Abstract
Cell growth and division require nutrients, and proliferating cells use a variety of sources to acquire the amino acids, lipids, and nucleotides that support macromolecule synthesis. Lipids are more reduced than other nutrients, whereas nucleotides and amino acids are typically more oxidized. Cells must therefore generate reducing and oxidizing (redox) equivalents to convert consumed nutrients into biosynthetic precursors. To that end, redox cofactor metabolism plays a central role in meeting cellular redox requirements. In this Minireview, we highlight the biosynthetic pathways that involve redox reactions and discuss their integration with metabolism in proliferating mammalian cells.
Keywords: cell metabolism, metabolism, mitochondrial metabolism, oxidation-reduction (redox), cell proliferation
Introduction
Proliferation imposes a biosynthetic requirement to support cell growth and division. To divide, a cell must replicate each of its components. All cells, whether proliferating or not, require a source of energy and reducing power to maintain homeostasis and combat stress. Proliferating cells must also duplicate their macromolecular contents: DNA, RNA, proteins, and lipids; to meet this demand, the proliferating cells carry out diverse metabolic reactions to generate the precursors for these molecules (1). Mammalian cells thrive in diverse environments where nutrient concentrations vary, so which nutrients are used and how these contribute to macromolecule precursors is not always evident from the metabolic network alone. Current studies seek to understand how metabolism supports proliferation in diverse cell types and environments. Many reactions involve the reduction or oxidation of substrates, and an understanding of cellular metabolism requires a careful consideration of redox reactions involved in biosynthesis.
Mammalian cells can proliferate in a variety of contexts. Regulated proliferation is found in developing embryos, stem cells, and immune cells, whereas cancer is a disease of uncontrolled proliferation. In all cases, proliferating cells have essentially the same macromolecular requirements, but the pathways different cells use to meet these metabolic demands differ, and there is growing interest in exploiting these differences for therapy (2–4). Drugs that alter redox status, such as mitochondrial inhibitors, can have significant effects on cellular metabolism and impact diverse cancers. For example, patients taking the mitochondrial inhibitor metformin have a lower risk of death across a variety of cancer types, and metformin is being investigated as a potential cancer therapy (5). Chemotherapies targeting nucleotide synthesis and DNA replication have been a mainstay of cancer therapy for decades, illustrating that inhibition of metabolic processes has the potential to stop proliferation.
In this Minireview, we describe the metabolic requirements of proliferating cells and examples of how they are met, discussing in particular which redox reactions support macromolecule synthesis. This provides a framework to understand how cellular redox status supports proliferation in a variety of contexts, including cancer.
Redox reactions for biosynthesis
Biosynthetic pathways often involve reduction and oxidation (redox) reactions, and pyridine nucleotide cofactors (NAD(H) and NADP(H)) serve as carriers to provide and remove electrons for most of these reactions (Fig. 1) (1, 6). NADPH is used in reduction reactions, and cells maintain a high NADPH/NADP+ ratio to ensure that these reactions are favorable (7, 8). Conversely, NAD+ is used as an electron acceptor to oxidize biosynthetic intermediates, and the NADH/NAD+ ratio is can be very low in proliferating cells (9, 10). The exact values of these ratios differ between cellular compartments and across growth conditions and impact which enzyme-catalyzed reactions are favorable in different contexts. Importantly, numerous shuttles link cytosolic redox metabolism to other compartments, so manipulating redox metabolism in one organelle can broadly impact cellular metabolism. Often a substrate is reduced in one compartment and transported to another compartment where it is oxidized, allowing redox equivalents to be shuttled throughout the cell.
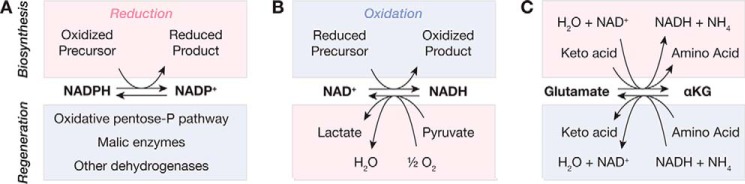
Figure 1.
Oxidation and reduction reactions and their relationship to biosynthesis. A, NADPH is used to reduce oxidized precursors, and NADPH can be regenerated from NADP+ by the oxidative pentose pathway, malic enzymes, and other reactions. B, NAD+ is used to oxidize reduced precursors in glycolysis, the TCA cycle, and biosynthetic pathways. NAD+ is regenerated from NADH by lactate dehydrogenase and by the electron transport chain, which reduce pyruvate and oxygen, respectively. C, transamination reactions involve a change in redox state for the substrates and products. Oxidation of glutamate to α-ketoglutarate reduces NAD+ to NADH or transaminates an α-keto acid to an amino acid. For all panels, the half-reactions in red are reduction reactions, and those in blue are oxidations. Abbreviations used are: αKG, α-ketoglutarate; pentose-P, pentose phosphate.
NADPH provides electrons for lipid, deoxyribonucleotide, and proline synthesis. Outside of biosynthesis, NADPH is also required to maintain a reduced intracellular environment and combat oxidative damage; consequently, proliferating and quiescent cells are reported to maintain similar NADPH/NADP+ ratios (11, 12). Several reactions can generate NADPH, and both the oxidative pentose phosphate pathway and mitochondrial reactions are important contributors to NADPH synthesis (Fig. 1A). In many cell types, the oxidative pentose phosphate pathway, which produces two molecules of NADPH per molecule of glucose 6-phosphate (Glc-6-P), is the largest contributor to the cytosolic NADPH pool, but cytosolic malic enzyme 1 may play a role in some contexts (13, 14). The first enzyme of the oxidative pentose phosphate pathway, Glc-6-P dehydrogenase, generates NADPH but is also strongly inhibited by NADPH, ensuring that pathway flux responds to changes in NADPH/NADP+ ratio (15). In the mitochondria, NADPH can be generated by malic enzyme 3, isocitrate dehydrogenase 2, and one-carbon cycle enzymes, and the relative contribution of each enzyme varies among cellular contexts (13, 16–18).
