Abstract
The systems integration of whole-body metabolism and immune signaling are central homeostatic mechanisms necessary for maintenance of normal physiology, and dysregulation of these processes leads to a variety of chronic disorders. However, the intracellular mechanisms responsible for cell-autonomous cross-talk between the inflammatory signaling pathways and metabolic flux have remained enigmatic. In this study, we discovered that the fructose-2,6-bisphosphatase TIGAR (Tp53-induced glycolysis and apoptosis regulator) critically regulates NF-κB activation. We found that TIGAR potently inhibits NF-κB–dependent gene expression by suppressing the upstream activation of IKKβ phosphorylation and kinase activation. This inhibition occurred through a direct binding competition between NEMO and TIGAR for association with the linear ubiquitination assembly complex (LUBAC). This competition prevented linear ubiquitination of NEMO, which is required for activation of IKKβ and other downstream targets. Furthermore, a TIGAR phosphatase activity–deficient mutant was equally effective as WT TIGAR in inhibiting NEMO linear ubiquitination, IKKβ phosphorylation/activation, and NF-κB signaling, indicating that TIGAR's effect on NF-κB signaling is due to its interaction with LUBAC. Physiologically, TIGAR knockout mice displayed enhanced adipose tissue NF-κB signaling, whereas adipocyte-specific overexpression of TIGAR suppressed adipose tissue NF-κB signaling. Together, these results demonstrate that TIGAR has a nonenzymatic molecular function that modulates the NF-κB signaling pathway by directly inhibiting the E3 ligase activity of LUBAC.
Keywords: NF-kappaB (NF-KB), adipocyte, ubiquitylation (ubiquitination), ubiquitin ligase, p53, adipocytes, linear ubiquitination, LUBAC, NF-κB, TIGAR, IKKbeta
Introduction
Tp53-induced glycolysis and apoptosis regulator (TIGAR)2 is a 270-amino acid protein that was originally identified as a p53-inducible protein that functions as a fructose-2,6-bisphosphatase but subsequently has been shown to have phosphatase activities for 2,3-bisphosphoglycerate (23BPG), 2-phosphoglycerate, phosphoglycolate, and phosphoenolpyruvate (1–5). Fructose 2,6-bisphosphate (F26P) is an allosteric activator of 6-phosphofructo-1-kinase (PFK1) and a negative regulator of fructose-1,6-bisphosphatase (FBP) that are key regulatory steps controlling glycolysis and gluconeogenesis, respectively (6, 7). By reducing the levels of F26P, TIGAR was found to suppress glycolysis, and the subsequent accumulation of glucose 6-phosphate was diverted into the pentose phosphate pathway to generate nucleotides, NADPH, and antioxidants, such as reduced GSH (5, 8). In contrast, TIGAR enzymatic activity was also reported to be 400-fold greater for 23BPG than F26P, generating 3-phosphoglycerate (9). Although an increase in 3-phosphoglycerate would be expected to increase glycolysis, the proximal substrate for the formation of phosphoenolpyruvate is 2-phosphoglycerate. 2-Phosphoglycerate is generated from 3-bisphosphoglycerate by phosphoglycerate mutase (PGM), which is allosterically activated by 23BPG. Thus, whether TIGAR functions to inhibit or activate glycolysis depends upon the relative contributions of PFK1, FBP, and PGM and their allosteric regulation by F26P and 23BPG. Through these mechanisms, TIGAR can modulate glucose metabolism for energy production and macromolecular synthesis as well as cellular redox state in different ways, depending on the combination of several cell context modulators.
Physiologically, consistent with an overall increase in glycolysis and decrease in pentose flux, TIGAR deficiency was found to increase oxidative stress during cardiac ischemic injury (10). TIGAR deficiency was also found to correlate with increased oxidative stress during Alzheimer's disease progression (11). Similarly, in some tumor cell models, TIGAR was found to contribute to the anti-tumor-promoting activity of p53 by suppressing aerobic glycolysis and cellular survival (5, 12). However, TIGAR was overexpressed in several other tumor cell types, suggesting that TIGAR may also function to promote rather than inhibit cancer development (4, 13, 14). For example, TIGAR deficiency in a mouse intestinal tumor model was shown to increase animal survival with decreased tumor burden, whereas increased TIGAR expression enhanced tumor progression (13). In these systems, TIGAR appears to promote cell survival and expansion by decreasing oxidative stress and increasing production of ribose for DNA and RNA synthesis. Moreover, TIGAR was reported to protect cells from genotoxic drug induced DNA damage partly through the regulation of pentose phosphate pathway products (NADPH and ribose) and reduction of reactive oxygen species (15). Thus, the growth-promoting and/or growth-inhibitory effects of TIGAR also appear to be cell context–dependent, suggesting the influence of additional signaling/metabolic events that determine the biological outcome of TIGAR function.
In this regard, there is mutual cross-talk and complex entanglement between metabolic flux, cellular oxidative stress, intracellular signal transduction, and in particular inflammatory signaling cascades, such that changes in one of these pathways can have multiple effects on any or all of these other pathways. During our studies of insulin resistance and inflammatory signaling of adipocytes, we examined the effect of TIGAR deficiency and overexpression in the differentiated 3T3-L1 adipocyte cell line. Surprisingly, independent of its role as a regulator of glucose flux, we have found that TIGAR is also a potent negative regulator of NF-κB signaling and inflammatory cytokine production. Molecular analyses of the NF-κB activation pathway revealed that TIGAR suppresses the activation of IKKβ and IKKβ-dependent phosphorylation of downstream substrate targets. Moreover, the ability of TIGAR to prevent NF-κB activation is independent of TIGAR's phosphatase activity that results from a direct binding interaction with the HOIP subunit of the linear ubiquitination assembly complex (LUBAC). TIGAR competes for NEMO binding to HOIP and thereby suppresses NEMO linear ubiquitination necessary for IKKβ activation and activation of downstream targets.
Results
TIGAR suppresses NF-κB signaling in a phosphatase activity–independent manner
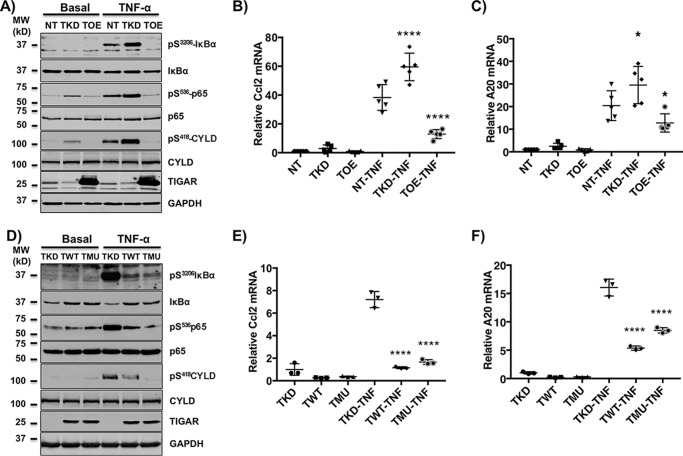
Adipocytes play a central role in the integrative normal and pathophysiologic regulation of appetite, adiposity, energy balance, and insulin responsiveness, including glucose and lipid metabolism (16–20). To assess the potential role of TIGAR in adipocytes, we generated control mammalian non-target shRNA lentivirus (NT)–, TIGAR short hairpin RNA (shRNA) knockdown lentivirus (TKD)–, and TIGAR cDNA-overexpressing lentivirus (TOE)–infected 3T3-L1 murine adipocyte cell lines. Examination of several signaling pathways in the differentiated adipocytes revealed that the TKD cells displayed increased TNFα-stimulated serine 32/36 IκBα and serine 536 p65 (RelA) phosphorylation (Fig. 1A). In contrast, the TOE cells had reduced IκBα and p65 phosphorylation following TNFα stimulation. In addition to IκBα and p65, CYLD is also a direct substrate of the IKKβ kinase, whose phosphorylation on serine residue 418 suppresses its deubiquitinase activity (21, 22). Similar to IκBα and p65, TNFα-stimulated CYLD phosphorylation was increased in the TKD and suppressed in the TOE adipocytes. Consistent with TIGAR regulating the NF-κB signal transduction pathway, TNFα increased Ccl2 and A20 gene expression in the TKD adipocytes that was suppressed in the TOE cells (Fig. 1, B and C). Essentially identical results were obtained from multiple independently generated NT, TKD, and TOE 3T3-L1 adipocyte cell lines (data not shown).
Figure 1.
TIGAR regulates canonical NF-κB signaling. NT, TKD, and TOE 3T3-L1 preadipocytes were generated as described under “Method details.” A, 3T3-L1 adipocytes were either left untreated (Basal) or stimulated with 10 ng/ml TNFα for 5 min (TNFα). Cell lysates were prepared and immunoblotted for the indicated proteins. These are representative immunoblots independently performed five times. B and C, the adipocytes were either left untreated or stimulated with 10 ng/ml TNFα for 4 h, and the expression of Ccl2 (B) and A20 (C) mRNAs was determined by qRT-PCR. These data represent the average of five independent determinations ± S.D. (error bars). D, the TKD 3T3-L1 cells were stably infected with TWT and TMU lentiviruses as described under “Method details.” The 3T3-L1 TKD, TWT, and TMU preadipocytes were differentiated into adipocytes either left untreated (Basal) or stimulated with 10 ng/ml TNFα for 5 min (TNFα). Cell lysates were prepared and immunoblotted for the indicated proteins. These are representative immunoblots independently performed three times. E and F, the TKD, TWT, and TMU adipocytes were treated with vehicle or 10 ng/ml TNFα for 4 h, and the expression of Ccl2 (E) and A20 (F) mRNAs was determined by qRT-PCR. These data represent the average of three independent determinations ± S.D. *, p < 0.05; ****, p < 0.0001.
