Abstract
3′ end cleavage of metazoan replication-dependent histone pre-mRNAs requires the multi-subunit holo-U7 snRNP and the stem–loop binding protein (SLBP). The exact composition of the U7 snRNP and details of SLBP function in processing remain unclear. To identify components of the U7 snRNP in an unbiased manner, we developed a novel approach for purifying processing complexes from Drosophila and mouse nuclear extracts. In this method, catalytically active processing complexes are assembled in vitro on a cleavage-resistant histone pre-mRNA containing biotin and a photo-sensitive linker, and eluted from streptavidin beads by UV irradiation for direct analysis by mass spectrometry. In the purified processing complexes, Drosophila and mouse U7 snRNP have a remarkably similar composition, always being associated with CPSF73, CPSF100, symplekin and CstF64. Many other proteins previously implicated in the U7-dependent processing are not present. Drosophila U7 snRNP bound to histone pre-mRNA in the absence of SLBP contains the same subset of polyadenylation factors but is catalytically inactive and addition of recombinant SLBP is sufficient to trigger cleavage. This result suggests that Drosophila SLBP promotes a structural rearrangement of the processing complex, resulting in juxtaposition of the CPSF73 endonuclease with the cleavage site in the pre-mRNA substrate.
INTRODUCTION
In metazoans, 3′ end processing of replication-dependent histone pre-mRNAs occurs through a single endonucleolytic cleavage, generating mature histone mRNAs that lack a poly(A) tail (1–3). This specialized 3′ end processing reaction depends on the U7 snRNP, the core of which consists of a ∼60-nt U7 snRNA (4–6) and a unique heptameric Sm ring. In the ring, the spliceosomal subunits SmD1 and SmD2 are replaced by the related Lsm10 and Lsm11 proteins (7,8), whereas the remaining subunits (SmB, SmD3, SmE, SmF and SmG) are shared with the spliceosomal snRNPs.
Lsm11 contains an extended N-terminal region that interacts with the N-terminal region of the 220 kDa protein FLASH (9,10). Together, they recruit a specific subset of the proteins that participate in 3′ end processing of canonical pre-mRNAs by cleavage and polyadenylation (11), resulting in formation of the holo-U7 snRNP (12). This subset of polyadenylation factors is referred to as the histone pre-mRNA cleavage complex (HCC) and in mammalian nuclear extracts includes symplekin, all subunits of CPSF (CPSF160, WDR33, CPSF100, CPSF73, Fip1 and CPSF30) and CstF64 as the only CstF subunit (13). The remaining components of the cleavage and polyadenylation machinery, including CstF50 and CstF77, the two CF Im subunits of 68 and 25 kDa, and the two subunits of CF IIm (Clp1 and Pcf11) were consistently absent in the HCC (13). A similar subset of polyadenylation factors is associated with the Drosophila holo-U7 snRNP (14).
The substrate specificity in the processing reaction is provided by the U7 snRNA, which through its 5′ terminal region base pairs with the histone downstream element (HDE), a sequence in histone pre-mRNA located downstream of the cleavage site (15). This interaction is assisted by the stem–loop binding protein (SLBP), which binds the highly conserved stem–loop structure located upstream of the cleavage site (16–18) and stabilizes the complex of U7 snRNP with histone pre-mRNA (19–21), likely by contacting FLASH and Lsm11 (12). In mammalian nuclear extracts, histone pre-mRNAs that form a strong duplex with the U7 snRNA are cleaved efficiently in the absence of SLBP (19–21). In contrast, Drosophila nuclear extracts lacking SLBP are inactive in cleaving histone pre-mRNAs, suggesting that Drosophila SLBP plays an essential role in processing in addition to stabilizing binding of the U7 snRNP to histone pre-mRNA (12,14,22,23).
Within the HCC, CPSF73 is the endonuclease (24,25), acting in a close partnership with its catalytically inactive homolog, CPSF100 (11,25,26), and the heat-labile scaffolding protein symplekin (27). RNAi-mediated depletion of these three HCC subunits in Drosophila cultured cells results in generation of polyadenylated histone mRNAs (14,28), an indication of their essential role in the U7-dependent processing. Depletion of the remaining components of the HCC had no effect on the 3′ end of histone mRNAs and their function in the U7 snRNP, if any, is less clear. Previous in vivo studies implicated multiple other proteins, in addition to SLBP and components of the U7 snRNP, in generation of correctly processed histone pre-mRNAs. These proteins include ZFP100 (29–31), CDC73/parafibromin (32,33), NELF E (34), Ars2 (35,36), CDK9 (37), CF Im68 (33) and RNA-binding protein FUS/TLS (Fused in Sarcoma/Translocated in Sarcoma) (38). ZFP100, CF Im68 and FUS were shown to interact with Lsm11, whereas Ars2 was shown to interact with FLASH, raising the possibility that they may be essential components of the cleavage machinery.
To determine which factors are required for the cleavage reaction, we developed a novel method for purification of in vitro assembled Drosophila and mouse processing complexes. In this method, histone pre-mRNAs containing biotin and a photo-cleavable linker in either cis or trans are incubated with a nuclear extract and the assembled processing complexes are immobilized on streptavidin beads, washed and released into solution by irradiation with long wave UV. This approach yielded remarkably pure processing complexes that were suitable for direct and unbiased analysis by mass spectrometry, providing a complete view of the holo-U7 snRNP and other proteins that associate with histone pre-mRNA for 3′ end processing.
MATERIALS AND METHODS
RNAs
RNA substrates and oligonucleotides were generated by T7 transcription or synthesized by GE Dharmacon (Lafayette, CO, USA), as listed below. All sequences are written in 5′-3′ orientation. The stem-loop and the purine-rich core of the HDE (GAGA or AGAG) are bold and underlined, the cleavage site is indicated with a semicolon and the 19-nt sequence complementary to the pcB/22-mer is underlined. Other symbols are as follows: pc (photo-cleavable linker), m (2′O-methyl modification), 18S (18 atom spacer).
-
- dH3 Ext pre-mRNA (T7 transcription), 125 nt:
GGCGAAUUCGAGCUCGGUACCAAAAAGGCUCUUUUCAGAGCCACCC;ACUGAAUUCAAUGAGAUAAAAUUUUCUGUUGCCGACUAUUUAUAACUUUAAAAAGCCGGAGUAGGCUCGAGUGUAAGCU
-
- dH3/21bp (T7 transcription), 125 nt:
GGCGAAUUCGAGCUCGGUACCAAAAAGGCUCUUUUCAGAGCCACCC;ACUGAAUUCAAAGAGAAUAAAAAUUUUCAAGCCGACUAUUUAUAACUUUAAAAAGCCGGAGUAGGCUCGAGUGUAAGCU
-
- 3′Biot-dH3/2m pre-mRNA(GE Dharmacon), 65-mer:
UAUAAUCGGUCCUUUUCAGGACCACAA;ACCAGAUUmCmAAUGAGAUAAAAUUUUCUGUUGCCAGCGA/18S/18S/3′Biot
-
- pcB-dH3/5m pre-mRNA (GE Dharmacon), 63 nt:
5′Biot/pc/18S/18S/CGUACUCUAUAAUCGGUCCUUUUCAGGACCACmAmAmAmCmCAGAUUCAAUGAGAUAAAAUUU UCUGU
-
- H2a-614 pre-RNA (T7 transcription), 85 nt,
GGCGAAUUCGAGCUCGGUACCAAAAAGGCUCUUUUCAGAGCCACCCA;CUGAAUCAGAUAAAGAGUUGUGUCACGGUAGCCAAGCU
-
- 3′Biot-mH2a/2m pre-mRNA (GE Dharmacon), 64 nt:
CAAAAGGCUCUUUUCAGAGCCACCCA;CUGAmAmUCAGAUAAAGAGCUGUGACACGGUAGCCGGUCU/18S/18S/3′Biot
-
- 3′Biot-mH2a/5m pre-mRNA (GE Dharmacon), 64 nt:
CAAAAGGCUCUUUUCAGAGCCACmCmCmAmCmUGAAUCAGAUAAAGAGUUGUGUCACGGUAGCCGGUCU/18S/18S/3′Biot
-
- pcB-mH2a/5m pre-mRNA (GE Dharmacon), 61 nt:
5′Biot/pc/18S/18S/CUCCCAAAAAGGCUCUUUUCAGAGCCACmCmCmAmCmUGAAUCAGAUAAAGAGCUGUGACACGGUA
-
- pcB/22mer adaptor oligonucleotide (GE Dharmacon), 22 nt:
5′Biot/pc/18S/18S/mAmGmUmAmGmCmUmUmAmCmAmCmUmCmGmAmGmCmCmUmAmC
SL RNA and oligonuclotides complementary to the 5′ end of mouse and Drosophila U7 snRNA were described previously (12,14,23).
Antibodies
Antibodies against human/mouse SLBP, Lsm11 and FLASH, and Drosophila FLASH, CstF64, symplekin, CPSF73 and CPSF100 were generated in rabbits using peptides or bacterially-expressed protein fragments, as described previously (9,14,16,39). Antibodies against Drosophila Lsm10 and Lsm11 (40,41) were kindly provided by J. Gall (Carnegie Institution for Science). Antibodies against mouse polyadenylation factors were from Bethyl Laboratories (Montgomery, TX, USA).
Cell culture and nuclear extract preparation
Mouse myeloma cells were grown in suspension to ∼1 × 106 cells/ml in Joklik medium containing 10% heat inactivated horse serum by Cell Culture Company (Minneapolis, MN, USA). The cells were shipped overnight on wet ice as a concentrated suspension in the same medium and used the following day for preparation of a nuclear extract, as described (42). Kc cells were grown in suspension to ∼2 × 106 cells/ml in Drosophila D-22 medium (US Biologicals), spun down and used for preparation of a nuclear extracts without prior freezing, as described (14,22).
RNA labeling and in vitro processing
Synthetic histone pre-mRNAs lacking phosphate at the 5′ end were labeled with 32P using T4 polynucleotide kinase (New England Biolabs). Polymerase T7-generated histone pre-mRNAs prior to labeling were treated with calf intestinal phosphatase (New England Biolabs) to remove the 5′ triphosphate. Processing in mouse and Drosophila nuclear extracts was carried out as previously described (10,22).
Purification of mouse and Drosophila processing complexes on histone pre-mRNAs containing covalently attached biotin and the photocleavable group (cis configuration)
Mouse and Drosophila processing complexes were assembled on pcB-mH2a/5m (61 nt) and pcB-dH3/5m (63 nt) pre-mRNAs, respectively. In each pre-mRNA, biotin and the photocleavable linker were placed at the 5′ end and the two groups were separated from the RNA sequence by two 18-atom spacers (18S). Both pre-mRNAs contain five 2′O-methyl-modified nucleotides around the cleavage site and hence are resistant to processing. The assembly of processing complexes and their immobilization on streptavidin beads (Sigma) were carried out as described previously (12–14), with minor modifications. Typically, 750 μl of mouse or Drosophila nuclear extracts supplemented with 250 μl of 80 mM ethylenediaminetetraacetic acid (EDTA) pH 8 (20 mM final EDTA concentration) were mixed with 75 ng of pcB-mH2a/5m or pcB-dH3/5m pre-mRNAs, respectively and the samples incubated for 5 min at either 32°C (mouse extracts) or 22°C (Drosophila extracts) to assemble processing complexes. All mouse nuclear extracts were supplemented with bacterially expressed N-terminal FLASH (amino acids 53–138) to a final concentration of 25 ng/μl (12). Note that in mouse and Drosophila nuclear extracts U7 snRNP is present at low concentrations and increasing the amount of the pre-mRNA substrate results in higher background of non-specific RNA binding proteins without significantly improving the yield of the U7-dependent processing complexes.