Cells use NAD+ to remove electrons from the nutrients they consume, such as sugars and lipids, which are reduced relative to many metabolic intermediates (Fig. 1B). The reactions of glycolysis and the tricarboxylic acid (TCA)2 cycle convert NAD+ to NADH, and this favorable oxidation of nutrients, ultimately yielding carbon dioxide, allows cells to derive energy from the nutrients they consume. Many intermediates of these NAD+-dependent pathways are also important precursors for biosynthesis, and NAD+ is directly used to oxidize precursors of some nucleotides and amino acids (1, 19, 20). Interestingly, the NADH/NAD+ ratio is lower in quiescent cells perhaps because of decreased consumption of NAD+ by biosynthetic pathways (11, 21). Regeneration of NAD+ is necessary to support the activities of a variety of metabolic pathways. In the cytosol, this is accomplished by lactate dehydrogenase (LDH), a highly expressed enzyme that converts the glycolytic product pyruvate into lactate (22). Proliferating cells generate large amounts of lactate, allowing high glycolytic flux to support the generation of ATP and biosynthetic precursors (23–26). The mitochondrial electron transport chain (ETC) also serves to regenerate NAD+ by converting oxygen to water, and when coupled to ATP synthesis, this process of oxidative phosphorylation can be a critical source of ATP for proliferating cells (27, 28). In producing pyruvate from glucose, glycolysis generates NADH, which can be converted back to NAD+ by LDH; therefore, conversion of glucose to lactate consumes and regenerates equal amounts of NAD+. As a result the conversion of glucose to lactate is unable to recycle the additional NADH generated by oxidative biosynthesis, and this must be accomplished by alternative reactions, many of which involve mitochondria. Consequently, reduced mitochondrial activity, resulting from either hypoxia or pharmacological inhibition of the ETC, can dramatically reduce the availability of NAD+ for biosynthesis. Cells lacking mitochondrial DNA, for example, are dependent on an exogenous source of pyruvate or another α-keto acid, which is reduced by LDH to regenerate the NAD+ consumed by biosynthesis (20, 29).
Although they do not use pyridine nucleotides, transamination reactions also involve carbon reduction-oxidation (Fig. 1C). Transamination reactions transfer an amino group between two molecules and are used to synthesize amino acids. In these reactions, glutamate is converted into α-ketoglutarate, and an α-keto acid is converted into an amino acid. Although no explicit redox reaction occurs, in each molecule a ketone and a primary amine are interconverted, changing the oxidation state of the adjacent carbon atom. Illustrating this fact, another reaction that generates α-ketoglutarate from glutamate is the glutamate dehydrogenase reaction. Glutamate dehydrogenase uses NAD+ to oxidize glutamate to produce α-ketoglutarate, showing that a change in oxidation state has occurred. In transamination reactions, an α-keto acid substitutes for NAD+ and is reduced to an amino acid. Transamination reactions are consequently linked to cellular redox status. For example, when alanine aminotransferase catalyzes the conversions of glutamate to α-ketoglutarate and pyruvate to alanine, it bypasses the immediate requirement for NAD+ to generate α-ketoglutarate, but ultimately this deprives the cell of pyruvate that could have otherwise been used by LDH to generate NAD+.
The reactions that allow cells to generate NADPH and NAD+ are critical for biosynthesis even though they do not directly provide new cell mass. Proliferating cells must abundantly regenerate these cofactors by catabolizing consumed nutrients, and on the whole, cellular metabolism must be balanced. Just as the amount of carbon consumed must equal the amounts excreted and incorporated into cell mass, so too must the reactions that reduce pyridine nucleotides equal those that oxidize them.
Nutrient environment, cell type, and growth signals can each influence how a cell generates the macromolecule precursors it requires to proliferate. Both in vivo and in vitro, mammalian cells are exposed to complex nutrient environments (30), and multiple pathways exist to generate amino acids, nucleotides, and lipids. The pathways that generate macromolecule precursors differ in both their use of nutrients and of redox cofactors, linking the use of a particular biosynthetic pathway to the availability of NAD+ and NADPH. The pathways proliferating cells use to generate amino acids, lipids, and nucleotides, which are the precursors of the vast majority of new cell mass, are described below.
Amino acids
Protein represents the largest fraction of cell mass, and acquisition of amino acids is a major biosynthetic requirement of cells. Cells can acquire amino acids through a variety of means, and cell type, environment, and genetics can all influence which are used. Mammalian cells can synthesize nonessential amino acids de novo but must obtain essential amino acids from the extracellular environment. Amino acid synthesis has a biosynthetic cost, requiring glycolytic or TCA cycle intermediates as well as redox cofactors and ATP. Thus, acquiring amino acids from the environment allows cells to spare these materials for other biosynthetic pathways. Notably, synthesis of many nonessential amino acids requires oxidation involving NAD+ and some require reduction by NADPH (Fig. 2). Human plasma contains each of the 20 amino acids at concentrations ranging from 10 to 600 μm. However, amino acid concentrations may be reduced in other fluids, limiting their availability to cells. In this context, acquiring amino acids from protein in the extracellular environment represents another source of amino acids (31–33).
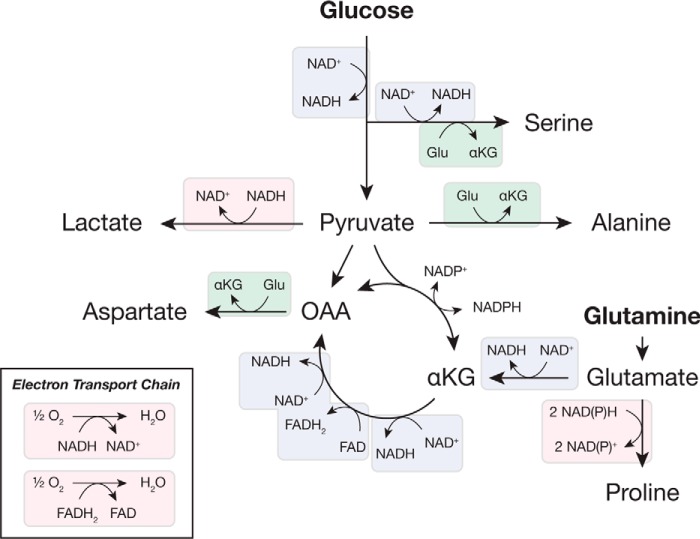
Figure 2.
Redox reactions used to synthesize nonessential amino acids from glucose and glutamine. Simplified pathways illustrating the interconversion of glycolytic and TCA cycle intermediates are shown, with redox reactions highlighted. Many of these reactions are oxidations and consume NAD+ (or FAD), although others are reductions and require NAD(P)H. As shown in Fig. 1, transamination reactions are also oxidation/reduction reactions. Major pathways involved in regenerating NAD+ and FAD are also shown for reference. Substrate oxidation is indicated in blue; substrate reduction is in pink; and transamination is in green. Abbreviations used are: αKG, α-ketoglutarate; OAA, oxaloacetate.