To further confirm specificity of the TIGAR knockdown shRNA and in parallel the necessity of the TIGAR phosphatase activity in the 3T3-L1 adipocyte cell context of TKD, we next re-expressed an shRNA-resistant WT TIGAR (TWT) and the phosphatase activity–defective TIGAR mutant (TMU) in which the three catalytic pocket residues (His11, Glu102, and His198) were mutated to alanine (5). As expected, re-expression of WT TIGAR in the cell context of TIGAR deficiency (TWT) resulted in suppression of TNFα-stimulated IκBα, p65, and CYLD phosphorylation (Fig. 1D) and target gene expression (Fig. 1, E and F). Surprisingly, expression of the phosphatase-defective mutant TIGAR (TMU) was just as effective in inhibiting TNFα-stimulated IκBα, p65, and CYLD phosphorylation (Fig. 1D). The ability of the phosphatase-defective mutant to inhibit these IKKβ-dependent phosphorylation events was also reflected in the suppression of TNFα-stimulated gene expression (Fig. 1, E and F). To directly demonstrate that the TIGAR inhibition of ligand-stimulated gene expression was specific to the NF-κB pathway and not due to potential regulation of other signaling pathways (i.e. mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and protein kinase C), human embryonic kidney 293T (HEK293T) cells were co-transfected with a specific NF-κB–driven luciferase gene reporter with either empty vector, TWT, or TMU cDNAs and then subsequently treated with TNFα (Fig. 2A). As is apparent, both TWT and TMU were effective suppressors of TNFα-stimulated luciferase activity. Together, these data demonstrate that TIGAR has a unique function to inhibit the canonical NF-κB signaling pathway that is independent of the TIGAR phosphatase activity.
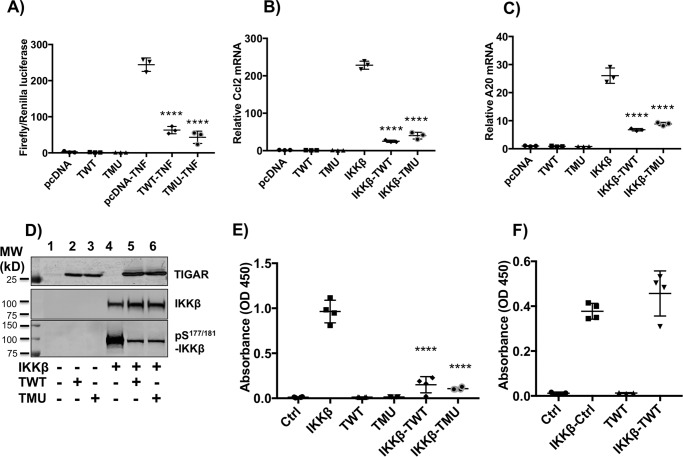
Figure 2.
TWT and TMU inhibit NF-κB pro-inflammatory gene expression, IKKβ phosphorylation, and IKKβ enzymatic activity. A, HEK293T cells were seeded in a 6-well plate (0.8 × 106 cells/well) in 2 ml of growth medium (10% fetal bovine serum with antibiotics) for 6–8 h. Following transfection with the NF-κB luciferase reporter gene, relative basal and TNFα-stimulated levels of luciferase activity were determined as described under “Method details.” These data are the average of three independent determinations ± S.D. (error bars). B and C, HEK293T cells were transfected with cDNA for IKKβ alone or in combination with TWT or TMU cDNAs, as described under “Method details.” Ccl2 (B) and A20 (C) mRNA levels were determined by qRT-PCR, and these data represent the average of three independent determinations ± S.D. D, the HEK293T cells were transfected with the indicated combinations of cDNAs (20 μg total/100-mm dish) for 24 h, followed by immunoblotting of cell lysates for the indicated proteins as described under “Method details.” Lane 1, 20 μg of pcDNA; lane 2, 10 μg of TWT; lane 3, 10 μg of TMU; lane 4, 5 μg of IKKβ; lane 5, 10 μg of IKKβ plus 10 μg of TWT; lane 6, 10 μg of IKKβ plus 10 μg of TMU cDNA plus various amounts of empty vector for a total of 20 μg of DNA. The extracts were then immunoblotted for the various proteins indicated. These are representative immunoblots independently performed three times. E, HEK293T cells (6-well plate, 0.8 × 106 cells/well) were transfected with 2.5 μg of empty vector (Ctrl), 1 μg of IKKβ, 1.5 μg of TWT, 1.5 μg of TMU, 2.5 μg of IKKβ plus TWT, and IKKβ plus TMU cDNAs for 24 h. Cell extracts were prepared, and IKKβ kinase activity in vitro was determined using IκBα as substrate as described under “Method details.” F, cell extracts were prepared, and equal amounts of IKKβ and TWT cell extracts were premixed (IKKβ + TWT) for 0.5 h before determination of IKKβ kinase activity. The data are the average ± S.E. from four independent experiments, each performed in duplicate.
TIGAR suppresses IKKβ activation by competing for NEMO
To address the mechanism(s) that could account for the apparent nonenzymatic function of TIGAR to suppress NF-κB signaling, we first examined the effect of TIGAR in cells overexpressing IKKβ. Previous studies have observed that overexpression of IKKβ results in its spontaneous phosphorylation and activation of NF-κB signaling (23, 24). Consistent with these findings, we observed that overexpression of IKKβ resulted in the marked increase of Ccl2 (Fig. 2B) and A20 (Fig. 2C) mRNA levels. However, co-expression with TWT or the TMU cDNAs also substantially suppressed the Ccl2 and A20 gene expression. Moreover, overexpression of IKKβ resulted in a robust level of IKKβ phosphorylation that was reduced when co-expressed with either TWT or TMU (Fig. 2D). These data suggest that TIGAR inhibits the function of IKKβ either directly or indirectly by inhibiting an upstream pathway.
To determine whether TIGAR directly inhibited IKKβ, we expressed IKKβ alone or in combination with TWT or TMU and examined the in vitro kinase activity of IKKβ in cell extracts. Consistent with the inhibition of IKKβ phosphorylation and NF-κB–dependent gene expression, co-expression of either TWT or TMU inhibited the in vitro kinase activity of IKKβ (Fig. 2E). We next mixed the cell extracts containing IKKβ with cell extracts containing TWT, followed by determination of in vitro IKKβ kinase activity (Fig. 2F). Under these conditions, TIGAR was completely ineffective in altering the IKKβ kinase activity. These data indicate that TIGAR blocks the activation of IKKβ indirectly and not through a direct interaction with IKKβ.
Because the in vitro kinase assay used IκBα as the substrate, usbit remained possible that TIGAR interacted with the IκBα–NF-κB complex, preventing IκBα accessibility as a substrate. To examine this, we co-expressed TIGAR with IKKβ and examined the phosphorylation of the IKKβ substrate CYLD (Fig. 3A). Overexpression of IKKβ resulted in the phosphorylation of the activation site serine residues 177/181 in IKKβ as well as the phosphorylation of serine 418 in CYLD (Fig. 3A, lane 3). It should be noted that unlike 3T3-L1 adipocytes, in which there is only a single band detected with the pS418-CYLD antibody (Fig. 1, A and D), in HEK293T cells, several bands are detected, with only one corresponding to the molecular weight of the CYLD protein (depicted by an arrow). The presence of these additional bands is probably due to the overexpression of IKKβ, as only a single band at the expected molecular weight is observed when HEK293T cells are stimulated with TNFα (data not shown). In any case, overexpression of TIGAR alone (Fig. 3A, lane 2) or NEMO alone (Fig. 3A, lane 4) had no effect on endogenous CYLD phosphorylation and was essentially identical to vector-transfected cells (Fig. 3A, lane 1). In contrast, co-expression of IKKβ with TIGAR markedly suppressed IKKβ and endogenous CYLD phosphorylation (Fig. 3A, compare lane 3 with lane 5), whereas co-expression of IKKβ with NEMO resulted in the full extent of substrate phosphorylation (Fig. 3A, lane 6). These data demonstrate that TIGAR suppresses IKKβ-mediated phosphorylation of both expressed IKKβ and endogenous CYLD, supporting an inhibition of IKKβ activation rather than an effect on substrate accessibility. Interestingly, expression of NEMO with TIGAR partially restored IKKβ and CYLD phosphorylation (Fig. 3A, compare lane 5 with lane 8). These data further suggest that TIGAR and NEMO compete at a common site responsible for IKKβ activation.
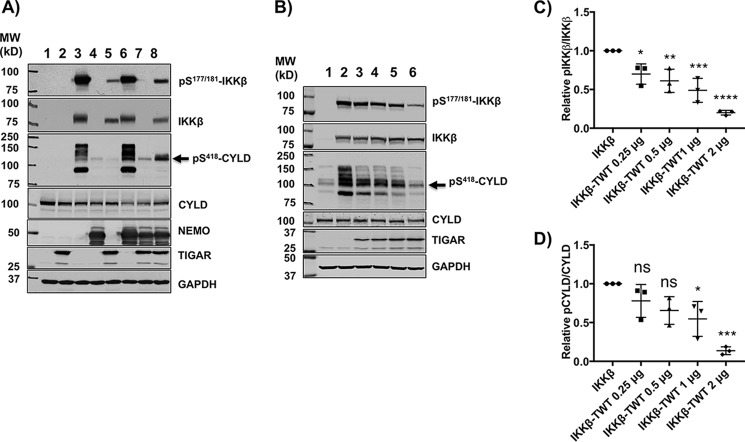
Figure 3.
