Abstract
Non-allelic homologous recombination events on chromosome 22q11.2 during meiosis can result in either the deletion (22q11.2DS) or duplication (22q11.2DupS) syndrome. Although the spectrum and frequency of congenital heart disease (CHD) are known for 22q11.2DS, there is less known for 22q11.2DupS. We now evaluated cardiac phenotypes in 235 subjects with 22q11.2DupS including 102 subjects we collected and 133 subjects that were previously reported as a confirmation and found 25% have CHD, mostly affecting the cardiac outflow tract (OFT). Previous studies have shown that global loss or gain of function (LOF; GOF) of mouse Tbx1, encoding a T-box transcription factor mapping to the region of synteny to 22q11.2, results in similar OFT defects. To further evaluate Tbx1 function in the progenitor cells forming the cardiac OFT, termed the anterior heart field, Tbx1 was overexpressed using the Mef2c-AHF-Cre driver (Tbx1 GOF). Here we found that all resulting conditional GOF embryos had a persistent truncus arteriosus (PTA), similar to what was previously reported for conditional Tbx1 LOF mutant embryos. To understand the basis for the PTA in the conditional GOF embryos, we found that proliferation in the Mef2c-AHF-Cre lineage cells before migrating to the heart, was reduced and critical genes were oppositely changed in this tissue in Tbx1 GOF embryos versus conditional LOF embryos. These results suggest that a major function of TBX1 in the AHF is to maintain the normal balance of expression of key cardiac developmental genes required to form the aorta and pulmonary trunk, which is disrupted in 22q11.2DS and 22q11.2DupS.
Introduction
The 22q11.2 deletion syndrome (22q11.2DS), also known as velo-cardio-facial syndrome (MIM #192430) and DiGeorge syndrome (MIM #188400), is a fairly common human congenital anomaly disorder occurring in 1:1000 fetuses (1) and 1:4000 live births (2). Most affected patients have a 3 million base pair (Mb) deletion, owing to non-allelic homologous recombination events between flanking low copy repeats [also referred to as segmental duplications (3–5)] during meiosis. Some have a nested distal deletion endpoint resulting in a smaller deletion of 1.5 or 2 Mb (3–5). Approximately 60–70% of 22q11.2DS individuals have congenital heart disease (CHD) irrespective of deletion size. The most serious defects include tetralogy of Fallot (TOF), persistent truncus arteriosus (PTA) and interrupted aortic arch (IAA) type B (6–9). When taken together, the majority of defects disrupt the conotruncal region of the heart, which is also referred to as the cardiac outflow tract (OFT).
Both intra-chromosomal or inter-chromosomal non-allelic homologous recombination events can result in the 22q11.2 deletion (3,10–12). The reciprocal product of an inter-chromosomal event is a duplication of the same region that is deleted in 22q11.2DS and this syndrome is referred to as 22q11.2DupS (13,14). It is thought to occur at half the frequency of 22q11.2DS (13,14), but it is not clinically as well characterized as 22q11.2DS. This may be in part because the anomalies seen in the duplication syndrome occur with reduced penetrance and the clinical presentation is more varied (13,15). According to the literature, the frequency of CHD in 22q11.2DupS is from 20% and above, with quite variable malformations (16–18). There is a gap in our understanding of the frequency and full spectrum of cardiovascular anomalies in 22q11.2DupS. To fill the gap, in this report, we examined cardiac phenotypes in a large number of affected 22q11.2DupS individuals. Second, we wanted to consider genes on 22q11.2 that might contribute to CHD in 22q11.2DS and 22q11.2DupS.
Since CHD occurs in individuals with the deletion and duplication, we examined the region for genes that may be sensitive to altered dosage. TBX1 (T-box transcription factor 1), which encodes a T-box transcription factor located within the nested 1.5 Mb hemizygously deleted region on chromosome 22q11.2. Heterozygous loss of function (LOF) mutations in TBX1 has been identified in a subset of non-deleted patients with related phenotypes, further supporting its role in the etiology of 22q11.2DS (19–22).
