Abstract
The biomechanical properties of extracellular matrices (ECMs) are critical to many biological processes, including cell–cell communication and cell migration and function. The correct balance between stiffness and elasticity is essential to the function of numerous tissues, including blood vessels and the lymphatic system, and depends on ECM constituents (the “matrisome”) and on their level of interconnection. However, despite its physiological relevance, the matrisome composition and organization remain poorly understood. Previously, we reported that the ADAMTS-like protein Lonely heart (Loh) is critical for recruiting the type IV collagen–like protein Pericardin to the cardiac ECM. Here, we utilized Drosophila as a simple and genetically amenable invertebrate model for studying Loh-mediated recruitment of tissue-specific ECM components such as Pericardin to the ECM. We focused on the functional relevance of distinct Loh domains to protein localization and Pericardin recruitment. Analysis of Loh deletion constructs revealed that one thrombospondin type 1 repeat (TSR1-1), which has an embedded WXXW motif, is critical for anchoring Loh to the ECM. Two other thrombospondin repeats, TSR1-2 and TSR1-4, the latter containing a CXXTCXXG motif, appeared to be dispensable for tethering Loh to the ECM but were crucial for proper interaction with and recruitment of Pericardin. Moreover, our results also suggested that Pericardin in the cardiac ECM primarily ensures the structural integrity of the heart, rather than increasing tissue flexibility. In conclusion, our work provides new insights into the roles of thrombospondin type 1 repeats and advances our understanding of cardiac ECM assembly and function.
Keywords: ADAMTS, cardiac development, Drosophila, Drosophila genetics, extracellular matrix, extracellular matrix protein, thrombospondin, cardiogenesis, ADAMTSL, cardiac matrix, TSR repeats, GAG, cardiogenesis, Drosophila melanogaster
Introduction
Extracellular matrices (ECMs),3 which support and protect cells and provide mechanical linkage between tissues like muscles and epidermis, are generally assembled in a similar manner. After incorporation of transmembrane receptors, such as integrins and dystroglycans (1, 2), meshwork-forming components like laminin and collagen IV are able to anchor. By interacting with each other, they form a complex network with distinct biomechanical properties, which is furthermore stabilized by nidogen (3, 4) and allows other proteins (e.g. perlecan) to bind to the matrix as well (5).
Whereas these steps can be found ubiquitously, the Drosophila cardiac ECM is different from the matrices of other tissues or organs in several ways. It forms a 3D meshwork that connects the contractile heart tube to the alary muscles and, thereby, to the epidermis (6, 7). Within this meshwork, embedded pericardial cells differentiate into a distinct population of cell types, such as nephrocytes (8–10) or wing hearts (11, 12).
In flies, the cardiac ECM combines two important biomechanical features: elasticity that accounts for a flexible connection between heart and alary muscle cells and a high tensile strength that withstands forces produced by lifelong heart contractions. One major difference between cardiac ECMs and matrices of other tissues is the presence of the ADAMTS-like adapter protein Lonely heart (Loh), which can be found exclusively at the surface of the heart and chordotonal organs. Here, Lonely heart is essential to proper recruitment of the type IV collagen–like protein Pericardin (13). Pericardin (Prc) is secreted into the hemolymph by pericardial nephrocytes and adipocytes, and, as soon as it becomes recruited to the cardiac matrix by Lonely heart, it starts to form a stable network (14). By this mechanism the heart is provided with an exceptional ECM that allows it to withstand the strong mechanical forces of a heartbeat. Lack of Pericardin or its anchor Lonely heart leads to a total collapse of the dorsal vessel and dissociation of the pericardial cells and alary muscles from the heart tube. Concomitants are severely impaired heartbeat and absence of heart-mediated hemolymph transport. Accordingly, corresponding mutant animals exhibit decreased fitness and shortened lifespan (13).
The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) superfamily consists of two classes of proteins: ADAMTS and ADAMTS-like proteins. Their main difference is that ADAMTS-like proteins, such as Loh, lack the proteolytically active motif within the ADAM spacer. Both classes share several domains, with most of them being poorly defined. In addition to a spacer region, a changing number of thrombospondin type 1 repeats (TSR1) can be found next to a protease and lacunin (PLAC) domain and a signal peptide (15). These ancillary domains apparently ensure proper substrate specificity as well as cell-surface or ECM tethering (16, 17).
TSR1 motifs were initially discovered in thrombospondins (TSPs), which belong to the family of calcium-binding glycoproteins that are secreted into the extracellular matrix of all complex organisms. TSPs have been shown to bind to fibronectin, laminin, collagen, and other matricellular proteins to form complex networks on the cell surface. TSP superfamily members are involved in regulation of spinal cord outgrowth (e.g. F-spondin) or act as specific anti-angiogenic factors in brain development (e.g. BAI-1). In addition, they can be critical to directed ECM proteolysis (16). TSPs are modular proteins containing several types of repetitive sequence motifs (Fig. 1). One of the most characteristic motifs is the evolutionarily conserved thrombospondin type 1 repeat (TSR1), which is ∼60 amino acids in length and supposed to form an antiparallel three-stranded structure that interacts with glycoproteins of the extracellular matrix. The human genome harbors ∼90 genes encoding TSR1-containing proteins (18), whereas ∼14 corresponding proteins are present in D. melanogaster (19). Among these, some have been shown to contribute to heart development during embryogenesis. These proteins are the transmembrane receptor Uncoordinated 5 (Unc5) (20) and the ADAMTS-like protein Lonely heart (Loh) (13).
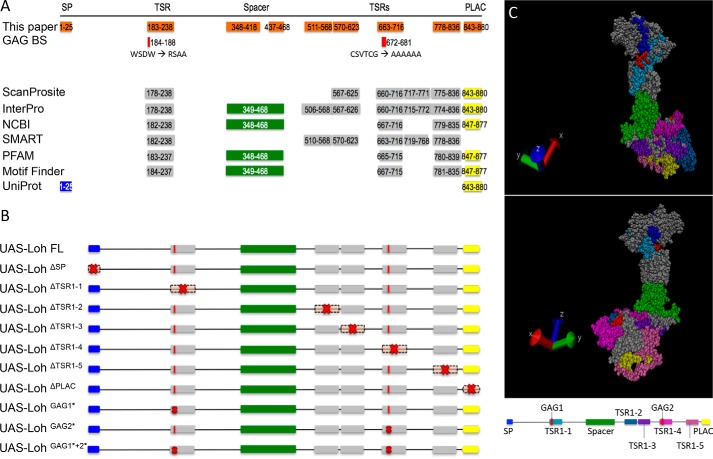
Figure 1.
Lonely heart domain structure and constructs used in this study. A, the Lonely heart isoform A sequence (GenBankTM entry AAF52956.3) was used to identify functional motifs. An N-terminal signal peptide, a variable number of thrombospondin repeats type I, an ADAM spacer region, and a single C-terminal PLAC domain were recognized by ScanProsite, Interpro, NCBI, SMART, PFAM, Motif Finder, and UniProt. Additionally, two putative glycosaminoglycan (GAG) binding sites were identified that locate within two thrombospondin repeat type I motifs (44, 45). Numbers depict the respective amino acid positions. B, Lonely heart constructs used in this study. Red crosses indicate mutated protein motifs. Nomenclature indicates deletions (Δ) or point mutations (*). Due to algorithm-dependent, inconsistent predictions of the TSR1 domains, constructs were generated based on the protein structure proposed previously (13). SP, signal peptide. C, 3D model of Lonely heart with color-coded protein domains. All thrombospondin domains locate to the surface of the protein. Modeling was performed using YASARA (42) and VMD version 1.9.3 (University of Illinois).
To understand the molecular mechanism by which Lonely heart ensures proper cardiac ECM formation in more detail, we analyzed a large set of individually mutated Loh proteins for their capability to incorporate into the ECM and recruit Pericardin. To allow quantitative measurements of Pericardin recruitment efficiency, we applied an in vivo recruitment assay established previously in our laboratory (13). In addition to the ECM of somatic muscles, we also investigated other types of matrices present in Drosophila for their capability to recruit Pericardin in a Loh-dependent manner. Furthermore, we analyzed whether Pericardin, once recruited to a target matrix, has an intrinsic capacity to self-assemble into a meshwork independent of Loh. To perform the analysis, we used imaginal discs to express full-length Loh in distinct compartments of the disc and evaluated potential spreading of the Pericardin meshwork over neighboring zones that lacked Loh.
Finally, to achieve an initial understanding of the biomechanical relevance of Pericardin, we searched for physiological consequences of ectopic Pericardin deposition. In this respect, body wall muscles represent an effective readout system to investigate, for example, altered animal locomotion or lifespan. We found that incorporation of Pericardin into the matrix of somatic muscles has no influence on lifespan but impairs contraction, thereby affecting the general locomotion performance.
Results
We have shown previously that Loh is essential and sufficient to recruit Pericardin from the hemolymph toward a given matrix (13). Here, we did a follow-up study aiming to understand the molecular mechanisms by which Lonely heart itself adheres to the cardiac matrix and, furthermore, how the protein recruits and assembles Pericardin, two processes that were not addressed until now. As depicted in Fig. 1, the Loh sequence contains an N-terminal signal peptide as well as five TSR1-like repeats. Of note, the number, size, and position of predicted TSR1 repeats differ, depending on the search and prediction algorithm used (Fig. 1A). Additionally, Loh contains two predicted glycosaminoglycan (GAG)-binding sites located within two of the TSR1 domains. Up to now, the functionality of these sites has not been confirmed experimentally.
It has been shown for ADAMTS-1 and other proteins that anchoring to the extracellular matrix requires the C-terminal presence of TSR motifs, suggesting an interaction with glycosaminoglycans, such as heparan sulfate, which are predeposited at the matrix (15). This work provides initial evidence that Lonely heart anchors to Drosophila matrices in a similar manner.
Functional analysis of distinct Lonely heart domains
In this study, a series of lonely heart mutations were generated to establish corresponding transgenic Drosophila lines as well as transiently transfected insect cell lines. Subsequently, a combined analysis of both systems was performed to investigate the molecular functions of individual domains present in the Loh protein in detail. Each of the established transgenic Drosophila lines carries a UAS-loh construct, consisting either of the full-length (FL) WT sequence (positive control) or of specifically mutated forms of loh lacking certain domains (Fig. 1B). Additionally, a construct lacking the ADAM spacer region, but retaining the antigenic region recognized by our anti-Loh antibody, was generated. However, after inducing expression by crossing to mef2-Gal4 or prc-Gal4 driver lines, this construct could not be detected by Western blot analysis (data not shown), indicating that the resulting protein is highly unstable and thus degraded. Consequently, this construct was excluded from further analysis. All other constructs were analyzed for their capability to recruit Pericardin by applying a previously established ectopic expression in vivo recruitment assay (13). Before performing these experiments, we verified that the individual Loh constructs were expressed properly and located to the cell surface, which was a prerequisite for the follow-up experiments. Corresponding analyses were done using a cell culture system (Sf21 cells; Fig. 2) as well as transgenic Drosophila (Fig. 3).