In some contexts, cells synthesize nonessential amino acids, and glucose is the source for several of these amino acids (Fig. 2). Most mammalian cells with high glycolytic flux excrete alanine as an alternative to lactate (34, 35). Importantly, the central carbon atom in both lactate and alanine is reduced relative to the corresponding atom in pyruvate, so synthesis of lactate or alanine from pyruvate regenerates NAD+. Most media formulations lack alanine, but alanine is not always synthesized in excess. Lymphocyte proliferation can be enhanced when alanine is included in media (36), and pancreatic cancer cells can consume alanine produced by pancreatic stellate cells in vitro, indicating the potential for a metabolic interaction in vivo (37). In that study, alanine utilization enhanced oxygen consumption, which might be necessary to recycle NADH produced indirectly by conversion of alanine to pyruvate. Another pathway supported by glucose is the serine biosynthesis pathway. Serine is generated from glycolytic intermediates through several reactions, the first of which is catalyzed by phosphoglycerate dehydrogenase, an enzyme that consumes NAD+. Glycolysis itself consumes NAD+, so synthesis of one molecule of serine from glucose requires two molecules of NAD+. As a result, mitochondrial inhibition, which prevents NAD+ regeneration, has been shown to inhibit serine synthesis from glucose, increasing the dependence on exogenous serine (38, 39). Serine can provide carbon for nucleotide biosynthesis and is further metabolized to produce cysteine and glycine, implying an NAD+ cost to synthesizing these amino acids from glucose.
Glutamine can supply carbon and nitrogen for synthesis of glutamate, aspartate, asparagine, and proline (39). Glutamine catabolism is sufficient to sustain biosynthesis of these amino acids, and they are often excluded from culture media (40, 41). Glutaminase activity produces glutamate, which can serve as a nitrogen donor for transamination reactions in the biosynthesis of other amino acids, such as serine and alanine (Fig. 2). Proline synthesis from glutamine is up-regulated by oncogenic signaling to promote proliferation of lymphoma cells, and this amino acid is synthesized even when provided exogenously (42). Of note, proline synthesis is unique in that it directly requires two molecules of NAD(P)H for reduction, linking its synthesis to NAD+ regeneration or NADPH consumption (43). Proline synthesis has been argued to consume NADPH to drive pentose phosphate pathway activity (13, 42), but loss of this activity is expected to impair proline synthesis only in cases where other NADPH-producing pathways cannot compensate (44).
Unlike most amino acids, which can be used by cells when supplied exogenously, aspartate is synthesized de novo: most cells (except in the prostate and nervous system) lack high-affinity transporters for aspartate, and typical plasma aspartate concentrations are insufficient to enable its consumption (19, 45). Aspartate synthesis is dependent on a supply of NAD+, and this amino acid becomes limiting when respiration is inhibited (19, 20). Conversely, enhanced aspartate synthesis has been observed in cells with an increased ability to regenerate NAD+ (46). Aspartate is synthesized from oxaloacetate in a reaction that consumes glutamate, and oxaloacetate itself can be synthesized from glutamine through several oxidation reactions. Oxaloacetate may also be synthesized by the reductive carboxylation of α-ketoglutarate, which requires less NAD+, and this pathway is favored during mitochondrial inhibition (20). In many cases in vivo, glutamine is not used, and glucose provides carbon for aspartate synthesis via pyruvate carboxylase (47–49).
Proliferating cells have a considerable requirement for protein synthesis and must meet the amino acid demand to grow and divide. Cell type and extracellular environment both influence how amino acids are acquired (33, 50), potentially in part by changing utilization and availability of redox cofactors, and this can affect protein synthesis as well as downstream biosynthetic reactions.
Lipids
Lipids are a diverse class of molecules, and acquiring lipids is necessary for proliferating cells (51). Lipids compose 50% of membrane mass, and they are also critical components of signaling pathways. Synthesizing membranes of the correct composition is critical for cell function (52, 53). In mammalian cells, cholesterol, phospholipids, and sphingolipids are the major lipid constituents of cell membranes. Fatty acids are core components of these latter two classes and are also present in cells as triglycerides. Each lipid species can be acquired exogenously or synthesized de novo, and nutrient supply and growth conditions influence how cells meet this demand.
A large fraction of cellular lipids are derived from the extracellular environment (39, 54). De novo lipid synthesis depends on a source of NADPH and potentially also on NAD+, but utilization of exogenous lipids allows cells to circumvent these costs (Fig. 3). Early work with proliferating lymphocytes demonstrated that exogenously supplied free fatty acids are incorporated into the lipid pool in these cells (55). Breast and ovarian cancer cells stimulate lipolysis in co-cultured adipocytes and can consume the released fatty acids (56, 57). Free fatty acids are not the only source of exogenous fatty acids. Hypoxic cells and cells with Ras activation utilize lysophospholipids present in serum as a source of unsaturated fatty acids (58). Lymphocytes and fibroblasts express lipoprotein lipase, allowing them to consume and hydrolyze triglycerides as a source of fatty acids for biosynthesis (59, 60), and some cancers overexpress other lipases to utilize exogenously esterified lipids (61).
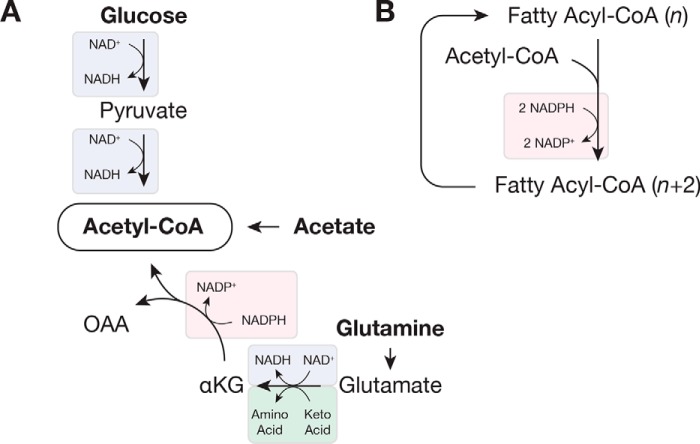
Figure 3.