TIGAR inhibits IKKβ-dependent phosphorylation of several direct IKKβ cellular substrate targets. A, HEK293T cells were transfected with the indicated combinations of cDNAs for 24 h followed by immunoblotting of cell lysates for the indicated proteins as described under “Method details.” Lane 1, 4 μg of empty vector; lane 2, 2 μg of TIGAR; lane 3, 1 μg of IKKβ; lane 4, 1 μg of NEMO; lane 5, 2 μg of TIGAR plus 1 μg of IKKβ; lane 6, 1 μg of IKKβ plus 1 μg of NEMO; lane 7, 2 μg of TIGAR plus 1 μg of NEMO; lane 8, 2 μg of TIGAR plus 1 μg of IKKβ plus 1 μg of NEMO cDNA plus various amounts of pcDNA for a total of 4 μg of DNA/60-mm dish for 24 h. These are representative immunoblots independently performed 3–5 times. B, HEK293T cells were transfected with 3 μg of empty vector (lane 1), 1 μg of IKKβ (lane 2), 1 μg of IKKβ (lane 3) plus increasing amounts of TIGAR cDNA (0.25 μg (lane 3), 0.5 μg (lane 4), 1 μg (lane 5), and 2 μg (lane 6)) plus various amounts of empty vector for a total of 3 μg/60-mm dish of DNA. 24 h later, cell extracts were prepared and immunoblotted for the indicated proteins. These are representative immunoblots independently performed 3–5 times. C, the TIGAR dose-dependent inhibition of IKKβ phosphorylation from the data obtained in B was quantified by ImageJ densitometry ± S.D. (error bars) as described under “Method details.” D, the TIGAR dose-dependent inhibition of CYLD phosphorylation (band indicated by arrow) from the data obtained in Fig. 3B was quantified by ImageJ densitometry ± S.D. (error bars) as described under “Method details.”
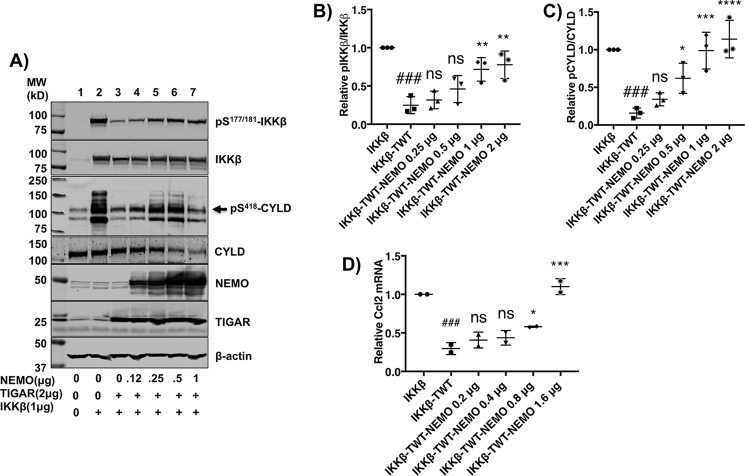
To test this prediction, we first determined the dose-dependent effect of TIGAR on IKKβ and CYLD phosphorylation (Fig. 3B). At a fixed IKKβ expression level (Fig. 3B, lane 2), TIGAR in a dose-dependent manner suppressed Ser177/181-IKKβ and Ser418-CYLD phosphorylation (Fig. 3B, lanes 3–6). Quantification of the TIGAR dose-dependent inhibition of IKKβ and CYLD phosphorylation is shown in Fig. 3 (C and D). Conversely, we determined the ability of NEMO to reverse the TIGAR inhibition of IKKβ activation (Fig. 4A). At a fixed concentration of IKKβ and TIGAR that results in a marked inhibition of IKKβ and CYLD phosphorylation (Fig. 4A, lane 2 versus lane 3), NEMO in a dose-dependent manner reversed the TIGAR inhibition of IKKβ and CYLD phosphorylation (Fig. 4A, lanes 4–7). It should be noted that with increasing amounts of expressed NEMO protein levels, the levels of the endogenous CYLD protein decreased. Whether this results from the activation of the MALT1-dependent proteolytic cleavage or ubiquitination and proteasome-mediated degradation of CYLD remains to be determined (25, 26). Nevertheless, quantification of the NEMO dose-dependent reversal of TIGAR-mediated inhibition of IKKβ and CYLD phosphorylation is shown in Fig. 4 (B and C). In parallel, we also demonstrated the ability of NEMO to reverse the TIGAR inhibition of IKKβ-stimulated Ccl2 gene expression (Fig. 4D). Together, these data are consistent with a direct competition of TIGAR and NEMO at a common upstream site necessary for IKKβ activation and NF-κB signaling.
Figure 4.
NEMO rescues the TIGAR inhibition of IKKβ-dependent signaling. A, HEK293T cells were transfected with 4 μg of empty vector (lane 1), 1 μg of IKKβ (lane 2), 1 μg of IKKβ plus 2 μg of TIGAR (lane 3) with increasing amounts (0.125 μg (lane 4), 0.25 μg (lane 5), 0.5 μg (lane 6), and 1 μg (lane 7)) of NEMO plus various amounts of empty vector for a total of 4 μg/60-mm dish of DNA. 24 h later, cell extracts were prepared and immunoblotted for the indicated proteins. This is a representative immunoblot independently performed 3–5 times. B, the NEMO dose-dependent rescue of TWT inhibition of IKKβ phosphorylation from the data obtained in A was quantified by ImageJ densitometry ± S.D. (error bars). C, the NEMO dose-dependent rescue of TWT inhibition of CYLD phosphorylation from the data obtained in A was quantified by ImageJ densitometry ± S.D. D, the NEMO dose-dependent rescue of TWT inhibition of Ccl2 gene expression from the cells transfected with cDNA was performed as in Fig. 4A. Ccl2 mRNA was determined by qRT-PCR from two independent experiments. The data are presented as the average ± S.D. ns, not significant; ##, p < 0.01; ###, p < 0.001 comparing IKKβ versus IKKβ + TIGAR; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 comparing IKKβ + TIGAR versus increasing amounts of NEMO.
TIGAR directly binds to the LUBAC and inhibits NEMO linear ubiquitination
Multiple analyses of endogenous and transfected TIGAR co-immunoprecipitation experiments failed to demonstrate any specific association of TIGAR with IKKα, IKKβ, or NEMO (data not shown). Although negative, this suggests that the site(s) of TIGAR action is not through direct interaction with the IKK complex, consistent with the data presented in Fig. 2. Furthermore, we have been unable to detect a specific interaction of TIGAR with the scaffolding protein TRAF2 (TNFα receptor) or TRAF6 (IL1β receptor) (data not shown). We then examined the interaction of TIGAR with the LUBAC in cells overexpressing TIGAR and three individual LUBAC subunit complex proteins, HOIP, HOIL-1L, and SHARPIN (Fig. 5A). Although neither HOIL-IL nor SHARPIN displayed any specific interaction with TIGAR, immunoprecipitation of HOIP clearly demonstrated a specific co-precipitation with TIGAR (Fig. 5B).
Figure 5.

TIGAR specifically co-immunoprecipitates with HOIP, the E3 ligase subunit of LUBAC. A, HEK293T cells were transfected with 3 μg of TIGAR or 3 μg of TIGAR plus 3 μg of FLAG-HOIP, 3 μg of TIGAR plus 3 μg of FLAG-HOIL-1L, or 3 μg of TIGAR plus 3 μg of FLAG-SHARPIN cDNAs on 100-mm plates for 18 h. Aliquots of the cell lysates were immunoblotted with the FLAG and TIGAR antibodies. B, the cell lysates in A were immunoprecipitated (IP) with the FLAG antibody and immunoblotted for FLAG and TIGAR. These are representative immunoblots independently performed two times.
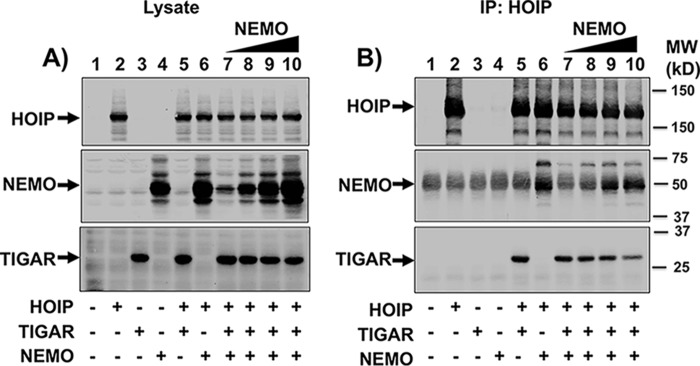
Based upon the finding that TIGAR and NEMO compete for IKKβ activation, we examined the NEMO competition for TIGAR binding to HOIP. Cells were transfected with empty vector, HOIP, TIGAR, and NEMO cDNAs separately or HOIP + TIGAR and HOIP + NEMO cDNAs together (Fig. 6A, lanes 1–6). In addition, HOIP and TIGAR were co-expressed with increasing amounts of NEMO cDNA (Fig. 6A, lanes 7–10). HOIP immunoprecipitation of the cells expressing HOIP + TIGAR demonstrated the co-immunoprecipitation of TIGAR, and similarly, cells expressing HOIP + NEMO demonstrated the co-immunoprecipitation of NEMO (Fig. 6B, lanes 5 and 6). Increasing levels of NEMO expression resulted in increased amounts of the NEMO protein co-immunoprecipitated with HOIP (Fig. 6B, lanes 7–10). In parallel, the amount of TIGAR co-immunoprecipitated with HOIP progressively decreased. These data directly demonstrate that TIGAR and NEMO compete for binding to the HOIP subunit of LUBAC.
Figure 6.