Studies in mouse models have provided genetic proof that Tbx1 is required for the formation of the cardiac OFT and its inactivation results in a PTA (23–25). A PTA occurs in approximately 10–35% of patients with 22q11.2DS (9,26). Tbx1 is highly expressed in the mesoderm within the embryonic pharyngeal apparatus and its expressed specifically in the anterior heart field (AHF) mesoderm (27). The Mef2c-AHF-Cre allele is the most commonly used driver to promote AHF specific recombination of genes containing loxP sites (28). Inactivation of Tbx1 in the AHF using the Mef2c-AHF-Cre driver resulted in a PTA with 100% penetrance, similar to that of Tbx1 null mutant embryos (29). This indicates that Tbx1 has a critical role in the AHF mesoderm in cardiac OFT formation and/or remodeling. Increased dosage of Tbx1 in its normal expression domain also results in similar types of cardiac OFT defects as in the global LOF mutant embryos (30–32). These studies implicate Tbx1 as sensitive to decreased and increased dosage for heart development. However, we do not know if the cardiac OFT defects in the gain of function (GOF) embryos result from increased expression of Tbx1 in the pharyngeal mesoderm or epithelia, where the gene is also expressed and required for heart development (33–35).
For this report, we compiled cardiac phenotype data from our cohort of 102 individuals with 22q11.2DupS and combined this with 133 subjects that were previously reported in the literature, as a confirmation. This greatly expanded our understanding of the cardiac phenotype in individuals with 22q11.2DupS. We also tested whether increase of Tbx1 expression in the AHF (32) can result in cardiac OFT defects and attempted to understand the basis of these defects.
Results
Congenital heart defects in individuals with 22q11.2DupS
The cardiac phenotypes in a total of 102 unrelated individuals with 22q11.2DupS were examined for the frequency and spectrum of CHD. The results are summarized in Table 1 and the defects in each subject are listed in Supplementary Material, Table S1. Overall, 24 of the 102 individuals in our cohort (24%) had CHD. We observed TOF (n = 3), PTA (n = 1), isolated subaortic ventricular septal defect (VSD; n = 4), subaortic VSD combined with an atrial septal defect (ASD) (n = 2), ASD (n = 5) and pulmonary valve (PA) or pulmonary artery stenosis (n = 5). Two individuals had hypoplastic left heart syndrome (HLHS; n = 2) and two had total anomalous pulmonary venous connection (TAPVC) with ASD (TAPVC w/ASD; n = 2) (Fig. 1).
Table 1.
Frequency and spectrum of CHD found in 22q11DupS patients
| 22q11.2 DupS | Total N | Cardiac anomalies N | % | Cardiac summarize findings: (N) and percentage |
|---|---|---|---|---|
| This report | 102 | 24 | 24 | OFT (13) 13% |
| HLHS (2) 2% | ||||
| ASD (7) 7% | ||||
| OFT/ASD (2) 2% | ||||
| Literature | 133 | 34 | 26 | OFT (21) 16% |
| HLHS (5) 4% | ||||
| ASD (3) 2% | ||||
| Valve Defects (2) 1.5% | ||||
| HLHS/OFT/ASD (1) 1% | ||||
| HLHS/OFT (1) 1% | ||||
| Complex Heart defect (1) 1% | ||||
| Total | 235 | 58 | 25 | OFT (34) 14% |
| HLHS (7) 3% | ||||
| ASD (10) 4% | ||||
| Valve Defects (2) 1% | ||||
| OFT/ASD (2) 1% | ||||
| HLHS/OFT/ASD (1) <1% | ||||
| HLHS/OFT (1) <1% | ||||
| Complex Heart defect (1) <1% |
Percentages of types of CHD in our cohort of 22q11DupS patients and from the literature are shown. Cardiac OFT defects include Tetralogy of Fallot (TOF); persistent truncus arteriosus (PTA); ventricular septum defect (VSD); pulmonary valve and artery stenosis; interrupted aortic arch (IAA); transposition of the great arteries (TGA); coarctation of aorta and aortic/mitral valve stenosis. Valve defects are mitral valve defects. Individuals with more than one category of defect were classified accordingly. The case with a complex heart defect was not specified in the publication so it could not be classified as another CHD category (52). For a more detailed analysis see Supplementary Material, Table S1. HLHS: hypoplastic left heart syndrome, ASD: atrial septal defect.
