Abstract
The extreme toxicity of botulinum neurotoxins (BoNTs) relies on their specific cleavage of SNARE proteins, which eventually leads to muscle paralysis. One newly identified mosaic toxin, BoNT/HA (aka H or FA), cleaves VAMP-2 at a unique position between residues L54 and E55, but the molecular basis underlying VAMP-2 recognition of BoNT/HA remains poorly characterized. Here, we report a ∼2.09 Å resolution crystal structure of the light chain protease domain of BoNT/HA (LC/HA). Structural comparison between LC/HA and LC of BoNT/F1 (LC/F1) reveals distinctive hydrophobic and electrostatic features near the active sites, which may explain their different VAMP-2 cleavage sites. When compared to BoNT/F5 that cleaves VAMP-2 at the same site as BoNT/HA, LC/HA displays higher affinity for VAMP-2, which could be caused by their different surface charge properties surrounding a VAMP-2 exosite-binding cleft. Furthermore, systematic mutagenesis studies on VAMP-2 and structural modeling demonstrate that residues R47 to K59 spanning the cleavage site in VAMP-2 may adopt a novel extended conformation when interacting with LC/HA and LC/F5. Taken together, our structure provides new insights into substrate recognition of BoNT/HA and paves the way for rational design of small molecule or peptide inhibitors against LC/HA.
Keywords: botulinum neurotoxin, VAMP-2, BoNT/HA, crystal structure, light chain
Crystal structure of the protease domain of the mosaic botulinum neurotoxin type HA.
INTRODUCTION
Botulinum neurotoxins (BoNTs) and tetanus neurotoxin (TeNT) are the most poisonous biological toxins for humans, which potently block neurotransmitter release (Rossetto, Pirazzini and Montecucco 2014). They adopt a similar architecture composed of a ∼50 kDa light chain (LC) and a ∼100 kDa C-terminal heavy chain (HC), which are linked by a disulfide bridge. HC could be further divided into a translocation domain (HN) that delivers LC to neuronal cytosol, and a receptor-binding domain (HC) that specifically recognizes motoneurons. LC is a Zn2+-containing protease that cleaves unique scissile peptide bonds in soluble N-ethylmaleimide-sensitive fusion protein attachment receptor (SNAREs) proteins, including synaptosome associated protein of 25 kDa (SNAP-25), vesicle associated membrane protein (VAMP) (also called synaptobrevin) and syntaxin. Cleavage of SNARE proteins inhibits the release of neurotransmitter acetylcholine and subsequently paralyzes the affected muscles (Turton, Chaddock and Acharya 2002; Brunger, Jin and Breidenbach 2008; Rossetto et al.2013; Pantano and Montecucco 2014).
There are seven established serotypes of BoNT, designated as BoNT/A to G, which are composed of more than 40 subtypes (Peck et al.2017). BoNT/A and E specifically hydrolyze SNAP-25; BoNT/B, D, F, G and TeNT cleave VAMP; BoNT/C uniquely cleaves both SNAP-25 and syntaxin (Schiavo, Matteoli and Montecucco 2000; Binz, Sikorra and Mahrhold 2010). Three new VAMP-specific BoNTs, namely HA (aka H or FA), X and eBoNT/J were identified in recent years (Dover et al.2014; Zhang et al.2017, 2018; Brunt et al.2018). BoNT/HA is a mosaic toxin, whose LC-HN domain and HC domain are similar to that of BoNT/F5 (∼73% amino acid identity) and BoNT/A1 (∼84% identity), respectively (Maslanka et al.2016). BoNT/HA cleaves VAMP-2 at L54 that is identical to BoNT/F5, but all other BoNT/F subtypes cleave VAMP-2 at Q58 (Kalb et al.2014). The structural mechanism underlying BoNT/HA’s unique substrate cleavage remains unknown.
BoNTs are categorized as Tier 1 select agents by the Centers for Disease Control and Prevention (CDC) and could be potentially misused for bioterrorism warfare (Burnett et al.2005). Therefore, there is an urgent need to develop countermeasures against BoNTs, especially the newly identified toxins (Pirazzini and Rossetto 2017). BoNT/HA could be neutralized by two BoNT/A-HC-targeting antibodies (RAZ1 and CR2), as well as a BoNT/F-HN-binding antibody (4E17.2) (Fan et al.2016; Yao et al.2017). However, no known antibody could cross-react with the LC of BoNT/HA (LC/HA). The unique enzymatic cleavage mechanism and the distinct immunological features of LC/HA thus motivated us to further explore its structure and function.
Here we present the first high-resolution crystal structure of LC/HA. We then carried out complementary mutagenesis studies and structural modeling, which reveal a novel conformation of VAMP-2 needed for LC/HA cleavage. Our data also provide new insights into the different VAMP-2 recognition and cleavage features displayed by LC/HA, LC/F1 and LC/F5, even though they have highly similar structures.
MATERIALS AND METHODS
Cloning, expression and purification
The plasmids for Escherichia coli expression of LC/F5 and LC/HA were generated by ligating PCR fragments encoding amino acids 1–438 of Clostridium botulinum strain CDC 54074 (Mendoza) and 1–434 of C. botulinum strain IBCA10–7060, respectively, into a derivative of pQE3 vector (Qiagen), and the E. coli codon optimized synthesized DNAs (Centic Biotec GmbH) were used as templates. Using the E. coli strain M15pREP4 (Qiagen GmbH, Hilden, Germany), the resulting plasmids allow production of LC carrying at the N-terminus a His6-tag followed by a triple FLAG-tag and a Strep-tag at the C-terminus. Briefly, cultures were induced for 15 h at 21°C and proteins were purified on Ni2+-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer’s instructions. Fractions containing the desired proteins were dialyzed against the toxin assay buffer (150 mM potassium glutamate, 10 mM HEPES, pH 7.2), and small aliquots were frozen in liquid nitrogen and kept at –70°C.
