Abstract
Rap1 proteins are members of the Ras subfamily of small GTPases involved in many biological responses, including adhesion, cell proliferation, and differentiation. Like all small GTPases, they work as molecular allosteric units that are active in signaling only when associated with the proper membrane compartment. Prenylation, occurring in the cytosol, is an enzymatic posttranslational event that anchors small GTPases at the membrane, and prenyl-binding proteins are needed to mask the cytoplasm-exposed lipid during transit to the target membrane. However, several of these proteins still await discovery. In this study, we report that cyclase-associated protein 1 (CAP1) binds Rap1. We found that this binding is GTP-independent, does not involve Rap1's effector domain, and is fully contained in its C-terminal hypervariable region (HVR). Furthermore, Rap1 prenylation was required for high-affinity interactions with CAP1 in a geranylgeranyl-specific manner. The prenyl binding specifically involved CAP1's C-terminal hydrophobic β-sheet domain. We present a combination of experimental and computational approaches, yielding a model whereby the high-affinity binding between Rap1 and CAP1 involves electrostatic and nonpolar side-chain interactions between Rap1's HVR residues, lipid, and CAP1 β-sheet domain. The binding was stabilized by the lipid insertion into the β-solenoid whose interior was occupied by nonpolar side chains. This model was reminiscent of the recently solved structure of the PDEδ–K-Ras complex; accordingly, disruptors of this complex, e.g. deltarasin, blocked the Rap1–CAP1 interaction. These findings indicate that CAP1 is a geranylgeranyl-binding partner of Rap1.
Keywords: protein isoprenylation, Ras-related protein 1 (Rap1), small GTPase, chaperone, lipid-binding protein, CAP1, cyclase-associated protein 1, prenyl-binding protein, Ras proteins, cell signaling, molecular modeling
Introduction
Rap1 proteins are members of the Ras subfamily of small GTPases involved in many biological responses, e.g. cell–cell (1) and cell–extracellular matrix attachment (2, 3), cytoskeletal dynamics (4–6), endocytosis/exocytosis (7–10), polarity (11), cell proliferation (12–16), apoptosis (17–19), differentiation (20–22), and migration/invasion (23). Like all small GTPases, they work as molecular allosteric units switching between inactive and active conformations in a regulated fashion with kinetic rate constants modulated by guanine-exchange factors and GTPase-activating proteins (24). In their GTP-bound active conformation, they bind and prompt the activation of effectors for signal propagation (25). All these nucleotide-dependent events are associated with the core G-domain (26), involving residues 1–166 in Ras/Rap1 proteins. Beyond this domain, the C terminus of these proteins is responsible for their trafficking and anchoring to a membrane-bound compartment and consists of ∼20 residues, the hypervariable region (HVR),5 and ends with a C-terminal CAAX motif (27).
Although at steady state members of the Ras subfamily of proteins reside at membrane environments, they are synthesized in the cytosol and require a series of posttranslational events for membrane association (28). Cytosolic prenyltransferases are first responsible for adding a prenyl group, i.e. C15 farnesyl (Far) in Ras and C20 geranylgeranyl (GerGer) in Rap1, as an irreversible thioether linkage to the cysteine residue in the C-terminal CAAX motif (29). Once prenylated, they move to the endoplasmic reticulum (ER) for further processing via ER-resident enzymes RceI (30, 31), a proteolytic activity responsible for releasing the terminal AAX, and Icmt (32, 33), a methyltransferase modifying the newly formed C-terminal isoprenylcysteine. However, prenylcysteines provide only a low-affinity interaction with membranes (34, 35), and it was recognized early on that a “second signal” was required for stable membrane interaction, i.e. palmitoylation in H-Ras/N-Ras, a reversible thioester linkage to upstream cysteine residue(s) that increases hydrophobicity, or a polybasic domain in the K-Ras/Rap1 HVR that provides electrostatic contacts with negatively charged phospholipids (36–38). Because of the differential localization of Golgi-resident acyltransferases and cytosolic esterases, dynamic acylation/deacylation allows vesicle-mediated transport of these isoforms via the secretory pathway to guarantee their steady-state localization in different membrane compartments (39, 40).
However, traversing the hydrophilic cytosol poses problems for pools of nascent prenylated proteins en route to the ER endomembrane, for retrograde transport of deacylated–prenylated proteins, and for prenylated polybasic domain–containing proteins on their way to plasma membrane. Recently, new chaperone proteins with prenyl-binding domains, able to mask the thermodynamically unfavorable exposure of the isoprenyl group to solvent, were described (e.g. Cdc42-Rho/RhoGDI (41, 42), Rab/RabGDI/REP (43), Rnd/14-3-3 (44), Ras/PDEδ/Arl2–3 (45), K-Ras/CaM (46), Rab/PRA1 (47), Ras/galectins (48), and N-Ras/VPS35 (49)). Most notably, newly identified SmgGDS variants differentially bind nonprenylated and prenylated forms, therefore facilitating processing and trafficking, respectively (50, 51), thus raising the possibility that prenylation of these polybasic domain–containing proteins could be subject to regulation.
In this study, we report the identification of cyclase-associated protein 1 (CAP1) (52) as a new partner of Rap1. Binding is nucleotide-independent and involves Rap1's C-terminal HVR. The C20 geranylgeranyl modification is required for high-affinity interaction with CAP1, involving specifically its C-terminal solenoid-like β-sheet domain, providing a hydrophobic tunnel. The structure is broadly reminiscent of the recently solved complex of PDEδ with K-Ras (53), and accordingly inhibitors of this complex, e.g. deltarasin (54), blocked the Rap1–CAP1 interaction. These studies indicate that CAP1 is a novel geranylgeranyl-binding partner of Rap1.
Results
Rap1b interacts with CAP1
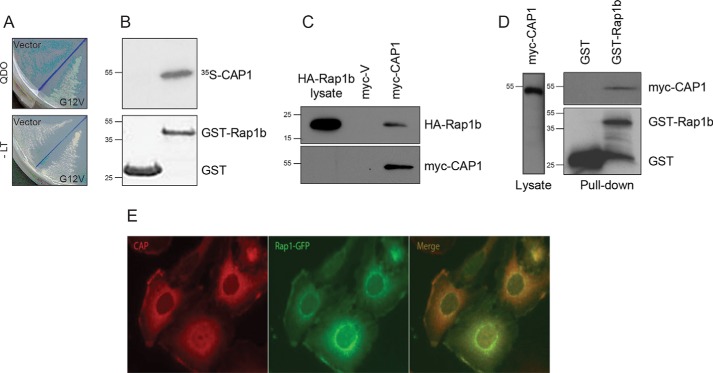
hCAP1 was isolated in a two-hybrid screening utilizing constitutively active Rap1b as bait against a human brain cDNA library. Along with the canonical effector RalGDS, CAP1 represented the most frequent isolate with multiple clones varying in size, all including the C-terminal 123 amino acid fragment (aa 353–475) (Fig. 1A). The N-terminal domain of CAP1 (aa 1–318), however, did not interact with Rap1b under similar conditions (data not shown). To address whether the interaction was direct, Escherichia coli–expressed GST or GST-Rap1b was immobilized on beads, and its ability to bind CAP1 was tested in vitro. As shown in Fig. 1B, [35S]methionine-labeled CAP1 associated specifically with GST-Rap1b, confirming a direct interaction. Similar studies were performed in cells upon cotransfection into HEK cells. Co-immunoprecipitation with HA-Rap1b (Fig. 1C) and pulldown assays of GST/GST-Rap1b (Fig. 1D) indicated a specific interaction with myc-CAP1. Additionally, fluorescence microscopy was used to assess intracellular Rap1b-CAP1 colocalization. PCCL3 thyroid follicular cells stably expressing or transiently transfected with GFP-Rap1b were stained with anti-CAP1 antibody to monitor endogenous CAP1 protein. Colocalization of CAP1 (red) and Rap1b (green) is observed with accumulation in the cytosol and at perinuclear sites as well as, with lower intensity, in peripheral actin-rich lamellipodia (Fig. 1E). Taken together, these results indicate that Rap1b physically interacts with CAP1.
Figure 1.
Rap1b interacts with CAP1. A, two-hybrid isolation of a C-terminal fragment of human CAP1. The activation domain fusion plasmid expressing the C terminus of hCAP1 was cotransformed with empty vector pGBKT7 (Vector) or bait plasmid expressing G12V-Rap1b (G12V) and plated on selection medium lacking Trp, Leu, His, and Ade (QDO; top) versus medium lacking only Trp and Leu (−LT; bottom). B, direct CAP1–Rap1b interaction. Immobilized GST or GST-Rap1b proteins (Coomassie Blue stain shown in lower panel) were incubated with [35S]methionine-labeled in vitro translated CAP1. After extensive washes, samples were resolved by SDS-PAGE. Once dried, radioactivity was visualized by fluorography using a phosphorimaging system (representative image of n = 2). C, in vivo Rap1b and CAP1 interaction. Empty vector (myc-V) or pCMV-myc-CAP1 was cotransfected with HA-Rap1b in HEK293T cells. After 48 h, cell lysates (normalized for equal HA-Rap1b signal) were immunoprecipitated with an anti-myc antibody (9E10), and the presence of Rap1b in the immunocomplex was assessed by Western blotting with an HA-specific antibody. D, in vivo Rap1b and CAP1 interaction. HEK293T cells were transfected with myc-CAP1 plasmid along with GST empty vector or GST-Rap1b mammalian expression plasmids. After 48 h, cell lysates (normalized for equal myc-CAP1 signal) were pulled down with GSH-Sepharose beads. The presence of CAP1 in the complex was assessed by Western blotting with a myc-specific antibody. A representative experiment (n = 3) is shown for C and D. E, colocalization of CAP1 and Rap1b. PCCL3 thyroid follicular cells stably expressing GFP-Rap1b were stained with anti-CAP1 antibodies, and fluorescence microscopy was used to assess intracellular colocalization of CAP1 (red) and Rap1b (green) (representative image; n = 3).
CAP1 is not a Rap1 effector
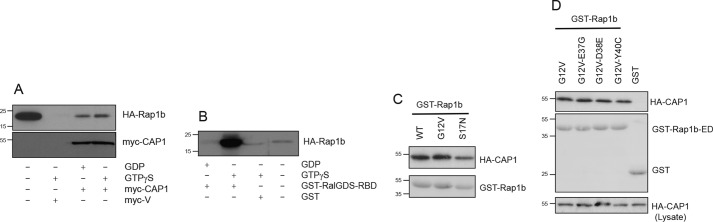
Different approaches were used to determine whether CAP1 binding uses the canonical Rap1 nucleotide-dependent switch. Lysates expressing HA-Rap1b were loaded in vitro with GDP or GTPγS, and binding was assessed upon incubation with myc-CAP1 immobilized on beads. Although the canonical RalGDS-RBD domain behaved as expected for an effector protein, i.e. Rap1-GTPγS–dependent interaction (Fig. 2B), CAP1 binds equally to Rap1-GDP and Rap1-GTPγS (Fig. 2A). Similarly, HA-CAP1 interacted with both constitutively active (G12V) and dominant negative (S17N) Rap1b proteins (Fig. 2C), consistent with a nucleotide-independent association. Moreover, the interaction was not disrupted by introduction of effector domain mutations in switch I (Fig. 2D), known to interfere with effector binding (55). Collectively, these results indicate that CAP1 binding to Rap1b does not involve the G-domain, i.e. the nucleotide-dependent switch and effector-binding domains. Thus, CAP1 is not a prototypical Rap1 effector protein.