TIGAR and NEMO compete for HOIP binding. A, HEK293T cells were transfected with the indicated combinations of cDNAs (8 μg total/100-mm dish) for 16 h, followed by immunoblotting of cell lysates for the indicated proteins. Lane 1, 8 μg of empty vector; lane 2, 3 μg of HOIP; lane 3, 2 μg of TIGAR; lane 4, 1 μg of the NEMO; lane 5, 3 μg of HOIP plus 2 μg of TIGAR; lane 6, 3 μg of HOIP plus 1 μg of NEMO; lanes 7–9, 3 μg of HOIP plus 2 μg of TIGAR with increasing amounts of NEMO (0.1, 0.3, 1, and 3 μg, respectively). B, the cell lysates from A were immunoprecipitated (IP) with a FLAG antibody and immunoblotted for the indicated proteins. These are representative immunoblots independently performed two times.
HOIP is composed of several modular domains, including a zinc finger (ZF), two Npl4 zinc finger (NZF), ubiquitin-associated (UBA), two R (ring fingers), ring between ring fingers (RBR), and linear ubiquitin chain–determining domain (LDD) (27–30), schematically represented in Fig. 7A. Previous studies have demonstrated that the NEMO binding specifically maps to the HOIP NZF1 domain (27). Consistent with these previous studies, immunoprecipitation of a HOIP NZF1 deletion mutant substantially reduced the amount of co-immunoprecipitated NEMO protein compared with WT HOIP (Fig. 7B, lanes 6 and 7). In contrast, deletion of the NZF1 domain had no effect on the ability of HOIP to co-immunoprecipitate TIGAR (Fig. 7B, lanes 8 and 9). In contrast, deletion of the ZF domain reduced TIGAR binding by 20%, whereas deletion of the NZF2 domain reduced TIGAR binding by ∼60% (Fig. 7C, lanes 3 and 5). Deletion of the NZF1 had no effect on HOIP binding to TIGAR (Fig. 7C, lane 4). Moreover, deletion of the UBA resulted in an apparent increase in TIGAR binding (Fig. 7C, lane 6).
Figure 7.
Identification of the HOIP amino acid sequences responsible for TIGAR binding. A, schematic representation of the HOIP linear amino acid sequence with known protein interaction domains and various deletion mutants generated. The values to the right of each model structure reflect the percentage of HOIP mutant binding to TIGAR compared with full-length HOIP determined as described below. The percentage of TIGAR binding was normalized for the relative levels of HOIP expression. B, HEK293T cells were transfected with TIGAR and HA-NEMO with full-length and NZF1 domain–deleted FLAG-HOIP cDNAs, as indicated. The cell lysates were immunoblotted for FLAG-HOIP, HA-NEMO, and TIGAR (left, lanes 1–9). The cell lysates were immunoprecipitated (IP) with the FLAG antibody, and these immunoprecipitates were immunoblotted for FLAG-HOIP, HA-NEMO, and TIGAR (right, lanes 1–9). These are representative immunoblots independently performed 2–3 times. C, HEK293T cells were transfected with TIGAR and the ZF, NZF1, NZF2, and UBA domain-deleted FLAG-HOIP cDNAs, as indicated. The cell lysates were immunoblotted for FLAG-HOIP and TIGAR (left, lanes 1–6). The cell lysates were immunoprecipitated with the FLAG antibody, and these immunoprecipitates were immunoblotted for FLAG-HOIP and TIGAR (right, lanes 1–6). These are representative immunoblots independently performed three times. D, cells were transfected with TIGAR and various N-terminal deletion FLAG-HOIP cDNAs, as indicated. The cell lysates were immunoblotted for FLAG-HOIP and TIGAR (left, lanes 1–5). The cell lysates were immunoprecipitated with the FLAG antibody, and these immunoprecipitates were immunoblotted for FLAG-HOIP and TIGAR (right, lanes 1–5). These are representative immunoblots independently performed three times. E, cells were transfected with TIGAR and various C-terminal deletion FLAG-HOIP cDNAs, as indicated. The cell lysates were immunoblotted for FLAG-HOIP and TIGAR (left, lanes 1–6). The cell lysates were immunoprecipitated with the FLAG antibody, and these immunoprecipitates were immunoblotted for FLAG-HOIP and TIGAR (right, lanes 1–6). These are representative immunoblots independently performed three times.
As the only domain that apparently resulted in a decrement in TIGAR binding, albeit relatively small, was the NZF2 domain, we next analyzed an N-terminal HOIP deletion set (Fig. 7D). Progressive deletions of the HOIP N terminus resulted in a decrease in the binding of TIGAR to HOIP when residues between 175 and 526 were deleted (Fig. 7D, lanes 2–5). Analyses of a carboxyl deletion set demonstrated that loss of residues from 679 to 1029 had little effect on HOIP binding to TIGAR (Fig. 7E, lanes 2–5). However, TIGAR binding was almost completely abrogated upon further deletion to eliminate the UBA domain (Fig. 7E, lane 6). These data suggest that the HOIP amino acid region required for TIGAR binding primarily lies between residues 175 and 679, which includes the ZF, NZF1, NZF2, and UBA domains.
Quantification of all of these binding data is presented as the percentage of TIGAR immunoprecipitated with full-length HOIP binding (denoted by the parenthesis) immediately next to the schematic HOIP structures shown in Fig. 7A. Taken together, these data indicate that TIGAR does not directly compete for NEMO at the NZF1 HOIP-binding site and therefore probably suppresses NEMO binding through steric hindrance and/or via induction of a mutually exclusive conformational change. In addition, TIGAR apparently interacts with HOIP through multiple contact sites most likely requiring the overall folding/conformational state of HOIP.
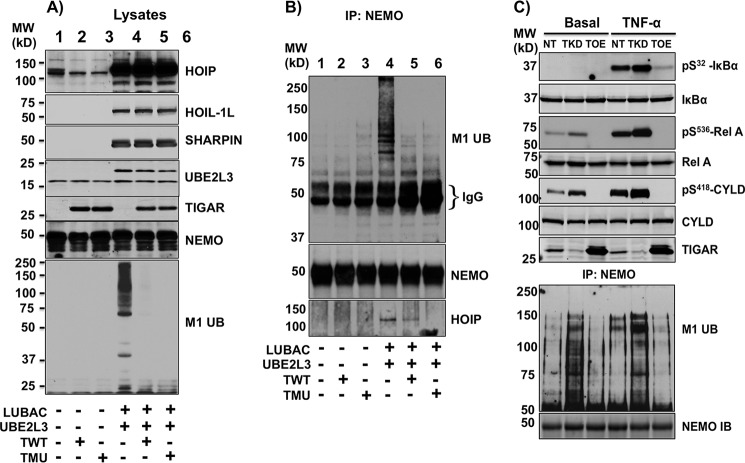
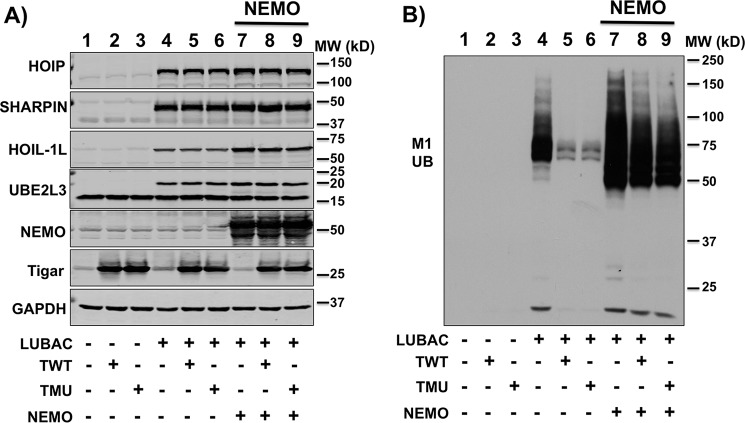
As HOIP is an essential component of the E3 linear ubiquitination activity of LUBAC, we hypothesized that TIGAR might suppress NF-κB activation by inhibiting NEMO M1 linear ubiquitination. As shown in Fig. 8A, we expressed the LUBAC components (HOIP and SHARPIN) along with the E2 ubiquitin-conjugating enzyme UBE2L3. This resulted in a substantial increase in total cellular protein linear ubiquitination that was prevented by the co-expression of both WT (TWT) and the phosphatase-defective (TMU) TIGAR proteins. Similarly, the expression of LUBAC with UBE2L3 resulted in a NEMO-specific increase in M1 linear ubiquitination, as observed in NEMO immunoprecipitates (Fig. 8B, lane 4). As observed in the whole-cell lysates, expression of TWT or TMU blocked NEMO-specific M1 linear ubiquitination (Fig. 8B, lanes 5 and 6). Furthermore, consistent with TIGAR and NEMO competing for IKKβ activation and HOIP binding, overexpression of NEMO reversed the TIGAR inhibition of NEMO linear ubiquitination (Fig. 9, A and B). In addition, immunoprecipitation of NEMO resulted in the co-immunoprecipitation of HOIP that was decreased in the presence of overexpressed TWT or TMU (Fig. 8B, lanes 4–6).
Figure 8.
TIGAR suppresses LUBAC-induced NEMO linear ubiquitination. A, HEK293T cells were transfected with HOIP-FLAG, HOIL-1L-FLAG, SHARPIN-FLAG, UBE2L3-FLAG (2.5 μg each), and either TWT or TMU (10 μg each) plus various amounts of empty vector for a total of 20 μg of cDNA/100-mm dish. 24 h later, cell lysates were prepared and immunoblotted for the indicated proteins and total M1 linear ubiquitinated proteins, as described under “Method details.” Lane 1, pcDNA; lane 2, TWT; lane 3, TMU; lane 4, LUBAC plus UBE2L3; lane 5, LUBAC plus UBE2L3 and TWT; lane 6, LUBAC plus UBE2L3 and TMU cDNAs. These are representative immunoblots independently performed three times. B, the lysates (500 μg) from A were immunoprecipitated (IP) with a NEMO-specific antibody (5 μg) and immunoblotted with the linear ubiquitination-specific (M1 Ub), NEMO, and HOIP antibodies. These are representative immunoblots independently performed three times. C, 3T3-L1 NT, TKD, and TOE adipocytes were either left untreated (Basal) or stimulated with 10 ng/ml TNFα for 5 min. Cell lysates were prepared and immunoblotted for the indicated proteins. The cell lysates (800 μg) were also immunoprecipitated with NEMO-specific antibody, and the NEMO immunoprecipitates were immunoblotted with the linear ubiquitination-specific (M1 Ub) and NEMO antibody. These are representative immunoblots independently performed three times.
