Abstract
Gluconeogenesis in the liver converts lipids and several other noncarbohydrate precursors into glucose, ensuring that blood sugar levels are maintained at healthy levels, especially during fasting. Effective regulation of gluconeogenesis is therefore critical for maintaining systemic metabolic homeostasis. Zhang and colleagues have discovered that the ubiquitous transcriptional regulator nuclear factor Y (NF-Y) confers cAMP responsiveness to key gluconeogenic genes and up-regulates hepatic glucose production. The study expands our understanding of transcriptional regulation of hepatic gluconeogenesis and also presents critical insights into the function of NF-Y in the liver.
Introduction
Besides its many other functions in the body, the liver is a major storage organ for glucose in its immobile form, glycogen. Whether the liver stores or releases glucose depends on the availability of this key source of energy, which is regulated by the interplay between the metabolic hormones insulin and glucagon. During short fasting periods, glucagon stimulates glycogen breakdown in the liver to release glucose. Prolonged fasting depletes the hepatic glycogen reserve, running the risk that general metabolism will begin to sputter. To counteract that potentially harmful scenario, the liver can switch to de novo glucose synthesis (gluconeogenesis) from pyruvate, glycerol, lactate, and other glucogenic amino acids. Gluconeogenesis thus maintains the body–glucose equilibrium. Because gluconeogenesis is important for maintaining healthy blood glucose levels (1, 2), it is tightly regulated at multiple levels, and a better understanding of this regulation could help inform the development of interventions for metabolic diseases such as type 2 diabetes.
Transcriptional regulation of gluconeogenic genes represents a major strategy for responding to metabolic conditions and needs. This regulatory network integrates many exogenous signaling events and relays them to regulate the activation or repression of specific sets of metabolically relevant genes (3). Understanding the interplay of the various transcription factors involved in gluconeogenesis, therefore, is crucial to deciphering the vast metabolic network that regulates glucose homeostasis. For instance, the key rate-limiting gluconeogenic genes G6pc (encoding glucose-6-phosphatase or G6Pase) and Pck1 (encoding phosphoenolpyruvate carboxykinase or PEPCK) are regulated by a host of hormone-sensing transcription factors and transcriptional co-activators. These transcriptional regulators include the cAMP response element–binding protein (CREB)2 (4) and CREB-binding protein (CBP)/p300 (5). Moreover, epigenetic regulators of glucose homeostasis also control gluconeogenic gene transcription (6). However, the full picture of this regulatory network is still incomplete.
Previous observations have indicated that cAMP signaling extensively controls gluconeogenesis via CREB, but other, yet unknown effectors besides CREB appear to be required for regulating gluconeogenesis. Nuclear factor Y (NF-Y) is a ubiquitous, evolutionarily conserved transcription factor that is also a cAMP target in other cells that respond to sugar, such as those in the pancreas. So, NF-Y appeared to also be a good candidate for a possible transcriptional regulator of gluconeogenesis in the liver. Zhang et al. (7) now report that NF-Y is a cAMP-induced, CREB-interacting positive regulator of hepatic gluconeogenesis. NF-Y comprises three different subunits, NF-YA, NF-YB, and NF-YC, that are required for binding to CCAAT boxes, canonical regulatory elements present in NF-Y–regulated genes. Traditionally, NF-Y has been primarily associated with regulation of cellular development and differentiation, but a previous report by the same group has provided strong hints that NF-Y is also regulated by endoplasmic reticulum stress in adipocytes (8). That NF-Y has now been shown to also stimulate the expression of gluconeogenic genes suggests that it may be a key metabolic stress–responsive transcription regulator.
Zhang et al. (7) used a multipronged approach to unravel NF-Y's role in gluconeogenesis. First, the authors found that expression of all three NF-Y subunits is up-regulated upon cAMP and glucagon treatments in mouse hepatoma cells and also during fasting conditions in vivo. Using ChIP assays, they confirmed that NF-Y activates the transcription of Pck1 or G6pc by binding to the canonical CCAAT regulatory elements in the promoters of these two genes. Moreover, lentivirus-mediated overexpression of all three NF-Y subunits induced G6pc and Pck1 expression in the hepatoma cells. The binding site of NF-Y at the promoter region of the gluconeogenic genes was further determined by luciferase reporter assays. The authors then used CRISPR-Cas9 to knock out the NF-Y subunits in hepatic cells or the Cre/lox system to obliterate the NF-YA subunit in the mouse liver (Nf-ya LKO), which attenuated expression of G6pc and Pck1 in both cases. More importantly, Nf-ya LKO mice had significantly reduced body weight, low serum glucose and glucagon levels, and low glucose production ability upon pyruvate challenge under starvation conditions. These are telltale signs of compromised metabolic function in these animals. Under fasting conditions, glucagon induces CREB phosphorylation to activate gluconeogenic genes. Intriguingly, Zhang et al. (7) observed that the Nf-ya LKO mice have reduced phosphorylation levels of CREB even upon glucagon induction, indicating that NF-Y is important for protein kinase A (PKA)–mediated CREB activation in hepatic tissue. The authors also found that NF-YA physically interacts with CREB, suggesting for the first time that NF-YA could be a co-activator of CREB and an integral part of the CREB–cAMP axis in mediating gluconeogenesis.