Following 5 min incubation, each sample was briefly cooled down on ice and spun 10 min at 10 000 rpm in a microcentrifuge to remove potential precipitates. The cleared supernatant was loaded over 35–40 μl of streptavidin beads (thoroughly rinsed with a processing buffer containing 15 mM HEPES-KOH pH 7.9, 75 mM KCl, 20 mM EDTA pH 8, 15% glycerol) and rotated at 4°C for 75 min. Streptavidin beads containing the immobilized complexes were gently spun down (3 min at 300 rpm in cold room), briefly rinsed twice with 1 ml of the processing buffer and left rotating in cold room in the same buffer for 75 min. The beads were transferred to a new 1.5 ml tube, rotated in the same buffer for an additional 75 min, transferred to a 0.5 ml tube and resuspended in 100 μl of the processing buffer. The tube containing the immobilized processing complexes was placed horizontally on ice and irradiated 30 min from a distance of 10 mm with a high intensity UV lamp (UVP Blak-Ray B-100AP) containing 100W spot bulb emitting 365 nm UV light (Cole-Parmer). The samples were frequently inverted and vortexed to ensure even exposure of the beads to UV and to avoid excessive heating. Following UV irradiation, the samples were spun down and the supernatant carefully collected and re-spun to remove residual beads.
Purification of Drosophila processing complexes on a duplex of histone pre-mRNA and pcB/22mer oligonucleotide (biotin and the photocleavable group provided in trans configuration)
Drosophila dH3 Ext pre-mRNA (125 nt, 38 kDa) was generated by T7 RNA polymerase and annealed to ∼5-fold molar excess of the chemically synthesized pcB/22mer (all 22 nt modified with 2′O-methyl groups, 8.5 kDa) containing biotin and the photocleavable group at the 5′ end. The duplex of uninterrupted 19 bp that forms at the 3′ end of the dH3 Ext pre-mRNA upon annealing with the pcB/22mer oligonucleotide is relatively strong and the 5-fold molar excess of the oligonucleotide proved sufficient to bind ∼70% of the pre-mRNA to streptavidin beads. Note that using too high molar excess of pcB/22mer may increase the background of non-specific RNA binding proteins that interact with the oligonucleotide. For other duplexes that might be used, optimal molar ratios of the RNA and the complementary oligonucleotide need to be determined experimentally. RNAs generated by T7 polymerase can be labeled at the 5′ end and the efficiency of annealing to complementary oligonucleotides can be conveniently monitored by measuring the amount of radioactivity that remains bound to streptavidin beads.
For a single assembly experiment with a Kc nuclear extract, 150 ng of dH3 Ext pre-mRNA and the same amount of pcB/22mer (∼5-fold molar excess) were combined in 100 μl of the processing buffer (see above) and the mixture was boiled for 5 min and slowly cooled down to room temperature (1.5–2 h). If multiple assembly experiments were carried out in parallel, the RNA, oligonucleotide and the processing buffer were scaled up appropriately. In the majority of cases, the duplex was first pre-selected on streptavidin. The annealing mixture was supplemented with the processing buffer to a final volume of 1 ml and rotated 75 min with 35–40 μl of streptavidin beads. Note that this step eliminated all dH3 substrate that failed to anneal to the oligonucleotide (which hence lacked biotin and the photocleavable group) but did not reduce the amount of the free oligonucleotide, which quantitatively binds to streptavidin. The beads containing the immobilized duplex and the oligonucleotide were washed twice with 1 ml of the processing buffer (75 min each time, with one tube change) and next rotated for 75 min with 1 ml of a pre-cleared (spun 10 min at 10 000 rpm) Kc nuclear extract containing 20 mM EDTA (prepared by adding 250 ml of 80 mM EDTA to 750 ml of the extract) to assemble processing complexes. Note that since the T7-generated dH3 Ext pre-mRNA lacks 2′O-methyl nucleotides, the incubation step at room temperature was omitted to avoid excessive cleavage of the substrate. The remaining steps, including extensive washes of the beads and UV elution of the immobilized complexes, were the same as described above for histone pre-mRNAs containing biotin and the photocleavable group in cis.
In addition to pre-binding of the duplex RNA to streptavidin, we tested an alternative approach in which the annealing mixture containing the duplex RNA was directly used for the assembly of processing complexes. In this variant, the annealing mixture was added to 750 μl of a Kc nuclear extract supplemented with 250 μl of 80 mM EDTA and the resulting sample was incubated 5 min on ice to assemble processing complexes. The sample was next pre-cleared by spinning 10 min at 10 000 rpm in a micro-centrifuge to remove any potential precipitates and loaded over 35–40 μl of streptavidin beads for 75 min rotation at 4°C. The remaining steps, including extensive washes and UV elution of the immobilized complexes, were the same as described above. This approach, compared to pre-selecting the duplex RNA on streptavidin, was less time consuming but yielded slightly lower amounts of the processing complexes (a fraction of them likely formed on pre-mRNA substrate that failed to anneal to the oligonucleotide and was lost) and was used rarely.
Analysis of the UV-eluted samples by mass spectrometry
A fraction (∼15%) of the UV-eluted supernatant was directly treated with trypsin and analyzed by LC- MS/MS using Nano-Acquity LC system (Waters) and Orbitrap Velos (Thermo Scientific) mass spectrometer. In some cases, 50% of the UV eluted sample was precipitated with 80% acetone, dissolved in a small volume of sodium dodecyl sulphate (SDS) dye and loaded on 4–12% SDS/polyacrylamide gel for a brief electrophoresis allowing the samples to enter 10 mm into the gel. A small section of the gel containing the sample was excised and in gel-treated with trypsin. Mass spectrometry analysis of both types of samples was carried out as described in Supplementary Materials.
Summary of various experiments using emPAI values
emPAI (exponentially modified protein abundance index) algorithm, as implemented in the Mascot search engine, was used to estimate abundance of proteins in each sample (43). Each protein’s emPAI was normalized to the sum of emPAI for all proteins in the sample and combined for all independent experiments. The combined normalized values were averaged for the 4 mouse experiments or 12 Drosophila experiments, and the average value for each protein in the negative samples (prepared in the presence of αU7 oligonucleotide and SL RNA) was subtracted from its average value in the positive samples (containing processing complexes), to calculate ΔAVE. ΔAVE was multiplied by 100 and expressed as a percentage (ΔAVE %).
Analysis of the catalytic activity of processing complexes
Mouse or Drosophila pre-mRNA substrates containing biotin at the 3′ end were labeled at the 5′ end with 32P and incubated with mouse or Drosophila nuclear extracts for 5 min on ice to assemble processing complexes but reduce cleavage to minimum. To assemble catalytically inactive Drosophila processing complexes lacking SLBP, Kc nuclear extract contained 10 ng/μl of SL RNA and the incubation step was carried out at room temperature. Each sample contained a total of 25 ng or ∼5 × 105 cpm (counts per minute) of radiolabeled pre-mRNA. The assembled processing complexes were immobilized on streptavidin beads, extensively washed with a processing buffer (see above), re-suspended in 200 μl of the same buffer additionally containing yeast tRNA at 0.25 μg/μl and bovine serum albumin at 0.1 μg/μl and divided into small aliquots containing equal amount of the radioactive substrate. To determine processing activity of the immobilized processing complexes, one aliquot was placed at 32°C (mouse) or 22°C (Drosophila) for 60–90 min and gently agitated or rotated. Drosophila processing complexes formed in the presence of SL RNA were supplemented with baculovirus-expressed SLBP to final a concentration of 25 ng/μl. The immobilized processing complexes were boiled in the presence of 2% SDS and 0.3 M sodium acetate and total RNA containing radioactive substrate and the upstream cleavage product was recovered by phenol extraction and ethanol precipitation and analyzed by gel electrophoresis and autoradiography.
RESULTS
Pre-mRNA substrates containing 2’O-methyl-modified nucleotides around the cleavage site are resistant to cleavage
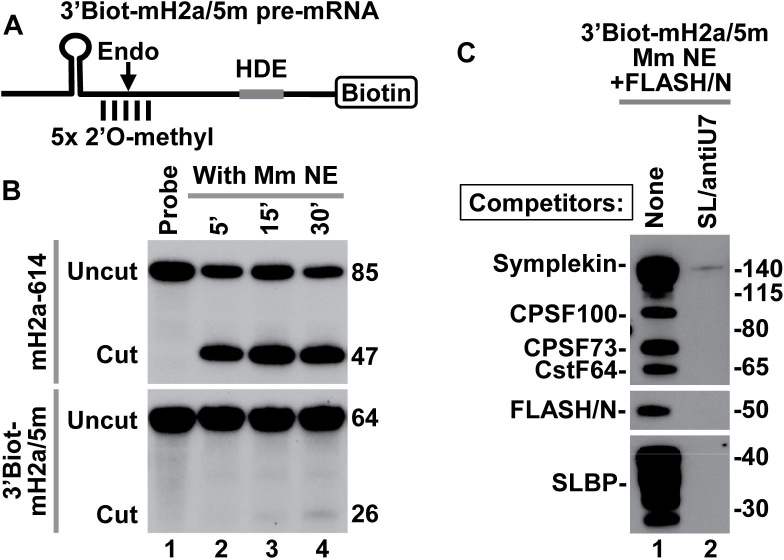
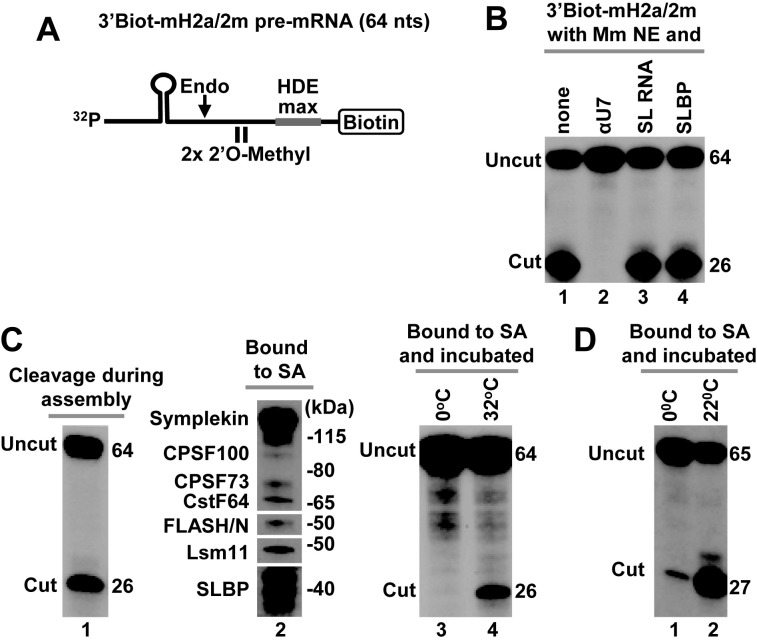
An important limitation in assembling processing complexes on histone pre-mRNAs in nuclear extracts active in processing is that even a brief incubation at low temperatures may result in significant cleavage of the substrate, reducing the amount of intact processing complexes recovered (12,14). In our previous purification experiments, we used 5′Biot-mH2a/5m pre-mRNA containing 5′ terminal biotin and five 2′O-methyl modified nucleotides (5m) around the cleavage site (12,13). A single modification of this type is sufficient to prevent 5′-3′ degradation of the downstream cleavage product of histone pre-mRNAs by CPSF73 (44). Whether a continuous cluster of 2′O-methyl-modified nucleotides in the 5′Biot-mH2a/5m pre-mRNA completely blocks endonucleolytic cleavage by CPSF73 has not been determined due to the lack of free 5′ end in this substrate for radioactive labeling. To test the effectiveness of 2′O-methyl modified nucleotides in rendering pre-mRNA substrates cleavage resistant, we used 64-nt 3′Biot-mH2a/5m pre-mRNA, a substrate similar in sequence to the 5′Biot-mH2a/5m pre-mRNA but containing biotin at the 3′ end (Figure 1A). The 3′Biot-mH2a/5m pre-mRNA was labeled at the 5′ end with 32P and incubated with a mouse nuclear extract under optimal processing conditions. After 30 min, only a trace of a 26-nt cleavage product was generated (Figure 1B, bottom, lane 4). During this incubation time, H2a-614 pre-mRNA (encoded by mouse Hist2H2aa2 gene) containing the same HDE but no modified nucleotide was processed with the efficiency exceeding 75% (Figure 1B, top, lane 4). Importantly, 3′Biot-mH2a/5m pre-mRNA readily associated with SLBP and components of the U7 snRNP (Figure 1C, lane 1), confirming that its resistance to cleavage is due to the presence of the modified groups rather than the inability to assemble into a processing complex. Note that in this mouse nuclear extract, SLBP is present as a number of N-terminal truncated proteolytic fragments that retain full activity in processing (12). As expected, processing complexes were not formed when the nuclear extract contained molar excess of stem–loop RNA (SL) to sequester SLBP, and an antisense oligonucleotide (αU7) to block U7 snRNA (Figure 1C, lane 2).