Figure 2.
CAP1 is not a Rap1b effector protein. A, nucleotide-independent Rap1–CAP1 interaction. Lysates expressing HA-Rap1b were loaded in vitro with GDP or GTPγS, and a binding assay was performed upon incubation with myc-CAP1 immobilized on beads. B, Rap1 activation was monitored by GST-RalGDS-RBD pulldown assay. C, HA-CAP1 interacted equally with WT, constitutively active (G12V), and dominant negative (S17N) Rap1 proteins. D, effector domain (ED) mutations in switch I did not affect the interaction between CAP1 and Rap1b. Representative experiments (n = 3) are shown. V, empty vector.
Rap1 binds CAP1's C-terminal domain
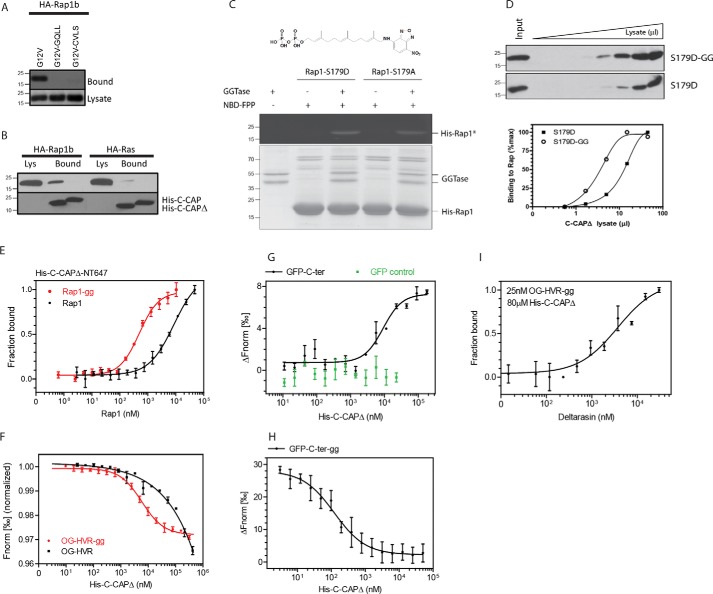
Full-length CAP1 can be divided in two main subdomains, a helical N-terminal and a β-sheet C-terminal subdomain (Fig. 3A). To identify the domain in CAP1 involved in the interaction with Rap1, we prepared constructs expressing both N-terminal (N-CAP; 1–318) and C-terminal fragments (C-CAP; 319–475). Unexpectedly, although full-length CAP1 interacted with Rap1, both N-CAP and C-CAP failed to do so (Fig. 3B). Prompted by the results from the original two-hybrid screen identifying the C-terminal fragment as a positive partner, our studies focused on this domain. The X-ray crystal structure of C-CAP1 (56) indicated the formation of a domain-swapped dimer, mediated by intertwined β-hairpins involving the last two β-strands of each monomer (Fig. 3C). To assess whether C-CAP dimerization could be responsible for masking the Rap1-binding site, those strands were deleted. As shown in Fig. 3D, C-CAPΔ (aa 319–448) strongly bound Rap1. Thus, consistent with the two-hybrid assays, these results confirmed that Rap1 interacts specifically with the C-terminal fragment of CAP1 encompassing the β-sheet domain.
Figure 3.
Rap1 interacts with the C-CAP domain. A, scheme of the CAP1 constructs used in this study. AC, adenylyl cyclase; PR, proline-rich; D, dimerization; FL, full length. B, both GST-N-CAP1 and GST-C-CAP1 failed to interact with Rap1b. The top panel shows purified GST-CAP1 fragments (Coomassie); the bottom panel shows associated Rap1b (HA) after GST-CAP1 pulldown from HA-Rap1b-G12V–expressing cells. C, C-CAP1 dimer formation from X-ray crystal structure (Protein Data Bank code 1K8F). D, C-terminal deletion, myc-C-CAPΔ (aa 319–448), restores binding to immobilized His-Rap1. Representative experiments (n = 3) are shown for B and D.
Rap1 isoprenylation modulates affinity interaction
Preliminary studies indicated no interaction between CAP1 and the deletion construct Rap(1–167) (not shown); therefore, in the absence of any evidence for the involvement of Rap1's effector domain in the interaction with CAP1, our efforts switched to the C terminus of the GTPase. Cys181 in the C-terminal CAAX domain is the target residue for isoprenylation, as noted above, a posttranslational enzymatic event required for small GTPases biology. To address whether Rap1 isoprenylation plays a role in the interaction with CAP1, a C181G mutation was introduced, and its effects on binding were assessed as described above. Fig. 4A shows that the lack of an isoprenyl moiety negatively impacts the interaction with CAP1. Based on the actual sequence of the CAAX box, isoprenylation results in the addition of a C15 farnesyl (i.e. CVLS in H-Ras) or C20 geranylgeranyl group (i.e. CQLL in Rap1). To assess whether the isoprenylation effect is lipid-specific, a farnesylated Rap1 construct (i.e. Rap-CVLS) was tested for CAP1 binding. As shown in Fig. 4A, the shorter C15 farnesyl moiety in Rap-CVLS is not able to replace the native C20 geranylgeranyl and rescue the interaction. Consistent with this observation, CAP1 does not interact with H-Ras (Fig. 4B).
Figure 4.
Rap1 isoprenylation modulates interaction with CAP1. A, Rap1 lysates from HEK293T cells (i.e. HA-Rap1b-G12V, HA-Rap1b-G12V-GGLL, and HA-Rap1b-G12V-CVLS) were incubated with Ni-NTA-agarose–prebound His-C-CAPΔ for 1 h at 4 °C. After washes, associated proteins were resolved by SDS-PAGE and blotted with anti-HA antibody. B, HA-Rap1b and HA-Ras lysates from HEK293T cells were incubated with Ni-NTA-agarose–prebound His-C-CAP and His-C-CAPΔ. His pulldown was performed as above. Associated proteins were revealed by anti-HA antibody (top panel). His-C-CAP and His-C-CAPΔ total proteins were revealed by anti-His tag antibody (bottom panel). Representative experiments (n = 3) are shown for A and B. C, purified Rap1 was in vitro isoprenylated with purified GGTase I utilizing NBD-FPP (top) as substrate. Total proteins were separated by SDS-PAGE (bottom panel; Coomassie), and fluorescent isoprenylated Rap1 proteins (His-Rap1*) were visualized under UV light (top panel). This experiment was repeated twice with similar results. D, immobilized in vitro isoprenylated (S179D-GG) and control His-Rap1-S179D (S179D) were incubated with normalized myc-C-CAPΔ lysates (Input lanes). The top panel shows the binding results as revealed by anti-myc antibody. The bottom panel shows quantitation of these blots upon densitometric analysis via ImageJ. Curves are expressed as a function of normalized lysate volume. A representative experiment (n = 3) is shown. E, MST analysis of in vitro labeled His-C-CAPΔ-NT647 with purified (Rap1) and in vitro prenylated Rap1 (Rap1-gg) used as titrants. F, MST analysis of labeled Rap1b HVR (OG-HVR) and in vitro prenylated (OG-HVR-gg) peptides and purified His-C-CAPΔ as titrant. G, MST analysis of purified GFP-Rap1b HVR (GFP-C-ter) and control GFP proteins used as probes against purified His-C-CAPΔ as titrant. H, MST analysis of in vitro prenylated (GFP-C-ter-gg) as probe against purified His-C-CAPΔ as titrant. I, MST analysis of the effect of deltarasin titrant on the binding of OG-HVR-GG to C-CAPΔ (conditions equivalent to ∼80% maximum based on F). Error bars for all MST studies represent mean ± S.E. (n = 3), and data analyses were performed using NanoTemper Analysis software.
The reduced CAP1–Rap1 interaction in the absence of a geranylgeranyl might represent a direct role for this lipid in binding or, alternatively, indicate that unprocessed cytosolic Rap1 is no longer colocalizing with CAP1 in cells. To directly assess a role for isoprenylation in binding, purified Rap1 was in vitro isoprenylated with purified geranylgeranyltransferase (GGTase I), and the reaction was monitored utilizing NBD-FPP, a fluorescent substrate (57), as shown in Fig. 4C. Moreover, as evidenced by phosphodeficient S179A and phosphomimetic S179D mutants, phosphorylation of Ser179 by PKA, just upstream of the substrate Cys181, does not interfere with the ability of GGTase I to isoprenylate Cys181 (see next section). Binding of CAP1 to lipidated and nonlipidated Rap1 was then assessed by His-affinity coprecipitation utilizing lysates from C-CAPΔ–transfected cells. Results shown in Fig. 4D indicate a left shift for the lipidated sample, qualitatively indicating a role for the isoprenyl group in binding. Microscale thermophoresis (MST) was then utilized for the quantitative determination of equilibrium dissociation constants; purified C-CAPΔ proteins were labeled in vitro (maleimide NT647) and titrated with increasing amounts of purified Rap1. Consistent with the pulldown assays (Fig. 4D), results from the MST assay (Fig. 4E) indicate a >10× increase in affinity upon isoprenylation (Rap1 KD, ∼5.6 ± 0.6 μm; Rap1-GG KD, ∼0.46 ± 0.07 μm). To directly test the role of Rap's C terminus, a peptide encompassing the HVR and CAAX domain (166NRKTPVPGKARKKSSCQLL184) was chemically synthesized and labeled at its N terminus with Oregon Green (OG) followed by in vitro isoprenylation with purified GGTase I. Labeled peptides (OG-HVR and OG-HVR-GG) were titrated with purified C-CAPΔ, and affinities were estimated by MST. Fig. 4F shows the results from this experiment: although, compared with the proteins, a much lower affinity is observed with the free peptides (OG-HVR KD, >156 ± 42 μm; OG-HVR-GG KD, ∼8.6 ± 0.8 μm), a similar effect of isoprenylation on affinity (>20×) is manifest. Moreover, when the same C-terminal sequence is expressed as a GFP fusion (GFP-C-ter) and isoprenylated in vitro (GFP-C-ter-GG), both a high-affinity interaction with C-CAPΔ and the effects of isoprenylation are reconstituted (Fig. 4, G and H; GFP-C-ter KD, ∼9 ± 1.3 μm; GFP-C-ter-GG KD, ∼0.17 ± 0.08 μm), approaching the values observed in full-length Rap1 protein. These results demonstrate a role for the geranylgeranyl group for the Rap1–CAP1 interaction affinity. To test whether the effect of the lipid depends on its ability to interact with the nonpolar environment of the β-sheet pocket as described above for other prenyl-binding proteins, we tested the effect of deltarasin, a recently identified inhibitor of the PDEδ–K-Ras interaction that binds with high affinity to the prenyl-binding pocket of PDEδ (58). As shown in Fig. 4I, deltarasin is able to inhibit the binding of OG-HVR-GG to C-CAPΔ (IC50, ∼3.6 ± 1.2 μm). Thus, the combined results show that the Rap1 C terminus is sufficient for CAP1 binding, and lipid modification increases affinity by interaction with the prenyl-binding pocket of CAP1.