Figure 1.
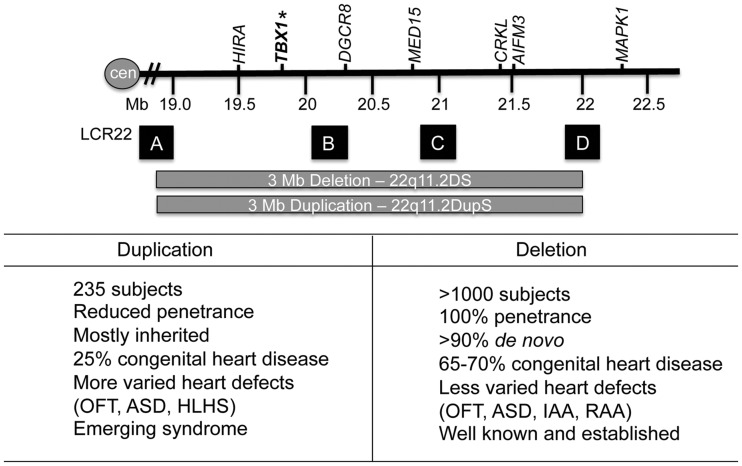
The 22q11.2 region and LCR22s. Top: a representation of the duplicated or deleted region on chromosome 22q11.2 shown with the coordinates on chromosome 22 (Mb is megabase; cen is centromere; hatched bars depicts a summary of the interval; GRCh37/hg19 assembly). A subset of the genes in the 22q11.2 region is shown. Low copy repeats on chromosome 22q11.2 (LCR22) are represented by letters A, B, C, D (sizes not at scale). Bottom: a comparison of some of the features of 22q11.2DupS and 22q11.2DS. Abbreviations: Outflow tract defect (OFT), atrial septal defect (ASD), hypoplastic left heart syndrome (HLHS), right-sided aortic arch (RAA).
We next performed a literature search and compiled cardiac findings in separate reports for a total of 133 reported individuals with 22q11.2DupS (Table 1; Supplementary Material, Table S1) to use as a confirmation of our results. For the 133 subjects, 34 of 133 (26%) had CHD and 21 of these had cardiac OFT defects alone or combined with other phenotype in two individuals (Table 1; Supplementary Material, Table S1). Some additional types of the cardiac OFT defects reported among the 133 subjects in the literature include transposition of the great arteries, double outlet right ventricle and IAA (Supplementary Material, Table S1). When taken together with our data, 25% (58/235) of individuals with 22q11.2DupS had a diagnosis of CHD, and a total of 66% among the ones with CHD had cardiac OFT defects alone or combined with other cardiac malformations (Fig. 2; Table 1; Supplementary Material, Table S1). Tbx1 is expressed in multiple tissues in the embryonic pharyngeal apparatus that forms the craniofacial region, thymus and parathyroid gland as well as the cardiac OFT. Since global loss and gain of Tbx1 expression in mouse models, including loss in the AHF, results in similar OFT malformations, such as a PTA (24,25,29–32,36,37), we wanted to determine if overexpression in the AHF would result in OFT defects.
Figure 2.