Figure 1.
3′Biot-mH2a/5m pre-mRNA is resistant to cleavage but assembles into processing complexes. (A) A schematic representation of chemically synthesized mouse-specific 3′Biot-mH2a/5m pre-mRNA (64-nt). The major cleavage site (located 5 nt downstream of the stem) and 2 nt on each side of the major cleavage site are modified with a 2′O-methyl group. Biotin is placed at the 3′ end. (B) In vitro processing of 3′Biot-mH2a/5m (bottom) and mH2a-614 (top) pre-mRNAs. mH2a-614 (85 nt) was generated by T7 transcription and contains the same HDE as 3′Biot-mH2a/5m but lacks biotin and modified nucleotides. Each pre-mRNA was labeled at the 5′ end with 32P and incubated in a mouse nuclear extract for 5, 15 and 30 min, as indicated. Probe alone is shown in lane 1. Numbers to the right indicate the length of the input pre-mRNA and the upstream cleavage product. (C) 3′Biot-mH2a/5m was incubated with a mouse myeloma nuclear extract (Mm NE) containing recombinant N-terminal FLASH (FLASH/N, amino acids 53–138) fused to GST. Assembled processing complexes were purified on streptavidin beads and analyzed by western blotting using specific antibodies (lane 1). In lane 2, formation of the processing complexes was blocked by excess SL RNA and αU7 oligonucleotide complementary to the 5′ end of mouse U7 snRNA.
Unbiased analysis of mouse U7-dependent processing complexes by mass spectrometry
In our previous studies, U7-dependent processing complexes formed in nuclear extracts were immobilized on streptavidin beads via biotin covalently attached to either 5′ or 3′ end of histone pre-mRNA (12–14). This method suffered from one major limitation: due to the unusually tight binding between biotin and streptavidin, components of the processing complexes could only be recovered from the beads by boiling in SDS, resulting in a significant contamination of the eluted samples with proteins that non-specifically bind to agarose beads. Some processing factors, particularly those with low molecular weight and/or existing in limiting amounts, could have been masked by this non-specific background, hence escaping previous detection by silver staining and mass spectrometry.
To overcome the virtually irreversible binding between biotin and streptavidin, an RNA of choice can be attached to biotin via a photo-cleavable linker and gently released into solution by irradiation with long wave UV (45). Our initial attempts with this approach that followed the recommended UV-elution protocol (5 min irradiation, 5W UV bulb, 20 mm distance) were largely unsuccessful, resulting in only a small amount of the RNA being released from streptavidin beads (not shown). However, increasing the energy of the UV source (100W bulbs) and time of irradiation (30 min) dramatically improved the yield of elution, leaving only 5–10% of the synthetic RNA on the beads (not shown).
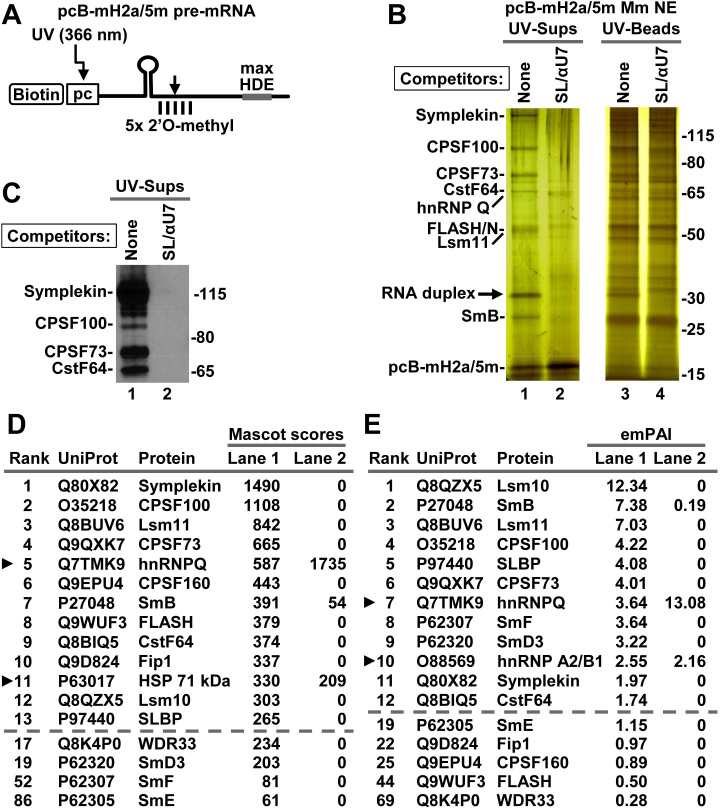
We designed a new mouse-specific histone pre-mRNA, pcB-mH2a/5m, which contained biotin (B) and the photo-cleavable (pc) linker at the 5′ end and five 2′O-methyl groups (5m) around the cleavage site (Figure 2A), and tested its suitability for purification of mouse processing complexes using the optimized UV-elution protocol. pcB-mH2a/5m pre-mRNA (75 ng) was incubated 5 min at 32°C in a mouse nuclear extract (750 μl) containing recombinant FLASH/N (9,12), and the assembled complexes were immobilized on streptavidin beads, washed and eluted from the beads by irradiation with long wave UV. In the negative control, formation of the processing complexes was blocked by addition of the αU7 and SL oligonucleotides to the nuclear extract. A fraction (∼15%) of each UV-eluted supernatant, representing only ∼100 μl of the nuclear extract, and the same fraction of the beads exposed to UV were first analyzed by silver staining. A small number of bands were visible in the UV-eluted supernatant of the sample prepared in the absence of the competitor oligonucleotides (Figure 2B, lane 1). These bands were not present in the negative sample (Figure 2B, lane 2), suggesting that they represent components of the processing complexes. The same bands were only weakly detectable in the material left on the beads, consistent with high efficiency of UV elution (Figure 2B, lane 3) and they were partially masked by multiple background proteins, underscoring the importance of the UV-elution step.
Figure 2.
Mouse nuclear proteins that bind pcB-mH2a/5m pre-mRNA. (A) A schematic representation of chemically synthesized mouse-specific pcB-mH2a/5m pre-mRNA (61 nt). Five nucleotides around the major cleavage site are modified with a 2′O-methyl group. 5′ biotin (B) is followed by a photo-cleavable (pc) linker sensitive to long wave UV (366 nm). The HDE was altered by two point mutations (max HDE) to increase its base pairing potential with the 5′ end of mouse U7 snRNA. (B) pcB-mH2a/5m was incubated with a mouse myeloma nuclear extract containing recombinant N-terminal FLASH to assemble processing complexes. In negative control, their formation was blocked by addition of SL RNA and αU7 oligonucleotide to the nuclear extract. Proteins bound to pcB-mH2a/5m pre-mRNA were immobilized on streptavidin beads and eluted by irradiation with long wave UV. Same fractions (15%) of the UV-eluted material (UV-sups) and the beads following UV-elution (UV-beads) were analyzed by silver staining. (C) A fraction of the UV-eluted material (15%) was analyzed by western blotting using selected antibodies. (D and E) A fraction (15%) of the UV-eluted material was directly analyzed by mass spectrometry and identified proteins were ranked based on their Mascot scores (panel D) or emPAI values (panel E). The top consecutive 13 and 12 hits are listed in panels D and E, respectively (shown above the dashed lines). Arrowheads indicate proteins that interacted with histone pre-mRNA in the presence of the SL RNA and αU7 oligonucleotide processing competitors. Below the dashed lines shown are the other proteins that fail to interact with histone pre-mRNA in the presence of the two competitors. Note that the emPAI value for FLASH is low since only a short fragment of the protein (amino acids 53–138) was added to the extract.
The proteins present in the processing complexes were readily identified by western blotting (Figure 2C, lane 1) and mass spectrometry of individual gel slices (not shown) as known subunits of the U7 snRNP. The identity of only one band migrating at ∼33 kDa (indicated with an arrow) was puzzling. This band was eliminated by treatment with RNase A, but unaffected by trypsin or phenol extraction (not shown), indicating that it represents an RNA. Further studies, including Northern blotting, demonstrated that the 33 kDa band represents a duplex formed between the endogenous mouse U7 snRNA and the pcB-mH2a/5m substrate (not shown).
For an unbiased and complete view of the proteome of the processing complexes, 15% of each UV-eluted supernatant was directly analyzed by mass spectrometry. The identified proteins were ranked based on the value of their Mascot scores (Figure 2D), which was calculated by adding accepted scores (higher than the threshold score) for all unique peptides matching a given protein in the searched database (46). Mascot score provides a simple and reliable way of identifying qualitative differences in protein composition between samples that contain and lack processing complexes. Among 10 highest scoring proteins identified in the sample containing processing complexes (i.e. assembled in the absence of αU7 oligonucleotide and SL RNA), 9 were known components of the holo-U7 snRNP: symplekin, CPSF100, Lsm11, CPSF73, CPSF160, SmB, FLASH, CstF64 and Fip1, and they all, with the exception of SmB, were undetectable in the sample lacking processing complexes (Figure 2D). The remaining protein among the top 10 scorers, hnRNP Q, was not eliminated by the presence of the αU7 oligonucleotide and SL RNA, indicating that its association with the pcB-mH2a/5m pre-mRNA is independent of U7 snRNA and SLBP, as previously demonstrated (12,39). Since mouse nuclear extracts were supplemented with the N-terminal FLASH to increase the amount of the holo-U7 snRNP, most FLASH peptides identified by mass spectrometry in the purified complexes were generated from this region. However, we also detected peptides from the central and C-terminal regions of the protein, consistent with a fraction of the bound U7 snRNP containing full length endogenous FLASH.
SLBP scored 13th and its binding to the pcB-mH2a/5m pre-mRNA was abolished by adding the SL RNA and αU7 oligonucleotide to the nuclear extract. Four other subunits of the U7-specifc Sm ring, Lsm10, SmD3, SmF and SmE, and one additional CPSF subunit, WDR33, were also detected only in the sample generated in the absence of the two processing competitors, and SmG was not detected in this experiment. Among the UV-eluted proteins, we also did not detect CPSF30, CstF50, CstF77 and other polyadenylation factors or proteins that were previously implicated in 3′ end processing of histone pre-mRNAs in vivo (see Supplementary Excel File 1 for Figure 2): ZFP100 (Q8BI67), Ars2 (Q99MR6), CDC73 (Q8JZM7), NELF E (P19426), CF Im68 (Q6NVF9) and FUS (P56959).
pcB-mH2a/5m pre-mRNA additionally associated with multiple other proteins but their binding to the substrate was not affected by the competitor oligonucleotides (Supplementary Excel File 1 for Figure 2). Many of these proteins, like the top scorer hnRNP Q, belong to the abundant class of hnRNPs and may non-specifically bind excess of histone pre-mRNA used in the experiment.
Since Mascot score is calculated based on the number of identified peptides, it favors large proteins and those that yield multiple peptides within the preferred mass range and amino acid composition. To eliminate this bias and to provide more quantitative information about proteins identified in the UV-eluted processing complexes, we used exponentially modified protein abundance index (emPAI), which corrects for the length and sequence variability among proteins in the analyzed sample (43,47). A hierarchy of proteins based on the emPAI values is shown for comparison in Figure 2E. All the subunits of the U7-specific ring, which are relatively small, are now ranked near the top of the list. Importantly, among the HCC subunits, CPSF73, CPSF100, CstF64 and symplekin, ranked substantially higher than CPSF160, WDR33 and Fip1, suggesting that they are more abundant in the purified U7 snRNP than the remaining CPSF subunits. Note that the FLASH is added to mouse nuclear extract as a short recombinant N-terminal protein of <10 kDa (amino acids 53–138) capable of yielding only a small number of peptides and its emPAI value (and ranking) is therefore underestimated.
