Abstract
Aerobactin, a citryl-hydroxamate siderophore, is produced by a number of pathogenic Gram-negative bacteria to aid in iron assimilation. Interest in this well-known siderophore was reignited by recent investigations suggesting that it plays a key role in mediating the enhanced virulence of a hypervirulent pathotype of Klebsiella pneumoniae (hvKP). In contrast to classical opportunistic strains of K. pneumoniae, hvKP causes serious life-threatening infections in previously healthy individuals in the community. Multiple contemporary reports have confirmed fears that the convergence of multidrug-resistant and hvKP pathotypes has led to the evolution of a highly transmissible, drug-resistant, and virulent “super bug.” Despite hvKP harboring four distinct siderophore operons, knocking out production of only aerobactin led to a significant attenuation of virulence. Herein, we continue our structural and functional studies on the biosynthesis of this crucial virulence factor. In vivo heterologous production and in vitro reconstitution of aerobactin biosynthesis from hvKP was carried out, demonstrating the specificity, stereoselectivity, and kinetic throughput of the complete pathway. Additionally, we present a steady-state kinetic analysis and the X-ray crystal structure of the second aerobactin synthetase IucC, as well as describe a surface entropy reduction strategy that was employed for structure determination. Finally, we show solution X-ray scattering data that support a unique dimeric quaternary structure for IucC. These new insights into aerobactin assembly will help inform potential antivirulence strategies and advance our understanding of siderophore biosynthesis.
Keywords: siderophore, natural product biosynthesis, structural biology, enzyme structure, Klebsiella pneumonia, protein structure, X-ray crystallography, Aerobactin, Surface Entropy Reduction
Introduction
Klebsiella pneumoniae (KP)3 is a Gram-negative bacillus ubiquitously found in surface water and soil (1). Besides this saprophytic existence, it is a common colonizer of the mucosal surfaces of the human gastrointestinal tract. However, KP has shown a propensity to transition from colonizer to opportunistic pathogen in susceptible individuals with underlying medical problems and compromised immune systems (2, 3). Historically described as a cause of community-acquired pneumonia, KP is now recognized as one of the most common causative agents of hospital-acquired infections globally, capable of infiltrating the lungs, urinary tract, surgical wounds, and bloodstream. This has been particularly problematic for the medical community as nosocomial KP isolates are often highly drug-resistant, making these healthcare-associated infections especially difficult to treat (4–6).
In addition to acquiring drug resistance, KP can also present with a novel hypervirulent pathotype (hvKP), which is now widely recognized by the medical community (7–9). In 1986, a seminal report was published linking KP to severe life-threatening infections in ambulatory individuals from the community (10). In the decades that followed, numerous reports from around the globe of KP-associated infections, including hepatic abscesses, endopthalmitis, meningitis, osteomyelitis, and necrotizing fasciitis, in otherwise healthy individuals, illustrated the emergence of this new hypervirulent pathotype (11–18). Amid recent reports suggesting the confluence of both drug-resistance and hypervirulence determinants in clinical isolates of KP, medical professionals fear a true “super bug” may be evolving (19, 20).
Investigations to understand the enhanced virulence of hvKP strains highlighted the importance of siderophore systems and iron acquisition (21, 22). Because iron is a vital mineral nutrient, and bioavailable iron is often below the micromolar concentration required to support growth, siderophores are often considered critical virulence factors for pathogenic bacteria (23, 24). hvKP is highly associated with a large virulence-conferring plasmid containing genes for the biosynthesis of aerobactin and the glycosylated enterobactin analogue salmochelin (25). Combined with chromosomally encoded enterobactin and yersiniabactin operons, most strains of hvKP have the ability to produce four distinct siderophores. However, in vitro and in vivo experiments have both demonstrated that of the four siderophores, knocking out the production of only aerobactin significantly diminished the virulence of hvKP (26, 27). Although the mechanistic basis underlying the relative importance of aerobactin is unknown, there is growing appreciation for the differing roles (e.g. colonization versus invasion) siderophores can perform, as well as numerous examples of individual siderophores contributing significantly to pathogenicity when redundant siderophores are available (24).
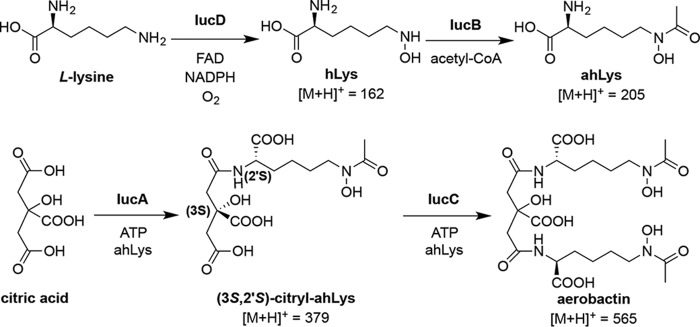
Based on the critical role of aerobactin in the evolution of a particularly worrisome new pathogen, we were motivated to revisit this early-described siderophore and study its biosynthesis in detail. Aerobactin, a mixed carboxylate-hydroxamate siderophore, was first isolated from the iron-deficient culture media of Aerobacter aerogenes by Gibson and Magrath in 1969 (28). Genetic experiments in the early 1980s using the Escherichia coli ColV-K30 plasmid identified the aerobactin operon containing genes for four enzymes (“iron uptake chelate” iucA–D) and an outer membrane receptor (“iron uptake transporter” iutA) (29). By selectively knocking out and re-complementing iuc gene function, and detecting the resultant biosynthetic intermediates in culture supernatants, Neilands and co-workers (30, 31) were able to decipher the aerobactin biosynthetic pathway, an effort they described as “ … the first entire gene-product-function analysis of any siderophore gene complex from any microbial species” (Fig. 1). Subsequent biochemical studies on isolated E. coli IucD investigated and confirmed its proposed l-lysine N6-monooxygenase activity (32). Similarly, early studies on isolated E. coli IucB were able to confirm its N6-hydroxy-l-lysine (hLys) acetyltransferase activity (33). However, functional characterization of the E. coli aerobactin NIS synthetases IucA or IucC, the enzymes proposed to ligate 2 eq of the N6-acetyl-N6-hydroxy-l-lysine (ahLys) intermediate with citrate, was never reported. IucA and IucC belong to a family of ligases that have been defined as the nonribosomal peptide synthetase (NRPS)-independent siderophore (NIS) synthetases (34, 35). IucA and IucC, respectively, represent the archetypal Type A (utilizing citrate as a substrate) and Type C (utilizing a citrate derivative) NIS synthetases. Furthermore, no one has described the in vitro reconstitution of aerobactin biosynthesis to functionally demonstrate the proposed pathway. Recent structural and functional studies of NIS synthetases, including hvKP IucA (36), achromobactin synthetase AcsD (37, 38), as well as petrobactin synthetases AsbA (39) and AsbB (40), have all advanced our understanding of the catalytic function of these closely-related enzymes.
Figure 1.
Aerobactin biosynthetic pathway. The model originally proposed by Neilands and co-workers (30) has been updated to reflect the biosynthesis functionally demonstrated herein. Also included are the [M + H]+ masses for aerobactin and its precursors.
Herein, we present structural and functional investigations of aerobactin biosynthesis from a hvKP clinical isolate. First, we produced aerobactin through heterologous expression of the four iucA–D genes in an aerobactin-deficient strain of E. coli. Second, using the four isolated and purified IucA–D enzymes, we were able to functionally reconstitute aerobactin biosynthesis in vitro. Additionally, we quantified pathway turnover frequency and demonstrated the high degree of stereoselectivity employed by the synthetases IucA and IucC. Finally, we report the steady-state kinetics, and structural characterization of IucC by X-ray crystallography and small-angle solution scattering. Together with our previous studies of IucA (36), hvKP IucA and IucC represent the first pair of Type A and Type C NIS synthetases from a single pathway to be structurally and functionally characterized. This work furthers our understanding of NIS biosynthesis and provides the biochemical and structural foundation to further explore this potential antivirulence target.
Results
Heterologous in vivo aerobactin production
Much of the prior investigation on aerobactin biosynthesis originated from a pathogenic strain of E. coli harboring the aerobactin operon in the ColV-K30 plasmid (29). Although Neilands and co-workers (31) studied aerobactin biosynthesis by knocking out and re-complementing aerobactin genes originating from this plasmid, we employed a parallel approach in which an aerobactin-deficient E. coli strain was engineered to produce the siderophore through heterologous co-expression of the four iucA–D genes. When protein expression was induced from a plasmid containing iucD and iucB, ahLys was prominently detected in the culture supernatant by HPLC-MS (Fig. 2A). However, as predicted, when expressing only the synthetase genes iucA and iucC, none of the aerobactin intermediates were detected (Fig. 2B). Finally, when the E. coli were transformed and induced to express all four iucA–D genes, a pronounced peak corresponding to aerobactin was observed alongside secondary peaks corresponding to the intermediates ahLys and citryl-ahLys (Fig. 2C). In addition to demonstrating aerobactin biosynthesis, this engineered system served as a relatively convenient/accessible source for isolating in vivo biosynthesized aerobactin and citryl-ahLys without using a pathogenic bacterial strain or requiring iron-deficient culture media. Although these metabolites were detected and isolated from the culture media supernatant, it is unclear whether they were actively secreted or if they accumulated via cell lysis or passive diffusion.