Rap1 Ser179 phosphorylation does not directly affect binding to CAP1
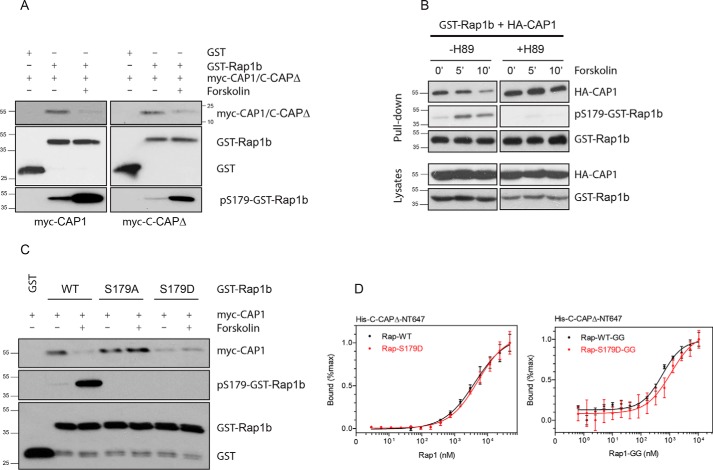
Rap1b is phosphorylated by PKA at residue Ser179 (59), just upstream of the isoprenylated Cys181. To assess potential effects of Rap1 Ser179 phosphorylation on the interaction with CAP1, we performed GST pulldown assays upon stimulation with forskolin, a cAMP-elevating and thus PKA-stimulating agent. Cells were cotransfected with GST-Rap1 or GST control and epitope-tagged myc-CAP1 or myc-C-CAPΔ. Upon forskolin stimulation, phosphorylation of pSer179-GST-Rap1b was confirmed by blotting, and a decrease in the amount of Rap1–CAP1/C-CAPΔ–associated protein was observed (Fig. 5A). Preincubation with the PKA inhibitor H89 blocked pSer179-GST-Rap1 phosphorylation and stabilized the GST-Rap1–CAP1 interaction (Fig. 5B), consistent with a PKA-dependent action of forskolin. Moreover, the negative effect of forskolin on the GST-Rap1–CAP1 interaction is lost in phosphodeficient GST-Rap1-S179A and fully mimicked by GST-Rap1-S179D (Fig. 5C) whose phosphomimetic properties were thoroughly characterized in our laboratory (60). These results indicate that forskolin/PKA–mediated Rap1 Ser179 phosphorylation negatively modulates its interaction with CAP1 in cells.
Figure 5.
Rap1 Ser179 phosphorylation modulates interaction in cells but not in vitro. A, HEK293T cells were co-transfected with GST-Rap1 or GST control and epitope-tagged myc-CAP1 or myc-C-CAPΔ. Upon forskolin stimulation (20 μm, 20min), GST pulldown assays were performed, and associated proteins assessed by Western blots with specific antibodies. B, PKA inhibitor H89 (20 μm) blocked pSer179-GST-Rap1 phosphorylation and stabilized the GST-Rap1–CAP1 interaction. C, The negative effect of forskolin on GST-Rap1–CAP1 interaction can be mimicked by phosphomimetic GST-Rap1-S179D and is lost in phospho-deficient GST-Rap1-S179A. Representative experiments (n = 3) shown in A–C. D, MST analysis of labeled His-C-CAPΔ-NT647, purified Rap1 (Rap-WT and Rap-S179D), and in vitro prenylated (Rap-WT-GG and Rap-S179D-GG) proteins as titrant. Error bars for all MST studies represent mean ± S.E. (n = 3), and data analyses were performed using NanoTemper Analysis software.
To directly quantitate the effect of Rap1 Ser179 phosphorylation on CAP1 binding, we utilized WT and Rap1-S179D as titrants on saturation binding MST assays with NT647-labeled C-CAPΔ as probe. Contrary to the effects observed in vivo, no significant phosphorylation-mediated differences are observed in the binding curves using either lipidated or nonlipidated Rap1 proteins (Fig. 5D). Thus, these results indicate that Ser179 phosphorylation does not directly act on the Rap1–CAP1 contacts, but rather other factor(s) are required to manifest its negative effect in cells.
CAP1 is required for proper Rap1 membrane localization
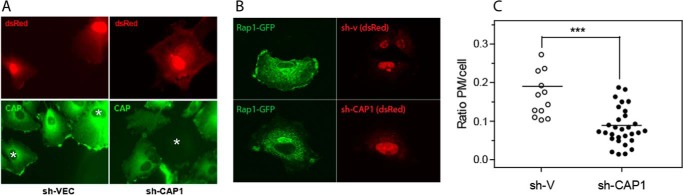
To investigate potential effects of CAP1 in Rap1b localization, an shCAP1 approach was utilized. Scrambled sequence and shCAP1-specific sequence against rat CAP1 were subcloned in a vector expressing an independent dsRed unit. As shown in Fig. 6A, almost complete down-regulation of CAP1 expression was observed in shCAP1- versus scrambled control (sh-V)–transfected (red) cells. To monitor Rap1 localization, experiments were repeated in the presence of GFP-Rap1, and green fluorescence was assessed in dsRed-transfected shCAP1 cells. As shown in Fig. 6, B and C, CAP1 down-regulation is accompanied by an almost complete loss of membrane GFP-Rap1. These results indicate a critical role for CAP1 in the proper plasma membrane localization of Rap1.
Figure 6.
CAP1 depletion alters Rap1 localization. A, CAP1 staining (green channel) in samples of PCCL3 cells transfected with control sh-V (sh-VEC) or sh-CAP1 for 72 h. Transfected cells (red channel) in the field are shown with * in the green channel. B, GFP-Rap1–expressing PCCL3 cells were transfected with sh-CAP1 or sh-V control plasmids. 72 h posttransfection GFP-Rap1 intracellular distribution was assessed by confocal microscopy. Representative fields are shown in A and B. C, quantification of GFP-Rap1 membrane delocalization upon CAP1 depletion. PM/total pixel intensity ratio (green channel) was determined in random red+ cells from experiment shown in B as described under “Experimental procedures.” Data were analyzed, and significance was tested using a two-tailed Student's t test (α level was defined as 0.05). ***, p < 0.0001. All data points are shown. Lines indicate the mean value.
Molecular modeling and dynamics simulations suggest differential binding affinities toward differently modified peptides
Currently no crystal structures of the C-CAP protein are available when it is complexed with Rap1b C-terminal peptides, and solution NMR awaits optimization of conditions to yield good spectra of the protein. Molecular modeling and dynamics simulations represent a suitable tool to make predictions of possible interactions. Because the farnesylated and geranylgeranylated peptides are highly hydrophobic due to attachment of the lipid group and the C-CAP domain presents a tunnel with an opening on both sides, we manually positioned the lipidated peptide as deep into the tunnel as possible, making favorable polar and charge interactions between the peptide RKKSSC residues at the N- or C-terminal opening of the tunnel (see “Experimental procedures”). The systems were then briefly minimized, solvated, and equilibrated for 5 ns using all-atom molecular dynamics using standard protocols. This was followed by a 95-ns production run. The equilibrated structures are shown in Fig. 7, A and B, for the N-terminal insertion and in Fig. S1 for the C-terminal insertion.
Figure 7.
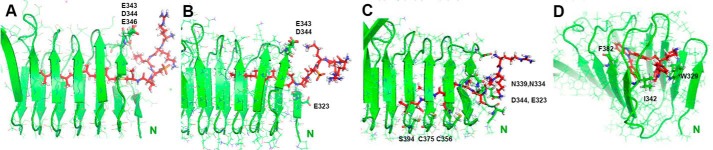
MD simulation and models for lipidated and unmodified peptides and for deltarasin inhibitor. Interactions of RKKSSC-Rap1b-GerGer (A) and -Far (B) prenylated peptides with the N-terminal opening of C-CAP after 5 ns of all-atom dynamics simulation are shown. Both peptides are phosphorylated on Ser179. Although the GG lipid tail is maintained along nearly the entire length of the tunnel, the Far-modified peptide substantially slides out of the tunnel, consistent with weaker binding experimentally (the Far group is initially ∼10 Å shorter than GerGer). Several salt bridges exist between peptide Lys/Arg residues and C-CAP Asp/Glu residues side chains at the N-terminal mouth. In the case of unprocessed peptide and deltarasin inhibitor, initial structures after minimization are shown (C and D). This peptide may be stabilized by the nonpolar side chains in the tunnel (labeled in C), whereas the carbon rings of the inhibitor deltarasin may be stabilized by Phe/Trp and Ile upon initial insertion into N-terminal C-CAP as shown (D). Note that D shows the CAP1 tunnel entrance from the N-terminal side, different from the side-on view of the other panels (A–C).
The behavior of the peptides in the tunnel and at the tunnel mouth was analyzed by root mean square deviation (r.m.s.d.) measurements using the position of lipid tail or the Cα atom of Cys181 in the initial structures as a reference, and the plots are shown in Fig. S2. It is clear that the behavior is dramatically different, depending on the nature of the peptide and direction of tunnel insertion. Briefly, peptides modified by farnesyl, especially the C-terminal inserted peptide, slide toward the mouth of the tunnel where they stay but are highly dynamic. By contrast, GerGer-modified peptides are relatively stable in the tunnel when inserted from the N-terminal side, whereas they slide slightly when inserted from the C-terminal side. Rapid transitions in sliding of the lipid/peptide in the tunnel (e.g. at 95 ns in Fig. S2, panel A3) arise from stochastic fluctuations in the simulations and are normal, especially in relatively long simulations. Because transitions can also revert back (e.g. at 90 ns in Fig. S2, panel A4), we need to consider the area under the r.m.s.d. curve to get an overall qualitative measure of structural stability. By those criteria, an N-terminal insertion of GerGer-Rap1 and Far-Rap1 is preferred. The persistence of contacts at either mouth of the tunnel was analyzed by the frequency key residues are in proximity of the peptide (with 5-Å cutoff between nonhydrogen atoms of residues). These data are summarized in Table S1 for all 10 simulations. In the case of the farnesyl-peptide simulation, they show relatively persistent contacts (>60% of time) between Ser179, Lys178, and Arg176 with C-CAP N-terminal residues Val341, Glu343, and Asp344; however, contacts on C-terminal insertion are only strong for Arg176 but not Lys178 or Ser179, also reflected in lesser contact frequency for residues at the C-terminal mouth of the tunnel (∼30% or less). The GerGer-modified peptide, inserted from the N-terminal side, behaves similarly to the Far-peptide but adds more contacts with Cys181 and the C-CAP protein residues Leu322, Trp329, and Glu346. The simulations with the GerGer peptides were also run with Ser179 when it is phosphorylated and given a −2 charge, showing contacts with N-terminal C-CAP residues 322–324 as well as 344 and 346 with higher frequency. At the C-terminal mouth, contacts with residue 436–438 and 450–452 are significantly more stable. Thus, although the data appear to show a preference of the GerGer peptide for N-terminal insertion, both modes of insertion are moderately stabilized by Ser179 phosphorylation.