Three additional mass spectrometry experiments with the pcB-mH2a/5m pre-mRNA were conducted using two different batches of mouse nuclear extract and they produced comparable results. To summarize the results from the four experiments, emPAI values for individual proteins in each experiment were first normalized, as explained in ‘Materials and Methods’. The normalized values were averaged for the four experiments (Supplementary Excel File 2), and the average score in the negative samples (prepared in the presence of αU7 oligonucleotide and SL RNA) was subtracted from the average score in the positive samples (containing processing complexes), to calculate ΔAVE. ΔAVE was multiplied by 100 and expressed as a percentage (ΔAVE %).
The summarized hierarchy of the most abundant proteins detected in the mouse processing complexes is shown in Table 1. With the exception of CPSF30, which was not detected in any of the four independent experiments, and WDR33, which ranked 22nd, all the remaining subunits of the holo-U7 snRNP and SLBP were among the top 16 proteins. The interaction of all these proteins with pcB-mH2a/5m pre-mRNA was either completely abolished or reduced by more than 95% in the presence of αU7 oligonucleotide and SL RNA (Supplementary Excel File 2). Fip1, CPSF160 and WDR33 have three to four times lower ΔAVE % values than CPSF73, CPSF100, symplekin and CstF64, suggesting that they, and CPSF30, are less abundant in the U7 snRNP and hence substoichiometric. As explained above, emPAI for FLASH is calculated for full length protein (∼2000 amino acids) and hence its ΔAVE % value is substantially underestimated.
Table 1. Summarized ranking of the most abundant proteins in mouse processing complexes.
| Rank | ID | Protein | Detected | ΔAVE (%) |
|---|---|---|---|---|
| 1 | Q8QZX5 | Lsm10 | 3 | 8.3 |
| 2 | P97440 | SLBP | 4 | 4.8 |
| 3 | Q8BUV6 | Lsm11 | 4 | 4.8 |
| 4 | P62307 | SmF | 4 | 3.6 |
| 5 | P27048 | SmB | 4 | 2.9 |
| 6 | O35218 | CPSF100 | 4 | 2.7 |
| 7 | Q9QXK7 | CPSF73 | 4 | 2.3 |
| 8 | Q8BIQ5 | CstF64 | 4 | 2.3 |
| 9 | Q80X82 | Symplekin | 4 | 2.1 |
| 10 | P62320 | SmD3 | 3 | 1.7 |
| 11 | P62309 | SmG | 2 | 1.4 |
| 12 | P62305 | SmE | 3 | 1.1 |
| 14 | Q9D824 | Fip1 | 4 | 0.7 |
| 15 | Q9EPU4 | CPSF160 | 3 | 0.5 |
| 16 | Q9WUF3 | *FLASH | 4 | 0.5 |
| 22 | Q8K4P0 | WDR33 | 4 | 0.2 |
The hierarchy is based on the value of ΔAVE (%) calculated for all four mouse experiments (see Supplementary Data for complete data and text for the method of calculation). Omitted from the list are proteins that were weakly competed (<25%) by SL RNA and anti-U7 oligonucleotide (proteins 17–20), and/or were detected in <3 experiments (proteins 13 and 21). *FLASH was added as a recombinant N-terminal fragment and its emPAI value is underestimated. CPSF30 was not detected.
Unbiased analysis of Drosophila processing complexes by mass spectrometry
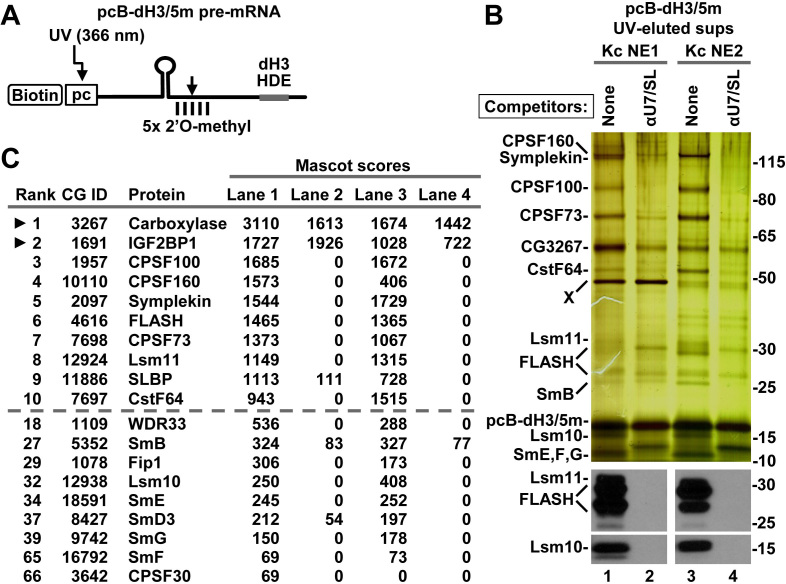
We designed a Drosophila-specific histone pre-mRNA, pcB-dH3/5m, containing both the photo-cleavable linker and five 2′O-methyl groups (Figure 3A) and used it for purification of Drosophila processing complexes. The pcB-dH3/5m pre-mRNA (75 ng) was incubated for 5 min at 22°C with two independent Kc nuclear extracts (750 μl each), both highly active in processing, and the assembled complexes were isolated and analyzed, as described above for the mouse substrate. The samples containing processing complexes were paired with matching negative controls prepared in the presence of excess SL RNA and Drosophila-specific αU7 oligonucleotide. FLASH is not limiting in Kc nuclear extracts, although in most of them it is degraded to short N-terminal fragments (detected by our N-terminal antibody) that retain full activity in processing.
Figure 3.
Drosophila nuclear proteins that bind pcB-dH3/5m pre-mRNA. (A) A schematic representation of chemically synthesized Drosophila-specific pcB-dH3/5m pre-mRNA (63 nt). (B) pcB-dH3/5m pre-mRNA was incubated with two different batches of Drosophila Kc nuclear extract in the absence or in the presence of SL RNA and αU7 oligonucleotide complementary to the 5′ end of Drosophila U7 snRNA. Proteins bound to pcB-dH3/5m pre-mRNA were immobilized on streptavidin beads and eluted by irradiation with long wave UV. A fraction of each UV-eluted supernatant was analyzed by silver staining (top panel) or western blotting (bottom panels). (C) The same fraction was also analyzed by mass spectrometry. Proteins with the 10 highest Mascot scores for Kc NE1 are shown above the dashed line, with arrowheads indicating proteins that interact with histone pre-mRNA in the presence of SL RNA and αU7 oligonucleotide. The remaining components of the U7 snRNP and their overall ranking among all identified proteins (numbering for Kc NE1) are shown below the dashed line. Note that carboxylase results from contamination of the UV eluate with a small amount of streptavidin beads.
As shown by silver staining, UV-eluted complexes formed on pcB-dH3/5m pre-mRNA contained several major bands, with only small differences being visible between the two nuclear extracts used in the experiment (Figure 3B, top panel, lanes 1 and 3). Most of these bands were either absent or barely detectable in the matching negative samples that were prepared in the presence of the competitor oligonucleotides (Figure 3B, top panel, lanes 2 and 4). Mass spectrometry of selected gel sections and western blotting (not shown) determined that the major bands represent Drosophila symplekin, CPSF100, CPSF73 and CstF64. The N-terminal degradation products of FLASH, Lsm11 and the remaining subunits of the Sm ring, including Lsm10, were visible as weakly stained bands near the bottom of the gel and their identity was confirmed by western blotting (Figure 3B, lanes 1 and 3). More complexes assembled in KC NE2, as indicated by stronger staining for the HCC subunits, FLASH and Lsm11. Incubation of the 5′pcB-dH3/5m pre-mRNA with Kc NE1 resulted in specific purification of a 50 kDa nuclear component (Figure 3B, top panel, band X), further highlighting differences between various Kc nuclear extracts. We failed to identify this component by mass spectrometry but the fact that it was present in both the positive and negative sample, and was undetectable in other Kc nuclear extract preparations suggests that it has no role in processing.
The same fraction of each UV-eluted Drosophila sample was directly analyzed by mass spectrometry (Supplementary Excel File 1 for Figure 3) and proteins with the top Mascot scores are shown in Figure 3C. Among the top 10 scoring proteins identified in the samples prepared in the absence of the competitor oligonucleotides, 6 or 7 (depending on the extract) were known components of the Drosophila U7 snRNP, including FLASH, Lsm11 and the major subunits of the HCC: symplekin, CPSF100, CPSF73 and CstF64. CPSF160 was detected with a much higher score in the complexes formed in Kc NE1. The remaining components of the HCC and the Sm ring scored among top 100 proteins, with CPSF30 being detected with a low score only in one of the two tested Kc extracts. In the UV-eluted supernatants, mass spectrometry also identified biotin-containing proteins (including the top scoring 3-methylcrotonyl-CoA carboxylase, CG3267), which are present due to contamination with a small amount of streptavidin beads, and a plethora of proteins containing known RNA binding domains (RBDs). The RNA binding proteins included insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1, CG1691), which contains four KH-type domains and was identified as a top scorer in all tested batches of the Drosophila nuclear extract. Binding of this protein to pcB-dH3/5m pre-mRNA occurs in the presence of αU7 oligonucleotide, indicating that it is not part of the Drosophila U7 snRNP and, as previously suggested (14), is unlikely to have an essential function in 3′ end processing. No peptides were identified for other components of the Drosophila cleavage and polyadenylation machinery, including orthologues of CF Im68 and the two remaining subunits of CstF, CstF50 and CstF77.
We also used silver staining and mass spectrometry analysis to compare UV-eluted processing complexes bound to pcB-mH2a/5m and pcB-dH3/5m pre-mRNAs side by side (Supplementary Figure S1). This comparison again demonstrated that mouse and Drosophila U7-dependent processing complexes have a remarkably similar composition and contain no additional, previously unidentified components (Supplementary Excel File 1 for Supplementary Figure S1).
Purification of Drosophila processing complexes via pre-mRNA attached to photo-cleavable moiety in trans
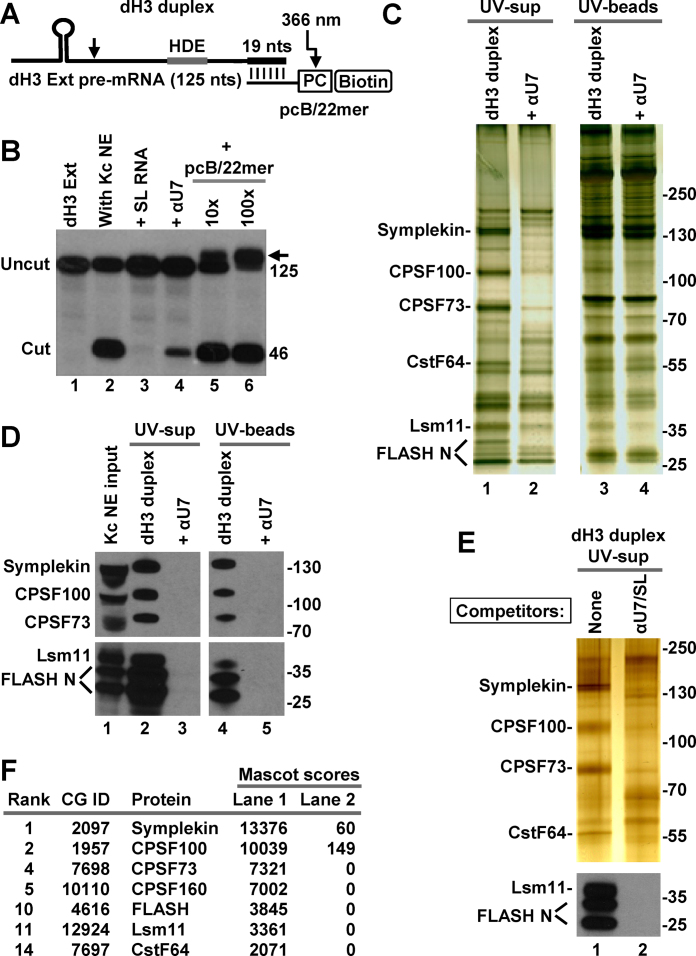
Due to limitations of chemical synthesis, biotin and the photo-cleavable group can be covalently attached only to the 5′ end of RNA (cis configuration). In addition, the synthesis is practical only with relatively short RNA sequences, not exceeding 60–65 nt. We tested whether the two groups can be attached to pre-mRNA in trans, as part of a short complementary adaptor oligonucleotide. This variation could potentially extend the usage of the UV-elution method to longer RNA species and larger RNA–protein complexes, improving at the same time its cost effectiveness.