Figure 2.
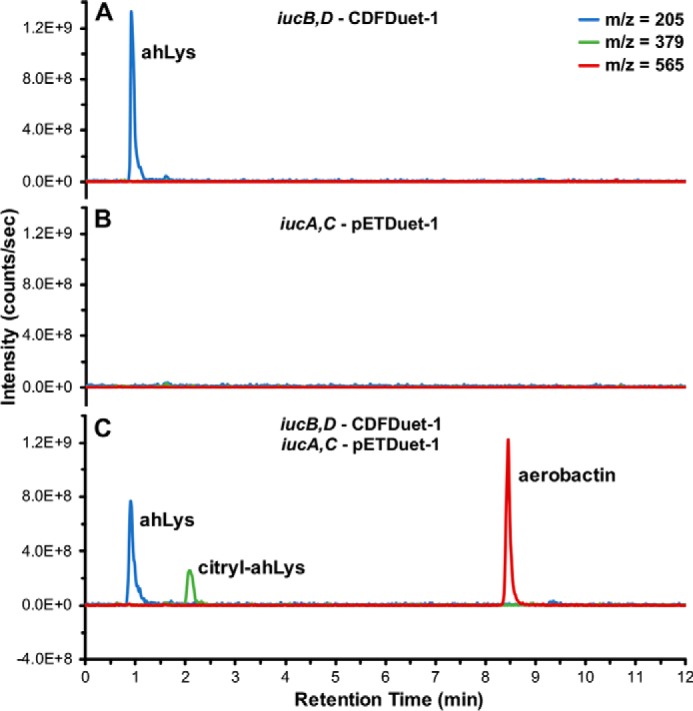
Heterologous in vivo aerobactin production. Shown are overlaid extracted ion chromatograms corresponding to the [M + H]+ masses of the metabolites ahLys, citryl-ahLys, and aerobactin from culture media supernatant of E. coli transformed and induced to express: A, iucB,D–CDFDuet-1; B, iucA,C–pETDuet-1; or C, both plasmids.
In vitro aerobactin reconstitution
After demonstrating expression of the four iucA–D genes was sufficient for aerobactin production in vivo, the pathway was investigated in vitro using purified enzymes (Fig. 3, Table S1 and Fig. S1). Despite a tendency to aggregate during expression, IucD, a putative membrane-associated enzyme, was purified as a monodisperse sample with the help of detergent. IucB, expressed as a cleavable chaperone chimera, had a broader size distribution and also had a tendency to aggregate at elevated concentrations and temperatures. However, when incubated together with substrates l-lysine, NADPH, acetyl-CoA, ATP, citrate, and the co-factor FAD, they were able to biosynthesize ahLys (Fig. 3B). When IucD, IucB, and IucA were combined, all of the ahLys was ligated to citrate, forming the citryl-ahLys intermediate, but no aerobactin was detected (Fig. 3C). In contrast, when IucD, IucB, and IucC were incubated together, no appreciable citryl-ahLys or aerobactin was detected (Fig. 3D). Finally, when substrates and all four purified IucA–D enzymes were included in the reaction mixture, a prominent peak corresponding to aerobactin was observed alongside lesser amounts of ahLys and citryl-ahLys remaining in the mixture (Fig. 3E). This in vitro reconstitution of aerobactin biosynthesis using purified enzymes provides direct functional evidence supporting the reported aerobactin pathway (30).
Figure 3.
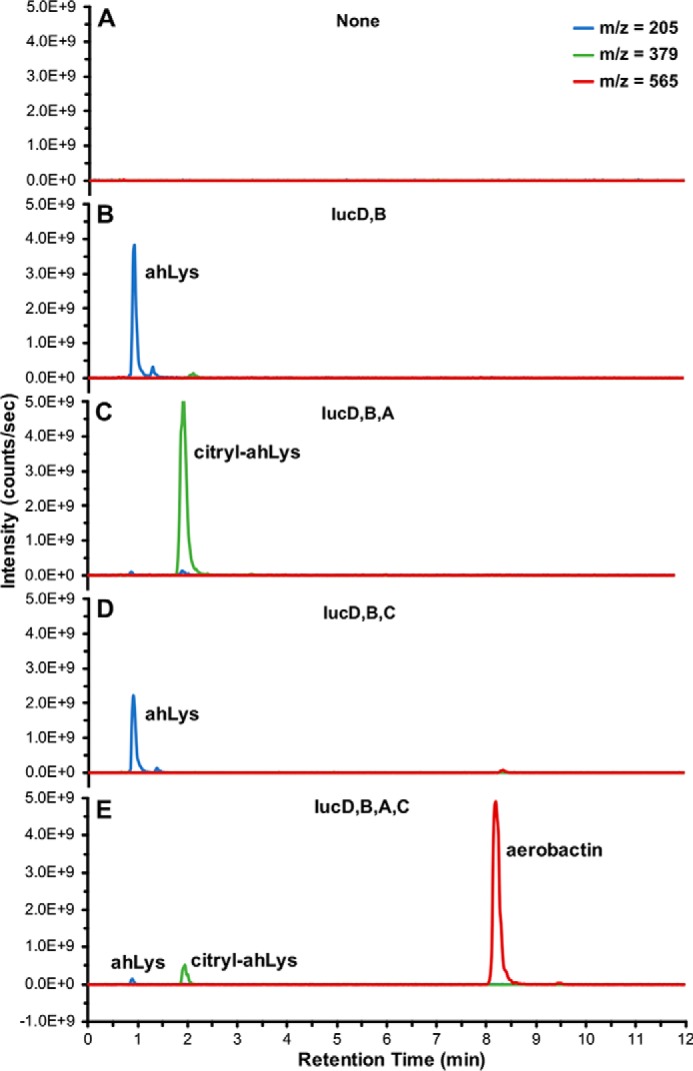
In vitro aerobactin reconstitution. Shown are overlaid extracted ion chromatograms corresponding to the [M + H]+ masses of metabolites ahLys, citryl-ahLys, and aerobactin from in vitro reaction mixtures containing (A) no enzymes; (B) IucD and IucB; (C) IucD, IucB, and IucA; (D) IucD, IucB, and IucC; or (E) IucA, IucB, IucC, and IucD.
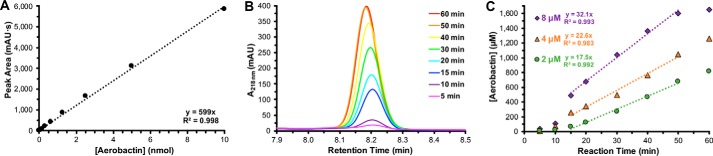
Aerobactin pathway turnover frequency
In addition to establishing the competency of the in vitro aerobactin biosynthetic system, the turnover of the pathway was also examined quantitatively. Aerobactin was purified from the culture supernatant of the engineered E. coli stain (Fig. S2, D–F) and used to construct a standard curve (Fig. 4A). The reconstitution reaction was sampled along its time course to monitor how aerobactin evolved over time (Fig. 4B). The linear time points on the reaction progress curves were fit with regression lines and used to estimate the pathway velocity (Fig. 4C). By employing a relatively high substrate concentration (5 mm) in the reconstitution, it was presumed that the resulting pathway kinetics would approximate saturating steady-state conditions. The reconstitution velocity was assessed at three different enzyme concentrations (2, 4, and 8 μm of each individual enzyme). When doubling the enzyme concentrations, the pathway velocity only increased by ≈1.4. It is unclear why reaction velocity was not directly proportional to enzyme concentration; however, it could be related to the limited stability of IucB and IucD. An aerobactin pathway turnover frequency of 6 ± 2 min−1 was estimated by averaging the turnover numbers at the three different enzyme concentrations.
Figure 4.
In vitro aerobactin pathway turnover frequency. A, standard curve relating HPLC A218 nm to aerobactin mass. The data points were fit with a linear regression best-fit line. B, overlaid A218 nm HPLC traces of the aerobactin peak at progressive in vitro reconstitution reaction times using 8 μm enzymes. C, progress curves of the aerobactin reconstitution at three enzyme concentrations (2, 4, and 8 μm). The linear regions were fit with regression lines to estimate pathway velocity.
Aerobactin synthetase stereoselectivity
The stereoselectivity of the aerobactin synthetases IucA and IucC has not been determined. Two distinct stereocenters are relevant in aerobactin biosynthesis. E. coli IucD was previously shown to only be capable of hydroxylating l-lysine, and had no activity with d-lysine (32). Therefore, it follows that the stereocenters derived from the α-carbon of lysine have the 2′S configuration in aerobactin and its precursors. Condensation of ahLys with one of the two pro-chiral carboxymethyl groups of citrate breaks the symmetry of the molecule and results in a second stereocenter at the citrate-derived C3 position in citryl-ahLys (Fig. 1). Citryl-ahLys purified from iucA–D co-expression eluted as a single peak by HPLC (Fig. 5A). Similarly, citryl-ahLys isolated from the in vitro IucA-catalyzed condensation of ahLys and citrate also eluted as a single peak (Fig. 5B). In contrast, a synthetic epimeric mixture of citryl-ahLys at the C3 position eluted as two distinct peaks, presumed to represent the two diastereomers (Fig. 5C). Elution of authentic 3R,2′S- and 3S,2′S-enriched citryl-ahLys produced through total synthesis, featuring enantioenrichment of a citrate intermediate by fractional recrystallization, supported this presumption and allowed the absolute configuration of the two peaks to be assigned (Fig. 5, D and E). The correlation of the retention times of the biosynthesized samples and the synthetic 3S,2′S-enriched sample indicated that only the 3S,2′S-diastereomer was appreciably produced by IucA. When biosynthesized citryl-ahLys was the substrate of IucC, all of it was turned over to make aerobactin (Fig. 5, A and B). Conversely, when the synthetic citryl-ahLys samples were incubated with IucC, only the peak corresponding to the 3S,2′S-diastereomer diminished, whereas the peak corresponding to the 3R,2′S-diastereomer remained unperturbed (Fig. 5, C–E). Taken together, these results demonstrate that the aerobactin synthetases are highly stereoselective; only 3S,2′S-citryl-ahLys is produced by IucA, and only 3S,2′S-citryl-ahLys is consumed by IucC in the production of aerobactin (Figs. 1 and 5).