Redox reactions involved in fatty acid synthesis. A, acetyl-CoA can be synthesized from glucose, glutamine, or free acetate, and each pathway relies on different redox cofactors. B, fatty acid synthesis and elongation use two molecules of NADPH per molecule of acetyl-CoA. In this figure, two carbon atoms from acetyl-CoA are added to a fatty acyl-CoA containing n carbon atoms. Substrate oxidation is indicated in blue; substrate reduction is in pink; and transamination is in green. Abbreviations used are: αKG, α-ketoglutarate; OAA, oxaloacetate.
Although a large portion of cellular lipids originates from exogenous sources (39, 56), glucose is a major carbon source for de novo fatty acid synthesis (58). Active lipogenesis was first observed in tumor tissue slices by Medes et al. (62), who noted that hepatomas incorporated more glucose into lipids than adjacent liver tissue. Pyruvate generated by glycolysis is decarboxylated by pyruvate dehydrogenase (PDH) in the mitochondria to produce acetyl-CoA, the basic building block of fatty acids. Synthesis of acetyl-CoA from glucose therefore requires two molecules of NAD+, one for glycolysis and one for PDH (Fig. 3A). In the cytosol, acetyl-CoA is used for de novo synthesis of palmitate (by fatty-acid synthase) and cholesterol. Synthesis of either lipid requires a large amount of reducing power, and two molecules of NADPH are needed for each acetyl-CoA that is incorporated into a saturated fatty acid (Fig. 3B). In fact, lipid synthesis is estimated to be a major user of NADPH (13). Fatty acids are diversified through elongation and desaturation reactions, and both require additional redox reactions. Sterol synthesis also requires redox reactions. In cells, fatty acids are esterified to glycerol 3-phosphate to produce phospholipids and triglycerides. Glycerol 3-phosphate is generated by the phosphorylation of exogenous glycerol or by the reduction of the glycolytic intermediate dihydroxyacetone phosphate, and proliferating cells obtain glycerol from both sources (63). Phospholipids also contain polar headgroups derived from serine, and their synthesis consequently also depends upon redox reactions. Many cancers up-regulate enzymes required for de novo lipid synthesis, and there is current interest in targeting these enzymes (64).
Glucose is not always the primary source of lipogenic acetyl-CoA. When cells are subject to mitochondrial inhibition, either as a result of hypoxia or genetic alterations, glutamine can serve as a source of acetyl-CoA for de novo lipid synthesis (Fig. 3A) (65–67). Lipid production from glutamine reduces the requirement for NAD+, whose regeneration is limited when mitochondrial activity is reduced. Reductive carboxylation of glutamine-derived α-ketoglutarate by isocitrate dehydrogenase (IDH) uses NADPH to ultimately produce citrate, which can be cleaved by ATP-citrate lysate to generate acetyl-CoA. In addition to decreased availability of NAD+, inhibitory phosphorylation reduces PDH flux in hypoxic cells, decreasing the availability of glucose-derived acetyl-CoA (68, 69). Mammalian cells express three IDH isoforms, and reductive carboxylation can be carried out by the NADPH-utilizing IDH1 and IDH2, which localize to the cytosol and mitochondria, respectively (65–67). The hypoxia-sensing pathway in mammals promotes this metabolic change, and the constitutive hypoxia-inducible factor activation in some cancers allows them to derive lipids from glutamine when oxygen is abundant (65). Cells proliferating with ample oxygen can derive acetyl-CoA from glutamine, but glutamine carbon is a minor contributor to lipid synthesis in this context (39, 65, 70).
Free acetate can serve as an additional lipogenic substrate for proliferating cells. Acetate can contribute to lipogenic acetyl-CoA when oxygen is abundant, but its contribution is increased in low oxygen (70–72). Acetyl-CoA synthetases (ACSS) are required to convert acetate to acetyl-CoA, and cytosolic ACSS2 is the primary enzyme that supplies acetate for biosynthesis (72). Incorporation of free acetate requires ATP but no redox cofactors (Fig. 3A), making it an attractive lipogenic substrate in hypoxia when NAD+ is limiting. Acetate concentration determines how much this substrate is used for lipogenesis (70), and acetate concentration varies in human plasma ranging from 50 to 200 μm (73). Free acetate in cells is predominantly generated by protein deacetylation, and a major role of ACSS is to salvage this acetate (74), raising the possibility that utilization of exogenous tracer acetate for biosynthesis is, in part, a readout of exchange between extracellular and intracellular acetate pools.
Lipids can be obtained from both exogenous sources and de novo synthesis, and each lipid synthesis pathway has different dependence upon redox cofactors. For some proliferating cells, lipid synthesis is sufficient to fully support proliferation, as these cells can multiply in lipid-free conditions. Conversely, lipids are abundant in the plasma, and proliferating cells are not thought to be lipid-deficient in fed animals. Therefore, which sources contribute to cellular lipid mass and how de novo synthesis supports proliferation remains an open question.
Nucleotides
Nucleic acids are another integral biosynthetic requirement of proliferating cells. DNA is necessarily synthesized during the cell division cycle, and RNA, the majority of which is ribosomal, synthesizes protein. Nucleotides are the precursors for nucleic acid synthesis but also serve as cofactors for many redox, energetic, and biosynthetic reactions. As with amino acids and lipids, nucleotides can be derived from de novo synthesis or salvaged from exogenous sources, and de novo nucleotide synthesis requires multiple oxidations by NAD+ and NADP+ (Fig. 4). Many extant cancer therapies target de novo nucleotide synthesis pathways, and their ability to impair the proliferation of both cancer and normal cells underscores the essentiality of de novo nucleotide synthesis for many cells (2, 75). All nucleotides are composed of a purine or pyrimidine base joined to ribose (or deoxyribose), which is derived from ribose 5-phosphate (Rib-5-P). Free ribose can be scavenged and phosphorylated by only a minority of cell types, and Rib-5-P is therefore synthesized from glucose by most cells (76, 77). Rib-5-P can be generated through either the oxidative or nonoxidative pentose phosphate pathways, and recent work has identified signaling pathways that can enhance flux through these pathways (78, 79). The oxidative pathway, which generates two molecules of NADPH per molecule of Rib-5-P produced, is proposed to be the primary source of Rib-5-P for biosynthesis in many proliferating cells (80, 81); however, the nonoxidative pathway may play a greater role in contexts where NADPH is abundant, as it is a potent inhibitor of the oxidative branch. Indeed, some cancers have been reported to preferentially obtain ribose from the nonoxidative pentose phosphate pathway (81–83), arguing the NADPH may not be limiting in those cells.