We used T7 RNA polymerase to generate 125-nt Drosophila dH3 Ext histone pre-mRNA and designed the 22-nt pcB/22mer for chemical synthesis. This oligonucleotide is uniformly modified with 2′O-methyl groups and contains the photo-cleavable group and biotin at the 5′ end followed by a 3-nt spacer and 19 nt complementary to the 3′ end of the substrate (Figure 4A). The 125-nt pre-mRNA was efficiently processed in Kc nuclear extracts and addition of 10- or 100-fold excess of the pcB/22mer oligonucleotide had no major effect (Figure 4B, lanes 5 and 6). Thus, the formation of a strong duplex at the 3′ end of histone pre-mRNA (note that the duplex RNA indicated with an arrow in Figure 4B persists in the denaturing conditions of the urea gel) does not interfere with binding of the U7 snRNP to the HDE. As expected, processing was abolished by excess of the SL RNA that sequesters SLBP, and was greatly reduced by the anti-U7 oligonucleotide (αU7) that binds Drosophila U7 snRNA (Figure 4B, lanes 3 and 4, respectively).
Figure 4.
Purification of Drosophila processing complexes with pre-mRNA attached to the photo-cleavable group in trans. (A) A schematic representation of the dH3 Ext duplex formed by annealing T7-generated dH3 Ext pre-mRNA and chemically synthesized 2′O-methyl pcB/22mer oligonucleotide. In pcB/22mer, biotin (B) is placed at the 5′ end and is followed by the photo-cleavable (pc) linker. The last 19 of 22 nt of pcB/22mer are complementary to the 3′ end of dH3 Ext pre-mRNA. (B) dH3 Ext pre-mRNA was labeled at the 5′ end and incubated at room temperature in a Kc nuclear extract either alone (lane 2) or in the presence of indicated oligonucleotides (lanes 3–6). SL RNA and αU7 oligonucleotide were added to a final concentration of 10 ng/μl, pcB/22mer was at either 10 ng/μl or 100 ng/μl, a 10 and 100 molar excess relative to dH3 Ext pre-mRNA, respectively. The arrow indicates an RNA duplex that survived denaturing conditions of the 7M urea gel. Lane 1 contains input dH3 Ext pre-mRNA. (C and D) dH3 Ext duplex was incubated with a Drosophila Kc nuclear extract to form processing complexes (lanes 1 and 3 in panel C, and lanes 2 and 4 in panel D). As a negative control, formation of the processing complexes was blocked by αU7 oligonucleotide complementary to the 5′ end of Drosophila U7 snRNA (lanes 2 and 4 in panel C, and lanes 3 and 5 in panel D). Proteins bound to the duplex RNA were purified on streptavidin beads and UV-eluted. A fraction of the UV-eluted material (UV-sups, lanes 1 and 2) and the beads following UV-elution (UV-beads, lanes 3 and 4) was analyzed by silver staining (panel C) or western blotting (panel D). (E and F) Drosophila processing complexes were assembled on the duplex RNA and purified, as described above with the difference that negative control contained both SL RNA and αU7 oligonucleotide. A fraction of the UV-eluted samples was analyzed by silver staining and western blotting for selected proteins (panel E). The remainder was concentrated by precipitation with acetone, subjected to a brief electrophoresis sufficient for proteins to enter the gel, in-gel digested with trypsin and analyzed by mass spectrometry (panel F). Only top scoring processing factors are listed.
The dH3 Ext histone pre-mRNA (150 ng) was annealed to a 5-fold molar excess of pcB/22mer and the resulting duplex pre-bound to streptavidin beads and incubated with a Kc nuclear extract (750 μl) either in the absence or presence of the Drosophila-specific αU7 oligonucleotide. Since the dH3 Ext histone pre-mRNA generated by T7 RNA polymerase contains no 2′O-methyl modified nucleotides, all incubation steps were carried out on ice to minimize cleavage. The UV-eluted supernatant (UV-sup) and the material that remained on the beads following UV-irradiation (UV-beads) were first analyzed by silver staining (Figure 4C). A characteristic pattern of bands was detected in the UV-eluted processing complexes, but not in the sample prepared in the presence of αU7 oligonucleotide (Figure 4C, lanes 1 and 2, respectively). The same bands were only weakly detectable in the material left on the beads, indicative of an efficient UV-elution (Figure 4C, lane 3). Western blotting (Figure 4D) and mass spectrometry of the excised gel fragments (not shown) identified them as symplekin, CPSF100, CPSF73, CstF64, Lsm11 and FLASH. The symplekin and CstF64 bands also contained CPSF160 and WDR33, respectively, although these two proteins yielded fewer peptides and lower scores, suggesting that they were present in substoichiometric amounts. In comparison with pcB-dH3/5m, the dH3 Ext duplex yielded more non-specific bands, likely representing abundant RNA binding proteins that interact with the longer substrate and/or excess of the pcB/22mer oligonucleotide used for annealing (Figure 4C, lanes 1 and 2). We conclude that attaching biotin and a photo-labile linker to pre-mRNA in trans via a complementary oligonucleotide can be used as an alternative method to using pre-mRNAs containing these two groups attached covalently. Note that 20 mM EDTA added to nuclear extracts in all of our experiments likely inhibited endogenous magnesium-dependent RNA helicases, hence preventing potential disruption of the duplex between the oligonucleotide and the pre-mRNA. It is possible that some unwinding of the duplex can occur in the absence of EDTA, making this method less effective than using pre-mRNAs with the photo-cleavable group in cis.
We also used this method with a different batch of the Kc nuclear extract. In the negative control of this experiment, formation of processing complexes was blocked by pre-incubating the extract with both the αU7 oligonucleotide and SL RNA. Analysis of a fraction (15%) of each UV-eluted sample by silver staining (Figure 4E, top panel) and western blotting (Figure 4E, bottom panel) showed that dH3 Ext duplex efficiently assembled into processing complexes only in the absence of the competitor oligonucleotides.
The remaining portion of each UV-eluted sample was precipitated with acetone, dissolved in a loading dye and subjected to a brief electrophoresis in an SDS/polyacrylamide gel. After the sample migrated ∼10 mm below the bottom of the well, a small fragment of the gel containing unresolved proteins was excised and subjected to in-gel digestion with trypsin. As a result of concentrating the eluted samples this approach yielded significantly more peptides, hence facilitating detection of less abundant proteins, but did not compromise the unbiased character of the analysis. Importantly, the mass spectrometry results were essentially identical to those obtained with the pcB-dH3/5m pre-mRNA (Figure 4F) (Supplementary Excel File 1 for Figure 4).
Altogether, we carried out 12 independent experiments using several different Drosophila nuclear extracts with either pcB-dH3/5m pre-mRNA (seven experiments) or dH3 duplex (five experiments). Mass spectrometry results from these experiments were used to calculate normalized emPAI values for all detected proteins (Supplementary Excel File 2) and to assemble a summarized hierarchy of the most abundant proteins in Drosophila processing complexes, as described for the mouse samples (Table 2). Among the HCC subunits, symplekin, CPSF100, CPSF73 and CstF64 ranked significantly higher than CPSF160, WDR33, Fip1 and CPSF30 (Table 1). Thus, in both organisms, symplekin, CPSF100, CPSF73 and CstF64 likely form the core of the HCC, and the remaining polyadenylation factors are substoichiometric, being associated with only a small fraction of the U7 snRNP.
Table 2. Summarized ranking of the most abundant proteins in Drosophila processing complexes.
| Rank | CG | Protein | Detected | ΔAVE (%) |
|---|---|---|---|---|
| 1 | 11 886 | SLBP | 12 | 17.2 |
| 2 | 12 924 | Lsm11 | 12 | 11.6 |
| 3 | 9742 | SmG | 10 | 6.4 |
| 4 | 2097 | Symplekin | 12 | 4.1 |
| 5 | 1957 | CPSF100 | 12 | 3.6 |
| 6 | 7698 | CPSF73 | 12 | 2.9 |
| 7 | 18 591 | SmE | 10 | 2.5 |
| 8 | 4616 | FLASH | 12 | 2.3 |
| 9 | 5352 | SmB | 11 | 2.1 |
| 10 | 7697 | CstF64 | 12 | 1.7 |
| 11 | 12 938 | Lsm10 | 10 | 1.2 |
| 12 | 8427 | SmD3 | 10 | 1.1 |
| 13 | 16 792 | SmF | 8 | 0.7 |
| 14 | 10 110 | CPSF160 | 11 | 0.8 |
| 17 | 1109 | WDR33 | 11 | 0.5 |
| 21 | 1078 | Fip1 | 7 | 0.08 |
| 24 | 3642 | CPSF30 | 5 | 0.06 |
The hierarchy is based on the value of ΔAVE (%) calculated for all 12 Drosophila experiments (see Supplementary Data for complete data and text for the method of calculation). Of the 12 experiments, 7 were carried out with pcB-dH3/5m and 5 with the dH3 duplex. Omitted from the list are proteins that were competed <50% by SL RNA and anti-U7 oligonucleotide (proteins 15, 18, 22 and 23), and/or were detected in <4 experiments (proteins 16, 19 and 20).
CstF50 and CstF77 (not shown in Table 2) were detected with low emPAI values in only two and three samples, respectively. Other Drosophila polyadenylation factors, including orthologues of the two subunits of CF Im, were either not detected, or if detected, their binding with dH3 pre-mRNA was not abolished by the αU7 oligonucleotide, indicative of a non-specific interaction (Supplementary Excel File 2).
Cleavage activity of the immobilized mouse and Drosophila processing complexes
To determine whether the mouse processing complexes immobilized on streptavidin beads contain all components required for catalysis, we tested their ability to support cleavage. For this purpose, we used 3′Biot-mH2a/2m pre-mRNA (Figure 5A), a mouse-specific substrate similar in design to the Drosophila-specific 3′Biot-dH3/2m pre-mRNA used in our previous study (12). 3′Biot-mH2a/2m pre-mRNA lacks the photo-cleavable group and biotin is directly attached to the 3′ end, leaving the 5′ end available for radioactive labeling. The two 2′O-methyl groups (2m) that are placed immediately upstream of the HDE do not interfere with endonucleolytic cleavage but prevent post-cleavage 5′-3′ degradation of the downstream product and displacement of the bound U7 snRNP (44). The HDE in 3′Biot-mH2a/2m has been altered by two point mutations (HDE max) to increase its base-pairing with U7 snRNA and enhance the recruitment of mouse U7 snRNP to the pre-mRNA. As a result of this alteration, processing of 3′Biot-mH2a/2m pre-mRNA is only weakly inhibited by excess SL RNA (which prevents binding of SLBP to the substrate), although it remains sensitive to αU7 oligonucleotide (Figure 5B, lanes 3 and 2, respectively). This almost complete independence of SLBP is further illustrated by the lack of any stimulatory effect of recombinant SLBP on processing efficiency (Figure 5B, lane 4).
Figure 5.