Figure 9.
NEMO overexpression rescues the TIGAR inhibition of linear ubiquitination. A, HEK293T cells were transfected with the various indicated cDNAs composed of HOIP, SHARPIN, HOIL-1L, UBEL3 cDNAs (LUBAC), TWT, TMU, and NEMO cDNAs (4 μg each) plus various amounts of empty vector for at a total of 20 μg/100-mm dish for 24 h. Cell lysates were immunoblotted for the indicated proteins as described under “Methods details.” B, the cell lysates in A were immunoblotted with the M1 linear ubiquitination (M1 Ub) antibody. These are representative immunoblots independently performed two times.
Because these data are based upon overexpression of LUBAC or IKKβ, we examined TNFα-stimulated NEMO M1 linear ubiquitination in TKD and TOE cells. As previously observed in Fig. 1, TNFα stimulation increased IκBα and p65 phosphorylation in control 3T3-L1 adipocytes (NT) (Fig. 8C). TKD cells displayed enhanced, whereas TOE cells had reduced, TNFα-stimulated IκBα, p65, and CYLD phosphorylation. In parallel, TNFα treatment of control cells increased NEMO M1 linear ubiquitination that was further increased in the TKD cells and suppressed in the TOE cells. These data demonstrate that endogenous TNFα receptor signal transduction results in NEMO M1 linear ubiquitination that is suppressed by the TIGAR protein.
TIGAR regulates NF-κB signaling in adipose tissue in vivo
To examine a potential role of TIGAR in adipocyte biology in vivo, we first determined the basal and TNFα stimulation of NF-κB signaling in epididymal adipose tissue from WT and whole-body TIGAR-deficient (TKO) mice. Intraperitoneal injection of TNFα for 15 min resulted in a small increase in in Ser32/36-IκB, Ser536-RelA, and Ser418-CYLD phosphorylation (Fig. 10A). In the TKO mice, the vehicle-treated adipose tissue displayed an increase in Ser32/36-IκB, Ser536-RelA, and Ser418-CYLD phosphorylation, similar to that of the TNFα-treated WT mice. However, TNFα stimulation in the TKO mice resulted in a significantly greater extent of IκB, RelA, and CYLD phosphorylation. It should be noted that as the extent of IκB phosphorylation increased, there was a concomitant decrease in IκB mobility, most likely due to an increase in IκB ubiquitination (31).
Figure 10.
TIGAR regulates adipose tissue NF-κB signaling in vivo. C57Bl6/J male WT, whole-body TKO, and TGRS mice at 12 weeks of age were maintained on a low-fat diet as described under “Experimental procedures.” A, two independent control WT and TKO male mice were given an intraperitoneal injection of vehicle or TNFα (10 μg/kg) for 15 min. The epididymal adipose tissue was extracted and immunoblotted for TIGAR, total IκB, pSer32/36-IκB, pSer536-RelA, total RelA, pSer418-CYLD, total CYLD, and actin as loading control. These are representative immunoblots independently performed three times. WT, wildtype mice from TKO inbreds. B, two independent control WT and TGRS mice were given an intraperitoneal injection of vehicle or TNFα for 15 min. The epididymal adipose tissue was extracted and immunoblotted for TIGAR, total IκB, pSer32/36-IκB, pSer536-RelA, total RelA, pSer418-CYLD, total CYLD, and actin as loading control. These are representative immunoblots independently performed two times. C, epididymal adipose tissue from control WT, TKO, and TGRS mice treated with and without TNFα for 15 min were subjected to qRT-PCR for Ccl2 mRNA expression. These data are the average of four independent determinations ± S.D. (error bars). ****, p < 0.0001, TKO-TNF versus WT-TNF and TGRS-TNF versus WT-TNF.
If TIGAR deficiency increased NF-κB signaling, then we would expect that increased TIGAR levels should decrease NF-κB signaling. To test this in vivo, we generated mice with TIGAR cDNA knockin Rosa26-floxed mice that were crossed with adiponectin-Cre (TGRS) mice to specifically increase TIGAR expression in adipocytes (32). Under these conditions, the TIGAR protein was increased ∼1.7 ± 0.3-fold (p < 0.01). In the basal state, there was a small but not significant decrease in the extent of adipose tissue CYLD phosphorylation and no discernible change in IκB or RelA phosphorylation (Fig. 10B). However, following acute TNFα stimulation, the adipocytes from the TIGAR-overexpressing (TGRS) mice displayed a marked blunting of IκB, RelA, and CYLD phosphorylation compared with adipose tissue of control mice. In parallel, the basal adipose tissue Ccl2 gene expression was elevated in the TKO mice and suppressed in the TGRS mice. Moreover, TIGAR deficiency enhanced, whereas TIGAR overexpression suppressed, TNFα stimulation of Ccl2 mRNA levels (Fig. 10C). It should be noted that acute (15-min) TNFα stimulation was insufficient to significantly increase the mRNA levels of Ccl5, IL-1β, IL-13, A20, or TNFα itself (data not shown). Together, these data demonstrate that TIGAR suppresses NF-κB signaling in adipocytes in vitro as well as adipose tissue in vivo.
Discussion
Detailed molecular and cellular analyses have defined a complex set of intracellular signal transduction events that controls the activation of the NF-κB signaling pathway. NF-κB is a transcription factor composed of homo- or heterodimers of Rel domain homology proteins, including p65/RelA, RelB, c-Rel, p105/p50 (NF-κB1), and p100/p52 (NF-κB2) (33, 34). The dimeric NF-κB complexes are localized to the cytosol through interaction with IκBα and, following canonical signaling activation (e.g. by TNFα), result in TNFα receptor activation of RIP1, TRAF2, and NEMO Lys63-linked ubiquitination (35, 36). Ubiquitinated TRAF2 also provides a scaffold for the recruitment to the TAK1-TAB1-TAB2/3 complex, and Lys63 ubiquitination recruits the IKK complex so that the TAK1 complex can phosphorylate and activate IKKβ, leading to subsequent IκBα phosphorylation and IκBα Lys48 ubiquitination and subsequent IκBα degradation (37, 38). The phosphorylation-dependent decrease in IκBα then releases the NF-κB dimer (i.e. p65/p50) that translocates to the nucleus and activates NF-κB–dependent target genes.
In addition to multisite ubiquitination, recent studies have demonstrated that NEMO undergoes M1 linear ubiquitination by LUBAC, composed of catalytically active HOIP associated with HOIL-1L and/or SHARPIN assembled as either HOIP–HOIL-IL or HOIP-SHARPIN dimeric and/or HOIL-IL–HOIP–SHARPIN trimeric complexes (35, 36, 39–42). Inhibition of LUBAC function substantially reduced NEMO linear ubiquitination and suppressed NF-κB activation and downstream signaling events in a cell context–dependent manner (27, 36, 39, 40).
Numerous studies have demonstrated an intimate and co-dependent relationship between metabolism and NF-κB–regulated inflammatory signaling. For example, endoplasmic reticulum and oxidative stress activate NF-κB signaling, and conversely, NF-κB activation has also been reported to induce oxidative stress (43–45). In both humans and animal models of insulin resistance, diet-induced obesity results in marked activation of adipose tissue inflammation, NF-κB signaling, and endoplasmic reticulum and oxidative stress (16, 17, 44, 46, 47). Relieving of any of these events has been shown to improve metabolic function and to restore insulin sensitivity.
One important pathway controlling cellular redox state is the bifurcation of glucose catabolism through the glycolytic or pentose phosphate pathways that increases or decreases the cellular oxidation state, respectively. As TIGAR was identified as an important control enzyme regulating carbon flux through the glycolytic and pentose phosphate pathways (5, 8), we undertook an analysis of TIGAR's metabolic function in cultured 3T3-L1 adipocytes. To our surprise, we noticed that TIGAR deficiency resulted in an enhanced TNFα activation of NF-κB signaling and NF-κB–dependent gene expression. In this regard, TIGAR was causally linked, albeit indirectly, to the regulation of the NF-κB inflammatory signaling pathways in aging and cancer (48–51). TIGAR was also found in the connectome associated with the E2 ubiquitin-conjugating enzyme UBE2L3 that provides ubiquitin to LUBAC for linear chain ubiquitination (52). As adipocytes express relatively high levels of the TIGAR protein, we speculated that the metabolic regulatory functions of TIGAR might also intersect with the NF-κB inflammatory signaling pathway. The data presented in this study demonstrate that TIGAR, independent of its phosphatase activity, suppresses NF-κB activation by preventing NEMO linear ubiquitination through a direct binding to the LUBAC subunit HOIP. Because TIGAR suppresses the E3-conjugating ligase activity of the LUBAC complex and co-immunoprecipitated with HOIP in the absence of either SHARPIN or HOIL-1L, it is unlikely that TIGAR binds to the HOIL-1L or SHARPIN subunits. Consistent with this conclusion, we were unable to detect any direct interaction of TIGAR with HOIL-1L or SHARPIN.