We also ran separate simulations with the nonlipidated peptide RKKSSCQLL and with deltarasin inhibitor molecules. Again, the manual positioning sought to optimize polar interactions with the C-CAP polar tunnel residues for the former (on both N-terminal (Fig. 7C) and C-terminal insertions) and C-CAP tunnel/mouth aromatic ring/nonpolar side-chain residues with the rings of deltarasin in the latter case (Fig. 7D). For the nonlipidated peptides, the peptide residues were not stable when inserted into the tunnel from either end and slid out within 30 ns, albeit slightly faster from the C-terminal side. Another difference is that, once it had slid out from the C-terminal side, the peptide actually detaches/interacts only with residue 452 at 50% frequency (others at <10%). By stark contrast, the N-terminal inserted peptide, although no longer resident near the center of the tunnel, maintains a stable conformation at the tunnel mouth by forming extensive contacts among residues Arg176, Lys177, Cys181–Leu184, and C-CAP residues 320/322/329 and 341/343/344/346. The fact that Leu182 and Leu183 can be accommodated is remarkable and suggests that the N-terminal mouth of the tunnel has considerable nonpolar character. A similar behavior is observed with the highly hydrophobic deltarasin inhibitor, which moves out very quickly from the CAP tunnel, either N- or C-terminally inserted, within less than 10 ns but then is maintained in the proximity of both N- and C-terminal mouths of C-CAP tunnel. However, the conformation is highly dynamic, and only a few contacts are formed (more so in the N-terminal than in the C-terminal case). In summary, the MD simulations are consistent with the deep insertion of GerGer-modified peptide into the C-CAP tunnel, especially from the N-terminal side. Far-modified peptides are not stable in the tunnel, and unlipidated peptides also slide out but are maintained at the tunnel mouth. A similar observation is made with deltarasin inhibitor. The latter two cases, unlipidated peptides and deltarasin, may highlight interactions to optimize for more tightly binding inhibitors/competitors to the natural Rap1b interaction with C-CAP.
Discussion
Here, we report the identification of CAP1 as a novel partner of the small GTPase Rap1b. The interaction does not involve the canonical nucleotide-dependent switch and is resistant to known effector domain mutants, indicating that CAP1 is not a Rap1 effector protein. The interaction instead involves Rap1's C-terminal HVR and its lipid moiety in a geranylgeranyl-specific manner. Thus, CAP1 represents a novel Rap1 prenyl-binding protein.
CAP1, which was originally isolated in yeasts (Srv2) (61, 62), is highly conserved in evolution (63). Mammalian CAP1 has 475 aa with several recognizable domains: an N-terminal oligomerization domain, a helical folded domain, a WH2 domain surrounded by two polyproline (P1 and P2) motifs, and a C-terminal β-solenoid domain (52, 63). Our studies indicate CAP1's C-terminal β-solenoid is responsible for Rap1 binding.
Although full-length CAP1 forms hexameric structures in cells (64, 65), its C-terminal β-solenoid domain is dimeric in vitro (56). X-ray crystallography shows each monomer presents a right-handed β-helical organization with six coils forming an elliptical solenoid whose interior is moderately well-packed with mostly nonpolar side chains, leaving a tunnel that runs through the entire protein. Distal to the solenoid, each monomer terminates with a β-hairpin (antiparallel strands β8-β9) responsible for intersubunit domain swapping and formation of an intertwined dimer. Contrary to full-length CAP1, we could not observe an interaction with the full-length C-CAP fragment isolated in the two-hybrid assay. We initially thought that the strand-exchanged C-CAP dimer might be responsible for capping and blocking the entry site to the hydrophobic solenoid, and deletion of the last ∼30 aa, including the β-hairpin, indeed unmasked a high-affinity Rap1 interaction domain.
The CAP1 C-terminal 27 aa are involved in binding monomeric G-actin (66, 67). Moreover, recent mutagenesis studies demonstrated a more extended interaction surface, including the C-terminal dimerization domain as well as conserved solvent-exposed hydrophobic residues in the solenoid domain of both units in the dimer (66, 68). These results suggest that Rap1 binding and G-actin binding to C-CAP might be mutually exclusive; however, pulldown of CAP1 from cell lysates shows an almost stoichiometric amount of bound G-actin that is not affected by overexpression of Rap1.6 Thus, we propose that either Rap1's effect on dimerization/oligomerization dynamics, the deletion of the C-terminal β-hairpin, and/or the presence of N-terminal sequences (e.g. full-length CAP1 or fusion two-hybrid library constructs) exposes a binding domain at the N-terminal opening of C-CAP, allowing prenylated Rap1 binding.
Molecular models, built with both Far- and GerGer-modified Rap1b C-terminal peptides, confirm these findings at the residue level. Farnesylated peptides are not stable in the tunnel and begin to slide out when inserted either from the N- or the C-terminal opening. In contrast, geranylgeranylated peptides are stably maintained in the tunnel over 100 ns of all-atom simulations, especially when inserted from the N terminus. Although the favorable entry site for inserted peptides from the N- versus C-terminal opening largely derives from the interactions that are possible between Rap1b residues and the tunnel mouth, the preference for GerGer- over Far-modified peptides likely originates from the extent of hydrophobic contacts that can be formed in the tunnel (probably proportional to tunnel length occupied). Although in principle shorter Far-modified peptides may insert from both sides to cover the entire tunnel, this is unlikely as it would require a very high concentration of Rap1b GTPases. Thus, consistent with the experimental results, in the presence of only one lipidated HVR per CAP, Far-lipidated peptide would not provide enough contacts, leaving a considerable nonpolar region of the tunnel unoccupied.
MD simulations also suggest how nonlipidated and unprocessed HVR peptide may interact with C-CAP. Although not stable near the center of the tunnel, the peptide is maintained at the N-terminal mouth of the tunnel by both charge–charge and nonpolar side-chain interactions. A similar number of interactions are not available at the C terminus of C-CAP, and the peptide dissociates. We also carried out simulations with the hydrophobic inhibitor deltarasin, inserted at either end into the tunnel after optimizing initial interactions. On both sides, the inhibitor quickly left its initial position (<10 ns) and moved to the mouth of the tunnel. However, the inhibitor then remained at the mouth of the tunnel for the 100-ns simulation. By contrast to the peptides, the deltarasin inhibitor is largely nonpolar except for a linking oxygen and two linking and two ring nitrogens. The only protonated nitrogen available for hydrogen bonding is at the most solvent-exposed part of the tunnel. The contacts between deltarasin and the CAP1 center are mostly hydrophobic, although ring stacking with a Phe and Trp at the N-terminal entrance would make this site more favorable and likely somewhat specific. The structure of deltarasin bound to PDEδ has not yet been solved, but from our model of the inhibitor bound to CAP1, it seems plausible that the inhibitor works by blocking the tunnel entrance, hence preventing the CAP1–Rap interaction.
It should be noted that, although the simulations are relatively lengthy at 100 ns, the positions of the peptides and inhibitor in the tunnel are first hand-modeled and then minimized. This treatment would need to be dramatically extended to obtain statistically significant results (four to eight repeat simulations) or even more computationally expensive, so-called umbrella-sampling simulations, to estimate free energies of binding. However, such calculations are beyond the scope of this report and our computational resources. The computational studies are, however, remarkably consistent with the experimental results across the series of peptides, and the predictions should be valuable in suggesting C-CAP mutants or HVR variants (e.g. testing GTPase isoforms with similar terminal residues) for future investigations.
CAP1 depletion results in Rap1 mislocalization from the plasma membrane, consistent with a role in stabilizing Rap1 at the membrane or as a chaperone for delivery of prenylated Rap1 to the membrane. Recently, several small GTPase chaperones sharing a common β-sandwich fold have been reported. Contrary to the GDP-bound specificity of RhoGDI (42) or the nonspecific prenyl–lipid interaction in PDEδ (69), our data showed that CAP1, with a different structure, binds in a nucleotide-independent manner specifically to only geranylgeranylated Rap1; no interaction was observed with native farnesylated H-Ras or farnesylated Rap1-CVLS. Rap1's G-domain by itself (aa 1–167) does not interact with CAP1 and the lipidated HVR peptide is able to fully recapitulate the affinity of the interaction with full-length Rap1. The different affinities observed for free peptide versus full-length Rap1b or GFP-C-ter reflects low-affinity contacts involving Rap1's G-domain or GFP or, most likely, an effect on decreased conformational entropy, i.e. the distribution of conformational states populated by the HVR tail. Whether other geranylgeranylated small GTPases can also bind CAP1 and/or the HVR sequence provides all specificity and/or it communicates allosterically with the G domain (70) remains to be investigated.
Rap1b is phosphorylated by PKA at Ser179 with cell-specific associated responses. We have shown in endocrine cells that Rap1-GTP and pSer179 are synergistically required for cAMP-dependent cell proliferation (71–73). In contrast, it was recently reported that in some cell types Rap1 phosphorylation might negatively impact its prenylation pathway, an effect involving a novel family of prenyl-binding proteins (74). SmgGDSs represent a family of alternatively spliced novel prenyl-binding proteins not involving a β-sandwich fold; although the largest isoform binds unprocessed Rap1 and seems to help in the prenylation pathway, the small isoform binds prenylated Rap1, facilitating its final translocation to membranes (51). Interestingly, phosphorylation of unprocessed Rap1 seems to inhibit its interaction with SmgGDS, affecting its prenylation and further trafficking (75). Although we observed a negative effect on the Rap1–CAP1 interaction in cells, pSer179 did not manifest any effect on binding when tested in vitro with either lipidated or nonlipidated Rap1 proteins or HVR-derived peptides. Overall, this is consistent with the molecular models and simulations where pSer179 showed only a slight effect, most likely organizing the neighboring charged Arg/Lys side chains of the HVR to point in the direction of the negative charged C-CAP residues at the mouth of the tunnel.
These results indicate that, for the effects observed in vivo, another cellular factor(s) might be required. Mammalian CAP1 itself can be phosphorylated in cells at many sites (76). However, all these events are PKA-independent and reside outside the C-CAP domain and are thus unlikely to affect Rap1b–CAP1 interaction. It has recently been reported that Rap1 phosphorylation creates a novel 14-3-3–binding site responsible for kinase suppressor of Ras (KSR) recruitment leading to B-Raf/ERK activation (77). Alternatively, a pSer179-mediated allosteric communication to switch regions (60) might be responsible for local conformational effects, resulting in effector coupling and activation. Even though the Rap1–CAP1 interaction is GTP-independent, the model predicts that Rap1-GTP might interact with effectors scaffolded in the CAP1 complex. This hypothesis is currently under investigation.