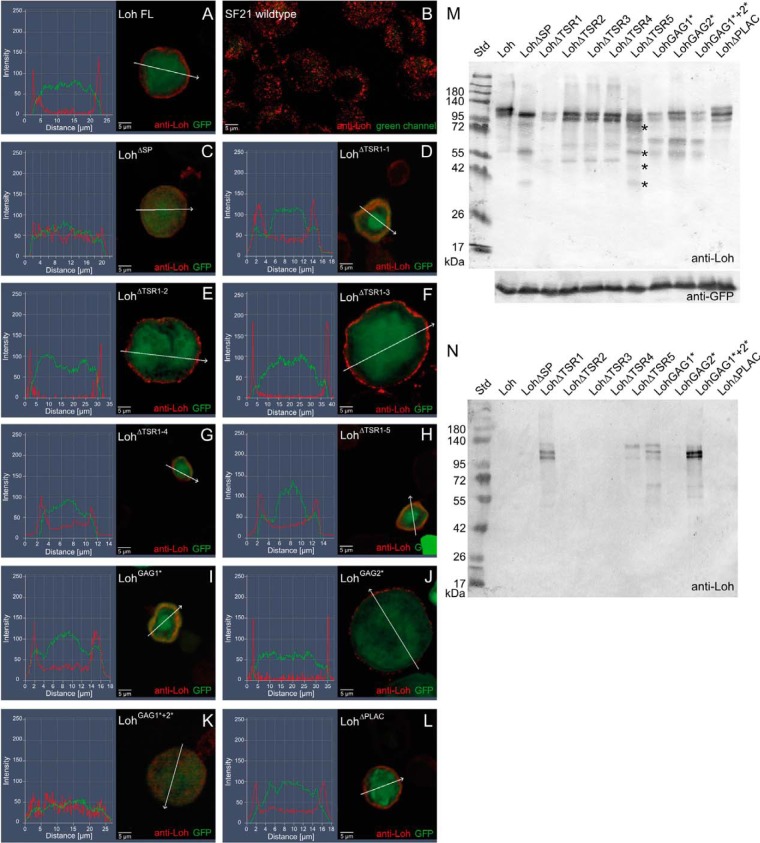
Figure 2.
Expression and localization of mutated Loh constructs in Sf21 cells. Sf21 cells were transfected with WT and individually mutated Loh constructs. Successful transfection was monitored by simultaneous expression of cytoplasmic GFP (green channel). WT as well as mutated Loh were visualized by anti-Loh antibody staining (red channel). Intensity profiling of stained cells was used to determine the subcellular localization of individual Loh constructs. The respective regions of evaluation are marked (arrows in A–L). Full-length Loh (A) is secreted and accumulates at the surface of the cell. Anti-Loh staining of untransfected cells results in a spotted distribution of low-intensity signals (overexposed to visualize the shape of the cells) (B). Loh lacking the signal peptide is not secreted and retained in the cytoplasm (C). The constructs LohΔTSR1-1 and LohGAG1* show no distinct surface location but accumulate in a patchy manner close to the plasma membrane (D and I). A similar behavior is observed for LohGAG1*+2*, yet with a broader distribution in the cytoplasm (K). Individual mutations in the remaining thrombospondin type I repeat domains (LohΔTSR1-2, LohΔTSR1-3, LohΔTSR1-4, and LohΔTSR1-5), as well as lack of the PLAC domain (LohΔPLAC) or a mutation in the second speculative GAG binding site (LohGAG2*), do not affect secretion or anchoring (E–H, J, and L). M, Western blotting of construct-specific cell lysates confirms expression and adequate molecular mass of all Loh constructs. Asterisks indicate putative degradation products that are unique to LohΔTSR1-5 and do not appear in the case of any other TSR1 deletion construct. Co-transfected GFP was used as a loading control. N, Western blotting of construct-specific cell culture media confirms the presence of LohΔTSR1-1, LohΔTSR1-5, LohGAG1*, and LohGAG1*+2*, indicating proper secretion but impaired binding of the respective constructs to the ECM. The depicted blots are representative of three individual biological replicates.
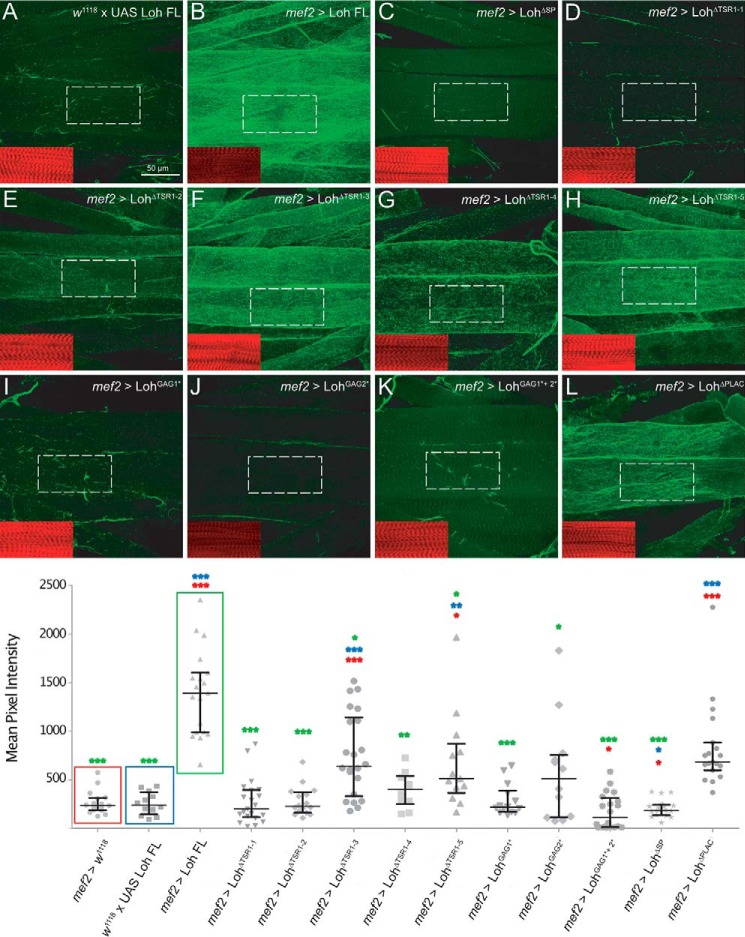
Figure 3.
Expression and localization of mutated Loh constructs in somatic muscles. All UAS constructs were inserted at the 86F8 landing site on the 3R chromosome. Expression was driven by crossing in mef2-Gal4, which induces expression of the target construct in the muscle lineage. Loh localization was visualized by anti-Loh antibody staining (red channel). Muscles were counterstained with anti-α-spectrin antibodies (green channel). Intensity profiling of stained cells was used to determine the subcellular localization of individual Loh constructs. The respective regions of evaluation are marked (arrows in A–L) Full-length Loh, driven by mef2-Gal4, is secreted and accumulates at the surface of the muscle (A). The UAS-Loh FL construct alone (negative control) shows no expression (B). Loh lacking the signal peptide is not secreted but appears to be directed to the nucleus (C). Loh lacking TSR1-2, TSR1-3, TSR1-4, or TSR1-5 is properly secreted and locates to the surface of the cells (E–H). A similar pattern is observed for the construct harboring a mutation in the second speculative GAG binding site (J). Loh that lacks TSR1–1 is not present at the muscle surface (D), which is also the case for the construct carrying a mutation in the first putative GAG-binding site (I). Mutating both predicted GAG-binding sites simultaneously results in complete loss of Loh incorporation into the muscle ECM (K). Deleting the PLAC domain (L) has no effect on secretion or localization of Loh.
Secretion and anchoring capability of Loh mutant constructs in transgenic animals and Sf21 cells
Pericardin recruitment to the ECM of a certain tissue depends on the presence of Loh at the target cell surface. This presence in turn requires successful processing, secretion, and anchoring of Loh. Therefore, we tested full-length WT Loh, as well as the individually mutated forms of the protein, for proper secretion and localization. In this context, Sf21 cells (expression driven by a constitutively active polyhedrin promoter) and somatic muscle cells (expression driven by mef2-Gal4) were analyzed. For visualizing Loh, we used a polyclonal anti-Loh antibody recognizing a peptide sequence within the ADAM spacer region of the protein. This antibody was described previously (13).
To ensure comparable transfection efficiencies of the Sf21 cells, eGFP was inserted into a second multiple cloning site present in the used expression vector; the resulting GFP signal was then used as a loading control for Western blot analysis (Fig. 2M) as well as a cytoplasmic counterstain for immunocytochemistry (Fig. 2, A–L). Captured images were processed for fluorescence intensity profiling to analyze Loh secretion and surface localization. As expected, full-length Loh was successfully secreted and anchored to the cell surface (Fig. 2A). As a negative control, UAS-LohΔSP that lacks the N-terminal signal sequence was used. The respective construct failed to become secreted and distributed uniformly within the cytoplasm (Fig. 2C). A similar localization was observed for LohGAG1*+2*, in which both speculative GAG-binding sites are mutated (Fig. 2K). The constructs LohΔTSR1-1 and LohGAG1* also showed impaired surface location and apparently accumulated in secretory vesicles close to the plasma membrane (Fig. 2, D and I). The respective localization patterns of these constructs may indicate that secretion still occurred, but anchoring to the ECM was compromised. Mutations in the remaining thrombospondin type I repeat domains (LohΔTSR1-2, LohΔTSR1-3, LohΔTSR1-4, LohΔTSR1-5), as well as lack of the PLAC domain (LohΔPLAC) or a mutation in the second speculative GAG binding site (LohGAG2*), did not affect secretion or anchoring (Fig. 2, E, F, G, H, J, and L). To address the question of whether the mislocalized constructs are expressed and secreted properly, but fail to incorporate into the ECM, Western blotting analyses of transfected Sf21 cells as well as of the respective construct-specific cell culture media were performed. If ECM incorporation fails, whereas expression and secretion are still proper, the respective Loh constructs should be detectable in the culture medium. As depicted in Fig. 2M, all constructs were expressed and exhibited molecular masses in line with expectations. To ensure comparable transfection rates, GFP was co-transfected and used as a loading control. Interestingly, in addition to the cell lysate (Fig. 2M), LohΔTSR1−1, LohGAG1*, and LohGAG1*+2* were also present in the cell culture medium (Fig. 2N), which strongly indicates that the respective constructs are not able to bind efficiently to the ECM. Thus, these data are in line with the results of the immunofluorescence stains (Fig. 2, D, I, and K). Of note, the LohΔTSR1-5 construct was also present in the culture medium, although immunocytochemistry detected it at the cell surface (Fig. 2H). One explanation for this discrepancy is a reduced stability of the construct, which is indicated by presence of additional bands on Western blots, probably degradation products that are unique to LohΔTSR1-5 and do not appear in the case of any other TSR1 deletion construct (Fig. 2M, asterisks). This reduced stability may be caused by impaired protein folding, which in turn could result in less efficient ECM incorporation. In this context, the essentially identical band composition that we observed for constructs holding similar mutations (e.g. TSR1 deletion constructs 1–4 and GAG substitutions 1, 2, and 1 + 2; Fig. 2M) indicates proper folding and stability of the respective proteins. Of note, the tryptophan residues present in the speculative GAG1 motif as well as the cysteine residues present in putative GAG2 (Fig. 1A) are probably of significance to the structure of the TSR domain in which they are embedded (19). However, as depicted above, substitution of the respective motifs apparently does not severely affect stability of the corresponding entire proteins (Fig. 2M).