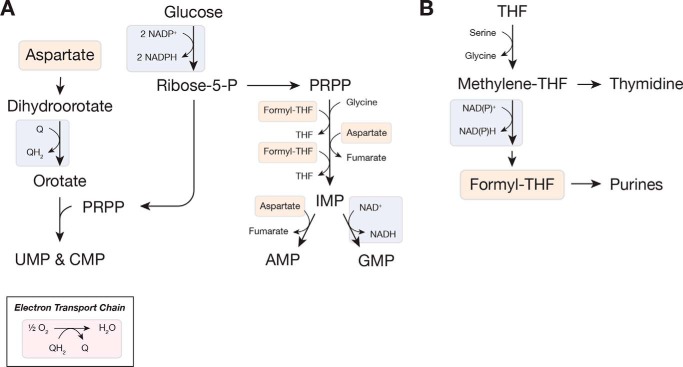
Figure 4.
Oxidation reactions are important for nucleotide synthesis. A, simplified schematic of de novo nucleotide biosynthesis highlighting the redox reactions involved. Ribose is produced via the pentose phosphate pathway, and synthesis of purine and pyrimidine nucleotides involve oxidation reactions. Both purine and pyrimidine synthesis use aspartate, which is generated from the oxidation of either glucose or glutamine (see Fig. 2). The use of oxygen to regenerate ubiquinone (Q) is also shown. B, formyl-THF, required for purine synthesis, is produced following the oxidation of one-carbon units derived from serine. Substrate oxidation is indicated in blue; substrate reduction is in pink; and products of oxidation reactions are in orange. Abbreviations used are: PRPP, phosphoribosyl pyrophosphate; Q, ubiquinone; QH2, ubiquinol; ribose-5-P, ribose-5-phosphate; THF, tetrahydrofolate.
Nucleotide base biosynthesis is closely linked to amino acid metabolism, requiring both amino acids and one-carbon units generated from amino acid catabolism (Fig. 4). Consequently, amino acid synthesis plays a direct role in nucleotide biosynthesis as well, dictating whether purine and pyrimidine bases derive from glucose or exogenous amino acids. One-carbon units, individual carbon atoms covalently linked to a folate cofactor, are required for both nucleotide biosynthesis pathways and are interconverted by redox reactions (Fig. 4B). Proliferating cells catabolize serine for one-carbon units to sustain nucleotide synthesis (84, 85). As with protein synthesis, this serine can be obtained by oxidizing glucose carbon or from an exogenous source (86). Recent work has highlighted the importance of mitochondrial function in one-carbon metabolism, and NAD(P)H can be derived from mitochondrial serine catabolism (13, 18). Mitochondria are also the major source of one-carbon units for nucleotide synthesis in the cytosol (16). When mitochondria are inhibited and the availability of mitochondrial NAD(P)+ is reduced, however, the equivalent reactions in the cytosol can supply one-carbon units (87).
In addition to serine and glycine, aspartate is essential to the synthesis of both purines and pyrimidines. As discussed above, aspartate must be synthesized de novo, and limitation for this amino acid impairs both nucleic acid and protein biosynthesis. Aspartate synthesis depends upon sustained mitochondrial activity, and cells become aspartate auxotrophs when mitochondria are inhibited (19, 20). Interestingly, aspartate can become limiting for nucleotides sooner than for protein synthesis, and cells unable to synthesize aspartate take longer to arrest growth when adenine and uracil are supplied exogenously (20). Beyond the substantial redox requirement for aspartate synthesis, nucleotide synthesis depends upon other redox reactions as well (Fig. 4A). Pyrimidine synthesis requires ETC activity to regenerate quinones required for orotate synthesis, and cells lacking mitochondrial DNA are pyrimidine auxotrophs (29). Guanine nucleotide synthesis requires NAD+, and reduction of nucleotides to deoxynucleotides requires NADPH. By regenerating NAD+ and quinones and by supporting aspartate synthesis, mitochondria play an integral role in nucleotide metabolism. Consistent with this relationship, both nucleotide depletion and mitochondrial inhibition block proliferation by inducing senescence (88, 89).
Some cells can bypass redox requirements by salvaging nucleosides or nucleobases if they are available in the extracellular environment. Tissues capable of salvage are resistant to nucleotide synthesis inhibitors (90). Nevertheless, the concentrations of nucleosides and their bases in human plasma are rarely greater than 10 μm (91), and circulating nucleosides are insufficient to rescue proliferation in many cases when cells in vivo are treated with nucleotide synthesis inhibitors. Consequently, de novo biosynthesis is an essential source of nucleotides for many proliferating cells, and inhibitors of this process might synergize with mitochondrial inhibitors that limit aspartate and NAD+ availability to further starve cells of nucleotides (6). Nucleotide synthesis inhibitors are currently used to treat cancer, demonstrating that there is a therapeutic window to target these pathways relative to normal proliferating cells. The preliminary evidence supporting the use of mitochondrial inhibitors to treat cancer and their potential to inhibit nucleotide synthesis suggests that altered redox metabolism could provide other opportunities to selectively target cancer.
Conclusion
Mammalian cells can acquire macromolecule precursors through multiple pathways. Cells can synthesize a large number of these compounds, and whether or not a cell synthesizes macromolecule precursors and which nutrients it uses are determined by a variety of factors, including cell type and the nutrient microenvironment. Most biosynthetic pathways depend upon reduction or oxidation reactions. Consequently biosynthesis depends on the continued generation of NADPH for reduction and NAD+ for oxidation. Several reactions in the cytosol and mitochondria produce NADPH; however, mitochondrial electron transport chain activity or an alternative source of electron acceptors is essential to regenerate NAD+. The conversion of glucose to lactate is net NAD+-neutral, so glycolysis and LDH activity alone are insufficient to support biosynthesis. Cells deficient for mitochondrial DNA cannot proliferate without exogenous pyruvate that is used to regenerate NAD+ (20, 29). Proliferation and biosynthesis cannot proceed without functional redox metabolism, and inhibition of these pathways renders cells dependent on exogenous macromolecule precursors. To exploit metabolic pathways for therapy, inhibitors of redox metabolism will be important tools to consider in conjunction with other agents that target metabolism.
This work was supported by NCI of the National Institutes of Health, the Lustgarten Foundation, the Ludwig Center at Massachusetts Institute of Technology, and SU2C. This work was also supported by an International Student Research Fellowship from the Howard Hughes Medical Institute (to A. M. H.) and a Howard Hughes Medical Institute Faculty Scholar Award (to M. V. H.). This is the first article in the Thematic Minireview Series “Redox metabolism and signaling.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCA
- tricarboxylic acid
- LDH
- lactate dehydrogenase
- ACSS
- acetyl-CoA synthetase
- ETC
- electron transport chain
- PDH
- pyruvate dehydrogenase
- IDH
- isocitrate dehydrogenase.