As discussed above, Rap1 has been proposed to be involved in several actin-dependent processes. It is localized in a Rab11+ perinuclear compartment and plasma membrane lamellipodia (78). Similarly, CAP1 localizes in F-actin–rich regions (67, 79–81), and it purifies from cells as a hexameric complex bound (1:1) to monomeric G-actin (82, 83). Its N-terminal domain is responsible for a cofilin-mediated actin-severing activity (64, 65, 84), suggesting a role in F-actin disassembly. Its C terminus is responsible for binding G-actin (66, 67) and in collaboration with profilin participates in nucleotide exchange on G-actin-ADP (85, 86), consequently indicating a role in F-actin assembly. Thus, like Rap1, the current model places CAP1 function in the context of cellular events requiring active actin dynamics, i.e. cell polarity, migration, and receptor-mediated endocytosis (63).
The role of Rap1 in tumor migration and invasion was confirmed in many tissues (23), and numerous recent reports demonstrate that CAP1 affects tumor migration and that its overexpression correlates with invasiveness in metastasis (88–100). Thus, it is tempting to speculate that the novel Rap1–CAP1 complex might provide a mechanistic link to integrate these biological responses.
Experimental procedures
Materials
TSH, forskolin, H89, and GTPγs were from Sigma. GSH-agarose was from GE Healthcare. Ni-NTA-agarose was from Qiagen. Antibodies against HA (HA.11) and myc (9E10) were from Covance. Anti-GST (A5800) antibody was from Invitrogen. Anti-phospho-(Ser/Thr) PKA substrate antibody was from Cell Signaling Technology. Anti-CAP1 antibody was from Proteintech. NBD-FPP and GGPP were from Jena Bioscience.
Cell lines and transfections
PCCL3, a normal TSH-dependent rat thyroid follicular cell line, was grown in 5% Coon's modified F-12 medium (Sigma) supplemented with 5% FBS and a combination of four hormones, TSH (1 mIU/ml), insulin (1 μg/ml), transferrin (5 μg/ml), and hydrocortisone (1 nm), as described before (73). HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 10% FBS. Cells were kept at 37 °C in a 5% CO2, 95% humidified air environment. Transfections were performed using a Lipofectamine 3000 transfection kit (Invitrogen) or X-tremeGENE HP DNA transfection reagent (Roche Applied Science), adjusting the total amount of DNA plasmid to 0.5–1 μg/well as directed by the manufacturers.
DNA constructs
pCGN-HA-Rap1, GST-Rap1, HA-Rap1-G12V, HA-Rap1-G12V/S179A, and GFP-Rap1 constructs were already described (59, 101). Effector domain mutants (provided by Dr. Stork) were introduced in HA-Rap1-G12V. Human CAP1 gene was amplified by PCR using human cDNA as template. The PCR products were digested with XbaI and BamHI and then inserted into pCGN-HA vector, generating HA-CAP1. myc-CAP1, myc-C-CAP (aa 319–475), and myc-C-CAPΔ (aa 319–448) were prepared by inserting SalI-XhoI PCR fragments into pCMV-myc vector using HA-CAP1 as template. PCR products from myc-C-CAP and myc-C-CAPΔ were digested with NdeI-XhoI and subcloned into pET-28c, generating His-C-CAP and His-C-CAPΔ. HA-Rap1-G12V-CVLS was subcloned as a PstI-BamHI PCR fragment. GFP-Rap1b-C-ter was prepared by digesting full-length GFP-Rap1b with BglII, removing most of Rap1b from its N terminus. The remaining fragment was blunted with Klenow and religated, generating GFP-Rap1b-C-ter. shCAP1 plasmid (targeting aa 1074–1092) was generated by subcloning the following annealed complementary oligos into BamHI-EcoRI–digested pSIREN-dsRed vector (Clontech): 5′-GATCCGCACGACATTGCAAATCAAGGAAGCTTGCTTGATTTGCAATGTCGTGCTTTTTTG-3′ and 5′-AATTCAAAAAAGCACGACATTGCAAATCAAGCAAGCTTCCTTGATTTGCAATGTCGTGCG-3′.
Yeast two-hybrid screening
MatchmakerTM GAL4 Two-Hybrid System 3 and pretransformed human brain cDNA library were purchased from Clontech. Rap1-G12V/S179D bait was released from pCGN-Rap1-G12V/S179D with SpeI-Klenow (blunt)/BamHI and subcloned into pGBKT7 at NcoI-Klenow (blunt)/BamHI sites. Approximately, 1.48 × 107 colonies were screened following the manufacturer's instructions.
Protein purification and in vitro binding
BL21/DE3 competent E. coli cells transformed with the appropriate pGEX or pET28c plasmids were grown until ∼0.8 A600 and induced for 16 h at 24 °C with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. After centrifugation, cells were lysed, and supernatants were subjected to affinity chromatography on GSH-Sepharose (GE Healthcare) or Ni-NTA-agarose (Qiagen), respectively. For in vitro binding, E. coli–expressed GST or GST-Rap1b was immobilized on beads, and binding was assessed with in vitro translated CAP1, generated from pET28c-CAP1 by TnT T7 Quick (Promega) and [35S]methionine (PerkinElmer Life Sciences) following the manufacturers' directions. Binding assays were performed in 50 mm Tris/Cl, pH 7.5, 50 mm NaCl, and 0.1%Tween 20 for 1 h at room temperature, and complexes were washed four times with binding buffer. Samples were resolved by 10% SDS-PAGE, transferred, dried, and exposed to film.
GST pulldowns
Cells were cotransfected with GST fusion and HA- or myc-tagged mammalian expression vectors. Upon lysis in immunoprecipitation buffer, GST proteins were pulled down with GSH-Sepharose beads, and associated proteins were resolved by SDS-PAGE and blotted with anti-myc or -HA antibodies.
His pulldowns
Ni-NTA-agarose–prebound His-tagged proteins were incubated with cell lysates at 4 °C for 1 h. Beads were spun down and washed four times, and associated proteins were resolved by SDS-PAGE and blotted with anti-myc or -HA antibodies.
Immunoprecipitation
pCMV-myc-CAP1 and HA-Rap1b were cotransfected in HEK293T or PCCL3 cells. After 48 h, cells were lysed with a buffer containing 50 mm Tris/Cl, pH 7.5, 50 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, and protease inhibitors. Lysates were incubated for 1 h at 4 °C with anti-c-myc-agarose (Thermo Scientific) beads followed by four washes with lysis buffer.
Rap1 activation assay using RalGDS-RBD
Cells transfected with a plasmid expressing HA-Rap1b were lysed with a buffer containing 25 mm Tris/Cl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 5% glycerol, 5 mm MgCl2, and protease inhibitors. Lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4 °C. Nucleotide loading was performed by incubation in the presence of 10 mm EDTA and either GTPγS (100 μm) or GDP (1 mm); the reaction was terminated after 30 min at 30 °C by addition of MgCl2 (60 mm). Purified GST-RalGDS-RBD precoupled to GSH-Sepharose beads (10 μg) was added to the supernatants and incubated at 4 °C for 60 min with agitation. Beads were washed four times in the same lysis buffer. After the final wash, Laemmli sample buffer was added to the samples. Proteins were fractionated by 12% SDS-PAGE and transferred to a polyvinylidene difluoride membrane for blotting.
Immunofluorescence staining and confocal microscopy
Stable PCCL3-GFP-Rap1b cells were cultured on glass coverslips in 6-well plates. For immunofluorescence staining, cells were rinsed with phosphate-buffered saline (PBS; pH 7.4), fixed in 3.7% formaldehyde in PBS for 10 min at room temperature, permeabilized in 0.5% Triton X-100 in PBS for 20 min, and washed in 0.1 m glycine in PBS for 10 min. The coverslips were washed five times for 5 min each with PBS and incubated in PBS containing 1% bovine serum albumin (BSA) and 1% goat serum for 30 min followed by anti-CAP1 and secondary antibodies. After five washes in PBS containing 1% BSA for 10 min, coverslips were mounted in PermaFluorTM (Thermo Fisher Scientific) and examined by confocal microscopy (Olympus Fluoview FV1000) utilizing the appropriate filters. Images were analyzed with the built-in software and ImageJ.
GGTase I expression and purification
Plasmids p316 (pGATEV-Ftase-α) and p310 (pET-28a-GGTase-I-β) were a kind gift from Dr. Kirill Alexandrov (University of Queensland, Australia). p316 and p310 were cotransformed in BL-21(DE3) competent cells. GGTase I expression and purification followed the protocol for His-tagged proteins mentioned above. Proteins were eluted in 1× PBS containing 200 mm imidazole. Imidazole was removed through dialysis against PBS buffer.
In vitro prenylation
In vitro prenylation of Rap1 proteins was performed in a buffer containing 5 mm MgCl2, 10 μm ZnCl2, 2 mm DTT, and 10 μm GDP in PBS. Typically, a 50-μl mixture contained 15 μm purified Rap1 protein, 2 μm GGTase I, and 15 μm GGPP or NBD-FPP. Rap1 proteins and GGTase I were independently prewarmed at 37 °C for 5 min. Mixed samples were incubated at 37 °C for 20 min. Free GGPP was removed through centrifugal filter units.
Microscale thermophoresis
Labeling of His-C-CAPΔ proteins with NT647-maleimide fluorescent dye followed the protocols provided by NanoTemper. Briefly, 20 μm purified proteins were mixed with 60 μm dye in a volume of 100 μl. Labeling mixture was incubated for 30 min at room temperature in the dark. Free dye was removed through the column from the kit. The thermophoresis measurements were performed in a Monolith NT.115 instrument with blue/red channels (NanoTemper) using Premium coated capillaries (NanoTemper, catalog number MO-K005). For experiments with OG-HVR peptide binding to His-C-CAPΔ, samples were prepared in MST binding buffer (50 mm Na-HEPPSO, pH 7.8, 5 mm TCEP, 0.05% Tween 20, 0.05% Pluronic F-127, and 0.1 mg/ml BSA). The peptides were used at a final concentration of 25 nm, and measurements were performed at 14% LED and 60% MST power. For experiments with NT647-maleimide–labeled His-C-CAPΔ binding to His-Rap1, samples were prepared in MST binding buffer, 1× PBS, 0.05% Tween 20, 0.05% Pluronic F-127, and 0.1 mg/ml BSA. The labeled His-C-CAPΔ was used at a final concentration of 100 nm, and measurements were performed at 95% LED and 60% MST power. For experiments with His-C-CAPΔ binding to GFP-Rap-C-ter, samples were prepared in the same MST binding buffer as labeled His-C-CAPΔ, GFP-Rap-C-ter was used at a final concentration of 15 nm, and measurements were performed at 40% LED and 60% MST power. Dose responses in triplicates (mean ± S.E.) were analyzed with the unit software or upon import into GraphPad Prism.
Rap1 localization upon CAP1 depletion
GFP-Rap1–expressing PCCL3 cells were transfected with a shCAP1 and sh-V control plasmids. The vector used, pSIREN, had an independently driven cassette for expression of a red fluorescent protein (dsRed), thus marking transfected cells. Cells were harvested 72 h posttransfection and processed for immunofluorescence detection of endogenous CAP1 expression and GFP-Rap1 intracellular distribution by confocal microscopy. Regions of interest were drawn (polygon tool in ImageJ) on plasma membrane (PM) and whole cells on random red+/green+ cells. Background-subtracted pixel intensity in the green channel was measured in the selected regions of interest, and the PM/total ratio was calculated for each cell. Data were analyzed, and significance was tested using a two-tailed Student's t test (α level was defined as 0.05).