To assess secretion and ECM incorporation in Drosophila, we expressed Loh and its mutated forms in somatic muscle cells using mef2-Gal4 as a driver (Fig. 3). Flat preparations of third instar larvae were fixed and immunostained for Loh. To visualize cell borders, anti-spectrin antibodies were used for counterstaining. This allowed us to perform intensity profiling to check for Loh secretion and surface localization in somatic muscles. In line with our results using cultured Sf21 cells, we found that most of the tested constructs were secreted and localized to the ECM. However, LohΔTSR1-1, LohGAG1*, and LohGAG1*+2* were again absent from the cell surface (Fig. 3, D, I, and K), which confirms the cell culture data and emphasizes the high relevance of these domains to proper ECM incorporation. The data of the localization and anchoring analyses as well as Pericardin recruitment efficiencies are summarized in Table S1.
Quantitative analysis of Pericardin recruitment efficiency
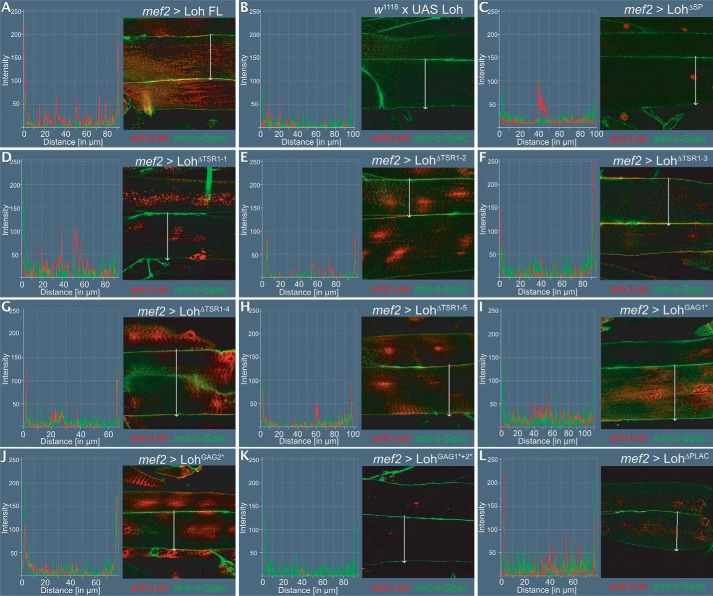
Utilizing the transgenic lines introduced above, we tested whether mutated forms of Loh still harbor the capability to recruit Pericardin to the ECM of muscle cells and whether Pericardin, if recruited, assembles into a network of ECM fibers. It has been shown previously that the full-length form of Loh, when expressed in muscles or adipocytes, is sufficient to recruit Pericardin from the circulating hemolymph and incorporate it into the surface meshwork of ECM fibers (13). We quantified the capacity of mutated forms of Loh to recruit Pericardin using a “sum pixel intensity” region of interest (ROI)-based approach. For normalization of the Pericardin signal in stained specimens of different genotypes, we utilized F-actin staining with phalloidin, a method that has recently been described and successfully used (21). As controls, we used animals that harbored the full-length UAS-Loh (UAS-Loh FL) construct but lacked Gal4 (negative control; Fig. 4A), UAS-Loh FL expressed by mef2-Gal4 (positive control; Fig. 4B), and UAS-LohΔSP driven by mef2-Gal4 (Fig. 4C). Our previous tests have shown that deleting the signal peptide completely inhibits secretion and Pericardin recruitment (Fig. 4C). We confirmed that full-length WT Loh is able to recruit Pericardin to the ECM of muscle cells. In addition, mef2-Gal4–driven expression of UAS-LohΔTSR1-3, UAS-LohΔTSR1-5, and UAS-LohΔPLAC results in considerable Pericardin recruitment (Fig. 4, F, H, and L), indicating that these protein domains play no major role in Pericardin assembly. By contrast, mef2-Gal4–driven expression of UAS-LohΔTSR1-1, UAS-LohΔTSR1-2, UAS-LohΔTSR1-4, UAS-LohGAG1*, UAS-LohGAG2*, and UAS-LohGAG1*+2* does not result in significant Pericardin accumulation (Fig. 4, D, E, G, I, J, and K), which suggests that these distinct domains of Lonely heart are responsible for efficient Pericardin recruitment. Thrombospondin repeat 1 (TSR1-1), with its embedded speculative GAG-binding site, appears to be most critical to Loh anchoring (Fig. 3D), whereas TSR1-2 and TSR1-4 are dispensable for anchoring but important for recruiting Pericardin toward a certain matrix (Figs. 3 (E and G) and 4 (E and G)). Furthermore, our results show that lack of the PLAC domain has no impact on Loh secretion, ECM adhesion, or Pericardin recruitment (Figs. 3L and 4L). We are aware of the fact that a slight reduction in Pericardin recruitment efficiency may not be measurable by our in vivo approach.
Figure 4.
Pericardin recruitment efficiency. To quantify Pericardin recruitment efficiency of individual Loh constructs, expression was driven by crossing in mef2-Gal4. Third instar larval offspring were prepared and stained for Pericardin (anti-Prc, green channel) and counterstained for F-actin (phalloidin, red channel, inset). Images were recorded with identical settings, and ROIs were measured using the “sum slices” method implemented into ImageJ. As a negative control, animals with the genotype w1118, +/UAS-Loh FL were used (A). Pixel intensity obtained for Pericardin staining (green channel) was normalized against F-actin staining (red channel). Quantification is presented as a scatter plot. Full-length Loh recruits Pericardin to the muscle matrix (B). Significant recruitment is also obvious for Loh constructs lacking the third TSR domain (UAS-LohΔTSR1–3) (F), the fifth TSR domain (UAS-LohΔTSR1–5) (H), or the PLAC domain (UAS- LohΔPLAC) (L). In Loh constructs lacking the signal peptide, no Pericardin signal above background is observed (C). Loh harboring a deleted first thrombospondin type 1 repeat (UAS-LohΔTSR1-1) (D) or a mutation of the embedded speculative GAG-binding site (UAS-LohGAG1*) (I) exhibits a considerably reduced capacity to recruit Pericardin. A strong reduction of Pericardin recruitment is also seen in LohΔTSR1-2, in LohΔTSR1-4, and in LohGAG2* mutants (E, G, and J). Loh proteins carrying mutations in both predicted GAG binding sites (LohGAG1*+2*) do also exhibit severely impaired Pericardin recruitment (K). Deleting the PLAC domain has no effect on Loh secretion on Pericardin recruitment (L). The box plot/scatter plot (bottom) depicts quantification of the construct-specific recruitment efficiencies. Colored asterisks indicate the respective significance levels (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001) as calculated for the individual controls (colored boxes).
We completed this set of experiments by testing all generated Loh constructs for their ability to recruit Pericardin to a tissue other than somatic muscles (Fig. S1). Pericardin-Gal4, as well as Cg-Gal4, mediates strong expression of the Gal4 transgene in adipocytes, and it has been shown that Loh is able to recruit Pericardin under these experimental conditions (13). Although not quantitatively proven in detail, our results using adipocytes as a second test tissue confirmed our observations from analyzing recruitment of Pericardin to somatic muscles. The analysis of adipocytes turned out to be more difficult because fat cells are a natural source of Pericardin production. Thus, there is always a certain amount of Pericardin at the cell surface, which is normally released into the hemolymph. Exposing Loh at the adipocyte surface apparently results in retention of the produced Pericardin rather than recruitment of it from the hemolymph. Nevertheless, in-line with our data from the muscle recruitment assay, mutations in the first or second thrombospondin type 1 repeat or in the first or second putative GAG-binding site result in loss of Pericardin recruitment to the adipocytes (Fig. S1, C, D, G, and H). This is also observed for the construct holding simultaneous mutations in both speculative GAG binding sites (Fig. S1E). Additionally, deleting the fourth TSR domain leads to considerably reduced Prc recruitment (Fig. S1J).
Tissue-specific analysis of Pericardin recruitment
Once recruited, Pericardin readily assembles into the preexisting or continuously forming meshwork of ECM fibers, indicating that not only the cardiac ECM, but also the ECM of muscles and adipocytes, harbor all the constituents needed for Pericardin incorporation. However, it is still unknown whether matrices in general harbor an intrinsic capacity to assemble and to incorporate Pericardin and whether Loh alone is necessary and sufficient or if other, yet unknown recruitment factors are also required. To answer this question, we expressed Loh in a variety of additional cells and tissues, including salivary glands, wing imaginal discs, and glial cells of the central nervous system, by crossing the UAS-Loh full-length transgenic line to the different tissue-specific Gal4 driver lines. Again, we used the anti-Loh antibody to confirm proper secretion and the presence of Loh at the respective cell surface. Subsequently, we monitored redistribution of Pericardin to the Loh-exposing tissues using the anti-Pericardin antibody (Fig. 5). We found that in addition to adipocytes and muscle cells (Fig. 5, A and B), also wing imaginal disc cells (Fig. 5E) and glial cells (Fig. 5C) are capable of recruiting Pericardin as long as Loh is exposed at the surface of the respective cells. This finding supports the hypothesis that the ability to recruit and incorporate Pericardin is a general property of a wide range of matrices. Interestingly, salivary glands are the only type of tissue that appears to be incapable of recruiting Pericardin. A careful microscopic analysis confirmed that Loh is produced and that it locates to the outer surface of the salivary gland cells (Fig. 5D). Thus, we can exclude the possibility that Loh itself is not stably present at the matrix and therefore fails to recruit Pericardin. It rather appears likely that a certain property of the salivary gland ECM or lack of a critical ECM constituent impedes Pericardin recruitment to this tissue.