Several studies have identified the structural domains of HOIP, HOIL-1L, and SHARPIN and their interactions responsible for LUBAC assembly and linear ubiquitination activity (27–30). Although the molecular basis for NEMO recognition has not been fully elucidated, NEMO has been reported to bind to the NZF1 domain of HOIP (27). The ability of NEMO in a dose-dependent manner to reverse TIGAR inhibition of IKKβ activation and conversely TIGAR in a dose-dependent manner to inhibit NEMO binding to HOIP strongly argues that TIGAR and NEMO compete with each other for binding to LUBAC. Although we confirmed the requirement for the HOIP NZF1 domain for NEMO binding, ablation of the NZF1 domain had no significant effect on TIGAR binding. Moreover, we were unable to detect any specific HOIP motif required for TIGAR binding, and both amino and C-terminal deletions suggest that the TIGAR binding requires multiple contacts across the conformational state of HOIP. This conclusion is consistent with an allosteric NEMO- and TIGAR-induced conformational change in HOIP that is responsible for the mutually exclusive binding properties rather than through a direct competition at a unique amino acid domain. Although the molecular structure of the HOIP ring between ring (UBA-RING1-IBR-RINF2-LDD) domain has recently been defined (53), structural analysis of full-length HOIP with TIGAR remains necessary to define our understanding of LUBAC-NEMO interactions.
Currently, it is generally thought that overexpressing IKKβ can result in the autophosphorylation and activation of IKKβ kinase activity independent of upstream activators, although it has also been reported that the TAK1 kinase complex (TAK1-TAB2/3) can phosphorylate IKKβ at a priming site (Ser177) that then allows for IKKβ autophosphorylation at Ser181 necessary for full activation (24). Nevertheless, our data clearly demonstrate that TIGAR expression inhibited the autophosphorylation of overexpressed IKKβ, proximal IKKβ downstream substrates, and NF-κB target gene expression. These findings further indicate that overexpressed IKKβ does not simply auto-activate itself but is also dependent upon LUBAC function for activation. In this regard, recent studies have suggested that the IKK complex, when recruited to LUBAC, results in the linear ubiquitination of NEMO (27, 36). In turn, NEMO also contains a ubiquitin-binding domain such that NEMO in a second IKK complex is then recruited to the previous ubiquitinated IKK complex bound to LUBAC (36, 37). The assembly of multimeric IKK complexes on LUBAC as proposed by Iwai and colleagues (54) then allows for the trans-autophosphorylation and activation of IKKβ.
In addition to these new mechanistic findings, our data also suggest an important physiologic role for TIGAR function in regulating adipose tissue inflammation. In vivo analysis of TIGAR knockout and adipocyte-specific TIGAR-overexpressing mice recapitulated the enhanced and suppressed TNFα stimulation of NF-κB signaling, respectively, that occurred in cultured 3T3-L1 adipocytes. We also observed that high-fat diet–induced obesity, a well-established inducer of adipose tissue inflammation (16), also results in the down-regulation of adipocyte TIGAR protein and mRNA levels.3 Although the major contribution to adipose tissue inflammation is the recruitment/activation of immune cell infiltrates, our data suggest that the enhancement of intrinsic adipocyte NF-κB signaling is, at least partly, due to the down-regulation of the TIGAR protein. Further studies are now needed to determine the mechanism(s) responsible for the adipocyte-specific diet-induced down-regulation of TIGAR.
In summary, the data presented in this paper demonstrate a novel cross-talk between an important metabolic regulator, TIGAR, and the major signaling pathway controlling innate and adaptive immune responses, NF-κB. TIGAR, independent of its phosphatase activity, inhibits NEMO linear ubiquitination through a direct binding interaction with LUBAC. This results in a suppression of NF-κB downstream inflammatory signaling and may provide novel targets for the development of agents to modulate these signaling events in states of dysregulated metabolism and inflammation.
Experimental procedures
TIGAR knockout/overexpressing and adiponectin-Cre mice
Adiponectin-Cre mice maintained on the C56Bl/6/J background for greater than 10 generations were a kind gift from Dr. Evan Rosen (Beth Israel Deaconess Medical Center). TIGAR conventional knockout (deletion) embryonic stem cells were purchased from the UC Davis KOMP Repository Knockout Mouse Project (Project ID VG19113). Embryonic stem cells were cultured, and the B6 chimera mice were produced by the Gene Targeting Facility, Albert Einstein College of Medicine (Bronx, NY). TIGAR-deficient mice (TKO) were back-crossed 8 generations in the C57BL/6J strain (000664, Jackson Laboratory). TIGAR tissue-specific overexpressing mice were generated by Applied StemCell, Inc. (Milpitas, CA). Briefly, the TIGAR mouse cDNA plasmid (9630033f20rik) was ligated into plasmid pCAG-loxP-stop-loxP-attB (pBT378-CAG-LSL). This plasmid and integrase mRNA for attP integration was injected into the pronucleus of zygotes, and the recombinant positive F1 germ line transmitted mice were shipped to Albert Einstein College of Medicine. These mice were then back-crossed with C57BL/6J more than seven times before mating with the adiponectin-Cre mice to generate adipocyte-specific TIGAR-overexpressing (TGRS) mice.
Mouse husbandry
WT, TKO, and TGRS mice were housed in a facility equipped with a 12-h light/dark cycle. Animals were fed a normal chow diet that contains 62.3% (kcal) carbohydrates, 24.5% protein, and 13.1% fat (5053, LabDiet). Male mice at 12 weeks of age were given an intraperitoneal injection of either 100 μl of saline or 100 μl of TNFα (10 μg of TNFα/kg body weight) in saline solution for 15 min. The mice were killed, and the tissues were collected and snap-frozen in liquid nitrogen and stored in a −80 °C freezer. All studies were approved by and performed in compliance with the guidelines of the Albert Einstein College of Medicine Institutional Animal Care and Use Committee.
Method details
Cell lines
Murine 3T3-L1 preadipocyte and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin. Cell lines were maintained in a 5% CO2 incubator at 37 °C. Cell lines were routinely tested to exclude Mycoplasma contaminations.
3T3-L1 adipocyte differentiation
3T3-L1 preadipocytes were obtained from the American Type Culture Collection repository and were cultured at 37 °C in an 8% CO2 atmosphere in DMEM containing 25 mm glucose and 10% calf serum. Confluent cultures were induced to differentiate by incubation of the cells with DMEM containing 25 mm glucose, 10% fetal bovine serum, 1 μg/ml insulin, 1 μm dexamethasone, 0.5 mm isobutyl-1-methylxanthine, and 1 μm rosiglitazone. After 3 days, the medium was changed to DMEM containing 25 mm glucose, 10% fetal bovine serum, and 1 μg/ml insulin, and the incubation was continued for an additional 2 days. The medium was then changed to DMEM containing 25 mm glucose and 10% fetal bovine serum. Under these conditions, more than 95% of the cell population morphologically differentiated into adipocytes. The adipocytes were maintained for an additional 3–5 days before use. Fully differentiated 3T3-L1 adipocytes were placed overnight in DMEM in the presence of 10% serum and then incubated with 10 ng/ml mouse TNFα for the time indicated.
Lentivirus short hairpin RNA knockdown in 3T3-L1 preadipocytes
MISSION lentiviral shRNA bacterial glycerol stocks for mouse TIGAR shRNA and non-target control shRNA plasmids were obtained from Sigma-Aldrich. The lentiviral human TIGAR plasmid DNA was obtained from GeneCopoeia (Rockville, MD). The plasmid DNAs were transformed and amplified in Mix & Go Competent Cells-Strain HB 101 (Zymo Research, catalog no. T3013, Irvine, CA) and purified using PowerPrep HP Plasmid Maxiprep kits with prefilters (Origene, Rockville, MD) and were transfected into human embryonic kidney 293T cells along with lentiviral packaging mix (Sigma-Aldrich) to produce lentiviruses per the manufacturer's instructions. 3T3-L1 preadipocytes (80% confluence) were infected with the non-target, mouse TIGAR shRNA, and human TIGAR overexpression lentivirus, respectively, selected by puromycin, and subjected to standard adipocyte differentiation. The TKD cells were further infected with human WT TIGAR or enzymatic inactive mutant TIGAR lentiviruses and selected by hygromycin to produce TKD rescue TWT and TMU cells.
Total RNA extraction and quantitative RT-PCR
Cellular total RNA was extracted using QIAzol lysis reagent and an RNeasy minikit (Qiagen, Germantown, MD). First-strand cDNA was synthesized using the SuperScript® VILO cDNA synthesis kit (Thermo Fisher Scientific Invitrogen). TaqMan RT-PCR was performed for measurement of mRNA using the ΔΔCt method. Gene expression was adjusted by comparison with Rpl7 expression. The quantitative RT-PCR results were analyzed by RG Manager version 1.2.1 (Applied Biosystems). Primer-probe mixture for Rpl7 was customized, and other primer-probe mixtures were obtained from Thermo Fisher Applied Biosystems.
WT and mutant human TIGAR lentiviral plasmid construction
The full-length human TIGAR cDNA (SC320794, Origene) was used as a template to produce DNA fragment with the addition of BamHI and XbaI sites, which were then cloned into a pCR-Blunt II-TOPO vector (Life Technologies, Inc.). The resulting constructs were sequenced and were used as templates to perform WT and site-directed mutagenesis to change His11, Glu102, and His198 to alanines simultaneously using the QuikChange II XL site-directed mutagenesis kit (Stratagene, San Diego, CA) according to the manufacturer's instructions. The primers used for specific histidine- and glutamic acid-to-alanine mutations are listed in Table S1. The fragments for WT and multiple mutated TIGAR were then subcloned into the lentiviral empty vector (CMV/TO promoter, hygromycin-selectable, Addgene plasmid 17484) using BamHI and XbaI restriction sites. The full-length WT and mutated human TIGAR in mammalian expression lentiviral vectors were confirmed by sequencing. qRT-PCR and Western blotting were also used to confirm the TWT and TMU for mRNA and protein expression. Plasmids used in this study are reported in Table S1.