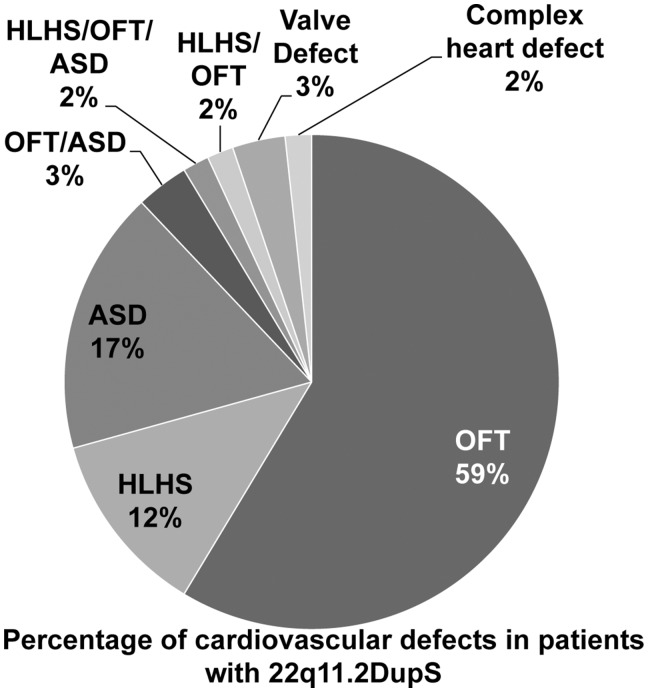
Percentage of cardiac anomalies in subjects with 22q11.2DupS. The pie chart indicates the relative composition of cardiac anomalies in the combined cohort. Abbreviations shown in the pie chart include: cardiac outflow tract defects [OFT: tetralogy of Fallot (TOF); persistent truncus arteriosus (PTA); ventricular septal defect (VSD); pulmonic valve (PV) stenosis; double outlet right ventricle (DORV); interrupted aortic arch (IAA); transposition of the great arteries (TGA)] and other malformations. We also indicate those with hypoplastic left heart syndrome (HLHS) and atrial septal defect (ASD). Valve malformations include mitral valve defects. Individuals with more than one defect were indicated accordingly. Complex heart defects are also indicated and this category includes defects that were not specified in the publication (52).
GOF of Tbx1 in the AHF results in a PTA
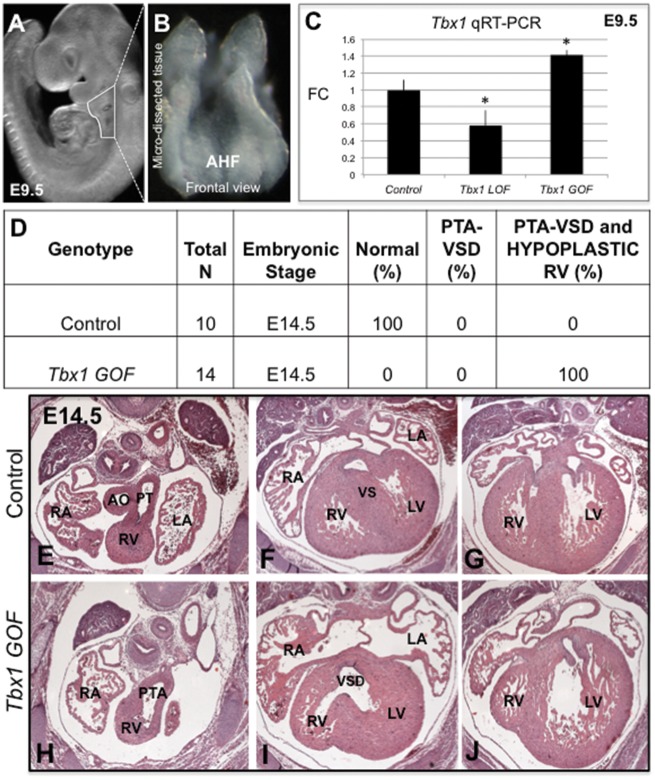
To determine the effect of constitutive activation of Tbx1 in the Mef2c-AHF-Cre domain, marking the AHF, we generated Tbx1 GOF conditional mutant mouse embryos that are of the genotype, Mef2c-AHF-Cre/+; Tbx1-GFP/+, in which the Tbx1-GFP allele is a fusion gene knocked into the ROSA26 locus (32). In these embryos, endogenous Tbx1 is expressed, and in addition, the Tbx1-GFP fusion gene is also expressed. We compared this with embryos in which we inactivated Tbx1 in the same domain, termed Tbx1 LOF (29). Tbx1 LOF embryos are of the genotype, Mef2c-AHF-Cre/+; Tbx1f/-, in which one allele of Tbx1 is completely inactivated, while the other is conditionally inactivated. We previously found that all Tbx1 LOF mutant mice died at birth with a PTA (29). The AHF is within the distal pharyngeal apparatus at E9.5 in developing embryos. The distal pharyngeal apparatus containing pharyngeal arches two, three and four, was micro-dissected (29) from Tbx1 GOF and LOF embryos at E9.5 (Fig. 3A and B). Wild-type, Tbx1-GFP/+ and Me2fc-AHF-Cre/+; Tbx1f/+ littermates were used as controls for the experiments. This stage was chosen because AHF progenitor cells within the distal pharyngeal apparatus are not yet differentiated. The dissected tissue contains the Mef2c-AHF-Cre lineage as well as adjacent non-mesodermal cells from the pharyngeal apparatus; however, it still is precise enough to obtain important data on gene expression changes (29). These cells are actively migrating into the cardiac OFT in order for it to elongate. We first determined the expression level of Tbx1 in the distal pharyngeal apparatus. As expected, Tbx1 expression in this tissue