Figure 5.
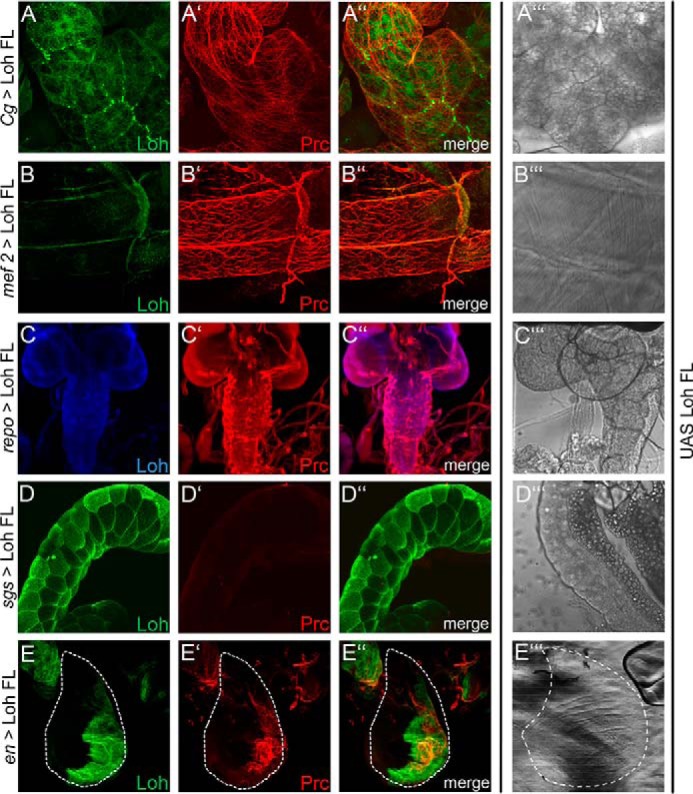
Tissue-specific recruitment of Pericardin. Lonely heart was ectopically expressed in different tissues to analyze the ability of the corresponding extracellular matrices to incorporate Pericardin. Drivers used are Cg-Gal4 for adipocytes (A), mef2-Gal4 for myocytes (B), repo-Gal4 for glial cells of the central nervous system (C), sgs-Gal4 for salivary glands (D), and en-Gal4 for imaginal discs (E). Anti-Loh staining (green (A, B, D, and E) or blue (C) channel) and anti-Pericardin staining (red channel (panels labeled with a prime symbol)) are shown individually and as merged images (panels labeled with a double prime symbol). A‴, B‴, C‴, D‴, and E‴ depict bright-field images of the individual tissues. Dashed lines (E) highlight the border of the wing imaginal disc. All panels show maximum intensity projections of confocal sections. With the exception of salivary glands (D), Pericardin is recruited to all tissues tested (A, B, C, and E).
Recruitment of Pericardin to distinct wing disc compartments
Previous work showed that Pericardin requires Loh for recruitment and proper incorporation into the ECM; however, proper Loh localization does not require Pericardin (13). To understand the molecular mechanisms by which Loh recruits and assembles Pericardin, we aimed to distinguish between two possible models. 1) Loh may act as a crystallization seed that recruits Pericardin from the hemolymph, either directly or via additional linker proteins. In a second step, the matrix-bound Pericardin molecules promote additional recruitment, assembly, and ECM incorporation of Pericardin from the hemolymph. Consequently, Pericardin matrices should spread over a tissue even if Loh is not present underneath. 2) Pericardin only adheres and incorporates if Loh localizes underneath; thus, coordinated self-assembly and spreading of Pericardin over a tissue that lacks Loh is not possible. To discriminate between these two possibilities, we expressed full-length Loh (UAS-Loh FL) in different compartments of the wing discs by utilizing dpp-Gal4, ci-Gal4, hh-Gal4, and sd-Gal4 as drivers (see Fig. 6). In each case, we found that Pericardin recruitment is restricted to those areas that express and display Loh. For example, hh-Gal4 drives Loh expression exclusively in cells of the posterior wing compartment (Fig. 6C, green channel). Consequently, Pericardin, synthesized and secreted by adipocytes of the fat body, adheres exclusively to this domain without spreading out into the anterior wing compartment (Fig. 6C, red channel). These results strongly support the second model proposed above, which postulates a necessity of underlying Loh for proper Pericardin recruitment and incorporation. Thus, the distinct localization of Loh allows for a temporally and spatially highly specific Pericardin incorporation into specialized extracellular matrices, such as the cardiac ECM.
Figure 6.
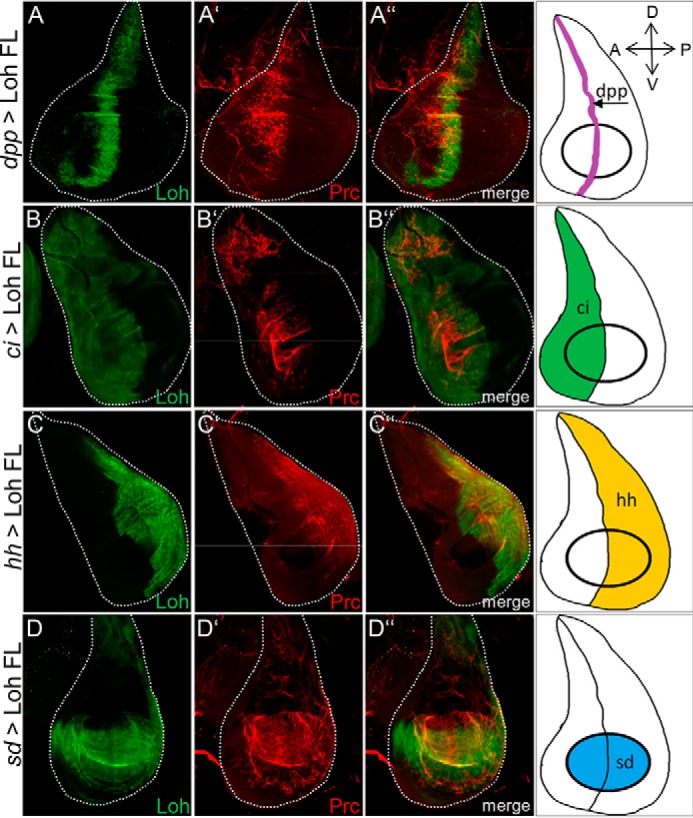
Pericardin recruitment to distinct compartments of the wing disc surface. Ectopic expression of UAS-Loh FL in different compartments of third instar larval wing imaginal discs using dpp-Gal4 (expressed along the anterior-posterior boundary; A–A″), ci-Gal4 (anterior compartment; B–B″), hh-Gal4 (posterior compartment; C–C″), and sd-Gal4 (expressed in the wing pouch; D–D″) as drivers. Loh is visualized using an anti-Loh antibody (green channel), and Pericardin is visualized using an anti-Pericardin antibody (red channel). The right column depicts a schematic illustration of the compartment-specific Gal4 expression for all driver lines used. Pericardin recruitment is restricted to areas that express and display Loh. Orientation of imaginal discs is indicated in the top right corner.
Analysis of the biochemical interactions in Pericardin-containing extracellular matrices
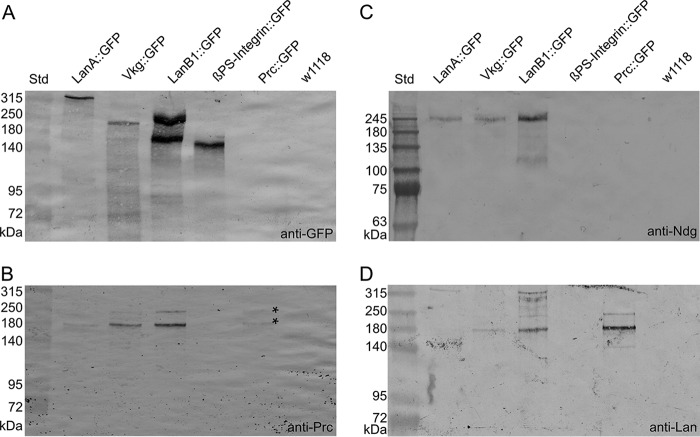
To understand the molecular mechanisms that facilitate Loh-dependent Pericardin incorporation into extracellular matrices in more detail, we analyzed the biochemical interactions characteristic to these matrices. In this effort, we utilized a series of GFP-tagged ECM proteins (LanA::GFP, LanB1::GFP, Vkg::GFP, βPS-integrin::GFP, and Pericardin::GFP) (22) and analyzed them for co-immunoprecipitation with Pericardin, nidogen, and laminin. Of note, co-immunoprecipitation of Loh and Pericardin has already been confirmed (13). As depicted in Fig. 7A, the GFP-tagged proteins exhibited a molecular mass consistent with expected values (including posttranslational modifications), thus indicating the presence of stable fusions. Furthermore, the respective proteins could be extracted from corresponding larval homogenates in rather convenient amounts. The only construct that could not be detected with anti-GFP antibodies was Pericardin::GFP. However, the fact that endogenous Pericardin as well as laminin was present in Pericardin::GFP pulldown fractions but absent in corresponding control preparations (w1118; Fig. 7 (B and D)) indicates that Pericardin::GFP can be isolated from corresponding transgenes, but in rather limited amounts. Further analyses revealed that Pericardin co-immunoprecipitates with LanA::GFP, LanB1::GFP, Vkg::GFP, and Pericardin::GFP, but not with βPS-integrin::GFP (Fig. 7B). The fact that the bands detected in the Pericardin::GFP sample correspond to the size of WT Pericardin (Fig. 7B, asterisks) confirms that endogenous Pericardin is labeled and not the GFP fusion protein. Nidogen co-immunoprecipitates with LanA::GFP, LanB1::GFP, and Vkg::GFP, but not with βPS-integrin::GFP and Pericardin::GFP (Fig. 7C), whereas laminin co-immunoprecipitates with Vkg::GFP and with Pericardin::GFP, but not with βPS-integrin::GFP (Fig. 7D). Despite a rather ineffective Pericardin::GFP isolation (Fig. 7A), a distinct laminin signal is present in the respective fraction (Fig. 7D), which indicates a strong interaction between Pericardin and laminin. Of note, we did not observe co-immunoprecipitation of βPS-integrin with laminin, although an interaction has been reported before (23). This discrepancy may be due to misfolding of the βPS-integrin::GFP fusion construct that we used in our study. Thus, our finding that Pericardin does not co-precipitate with integrin must be considered preliminary. The data of the immunoprecipitation assay are summarized in the assembly model depicted in Fig. 10.
Figure 7.