WT and mutant human TIGAR plasmid cDNA construction
The lentiviral WT and mutant TIGAR plasmid DNAs were then used as templates to produce blunt PCR fragments with forward primer containing CACC sequence at the 5′-end and with reverse primer containing TAA stop codon at the 5′-end. The WT and mutant human TIGAR plasmids were then produced using the pcDNA3.1 directional TOPO expression kit (Life Technologies) according to the manufacturer's protocol.
Immunoblotting
Culture cells were washed with cold PBS and scraped and homogenized using Ceria stabilized zirconium oxide beads (MidSci, Valley Park, MO) in a radioimmune precipitation assay lysis buffer (sc-24948, Santa Cruz Biotechnology) containing Halt protease and phosphatase inhibitor mixture (catalog no. 78442, Thermo Fisher Scientific), 50 μm MG132, 50 μm ALLN, and 50 μm PR-619 (EMD Millipore, Darmstadt, Germany). Homogenates were centrifuged for 15 min at 21,000 × g at 4 °C, and supernatants were collected for the protein assay. Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membrane using iBlot Blotting System (Thermo Fisher Scientific). The immunoblot membrane was blocked with Pierce Protein-Free T20 (TBS) blocking buffer (product no. 37571, Thermo Fisher Scientific) and incubated with the first antibody indicated in the blocking buffer. Blots were washed in TBS with Tween 20 (TBST) and incubated with either IRDye 800CW secondary antibody (LI-COR, Lincoln, NE) or horseradish peroxidase–conjugated secondary antibody in blocking buffer (listed in Table S1). The Membrane was washed with TBST and visualized either by the Odyssey Imaging System (LI-COR) or enhanced chemiluminescence (ECL) (Thermo Fisher Scientific Pierce) method. ImageJ was used to quantify protein bands on the membrane.
Co-immunoprecipitation
The antibody for immunoprecipitation was diluted in 250 μl of Pierce protein-free T20 (TBS) blocking buffer and incubated with magnetic Dynabeads Protein G (catalog no. 10004D, Thermo Fisher Scientific Novex) for 10 min at room temperature. The cleared cellular lysate was then incubated with the antibody-beads complex for 15 min at room temperature and washed five times with immunoprecipitation lysis buffer (catalog no. 87788, Thermo Fisher Scientific Pierce). Immunoprecipitated proteins were eluted from beads by boiling samples for 5 min at 95 °C in 2× SDS loading sample buffer and analyzed by Western blot. For linear ubiquitinated NEMO blot, 1.2× SDS loading sample buffer was used to elute the denatured immunoprecipitated proteins, and 7.5% polyacrylamide gel was used for protein separation.
Plasmid DNA transfection
HEK293T cells were transfected when 80% confluent in 60-mm dishes except where otherwise indicated with the various plasmid DNAs with transfection reagents Lipofectamine 2000 (Thermo Fisher), GenJet II (SignaGen Laboratories, Rockville, MD), or BioT (Morganville Scientific, Morganville, NJ) per the manufacturer's instructions. The transfected cells were in culture for 24 h and were used for further experiments.
HOIP deletion constructs
HOIP deletion mutants were generated from N-terminal FLAG-tagged human HOIP (EX-Z1067-M12, GeneCopoeia). Deletions of N-terminal FLAG-tagged human HOIP were made using a QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol and verified by sequencing.
Luciferase activity assay
The NF-κB–specific luciferase reporter construct (0.8 μg) plus Renilla luciferase (0.2 μg) and either empty vector (2 μg), TWT (2 μg), or TMU (2 μg) plasmid cDNAs were mixed in 100 μl of DMEM, and 3 μl of BioT were added into the DNA/DMEM solution to make the DNA-transfection reagent complex by standing at room temperature for 5 min. The entire complex was added directly to the HEK293T cells carefully, the mixture was tilted back and forth a few times to mix the complex into the medium, and the cells were returned to the CO2 incubator for 24 h. The transfected cells were either left untreated or stimulated with 20 ng/ml human TNFα (PeproTech, Rocky Hill, NJ) for 24 h. The cells were lysed in 0.5 ml of passive lysis buffer, and the cleared lysates were used for a luciferase assay using the Dual-Luciferase reporter assay system and a Glomax 96-microplate Luminometer (Promega, Madison, WI) with the company's protocol.
IKKβ in vitro kinase assay
HEK293T cells were cultured on 6-well plates for 24 h and transfected with IKKβ and TIGAR expression plasmids. The cells were collected and lysed with 150 μl of lysis buffer (50 mm Tris, pH 7.4, 250 mm NaCl, 5 mm EDTA, 50 mm NaF, 1 mm Na3VO4, 1% Nonidet P-40, and protease inhibitor mixtures) after 18 h transfection. The cell extracts were prepared and used (10 μl) for the IKKβ kinase assay according to manufacturer's protocol. IκBα was used as a substrate.
Quantification and statistical analyses
The number of independent experimental replications and the average with S.D. are reported in the figure legends. The data were analyzed by one-way analysis of variance using Prism version 7 software (GraphPad Software, Inc., La Jolla, CA) for comparison of multiple groups or unpaired t test for two groups. The statistical analyses were made at significance levels as follows: ns, not statistically significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Author contributions
Y. T., H. K., and B. A. N. designed and conducted experiments, compiled/analyzed and interpreted data, prepared figures, and wrote and edited the manuscript. M. K.-M. conducted experiments that provided the basis for the data presented and assisted in experimental design and editing the manuscript. J. B. P. assisted in experimental design, compiled/analyzed and interpreted data, and wrote and edited the manuscript. E. Y. prepared plasmids, assisted in experimental design, compiled/analyzed data, and wrote and edited the manuscript. J. E. P. was responsible for the overall direction, experimental design, analyses and interpretation of data, and writing and editing of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Anjana Rao (FLAG-IKKβ), Kunliang Guan (HA-NEMO), Martin Dorf (FLAG-HOIP, FLAG-HOIL-1L, FLAG-Sharpin), Shao-Cong Sun (HA-UBE2L3), and Eric Campeau (pLenti CMV/TO Hygro) for providing the indicated cDNAs.
This work was supported by National Institutes of Health Grants DK033823 and DK020541. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
Y. Tang, H. Kwon, B. A. Neel, M. Kasher-Meron, J. B. Pessin, E. Yamada, and J. E. Pessin, unpublished results.
- TIGAR
- Tp53-induced glycolysis and apoptosis regulator
- 23BPG
- 2,3-bisphosphoglycerate
- F26P
- fructose-2,6-bisphosphate
- PFK1
- 6-phosphofructo-1-kinase
- FBP
- fructose-1,6-bisphosphatase
- PGM
- phosphoglycerate mutase
- LUBAC
- linear ubiquitination assembly complex
- NT
- mammalian Non-Target shRNA lentivirus
- TKD
- TIGAR shRNA knockdown lentivirus
- TOE
- TIGAR cDNA-overexpressing lentivirus
- TWT
- shRNA-resistant WT TIGAR
- TMU
- phosphatase activity–defective TIGAR mutant
- ZF
- zinc finger
- NZF
- Npl4 zinc finger
- UBA
- ubiquitin-associated
- R
- ring fingers
- RBR
- ring between ring fingers
- LDD
- linear ubiquitin chain–determining domain
- TNF
- tumor necrosis factor
- HEK
- human embryonic kidney
- DMEM
- Dulbecco's modified Eagle's medium
- shRNA
- short hairpin RNA
- qRT-PCR
- quantitative RT-PCR
- TKO
- TIGAR-deficient
- TGRS
- adipocyte-specific TIGAR-overexpressing.