Cleavage activity of immobilized mouse and Drosophila processing complexes. (A) A schematic representation of chemically synthesized mouse-specific 3′Biot-mH2a/2m pre-mRNA (64 nt). The two 2′O-methyl groups placed immediately upstream of the HDE have no effect on endonucleolytic cleavage but prevent subsequent 5′-3′ degradation of the downstream cleavage product. Biotin is placed at the 3′ end and the HDE is modified by two point mutations to maximize its base pair interaction with U7 snRNA (HDE max). (B) In vitro processing of 3′Biot-mH2a/2m pre-mRNA in a mouse nuclear extract alone (lane 1) or containing indicated reagents: αU7 oligonucleotide (10 ng/μl), SL RNA (10 ng/μl) or human baculovirus-expressed SLBP (25 ng/μl). (C) Mouse processing complexes (lanes 1–4) were assembled on ice by incubating 32P-labeled 3′Biot-mH2a/2m pre-mRNA with a mouse nuclear extract containing recombinant N-terminal FLASH. During this step, significant cleavage occurred reducing the amount of fully assembled processing complexes (lane 1). Length in nucleotides of the input 3′Biot-mH2a/2m pre-mRNA and of the upstream cleavage product in nucleotides is shown to the right. The assembled complexes were immobilized on streptavidin beads, thoroughly washed and analyzed by western blotting for the presence of SLBP and selected subunits of the U7 snRNP (lane 2) or incubated with a gentle agitation at indicated temperatures to determine their ability to support cleavage (lanes 3 and 4). As explained above, in mouse nuclear extracts SLBP is partially degraded and in SDS gels migrates as a group of closely spaced bands rather than a single full length species of ∼45 kDa. (D) Drosophila processing complexes were assembled on Drosophila-specific 3′Biot-dH3/2m pre-mRNA. The complexes were immobilized on streptavidin beads, washed and incubated with a gentle agitation at 0°C (lane 1) or 22°C (lane 2) to determine their ability to support cleavage.
The 3′Biot-mH2a/2m pre-mRNA labeled at the 5′ end was briefly incubated on ice with mouse nuclear extract and the assembled processing complexes were subsequently bound to streptavidin beads. In spite of conducting this step in cold, more than 25% of the pre-mRNA was cleaved (Figure 5C, lane 1). Additional cleavage occurred during long washes (not shown). Importantly, at least a fraction of the intact processing complexes, as judged by the presence of SLBP (Figure 5C, lane 2), remained bound to streptavidin beads and was used for functional analysis. The beads were resuspended in a small amount of a processing buffer and gently agitated for 60 min at either 0°C or 32°C to monitor the ability of the immobilized processing complexes to support cleavage. While no processing occurred when the complexes were constantly kept on ice, a small amount of the upstream cleavage product was generated during the same time at 32°C (Figure 5C, lanes 3 and 4, respectively). We conclude that at least a fraction of the immobilized mouse processing complexes contains all factors required for catalysis. Note that compared to the 3′Biot-mH2a/2m pre-mRNA, the pcB-mH2a/5m pre-mRNA due to its resistance to cleavage is expected to be more efficient in capturing complete and catalytically active processing complexes and revealing their proteome by mass spectrometry.
We used the same approach to test cleavage activity of purified Drosophila processing complexes assembled on Drosophila-specific 3′Biot-dH3/2m histone pre-mRNA (12). During the assembly and purification on streptavidin beads, more than 50% of the substrate was cleaved (not shown). Immobilized processing complexes assembled on the remaining intact substrate were divided into two equal aliquots and incubated for 90 min at either 0°C or at 22°C. Only a small amount of histone pre-mRNA was cleaved at 0°C (Figure 5D, lane 1). Importantly, the efficiency of cleavage increased at room temperature to 60% (Figure 5D, lane 2), demonstrating that most of the immobilized Drosophila processing complexes are catalytically active.
Cleavage activity of Drosophila processing complexes assembled in the absence of SLBP
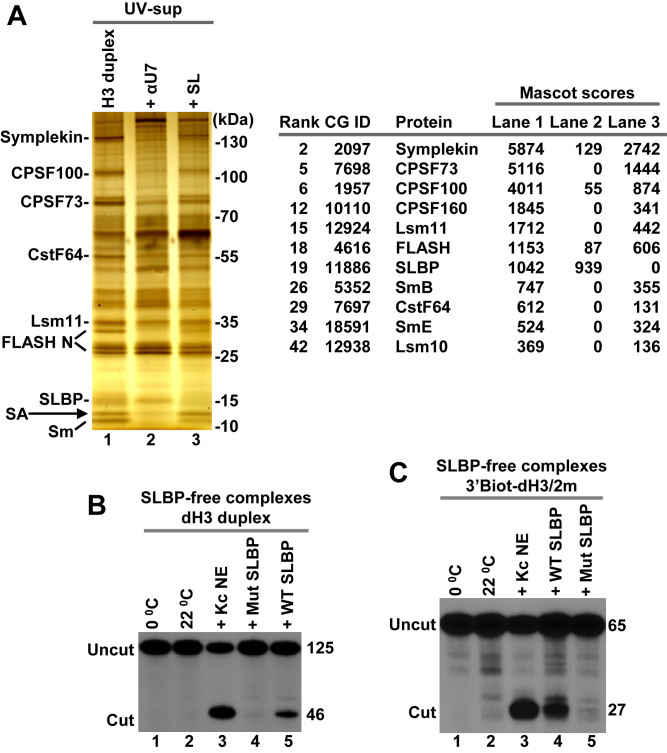
As explained above, nuclear extracts can be readily depleted of free SLBP by a brief incubation with excess SL RNA, which forms a tight complex with SLBP and prevents its interaction with histone pre-mRNA. In these extracts, Drosophila U7 snRNP binds histone pre-mRNA less efficiently than in the presence of free SLBP (12,14). Intriguingly, the bound U7 snRNP in spite of containing all subunits of the HCC, including the CPSF73 endonuclease, is inactive in processing (12,14). Thus, Drosophila SLBP in addition to promoting the recruitment of U7 snRNP to histone pre-mRNA plays a second, indispensable role in processing. Since one possibility was that Drosophila SLBP recruits an unknown factor critical for processing, we used the UV-elution method and unbiased analysis by mass spectrometry to compare the proteome of processing complexes formed in a Kc nuclear extract in the absence and the presence of SL RNA. As a negative control, we also generated a sample lacking processing complexes by pre-incubating the same Kc nuclear extract with the αU7 oligonucleotide. dH3 Ext pre-mRNA was annealed to pcB/22mer and the resultant duplex incubated in a Kc nuclear extract without or with the competitors. UV-eluted proteins were precipitated and resolved in an SDS gel for silver staining (Figure 6A, left) or subjected to only a brief electrophoresis for global mass spectrometry analysis (Figure 6A, right), as described above. Silver staining revealed the presence of all major subunits of the U7 snRNP in the processing complexes containing SLBP (Figure 6A, left, lane 1). Addition of SL RNA to the extract significantly reduced their amounts in the complex (Figure 6A, left, lane 3), consistent with the role of SLBP in recruiting U7 snRNP. The αU7 oligonucleotide eliminated U7 snRNP from the complex, but as expected SLBP remained bound to the pre-mRNA (Figure 6A, left, lane 2). The same results were evident from the mass spectrometry analysis (Figure 6A, right) (Supplementary Excel File 1 for Figure 6). Importantly, this analysis also demonstrated that SLBP was the only major protein eliminated from the processing complexes by excess SL RNA.
Figure 6.
Cleavage activity of Drosophila processing complexes assembled in the absence of SLBP. (A) dH3 Ext duplex was incubated in a Kc nuclear extract in the absence or in the presence of indicated competitors and the UV-eluted samples were visualized by silver staining (left) and analyzed by mass spectrometry following a brief electrophoresis into 4–12 gel SDS/polyacrylamide gel (right). Only SLBP and components of the U7 snRNP are listed. They all fail to interact with dH3 pre-mRNA in the presence of SL RNA or αU7 oligonucleotide. A small amount of contaminating streptavidin (SA) in lanes 1 and 3 is indicated with an arrow. (B and C) SLBP-free Drosophila processing complexes assembled on dH3 Ext duplex (panel B) or 3′Biot-dH3/2m (panel C) in a nuclear extract containing the SL RNA (as in lane 3 of panel A) were immobilized on streptavidin beads, washed and tested for the ability to support cleavage at 22°C (lanes 2–5 in both panels) either alone or in the presence of Kc nuclear extract or indicated SLBP variants. The aliquots incubated on ice are shown in lane 1 in both panels.
It remained possible that SLBP recruits for processing a weakly bound factor(s) that is either absent in the immobilized processing complexes or substoichiometric and hence undetectable by mass spectrometry. To address this possibility, we tested whether the immobilized complexes that lack SLBP can be activated for processing by addition of highly purified recombinant SLBP expressed in baculovirus (12,22). The dH3 Ext duplex labeled at the 5′ end of the pre-mRNA with 32P was incubated in a Kc nuclear extract in the presence of SL RNA to assemble processing complexes without SLBP. Since cleavage under these conditions is inhibited, this step was carried at room temperature rather than on ice to maximize binding of the U7 snRNP to the pre-mRNA. Following thorough washes, the immobilized processing complexes were tested for the ability to support cleavage. In the absence of exogenous SLBP no cleavage occurred at either 0 or 22°C (Figure 6B, lanes 1 and 2), consistent with the essential role of SLBP in processing. After addition of a small amount of Kc nuclear extract to the suspension of the beads, as much as 50% of the bound dH3 Ext was cleaved, confirming that the immobilized substrate is competent for cleavage in the presence of all essential processing components provided by the extract (Figure 6B, lane 3). Importantly, 5–10% of the input pre-mRNA was cleaved after addition of a highly purified preparation of baculovirus-expressed WT Drosophila SLBP instead of the extract (Figure 6B, lane 5). Note that this relatively low efficiency of processing results at least in part from the very inefficient binding of the U7 snRNP to histone pre-mRNA in the absence of SLBP (Figure 6A, left, lane 3). Addition of recombinant ER/Hs17C SLBP that is inactive in processing had no effect (Figure 6B, lane 4). This mutant SLBP binds the stem–loop in histone pre-mRNA but is unable to interact with the U7 snRNP due to mutations in helix B of the RBD and in the C-terminal region (12). We conclude that SLBP is the only factor missing when processing complexes are assembled in the nuclear extract containing SL RNA, and that the activation of the U7 snRNP bound to histone pre-mRNA by SLBP requires direct contact(s) between these two processing factors.
We repeated the same experiment with 32P-labeled 3′Biot-dH3/2m pre-mRNA. Again, addition of WT recombinant SLBP, but not the mutant SLBP, to the immobilized SLBP-free complexes was sufficient to activate the bound U7 snRNP for cleavage (Figure 6C, lanes 4 and 5, respectively), confirming that besides SLBP they contain all other essential factors.