Pericardin co-immunoprecipitates with distinct ECM components. GFP-tagged ECM proteins (laminin A, LanA::GFP; laminin B1, LanB1::GFP; Viking, Vkg::GFP; βPS-integrin, βPS-integrin::GFP; Pericardin, Pericardin::GFP) were purified from third instar larvae and subjected to Western blot analysis. Anti-GFP antibodies were applied to estimate the protein-specific purification efficiency (A). Anti-Pericardin (Prc; B), anti-nidogen (Ndg; C), and anti-laminin (Lan; D) antibodies were used to assess co-immunoprecipitation of the individual ECM components with the purified GFP fusion proteins. The rather weak detection of endogenous Pericardin in the Pericardin::GFP sample is indicated (B; asterisks). The observation that nidogen migrates significantly higher than expected (predicted molecular mass, 149.1 kDa) presumably reflects extensive posttranslational modification of the protein. Similar mass shifts, predominantly caused by N- and O-glycosylation, have been reported for nidogens from other species (4). The depicted blots are representative of two individual biological replicates.
Figure 10.
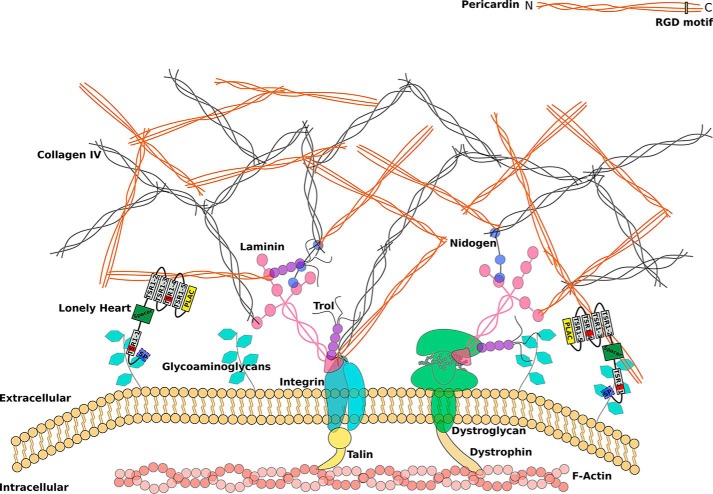
Schematic representation of an extracellular matrix with incorporated Pericardin. The ECM is coupled to the actin cytoskeleton via integrins or dystroglycans. Laminins form the connection between the large structural ECM constituents, including collagen IV, Pericardin, and integrin/dystroglycan. Based on our results, we postulate that Lonely heart binds to glycosaminoglycans of membrane-associated proteins via its first speculative GAG-binding site and to Pericardin (directly or indirectly) via its second and fourth thrombospondin type 1 repeat. Loh itself is shown as a protein domain scheme with a signal peptide indicated in light blue, the ADAMTS spacer in green, the PLAC domain in yellow, and the remaining thrombospondin type 1 repeats in gray, of which two harbor a predicted GAG binding site (red bars). The form of the scheme is based on the protein model shown in Fig. 1C. The depicted interaction between Pericardin and integrin is based solely on the presence of an RGD motif at the C terminus of Pericardin and is not experimentally proven.
Incorporation of Pericardin into the matrix of somatic muscles affects contraction performance
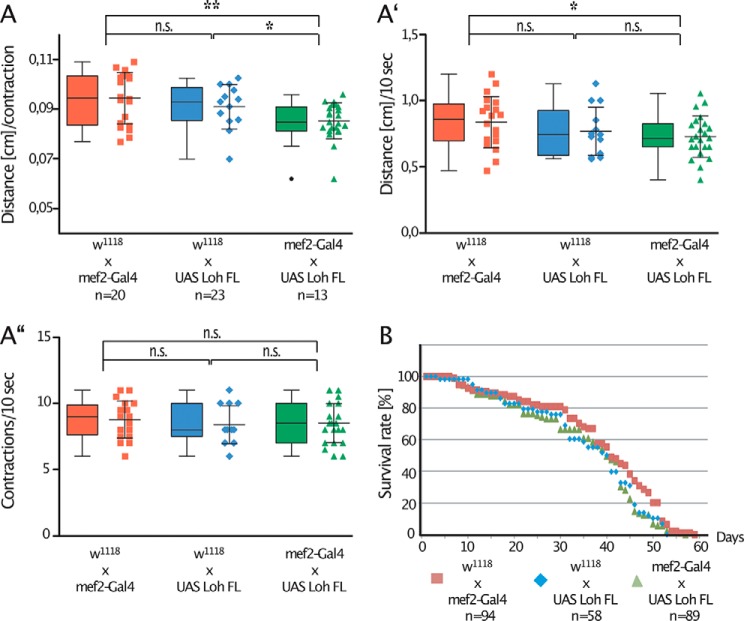
To achieve an initial understanding of the physiological relevance of Pericardin, we analyzed whether recruitment and assembly of the protein into ectopic matrices affected the biomechanical properties of the corresponding matrix. We considered recruitment of Pericardin to muscles to be a useful readout model because biomechanical modulations of the muscle matrix probably have a direct influence on contraction efficiency and, thereby, on locomotion in general. To measure muscle performance, we analyzed the distance a larva covers within a straight run of 10 s (Fig. 8A′), as well as the number of contractions that occur during this time frame (Fig. 8A″). Based on these data, we also calculated the distance covered by a single contraction (Fig. 8A). As depicted, specimens with Pericardin ectopically incorporated into the muscle matrix exhibit reduced movement speed. Interestingly, the effect is not caused by a reduced contraction rate in corresponding animals, which is comparable with that of controls (Fig. 8A″), but by a reduced distance covered per contraction (Fig. 8A). This result may indicate that artificial incorporation of Pericardin into the ECM of muscle cells increases stiffness of the matrix, which eventually impairs proper contraction of the tissue. We also assessed possible effects on lifespan and found no significant influence of ectopic Pericardin deposition under laboratory conditions (Fig. 8B).
Figure 8.
Artificial recruitment of Pericardin affects muscle function. A, ectopic expression of full-length Loh in muscles (mef2-Gal4 × UAS Loh FL; green triangles) significantly impairs crawling performance. As controls, F1 animals from either a crossing of mef2-Gal4 × w1118 (red squares) or from w1118 × UAS-Loh FL (blue diamonds) were used. Crawling performance was measured as crawling distance in cm/contraction (A) or crawling distance/10 s (A′). Crawling performance is significantly reduced if Pericardin is recruited to the muscle ECM (green triangles in A and A′). The contraction frequency is not significantly altered in corresponding animals (A″). Ectopic expression of full-length Loh (mef2-Gal4 × UAS Loh FL; green triangles) has no effect on lifespan, compared with controls (mef2-Gal4 × w1118 (red squares) and w1118 × UAS Loh FL (blue diamonds)) (B). Depicted are mean values ± S.D. (error bars). Corresponding significance levels are indicated (asterisks, Student's t test; *, p < 0.05; **, p < 0.01; n.s., not significant). Data are presented as box plots and scatter plots.
Overexpression of Lonely heart in cardiomyocytes leads to matrix phenotypes and affects heart performance
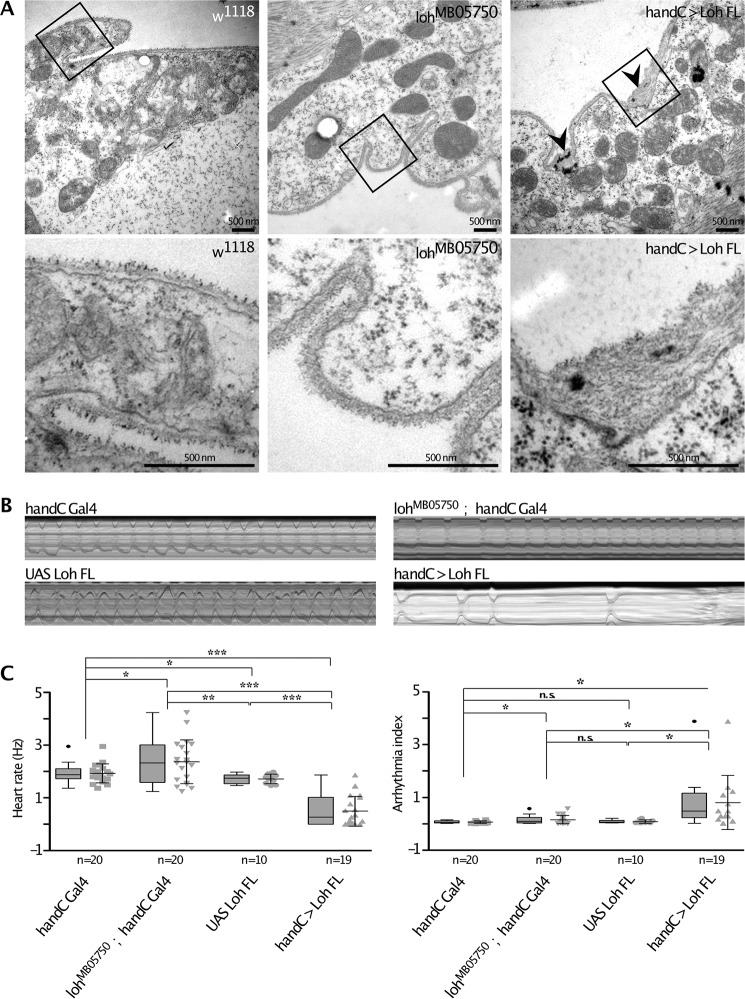
The primary function of Loh is the recruitment of Pericardin toward the cardiac matrix. Pericardin requires the presence of Loh to become properly incorporated into the ECM meshwork and for full functionality (13, 14). Therefore, we tested whether the amount of Loh present at the surface of cardiac cells directly affects ECM architecture (e.g. by recruiting increased or reduced quantities of Pericardin or other, yet unknown Loh interactors). An ultrastructural analysis of the cardiac matrix of homozygous lohMB05750 mutant wandering third instar larvae, compared with WT, revealed no visible difference with respect to, for example, thickness of the matrix (Fig. 9A). By contrast, overexpression of full-length Loh (UAS-Loh FL) in cardiac cells results in irregular matrix deposition characterized by a dramatic increase in the width of the matrix (Fig. 9A, arrowheads). Heart performance, measured by high-speed video microscopy in semi-intact third instar larvae, is also affected when full-length Loh is overexpressed in cardiac cells. Corresponding animals display a considerably reduced heart rate, concomitant with severe arrhythmia (Fig. 9, B and C). Of note, arrhythmia includes long periods of heartbeat arrest, which emphasizes the severity of the phenotype. Representative examples are shown in Fig. 9B and in Movies S1–S4.
Figure 9.