Molecular modeling and dynamics simulations
Simulations were performed on the human C-CAP in complex with the processed and unprocessed C terminus of human Rap1b. Specifically, for the processed peptide, amino acids 176RKKSSC181 were modeled in an extended conformation where the C-terminal carboxylic acid group is methylated and the cysteine is linked to a farnesyl or geranylgeranyl lipid group. The structures of the complex were built by placing the farnesyl or geranylgeranyl into the higher resolution (lower B-factor) monomer unit of the C-CAP protein, taking residues 319–452 of the asymmetric dimer crystal structure (Protein Data Bank code 1K8F, residues 319–475; this removes the C-terminal β-strands of C-CAP that are domain-swapped in the dimer). The lipid group was inserted either in the N- or the C-terminal opening of the C-CAP β-solenoid to the farthest extent possible and oriented so that the charged or polar groups of RKKSS point in the direction of oppositely charged groups or polar groups that surround the tunnel opening. Simulations were also run with lipidated peptides in which the Ser179 side-chain hydroxyl was modified with a phosphorous, PO3−2 group. In the case of the unprocessed Rap1b C-terminal peptide, amino acids 176RKKSSCQLL184 were also inserted into the N- or C-terminal tunnel opening as deeply as possible. However, because the C-terminal group (treated as protonated), Cys–SH, and Gln and Ser side chains are polar, the peptide was placed near the side of the tunnel that contains polar residues, for example, making initial contacts with Ser394, Cys375, and Cys356 for N-terminal insertion. Similarly, no structure for a CAP1 protein (of PDE)–inserted deltarasin is available, and we placed the molecule into the tunnel to initially make contacts between its rings and C-CAP residues Phe382, Ile342, and Trp329. The potential function parameters for the farnesyl and geranylgeranyl groups as well as for deltarasin (PubChem CID 73292904) were generated by the CHARMM generalized force field (CGenFF) online tool (102). Otherwise the CHARMM36 potential function, including the CMAP correction, which also includes phosphoserine, was applied to the system (103–105). The TIP3P model (106) was used for water. Each system was first briefly minimized to remove possible clashes and then solvated in water within a box of pre-equilibrated solvent so that no protein atom was closer than 1.0 nm to the box edge. The system was then neutralized and maintained at a near-physiological ion concentration of 0.15 m NaCl by randomly replacing water molecules. In these all-atom simulations, the electrostatic interaction was treated by the particle-mesh Ewald method (107) for a rectangular periodic boundary box. The van der Waals interaction was truncated at 1.2 nm. The time step was set as 2 fs. Temperature was coupled by using a Langevin thermostat at 300 K, whereas pressure was 1 bar controlled by a semi-isotropic Langevin scheme. Energy minimization was done on the protein complex and waters, respectively, and on the entire system after solvation. All simulations of C-CAP in complex with peptides were run initially for 5 ns using the NAMD 2.10 package (87). The simulations were transferred to the Ohio Supercomputer Center and run for another 95 ns. The simulations of C-CAP in complex with deltarasin were run on the Ohio Supercomputer Center for 50 ns. The trajectories were analyzed using distance measurements between nonhydrogen atoms of C-CAP to the nearest nonhydrogen atoms of peptide or inhibitor. Distances within a cutoff of 5 Å were counted in each trajectory frame, examined at 100-ps intervals. After superposition on Cα atoms of C-CAP, we calculated the distance between the C-terminal peptide or lipid carbon atom inserted initially into the tunnel and its subsequent position, measured as distance as a function of simulation time (Fig. S2). To remove directionality and to calculate the distance between two points at time t = 0 and at a later time in x,y,z coordinate space, the deviation distance is the r.m.s.d.
Author contributions
X. Z., S. C., G. B., M. B., and D. L. A. conceptualization; X. Z., S. C., G. B., M. B., and D. L. A. formal analysis; X. Z., M. B., and D. L. A. supervision; X. Z., M. B., and D. L. A. funding acquisition; X. Z., S. C., G. B., M. M. E., M. W., N. N., and M. B. methodology; X. Z., S. C., G. B., M. B., and D. L. A. writing-original draft; X. Z., M. B., and D. L. A. writing-review and editing; S. C. and G. B. visualization; G. B., M. M. E., M. W., N. N., and M. B. resources.
Supplementary Material
Acknowledgments
We thank Drs. Pat Casey (Duke, North Carolina) and Kirill Alexandrov (Queensland, Australia) for GGTase I protein and GGTase I expression plasmids, respectively, and Dr. Phil Stork (Vollum Institute, Oregon) for the effector domain constructs. We also thank Dr. Zhen Lu Li (Case Western Reserve University, Ohio) for help with the simulation setup and analysis. This project used the University of Pittsburgh Peptide and Peptoid Synthesis Core for peptide synthesis and fluorophore conjugation services. Computational resources at the Case High Performance Computing Cluster and Ohio Supercomputing Center are acknowledged as well.
This work was supported by National Institutes of Health Grant R01 GM099775 (to D. L. A. and partially to M. B). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Table S1.
X. Zhang, S. Cao, G. Barila, M. M. Edreira, M. Wankhede, N. Naim, M. Buck, and D. L. Altschuler, unpublished results.
- HVR
- hypervariable region
- CAP1
- cyclase-associated protein 1
- Far
- farnesyl
- GerGer or GG
- geranylgeranyl
- ER
- endoplasmic reticulum
- GDI
- GDP-dissociation inhibitor
- PDE
- phosphodiesterase
- CaM
- calmodulin
- REP
- Rab escort protein
- GDS
- GDP-dissociation stimulator
- C-ter
- C-terminal sequence
- hCAP1
- human CAP1
- aa
- amino acids
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- RBD
- Rap-binding domain
- GGTase I
- geranylgeranyltransferase
- NBD-FPP
- {3,7-dimethyl-8-(7-nitro-benzo[1,2,5]oxadiazol-4-ylamino)-octa-2,6-diene-1}pyrophosphate
- MST
- microscale thermophoresis
- OG
- Oregon Green
- sh-V
- scrambled control
- r.m.s.d.
- root mean square deviation
- MD
- molecular dynamics
- TSH
- thyrotropin
- Ni-NTA
- nickel-nitrilotriacetic acid
- GGPP
- geranylgeranyl diphosphate
- HEPPSO
- β-hydroxy-4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid monohydrate
- TCEP
- tris(2-carboxyethyl)phosphine
- LED
- light-emitting diode
- PM
- plasma membrane.
References
- 1. Pannekoek W. J., Kooistra M. R., Zwartkruis F. J., and Bos J. L. (2009) Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim. Biophys. Acta 1788, 790–796 10.1016/j.bbamem.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 2. Lagarrigue F., Kim C., and Ginsberg M. H. (2016) The Rap1-RIAM-talin axis of integrin activation and blood cell function. Blood 128, 479–487 10.1182/blood-2015-12-638700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu L., Yang J., Bromberger T., Holly A., Lu F., Liu H., Sun K., Klapproth S., Hirbawi J., Byzova T. V., Plow E. F., Moser M., and Qin J. (2017) Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation. Nat. Commun. 8, 1744 10.1038/s41467-017-01822-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freeman S. A., Lei V., Dang-Lawson M., Mizuno K., Roskelley C. D., and Gold M. R. (2011) Cofilin-mediated F-actin severing is regulated by the Rap GTPase and controls the cytoskeletal dynamics that drive lymphocyte spreading and BCR microcluster formation. J. Immunol. 187, 5887–5900 10.4049/jimmunol.1102233 [DOI] [PubMed] [Google Scholar]
- 5. Lin K. B., Freeman S. A., Zabetian S., Brugger H., Weber M., Lei V., Dang-Lawson M., Tse K. W., Santamaria R., Batista F. D., and Gold M. R. (2008) The rap GTPases regulate B cell morphology, immune-synapse formation, and signaling by particulate B cell receptor ligands. Immunity 28, 75–87 10.1016/j.immuni.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 6. Wang J. C., Lee J. Y., Christian S., Dang-Lawson M., Pritchard C., Freeman S. A., and Gold M. R. (2017) The Rap1-cofilin-1 pathway coordinates actin reorganization and MTOC polarization at the B cell immune synapse. J. Cell Sci. 130, 1094–1109 10.1242/jcs.191858 [DOI] [PubMed] [Google Scholar]
- 7. Chung J., Serezani C. H., Huang S. K., Stern J. N., Keskin D. B., Jagirdar R., Brock T. G., Aronoff D. M., and Peters-Golden M. (2008) Rap1 activation is required for Fc gamma receptor-dependent phagocytosis. J. Immunol. 181, 5501–5509 10.4049/jimmunol.181.8.5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoshino T., Sakisaka T., Baba T., Yamada T., Kimura T., and Takai Y. (2005) Regulation of E-cadherin endocytosis by nectin through afadin, Rap1, and p120ctn. J. Biol. Chem. 280, 24095–24103 10.1074/jbc.M414447200 [DOI] [PubMed] [Google Scholar]
- 9. van Hooren K. W., van Agtmaal E. L., Fernandez-Borja M., van Mourik J. A., Voorberg J., and Bierings R. (2012) The Epac-Rap1 signaling pathway controls cAMP-mediated exocytosis of Weibel-Palade bodies in endothelial cells. J. Biol. Chem. 287, 24713–24720 10.1074/jbc.M111.321976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. York R. D., Molliver D. C., Grewal S. S., Stenberg P. E., McCleskey E. W., and Stork P. J. (2000) Role of phosphoinositide 3-kinase and endocytosis in nerve growth factor-induced extracellular signal-regulated kinase activation via Ras and Rap1. Mol. Cell. Biol. 20, 8069–8083 10.1128/MCB.20.21.8069-8083.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shah B., Lutter D., Tsytsyura Y., Glyvuk N., Sakakibara A., Klingauf J., and Püschel A. W. (2017) Rap1 GTPases are master regulators of neural cell polarity in the developing neocortex. Cereb. Cortex 27, 1253–1269 10.1093/cercor/bhv341 [DOI] [PubMed] [Google Scholar]
- 12. Altschuler D. L., and Ribeiro-Neto F. (1998) Mitogenic and oncogenic properties of the small G protein Rap1b. Proc. Natl. Acad. Sci. U.S.A. 95, 7475–7479 10.1073/pnas.95.13.7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flacke J. P., Flacke H., Appukuttan A., Palisaar R. J., Noldus J., Robinson B. D., Reusch H. P., Zippin J. H., and Ladilov Y. (2013) Type 10 soluble adenylyl cyclase is overexpressed in prostate carcinoma and controls proliferation of prostate cancer cells. J. Biol. Chem. 288, 3126–3135 10.1074/jbc.M112.403279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Misra U. K., and Pizzo S. V. (2009) Epac1-induced cellular proliferation in prostate cancer cells is mediated by B-Raf/ERK and mTOR signaling cascades. J. Cell. Biochem. 108, 998–1011 10.1002/jcb.22333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ribeiro-Neto F., Leon A., Urbani-Brocard J., Lou L., Nyska A., and Altschuler D. L. (2004) cAMP-dependent oncogenic action of Rap1b in the thyroid gland. J. Biol. Chem. 279, 46868–46875 10.1074/jbc.M406858200 [DOI] [PubMed] [Google Scholar]