For crystallographic study, LC/HA (M1 – K434) was subcloned into pGEX-4T-2 expression vector (GE Healthcare). The recombinant LC/HA was expressed in E. coli strain BL21-star (DE3) (ThermoFisher). Bacteria were grown at 37°C in LB (Luria-Bertani) medium containing 100 μg/ml ampicillin. The temperature was decreased to 18°C when OD600 reached 0.4–0.6. Expression was induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 16 h. The cells were harvested by centrifugation and stored at –20°C until use.
LC/HA was purified using a GST-affinity column in a buffer containing 400 mM NaCl, 4 mM DTT, 50 mM Tris, pH 8.0. The GST-tag was subsequently removed by incubating resins with thrombin at 8°C for 16 h. The eluted proteins were further purified by Superdex 200 size-exclusion chromatography (SEC) in a buffer containing 150 mM NaCl, 10 mM HEPES, pH 7.0, concentrated to ∼5 mg/ml using Amicon Ultra centrifugal filters (Millipore) and stored at –80°C until use.
Cleavage assays
Cleavage assays were performed as previously described (Sikorra et al.2008). Briefly, rat VAMP-2 and its mutants were generated by in vitro transcription/translation. Cleavage assays contained 1 μl of the transcription/translation mixture of [35S]methionine-labeled wild-type or mutated VAMP-2 and purified LC (LC/F5 and LC/HA at 100 and 50 nM final concentrations, respectively) and were incubated for 60 min at 37°C in a total volume of 10 μl of toxin assay buffer.
For determination of the enzyme kinetic parameters, VAMP-2 concentration was varied between 3 and 130 μM using E. coli expressed His6-VAMP-2. Each of the various substrate concentrations was endowed by the addition of 1 μl of radiolabeled His6-VAMP-2 generated by in vitro transcription/translation. LC/F5 and LC/HA were used at final concentrations of 2 and 10 nM, respectively. Incubation was done in a final volume of 25 μl of toxin assay buffer. After 2 and 4 min of incubation at 37°C, aliquots of 10 μl were taken.
Enzymatic reactions were stopped by mixing with 10 μl of pre-chilled double-concentrated SDS-PAGE sample buffer. VAMP-2 and its cleavage products were separated by SDS-PAGE, and radiolabeled protein was visualized using a FLA-9000 image scanner (Fuji Photo Film, Co., Ltd., Tokyo, Japan). The percentage of hydrolyzed VAMP-2 was determined from the turnover of the radiolabeled substrate applying the Multigauge 3.2 software (Fuji Photo Film) and used to calculate the initial velocity of substrate hydrolysis. KM and Vmax values were calculated by non-linear regression using the GraphPad Prism 4.03 program (GraphPad Software Inc., San Diego, CA).
Crystallization and X-ray diffraction data collection
Initial crystallization screens for LC/HA were carried out using a Gryphon crystallization robot (Art Robbins Instrument, Sunnyvale, CA, USA) and commercial high-throughput crystallization screening kits (Hampton Research, Aliso Viejo, CA, USA and Qiagen, Germantown, MD, USA). After extensive manual optimizations, the best LC/HA crystals were grown by hanging-drop vapor-diffusion at 18°C, in which the protein (5 mg/ml) was mixed in 1:1 (v/v) ratio with a reservoir solution containing 100 mM calcium acetate, 100 mM sodium cacodylate, pH 5.5 and 4% polyethylene glycol (PEG) 8000 (Hampton Research).
The LC/HA crystals were cryo-protected in the original mother liquor supplemented with 20% (v/v) glycerol and flash-frozen in liquid nitrogen. The crystals belong to space group C2221, with unit cell dimensions of a = 76.2 Å, b = 160.7 Å, c = 219.1 Å; α = β = γ = 90o. The best data set was collected at 2.09 Å resolution. The X-ray diffraction data were collected at 100 K at beam line 24-ID-E, Advanced Photon Source (APS), using detector ADSC Q315. All data sets were processed and scaled using XDS (Kabsch 2010) and CCP4i (Potterton et al.2003). Data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| Data collection | LC/HA |
|---|---|
| Wavelength (Å) | 0.97918 |
| Resolution (Å) | 109.55–2.09 (2.13–2.09) |
| Space group | C 2 2 21 |
| Cell dimensions | |
| a, b, c (Å) | 76.2, 160.7, 219.1 |
| α, β, γ (°) | 90, 90, 90 |
| Completeness (%) | 96.0 (96.7) |
| Redundancy | 3.3 (3.3) |
| R merge (%) | 10.0 (39.3) |
| Mean I/σ(I) | 8.2 (2.0) |
| Refinement | |
| Resolution (Å) | 54.05–2.09 |
| No. reflections | 76 565 |
| R work/Rfree (%) | 17.61/19.12 |
| No. atoms | |
| Protein | 6924 |
| Ligand/ion | 19 |
| Water | 584 |
| B-factors (Å2) | |
| Protein | 36.2 |
| Ligand/ion | 42.9 |
| Water | 41.9 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.24 |
Values in parentheses are for the highest resolution shell.
Structure determination
The LC/HA structure was determined with Phaser molecular replacement using the structure of LC/F1 (PDB code 2A8A) (Agarwal, Binz and Swaminathan 2005) as the search model. The structural modeling and refinement were carried out iteratively using COOT (Emsley and Cowtan 2004) and PHENIX (Adams et al.2010) in an iterative manner. The refinement progress was monitored with the Rfree value with a 5% randomly selected test set (Brünger 1992). LC/HA structure was refined to 2.09 Å with Rwork/Rfree = 17.61/19.12. The structure was validated by the MolProbity web server and showed excellent stereochemistry (Chen et al.2010). Structural refinement statistics are listed in Table 1. Coordinates and structure factors for LC/HA have been deposited in the Protein Data Bank under accession code 6BVD. All structure figures were prepared with PyMOL (http://www.pymol.org/).
