieria sulphuraria co-assemble into dynamic filaments. J. Biol. Chem. 292, 5207–5215 10.1074/jbc.M116.767715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.