Figure 5.
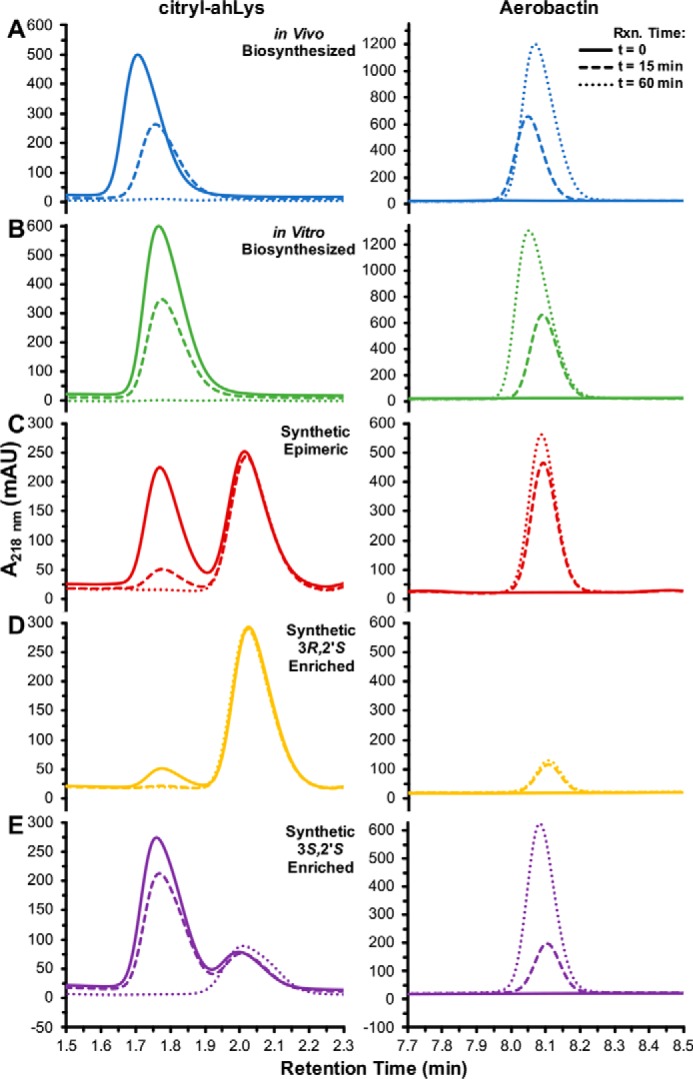
Stereoselectivity of aerobactin synthetases IucA and IucC. Shown are overlaid HPLC peaks corresponding to citryl-ahLys and aerobactin during the IucC-catalyzed condensation of ahLys with (A) in vivo biosynthesized, (B) in vitro biosynthesized, (C) synthetic epimeric mixture at C3 (D) synthetic 3R,2′S-enriched, or (E) synthetic 3S,2′S-enriched citryl-ahLys.
Steady-state kinetics of IucC
By using IucA to biosynthesize citryl-ahLys from synthetic ahLys, all three substrates were available to evaluate the steady-state kinetics of IucC (Table 1, Fig. S3). To conserve limited quantities of ahLys, hydroxylamine was employed as a saturating surrogate nucleophile when evaluating the activity of ATP and citryl-ahLys. IucC displayed relatively high-affinity for ATP (Km = 170 ± 20 μm) compared with its other two substrates citryl-ahLys (Km = 1.32 ± 0.08 mm) and ahLys (Km = 1.9 ± 0.2 mm). IucC showed similar turnover frequencies (kcat = 25–35 min−1) for all three substrates and, unsurprisingly, exhibited the highest velocity when consuming only native substrates. Overall, IucC displayed a relatively modest catalytic efficiency (kcat/Km = 300–2,500 m−1 s−1) in the production of aerobactin.
Table 1.
Apparent steady-state kinetic constants of IucC
| Substrate | Kma | kcata | kcat/Km |
|---|---|---|---|
| μm | min−1 | m−1 s−1 | |
| ATPb | 170 ± 20 | 25 ± 1 | 2,500 |
| citryl-ahLysb | 1,320 ± 80 | 29.8 ± 0.6 | 380 |
| ahLys | 1,900 ± 200 | 35 ± 1 | 300 |
a Non-linear regression best-fit values ± S.E.
b Substrates evaluated with saturating hydroxylamine as surrogate nucleophile.
X-ray crystal structure of IucC
Crystals of WT IucC were grown, but despite extensive optimization efforts, they were of insufficient quality to solve its X-ray crystal structure. To overcome this hurdle and grow better diffracting crystals, a surface entropy reduction strategy was employed (Fig. S4). Using one of 12 engineered IucC surface mutants (designated M1–M12), the X-ray crystal structure of IucC surface mutant M5 (containing five substitutions, S182G, E183S, D185T, Q187G, and Q188T) was solved to 2.45-Å resolution by molecular replacement (Table 2, Fig. 6).
Table 2.
X-ray data collection and refinement statistics of IucC M5
Values in parentheses reflect the highest resolution shell.
| Data collection | |
| Space group | P21 |
| Unit cell (a, b, c, Å) | 124.0, 197.7, 130.4 |
| (α, β, ɣ, °) | 90, 109.6, 90 |
| IucC molecules/ASU | 8 |
| Resolution range (Å) | 57.4–2.45 (2.54–2.45) |
| Completeness (%) | 99.6 (98.8) |
| Multiplicity | 6.7 (6.7) |
| No. unique reflections | 215,668 (21,423) |
| Wilson B-Factor (Å2) | 59.6 |
| I/σI | 10.0 (1.8) |
| Rmerge | 0.099 (1.067) |
| CC1/2 | 0.995 (0.636) |
| Refinement | |
| Rwork/Rfree | 0.213 (0.322)/0.241 (0.351) |
| No. protein/water atoms | 33,330/248 |
| No. protein residues | 4,344 |
| RMSDs | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.55 |
| Average protein/water B-factors (Å2) | 73.6/53.9 |
| Ramachandran analysis (%) | |
| Favored | 96.8 |
| Allowed | 2.8 |
| Outliers | 0.4 |
| Rotamer outliers (%) | 1.1 |
| MolProbity Clashscore | 2.6 |
| PDB code | 6CN7 |
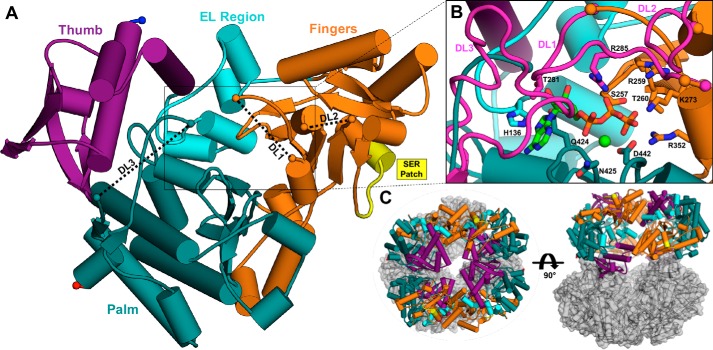
Figure 6.
The X-ray crystal structure of IucC. A, the monomeric tertiary structure of IucC surface mutant M5 colored by domain. Gaps in the model corresponding to disordered loops (DL1–3) are connected by black dashed lines for clarity. Protein chain termini are highlighted by spheres, with the N and C termini colored blue and red, respectively. The SER patch is emphasized in yellow. B, close-up view of the modeled active site. The disordered loops (DL1–3, magenta) were modeled by overlaying corresponding regions of a homology model (Fig. S4A). ATP (green carbons) and Mg2+ (light green sphere) from the crystal structure of IucA (PDB code 5JM8) are also modeled into the active site with IucC the side chains of several predicted binding residues labeled and depicted as sticks. C, the crystallographic asymmetric unit containing eight copies of IucC illustrated in two orthogonal views. Potential tetrameric (left) and dimeric (right) assemblies are highlighted using the same color scheme as panel A.