Molecular modeling
The structure model of LC/F5 was processed in UCSF Chimera using Modeller (Yang et al.2012) with LC/HA as the starting template. For structural model of LC/HA-VAMP-2 peptide, VAMP-2 was modeled based on the LC/F1-inh1 inhibitor (PDB code 3FIE) structure (Agarwal et al.2009) and then modified manually using COOT (Emsley and Cowtan 2004). The peptide docking was initiated using the Rosetta FlexPepDock web server (London et al.2011). The top 10 models were analyzed and the model that is consistent with our biochemical enzymatic cleavage data was selected for further analysis.
RESULTS AND DISCUSSION
Crystal structure of LC/HA
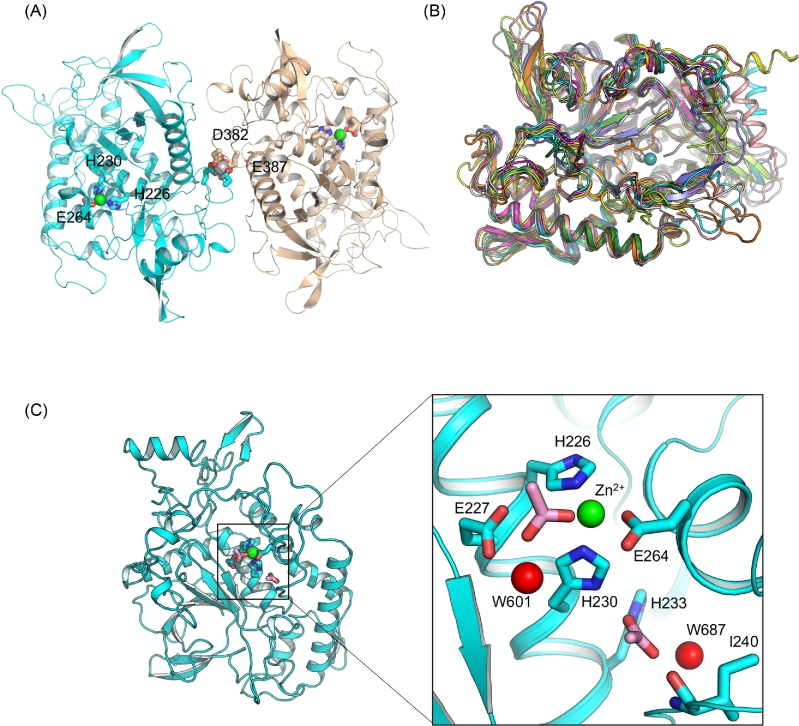
The crystal structure of a recombinant LC/HA (residues 1–434) was determined at 2.09 Å (Table 1). Two molecules of LC/HA were identified in an asymmetric unit (AU), which are nearly identical with root-mean-square deviation (RMSD) of 0.4 Å over 422 Cα atoms (Pymol, www.pymol.org). Nearly all LC/HA residues are visible except for a few solvent-exposed loops in chain B including residues I208 – H211, and N430 – K434, which is likely due to high structural flexibility. Interestingly, a Ca2+ was found between the two LC/HA molecules, which interacts with D382 and E387 in each LC/HA (Fig. 1A). The high Ca2+ concentration in the crystallization buffer (100 mM calcium acetate) likely facilitated LC/HA dimerization at high protein concentration during crystallization. LC/HA shares a high structural homology with LC of other BoNTs as evidenced by the high DALI Z-scores (Holm and Rosenström 2010) during pairwise structure alignment (LC/A, 44.5; LC/B, 50.6; LC/C, 45.7; LC/D, 45.4; LC/E, 50.5; LC/F1, 53.0; LC/G, 48.2; TeNT-LC, 54.5) (Fig. 1B). The structures most similar to LC/HA are TeNT-LC and LC/F1, which have a RMSD of 1.2 Å over 387 amino acids and 1.7 Å over 395 amino acids, respectively.
Figure 1.
Overall structure of LC/HA. (A) Cartoon representation of an LC/HA crystallographic dimer. The Zn2+- and Ca2+-coordinating residues are drawn as sticks and labeled. Ca2+ and Zn2+ are shown as silver and green spheres, respectively. (B) Structural superposition of LC/HA and the LCs of seven established BoNT serotypes and TeNT: LC/A (PDB code: 1XTF) (Breidenbach and Brunger 2004), white; LC/B (2ETF), salmon; LC/C (2QN0) (Jin et al.2007), slate; LC/D (2FPQ) (Arndt et al.2006), orange; LC/E (1T3A) (Agarwal et al.2004), yellow; LC/F1 (2A97) (Agarwal, Binz and Swaminathan 2005), magenta; LC/G (1ZB7) (Arndt et al.2005), lime; TeNT-LC (1YVG) (Rao et al.2005), forest, LC/HA, cyan. (C) The active site of LC/HA. Two acetate ions identified near the active site are drawn as pink sticks and two interacting water molecules are shown as red spheres.
The conserved Zn2+-binding HEXXH motif in LC/HA adopts a geometry that is highly similar to LCs of other BoNTs. In LC/HA, the Zn2+ is coordinated tetrahedrally by nitrogen atoms of H226 and H230, carboxylate oxygens of E264 and an acetate ion. The acetate ion replaces the nucleophilic water and further interacts with the carboxylate oxygen of E227 and a water molecule (W601) (Fig. 1C). Similar observations were reported before, in which the nucleophilic water was replaced by an acetate ion in a LC/A structure (Kumaran et al.2008) or by a sulfate ion in BoNT/B structure (Swaminathan and Eswaramoorthy 2000). Moreover, another acetate ion was identified adjacent to the active site, which is buried deeply in an electropositive pocket and forms several hydrogen bonds with nitrogen atoms of H230 and H233, the main-chain nitrogen atom of I240 and a water molecule (W687).