Ultrastructural and physiological effects of modulated Loh levels in cardiac tissue. A, transsections of the heart chamber of wandering third instar larvae analyzed by transmission EM. Images in the top panels represent overviews (×20,000), whereas the bottom panels depicts close-ups of the marked areas. Black arrowheads indicate ECM accumulations that mainly occur at membrane invaginations of cardiomyocytes in loh overexpression animals (handC > Loh FL) but are absent in control animals (w1118) or loh null mutants (lohMB05750). B, representative M-modes (10-s videos) of third instar larvae of the depicted genotypes indicate heart rate over time and occurrence of arrhythmia. C, quantification of B. Both heart rate and rhythmicity (arrhythmia index) are significantly affected in loh knockout animals (lohMB05750; handC-Gal4) and loh overexpression larvae (handC > Loh FL), compared with controls (handC-Gal4 and UAS-Loh FL). Depicted are mean values ± S.D. (error bars). Corresponding significance levels are indicated (asterisks, Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant). Data are presented as box plots and scatter plots.
Discussion
In contrast to matrices present at the surface of most other tissues, the cardiac ECM is exposed to permanent mechanical stress generated by the regular and repetitive contraction cycles of the heart. These unique biomechanical conditions require ECM adaptation, which is achieved predominantly by incorporating specific structural components into the respective matrices. In Drosophila, one of these components is the type IV collagen–like protein Pericardin, which is recruited specifically to cardiac tissue by its adaptor protein Lonely heart (13). However, until now, neither the recruitment process itself nor the relevance of Pericardin to the biomechanical properties of the cardiac ECM have been studied in detail. By conducting a recruitment assay based on systematically generated domain-specific Loh mutants, we found that presence of the first TSR1 domain is critical to localizing Loh to the ECM (Fig. 3D). Interestingly, mutating only the speculative GAG-binding site embedded within the first TSR1 domain is sufficient to abrogate Loh anchoring, indicating a high functional relevance of this distinct sequence motif (Fig. 3I). This result was confirmed by expressing the same constructs in Sf21 cells. Also in this system, deletion of the first TSR1 domain or mutation of the embedded putative GAG site resulted in considerably reduced surface localization of the respective Loh constructs (Fig. 2, D and I). Significantly, Western blot analysis detected the proteins in the culture medium (Fig. 2N), which indicates that production and secretion still occurred, whereas incorporation into the ECM was impaired. Thus, our data suggest that TSR1-1, with its embedded putative GAG-binding site, is crucial for anchoring Loh to the ECM, which represents a prerequisite for the subsequent recruitment of Pericardin. On the other hand, the second speculative GAG binding site, embedded within the TSR1-4 domain, appears to be dispensable for localizing Loh (Figs. 2 (G, J, and N) and 3 (G and J)) but is required for efficient Pericardin recruitment (Fig. 4J). Of note, previous work identified the respective CXXTCXXG motif as a consensus site for O-fucosylation and showed that mutating this motif results in impaired protein secretion (24). Because the substitution in UAS-LohGAG2* covers this motif (Fig. 1A), slightly impaired secretion of this construct appears possible. However, its complete inability to recruit Pericardin (Fig. 4J) cannot be attributed to minor deficiencies in secretion. Thus, our findings indicate that both speculative GAG-binding sites are of high functional relevance, with the first site being essential to proper anchoring of Loh, whereas the second one appears to be required for Pericardin recruitment. Interestingly, also lack of the second TSR1 domain results in failure to recruit Pericardin (Fig. 4), whereas localization of Loh is not affected (Figs. 2 (E and N) and 3E). Thus, the TSR1-2 and TSR1-4 domains as well as the putative GAG2-binding site seem to be dispensable for localizing Loh but crucial to proper Pericardin interaction and recruitment. In this context, the distinct position of the respective domains within Loh is probably decisive. According to structural modeling, TSR1-2 and TSR1-4, the latter containing the predicted GAG2 site, exhibit close spatial proximity (Fig. 1C). The fact that lack of either domain completely abolishes the capacity of Loh to recruit Pericardin (Fig. 4) indicates that these two domains constitute the interaction site between Loh and Prc, with the embedded speculative GAG binding site being of critical relevance. Subsequent to the initial binding, the nearby TSR1-3 and TSR1-5 domains may support interaction; however, their functional relevance is minor compared with TSR1-2 and TSR1-4 (Fig. 4). Taking these data into account, it appears likely that the N-terminal part of Loh, including the first TSR1 repeat and the embedded predicted GAG binding site, is facing the plasma membrane and anchors the protein to the cell surface, probably via glycosaminoglycan binding. The C-terminal part of Loh would then be available for interaction with Pericardin, and possibly also with other ECM components, via the second and fourth TSR1 repeats. Of note, a function of the PLAC domain, which is present in several enzymes and ECM proteins, such as ADAMTS-2, -3, -10, and others, was not uncovered by our approach. Deleting the C-terminal PLAC domain in Loh has no distinct consequences, either for Loh secretion or for Pericardin recruitment efficiency, as far as we can state in view of the sensitivity limitations of our test system. Results are summarized in Table S1.
Regarding the question of whether ECMs are generally capable of recruiting and incorporating Pericardin, we found that this is not the case. Whereas Loh-dependent recruitment was observed for fat body cells, somatic muscles, glial cells of the central nervous system, and wing discs, salivary gland cells did not incorporate Pericardin into the ECM, although Loh was present at the surface (Fig. 5). This result indicates that other, yet unknown ECM components are required, in addition to Loh, for proper recruitment of Pericardin and that at least one of these factors is not present in salivary gland cells. Identification of the respective constituents represents an important objective of future studies because it would complement the current understanding of the interconnections that form the cardiac extracellular matrix (Fig. 10). The alternative explanation, the presence of an inhibitory protein that prevents Pericardin incorporation into the ECM of salivary glands, appears unlikely, although we cannot rule out this possibility for sure.
When dissecting third instar larvae, Pericardin fibers randomly associated with various tissues are constantly observed (not shown). However, significant amounts of Pericardin, organized and incorporated into the meshwork of a matrix, are only present at the surface of the heart and chordotonal organs. This tissue specificity essentially depends on the presence of Loh (13), and this paper. The dispersed and random adhesion of Pericardin, synthesized and secreted by adipocytes, to other tissues presumably reflects an intrinsic property of the protein to interact, with low affinity, with matrix components other than Loh (e.g. with collagen IV or laminin) (Fig. 7). Thus, Pericardin is able to adhere to an existing matrix, yet efficient incorporation requires Loh presence. This hypothesis is also supported by the results of experiments expressing Loh at distinct surface areas of wing discs (Fig. 6). Therefore, we speculate that Loh is a primary anchor for Pericardin. Whether anchoring is mediated by direct or indirect interaction remains to be determined.
To understand the functional impact of Pericardin incorporation into extracellular matrices in more detail, we directed the protein to the surface of somatic muscles by ectopically expressing Loh in this tissue. Because neither Loh nor Pericardin are present at the surface of WT somatic muscles, the setup allows distinct evaluation of the resulting biomechanical effects in corresponding transgenic animals. As depicted (Fig. 8), specimens with Pericardin incorporated into the muscle matrix exhibit a considerably reduced movement speed, which is caused by a reduction in the distance covered by a single contraction wave (Fig. 8A); the contraction rate is not altered in corresponding animals (Fig. 8A″). These effects are consistent with an increase in ECM stiffness as a result of Pericardin incorporation, which, in the case of body wall muscles, impairs flexibility and thus contraction efficiency. Of note, as has been shown in the aging mouse myocardium, an increase in cardiac stiffness correlates with the accumulation of collagen (25). Furthermore, fly hearts also exhibit reduced contraction efficiency with age, which manifests in decreased diastolic and systolic dimensions (26–28). The fact that Pericardin knockdown flies do not show this age-dependent decline (21) indicates that progressive Pericardin incorporation increases stiffness of the cardiac ECM in a similar way. Our data indicating that elevated Loh levels at the surface of cardiac cells result in increased matrix deposition and severely impaired heart function (Fig. 9) support this indication and emphasize the relevance of an adequate cardiac ECM composition to proper heart function. Aside from simply reflecting increased cardiac stiffness, the observed effects on heart rate and rhythmicity may also be indicative of altered chronotropy, caused by impaired activity of certain ion channels that in turn affects pacemaker function. This possibility is supported by recent data on Drosophila cardiac muscles, suggesting that basement membrane (BM) proteins are more than simple structural entities. Based on the fact that knockdown of Pericardin, laminin A, and viking increased cardiac contractility, whereas BM stiffness was apparently not altered, it was postulated that BM proteins are capable of regulating cardiac function by modulating interaction of the heart tube with adjacent muscle layers (21). By altering Loh abundance, and thus matrix composition, this interaction may be impaired, which could affect activity of corresponding mechanosensitive channels, eventually resulting in the observed phenotypes.
Whereas an in-depth understanding of Pericardin functionality clearly requires further studies, our current data suggest that the presence of the protein within the cardiac ECM is primarily required to ensure the structural integrity of the beating heart, rather than representing a means to increase flexibility of the tissue.
Experimental procedures
Drosophila stocks/fly lines
All UAS-Loh constructs were generated in this laboratory with the assistance of a commercial injection service (BestGene, Chino Hills, CA). Drosophila stocks labeled with BL numbers were obtained from the Bloomington stock center. For the WT, we used w1118. Gal4 driver lines used were Cg-Gal4 (29); ci-Gal4, hh-Gal4, sd-Gal4, en-Gal4 received from T. Klein (Düsseldorf, Germany); dpp-Gal4 received from H. Aberle (Düsseldorf, Germany); mef2-Gal4 received from H. Nguyen (Erlangen, Germany); pericardin-Gal4 received from L. Perrin (Marseille, France); repo-Gal4 received from G. Technau (Mainz, Germany); and sgs4-58-Gal4 received from A. Hofmann (Berlin, Germany). Fly husbandry was carried out as described previously (30).
Generation of Loh constructs
Loh constructs used to generate transgenic Drosophila were cloned into a yeast/Escherichia coli/Saccharomyces cerevisiae triple-shuttle vector (pJJH1784) based on vectors described previously (31). The vector pJJH1784 allows cloning via homologous recombination in yeast and site-specific integration into the Drosophila genome. All Loh constructs were introduced into the germ line by injections in the presence of the PhiC31 integrase (using line BL24749, Bloomington Drosophila Stock Center), which resulted in insertion into the 86F8 landing site on the 3R chromosome. Designs of the Loh constructs are depicted in Fig. 1B. All mutations were established by PCR and an appropriate primer design. Transgenic flies were generated using commercial services (BestGene). Complete sequences are available on request.