References
- 1. Lunt S. Y., and Vander Heiden M. G. (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464 10.1146/annurev-cellbio-092910-154237 [DOI] [PubMed] [Google Scholar]
- 2. Vander Heiden M. G. (2011) Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684 10.1038/nrd3504 [DOI] [PubMed] [Google Scholar]
- 3. Weinberg S. E., and Chandel N. S. (2015) Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11, 9–15 10.1038/nchembio.1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tennant D. A., Durán R. V., and Gottlieb E. (2010) Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 10.1038/nrc2817 [DOI] [PubMed] [Google Scholar]
- 5. Wheaton W. W., Weinberg S. E., Hamanaka R. B., Soberanes S., Sullivan L. B., Anso E., Glasauer A., Dufour E., Mutlu G. M., Budigner G. S., and Chandel N. S. (2014) Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife 3, e02242 10.7554/eLife.02242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vander Heiden M. G., and DeBerardinis R. J. (2017) Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Veech R. L., Eggleston L. V., and Krebs H. A. (1969) The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 115, 609–619 10.1042/bj1150609a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pollak N., Dölle C., and Ziegler M. (2007) The power to reduce: pyridine nucleotides–small molecules with a multitude of functions. Biochem. J. 402, 205–218 10.1042/BJ20061638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Y., Wang A., Zou Y., Su N., Loscalzo J., and Yang Y. (2016) In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD+/NADH redox state. Nat. Protoc. 11, 1345–1359 10.1038/nprot.2016.074 [DOI] [PubMed] [Google Scholar]
- 10. Schwartz J. P., Passonneau J. V., Johnson G. S., and Pastan I. (1974) The effect of growth conditions on NAD+ and NADH concentrations and the NAD+:NADH ratio in normal and transformed fibroblasts. J. Biol. Chem. 249, 4138–4143 [PubMed] [Google Scholar]
- 11. Coloff J. L., Murphy J. P., Braun C. R., Harris I. S., Shelton L. M., Kami K., Gygi S. P., Selfors L. M., and Brugge J. S. (2016) Differential glutamate metabolism in proliferating and quiescent mammary epithelial cells. Cell Metab. 23, 867–880 10.1016/j.cmet.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 12. Lemons J. M., Feng X. J., Bennett B. D., Legesse-Miller A., Johnson E. L., Raitman I., Pollina E. A., Rabitz H. A., Rabinowitz J. D., and Coller H. A. (2010) Quiescent fibroblasts exhibit high metabolic activity. PLos Biol. 8, e1000514 10.1371/journal.pbio.1000514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., and Rabinowitz J. D. (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 10.1038/nature13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu L., Shah S., Fan J., Park J. O., Wellen K. E., and Rabinowitz J. D. (2016) Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat. Chem. Biol. 12, 345–352 10.1038/nchembio.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holten D., Procsal D., and Chang H. L. (1976) Regulation of pentose phosphate pathway dehydrogenases by NADP+/NADPH ratios. Biochem. Biophys. Res. Commun. 68, 436–441 10.1016/0006-291X(76)91164-5 [DOI] [PubMed] [Google Scholar]
- 16. Lewis C. A., Parker S. J., Fiske B. P., McCloskey D., Gui D. Y., Green C. R., Vokes N. I., Feist A. M., Vander Heiden M. G., and Metallo C. M. (2014) Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 10.1016/j.molcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang L., Shestov A. A., Swain P., Yang C., Parker S. J., Wang Q. A., Terada L. S., Adams N. D., McCabe M. T., Pietrak B., Schmidt S., Metallo C. M., Dranka B. P., Schwartz B., and DeBerardinis R. J. (2016) Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258 10.1038/nature17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye J., Fan J., Venneti S., Wan Y. W., Pawel B. R., Zhang J., Finley L. W., Lu C., Lindsten T., Cross J. R., Qing G., Liu Z., Simon M. C., Rabinowitz J. D., and Thompson C. B. (2014) Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4, 1406–1417 10.1158/2159-8290.CD-14-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birsoy K., Wang T., Chen W. W., Freinkman E., Abu-Remaileh M., and Sabatini D. M. (2015) An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 10.1016/j.cell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sullivan L. B., Gui D. Y., Hosios A. M., Bush L. N., Freinkman E., and Vander Heiden M. G. (2015) Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 10.1016/j.cell.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olenchock B. A., Rathmell J. C., and Vander Heiden M. G. (2017) Biochemical underpinnings of immune cell metabolic phenotypes. Immunity 46, 703–713 10.1016/j.immuni.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wuntch T., Chen R. F., and Vesell E. S. (1970) Lactate dehydrogenase isozymes: kinetic properties at high enzyme concentrations. Science 167, 63–65 10.1126/science.167.3914.63 [DOI] [PubMed] [Google Scholar]
- 23. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hume D. A., and Weidemann M. J. (1979) Role and regulation of glucose metabolism in proliferating cells. J. Natl. Cancer Inst. 62, 3–8 [PubMed] [Google Scholar]
- 25. Pavlova N. N., and Thompson C. B. (2016) The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu P. P., and Sabatini D. M. (2008) Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 10.1016/j.cell.2008.08.021 [DOI] [PubMed] [Google Scholar]
- 27. Fan J., Kamphorst J. J., Mathew R., Chung M. K., White E., Shlomi T., and Rabinowitz J. D. (2013) Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 9, 712 10.1038/msb.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zu X. L., and Guppy M. (2004) Cancer metabolism: facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 313, 459–465 10.1016/j.bbrc.2003.11.136 [DOI] [PubMed] [Google Scholar]
- 29. King M. P., and Attardi G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503 10.1126/science.2814477 [DOI] [PubMed] [Google Scholar]