at E9.5 was significantly increased in Tbx1 GOF embryos versus controls, especially when compared with conditional Tbx1 LOF embryos, as shown by quantitative real-time PCR (qRT-PCR) at E9.5 (Fig. 3C). Expression in Tbx1 LOF embryos was not completely ablated, as can be seen in Figure 3C, since the microdissected tissue also contains the pharyngeal epithelia that expresses Tbx1. Histological analysis of tissue sections from Tbx1 GOF embryos at E14.5 showed that all (n = 14) had a PTA with complete penetrance (Fig. 3D, H–J) when compared with controls (Fig. 3D, E–G). A PTA is usually accompanied by a VSD, as what occurred in these mutant embryos (Fig. 3I). Surprisingly, they also had a hypoplastic right ventricle (Fig. 3J), which was not observed in Tbx1 mutant mouse models. For example, Tbx1 LOF embryos had a PTA (with a VSD) but had a normal right ventricle at the same stage (29). The hypoplastic right ventricle might result from overexpression of Tbx1 in the progenitor cells located in the first pharyngeal arch that form the right ventricle (38) resulting in ectopic expression of Tbx1 in the heart. Since Tbx1 is not normally expressed in the right ventricle, we decided not to further examine this malformation. We therefore focused on the tissue within the distal pharyngeal apparatus that forms the OFT, and not the right ventricle.
Figure 3.
Characterization of Tbx1 GOF mutant embryos. (A) Whole mount image of a mouse embryo at E9.5 showing the region that has been dissected for quantitative RT-PCR (qRT-PCR) experiments. (B) The micro-dissected region containing the Mef2c-AHF-Cre lineage is shown as a frontal view. (C) Quantification of Tbx1 expression levels in Tbx1 GOF and Tbx1 LOF mutant embryos as well as control embryos at E9.5 by qRT-PCR analysis. Statistical significance of the difference in gene expression was estimated using a two-tailed t-test; FC is fold change, P values <0.05. Error bars are standard deviation (SD). D) PTA phenotype observed in Tbx1 GOF embryos at E14.5. N is total number of hearts observed per group. Hematoxylin and eosin (H&E) stained histological sections of the heart of a control embryo at E14.5 with a normal cardiac OFT and ventricular septum (VS) (E–G). A Tbx1 GOF mutant embryo heart showing a PTA and VSD (H, I) as well as a hypoplastic right ventricle [RV] (J). Abbreviations: aorta (Ao), pulmonary trunk (PT), left atrium (LA), right atrium (RA), left ventricle (LV), right ventricle (RV), pharyngeal arch (PA). Control: Tbx1-GFPf/+ and Mef2c-AHF-Cre/+; Tbx1 f/+, Tbx1 GOF: Mef2c-AHF-Cre/+; Tbx1-GFPf/+, Tbx1 LOF: Mef2c-AHF-Cre/+; Tbx1 f/f.