The IucC monomeric tertiary structure is described using a cupped hand analogy (Fig. 6A), following the convention employed for other structurally-characterized NIS synthetases (38, 40). The thumb domain (residues 2–113) consists of a 3-helix bundle adjoined by a distorted 4-stranded antiparallel β-sheet. The extended loop region (114–163) contains a loop that is directed into the interior of the protein followed by four short α-helices. The fingers domain (164–362) comprises a 6-stranded and a 3-stranded anti-parallel β-sheet decorated with loops and α-helices. This domain also includes the mutated surface patch (182–188) that helped mediate crystallization. Finally, the palm domain (363–576) contains a prominent 3-helix bundle and forms the base of the central active site depression. The tertiary fold is homologous to the other structurally characterized NIS synthetases, including Type C AlcC (RMSD = 1.5 Å, 392 Cα), and AsbB (RMSD = 2.3 Å, 386 Cα), as well as Type A IucA (r.m.s. displacement = 4.1 Å, 361 Cα), and AcsD (RMSD = 5.1 Å, 315 Cα).
The IucC model has three gaps where disordered loops (DL1–3) were unresolved in the electron density map (Fig. 6A). In Fig. 6B, the three disordered loops were modeled onto the crystal structure to better appreciate their proximity to the putative active site. The first gap (DL1, ≈279–288) in the fingers domain encompasses a lid loop extending over the putative active site and has been shown to be important in binding ATP and citrate in the other structurally-characterized NIS synthetases. The second gap (DL2, ≈331–341) in the fingers domain is a surface loop connecting a β-sheet and a short α-helix. Finally, the third disordered loop (DL3, ≈550–562) occurred toward the C terminus in the palm domain. This loop is in close proximity to the putative active site and appears to have inherent flexibility, with gaps occurring in the corresponding regions of the homologous AcsD, AlcC, and IucA crystal structures. The IucC surface mutant M12 has an engineered patch (551–558) within this disordered loop region. The negligible catalytic activity of this mutant enzyme provides evidence for a potential functional role of this flexible region (Fig. S5). ATP and Mg2+ were also modeled into the putative active site to highlight the conserved binding mode that IucC and the other NIS synthetases appear to employ for this common substrate (Fig. 6B). In this binding pose, several conserved residues including His-136, Ser-257, Arg-259, Thr-260, Lys-273, Thr-281, Arg-285, Arg-352, and His-422 are available in IucC to make specific interactions with the adenine, ribose, and phosphate portions of the nucleotide. Conserved residues Gln-424, Asn-425, and Asp-442 are also available to coordinate the Mg2+ ion intimately contacting the phosphate groups of ATP (Fig. S6). The unliganded active site and proximally-located disordered loops limit our ability to infer how ahLys and citryl-ahLys bind in the active site, as well as how IucC catalyzes their condensation.
The quaternary structure of IucC
Features from the crystal lattice were combined with biochemical and solution-phase small-angle X-ray scattering (SAXS) data to investigate the oligomeric state and quaternary structure of IucC. Despite migrating a bit lower than expected by SDS-PAGE (Fig. S1A), MS indicated that the full construct was expressed, and had a monomeric mass of 66.1 kDa (Table S1). During purification, IucC eluted from size exclusion chromatography primarily as a tetramer, partially overlapping a secondary dimer peak (Fig. S1D). After freezing, storing, and thawing tetrameric fractions, the tetrameric oligomer was observed to be stable (Fig. S1, B and C) and appeared the most crystallizable oligomer in screening experiments. Molecular weight calculations from SAXS data of IucC in standard storage buffer (≈230–240 kDa) slightly underestimated its theoretical tetrameric mass (264 kDa) (Table S2). However, when SAXS data were collected on IucC in the presence of ATP, the calculated molecular mass was reduced significantly to ≈155 kDa, a value marginally overestimating a dimer (132 kDa). Size exclusion chromatography also illustrated the propensity of IucC to transition to a dimer upon exposure to its nucleotide substrate (Fig. S1E).
Inspection of the asymmetric unit containing eight IucC molecules revealed both plausible tetrameric and dimeric assemblies in crystallo (Fig. 6C). The crystalline asymmetric unit can be reduced into two abutting tetramers adopting a circular configuration. This tetramer can be further divided into two identical dimers bearing resemblance to the dimeric assembly reported for the Type C NIS synthetases AlcC and AsbB (Fig. S7) (40). Comparison of the experimental IucC scattering curve to scattering curves calculated from potential crystalline assemblies revealed the strongest correlation with the tetramer (Fig. 7A). Likewise, the size and shape of an ab initio molecular envelope calculated from the SAXS data were consistent with the general size and shape of the in crystallo tetramer (Fig. 7A, inset). SAXS data collected on IucC in the presence of 1 mm ATP and MgCl2 showed a marked reduction in the near zero-angle scatting intensity, indicating a reduction in particle mass (Fig. 7B). In the conditions containing ATP, the SAXS data were most closely approximated by the curve calculated from the dimer, particularly in the low q region. Again, there is reasonable correlation between the crystalline dimer and an ab inito molecular envelope calculated from IucC in the presence of ATP (Fig. 7B, inset). Nevertheless, inspection of the molecular envelopes in Fig. 7 reveals that they are a bit too “snug” for the tetramer and a bit too “loose” for the dimer. Moreover, the molecular weight estimations seemed to underestimate a tetramer and overestimate a dimer (Table S2). Both of these observations suggest IucC likely exists as a mixture of both tetramer and dimer under both conditions. Modeling the scattering curves as a mixture of the two oligomers revealed that the experimental data were best fit by a 91:9% mixture of tetramer:dimer in storage buffer, whereas it was best approximated as a 28:72% tetramer:dimer ratio in the presence of its substrate ATP.
Figure 7.
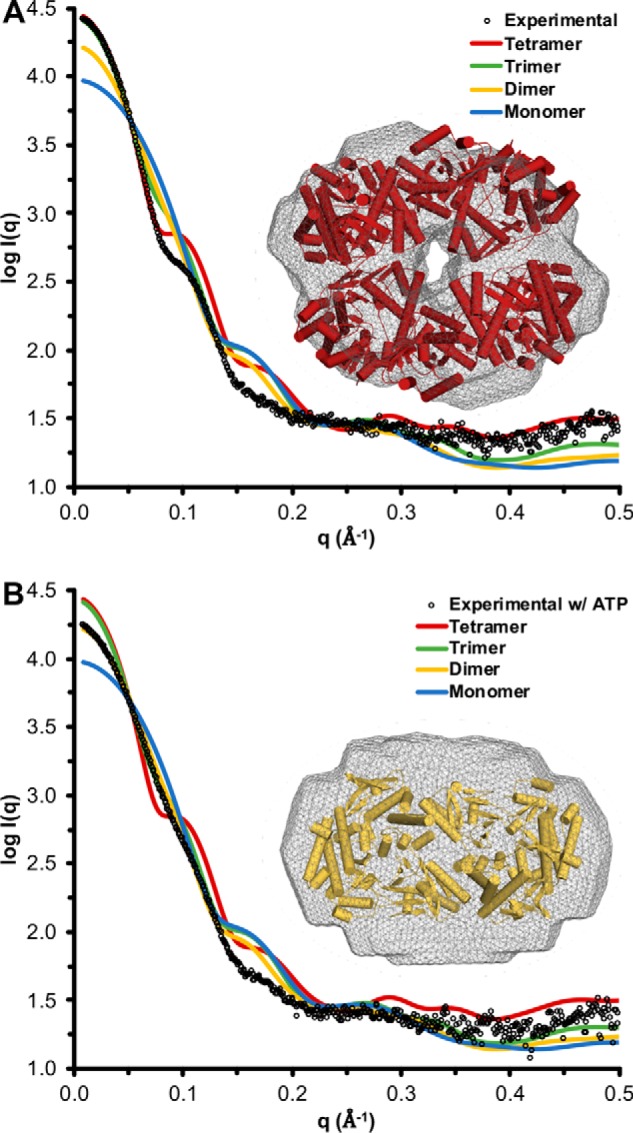
IucC SAXS. A, the experimental scattering curve of IucC in storage buffer extrapolated to infinite dilution overlaid with scattering curves calculated from potential oligomeric assemblies found in the crystalline asymmetric unit. Inset, a crystallographic tetramer docked inside an ab initio molecular envelope (wireframe) calculated from the experimental curve. B, the experimental scattering curve of IucC in storage buffer plus 1 mm ATP and MgCl2 extrapolated to infinite dilution overlaid with scattering curves calculated from potential oligomeric assemblies found in the crystal structure. Inset, a crystallographic dimer docked inside an ab initio molecular envelope (wireframe) calculated from the experimental curve.
Computational analysis of the crystal lattice (PISA (41) and EPPIC (42)) predicted the dimer was the only biologically relevant oligomeric assembly, burying ≈1,080 Å2 of surface area between the two protomers. This prediction is also supported by the observation that all four dimers within the asymmetric unit are identical, whereas the structure of the two tetramers are marginally different (Fig. S7, A and B). Taken together, it appears IucC is capable of forming both a tetramer and dimer in solution. However, although the tetrameric assembly appears to promote crystallization, it is more likely an artifactual consequence of heterologous expression and purification, with the biologically relevant dimer predominating in the presence of its substrate ATP.