Mutagenesis studies revealed VAMP-2 residues that interact with LC/HA and LC/F5
Earlier studies on BoNT-substrate recognition suggest that LC exploits an exosite-based mechanism to bind and cleave SNARE proteins: (1) LC first contacts its substrate peptide at the exosites that are remote from the cleavage site; (2) the unusually extended substrate-binding groove of LC provides additional anchoring points that bend the substrate so it wraps around the LC and has its cleavage site move closer to the LC active site; (3) LC binds selective cleavage site residues in substrate that ensure proper register of the scissile peptide bond for hydrolysis. This mechanism is supported by the crystal structures of the LC/A-SNAP-25 and LC/F1-VAMP-2 inhibitor complexes, as well as a wealth of biochemical studies of many other BoNT serotypes (Breidenbach and Brunger 2004; Sikorra et al.2008; Agarwal et al.2009). But how LC/HA and LC/F5 cut VAMP-2 at a unique position between L54 and E55 remains elusive.
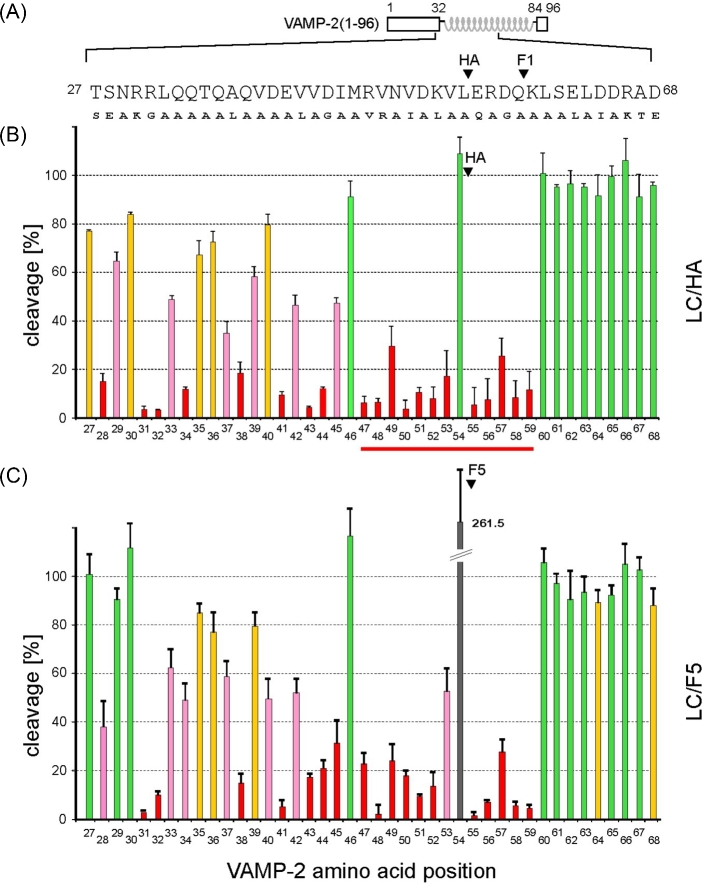
In order to identify VAMP-2 residues that are important for interactions with LC/HA or LC/F5, we performed a systematic mutagenesis study of VAMP-2 focusing on residues 27 to 68. We used the TeNT-insensitive VAMP (TI-VAMP; also known as VAMP-7), which is not hydrolyzed by clostridial neurotoxins, as a guide when designing VAMP-2 mutants (Galli et al.1998). VAMP-2 residues that are identical to TI-VAMP were substituted by alanine; residues Q33, Q34, V43, Q58, K59 and S61 were mutated to alanine; A37 was mutated to leucine, and all other VAMP-2 residues in this region were mutated to the corresponding residues in TI-VAMP. The same mutagenesis strategy has been successfully applied to study other VAMP-2-cleaving BoNT serotypes such as BoNT/B, D, F1 and TeNT and reported in our earlier work (Sikorra et al. 2006, 2008).
The VAMP-2 variants were produced as radiolabeled molecules by in vitro transcription/translation and incubated for 1 h with LC/HA or LC/F5. The percentage of cleavage of the VAMP-2 mutants versus that of the wild-type VAMP-2 was calculated. The amino acid substitutions that resulted in at least 33% diminished cleavage rate were considered critical for substrate cleavage. We observed significant effects when VAMP-2 residues between S28 and K59 were mutated, which include residues around the SNARE secondary recognition motif (SSR motif) V1 (residues 39–47), but not the V2 (residues 62–71) (Rossetto et al.1994). This finding suggests that LC/HA and LC/F5 require a substrate segment spanning 32 amino acids surrounding the cleavage site for optimal cleavage.
Within this 32 amino acid segment in VAMP-2, 26 or 25 amino acids were found to be crucial for LC/HA or LC/F5 cleavage, respectively, as mutating these residues led to at least 33% decreased VAMP-2 cleavage (Fig. 2). In contrast to these findings on LC/HA and LC/F5, other VAMP-2 cleaving BoNTs seem to have higher tolerance to side-chain mutations in their equivalent VAMP-2 segments. For example, the numbers of amino acids in this segment that, if mutated, caused more than 33% in VAMP-2 cleavage are 14 for LC/F1 (Sikorra et al.2008; Chen and Wan 2011), 7 for LC/B, 9 for LC/D and 11 for TeNT-LC (Chen, Hall and Barbieri 2008; Sikorra et al.2008). These findings suggest that LC/HA and LC/F5 require more on side chain-mediated interactions to achieve optimal VAMP-2 binding and specific cleavage.