Sf21 expression constructs and cell culture
Heterologous expression was performed in Sf21 cells (RRID:CVCL_0518) using the Bac-to-Bac® baculovirus expression system (Life Technologies, Inc.). Full-length WT and mutated forms of Loh were cloned into an E. coli/S. cerevisiae/baculovirus triple-shuttle derivative of the pFastBacTM Dual vector. The respective vector (pJJH1460) was constructed similarly to the vectors described previously (31). Expression was induced as described previously (32). To track transfection efficiency, an eGFP reporter gene was inserted into the same vector under control of the p10 promoter. Transfected Sf21 cells were grown on coverslips in 6-well plates for 72 h. Subsequently, the coverslips were removed from the wells, and adherent cells were fixed in 3% paraformaldehyde in PBS for 30 min. After washing (3 × 5 min in PBS), cells were blocked and permeabilized in 2% BSA, 0.01% Triton X-100 (in PBS) for 20 min and incubated with primary antibodies (anti-Loh; 1:500) overnight (4 °C). Unbound antibodies were removed by washing (3 × 5 min in PBS), and cells were blocked again for 60 min (Roti Immuno-Block, Carl Roth, Karlsruhe, Germany), followed by additional washing (3 × 5 min in PBS). Secondary antibodies (anti-guinea pig, Cy3 conjugate, Dianova) were diluted in PBS (1:200) and applied for 90 min at room temperature. Finally, cells were washed again (as described above) and mounted in Fluoromount-G (Southern Biotech, Birmingham, AL). Confocal images were captured with an LSM 5 Pascal confocal microscope (Zeiss, Jena, Germany).
Immunoprecipitation
For each sample, 100 wandering third instar larvae were snap-frozen in liquid nitrogen and ground to powder. The powder was resuspended in 1 ml of lysis buffer (150 mm NaCl, 5% glycerol, 1% IGEPAL-CA-630, 1 mm MgCl2, 50 mm Tris, pH 7.5, 1× protease inhibitor mix from Sigma-Aldrich, Heidelberg, Germany) and incubated for 30 min at room temperature. Subsequent to centrifugation (15 min, 4000 × g), 800 μl of the supernatant were mixed with paramagnetic beads precoupled to anti-GFP antibodies (μMACS GFP Isolation Kit, Miltenyi Biotec, Auburn, CA) and run over magnetic μ-columns according to the manufacturer's instructions (Miltenyi Biotec). Elution fractions were subjected to SDS-PAGE and Western blotting as described previously (33). Primary antibodies were anti-GFP (made in goat, 1:2000; Abcam, Cambridge, UK), anti-Pericardin (made in mouse, 1:100; Developmental Studios Hybridoma Bank, Iowa City, IA), anti-nidogen (made in rabbit, 1:3000; gift from Stefan Baumgartner), and anti-KcLaminin (made in rabbit, 1:2000; gift from John Fessler). Secondary antibodies were anti-goat-AP (1:10,000; Sigma), anti-mouse-AP (1:10,000; Sigma), and anti-rabbit-AP (1:10,000; Sigma).
Immunohistochemistry, Western blotting analyses, and confocal microscopy
Immunostainings were carried out as described previously (33). Antibodies used in this study were as follows: guinea pig anti-Loh (1:500; this laboratory), mouse anti-Pericardin/EC11 (1:5; Developmental Studios Hybridoma Bank), mouse anti-α-spectrin/3A9 (1:3; Developmental Studios Hybridoma Bank), rabbit anti-nidogen/entactin (1:1000; a gift from S. Baumgartner), rabbit anti-laminin (detects only secreted laminin trimers; a gift from J. Fessler), mouse anti-β-Tubulin (1:5 to 1:200, Developmental Studios Hybridoma Bank), and rabbit anti-GFP (1:1000; Abcam). Secondary antibodies used were anti-mouse Cy2/Cy3 (1:100/1:200; Dianova), anti-rabbit-Cy2/Cy3 (1:100/1:200; Dianova), and anti-guinea pig Cy2/Cy3/Alexa633 (1:100/1:200/1:200; Thermo Fisher Scientific, Dianova, and Abcam). F-Actin was visualized by staining fixed tissues using TRITC coupled with phalloidin (Sigma) at a concentration of 0.4 μg/ml (1:200) in 1× PBS, for 2 h at room temperature. Confocal images were captured with a Zeiss LSM 5 Pascal cLSM. Z-stacks were acquired using standard settings and objectives. If not otherwise noted, Z-stacks are depicted as maximum projections. Image processing was done with ImageJ and Affinity Photo.
Transmission EM
Larvae were processed as described previously with minor modifications (34). Briefly, abdomens of larvae were prepared and fixed for 4 h at room temperature in 2% glutaraldehyde (Sigma) in artificial hemolymph, subsequently washed in 0.5 m cacodylate buffer, pH 7.4, post-fixed for 2 h at room temperature in 1% osmium tetroxide in 0.5 m cacodylate buffer, pH 7.4 (Science Services, Munich, Germany), dehydrated stepwise in a graded ethanol series, and embedded in Epon 812. Ultrathin sections (70 nm) were assembled on an ultramicrotome (UC6 and UC7 Leica, Wetzlar, Germany) and mounted on copper slot grids. Sections were stained for 30 min in 2% uranyl acetate (Science Services) and for 20 min in 3% lead citrate (Carl Roth, Karlsruhe, Germany). TEM images were acquired with a Zeiss 902 or a Zeiss 120-kV transmission electron microscope. For all genotypes, n = 3.
Recruitment assay and pixel intensity analysis
For the Pericardin recruitment assay, UAS-loh constructs (full-length WT or mutated forms of Loh) were expressed under the control of mef2-Gal4 (muscle cell lineage); pericardin-Gal4 (adipocytes and pericardial cells); Cg-Gal4 (adipocytes); repo-Gal4 (central nervous system); sgs-Gal4 (salivary glands); or dpp-Gal4, ci-Gal4, hh-Gal4, sd-Gal4, or en-Gal4 (imaginal disc). All crossings were set up, and offspring were allowed to grow up at 27 °C until the third instar wandering stage. Subsequently, animals were dissected, and tissues of interest were fixed for further processing (13, 35).
Confocal images of stained third instar larvae were obtained with an LSM5 Pascal confocal microscope (Zeiss). The Cy2 signal, depicting the distribution of Pericardin, was normalized to the TRITC signal of the phalloidin staining to adapt to variable parameters in the staining procedure, such as fixation or permeabilization efficiencies. Captured Z-stacks were further analyzed using ImageJ. After a reduction of the background, a maximum projection using the “sum slices” option was created. Subsequently, the pixel intensity within an ROI was measured (at least 10 animals/genotype). Further data analysis was conducted with GraphPad Prism (GraphPad Software, La Jolla, CA).
Lifespan assay
Following an approved protocol (36), 2–10 male or female flies, collected within 1 day of eclosion, were transferred twice per week to fresh vials with standard food. A minimum of 55 flies were analyzed per genotype. Vials were kept at 27 °C for the duration of the lifespan assay. Surviving flies were counted after vial changes. All animals were maintained under constant 12-h/12-h, light/dark cycles. Data analysis was conducted with GraphPad Prism (GraphPad Software).
Crawling assay and data analysis
Wandering third instar larvae were collected, and their locomotion activity was recorded with a standard digital camera under a Zeiss binocular (37). The larval linear forward crawling distance and the number of body wall contractions within a time frame of 10 s were assessed for each animal. Corresponding measurements were done for at least 13 animals per crossing. As negative controls, third instar offspring of mef2-Gal4 × w1118, as well as UAS-Loh FL × w1118 crossings, were analyzed. All distance measurements were performed using ImageJ, whereas further data analysis was conducted with GraphPad Prism (GraphPad Software).
Animal preparation and video analysis
Wandering third instar larvae were pinned down with the dorsal side downward onto Sylgard 184 silicone elastomer plates, which were filled with artificial hemolymph. Artificial hemolymph contains 108 mm NaCl, 5 mm KCl, 2 mm CaCl2, 8 mm MgCl2, 1 mm NaH2PO4, 4 mm NaHCO3, and 5 mm HEPES, pH 7.1. Before use, the buffer was supplemented with sucrose (final concentration, 10 mm) and trehalose (final concentration, 10 mm) (38). After dissecting the animals, the specimens were allowed to rest for 10 min. To record heartbeat, a high-speed video camera (Basler piA-640) was mounted on an upright microscope (Leica DMLB), equipped with a ×10 Leica Fluotar. Movies were captured with the software Firecapture® (freeware by Torsten Edelmann) and processed with ImageJ (39). Heart parameters were analyzed using SOHA (Semi-Automated Optical Heartbeat Analysis), a MATLAB application first introduced by Fink et al. (40, 41). Additional data analysis was done using Microsoft Excel and GraphPad Prism (GraphPad Software).
Structural modeling
Yasara Structure 15.7.25 (42) was used to calculate a 3D homology model of Lonely heart. Templates for the homology modeling process were identified by performing a BLAST search of the lonely heart target sequence against ExPDB (43). 16 template structures were provided for building 56 homology models. Finally, one hybrid model of the 56 homology models was generated.
Author contributions
A. P. conceptualization; B. R., Y. P., M. R., K. L., A. B., H. M., and J. J. H. investigation; A. P. and H. M. writing-original draft; B. R., Y. P., H. M., and M. R. writing-review and editing; A. P. funding acquisition; B. R., H. M., and A. P. supervision.
Supplementary Material
Acknowledgments
We thank Mechthild Krabusch, Martina Biedermann, and Kerstin Etzold for excellent technical assistance and fly-keeping. We thank Markus Schneider for providing the Loh 3D model in Fig. 1C and Maik Drechsler, Ariane Wilmes, Philipp Merling, and Patrick Tiltmann for initial experiments. We thank Thomas Klein and Hermann Aberle for sharing fly stocks and reagents. We also thank the Bloomington Drosophila Stock Center for providing fly stocks and Flybase for providing genetic, genomic, and functional information.
This work was supported by funding from the German Research Foundation (SFB 944: Physiology and Dynamics of Cellular Microcompartments) (to A. P.) and by State of Lower-Saxony, Hannover, Germany, Grant 11-76251-99-15/12 (ZN2832). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1, Fig. S1, and Movies S1–S4.
- ECM
- extracellular matrix
- PLAC
- protease and lacunin
- TSP
- thrombospondin
- FL
- full-length
- eGFP
- enhanced GFP
- ROI
- region of interest
- UAS
- upstream activation sequence
- BM
- basement membrane
- TRITC
- tetramethylrhodamine isothiocyanate
- GAG
- glycosaminoglycan.