Change of Tbx1 dosage affects the Mef2c-AHF-Cre cell lineage
To understand the role of Tbx1 in the AHF within the distal pharyngeal apparatus, we performed lineage tracing of the Mef2c-AHF-Cre, GFP+ [green fluorescent protein (GFP) expressing] cell population in conditional gain and LOF embryos at E9.5 and E10.5 (Fig. 4A–K). In each comparison, we will describe the results for the Tbx1 GOF mutant embryos first, followed by results for the Tbx1 LOF mutant embryos.
Figure 4.
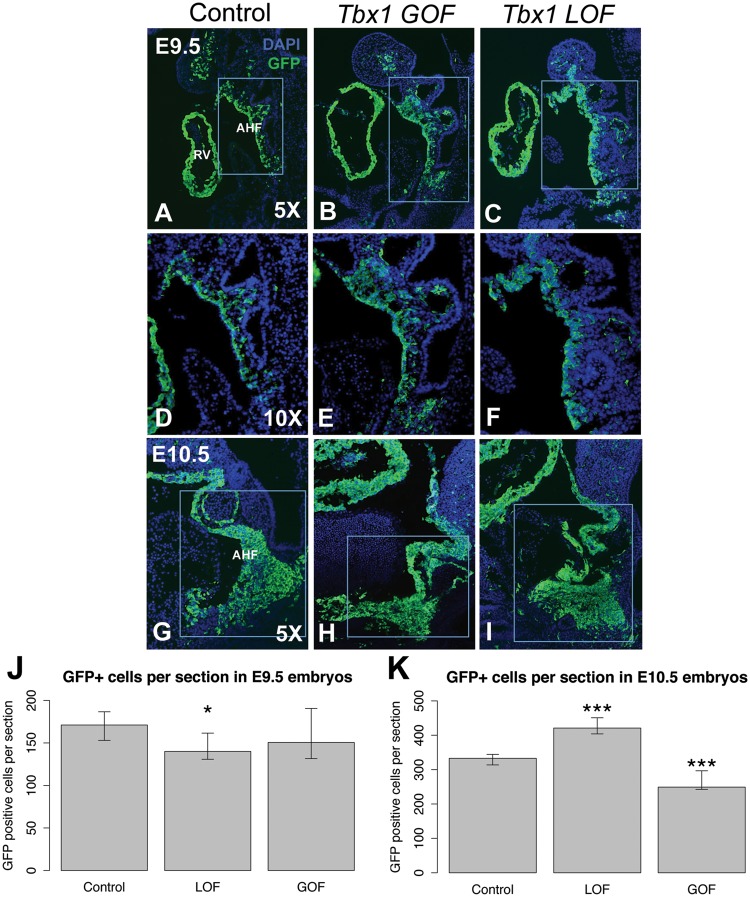
Changing the Tbx1 dosage in both directions has opposite affects on the Mef2c-AHF-Cre lineage. Representative sagittal sections showing the Mef2c-AHF-Cre lineage using a GFP reporter allele as well as DAPI fluorescent stain to visualize nuclei and identify the tissue is shown in blue. (A) E9.5 control, (B) Tbx1 GOF and (C) Tbx1 LOF embryo sections. (D–F) Higher magnification images of the rectangular areas shown in A, B and C, respectively. (G) E10.5 control, (H) Tbx1 GOF and (I) Tbx1 LOF embryo sections. Quantification of the Mef2c-AHF-Cre lineage from the area shown in the inset for each type of embryo at E9.5 (J) and E10.5 (K). Only one type of control is shown. Control for Tbx1 GOF: Mef2c-AHF-Cre/+; Rosa26GFP/+, Tbx1 GOF: Mef2c-AHF-Cre/+; Tbx1-GFPf/+, control for Tbx1 LOF: Mef2c-AHF-Cre/+; Tbx1 f/+; Rosa26GFP/+, Tbx1 LOF: Mef2c-AHF-Cre/+; Tbx1 f/f; Rosa26GFP/+. Asterisks indicate P value <0.05.