Figure 2.
Cleavage analysis of various VAMP-2 point mutants. (A) Schematic representation of VAMP-2 and the mutations introduced in a segment between residues T27 to D68. The scissile peptide bonds are marked for LC/HA and LC/F1. (B, C) Cleavage assays. His6-VAMP-2 (1–96) variants were radiolabeled by in vitro transcription/translation and incubated for 1 h in the presence of 50 nM LC/HA, 100 nM LC/F5. Samples were analyzed by Tris/Tricine-PAGE using 15% gels. Columns represent percentages of cleavage caused by the mutants versus the wild-type VAMP-2. Data represent means ± SD of at least four independent experiments. The color code applied to the columns is as follows: green, no or less than 10% reduction of cleavability; yellow, >10%; pink, >33%; red, >66%. A peptide segment between R47 and K59 that may adopt an extended conformation is highlighted with a red line.
Notably, except for the P1 residue L54, residues R47 to K59 spanning the cleavage site are all essential for LC/HA and LC/F5 hydrolysis, as any mutations in this segment strongly inhibited the cleavage of VAMP-2 (Fig. 2). This finding argues against an earlier model suggesting that residues 47–50 in VAMP-2 adopted a helical conformation when bound by LC/F5 (Guo, Chan and Chen 2016). If this region adopts a helical conformation, one would expect that VAMP-2 residues facing away from LC/F5 would not have much effect on VAMP-2 cleavage when mutated. Taken together, we suggest that residues R47 to K59 spanning the cleavage site in VAMP-2 likely adopt an extended conformation when interacting with LC/HA and LC/F5 (Fig. 4B and C).
Figure 4.
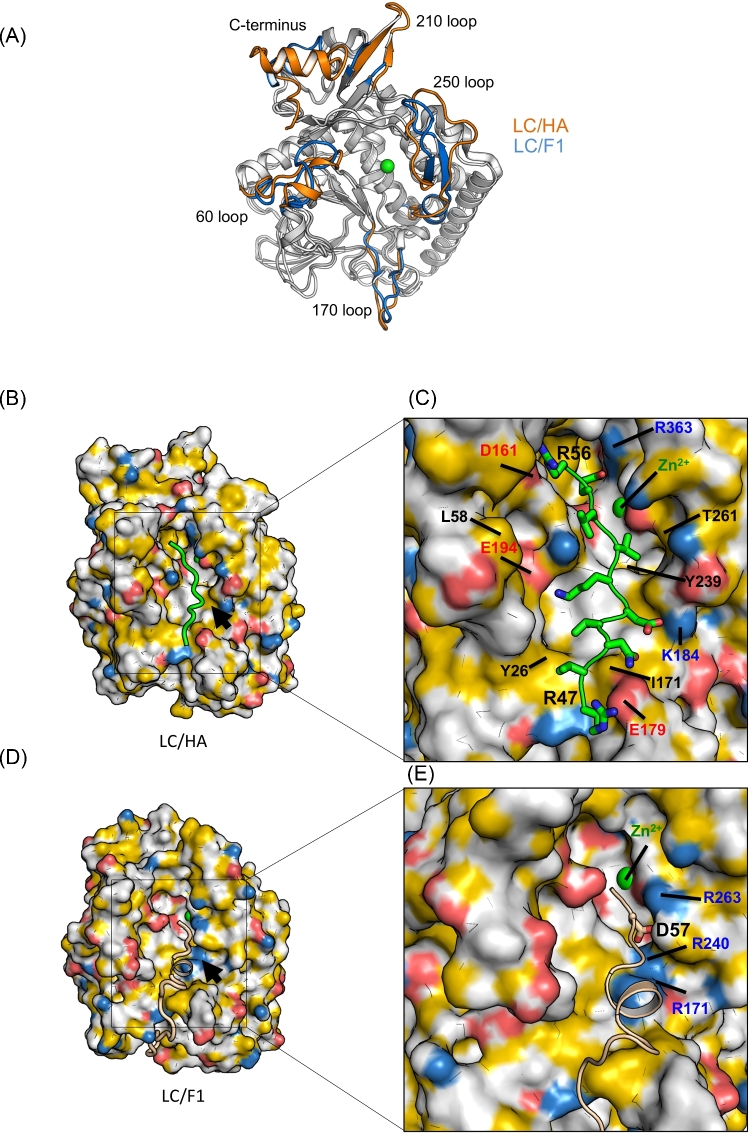
Structural comparison between LC/HA and LC/F1. (A) Structural superposition between LC/HA and LC/F1, and areas that show different conformations are colored in orange for LC/HA and blue for LC/F1. (B) A structure model of the LC/HA-VAMP-2 (R47 – K59) complex. Atoms are colored to highlight hydrophobicity features (Hagemans et al.2015): carbon atoms not bound to oxygen or nitrogen atoms are colored orange, nitrogen atoms carrying positive charges in arginine and lysine are blue, oxygen atoms carrying negative charges in glutamate and aspartate are red, and all remaining atoms are white. A structural model of VAMP-2 is colored green. (C) Close-up view of the LC/HA-VAMP-2 binding surface. Key residues of LC/HA predicted to interact with VAMP-2 are labeled. (D) The structure of LC/F1-VAMP-2 complex (PDB code: 3FIE). The color scheme for LC/F1 is the same as that shown in panel (B), and the VAMP-2 peptide is shown as a wheat cartoon loop. Black arrows mark an electropositive VAMP-2-binding pocket in LC/F1, whereas the corresponding surface in LC/HA displayed different charge property. (E) Close-up view of the LC/F1-VAMP-2 binding surface. Residues that contribute to the unique electropositive surface patch in LC/F1 are labeled.