The effect of improving the HDE on processing
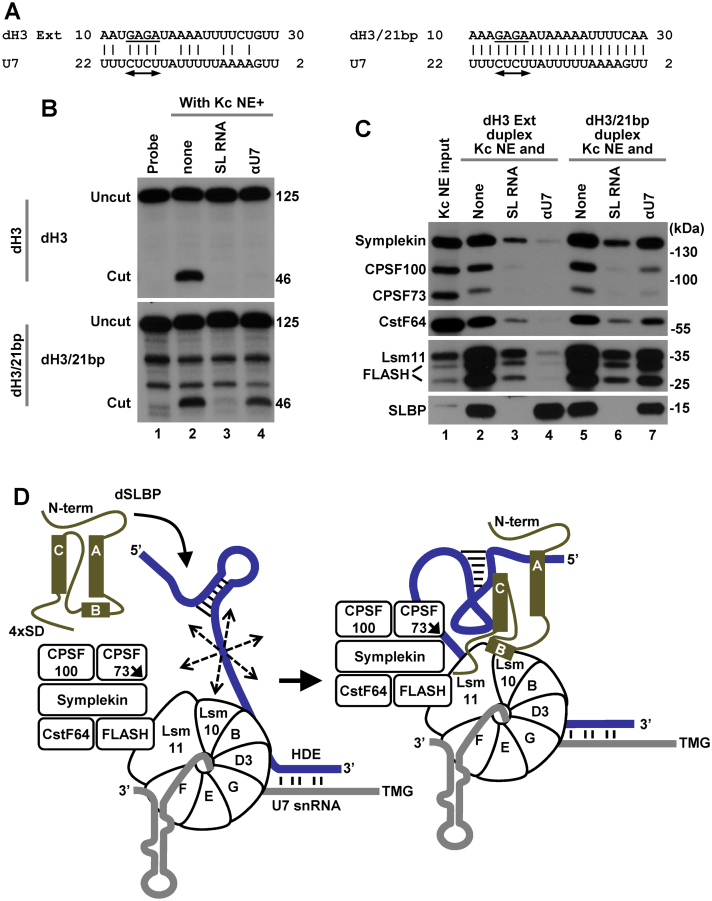
In mouse and human nuclear extracts, histone pre-mRNAs that form strong duplexes with the 5′ end of the mammalian U7 snRNA are cleaved even in the absence of SLBP (19–21) (Figure 5B, lane 3). The HDEs of all five Drosophila histone pre-mRNAs are AU-rich and have relatively poor complementarity with the 5′ end of Drosophila U7 snRNA, which is also AU-rich (23). We tested whether the requirement for Drosophila SLBP during processing in Drosophila nuclear extracts can be alleviated by improving this weak base pair potential.
The HDE of the dH3 pre-mRNA and the 5′ end of the U7 snRNA can form 5 alternative duplexes containing 10 to 15 bp, interrupted by a various number of mismatches (23). As a template for improving the complementarity between the two RNAs, we selected the alignment that results in a duplex containing only 12 bp (ΔG, −4.75 kcal/mol) but engages the entire GAGA purine core in the base pair interaction with the U7 snRNA (Figure 7A). Substitution of these 4 nt, in contrast to mutations in other parts of the HDE, prevented binding of the U7 snRNP to histone pre-mRNA and abolished processing in vitro (22,48). By changing 9 nt in dH3 Ext outside this motif, we created dH3/21bp pre-mRNA that can form a continuous duplex of 21 bp with the 5′ end of the U7 snRNA (Figure 7A), drastically reducing ΔG to −35.17 kcal/mol.
Figure 7.
Effects of increasing the base pairing potential between histone pre-mRNA and Drosophila U7 snRNA. (A) A proposed base pair alignment between the HDE of dH3 Ext (left) and dH3/21bp (right) pre-mRNAs and Drosophila U7 snRNA. The HDE starts 10 nt 3′ of the cleavage site and contains the GAGA sequence (underlined) that is essential for processing and is predicted to form an obligatory duplex with the UCUC motif (indicated with a double-headed arrow) conserved in most U7 snRNAs. (B) Processing of 5′-labeled dH3 Ext (top) and dH3/21bp (bottom) pre-mRNAs in a Kc nuclear extract either lacking (lane 2) or containing indicated competitors (lanes 3 and 4). The input RNA is shown in lane 1. Length of the uncut pre-mRNAs and the upstream cleavage products (cut) in nucleotides is shown to the right (C). dH3 Ext and dH3/21bp pre-mRNAs were annealed to pcB/21mer and the resultant duplexes incubated with a Kc nuclear extract in the absence or presence of oligonucleotide competitors, as indicated. The assembled processing complexes were immobilized on streptavidin beads and analyzed by western blotting for the presence of SLBP and major subunits of the U7 snRNP. (D) A hypothetical model explaining essential role of Drosophila in processing. The three α helices of the RBD of Drosophila SLBP are depicted as rectangles and the repeated SD motif is shown at the C-terminal region. In the absence of Drosophila SLBP, U7 snRNP inefficiently binds histone pre-mRNA (blue line) via limited base paring between U7 snRNA (gray line) and the HDE. The bound U7 snRNP in spite of containing all necessary subunits is unable to carry out the cleavage reaction, possibly due to the excessive flexibility of the pre-mRNA substrate (illustrated by multiple arrows). SLBP bound to the stem-loop uses helix B and the C-terminal region to promote the recruitment of the U7 snRNP to the HDE, likely by contacting Lsm11 bound to FLASH and possibly Lsm10. The same network of interactions may result in a major structural rearrangement of the processing complex and stable alignment of the active site of CPSF73 (indicated with an arrow) with the pre-mRNA.
The efficiency of processing of the dH3/21bp pre-mRNA in a Kc nuclear extract did not significantly differ from that observed for the wild-type dH3 Ext pre-mRNA (Figure 7B, compare lane 2 in top and bottom panels) and excess of the SL RNA had the same inhibitory effect, abolishing processing of both pre-mRNAs as a result of sequestering the entire pool of SLBP present in the extract (Figure 7B, compare lane 3 in both panels). Thus, in vitro processing of the dH3/21bp pre-mRNA that is capable of forming a perfect duplex with the 5′ end of the Drosophila U7 snRNA remains fully dependent on SLBP, strongly contrasting with the situation observed in mammalian nuclear extracts (19–21). Interestingly, the αU7 oligonucleotide abolished processing of dH3 Ext pre-mRNA but was only partially inhibitory on processing of dH3/21bp pre-mRNA (Figure 7B, compare lane 4 in both panels). Note that the αU7 oligonucleotide forms only a 16-bp duplex with the U7 snRNA but it is added to the nuclear extract in a large molar excess prior to adding the pre-mRNA substrate.
We compared the ability of the dH3 Ext and dH3/21bp pre-mRNAs to assemble processing complexes. Each pre-mRNA was annealed to the pcB/22mer oligonucleotide and the resulting duplexes incubated with a Kc nuclear extract either lacking or containing appropriate competitors (SL RNA or αU7 oligonucleotide), as described above. dH3/21bp pre-mRNA, in spite of containing the improved HDE, was only slightly more efficient than dH3 Ext in binding U7 snRNP (Figure 7C, lanes 2 and 5), and the same small difference between the two pre-mRNAs was observed when processing complexes were formed in the presence of SL RNA (Figure 7C, lanes 3 and 6). Consistent with the processing data, binding of U7 snRNP to dH3/21bp pre-mRNA was only partially blocked by the αU7 oligonucleotide (Figure 7C, lanes 4 and 7). Thus, the improved HDE in this pre-mRNA does not recruit significantly more U7 snRNP into a stable complex with the pre-mRNA, although it is sufficiently strong to compete for U7 snRNP with the αU7 oligonucleotide.
We conclude that creation of a perfect HDE in histone pre-mRNA by improving the base pairing potential with the Drosophila U7 snRNA is not sufficient to relieve the absolute dependence of processing in vitro on Drosophila SLBP.
DISCUSSION
Purification of soluble U7-dependent processing complexes
In animal cells, 3′ end cleavage of replication-dependent histone pre-mRNAs requires at least two components: SLBP and the holo-U7 snRNP; a multi-subunit factor that in addition to the U7 snRNA and the U7-specific Sm ring contains FLASH and a subset of polyadenylation factors that we collectively refer to as the HCC (12,13). To identify all subunits of the HCC and to determine whether processing requires any additional factor(s), we developed an efficient single-step method of purifying in vitro assembled mouse and Drosophila processing complexes.
In this method, processing complexes were assembled in a nuclear extract on a synthetic histone pre-mRNA containing biotin and a photo-cleavable linker at the 5′ end. The major cleavage site and the two neighboring nucleotides on each side were modified with a 2′O-methyl group, hence preventing endonucleolytic cleavage of the pre-mRNA and increasing the efficiency of capturing intact processing complexes. Following immobilization on streptavidin beads, the pre-mRNA and the bound proteins were washed and released to solution by irradiation with long wave UV. This UV-elution step, by eliminating all background proteins non-specifically bound to streptavidin beads, resulted in isolation of remarkably pure processing complexes that were suitable for direct analysis by mass spectrometry. To our knowledge, this is the first successful use of the photo-cleavable linker and the UV-elution step for purification of an in vitro assembled RNA/protein complex. Parallel experiments with pre-mRNA substrates lacking 2′O-methyl nucleotides at the cleavage site demonstrated that the immobilized processing complexes retain catalytic activity. Thus, the mass spectrometry analysis of the UV-eluted material is likely to provide a global and unbiased view of all essential proteins that associate with histone pre-mRNA for 3′ end processing.
Since chemical synthesis of RNAs containing covalently attached biotin and the photo-cleavable linker (cis configuration) is both expensive and limited to sequences not exceeding 60–70 nt, we also tested longer histone pre-mRNAs generated by T7 transcription. Biotin and the photo-cleavable linker can be attached to the 3′ end of these pre-mRNAs in trans via a short complementary oligonucleotide. This modification makes the UV-elution method more cost effective and potentially applicable for purification of RNA–protein complexes that require longer RNA binding targets, including spliceosomes and complexes involved in cleavage and polyadenylation.
Components of mouse and Drosophila processing complexes
In the UV-eluted mouse and Drosophila processing complexes, mass spectrometry identified SLBP and all known subunits of the U7-specific Sm ring, including Lsm10 and Lsm11. Readily detectable in mouse and Drosophila processing complexes were also FLASH and subunits of the HCC. The HCC is remarkably similar in composition between the two species, with symplekin, CPSF100, CPSF73 and CstF64 being most abundant and present in close to stoichiometric amounts, as determined by both silver staining and emPAI value analysis. The remaining CPSF subunits (CPSF160, WDR33, Fip1 and CPSF30) are present in lower amounts, suggesting that they are substoichiometric, being stably associated only with a fraction of the U7 snRNP.
In both mouse and Drosophila experiments, SLBP and the components of the U7 snRNP were the only proteins that consistently failed to bind histone pre-mRNAs in the presence of processing competitors: SL RNA and αU7 oligonucleotide. Other proteins were detected both in the samples containing processing complexes and in the matching negative controls, where formation of processing complexes was blocked. Among them, the most prevalent were non-specific RNA binding proteins, including hnRNP Q in mouse nuclear extracts, and IGF2BP1 in Drosophila nuclear extracts. All these proteins likely bind to sites in histone pre-mRNAs unoccupied by SLBP and U7 snRNP, and play no essential role in processing.
Proteins not detected in mouse and Drosophila processing complexes
CstF50 and CstF77 were not detected in the UV-eluted mouse processing complexes and were present only in some Drosophila complexes, always with low scores, consistent with our previous conclusion that of the three CstF subunits only CstF64 stably associates with the U7 snRNP (13). No peptides were detected for CF Im (68 and 25 kDa) and CF IIm (Clp1 and Pcf11) in any of the mouse experiments, suggesting that these factors are also uniquely involved in cleavage and polyadenylation. Mass spectrometry identified the orthologues of the 68 and 25 kDa subunits in some Drosophila experiments, but they were clearly contaminants, persisting in the presence of the SL RNA and αU7 oligonucleotide (Supplementary Excel File 2). CF Im68 was previously reported to interact with Lsm11 and to co-purify with U7 snRNP (33). Based on our analysis, this subunit is unlikely to interact with Lsm11 in the processing complex.
Catalytically active mouse processing complexes also lacked ZFP100 (ZN473), a zinc finger protein that co-localizes with Lsm11 and stimulates expression of a reporter gene containing U7-dependent processing signals (30). ZFP100 was initially identified by the yeast two-hybrid system as a protein interacting with SLBP bound to the SL RNA (29) and suggested to function as a bridging factor in the SLBP-mediated recruitment of the U7 snRNP to histone pre-mRNA (29,31,49). However, the absence of ZFP100 in the UV-eluted mouse processing complexes containing both SLBP and U7 snRNP strongly argues against this function. ZFP100 may instead participate in a different aspect of histone gene expression in vivo, perhaps acting as a coupling factor that integrates transcription of histone genes with 3′ end processing of the nascent histone pre-mRNAs.