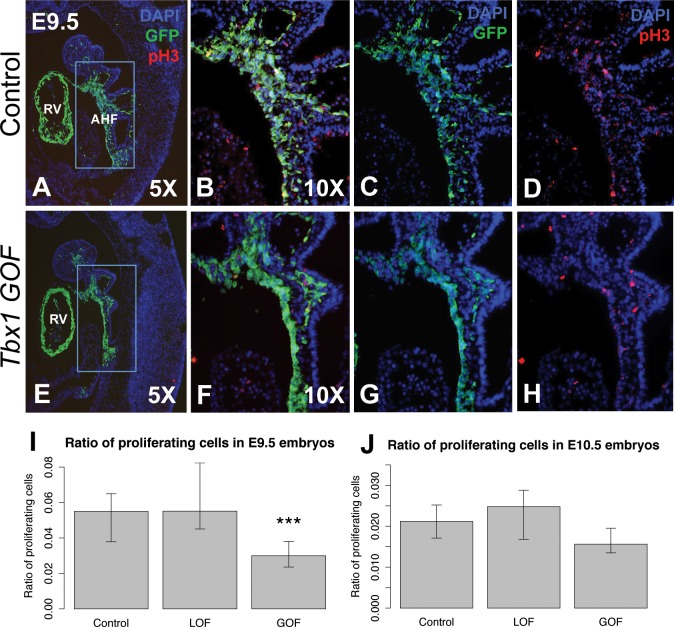
We first analyzed the GFP+ cell number in the AHF in Tbx1 GOF embryos. In Tbx1 GOF embryos, there was no change in number of GFP+ cells in the distal pharyngeal apparatus, when compared with controls at E9.5 (Fig. 4A, B, D, E and J). In contrast to what was observed at E9.5, we found at E10.5, there was a reduction of the number of GFP+ cells in the distal pharyngeal apparatus in the Tbx1 GOF embryos in comparison to littermate controls (Fig. 4G, H and K). To explain the loss of cell number in the GOF embryos at E9.5, proliferation and apoptosis assays were performed (Fig. 5). At E9.5, there was a significant reduction of cell proliferation in the GFP+ domain in the distal pharyngeal apparatus in the Tbx1 GOF embryos in comparison to controls (Fig. 5A–H and I). This is consistent with the reduction of the size of the Mef2c-AHF-Cre lineage population marked by GFP expression at E10.5 (Fig. 4H and K). At E10.5 there was no change in cell proliferation in the Tbx1 GOF embryos in comparison to the controls (Fig. 5J).
Figure 5.
Proliferation of AHF. (A–J) Proliferation of AHF in controls, Tbx1 GOF and Tbx1 LOF embryos. Immunofluorescence images of sagittal sections to visualize the AHF lineage (GFP, green) and cell proliferation [anti-phospho Histone H3 (Ser10); red]; in control and Tbx1 GOF embryos are shown. DAPI fluorescent stain to visualize nuclei and identify the tissue is shown in blue. (D–F) Higher resolution images provide a snapshot of how the cells were counted for statistical analysis. Statistical analysis was performed to determine whether cell proliferation was the same or different between groups of embryos by two-tailed t-test, P value <0.05. Error bars = standard deviation (SD). Abbreviations: AHF (anterior heart field), right ventricle (RV). Control for Tbx1 GOF: Mef2c-AHF-Cre/+; Rosa26GFP/+, Tbx1 GOF: Mef2c-AHF-Cre/+; Tbx1-GFPf/+, control for Tbx1 LOF: Mef2c-AHF-Cre/+; Tbx1f/+; Rosa26GFP/+, Tbx1 LOF: Mef2c-AHF-Cre/+; Tbx1 f/f; Rosa26GFP/+. Asterisks indicate P value <0.05.