LC/HA and LC/F5 share a high sequence identity (∼80%) and cleave the same scissile peptide bond in VAMP-2 (Maslanka et al.2016). Interestingly, LC/HA displays a ∼4-fold higher substrate hydrolysis efficiency (kcat/KM) than LC/F5, even though they have a similar turnover number (kcat) towards VAMP-2 (Table 2). Our mutagenesis data reveal that several VAMP-2 residues that are distant to the cleavage site (e.g. T27-R30, Q34) showed stronger effects on LC/HA cleavage than LC/F5 when mutated (Fig. 2). For instance, S28E and Q34A mutants reduced the VAMP-2 cleavability by ∼2-fold and ∼3-fold, respectively, in LC/HA comparing with LC/F5. These exosite residues may contribute to the higher affinity (lower KM) of LC/HA towards VAMP-2, because earlier studies suggest that mutations at the exosite affect substrate-binding affinity (Sikorra et al.2008).
Table 2.
Kinetic constants of LC/HA and LC/F5.
| KM (μM) | kcat (1/min) | kcat/KM (1/μM min) | |
|---|---|---|---|
| LC/HA | 253.5 ± 15.4 | 1038 ± 114 | 4.1 |
| LC/F5 | 883 ± 109 | 792 ± 258 | 0.90 |
Since the crystal structure of LC/F5 is not available, we generated a model of LC/F5 using LC/HA as a template for further structural analysis. As expected, the overall electrostatic features of these two LCs around the active site are very similar (Fig. 3A). However, they exhibit distinct surface charge distribution in a potential VAMP-2 exosite-binding pocket, which is predicted based on the structure of the LC/F1-VAMP-2 complex (Agarwal et al.2009). This area bears more negative charges in LC/HA, but more positive charge in LC/F5 (Fig. 3B). We speculate that this difference may contribute to the different affinity of LC/HA and LC/F5 towards VAMP-2.
Figure 3.
Electrostatic surfaces of LC/HA, LC/F1 (PDB code: 3FIE) and a model of LC/F5. The molecular surfaces are shown and colored by the electrostatic potential with a contour +3 kBT/e (blue) and –3 kBT/e (red). Electrostatic surfaces were calculated using the software APBS (Baker et al.2001). A VAMP-2-based peptide inhibitor in complex with LC/F1 is shown as a wheat cartoon loop (Agarwal et al.2009). The top panel (A) and bottom panel (B) are related by a rotation of 180° along the y-axis. Arrows mark a predicted exosite-binding pocket (green) and the cleavage sites pockets that accommodate VAMP-2 residues N-terminal (yellow) or C-terminal (cyan) to the cleavage site.
Remarkably, we found that mutating the P1 residue L54 to Ala enhanced the VAMP-2 cleavability by ∼2.5 fold in LC/F5, but had no effect on LC/HA. The increased VAMP-2 cleavage by LC/F5 is unexpected because earlier studies show that mutations at P1 position usually do not significantly change the VAMP-2 cleavability of other BoNT LCs (Vaidyanathan et al.1999; Jin et al.2007; Sikorra et al.2008). This finding thus suggests that the P1 residue L54 probably is not directly involved in positioning the scissile peptide bond in the active sites of LC/F5, and it might even impose a steric hindrance for VAMP-2 binding to LC/F5.
Structural comparison between LC/HA and LC/F1
LC/HA shows a high-sequence identity to LC/F1 (∼46%), but they cleave VAMP-2 at two distinct sites four residues away from each other. To better understand the unique VAMP-2 cleavage property of LC/HA, we carried out detailed structural comparison between LC/HA and a structure of LC/F1 in complex with a VAMP-2-based peptide inhibitor (VAMP 22–58/Q58D-cysteine) (Agarwal et al.2009).
The overall structures of LC/HA and LC/F1 are very similar, but LC/HA adopts different conformations in several surface loops, including the 60, 170, 210 and 250 loops and the C-terminus (Fig. 4A). The conformations of the 60, 170 and 250 loops in LC/HA may be affected by crystal packing because they interact with a neighboring symmetric LC/HA molecule, but the 210 loop and the C-terminal helix of LC/HA do not. In LC/HA, the 60 loop, 210 loop and the C-terminal helix are more structured, while the 170 loop and 250 loop show higher structure flexibility. In particular, the 250 loop of LC/HA is largely unstructured while the corresponding loop forms an anti-parallel β-hairpin in LC/F1, LC/A and LC/C (Breidenbach and Brunger 2004; Jin et al.2007; Agarwal et al.2009). It is known that the 60 and 250 loops in LC/A1 bind SNAP-25 at a site C-terminal to the cleavage site, and the 170 loop in LC/F1 binds VAMP-2 at a site N-terminal to the cleavage site (Agarwal et al.2009). Therefore, these unique conformations observed in LC/HA may indicate different VAMP-2 binding modes between LC/HA and LC/F1, which may partly contribute to their preference for different VAMP-2 cleavage sites.
We next examined the surface electrostatic properties of BoNT/HA and BoNT/F1. One of the major differences is that LC/F1 has a more electropositive surface around the active site pocket that accommodates VAMP-2 residues located N-terminal to the cleavage site, while the corresponding pocket in LC/HA is hydrophobic (indicated by yellow arrows in Fig. 3A and black arrows in Fig. 4B and D). Structure-based sequence alignment between LC/F1 and LC/HA revealed that several charged residues of LC/F1, such as R171, R240 and R263, are replaced by I171, Y239 and T261 in LC/HA, respectively (Fig. 4B–E). It is worth noting that R240 and R263 in LC/F1 form key salt bridges with the P2 residue D57 of VAMP-2, which play a crucial role in LC/F1 catalyzed hydrolysis (Agarwal et al.2009). Therefore, these amino acid variations around the active site may contribute to the different scissile bond specificities between LC/HA and LC/F1 (Chen and Barbieri 2009).