Discussion
Heterologous expression of the four iucA–D genes in an aerobactin-deficient strain of E. coli enabled the production of the siderophore. This implies that all the necessary substrates can be harvested from the cellular metabolic pool, not requiring any additional enzymatic activity to supply the pathway. It is interesting to note that the operon of the closely related, and virulence-promoting, NIS staphyloferrin B contains specialized enzymes dedicated to citrate and amino acid precursor production (43). The relatively low burden of the 5-gene, 8-kb aerobactin operon likely contributes to its relatively wide distribution in pathogenic members of the Enterobacteriaceae family, including Escherichia, Klebsiella, Salmonella, Yersinia, Enterobacter, Shigella, Serratia, and Citrobacter, as well as marine Vibrio (44) species.
The four purified IucA–D enzymes were also able to reconstitute aerobactin biosynthesis in vitro. Although hLys was not detected in the assay when only IucD was included, the three downstream metabolites ahLys, citryl-ahLys, and aerobactin were all clearly identified when additional Iuc enzymes were added. Enzyme combinations in which either IucD or IucB were omitted did not lead to the identification of nonhydroxylated or nonacetylated aerobactin analogues, respectively (data not shown). This observation demonstrates the aerobactin secondary metabolic pathway is engrained with a high-degree of substrate selectivity to limit production of nonfunctional analogues. Both of the synthetases are required to synthesize aerobactin; neither was capable of compensating for the activity of the other to any appreciable extent. This is in contrast to the Type C NIS synthetase AsbB in petrobactin biosynthesis, which was reported to partially compensate for the absence of its Type A counterpart AsbA (40). Although having one synthetase capable of catalyzing both condensations would be ideal from a metabolic efficiency perspective, the presence of two distinct synthetases in aerobactin biosynthesis, as well as many of the other NIS pathways, likely implies a difficult evolutionary challenge of designing a single stereoselective active site to accommodate both sets of substrates. One unique way of addressing this challenge would be by combining two synthetase active sites into a single polypeptide, an approach that may be adopted by the putative NIS synthetase CiuE in the biosynthesis of corynebactin, a lysine and citrate-based siderophore closely-related to aerobactin (45, 46).
Functional characterization of the Type A NIS synthetase AcsD described a highly stereoselective mechanism for the condensation of citrate and l-serine (38). Furthermore, structural models of the AcsD and IucA active sites predicted a mechanism in which only one of the two pro-chiral carboxymethyl groups of citrate is activated by ATP to form a citryl-adenylate intermediate (36, 38). Experiments presented herein confirmed the structural predictions by functionally demonstrating the stereoselective mechanisms of IucA and IucC. Although the final symmetry of aerobactin renders the citrate-derived C3 position achiral, stereochemistry at the C3 position has been shown to play a critical role in influencing siderophore potency and iron affinity in the NIS staphyloferrin B (43).
In addition to demonstrating aerobactin biosynthesis in vitro, the velocity of the pathway was determined to more quantitatively evaluate aerobactin production and compare it to other reconstituted pathways. The aerobactin turnover frequency (6 ± 2 min−1) is similar to values reported for the in vitro production of the siderophores pyochelin (2 min−1) (47) and yersiniabactin (1.4 min−1) (48), despite employing contrasting NRPS and hybrid NRPS/polyketide synthetase enzymatic mechanisms, respectively. The apparent steady-state kinetics of the individual aerobactin synthetase IucC was also examined quantitatively. IucC showed an affinity for ATP similar to IucA (Km = 130 ± 30 μm) (36), a result supporting the common ATP binding mechanism that NIS synthetases are predicted to share. The relatively low catalytic efficiency of IucC also appeared comparable with values reported for the Type C NIS synthetase homologue AsbB (240 m−1 s−1) (40). These seemingly modest pathway turnover frequencies and enzyme catalytic efficiencies support the premise that high production may not be essential for these siderophore pathways to meet the demand for iron.
Although WT hvKP IucC was a stable enzyme and expressed robustly, a surface entropy reduction mutant (M5) was required to grow high-quality crystals. Computational analysis of the resulting crystal lattice using EPPIC (42) revealed that residues within the mutated surface patch (182–188) contributed to one-third (5 of 15) of the distinct crystal lattice interfaces, while not disturbing the predicted biologically relevant dimer interface or catalytic competency (Fig. S5). This significant contribution to crystal lattice contacts helps rationalize how this particular mutant patch was able to promote higher quality crystal growth. Additionally, this most successful surface mutant had the highest melting temperature off all the evaluated IucC constructs, consistent with the commonly-held assertion that maximizing protein stability promotes successful crystallization (Fig. S4C) (49).
Attempts to obtain a co-crystal structure with substrates, especially with ATP, were surprisingly unsuccessful, given the co-crystallization success of the other NIS synthetases. Although the underlying reasons behind this phenomenon are uncertain, it was observed that the disordered loops (DL1–3) in proximity to the active site all faced toward the interior of the asymmetric unit. Given the presumed active site flexibility required to bind the relatively complex substrates ahLys and citryl-ahLys, it is possible that inter-particle interactions from higher-order oligomerization, especially in crystallo, could have disrupted stable substrate binding. The dimeric quaternary structure predicted to be biologically relevant for IucC is similar, but clearly distinct from the assembly shared by its Type C homologues AlcC and AsbB (Fig. S7C). This represents the fourth distinct quaternary structure among the five structurally characterized NIS synthetases. Considering their high monomeric structural homology, their varied quaternary structure is noteworthy, despite its biological relevance remaining uncertain.
Motivated by the role aerobactin is reported to play in the newly recognized hvKP, the biosynthesis of the siderophore was revisited. Building upon pioneering studies in the 1980s, the aerobactin biosynthetic pathway was functionally demonstrated while attempting to further our understanding of a key virulence factor and of NIS biosynthesis in general. Even with the results of the current study, a number of inquiries related to aerobactin biosynthesis remain open: the detailed active site architecture and substrate binding geometry of IucC, what, if any, cellular localization or multienzyme assembly exists for the pathway, why it appears to be so important for mediating virulence in hvKP, and if it can be inhibited in an anti-virulence therapeutic approach.
Experimental procedures
Cloning, expression, and purification of hvKP Iuc enzymes
Details of Iuc enzyme expression and purification are outlined in Table S1; a general expression and purification procedure is described below. The four iucA–D genes were individually amplified from the hvKP1 clinical isolate (50) genomic DNA using primers to incorporate restriction sites at the 5′ and 3′ ends of the gene. The genes were subcloned into modified expression vectors with N-terminal His tags and protease recognition sites for tag removal. The vectors were transformed into E. coli BL21(DE3) for protein production. Cells were grown in LB media (250 rpm) to an A600 nm of ≈0.5–0.7. Expression was induced with the addition of isopropyl 1-thio-β-d-galactopyranoside, followed by incubation for ≈18 h overnight at 16 °C. Cells were harvested by centrifugation at 6,000 × g for 15 min at 4 °C. After decanting off the supernatant media, the cell pellets were flash frozen in liquid nitrogen and stored at −80 °C for later use.
Frozen cell pellets were re-suspended in lysis buffer and lysed by sonication. The resulting slurry was separated by ultracentrifugation at 185,000 × g. The supernatant was filtered over a 0.45-μm polysulfone membrane before being subjected to immobilized metal affinity chromatography. The lysate supernatant was passed over a 5-ml Ni2+- nitrilotriacetic acid column. Bound proteins were eluted from the column using lysis buffer plus 300 mm imidazole. Fractions that contained His-tagged Iuc enzymes by SDS-PAGE were combined and dialyzed overnight at 4 °C or room temperature with the appropriate protease in dialysis buffer for His/chaperone tag removal. After spiking with imidazole to 20 mm, the dialyzed sample was passed over the Ni2+- nitrilotriacetic acid column for a second time. The flow-through fractions containing the desired protein without the His tag were combined and concentrated using an appropriate centrifugal filter before being subjected to size exclusion chromatography (HiLoadTM 16/60 SuperdexTM 200, GE Healthcare Life Sciences). The desired fractions were combined, concentrated, and flash frozen in liquid nitrogen before being stored at −80 °C for subsequent use.
Heterologous in vivo aerobactin production
The four iucA–D genes were subcloned into duet expression vectors for simultaneous co-expression. The iucB and iucD genes were subcloned into MCS1 (NcoI and BamHI) and MCS2 (NdeI and XhoI) of the CDFDuet-1 expression plasmid (StrepR/SpecR), respectively. Similarly, the iucA and iucC genes were subcloned into MCS1 (NcoI and BamHI) and MCS2 (NdeI and BglII) of the pETDuet-1 expression plasmid (AmpR), respectively. The iucA gene construct included a N-terminal His tag, whereas the other three enzymes were untagged. E. coli BL21(DE3) were then transformed with either CDFDuet-1, pETDuet-1, or both of the plasmids. The transformed E. coli strains were grown in 60 ml of M9 minimal media with appropriate antibiotics to an A600 nm of ≈0.6–0.8 (37 °C, 250 rpm). Protein expression was induced with the addition of 750 μm isopropyl 1-thio-β-d-galactopyranoside, and cells were incubated for ≈18 h overnight. The cells were pelleted by centrifugation at 4,500 rpm for 10 min, and the culture supernatant was decanted and frozen at −20 °C for subsequent analysis.