- 30. Mayers J. R., and Vander Heiden M. G. (2015) Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 40, 130–140 10.1016/j.tibs.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirayama A., Kami K., Sugimoto M., Sugawara M., Toki N., Onozuka H., Kinoshita T., Saito N., Ochiai A., Tomita M., Esumi H., and Soga T. (2009) Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 69, 4918–4925 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- 32. Kamphorst J. J., Nofal M., Commisso C., Hackett S. R., Lu W., Grabocka E., Vander Heiden M. G., Miller G., Drebin J. A., Bar-Sagi D., Thompson C. B., and Rabinowitz J. D. (2015) Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 10.1158/0008-5472.CAN-14-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Commisso C., Davidson S. M., Soydaner-Azeloglu R. G., Parker S. J., Kamphorst J. J., Hackett S., Grabocka E., Nofal M., Drebin J. A., Thompson C. B., Rabinowitz J. D., Metallo C. M., Vander Heiden M. G., and Bar-Sagi D. (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., and Thompson C. B. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 10.1073/pnas.0709747104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A. L., Kafri R., Kirschner M. W., Clish C. B., and Mootha V. K. (2012) Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 10.1126/science.1218595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chuang J. C., Yu C. L., and Wang S. R. (1990) Modulation of human lymphocyte proliferation by amino acids. Clin. Exp. Immunol. 81, 173–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sousa C. M., Biancur D. E., Wang X., Halbrook C. J., Sherman M. H., Zhang L., Kremer D., Hwang R. F., Witkiewicz A. K., Ying H., Asara J. M., Evans R. M., Cantley L. C., Lyssiotis C. A., and Kimmelman A. C. (2016) Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 10.1038/nature19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meiser J., Tumanov S., Maddocks O., Labuschagne C. F., Athineos D., Van Den Broek N., Mackay G. M., Gottlieb E., Blyth K., Vousden K., Kamphorst J. J., and Vazquez A. (2016) Serine one-carbon catabolism with formate overflow. Sci. Adv. 2, e1601273 10.1126/sciadv.1601273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hosios A. M., Hecht V. C., Danai L. V., Johnson M. O., Rathmell J. C., Steinhauser M. L., Manalis S. R., and Vander Heiden M. G. (2016) Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell 36, 540–549 10.1016/j.devcel.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeBerardinis R. J., and Cheng T. (2010) Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 10.1038/onc.2009.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eagle H., Oyama V. I., Levy M., Horton C. L., and Fleischman R. (1956) The growth response of mammalian cells in tissue culture to l-glutamine and l-glutamic acid. J. Biol. Chem. 218, 607–616 [PubMed] [Google Scholar]
- 42. Liu W., Hancock C. N., Fischer J. W., Harman M., and Phang J. M. (2015) Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci. Rep. 5, 17206 10.1038/srep17206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Phang J. M., Liu W., Hancock C., and Christian K. J. (2012) The proline regulatory axis and cancer. Front. Oncol. 2, 60 10.3389/fonc.2012.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu G. (1996) An important role for pentose cycle in the synthesis of citrulline and proline from glutamine in porcine enterocytes. Arch. Biochem. Biophys. 336, 224–230 10.1006/abbi.1996.0552 [DOI] [PubMed] [Google Scholar]
- 45. Franklin R. B., Zou J., Yu Z., and Costello L. C. (2006) EAAC1 is expressed in rat and human prostate epithelial cells; functions as a high-affinity l-aspartate transporter; and is regulated by prolactin and testosterone. BMC Biochem. 7, 10 10.1186/1471-2091-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Titov D. V., Cracan V., Goodman R. P., Peng J., Grabarek Z., and Mootha V. K. (2016) Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235 10.1126/science.aad4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davidson S. M., Papagiannakopoulos T., Olenchock B. A., Heyman J. E., Keibler M. A., Luengo A., Bauer M. R., Jha A. K., O'Brien J. P., Pierce K. A., Gui D. Y., Sullivan L. B., Wasylenko T. M., Subbaraj L., Chin C. R., et al. (2016) Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 10.1016/j.cmet.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sellers K., Fox M. P., Bousamra M. 2nd., Slone S. P., Higashi R. M., Miller D. M., Wang Y., Yan J., Yuneva M. O., Deshpande R., Lane A. N., and Fan T. W. (2015) Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 125, 687–698 10.1172/JCI72873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hensley C. T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L., Wodzak M., Klimko C., McMillan E., Butt Y., Ni M., et al. (2016) Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 10.1016/j.cell.2015.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mayers J. R., Torrence M. E., Danai L. V., Papagiannakopoulos T., Davidson S. M., Bauer M. R., Lau A. N., Ji B. W., Dixit P. D., Hosios A. M., Muir A., Chin C. R., Freinkman E., Jacks T., Wolpin B. M., et al. (2016) Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 10.1126/science.aaf5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baenke F., Peck B., Miess H., and Schulze A. (2013) Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6, 1353–1363 10.1242/dmm.011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simons K., and Sampaio J. L. (2011) Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697 10.1101/cshperspect.a004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thibault G., Shui G., Kim W., McAlister G. C., Ismail N., Gygi S. P., Wenk M. R., and Ng D. T. (2012) The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell 48, 16–27 10.1016/j.molcel.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yao C. H., Fowle-Grider R., Mahieu N. G., Liu G. Y., Chen Y. J., Wang R., Singh M., Potter G. S., Gross R. W., Schaefer J., Johnson S. L., and Patti G. J. (2016) Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem. Biol. 23, 483–493 10.1016/j.chembiol.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ardawi M. S., and Newsholme E. A. (1984) Metabolism of ketone bodies, oleate and glucose in lymphocytes of the rat. Biochem. J. 221, 255–260 10.1042/bj2210255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Balaban S., Shearer R. F., Lee L. S., van Geldermalsen M., Schreuder M., Shtein H. C., Cairns R., Thomas K. C., Fazakerley D. J., Grewal T., Holst J., Saunders D. N., and Hoy A. J. (2017) Adipocyte lipolysis links obesity to breast cancer growth: adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer Metab. 5, 1 10.1186/s40170-016-0163-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nieman K. M., Kenny H. A., Penicka C. V., Ladanyi A., Buell-Gutbrod R., Zillhardt M. R., Romero I. L., Carey M. S., Mills G. B., Hotamisligil G. S., Yamada S. D., Peter M. E., Gwin K., and Lengyel E. (2011) Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503 10.1038/nm.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kamphorst J. J., Cross J. R., Fan J., de Stanchina E., Mathew R., White E. P., Thompson C. B., and Rabinowitz J. D. (2013) Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U.S.A. 110, 8882–8887 10.1073/pnas.1307237110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oram J. F., Shafrir E., and Bierman E. L. (1980) Triacylglycerol metabolism and triacylglycerol lipase activities of cultured human skin fibroblasts. Biochim. Biophys. Acta 619, 214–227 10.1016/0005-2760(80)90070-3 [DOI] [PubMed] [Google Scholar]