Another major difference is observed at a VAMP-2-binding pocket that interacts with VAMP-2 residues located C-terminal to the cleavage site, which is electronegative in LC/HA but more hydrophobic in LC/F1 (indicated by cyan arrows in Fig. 3A). Indeed, this charged pocket in LC/HA seems to match well with P1’-P5’ residues of VAMP-2, which are largely charged/polar and all are essential for optimal LC/HA activity (Fig. 2). A subtle mutation of P1’ residue E55 of VAMP-2 to Gln was sufficient to cause a >90% decrease in cleavability by LC/HA. On the other hand, LC/F1 prefers a hydrophobic P2’ residue (L60), which is needed for optimal interactions with the corresponding hydrophobic surface in LC/F1 (Chen and Wan 2011). Interestingly, we noticed that this specific substrate-binding region in LCs that accommodates amino acids C-terminal to the cleavage site usually forms a deep pocket in almost all known LC structures. It therefore would be a good binding site for a small molecule or a peptide inhibitor (Fig. 3A). For example, a group of peptide inhibitors that bind to a similar site in LC/A1 (termed the β-exosite) have been previously reported (Zuniga et al.2008; Kumaran et al.2015).
Based on our biochemical and structural analyses, we propose a model on how VAMP-2 (P8 – P2’) might bind LC/HA near the active site pocket. We used the structure of VAMP-2 (22–58) in complex with LC/F1 as a starting model (Agarwal et al.2009). In the LC/F1-VAMP-2 complex, the P8 – P5 residues (D51 – L54) of VAMP-2 adopt a helical conformation, but our mutagenesis data suggest a more extended conformation in this segment when it interacts with LC/HA (Fig. 2). Therefore, we manually modified P8–P5 residues (R47 – V50) into an extended conformation. The modified VAMP-2 (P8 – P2’) was submitted to Rosetta FlexPepDock web server for high-resolution peptide docking, which optimizes the peptide backbones and rigid body orientation using the Monte-Carlo with the minimization approach (London et al.2011). In the optimized structure model (Fig. 4B and C), VAMP-2 V53 interacts with Y239 of LC/HA; V48 and V50 are pointing to Y26; and D51, K52, R47 and N49 of VAMP-2 interact electrostatically with K184, E194 and E179 of LC/HA. C-terminal to the active site, E55 and R56 of VAMP-2 form salt bridges with R363 and D161 of LC/HA, respectively. This novel structural model will guide our future studies of the VAMP-2 cleavage mechanism for LC/HA.
CONCLUSION
In summary, we present here the first comprehensive structural and biochemical analysis of the substrate-binding mechanism of LC/HA. Our findings demonstrate that LC/HA recognizes VAMP-2 in a way that is distinct from other VAMP-2-specific BoNTs. Most notably, VAMP-2 may adopt an extended conformation in a segment surrounding the scissile peptide bond, which involves more side-chain mediated interactions with the active site of LC/HA to ensure a proper positioning of the scissile peptide bond. Furthermore, the different surface electrostatic properties of LC/HA and LC/F1 in the VAMP-2 binding groove might account for their different cleavage sites. Our structural model could facilitate the development of peptide or small molecule inhibitors targeting LC/HA. Such inhibitors against the LC of BoNT will be important for clinically relevant post-intoxication treatment, which will greatly complement the currently available treatments that use neutralizing antibodies to clear the toxins in the blood stream.
FUNDING
This work was partly supported by National Institute of Health (NIH) grants R01AI091823, R01 AI125704 and R21AI123920 to RJ and by grant BI 660/3–1 from the Deutsche Forschungsgemeinschaft to TB. NE-CAT at the Advanced Photon Source (APS) is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403). Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02–06CH11357. Coordinates and structure factors of LC/HA have been deposited in the Protein Data Bank under accession code 6BVD.