The supernatant was diluted 1:5 in 5% acetonitrile in water before being analyzed by HPLC-MS. Ten microliters of sample was injected over a 3 × 50-mm Agilent InfinityLab Poroshell 120 EC-C18 2.7 μm column held at 30 °C with a flow rate of 0.4 ml/min. The mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The elution gradient started with isocratic 5% B for 2 min, followed by a linear gradient to 20% B over 10 min. The eluant was monitored for UV absorbance at 218 nm and by an in-line Advion ExpressionL compact mass spectrometer configured for positive mode electrospray ionization. The mass spectrometer was further configured with the following parameters: scan range: 100–1,000 m/z; capillary voltage: 150 V; ESI voltage +3,500 V; capillary temperature: 150 °C; gas temperature: 350 °C; gas flow: 4 liter/min.
In vitro aerobactin reconstitution
A 100-μl reaction mixture was prepared containing 50 mm HEPES pH 7.5, 5 mm MgCl2, 50 mm NaCl, 0.2 mm tris(2-carboxyethyl)phosphine, 5 mm l-lysine, 5 mm NADPH, 0.5 mm FAD, 5 mm acetyl-CoA, 5 mm ATP, 5 mm citrate, and various combinations of the Iuc enzymes at 10 μm concentration. After incubating at 37 °C, 10-μl aliquots of the reaction mixture were removed at specified time points (up to 1 h) and quenched by the addition of 40 μl of 0.1% formic acid and 5% acetonitrile in water. The sample was briefly centrifuged to remove any precipitated protein before being analyzed using the same HPLC-MS procedure outlined above.
Purification of biosynthesized citryl-ahLys and aerobactin
The general purification strategy was informed by the work of Butler and colleagues (44, 51). After acidifying to pH 2.5 using concentrated HCl, 250 ml of M9 minimal media culture supernatant from iucA–D co-expression was stirred with 10 g of AmberLite XAD-4 resin overnight at 4 °C. The resin was filtered from the supernatant and rinsed with 30 ml of water before being eluted with 100% methanol. Fractions evidenced to contain iron chelators by the liquid chrome azurol S assay (52) were combined and concentrated to a residue (≈120 mg) under vacuum. This crude residue was re-dissolved in 1 ml of 5% acetonitrile in water, centrifuged to remove any remaining solids, and 50–100 μl was eluted over an Agilent ZORBAX SB-C18 9.4 × 250 mm 5-μm semi-preparative HPLC column held at 30 °C with a flow rate of 4 ml/min. The same mobile phase composition and elution gradient was employed as described above. Fractions from multiple injections shown to contain the iron chelators citryl-ahLys and aerobactin by the chrome azurol S assay and analytical HPLC-MS analysis were combined and reduced to dryness by rotary evaporation. From ≈250 ml of culture supernatant, 3.5 mg of citryl-ahLys and 7.2 mg of aerobactin was isolated with high purity (Fig. S2). A standard curve for the quantitation of aerobactin production was constructed by analyzing known quantities of this aerobactin standard using the same HPLC method described above to quantify peak area.
In vitro biosynthesized citryl-ahLys was isolated by scaling-up the IucA-catalyzed reaction. A 2.5-ml reaction mixture was prepared containing 10 mm HEPES pH 7.5, 5 mm MgCl2, 50 mm NaCl, 27.5 mm ATP, 27.5 mm citrate, 25 mm ahLys, and 10 μm IucA, and incubated at 37 °C for ≈15 h overnight. The enzyme was removed by filtration through a centrifugal filter and the filtrate was reduced to dryness under vacuum. The crude residue was re-dissolved in 5% acetonitrile in water before being purified by the identical semi-preparative HPLC procedure outlined above. The combined yield of the reaction and purification was 65%.
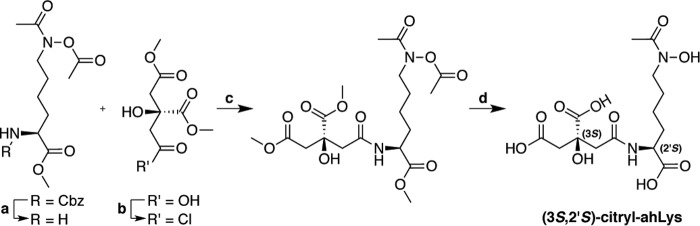
Preparation of synthetic ahLys and citryl-ahLys
The detailed synthesis of aerobactin precursors ahLys and citryl-ahLys is presented in the supporting information (Data S10–S25). Briefly, the N6-oxidized and acetylated l-lysine derivative, ahLys was synthesized in accordance with previously described methods reported by Hu and Miller (53). Late stage divergence from the reported route allowed for functionalization of the α-amino moiety of the fully protected ahLys derivative (Fig. 8). Enantiopure (S)-1,3-dimethyl citrate was prepared by co-crystallization with (R)-methylbenzylamine (54, 55). Subsequent activation to the corresponding citryl-chloride and coupling to the α-amine of the protected ahLys derivative yielded methyl-N6-acetoxy-N6-acetyl-N2-[(S)-1,3-dimethylcitryl]-l-lysinate. Global saponification with lithium hydroxide afforded the penultimate aerobactin intermediate (3S,2′S)-citryl-ahLys with minor epimerization.
Figure 8.
Synthesis of aerobactin precursor (3S,2′S)-citryl-ahLys. Reaction conditions: (a) Pd/C, H2, EtOH, room temperature, 1 h; (b) SOCl2, 85 °C, 1 h; (c) Et3N, 0 °C to room temperature, 2 h; (d) LiOH, THF:H2O.
IucC steady-state kinetics
A coupled NADH oxidation assay was employed to evaluate the steady-state kinetics of IucC (56). Reaction mixtures (50 μl) were prepared containing 50 mm HEPES pH 7.5, 15 mm MgCl2, 3 mm PEP, 500 μm NADH, 10 units/ml of pyruvate kinase, lactate dehydrogenase, and adenylate kinase, 2 μm IucC, and varied concentrations of substrates. The concentration of a single substrate was varied while the other two were at saturating/near saturating concentrations. When not being varied, ATP and citryl-ahLys were at 5 and 6.5 mm, respectively. When hydroxylamine was employed as a surrogate nucleophile, it was at 75 mm. Reactions were carried out in triplicate in 96-well half-area clear polystyrene plates at 37 °C; the absorbance (340 nm, ϵ = 6,220 m−1 cm−1) was measured every 10–15 s using a Biotek Synergy 4 plate reader for 10 min. The maximum initial absorbance slopes were converted to enzyme activity and plotted versus varying substrate concentrations. The plots were then fit with nonlinear Michaelis-Menten regression in GraphPad Prism to calculate Vmax and Km values.
IucC surface entropy reduction (SER) and crystallization
The general SER approach was informed by numerous literature reports, two of which are cited here (57, 58). A homology model of hvKP IucC was created based on the crystal structures of the two Type C NIS synthetases AsbB and AlcC using YASARA software. The homology model was visually inspected to identify sequences on the surface of the enzyme enriched in high-entropy charged/polar residues, including Lys, Glu, Gln, Arg, Asp, and Asn. Mutations were then designed to introduce lower entropy side chains (Ala, Gly, Ser, Thr, Tyr, and His) to promote the formation of stable crystal lattice contacts, while trying to avoid making the surface of the enzyme too hydrophobic. In total, 12 constructs were designed with a single mutated surface patch (Fig. S4B). The hvKP iucC plasmid was sent to GENEWIZ, Inc. (South Plainfield, NJ) to synthesize and sequence the 12 mutant genes. E. coli BL21(DE3) were transformed with the vectors containing the mutant IucC genes, and protein expression and purification was carried out using the same general procedure described for WT IucC.
Of the 12 surface mutant proteins designed, seven were successfully purified in sufficient quantity for crystallization screening using a 120-condition in-house screen and the 1,536-condition high-throughput crystallization screening service available at the Hauptman-Woodward Medical Research Institute (59). The thermal stability of WT IucC and the seven successfully purified SER mutants was assayed using a fluorescence-based thermal shift assay (60). Although crystallization leads were identified for all seven of the surface mutants, initial crystals from surface mutant M5 appeared most promising. Conditions were optimized to ≈5.5 mg/ml of IucC M5 combined 1:1 with a mixture of 10–11% PEG 20,000 and 100 mm MES, pH 6.5, in hanging-drop vapor-diffusion format at 20 °C. Microseeding was employed to control nucleation and promote the growth of single crystals. Crystals were cryo-protected by serial transfer through mixture plus 8, 16, and 24% ethylene glycol before being flash frozen in liquid nitrogen.
X-ray data collection, structure determination, and refinement
Frozen crystals were shipped to GM/CA ID-B at the Advanced Photon Source (APS) at the Argonne National Laboratory (Lemont, IL) for remote X-ray diffraction data collection. A total of 400 images were collected on a single crystal using a 100 × 45 μm beam (no attenuation) over 360° (0.9°, 0.5 s) on a Dectris Eiger-16m detector at a distance of 350 mm. The diffraction images were processed using iMosflm (61) and the structure was solved by molecular replacement using Phaser (62) software and employing monomeric AlcC (PDB code 3X0Q, 29.9% identity) as the search model. Automated model building was performed using PHENIX.Autobuild (63), manual model building and refinement was carried out using Coot (64), and automated structure refinement was executed using PHENIX.Refine (65). Data collection and refinement statistics are presented in Table 2. In addition to the disordered loops discussed in the text, the protein chain was modeled for residues 542–545 despite their weak electron density. Structure factors and coordinates for hvKP IucC surface mutant M5 have been deposited in the Protein Data Bank (code 6CN7).