The study by Zhang et al. (7) provides compelling evidence for a previously unrecognized role of the transcriptional regulator NF-Y in controlling hepatic gluconeogenesis, and that it is an integral part of the CREB/cAMP signaling pathway (Fig. 1). These observations point to the fact that besides its known roles, mainly in cellular developmental processes, NF-Y also regulates a pivotal metabolic pathway in the liver. The data presented by Zhang et al. (7) have significantly opened up several new avenues for future research on NF-Y. For example, it will be interesting to see how NF-Y or its post-translationally modified form could be influencing the epigenetic landscape of chromatin and consequent metabolic gene expression programs. Further, since NF-Y is known to associate with a myriad of transcription factors, future studies into the relative contribution of these associations influencing transcription of metabolic genes will expand our knowledge in understanding the intricacies of cellular metabolic homeostasis. The old paradigm of considering metabolic processes as a set of energy-supplying housekeeping reactions is beginning to change. The vast amount of research data in the last decade has pointed to the association of metabolic flux regulation to developmental disorders like cancer. The newfound role of NF-Y, a well-established player in the cellular developmental program, in metabolic regulation is an example of biological cross-talk among functionally different processes in the cell (9). Additionally, the newly discovered role of NF-Y in the regulation of hepatic glucose production also has important therapeutic implications, as NF-Y may represent a useful drug target for future approaches to manage type 2 diabetes.
Figure 1.
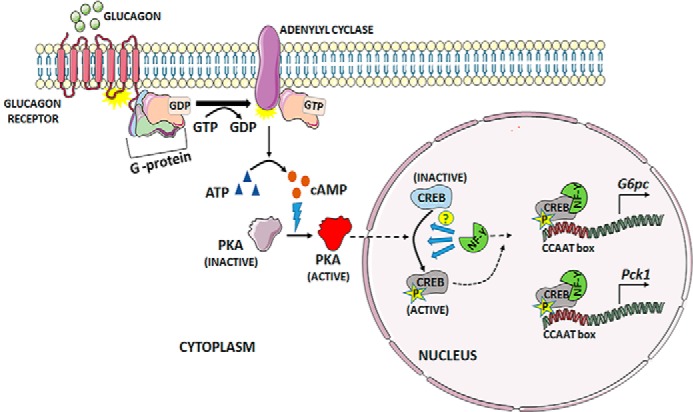
Role of NF-Y in glucagon-mediated activation of the gluconeogenic genes G6pc and Pck1 through the cAMP–CREB axis. Binding of glucagon to its receptor in hepatocytes triggers the guanylyl nucleotide exchange, leading to dissociation of the activated, GTP-bound Gα subunit from the heterotrimeric G-protein complex. Binding of the active Gα subunit to adenylyl cyclase triggers the conversion of ATP to the cyclic AMP. This, in turn, activates protein kinase A (PKA), which then translocates to the nucleus and phosphorylates CREB to make it transcriptionally active. Zhang et al. (7) show that NF-Y is key in mediating this phosphorylation step, although the exact mechanism is yet to be explored. The authors also show that activated CREB binds to NF-Y, ultimately up-regulating expression of G6pc and Pck1 by binding to the CCAAT regulatory region (7).
Acknowledgment
We thank Payel Mondal for her help with the preparation of the figure used in this highlight.
This work was supported by Biomolecular Assembly, Recognition, and Dynamics (BARD) Research Grant 12-R&D-SIN-5.04-0103 from the Department of Atomic Energy (DAE), Government of India. The authors declare that they have no conflicts of interest with the contents of this article.
- CREB
- cAMP-responsive element–binding protein
- NF-Y
- nuclear factor Y.
References
- 1. Magnusson I., Rothman D. L., Katz L. D., Shulman R. G., and Shulman G. I. (1992) Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J. Clin. Invest. 90, 1323–1327 10.1172/JCI115997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tappy L. (1995) Regulation of hepatic glucose production in healthy subjects and patients with non-insulin-dependent diabetes mellitus. Diabetes Metab. 21, 233–240 [PubMed] [Google Scholar]
- 3. Desvergne B., Michalik L., and Wahli W. (2006) Transcriptional regulation of metabolism. Physiol. Rev. 86, 465–514 10.1152/physrev.00025.2005 [DOI] [PubMed] [Google Scholar]
- 4. Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., and Montminy M. (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 10.1038/35093131 [DOI] [PubMed] [Google Scholar]
- 5. Bedford D. C., Kasper L. H., Wang R., Chang Y., Green D. R., and Brindle P. K. (2011) Disrupting the CH1 domain structure in the acetyltransferases CBP and p300 results in lean mice with increased metabolic control. Cell Metab. 14, 219–230 10.1016/j.cmet.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sen S., Sanyal S., Srivastava D. K., Dasgupta D., Roy S., and Das C. (2017) Transcription factor 19 interacts with histone 3 lysine 4 trimethylation and controls gluconeogenesis via the nucleosome-remodeling-deacetylase complex. J. Biol. Chem. 292, 20362–20378 10.1074/jbc.M117.786863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Guan Q., Liu Y., Zhang Y., Chen Y., Chen J., Liu Y., and Su Z. (2018) Regulation of hepatic gluconeogenesis by nuclear factor Y transcription factor in mice. J. Biol. Chem. 293, 7894–7904 10.1074/jbc.RA117.000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y., Zhang Y., Zhang Y., Zhang J., Liu Y., Feng P., and Su Z. (2017) Obesity-induced endoplasmic reticulum stress suppresses nuclear factor-Y expression. Mol. Cell. Biochem. 426, 47–54 10.1007/s11010-016-2879-7 [DOI] [PubMed] [Google Scholar]
- 9. Metallo C. M., and Vander Heiden M. G. (2013) Understanding metabolic regulation and its influence on cell physiology. Mol. Cell. 49, 388–398 10.1016/j.molcel.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]