References
- 1. Hynes R. O. (1992) Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69, 11–25 10.1016/0092-8674(92)90115-S [DOI] [PubMed] [Google Scholar]
- 2. Hynes R. O., and Yamada K. M. (2012) Extracellular Matrix Biology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 3. Timpl R. (1996) Macromolecular organization of basement membranes. Curr. Opin. Cell Biol. 8, 618–624 10.1016/S0955-0674(96)80102-5 [DOI] [PubMed] [Google Scholar]
- 4. Kohfeldt E., Sasaki T., Göhring W., and Timpl R. (1998) Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 282, 99–109 10.1006/jmbi.1998.2004 [DOI] [PubMed] [Google Scholar]
- 5. Pastor-Pareja J. C., and Xu T. (2011) Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and Perlecan. Dev. Cell 21, 245–256 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rotstein B., and Paululat A. (2016) On the morphology of the Drosophila heart. J. Cardiovasc. Dev. Dis. 3, E15 10.3390/jcdd3020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Volk T., Wang S., Rotstein B., and Paululat A. (2014) Matricellular proteins in development: perspectives from the Drosophila heart. Matrix Biol. 37, 162–166 10.1016/j.matbio.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 8. Das D., Aradhya R., Ashoka D., and Inamdar M. (2008) Post-embryonic pericardial cells of Drosophila are required for overcoming toxic stress but not for cardiac function or adult development. Cell Tissue Res. 331, 565–570 10.1007/s00441-007-0518-z [DOI] [PubMed] [Google Scholar]
- 9. Ivy J. R., Drechsler M., Catterson J. H., Bodmer R., Ocorr K., Paululat A., and Hartley P. S. (2015) Klf15 is critical for the development and differentiation of Drosophila nephrocytes. PLoS One 10, e0134620 10.1371/journal.pone.0134620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tutor A. S., Prieto-Sánchez S., and Ruiz-Gómez M. (2014) Src64B phosphorylates Dumbfounded and regulates slit diaphragm dynamics: Drosophila as a model to study nephropathies. Development 141, 367–376 10.1242/dev.099408 [DOI] [PubMed] [Google Scholar]
- 11. Tögel M., Pass G., and Paululat A. (2008) The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev. Biol. 318, 29–37 10.1016/j.ydbio.2008.02.043 [DOI] [PubMed] [Google Scholar]
- 12. Tögel M., Pass G., and Paululat A. (2013) In vivo imaging of Drosophila wing heart development during pupal stages. Int. J. Dev. Biol. 57, 13–24 10.1387/ijdb.120111ap [DOI] [PubMed] [Google Scholar]
- 13. Drechsler M., Schmidt A. C., Meyer H., and Paululat A. (2013) The conserved ADAMTS-like protein Lonely heart mediates matrix formation and cardiac tissue integrity. PLoS Genet. 9, e1003616 10.1371/journal.pgen.1003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilmes A. C., Klinke N., Rotstein B., Meyer H., and Paululat A. (2018) Biosynthesis and assembly of the Collagen IV-like protein Pericardin in Drosophila melanogaster. Biol. Open 7, bio030361 10.1242/bio.030361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuno K., and Matsushima K. (1998) ADAMTS-1 protein anchors at the extracellular matrix through the thrombospondin type I motifs and its spacing region. J. Biol. Chem. 273, 13912–13917 10.1074/jbc.273.22.13912 [DOI] [PubMed] [Google Scholar]
- 16. Apte S. S. (2009) A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 284, 31493–31497 10.1074/jbc.R109.052340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tortorella M. D., Malfait F., Barve R. A., Shieh H. S., and Malfait A. M. (2009) A review of the ADAMTS family, pharmaceutical targets of the future. Curr. Pharm. Des. 15, 2359–2374 10.2174/138161209788682433 [DOI] [PubMed] [Google Scholar]
- 18. Adams J. C., and Lawler J. (2011) The thrombospondins. Cold Spring Harb. Perspect. Biol. 3, a009712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan K., Duquette M., Liu J.-H., Dong Y., Zhang R., Joachimiak A., Lawler J., and Wang J.-H. (2002) Crystal structure of the TSP-1 type 1 repeats: A novel layered fold and its biological implication. J. Cell Biol. 159, 373–382 10.1083/jcb.200206062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albrecht S., Altenhein B., and Paululat A. (2011) The transmembrane receptor Uncoordinated5 (Unc5) is essential for heart lumen formation in Drosophila melanogaster. Dev. Biol. 350, 89–100 10.1016/j.ydbio.2010.11.016 [DOI] [PubMed] [Google Scholar]
- 21. Sessions A. O., Kaushik G., Parker S., Raedschelders K., Bodmer R., Van Eyk J. E., and Engler A. J. (2017) Extracellular matrix downregulation in the Drosophila heart preserves contractile function and improves lifespan. Matrix Biol. 62, 15–27 10.1016/j.matbio.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarov M., Barz C., Jambor H., Hein M. Y., Schmied C., Suchold D., Stender B., Janosch S., KJ V. V., Krishnan R. T., Krishnamoorthy A., Ferreira I. R., Ejsmont R. K., Finkl K., Hasse S., et al. (2016) A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gotwals P. J., Paine-Saunders S. E., Stark K. A., and Hynes R. O. (1994) Drosophila integrins and their ligands. Curr. Opin. Cell Biol. 6, 734–739 10.1016/0955-0674(94)90101-5 [DOI] [PubMed] [Google Scholar]
- 24. Wang L. W., Dlugosz M., Somerville R. P., Raed M., Haltiwanger R. S., and Apte S. S. (2007) O-Fucosylation of thrombospondin type 1 repeats in ADAMTS-like-1/punctin-1 regulates secretion: implications for the ADAMTS superfamily. J. Biol. Chem. 282, 17024–17031 10.1074/jbc.M701065200 [DOI] [PubMed] [Google Scholar]
- 25. Bradshaw A. D., Baicu C. F., Rentz T. J., Van Laer A. O., Bonnema D. D., and Zile M. R. (2010) Age-dependent alterations in fibrillar collagen content and myocardial diastolic function: role of SPARC in post-synthetic procollagen processing. Am. J. Physiol. Heart Circ. Physiol. 298, H614–H622 10.1152/ajpheart.00474.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cammarato A., Dambacher C. M., Knowles A. F., Kronert W. A., Bodmer R., Ocorr K., and Bernstein S. I. (2008) Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol. Biol. Cell 19, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishimura M., Kumsta C., Kaushik G., Diop S. B., Ding Y., Bisharat-Kernizan J., Catan H., Cammarato A., Ross R. S., Engler A. J., Bodmer R., Hansen M., and Ocorr K. (2014) A dual role for integrin-linked kinase and β1-integrin in modulating cardiac aging. Aging Cell 13, 431–440 10.1111/acel.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaushik G., Spenlehauer A., Sessions A. O., Trujillo A. S., Fuhrmann A., Fu Z., Venkatraman V., Pohl D., Tuler J., Wang M., Lakatta E. G., Ocorr K., Bodmer R., Bernstein S. I., Van Eyk J. E., et al. (2015) Vinculin network-mediated cytoskeletal remodeling regulates contractile function in the aging heart. Sci. Transl. Med. 7, 292ra99 10.1126/scitranslmed.aaa5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Asha H., Nagy I., Kovacs G., Stetson D., Ando I., and Dearolf C. R. (2003) Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kölsch V., and Paululat A. (2002) The highly conserved cardiogenic bHLH factor Hand is specifically expressed in circular visceral muscle progenitor cells and in all cell types of the dorsal vessel during Drosophila embryogenesis. Dev. Genes Evol. 212, 473–485 10.1007/s00427-002-0268-6 [DOI] [PubMed] [Google Scholar]
- 31. Paululat A., and Heinisch J. J. (2012) New yeast/E. coli/Drosophila triple shuttle vectors for efficient generation of Drosophila P element transformation constructs. Gene 511, 300–305 10.1016/j.gene.2012.09.058 [DOI] [PubMed] [Google Scholar]
- 32. Hallier B., Schiemann R., Cordes E., Vitos-Faleato J., Walter S., Heinisch J. J., Malmendal A., Paululat A., and Meyer H. (2016) Drosophila neprilysins control insulin signaling and food intake via cleavage of regulatory peptides. eLife 5, e19430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meyer H., Panz M., Zmojdzian M., Jagla K., and Paululat A. (2009) Neprilysin 4, a novel endopeptidase from Drosophila melanogaster, displays distinct substrate specificities and exceptional solubility states. J. Exp. Biol. 212, 3673–3683 10.1242/jeb.034272 [DOI] [PubMed] [Google Scholar]
- 34. Lehmacher C., Abeln B., and Paululat A. (2012) The ultrastructure of Drosophila heart cells. Arthropod. Struct. Dev. 41, 459–474 10.1016/j.asd.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 35. Lammers K., Abeln B., Hüsken M., Lehmacher C., Psathaki O. E., Alcorta E., Meyer H., and Paululat A. (2017) Formation and function of intracardiac valve cells in the Drosophila heart. J. Exp. Biol. 220, 1852–1863 10.1242/jeb.156265 [DOI] [PubMed] [Google Scholar]
- 36. Linford N. J., Bilgir C., Ro J., and Pletcher S. D. (2013) Measurement of lifespan in Drosophila melanogaster. J. Vis. Exp. 10.3791/50068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nichols C. D., Becnel J., and Pandey U. B. (2012) Methods to assay Drosophila behavior. J. Vis. Exp. 10.3791/3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogler G., and Ocorr K. (2009) Visualizing the beating heart in Drosophila. J. Vis. Exp. 10.3791/1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rasband W. S. (2016) ImageJ, National Institutes of Health, Bethesda, MD [Google Scholar]
- 40. Fink M., Callol-Massot C., Chu A., Ruiz-Lozano P., Izpisua Belmonte J. C., Giles W., Bodmer R., and Ocorr K. (2009) A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. BioTechniques 46, 101–113 10.2144/000113078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ocorr K., Vogler G., and Bodmer R. (2014) Methods to assess Drosophila heart development, function and aging. Methods 68, 265–272 10.1016/j.ymeth.2014.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krieger E., and Vriend G. (2014) View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 30, 2981–2982 10.1093/bioinformatics/btu426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwede T., Diemand A., Guex N., and Peitsch M. C. (2000) Protein structure computing in the genomic era. Res. Microbiol. 151, 107–112 10.1016/S0923-2508(00)00121-2 [DOI] [PubMed] [Google Scholar]
- 44. Guo N. H., Krutzsch H. C., Nègre E., Zabrenetzky V. S., and Roberts D. D. (1992) Heparin-binding peptides from the type I repeats of thrombospondin: structural requirements for heparin binding and promotion of melanoma cell adhesion and chemotaxis. J. Biol. Chem. 267, 19349–19355 [PubMed] [Google Scholar]
- 45. Tuszynski G. P., Rothman V. L., Deutch A. H., Hamilton B. K., and Eyal J. (1992) Biological activities of peptides and peptide analogues derived from common sequences present in thrombospondin, properdin, and malarial proteins. J. Cell Biol. 116, 209–217 10.1083/jcb.116.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.