SAXS
SAXS data on WT hvKP IucC were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 4–2 (66, 67). Protein samples (40 μl) were prepared in a dilution series of 5.3, 4.0, 2.7, and 1.3 mg/ml in a buffer of 50 mm HEPES, 150 mm NaCl, and 0.2 mm tris(2-carboxyethyl)phosphine. To investigate the effect of ATP on the conformation of IucC, a second dilution series (5.1, 3.8, 2.5, and 1.3 mg/ml) was prepared in an identical buffer system plus 1 mm ATP and MgCl2. Each sample was held at 10 °C and data were collected with 20 exposures of 1-s duration. Scattering from a buffer blank was subtracted from the data. The scattering curves were produced using SASTool software (67) and the scattering curves were analyzed using software from the ATSAS program package (68): PRIMUSQT, DAMMIF, DAMAVER, CRYSOL, SUPCOMB, OLIGOMER, and DATVC. Guinier analysis (0.4 ≤ q·RG ≤ 1.3) was used to determine the zero-angle scattering intensity (I0). This value was then used to calculate the molecular mass by employing lysozyme (14.3 kDa, 16.3 mg/ml) as a reference standard. For molecular weight calculations based on Porod volume, the pair distance distribution (P(r)) function (q ≤ 0.3) was employed to calculate the Porod volume, which was then divided by 1.66 to yield the molecular mass in Da (69). The DATVC software was used to calculate the molecular weight by the volume of correlation (VC) including data for q ≤ 0.3. The IucC ab initio molecular envelopes were calculated using P222 (without ATP) and P2 (with ATP) symmetry restraints in DAMMIF and including data for q ≤ 0.3. The CRYSOL scattering curves were calculated using the solvent electron density of water (0.334 e−/Å3). To correct for a slight concentration dependence (minor interparticle repulsion) in the SAXS data, PRIMUSQT was used to generate scattering curves extrapolated to infinite dilution.
Author contributions
D. C. B., C. C. A., and A. M. G. conceptualization; D. C. B. and A. M. G. data curation; D. C. B., E. A., and A. M. G. validation; D. C. B., E. A., M. R. R., E. J. D., L. S. M., C. C. A., and A. M. G. investigation; D. C. B., E. A., M. R. R., E. J. D., L. S. M., C. C. A., and A. M. G. methodology; D. C. B., E. A., C. C. A., and A. M. G. writing-original draft; D. C. B., C. C. A., and A. M. G. writing-review and editing; E. A. visualization; C. C. A. and A. M. G. supervision; C. C. A. and A. M. G. project administration; A. M. G. resources; A. M. G. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Edward Snell for collecting the SAXS data on IucC, and Dr. Thomas Grant for guidance analyzing it. We thank Dr. Artem Evdokimov for invaluable assistance in SER mutant design. We thank Dr. Thomas Russo for insightful discussions regarding hvKP. The compact mass spectrometer was purchased with funds from the Richard W. and Mae Stone Goode Foundation. Diffraction data were collected at the GM/CA ID-B beamline of APS, which is supported by the National Cancer Institute Grant ACB-12002, National Institute of General Medical Sciences Grant AGM-12006 under Department of Energy Contract Number DE-AC02-06CH11357 to APS, and the National Institutes of Health Office of Research Infrastructure Program Grant 1S10OD012289-01A1.
This work was supported in part by the National Institutes of Health Grant AI116998 (to A. M. G.) and pilot studies supported by The Buffalo Clinical and Translational Research Center, National Institutes of Health Grant UL1TR001412 (to Dr. Timothy F. Murphy). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7, Tables S1 and S2, Schemes S1–S3, and Data S10–S25.
The atomic coordinates and structure factors (code 6CN7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- KP
- Klebsiella pneumoniae
- hvKP
- hypervirulent pathotype KP
- NRPS
- nonribosomal peptide synthetase
- NIS
- NRPS-independent siderophore
- RMSD
- root mean square displacement
- DL
- disordered loop
- SER
- surface entropy reduction
- hLys
- N6-hydroxy-l-lysine
- ahLys
- N6-acetyl-N6-hydroxy-l-lysine
- SAXS
- small-angle X-ray scattering.
References
- 1. Podschun R., and Ullmann U. (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keynan Y., and Rubinstein E. (2007) The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents 30, 385–389 10.1016/j.ijantimicag.2007.06.019 [DOI] [PubMed] [Google Scholar]
- 3. Li B., Zhao Y., Liu C., Chen Z., and Zhou D. (2014) Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 9, 1071–1081 10.2217/fmb.14.48 [DOI] [PubMed] [Google Scholar]
- 4. Kuehn B. M. (2013) “Nightmare” bacteria on the rise in US hospitals, long-term care facilities. JAMA 309, 1573–1574 10.1001/jama.2013.2922 [DOI] [PubMed] [Google Scholar]
- 5. Guillermo V. S., Ronald N. M., Richard B. C., Madiha F., Padmaraj D., Gupta E., and Jose B. (2013) Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998–2010. Emerg. Infect. Dis. J. 19, 133 10.3201/eid1901.120310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braykov N. P., Eber M. R., Klein E. Y., Morgan D. J., and Laxminarayan R. (2013) Trends in resistance to carbapenems and third-generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect. Control Hosp. Epidemiol. 34, 259–268 10.1086/669523 [DOI] [PubMed] [Google Scholar]
- 7. Patel P. K., Russo T. A., and Karchmer A. W. (2014) Hypervirulent Klebsiella pneumoniae. Open Forum Infect. Dis. 1, ufo028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shon A. S., Bajwa R. P., and Russo T. A. (2013) Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shon A. S., and Russo T. A. (2012) Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 7, 669–671 10.2217/fmb.12.43 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Cheng D., and Lin C. L. (1986) Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916 10.1001/archinte.146.10.1913,10.1001/archinte.1986.00360220057011 [DOI] [PubMed] [Google Scholar]
- 11. Iwasaki Y., Inokuchi R., Harada S., Aoki K., Ishii Y., and Shinohara K. (2017) Bacterial meningitis caused by hypervirulent Klebsiella pneumoniae capsular genotype K54 with development of granuloma-like nodal enhancement in the brain during the subacute phase. Intern. Med. 56, 373–376 10.2169/internalmedicine.56.7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prokesch B. C., TeKippe M., Kim J., Raj P., TeKippe E. M., and Greenberg D. E. (2016) Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16, e190–e195 10.1016/S1473-3099(16)30021-4 [DOI] [PubMed] [Google Scholar]
- 13. Takahashi K., Miura A., Yamaguchi T., and Kanematsu M. (2015) Novel cord-like structures on MRI in a case of hypervirulent Klebsiella pneumoniae. Intern. Med. 54, 355–356 10.2169/internalmedicine.54.3485 [DOI] [PubMed] [Google Scholar]
- 14. Ng D., and Frazee B. (2015) Necrotizing fasciitis caused by hypermucoviscous Klebsiella pneumoniae in a Filipino female in North America. West. J. Emerg. Med. 16, 165–168 10.5811/westjem.2014.11.23599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vandevelde A., and Stepanovic B. (2014) On a boat: a case in Australia of endophthalmitis and pyogenic liver, prostatic, and lung abscesses in a previously well patient due to Klebsiella pneumoniae. Case Rep. Infect. Dis. 2014, 5 10.1155/2014/137248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kashani A. H., and Eliott D. (2013) The emergence of Klebsiella pneumoniae endogenous endophthalmitis in the U.S.A.: basic and clinical advances. J. Ophthal. Inflamm. Infect. 3, 28 10.1186/1869-5760-3-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin S. S., Kim Y. J., Kim S. I., Hu J. Y., Kim Y. S., Cho S. Y., and Kang M. W. (2013) Necrotizing fasciitis and psoas abscess caused by hypervirulent Klebsiella pneumoniae. Korean J. Med. 85, 540–544 10.3904/kjm.2013.85.5.540 [DOI] [Google Scholar]
- 18. Kwon J. M., Jung H. L., Shim J. W., Kim D. S., Shim J. Y., and Park M. S. (2013) Klebsiella pneumoniae liver abscess in an immunocompetent child. Korean J. Pediatr. 56, 407–410 10.3345/kjp.2013.56.9.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., Chan E. W., Shu L., Yu J., Zhang R., and Chen S. (2018) A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46 [DOI] [PubMed] [Google Scholar]
- 20. Chen L., and Kreiswirth B. N. (2018) Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet. Infect. Dis. 18, 2–3 [DOI] [PubMed] [Google Scholar]
- 21. Russo T. A., Shon A. S., Beanan J. M., Olson R., MacDonald U., Pomakov A. O., and Visitacion M. P. (2011) Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PloS One 6, e26734 10.1371/journal.pone.0026734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nassif X., and Sansonetti P. J. (1986) Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fischbach M. A., Lin H., Liu D. R., and Walsh C. T. (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138 10.1038/nchembio771 [DOI] [PubMed] [Google Scholar]