- 16. Sun W., Jiao W., Huang Y., Li R., Zhang Z., Wang J., and Lei T. (2017) Exchange proteins directly activated by cAMP induce the proliferation of rat anterior pituitary GH3 cells via the activation of extracellular signal-regulated kinase. Biochem. Biophys. Res. Commun. 485, 355–359 10.1016/j.bbrc.2017.02.075 [DOI] [PubMed] [Google Scholar]
- 17. Insel P. A., Zhang L., Murray F., Yokouchi H., and Zambon A. C. (2012) Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol. 204, 277–287 10.1111/j.1748-1716.2011.02273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mangmool S., Hemplueksa P., Parichatikanond W., and Chattipakorn N. (2015) Epac is required for GLP-1R-mediated inhibition of oxidative stress and apoptosis in cardiomyocytes. Mol. Endocrinol. 29, 583–596 10.1210/me.2014-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki S., Yokoyama U., Abe T., Kiyonari H., Yamashita N., Kato Y., Kurotani R., Sato M., Okumura S., and Ishikawa Y. (2010) Differential roles of Epac in regulating cell death in neuronal and myocardial cells. J. Biol. Chem. 285, 24248–24259 10.1074/jbc.M109.094581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khan I., Ali A., Akhter M. A., Naeem N., Chotani M. A., Iqbal H., Kabir N., Atiq M., and Salim A. (2017) Epac-Rap1-activated mesenchymal stem cells improve cardiac function in rat model of myocardial infarction. Cardiovasc. Ther. 35, e12248 10.1111/1755-5922.12248 [DOI] [PubMed] [Google Scholar]
- 21. Marada S., Truong A., and Ogden S. K. (2016) The small GTPase Rap1 is a modulator of Hedgehog signaling. Dev. Biol. 409, 84–94 10.1016/j.ydbio.2015.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu Y., Zhou J., Li Y., Zhou Y., Cui Y., Yang G., and Hong Y. (2015) Rap1A regulates osteoblastic differentiation via the ERK and p38 mediated signaling. PLoS One 10, e0143777 10.1371/journal.pone.0143777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y. L., Wang R. C., Cheng K., Ring B. Z., and Su L. (2017) Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol. Med. 14, 90–99 10.20892/j.issn.2095-3941.2016.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 25. Guo X. X., An S., Yang Y., Liu Y., Hao Q., and Xu T. R. (2016) Rap-interacting proteins are key players in the Rap symphony orchestra. Cell. Physiol. Biochem. 39, 137–156 10.1159/000445612 [DOI] [PubMed] [Google Scholar]
- 26. Wittinghofer A., and Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 10.1146/annurev-biochem-062708-134043 [DOI] [PubMed] [Google Scholar]
- 27. Prior I. A., and Hancock J. F. (2012) Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 23, 145–153 10.1016/j.semcdb.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang M., and Casey P. J. (2016) Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17, 110–122 10.1038/nrm.2015.11 [DOI] [PubMed] [Google Scholar]
- 29. Taylor J. S., Reid T. S., Terry K. L., Casey P. J., and Beese L. S. (2003) Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 22, 5963–5974 10.1093/emboj/cdg571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyartchuk V. L., Ashby M. N., and Rine J. (1997) Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275, 1796–1800 10.1126/science.275.5307.1796 [DOI] [PubMed] [Google Scholar]
- 31. Otto J. C., Kim E., Young S. G., and Casey P. J. (1999) Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 274, 8379–8382 10.1074/jbc.274.13.8379 [DOI] [PubMed] [Google Scholar]
- 32. Hrycyna C. A., Sapperstein S. K., Clarke S., and Michaelis S. (1991) The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 10, 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pillinger M. H., Volker C., Stock J. B., Weissmann G., and Philips M. R. (1994) Characterization of a plasma membrane-associated prenylcysteine-directed α carboxyl methyltransferase in human neutrophils. J. Biol. Chem. 269, 1486–1492 [PubMed] [Google Scholar]
- 34. Brunsveld L., Waldmann H., and Huster D. (2009) Membrane binding of lipidated Ras peptides and proteins—the structural point of view. Biochim. Biophys. Acta 1788, 273–288 10.1016/j.bbamem.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 35. Hancock J. F., Cadwallader K., Paterson H., and Marshall C. J. (1991) A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10, 4033–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hancock J. F., Paterson H., and Marshall C. J. (1990) A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–139 10.1016/0092-8674(90)90294-O [DOI] [PubMed] [Google Scholar]
- 37. Zhou Y., Prakash P., Liang H., Cho K. J., Gorfe A. A., and Hancock J. F. (2017) Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168, 239–251.e216 10.1016/j.cell.2016.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hancock J. F., Magee A. I., Childs J. E., and Marshall C. J. (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177 10.1016/0092-8674(89)90054-8 [DOI] [PubMed] [Google Scholar]
- 39. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., and Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 10.1126/science.1105654 [DOI] [PubMed] [Google Scholar]
- 40. Pedro M. P., Vilcaes A. A., Gomez G. A., and Daniotti J. L. (2017) Individual S-acylated cysteines differentially contribute to H-Ras endomembrane trafficking and acylation/deacylation cycles. Mol. Biol. Cell 28, 962–974 10.1091/mbc.E16-08-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoffman G. R., Nassar N., and Cerione R. A. (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 10.1016/S0092-8674(00)80670-4 [DOI] [PubMed] [Google Scholar]
- 42. Tnimov Z., Guo Z., Gambin Y., Nguyen U. T., Wu Y. W., Abankwa D., Stigter A., Collins B. M., Waldmann H., Goody R. S., and Alexandrov K. (2012) Quantitative analysis of prenylated RhoA interaction with its chaperone, RhoGDI. J. Biol. Chem. 287, 26549–26562 10.1074/jbc.M112.371294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu Y. W., Tan K. T., Waldmann H., Goody R. S., and Alexandrov K. (2007) Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc. Natl. Acad. Sci. U.S.A. 104, 12294–12299 10.1073/pnas.0701817104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riou P., Kjær S., Garg R., Purkiss A., George R., Cain R. J., Bineva G., Reymond N., McColl B., Thompson A. J., O'Reilly N., McDonald N. Q., Parker P. J., and Ridley A. J. (2013) 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell 153, 640–653 10.1016/j.cell.2013.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ismail S. A., Chen Y. X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P. I., and Wittinghofer A. (2011) Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942–949 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- 46. Wu L. J., Xu L. R., Liao J. M., Chen J., and Liang Y. (2011) Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS One 6, e21929 10.1371/journal.pone.0021929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Figueroa C., Taylor J., and Vojtek A. B. (2001) Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 276, 28219–28225 10.1074/jbc.M101763200 [DOI] [PubMed] [Google Scholar]
- 48. Rotblat B., Niv H., André S., Kaltner H., Gabius H. J., and Kloog Y. (2004) Galectin-1(L11A) predicted from a computed galectin-1 farnesyl-binding pocket selectively inhibits Ras-GTP. Cancer Res. 64, 3112–3118 10.1158/0008-5472.CAN-04-0026 [DOI] [PubMed] [Google Scholar]
- 49. Zhou M., Wiener H., Su W., Zhou Y., Liot C., Ahearn I., Hancock J. F., and Philips M. R. (2016) VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J. Cell Biol. 214, 445–458 10.1083/jcb.201604061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schuld N. J., Vervacke J. S., Lorimer E. L., Simon N. C., Hauser A. D., Barbieri J. T., Distefano M. D., and Williams C. L. (2014) The chaperone protein SmgGDS interacts with small GTPases entering the prenylation pathway by recognizing the last amino acid in the CAAX motif. J. Biol. Chem. 289, 6862–6876 10.1074/jbc.M113.527192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berg T. J., Gastonguay A. J., Lorimer E. L., Kuhnmuench J. R., Li R., Fields A. P., and Williams C. L. (2010) Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 10.1074/jbc.M110.129916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hubberstey A. V., and Mottillo E. P. (2002) Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16, 487–499 10.1096/fj.01-0659rev [DOI] [PubMed] [Google Scholar]
- 53. Dharmaiah S., Bindu L., Tran T. H., Gillette W. K., Frank P. H., Ghirlando R., Nissley D. V., Esposito D., McCormick F., Stephen A. G., and Simanshu D. K. (2016) Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. U.S.A. 113, E6766–E6775 10.1073/pnas.1615316113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M., Hahn S. A., Triola G., Wittinghofer A., Bastiaens P. I., and Waldmann H. (2013) Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 10.1038/nature12205 [DOI] [PubMed] [Google Scholar]
- 55. Rodriguez-Viciana P., Sabatier C., and McCormick F. (2004) Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 24, 4943–4954 10.1128/MCB.24.11.4943-4954.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dodatko T., Fedorov A. A., Grynberg M., Patskovsky Y., Rozwarski D. A., Jaroszewski L., Aronoff-Spencer E., Kondraskina E., Irving T., Godzik A., and Almo S. C. (2004) Crystal structure of the actin binding domain of the cyclase-associated protein. Biochemistry 43, 10628–10641 10.1021/bi049071r [DOI] [PubMed] [Google Scholar]
- 57. Dursina B., Reents R., Delon C., Wu Y., Kulharia M., Thutewohl M., Veligodsky A., Kalinin A., Evstifeev V., Ciobanu D., Szedlacsek S. E., Waldmann H., Goody R. S., and Alexandrov K. (2006) Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J. Am. Chem. Soc. 128, 2822–2835 10.1021/ja052196e [DOI] [PubMed] [Google Scholar]
- 58. Papke B., Murarka S., Vogel H. A., Martín-Gago P., Kovacevic M., Truxius D. C., Fansa E. K., Ismail S., Zimmermann G., Heinelt K., Schultz-Fademrecht C., Al Saabi A., Baumann M., Nussbaumer P., Wittinghofer A., et al. (2016) Identification of pyrazolopyridazinones as PDEδ inhibitors. Nat. Commun. 7, 11360 10.1038/ncomms11360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Altschuler D., and Lapetina E. G. (1993) Mutational analysis of the cAMP-dependent protein kinase-mediated phosphorylation site of Rap1b. J. Biol. Chem. 268, 7527–7531 [PubMed] [Google Scholar]
- 60. Edreira M. M., Li S., Hochbaum D., Wong S., Gorfe A. A., Ribeiro-Neto F., Woods V. L. Jr., and Altschuler D. L. (2009) Phosphorylation-induced conformational changes in Rap1b: allosteric effects on switch domains and effector loop. J. Biol. Chem. 284, 27480–27486 10.1074/jbc.M109.011312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fedor-Chaiken M., Deschenes R. J., and Broach J. R. (1990) SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340 10.1016/0092-8674(90)90813-T [DOI] [PubMed] [Google Scholar]