- 60. Calder P. C., Yaqoob P., and Newsholme E. A. (1994) Triacylglycerol metabolism by lymphocytes and the effect of triacylglycerols on lymphocyte proliferation. Biochem. J. 298, 605–611 10.1042/bj2980605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zaidi N., Lupien L., Kuemmerle N. B., Kinlaw W. B., Swinnen J. V., and Smans K. (2013) Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 52, 585–589 10.1016/j.plipres.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Medes G., Thomas A., and Weinhouse S. (1953) Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 13, 27–29 [PubMed] [Google Scholar]
- 63. Harding J. W. Jr., Pyeritz E. A., Morris H. P., and White H. B. 3rd. (1975) Proportional activities of glycerol kinase and glycerol 3-phosphate dehydrogenase in rat hepatomas. Biochem. J. 148, 545–550 10.1042/bj1480545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Svensson R. U., and Shaw R. J. (2016) Lipid synthesis is a metabolic liability of non-small cell lung cancer. Cold Spring Harb. Symp. Quant. Biol. 81, 93–103 10.1101/sqb.2016.81.030874 [DOI] [PubMed] [Google Scholar]
- 65. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., and Stephanopoulos G. (2011) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 10.1038/nature10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., and DeBerardinis R. J. (2011) Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 10.1038/nature10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., and Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 10.1073/pnas.1117773108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim J. W., Tchernyshyov I., Semenza G. L., and Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 10.1016/j.cmet.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 69. Fendt S. M., Bell E. L., Keibler M. A., Olenchock B. A., Mayers J. R., Wasylenko T. M., Vokes N. I., Guarente L., Vander Heiden M. G., and Stephanopoulos G. (2013) Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236 10.1038/ncomms3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kamphorst J. J., Chung M. K., Fan J., and Rabinowitz J. D. (2014) Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2, 23 10.1186/2049-3002-2-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schug Z. T., Peck B., Jones D. T., Zhang Q., Grosskurth S., Alam I. S., Goodwin L. M., Smethurst E., Mason S., Blyth K., McGarry L., James D., Shanks E., Kalna G., Saunders R. E., et al. (2015) Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 10.1016/j.ccell.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Comerford S. A., Huang Z., Du X., Wang Y., Cai L., Witkiewicz A. K., Walters H., Tantawy M. N., Fu A., Manning H. C., Horton J. D., Hammer R. E., McKnight S. L., and Tu B. P. (2014) Acetate dependence of tumors. Cell 159, 1591–1602 10.1016/j.cell.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hosios A. M., and Vander Heiden M. G. (2014) Acetate metabolism in cancer cells. Cancer Metab. 2, 27 10.1186/s40170-014-0027-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bulusu V., Tumanov S., Michalopoulou E., van den Broek N. J., MacKay G., Nixon C., Dhayade S., Schug Z. T., Vande Voorde J., Blyth K., Gottlieb E., Vazquez A., and Kamphorst J. J. (2017) Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 18, 647–658 10.1016/j.celrep.2016.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Martinez-Outschoorn U. E., Peiris-Pagés M., Pestell R. G., Sotgia F., and Lisanti M. P. (2017) Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 10.1038/nrclinonc.2016.60 [DOI] [PubMed] [Google Scholar]
- 76. Agranoff B. W., and Brady R. O. (1956) Purification and properties of calf liver ribokinase. J. Biol. Chem. 219, 221–229 [PubMed] [Google Scholar]
- 77. Eagle H., Barban S., Levy M., and Schulze H. O. (1958) The utilization of carbohydrates by human cell cultures. J. Biol. Chem. 233, 551–558 [PubMed] [Google Scholar]
- 78. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., and Vousden K. H. (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- 79. Düvel K., Yecies J. L., Menon S., Raman P., Lipovsky A. I., Souza A. L., Triantafellow E., Ma Q., Gorski R., Cleaver S., Vander Heiden M. G., MacKeigan J. P., Finan P. M., Clish C. B., Murphy L. O., and Manning B. D. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reitzer L. J., Wice B. M., and Kennell D. (1980) The pentose cycle. Control and essential function in HeLa cell nucleic acid synthesis. J. Biol. Chem. 255, 5616–5626 [PubMed] [Google Scholar]
- 81. Lee W. N., Boros L. G., Puigjaner J., Bassilian S., Lim S., and Cascante M. (1998) Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. Am. J. Physiol. 274, E843–E851 [DOI] [PubMed] [Google Scholar]
- 82. Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 10.1016/j.cell.2012.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Boros L. G., Lee P. W., Brandes J. L., Cascante M., Muscarella P., Schirmer W. J., Melvin W. S., and Ellison E. C. (1998) Nonoxidative pentose phosphate pathways and their direct role in ribose synthesis in tumors: is cancer a disease of cellular glucose metabolism? Med. Hypotheses 50, 55–59 10.1016/S0306-9877(98)90178-5 [DOI] [PubMed] [Google Scholar]
- 84. Mattaini K. R., Sullivan M. R., and Vander Heiden M. G. (2016) The importance of serine metabolism in cancer. J. Cell Biol. 214, 249–257 10.1083/jcb.201604085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Labuschagne C. F., van den Broek N. J., Mackay G. M., Vousden K. H., and Maddocks O. D. (2014) Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258 10.1016/j.celrep.2014.04.045 [DOI] [PubMed] [Google Scholar]
- 86. Maddocks O. D., Berkers C. R., Mason S. M., Zheng L., Blyth K., Gottlieb E., and Vousden K. H. (2013) Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 10.1038/nature11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bao X. R., Ong S. E., Goldberger O., Peng J., Sharma R., Thompson D. A., Vafai S. B., Cox A. G., Marutani E., Ichinose F., Goessling W., Regev A., Carr S. A., Clish C. B., and Mootha V. K. (2016) Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 10.7554/eLife.10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Aird K. M., Zhang G., Li H., Tu Z., Bitler B. G., Garipov A., Wu H., Wei Z., Wagner S. N., Herlyn M., and Zhang R. (2013) Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 3, 1252–1265 10.1016/j.celrep.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wiley C. D., Velarde M. C., Lecot P., Liu S., Sarnoski E. A., Freund A., Shirakawa K., Lim H. W., Davis S. S., Ramanathan A., Gerencser A. A., Verdin E., and Campisi J. (2016) Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 10.1016/j.cmet.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. DeLapp N. W., and Karasek M. A. (1976) Importance of pyrimidine nucleotide salvage pathways for DNA synthesis in skin. J. Invest. Dermatol. 66, 306–312 10.1111/1523-1747.ep12482292 [DOI] [PubMed] [Google Scholar]
- 91. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]