Conflict of interest. None declared.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Binz T, Swaminathan S. Structural analysis of botulinum neurotoxin serotype F light chain: implications on substrate binding and inhibitor design. Biochemistry 2005;44:11758–65. [DOI] [PubMed] [Google Scholar]
- Agarwal R, Eswaramoorthy S, Kumaran D et al. . Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212→Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway. Biochemistry 2004;43:6637–44. [DOI] [PubMed] [Google Scholar]
- Agarwal R, Schmidt JJ, Stafford RG et al. . Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat Struct Mol Biol 2009;16:789–94. [DOI] [PubMed] [Google Scholar]
- Arndt JW, Chai Q, Christian T et al. . Structure of botulinum neurotoxin type D light chain at 1.65 A resolution: repercussions for VAMP-2 substrate specificity. Biochemistry 2006;45:3255–62. [DOI] [PubMed] [Google Scholar]
- Arndt JW, Yu W, Bi F et al. . Crystal structure of botulinum neurotoxin type G light chain: serotype divergence in substrate recognition. Biochemistry 2005;44:9574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S et al. . Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 2001;98:10037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz T, Sikorra S, Mahrhold S. Clostridial neurotoxins: mechanism of SNARE cleavage and outlook on potential substrate specificity reengineering. Toxins 2010;2:665–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 2004;432:925–9. [DOI] [PubMed] [Google Scholar]
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992;355:472–5. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Jin R, Breidenbach MA. Highly specific interactions between botulinum neurotoxins and synaptic vesicle proteins. Cell Mol Life Sci 2008;65:2296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt J, Carter AT, Stringer SC et al. . Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS Lett 2018;592:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Henchal EA, Schmaljohn AL et al. . The evolving field of biodefence: therapeutic developments and diagnostics. Nat Rev Drug Discov 2005;4:281–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Barbieri JT. Engineering botulinum neurotoxin to extend therapeutic intervention. Proc Natl Acad Sci USA 2009;106:9180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Hall C, Barbieri JT. Substrate recognition of VAMP-2 by botulinum neurotoxin B and tetanus neurotoxin. J Biol Chem 2008;283:21153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wan HY. Molecular mechanisms of substrate recognition and specificity of botulinum neurotoxin serotype F. Biochem J 2011;433:277–84. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ et al. . MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 2010;66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover N, Barash JR, Hill KK et al. . Molecular characterization of a novel botulinum neurotoxin type H gene. J Infect Dis 2014;209:192–202. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- Fan Y, Barash JR, Lou J et al. . Immunological characterization and neutralizing ability of monoclonal antibodies directed against botulinum neurotoxin type H. J Infect Dis 2016;213:1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T, Zahraoui A, Vaidyanathan VV et al. . A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol Biol Cell 1998;9:1437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chan EWC, Chen S. Mechanism of substrate recognition by the novel botulinum neurotoxin subtype F5. Sci Rep 2016;6:19875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemans D, van Belzen IAEM, Morán Luengo T et al. . A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci 2015;2:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010;38:W545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Sikorra S, Stegmann CM et al. . Structural and biochemical studies of botulinum neurotoxin serotype C1 light chain protease: implications for dual substrate specificity. Biochemistry 2007;46:10685–93. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb SR, Baudys J, Smith TJ et al. . Three enzymatically active neurotoxins of Clostridium botulinum strain Af84: BoNT/A2, /F4, and /F5. Anal Chem 2014;86:3254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran D, Adler M, Levit M et al. . Interactions of a potent cyclic peptide inhibitor with the light chain of botulinum neurotoxin A: Insights from X-ray crystallography. Bioorg Med Chem 2015;23:7264–73. [DOI] [PubMed] [Google Scholar]
- Kumaran D, Rawat R, Ludivico ML et al. . Structure- and substrate-based inhibitor design for Clostridium botulinum neurotoxin serotype A. J Biol Chem 2008;283:18883–91. [DOI] [PubMed] [Google Scholar]
- London N, Raveh B, Cohen E et al. . Rosetta FlexPepDock web server—high resolution modeling of peptide–protein interactions. Nucleic Acids Res 2011;39:W249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslanka SE, Lúquez C, Dykes JK et al. . A novel botulinum neurotoxin, previously reported as serotype H, has a hybrid-like structure with regions of similarity to the structures of serotypes A and F and is neutralized with serotype A antitoxin. J Infect Dis 2016;213:379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano S, Montecucco C. The blockade of the neurotransmitter release apparatus by botulinum neurotoxins. Cell Mol Life Sci 2014;71:793–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck M, Smith T, Anniballi F et al. . Historical perspectives and guidelines for botulinum neurotoxin subtype nomenclature. Toxins 2017;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzini M, Rossetto O. Challenges in searching for therapeutics against botulinum neurotoxins. Expert Opin Drug Dis 2017;12:497–510. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M et al. . A graphical user interface to the CCP4 program suite. Acta Crystallogr D 2003;59:1131–7. [DOI] [PubMed] [Google Scholar]
- Rao KN, Kumaran D, Binz T et al. . Structural analysis of the catalytic domain of tetanus neurotoxin. Toxicon 2005;45:929–39. [DOI] [PubMed] [Google Scholar]
- Rossetto O, Megighian A, Scorzeto M et al. . Botulinum neurotoxins. Toxicon 2013;67:31–36. [DOI] [PubMed] [Google Scholar]
- Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat Rev Microbiol 2014;12:535–49. [DOI] [PubMed] [Google Scholar]
- Rossetto O, Schiavo G, Montecucco C et al. . SNARE motif and neurotoxins. Nature 1994;372:415–6. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev 2000;80:717–66. [DOI] [PubMed] [Google Scholar]
- Sikorra S, Henke T, Galli T et al. . Substrate recognition mechanism of VAMP/synaptobrevin-cleaving clostridial neurotoxins. J Biol Chem 2008;283:21145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorra S, Henke T, Swaminathan S et al. . Identification of the amino acid residues rendering TI-VAMP insensitive toward botulinum neurotoxin B. J Mol Biol 2006;357:574–82. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol 2000;7:693–9. [DOI] [PubMed] [Google Scholar]
- Turton K, Chaddock JA, Acharya KR. Botulinum and tetanus neurotoxins: structure, function and therapeutic utility. Trends Biochem Sci 2002;27:552–8. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan VV, Yoshino K, Jahnz M et al. . Proteolysis of SNAP-25 isoforms by botulinum neurotoxin types A, C, and E. J Neurochem 1999;72:327–37. [DOI] [PubMed] [Google Scholar]
- Yang Z, Lasker K, Schneidman-Duhovny D et al. . UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol 2012;179:269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Lam K, Perry K et al. . Crystal structure of the receptor-binding domain of botulinum neurotoxin type HA, also known as type FA or H. Toxins 2017;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lebreton F, Mansfield MJ et al. . Identification of a botulinum neurotoxin-like toxin in a commensal strain of Enterococcus faecium. Cell Host Microbe 2018;23:169–176.e6e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Masuyer G, Zhang J et al. . Identification and characterization of a novel botulinum neurotoxin. Nat Commun 2017;8:14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga JE, Schmidt JJ, Fenn T et al. . A potent peptidomimetic inhibitor of botulinum neurotoxin serotype A has a very different conformation than SNAP-25 substrate. Structure 2008;16:1588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]