References
- 1. Mor I., Cheung E. C., and Vousden K. H. (2011) Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold. Spring Harb. Symp. Quant. Biol. 76, 211–216 10.1101/sqb.2011.76.010868 [DOI] [PubMed] [Google Scholar]
- 2. Rigden D. J. (2008) The histidine phosphatase superfamily: structure and function. Biochem. J. 409, 333–348 10.1042/BJ20071097 [DOI] [PubMed] [Google Scholar]
- 3. Wang S. J., and Gu W. (2014) To be, or not to be: functional dilemma of p53 metabolic regulation. Curr. Opin. Oncol. 26, 78–85 10.1097/CCO.0000000000000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao L., Mao Y., Zhao Y., Cao Y., and Chen X. (2016) Role of multifaceted regulators in cancer glucose metabolism and their clinical significance. Oncotarget 7, 31572–31585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., and Vousden K. H. (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- 6. Rider M. H., Bertrand L., Vertommen D., Michels P. A., Rousseau G. G., and Hue L. (2004) 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 381, 561–579 10.1042/BJ20040752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okar D. A., Manzano A., Navarro-Sabatè A., Riera L., Bartrons R., and Lange A. J. (2001) PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem. Sci. 26, 30–35 10.1016/S0968-0004(00)01699-6 [DOI] [PubMed] [Google Scholar]
- 8. Li H., and Jogl G. (2009) Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator). J. Biol. Chem. 284, 1748–1754 10.1074/jbc.M807821200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerin I., Noël G., Bolsée J., Haumont O., Van Schaftingen E., and Bommer G. T. (2014) Identification of TP53-induced glycolysis and apoptosis regulator (TIGAR) as the phosphoglycolate-independent 2,3-bisphosphoglycerate phosphatase. Biochem. J. 458, 439–448 10.1042/BJ20130841 [DOI] [PubMed] [Google Scholar]
- 10. Hoshino A., Matoba S., Iwai-Kanai E., Nakamura H., Kimata M., Nakaoka M., Katamura M., Okawa Y., Ariyoshi M., Mita Y., Ikeda K., Ueyama T., Okigaki M., and Matsubara H. (2012) p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J. Mol. Cell. Cardiol. 52, 175–184 10.1016/j.yjmcc.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 11. Katsel P., Tan W., Fam P., Purohit D. P., and Haroutunian V. (2013) Cell cycle checkpoint abnormalities during dementia: a plausible association with the loss of protection against oxidative stress in Alzheimer's disease [corrected]. PLoS One 8, e68361 10.1371/journal.pone.0068361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lui V. W., Wong E. Y., Ho K., Ng P. K., Lau C. P., Tsui S. K., Tsang C. M., Tsao S. W., Cheng S. H., Ng M. H., Ng Y. K., Lam E. K., Hong B., Lo K. W., Mok T. S., et al. (2011) Inhibition of c-Met downregulates TIGAR expression and reduces NADPH production leading to cell death. Oncogene 30, 1127–1134 10.1038/onc.2010.490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung E. C., Athineos D., Lee P., Ridgway R. A., Lambie W., Nixon C., Strathdee D., Blyth K., Sansom O. J., and Vousden K. H. (2013) TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev. Cell 25, 463–477 10.1016/j.devcel.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wanka C., Steinbach J. P., and Rieger J. (2012) Tp53-induced glycolysis and apoptosis regulator (TIGAR) protects glioma cells from starvation-induced cell death by up-regulating respiration and improving cellular redox homeostasis. J. Biol. Chem. 287, 33436–33446 10.1074/jbc.M112.384578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu H. P., Xie J. M., Li B., Sun Y. H., Gao Q. G., Ding Z. H., Wu H. R., and Qin Z. H. (2015) TIGAR regulates DNA damage and repair through pentosephosphate pathway and Cdk5-ATM pathway. Sci. Rep. 5, 9853 10.1038/srep09853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hotamisligil G. S. (2006) Inflammation and metabolic disorders. Nature 444, 860–867 10.1038/nature05485 [DOI] [PubMed] [Google Scholar]
- 17. Kwon H., and Pessin J. E. (2013) Adipokines mediate inflammation and insulin resistance. Front. Endocrinol. (Lausanne) 4, 71 10.3389/fendo.2013.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patni N., and Garg A. (2015) Congenital generalized lipodystrophies– new insights into metabolic dysfunction. Nat. Rev. Endocrinol. 11, 522–534 10.1038/nrendo.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samuel V. T., and Shulman G. I. (2016) The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J. Clin. Invest. 126, 12–22 10.1172/JCI77812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shoelson S. E., Lee J., and Goldfine A. B. (2006) Inflammation and insulin resistance. J. Clin. Invest. 116, 1793–1801 10.1172/JCI29069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reiley W., Zhang M., Wu X., Granger E., and Sun S. C. (2005) Regulation of the deubiquitinating enzyme CYLD by IκB kinase γ-dependent phosphorylation. Mol. Cell. Biol. 25, 3886–3895 10.1128/MCB.25.10.3886-3895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun S. C. (2010) CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ. 17, 25–34 10.1038/cdd.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Polley S., Huang D. B., Hauenstein A. V., Fusco A. J., Zhong X., Vu D., Schröfelbauer B., Kim Y., Hoffmann A., Verma I. M., Ghosh G., and Huxford T. (2013) A structural basis for IκB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol. 11, e1001581 10.1371/journal.pbio.1001581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang J., Clark K., Lawrence T., Peggie M. W., and Cohen P. (2014) An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem. J. 461, 531–537 10.1042/BJ20140444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meininger I., Griesbach R. A., Hu D., Gehring T., Seeholzer T., Bertossi A., Kranich J., Oeckinghaus A., Eitelhuber A. C., Greczmiel U., Gewies A., Schmidt-Supprian M., Ruland J., Brocker T., Heissmeyer V., et al. (2016) Alternative splicing of MALT1 controls signalling and activation of CD4+ T cells. Nat. Commun. 7, 11292 10.1038/ncomms11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Staal J., Driege Y., Bekaert T., Demeyer A., Muyllaert D., Van Damme P., Gevaert K., and Beyaert R. (2011) T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 30, 1742–1752 10.1038/emboj.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., and Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 10.1038/ncb1821 [DOI] [PubMed] [Google Scholar]
- 28. Sasaki K., and Iwai K. (2015) Roles of linear ubiquitinylation, a crucial regulator of NF-κB and cell death, in the immune system. Immunol. Rev. 266, 175–189 10.1111/imr.12308 [DOI] [PubMed] [Google Scholar]
- 29. Schaeffer V., Akutsu M., Olma M. H., Gomes L. C., Kawasaki M., and Dikic I. (2014) Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell 54, 349–361 10.1016/j.molcel.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 30. Shimizu Y., Taraborrelli L., and Walczak H. (2015) Linear ubiquitination in immunity. Immunol. Rev. 266, 190–207 10.1111/imr.12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gonen H., Bercovich B., Orian A., Carrano A., Takizawa C., Yamanaka K., Pagano M., Iwai K., and Ciechanover A. (1999) Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J. Biol. Chem. 274, 14823–14830 10.1074/jbc.274.21.14823 [DOI] [PubMed] [Google Scholar]
- 32. Kong X., Williams K. W., and Liu T. (2017) Genetic mouse models: the powerful tools to study fat tissues. Methods Mol. Biol. 1566, 99–107 10.1007/978-1-4939-6820-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayden M. S., and Ghosh S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 10.1101/gad.183434.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vallabhapurapu S., and Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- 35. Emmerich C. H., Ordureau A., Strickson S., Arthur J. S., Pedrioli P. G., Komander D., and Cohen P. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 110, 15247–15252 10.1073/pnas.1314715110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., and Walczak H. (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 10.1016/j.molcel.2009.10.013 [DOI] [PubMed] [Google Scholar]
- 37. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., and Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 10.1016/j.cell.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 38. Ruland J. (2011) Return to homeostasis: downregulation of NF-κB responses. Nat. Immunol. 12, 709–714 10.1038/ni.2055 [DOI] [PubMed] [Google Scholar]
- 39. Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S. J., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., et al. (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 10.1038/nature09814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., and Iwai K. (2011) SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 10.1038/nature09815 [DOI] [PubMed] [Google Scholar]
- 41. Niu J., Shi Y., Iwai K., and Wu Z. H. (2011) LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 30, 3741–3753 10.1038/emboj.2011.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., and Walczak H. (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 10.1038/nature09816 [DOI] [PubMed] [Google Scholar]
- 43. Nakajima S., and Kitamura M. (2013) Bidirectional regulation of NF-κB by reactive oxygen species: a role of unfolded protein response. Free Radic. Biol. Med. 65, 162–174 10.1016/j.freeradbiomed.2013.06.020 [DOI] [PubMed] [Google Scholar]
- 44. Meyerovich K., Ortis F., Allagnat F., and Cardozo A. K. (2016) Endoplasmic reticulum stress and the unfolded protein response in pancreatic islet inflammation. J. Mol. Endocrinol. 57, R1–R17 10.1530/JME-15-0306 [DOI] [PubMed] [Google Scholar]
- 45. Cai D., and Liu T. (2012) Inflammatory cause of metabolic syndrome via brain stress and NF-κB. Aging 4, 98–115 10.18632/aging.100431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwon H., Laurent S., Tang Y., Zong H., Vemulapalli P., and Pessin J. E. (2014) Adipocyte-specific IKKβ signaling suppresses adipose tissue inflammation through an IL-13-dependent paracrine feedback pathway. Cell Rep. 9, 1574–1583 10.1016/j.celrep.2014.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Yamada E., Zong H., and Pessin J. E. (2015) Fyn activation of mTORC1 stimulates the IRE1α-JNK pathway, leading to cell death. J. Biol. Chem. 290, 24772–24783 10.1074/jbc.M115.687020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salminen A., and Kaarniranta K. (2010) Glycolysis links p53 function with NF-κB signaling: impact on cancer and aging process. J. Cell. Physiol. 224, 1–6 [DOI] [PubMed] [Google Scholar]
- 49. Sinha S., Ghildiyal R., Mehta V. S., and Sen E. (2013) ATM-NFκB axis-driven TIGAR regulates sensitivity of glioma cells to radiomimetics in the presence of TNFα. Cell Death Dis. 4, e615 10.1038/cddis.2013.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kawauchi K., Araki K., Tobiume K., and Tanaka N. (2008) p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat. Cell Biol. 10, 611–618 10.1038/ncb1724 [DOI] [PubMed] [Google Scholar]
- 51. Martinez-Outschoorn U. E., Trimmer C., Lin Z., Whitaker-Menezes D., Chiavarina B., Zhou J., Wang C., Pavlides S., Martinez-Cantarin M. P., Capozza F., Witkiewicz A. K., Flomenberg N., Howell A., Pestell R. G., Caro J., et al. (2010) Autophagy in cancer associated fibroblasts promotes tumor cell survival: role of hypoxia, HIF1 induction and NFκB activation in the tumor stromal microenvironment. Cell Cycle 9, 3515–3533 10.4161/cc.9.17.12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Havugimana P. C., Hart G. T., Nepusz T., Yang H., Turinsky A. L., Li Z., Wang P. I., Boutz D. R., Fong V., Phanse S., Babu M., Craig S. A., Hu P., Wan C., Vlasblom J., et al. (2012) A census of human soluble protein complexes. Cell 150, 1068–1081 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lechtenberg B. C., Rajput A., Sanishvili R., Dobaczewska M. K., Ware C. F., Mace P. D., and Riedl S. J. (2016) Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550 10.1038/nature16511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fujita H., Rahighi S., Akita M., Kato R., Sasaki Y., Wakatsuki S., and Iwai K. (2014) Mechanism underlying IκB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol. Cell. Biol. 34, 1322–1335 10.1128/MCB.01538-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.