- 24. Lamb A. L. (2015) Breaking a pathogen's iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim. Biophys. acta 1854, 1054–1070 10.1016/j.bbapap.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Struve C., Roe C. C., Stegger M., Stahlhut S. G., Hansen D. S., Engelthaler D. M., Andersen P. S., Driebe E. M., Keim P., and Krogfelt K. A. (2015) Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6, e00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Russo T. A., Olson R., MacDonald U., Beanan J., and Davidson B. A. (2015) Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83, 3325–3333 10.1128/IAI.00430-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russo T. A., Olson R., Macdonald U., Metzger D., Maltese L. M., Drake E. J., and Gulick A. M. (2014) Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367 10.1128/IAI.01667-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibson F., and Magrath D. I. (1969) The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta 192, 175–184 10.1016/0304-4165(69)90353-5 [DOI] [PubMed] [Google Scholar]
- 29. Carbonetti N. H., and Williams P. H. (1984) A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect. Immun. 46, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lorenzo V., and Neilands J. B. (1986) Characterization of iucA and iucC genes of the aerobactin system of plasmid ColV-K30 in Escherichia coli. J. Bacteriol. 167, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Lorenzo V., Bindereif A., Paw B. H., and Neilands J. B. (1986) Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165, 570–578 10.1128/jb.165.2.570-578.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macheroux P., Plattner H. J., Romaguera A., and Diekmann H. (1993) FAD and substrate analogs as probes for lysine N6-hydroxylase from Escherichia coli EN 222. Eur. J. Biochem. 213, 995–1002 10.1111/j.1432-1033.1993.tb17846.x [DOI] [PubMed] [Google Scholar]
- 33. Coy M., Paw B. H., Bindereif A., and Neilands J. B. (1986) Isolation and properties of Nϵ-hydroxylysine:acetyl coenzyme A Nϵ-transacetylase from Escherichia coli pABN11. Biochemistry 25, 2485–2489 10.1021/bi00357a030 [DOI] [PubMed] [Google Scholar]
- 34. Oves-Costales D., Kadi N., and Challis G. L. (2009) The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem. Commun. 2009, 6530–6541 10.1039/B913092F [DOI] [PubMed] [Google Scholar]
- 35. Challis G. L. (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6, 601–611 10.1002/cbic.200400283 [DOI] [PubMed] [Google Scholar]
- 36. Bailey D. C., Drake E. J., Grant T. D., and Gulick A. M. (2016) Structural and functional characterization of aerobactin synthetase IucA from a hypervirulent pathotype of Klebsiella pneumoniae. Biochemistry 55, 3559–3570 10.1021/acs.biochem.6b00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmelz S., Botting C. H., Song L., Kadi N. F., Challis G. L., and Naismith J. H. (2011) Structural basis for acyl acceptor specificity in the achromobactin biosynthetic enzyme AcsD. J. Mol. Biol. 412, 495–504 10.1016/j.jmb.2011.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmelz S., Kadi N., McMahon S. A., Song L., Oves-Costales D., Oke M., Liu H., Johnson K. A., Carter L. G., Botting C. H., White M. F., Challis G. L., and Naismith J. H. (2009) AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nat. Chem. Biol. 5, 174–182 10.1038/nchembio.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oves-Costales D., Song L., and Challis G. L. (2009) Enantioselective desymmetrisation of citric acid catalyzed by the substrate-tolerant petrobactin biosynthetic enzyme AsbA. Chem. Commun. 2009, 1389–1391 [DOI] [PubMed] [Google Scholar]
- 40. Nusca T. D., Kim Y., Maltseva N., Lee J. Y., Eschenfeldt W., Stols L., Schofield M. M., Scaglione J. B., Dixon S. D., Oves-Costales D., Challis G. L., Hanna P. C., Pfleger B. F., Joachimiak A., and Sherman D. H. (2012) Functional and structural analysis of the siderophore synthetase AsbB through reconstitution of the petrobactin biosynthetic pathway from Bacillus anthracis. J. Biol. Chem. 287, 16058–16072 10.1074/jbc.M112.359349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 42. Duarte J. M., Srebniak A., Schärer M. A., and Capitani G. (2012) Protein interface classification by evolutionary analysis. BMC Bioinformatics 13, 334 10.1186/1471-2105-13-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madsen J. L., Johnstone T. C., and Nolan E. M. (2015) Chemical synthesis of staphyloferrin B affords insight into the molecular structure, iron chelation, and biological activity of a polycarboxylate siderophore deployed by the human pathogen Staphylococcus aureus. J. Am. Chem. Soc. 137, 9117–9127 10.1021/jacs.5b04557 [DOI] [PubMed] [Google Scholar]
- 44. Haygood M. G., Holt P. D., and Butler A. (1993) Aerobactin production by a planktonic marine Vibrio sp. Limnol. Oceanogr. 38, 1091–1097 10.4319/lo.1993.38.5.1091 [DOI] [Google Scholar]
- 45. Zajdowicz S., Haller J. C., Krafft A. E., Hunsucker S. W., Mant C. T., Duncan M. W., Hodges R. S., Jones D. N., and Holmes R. K. (2012) Purification and structural characterization of siderophore (corynebactin) from Corynebacterium diphtheriae. PloS One 7, e34591 10.1371/journal.pone.0034591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kunkle C. A., and Schmitt M. P. (2005) Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 187, 422–433 10.1128/JB.187.2.422-433.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Patel H. M., and Walsh C. T. (2001) In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry 40, 9023–9031 10.1021/bi010519n [DOI] [PubMed] [Google Scholar]
- 48. Miller D. A., Luo L., Hillson N., Keating T. A., and Walsh C. T. (2002) Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9, 333–344 [DOI] [PubMed] [Google Scholar]
- 49. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., and Nordlund P. (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 10.1016/j.ab.2006.07.027 [DOI] [PubMed] [Google Scholar]
- 50. Russo T. A., and Gill S. R. (2013) Draft genome sequence of the hypervirulent Klebsiella pneumoniae strain hvKP1, isolated in Buffalo, New York. Genome Announc. 1, e0006513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Küpper F. C., Carrano C. J., Kuhn J. U., and Butler A. (2006) Photoreactivity of iron(III)-aerobactin: photoproduct structure and iron(III) coordination. Inorg. Chem. 45, 6028–6033 10.1021/ic0604967 [DOI] [PubMed] [Google Scholar]
- 52. Schwyn B., and Neilands J. B. (1987) Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 53. Hu J., and Miller M. J. (1994) A new method for the synthesis of Nϵ-acetyl-Nϵ-hydroxy-l-lysine, the iron-binding constituent of several important siderophores. J. Org. Chem. 59, 4858–4861 10.1021/jo00096a030 [DOI] [Google Scholar]
- 54. Weaver R., and Gilbert I. H. (1997) The design and synthesis of nucleoside triphosphate isosteres as potential inhibitors of HIV reverse transcriptase. Tetrahedron 53, 5537–5562 10.1016/S0040-4020(97)00210-X [DOI] [Google Scholar]
- 55. Ancliff R. A., Russell A. T., and Sanderson A. J. (1997) Resolution of a citric acid derivative: synthesis of (R)-(−)-homocitric acid-γ-lactone. Tetrahedron Asymmetry 8, 3379–3382 10.1016/S0957-4166(97)00435-7 [DOI] [Google Scholar]
- 56. Wu M. X., and Hill K. A. (1993) A continuous spectrophotometric assay for the aminoacylation of transfer RNA by alanyl-transfer RNA synthetase. Anal. Biochem. 211, 320–323 10.1006/abio.1993.1276 [DOI] [PubMed] [Google Scholar]
- 57. Goldschmidt L., Eisenberg D., and Derewenda Z. S. (2014) Salvage or recovery of failed targets by mutagenesis to reduce surface entropy. Methods Mol. Biol. 1140, 201–209 10.1007/978-1-4939-0354-2_16 [DOI] [PubMed] [Google Scholar]
- 58. Cooper D. R., Boczek T., Grelewska K., Pinkowska M., Sikorska M., Zawadzki M., and Derewenda Z. (2007) Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr. Sect. D Biol. Crystallogr. 63, 636–645 10.1107/S0907444907010931 [DOI] [PubMed] [Google Scholar]
- 59. Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., and DeTitta G. T. (2003) A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 142, 170–179 10.1016/S1047-8477(03)00048-0 [DOI] [PubMed] [Google Scholar]
- 60. Boivin S., Kozak S., and Meijers R. (2013) Optimization of protein purification and characterization using Thermofluor screens. Protein Exp. Purif. 91, 192–206 10.1016/j.pep.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 61. Leslie A. G. W., and Powell H. R. (2007) Processing diffraction data with mosflm. in Evolving Methods for Macromolecular Crystallography: The Structural Path to the Understanding of the Mechanism of Action of CBRN Agents (Read R. J., and Sussman J. L., eds) pp. 41–51, Springer, Dordrecht, Netherlands [Google Scholar]
- 62. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L.-W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 65. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martel A., Liu P., Weiss T. M., Niebuhr M., and Tsuruta H. (2012) An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J. Synchrotron Radiat. 19, 431–434 10.1107/S0909049512008072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smolsky I. L., Liu P., Niebuhr M., Ito K., Weiss T. M., and Tsuruta H. (2007) Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Crystallogr. 40, s453–s458 10.1107/S0021889807009624 [DOI] [Google Scholar]
- 68. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grant T. D., Luft J. R., Carter L. G., Matsui T., Weiss T. M., Martel A., and Snell E. H. (2015) The accurate assessment of small-angle X-ray scattering data. Acta Crystallogr. Sect. D Biol. Crystallogr. 71, 45–56 10.1107/S1399004714010876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.