A similar role in vivo may be played by the multi-functional protein FUS and other proteins previously linked to 3′ end processing of histone pre-mRNAs in mammalian cells, including Ars2 (35,36), CDC73/parafibromin (32), NELF E (34) and CDK9 (50). These factors were never specifically detected in the UV-eluted mouse processing complexes, suggesting that they have no direct role in processing in vitro. Their downregulation by RNAi results in production of a small amount of polyadenylated histone mRNAs, which may be due to a defect in coupling of histone gene transcription with processing and/or cell-cycle progression.
What components are essential for processing?
Although we identified several polyadenylation subunits in a stable association with the U7 snRNP, our experiments do not directly address which of them are essential for processing of histone pre-mRNAs. In Drosophila cultured cells, RNAi-mediated depletion of each of only three U7-associated polyadenylation subunits, symplekin, CPSF100 and CPSF73, consistently resulted in accumulation of histone mRNAs terminated with a poly(A) tail, an indication of a defect in the U7-dependent processing mechanism. Depletion of the remaining HCC subunits had no effect, suggesting that their association with the U7 snRNP is not essential for 3′ end processing of histone pre-mRNAs (14,28). Symplekin, CPSF100 and CPSF73 are present in Drosophila cells as a stable sub-complex (51) and likely act together as an autonomous cleavage module recruited for processing to either histone or canonical pre-mRNAs by specialized RNA recognition sub-complexes. For canonical pre-mRNAs, this role is played by the remaining CPSF subunits, CPSF160, WDR33, Fip1 and CPSF30, recently shown to co-operate in recognizing the AAUAAA signal during the polyadenylation step (52–54). In 3′ end processing of histone pre-mRNAs, the recruitment of the cleavage sub-complex is mediated by the U7 snRNA, which recognizes the substrate by the base pairing interaction, further arguing that CPSF160, WDR33, Fip1 and CPSF30 are likely non-essential bystanders in the U7 snRNP.
A less clear role in 3′ end processing of histone pre-mRNAs is played by CstF64, which in spite of being relatively abundant in Drosophila U7 snRNP can be depleted from Drosophila cells without causing a detectable misprocessing of histone pre-mRNAs (14,28). A defect in the U7-dependent processing was however observed in human cells partially depleted of CstF64 (55,56), suggesting that in mammalian cells this subunit may play a more critical role, perhaps helping to stabilize the three essential subunits of the HCC on the FLASH/Lsm11 complex. Clearly, determining which subunits are essential for cleavage will require reconstitution of a catalytically active processing complex from recombinant components.
Function of Drosophila SLBP in processing
Our study brings a new perspective on the essential role of Drosophila SLBP in processing. We recently demonstrated that Drosophila SLBP, like its mammalian counterpart, enhances the recruitment of U7 snRNP to histone pre-mRNA (12). A small amount of U7 snRNP binds to histone pre-mRNA in the absence of Drosophila SLBP but the bound U7 snRNP in spite of containing all major HCC subunits is catalytically inactive. We now showed that processing complexes assembled in the absence of SLBP can be activated for cleavage by simply adding recombinant WT SLBP, providing evidence that SLBP is the only missing factor in the assembled complexes. A mutant Drosophila SLBP that is deficient in recruiting U7 snRNP to histone pre-mRNA is also unable to activate the assembled complex for cleavage. Based on these results, we propose that the interaction of Drosophila SLBP with the U7 snRNP promotes an essential structural rearrangement of the entire processing complexes that juxtaposes the catalytic site of CPSF73 with the pre-mRNA (Figure 7D). It is possible that higher metazoans developed an additional positioning mechanism for the CPSF73 endonuclease, resulting in efficient cleavage in the absence of SLBP.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (57) partner repository with the dataset identifier PXD008917.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Gall (Carnegie Institution for Science) for antibodies against Drosophila Lsm10 and Lsm11. We also thank J. Oledzki and A. Fabijanska (Institute of Biochemistry and Biophysics, Polish Academy of Sciences) for mass spectrometry analysis and Y. Shi (University of California, Irvine), M. Jurica (Montana State University) and J. Vilardell (IBMB, Spain) for reagents and helpful suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [GM 29832 to W.F.M., Z.D., GM 58921 to W.F.M.]; Polish National Science Centre, MAESTRO Grant [UMO-2014/14/A/NZ1/00306 to A.S., M.D.]. Funding for open access charge: NIH [GM 29832, GM 58921].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dominski Z., Marzluff W.F.. Three-hybrid screens for RNA-binding proteins: proteins binding the 3′ end of histone mRNA. Methods Mol. Biol. 2001; 177:291–318. [DOI] [PubMed] [Google Scholar]
- 2. Marzluff W.F. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005; 17:274–280. [DOI] [PubMed] [Google Scholar]
- 3. Dominski Z., Carpousis A.J., Clouet-d’Orval B.. Emergence of the beta-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta. 2013; 1829:532–551. [DOI] [PubMed] [Google Scholar]
- 4. Birchmeier C., Schümperli D., Sconzo G., Birnstiel M.L.. 3′ editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:1057–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galli G., Hofstetter H., Stunnenberg H.G., Birnstiel M.L.. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3′ histone mRNA termini. Cell. 1983; 34:823–828. [DOI] [PubMed] [Google Scholar]
- 6. Mowry K.L., Steitz J.A.. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987; 238:1682–1687. [DOI] [PubMed] [Google Scholar]
- 7. Pillai R.S., Grimmler M., Meister G., Will C.L., Luhrmann R., Fischer U., Schumperli D.. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003; 17:2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B.. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001; 20:5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X.C., Burch B.D., Yan Y., Marzluff W.F., Dominski Z.. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 2009; 36:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang X.C., Xu B., Sabath I., Kunduru L., Burch B.D., Marzluff W.F., Dominski Z.. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 2011; 31:1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mandel C.R., Bai Y., Tong L.. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008; 65:1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skrajna A., Yang X.C., Bucholc K., Zhang J., Hall T.M.T., Dadlez M., Marzluff W.F., Dominski Z.. U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein. RNA. 2017; 23:938–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X.C., Sabath I., Debski J., Kaus-Drobek M., Dadlez M., Marzluff W.F., Dominski Z.. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell Biol. 2013; 33:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabath I., Skrajna A., Yang X.C., Dadlez M., Marzluff W.F., Dominski Z.. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA. 2013; 19:1726–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schaufele F., Gilmartin G.M., Bannwarth W., Birnstiel M.L.. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986; 323:777–781. [DOI] [PubMed] [Google Scholar]
- 16. Wang Z.-F., Whitfield M.L., Ingledue T.I., Dominski Z., Marzluff W.F.. The protein which binds the 3′ end of histone mRNA: a novel RNA- binding protein required for histone pre-mRNA processing. Genes Dev. 1996; 10:3028–3040. [DOI] [PubMed] [Google Scholar]
- 17. Martin F., Schaller A., Eglite S., Schümperli D., Müller B.. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997; 16:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan D., Marzluff W.F., Dominski Z., Tong L.. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013; 339:318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Streit A., Koning T.W., Soldati D., Melin L., Schümperli D.. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993; 21:1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spycher C., Streit A., Stefanovic B., Albrecht D., Koning T.H.W., Schümperli D.. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994; 22:4023–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dominski Z., Zheng L.X., Sanchez R., Marzluff W.F.. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 1999; 19:3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dominski Z., Yang X., Raska C.S., Santiago C.S., Borchers C.H., Duronio R.J., Marzluff W.F.. 3′ end processing of Drosophila histone pre-mRNAs: Requirement for phosphorylated dSLBP and co-evolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 2002; 22:6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dominski Z., Yang X.C., Purdy M., Marzluff W.F.. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. RNA. 2005; 11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominski Z., Yang X.C., Marzluff W.F.. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005; 123:37–48. [DOI] [PubMed] [Google Scholar]
- 25. Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., Tong L.. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006; 444:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dominski Z., Yang X.C., Purdy M., Wagner E.J., Marzluff W.F.. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 2005; 25:1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kolev N.G., Steitz J.A.. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005; 19:2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wagner E.J., Burch B.D., Godfrey A.C., Salzler H.R., Duronio R.J., Marzluff W.F.. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell. 2007; 28:692–699. [DOI] [PubMed] [Google Scholar]
- 29. Dominski Z., Erkmann J.A., Yang X., Sanchez R., Marzluff W.F.. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 2002; 16:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagner E.J., Marzluff W.F.. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol. Cell. Biol. 2006; 26:6702–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Azzouz T.N., Gruber A., Schumperli D.. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 2005; 33:2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farber L.J., Kort E.J., Wang P., Chen J., Teh B.T.. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol. Carcinog. 2010; 49:215–223. [DOI] [PubMed] [Google Scholar]
- 33. Ruepp M.D., Vivarelli S., Pillai R.S., Kleinschmidt N., Azzouz T.N., Barabino S.M., Schumperli D.. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′-end processing of animal histone mRNAs. Nucleic Acids Res. 2010; 38:7637–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H.. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007; 26:349–365. [DOI] [PubMed] [Google Scholar]
- 35. Kiriyama M., Kobayashi Y., Saito M., Ishikawa F., Yonehara S.. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 2009; 29:4729–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gruber J.J., Olejniczak S.H., Yong J., La R.G., Dreyfuss G., Thompson C.B.. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012; 45:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pirngruber J., Shchebet A., Johnsen S.A.. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009; 8:3636–3642. [DOI] [PubMed] [Google Scholar]
- 38. Raczynska K.D., Ruepp M.D., Brzek A., Reber S., Romeo V., Rindlisbacher B., Heller M., Szweykowska-Kulinska Z., Jarmolowski A., Schumperli D.. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 2015; 43:9711–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang X.C., Torres M.P., Marzluff W.F., Dominski Z.. Three proteins of the U7-specific Sm ring function as the molecular ruler to determine the site of 3′-end processing in mammalian histone pre-mRNA. Mol. Cell. Biol. 2009; 29:4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nizami Z., Deryusheva S., Gall J.G.. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010; 2:a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu J.L., Buszczak M., Gall J.G.. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 2006; 14:465–475. [DOI] [PubMed] [Google Scholar]
- 42. Dominski Z., Sumerel J., Hanson R.J., Marzluff W.F.. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995; 1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 43. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M.. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics. 2005; 4:1265–1272. [DOI] [PubMed] [Google Scholar]
- 44. Yang X.C., Sullivan K.D., Marzluff W.F., Dominski Z.. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 2009; 29:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Olejnik J., Sonar S., Krzymanska-Olejnik E., Rothschild K.J.. Photocleavable biotin derivatives: a versatile approach for the isolation of biomolecules. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:7590–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S.. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- 47. Rappsilber J., Ryder U., Lamond A.I., Mann M.. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002; 12:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dominski Z., Yang X.C., Purdy M., Marzluff W.F.. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9422–9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagner E.J., Ospina J.K., Hu Y., Dundr M., Matera A.G., Marzluff W.F.. Conserved zinc fingers mediate multiple functions of ZFP100, a U7snRNP associated protein. RNA. 2006; 12:1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pirngruber J., Johnsen S.A.. Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3′ end processing through p21, NPAT and CDK9. Oncogene. 2010; 29:2853–2863. [DOI] [PubMed] [Google Scholar]
- 51. Sullivan K.D., Steiniger M., Marzluff W.F.. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell. 2009; 34:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun Y., Zhang Y., Hamilton K., Manley J.L., Shi Y., Walz T., Tong L.. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl. Acad. Sci. U.S.A. 2017; E1419–E1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan S.L., Huppertz I., Yao C., Weng L., Moresco J.J., Yates J.R. 3rd, Ule J., Manley J.L., Shi Y.. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 2014; 28:2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schonemann L., Kuhn U., Martin G., Schafer P., Gruber A.R., Keller W., Zavolan M., Wahle E.. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014; 28:2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Romeo V., Griesbach E., Schumperli D.. CstF64: cell cycle regulation and functional role in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. Biol. 2014; 34:4272–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruepp M.D., Schweingruber C., Kleinschmidt N., Schumperli D.. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell. 2011; 22:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (57) partner repository with the dataset identifier PXD008917.