- 62. Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S., Riggs S., Rodgers L., Wieland I., Wheland B., and Wigler M. (1990) Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 61, 319–327 10.1016/0092-8674(90)90812-S [DOI] [PubMed] [Google Scholar]
- 63. Ono S. (2013) The role of cyclase-associated protein in regulating actin filament dynamics—more than a monomer-sequestration factor. J. Cell Sci. 126, 3249–3258 10.1242/jcs.128231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chaudhry F., Breitsprecher D., Little K., Sharov G., Sokolova O., and Goode B. L. (2013) Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol. Biol. Cell 24, 31–41 10.1091/mbc.E12-08-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jansen S., Collins A., Golden L., Sokolova O., and Goode B. L. (2014) Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J. Biol. Chem. 289, 30732–30742 10.1074/jbc.M114.601765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Iwase S., and Ono S. (2016) The C-terminal dimerization motif of cyclase-associated protein is essential for actin monomer regulation. Biochem. J. 473, 4427–4441 10.1042/BCJ20160329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zelicof A., Protopopov V., David D., Lin X. Y., Lustgarten V., and Gerst J. E. (1996) Two separate functions are encoded by the carboxyl-terminal domains of the yeast cyclase-associated protein and its mammalian homologs. Dimerization and actin binding. J. Biol. Chem. 271, 18243–18252 10.1074/jbc.271.30.18243 [DOI] [PubMed] [Google Scholar]
- 68. Iwase S., and Ono S. (2017) Conserved hydrophobic residues in the CARP/β-sheet domain of cyclase-associated protein are involved in actin monomer regulation. Cytoskeleton 74, 343–355 10.1002/cm.21385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang H., Constantine R., Frederick J. M., and Baehr W. (2012) The prenyl-binding protein PrBP/δ: a chaperone participating in intracellular trafficking. Vision Res. 75, 19–25 10.1016/j.visres.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Banerjee A., Jang H., Nussinov R., and Gaponenko V. (2016) The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr. Opin. Struct. Biol. 36, 10–17 10.1016/j.sbi.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hochbaum D., Barila G., Ribeiro-Neto F., and Altschuler D. L. (2011) Radixin assembles cAMP effectors Epac and PKA into a functional cAMP compartment: role in cAMP-dependent cell proliferation. J. Biol. Chem. 286, 859–866 10.1074/jbc.M110.163816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hochbaum D., Hong K., Barila G., Ribeiro-Neto F., and Altschuler D. L. (2008) Epac, in synergy with cAMP-dependent protein kinase (PKA), is required for cAMP-mediated mitogenesis. J. Biol. Chem. 283, 4464–4468 10.1074/jbc.C700171200 [DOI] [PubMed] [Google Scholar]
- 73. Ribeiro-Neto F., Urbani J., Lemee N., Lou L., and Altschuler D. L. (2002) On the mitogenic properties of Rap1b: cAMP-induced G1/S entry requires activated and phosphorylated Rap1b. Proc. Natl. Acad. Sci. U.S.A. 99, 5418–5423 10.1073/pnas.082122499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson J. M., Lorimer E., Tyburski M. D., and Williams C. L. (2015) β-Adrenergic receptors suppress Rap1B prenylation and promote the metastatic phenotype in breast cancer cells. Cancer Biol. Ther. 16, 1364–1374 10.1080/15384047.2015.1070988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wilson J. M., Prokop J. W., Lorimer E., Ntantie E., and Williams C. L. (2016) Differences in the phosphorylation-dependent regulation of prenylation of Rap1A and Rap1B. J. Mol. Biol. 428, 4929–4945 10.1016/j.jmb.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou G. L., Zhang H., Wu H., Ghai P., and Field J. (2014) Phosphorylation of the cytoskeletal protein CAP1 controls its association with cofilin and actin. J. Cell Sci. 127, 5052–5065 10.1242/jcs.156059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Takahashi M., Li Y., Dillon T. J., and Stork P. J. (2017) Phosphorylation of Rap1 by cAMP-dependent protein kinase (PKA) creates a binding site for KSR to sustain ERK activation by cAMP. J. Biol. Chem. 292, 1449–1461 10.1074/jbc.M116.768986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Goitre L., Cutano V., and Retta S. F. (2014) Fluorescence microscopy study of Rap1 subcellular localization. Methods Mol. Biol. 1120, 197–205 10.1007/978-1-62703-791-4_13 [DOI] [PubMed] [Google Scholar]
- 79. Bertling E., Hotulainen P., Mattila P. K., Matilainen T., Salminen M., and Lappalainen P. (2004) Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell 15, 2324–2334 10.1091/mbc.E04-01-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Freeman N. L., and Field J. (2000) Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil. Cytoskeleton 45, 106–120 [DOI] [PubMed] [Google Scholar]
- 81. Moriyama K., and Yahara I. (2002) Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115, 1591–1601 [DOI] [PubMed] [Google Scholar]
- 82. Balcer H. I., Goodman A. L., Rodal A. A., Smith E., Kugler J., Heuser J. E., and Goode B. L. (2003) Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159–2169 10.1016/j.cub.2003.11.051 [DOI] [PubMed] [Google Scholar]
- 83. Quintero-Monzon O., Jonasson E. M., Bertling E., Talarico L., Chaudhry F., Sihvo M., Lappalainen P., and Goode B. L. (2009) Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J. Biol. Chem. 284, 10923–10934 10.1074/jbc.M808760200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Normoyle K. P., and Brieher W. M. (2012) Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J. Biol. Chem. 287, 35722–35732 10.1074/jbc.M112.396051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bertling E., Quintero-Monzon O., Mattila P. K., Goode B. L., and Lappalainen P. (2007) Mechanism and biological role of profilin-Srv2/CAP interaction. J. Cell Sci. 120, 1225–1234 10.1242/jcs.000158 [DOI] [PubMed] [Google Scholar]
- 86. Makkonen M., Bertling E., Chebotareva N. A., Baum J., and Lappalainen P. (2013) Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin through a conserved mechanism. J. Biol. Chem. 288, 984–994 10.1074/jbc.M112.435719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bao Z., Qiu X., Wang D., Ban N., Fan S., Chen W., Sun J., Xing W., Wang Y., and Cui G. (2016) High expression of adenylate cyclase-associated protein 1 accelerates the proliferation, migration and invasion of neural glioma cells. Pathol. Res. Pract. 212, 264–273 10.1016/j.prp.2015.12.017 [DOI] [PubMed] [Google Scholar]
- 89. Fan Y. C., Cui C. C., Zhu Y. S., Zhang L., Shi M., Yu J. S., Bai J., and Zheng J. N. (2016) Overexpression of CAP1 and its significance in tumor cell proliferation, migration and invasion in glioma. Oncol. Rep. 36, 1619–1625 10.3892/or.2016.4936 [DOI] [PubMed] [Google Scholar]
- 90. Hua M., Yan S., Deng Y., Xi Q., Liu R., Yang S., Liu J., Tang C., Wang Y., and Zhong J. (2015) CAP1 is overexpressed in human epithelial ovarian cancer and promotes cell proliferation. Int. J. Mol. Med. 35, 941–949 10.3892/ijmm.2015.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kakurina G. V., Kondakova I. V., Cheremisina O. V., Shishkin D. A., and Choinzonov E. L. (2016) Adenylyl cyclase-associated protein 1 in the development of head and neck squamous cell carcinomas. Bull. Exp. Biol. Med. 160, 695–697 10.1007/s10517-016-3252-2 [DOI] [PubMed] [Google Scholar]
- 92. Liu X., Yao N., Qian J., and Huang H. (2014) High expression and prognostic role of CAP1 and CtBP2 in breast carcinoma: associated with E-cadherin and cell proliferation. Med. Oncol. 31, 878 10.1007/s12032-014-0878-7 [DOI] [PubMed] [Google Scholar]
- 93. Liu Y., Cui X., Hu B., Lu C., Huang X., Cai J., He S., Lv L., Cong X., Liu G., Zhang Y., and Ni R. (2014) Upregulated expression of CAP1 is associated with tumor migration and metastasis in hepatocellular carcinoma. Pathol. Res. Pract. 210, 169–175 10.1016/j.prp.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 94. Masugi Y., Tanese K., Emoto K., Yamazaki K., Effendi K., Funakoshi T., Mori M., and Sakamoto M. (2015) Overexpression of adenylate cyclase-associated protein 2 is a novel prognostic marker in malignant melanoma. Pathol. Int. 65, 627–634 10.1111/pin.12351 [DOI] [PubMed] [Google Scholar]
- 95. Tan M., Song X., Zhang G., Peng A., Li X., Li M., Liu Y., and Wang C. (2013) Overexpression of adenylate cyclase-associated protein 1 is associated with metastasis of lung cancer. Oncol. Rep. 30, 1639–1644 10.3892/or.2013.2607 [DOI] [PubMed] [Google Scholar]
- 96. Xie S. S., Tan M., Lin H. Y., Xu L., Shen C. X., Yuan Q., Song X. L., and Wang C. H. (2015) Overexpression of adenylate cyclase-associated protein 1 may predict brain metastasis in non-small cell lung cancer. Oncol. Rep. 33, 363–371 10.3892/or.2014.3577 [DOI] [PubMed] [Google Scholar]
- 97. Yamazaki K., Takamura M., Masugi Y., Mori T., Du W., Hibi T., Hiraoka N., Ohta T., Ohki M., Hirohashi S., and Sakamoto M. (2009) Adenylate cyclase-associated protein 1 overexpressed in pancreatic cancers is involved in cancer cell motility. Lab. Invest. 89, 425–432 10.1038/labinvest.2009.5 [DOI] [PubMed] [Google Scholar]
- 98. Yu X. F., Ni Q. C., Chen J. P., Xu J. F., Jiang Y., Yang S. Y., Ma J., Gu X. L., Wang H., and Wang Y. Y. (2014) Knocking down the expression of adenylate cyclase-associated protein 1 inhibits the proliferation and migration of breast cancer cells. Exp. Mol. Pathol. 96, 188–194 10.1016/j.yexmp.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 99. Zhang H., and Zhou G. L. (2016) CAP1 (cyclase-associated protein 1) exerts distinct functions in the proliferation and metastatic potential of breast cancer cells mediated by ERK. Sci. Rep. 6, 25933 10.1038/srep25933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Xie S., Shen C., Tan M., Li M., Song X., and Wang C. (2017) Systematic analysis of gene expression alterations and clinical outcomes of adenylate cyclase-associated protein in cancer. Oncotarget 8, 27216–27239 10.18632/oncotarget.16111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Altschuler D. L., Peterson S. N., Ostrowski M. C., and Lapetina E. G. (1995) Cyclic AMP-dependent activation of Rap1b. J. Biol. Chem. 270, 10373–10376 10.1074/jbc.270.18.10373 [DOI] [PubMed] [Google Scholar]
- 102. Vanommeslaeghe K., Raman E. P., and MacKerell A. D. Jr. (2012) Automation of the CHARMM general force field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 10.1021/ci3003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Buck M., Bouguet-Bonnet S., Pastor R. W., and MacKerell A. D. Jr. (2006) Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 90, L36–38 10.1529/biophysj.105.078154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Huang J., and MacKerell A. D. Jr. (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 106. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 107. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 10.1063/1.470117 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.