We next examined the number of GFP+ AHF cells in Tbx1 LOF embryos. We previously counted the number of such cells at E9.5 (29), shown here as a comparison (Fig. 4C and F) and at E10.5 (Fig. 4I). In Tbx1 LOF embryos, the cell number was normal at E9.5 (29). In contrast, we found an increase of the number of GFP+ cells in Tbx1 LOF embryos at E10.5 (Fig. 4I and K). This may be owing to an expansion or accumulation of the GFP+ cell population in the caudal part of the pharyngeal apparatus in Tbx1 LOF embryos at this stage (Fig. 4I). Data supports an accumulation of GFP+ cells since there were no changes in proliferation in the GFP+ cells in the distal pharyngeal apparatus (Fig. 5I and J).
In both GOF and LOF mutants, there was no change in apoptosis at either stage (data not shown). Based upon the analysis of cell number and proliferation assays, there was a change in proliferation in Tbx1 GOF embryos and an accumulation of cells in Tbx1 LOF embryos, but not visa versa.
Opposite changes in expression of downstream genes when Tbx1 dosage is altered
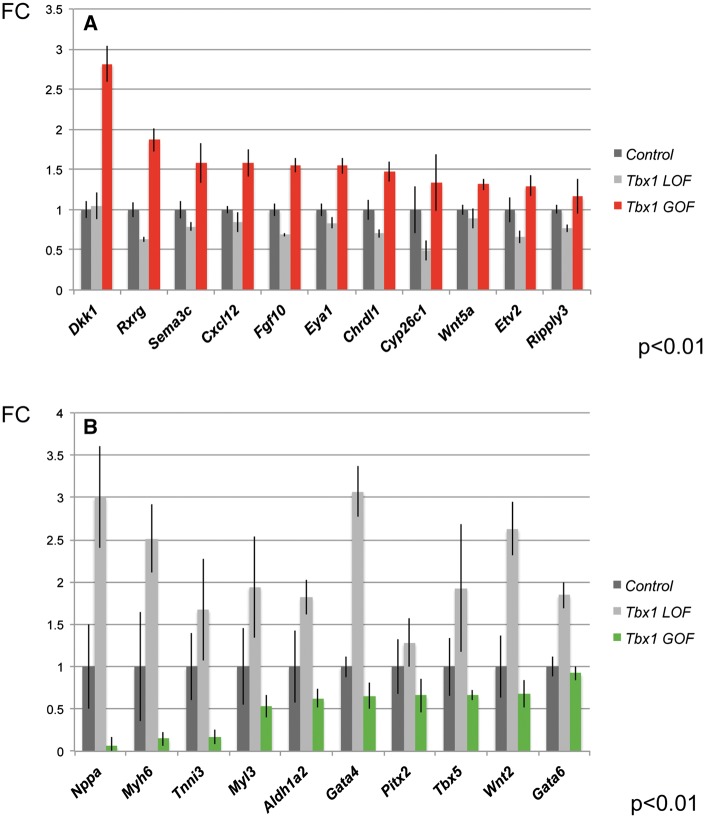
In this section, we will first describe previous gene expression profiling results for the Tbx1 LOF embryos versus controls and then the Tbx1 GOF embryos versus controls. This is to provide a rationale for choosing genes to test in the Tbx1 GOF embryos. Global (39,40), mesodermal (27) or AHF specific loss (29) of Tbx1 demonstrates that it is required in the AHF to promote expression of genes responsible for morphogenesis of the cardiac OFT but at the same time, to prevent premature differentiation while progenitor cells are still located within the pharyngeal apparatus. We previously performed gene expression profiling of distal pharyngeal tissue at E9.5 from AHF conditional Tbx1 LOF embryos and found similar genes were altered, and in the same direction when compared with that of controls, in global Tbx1-/- embryos at E9.5 (29,40). We next wanted to test whether the same genes found in Tbx1 LOF embryos would be found changed, and if so, whether they would be changed in the same or opposite direction in Tbx1 GOF embryos. We selected genes based on gene expression results from microarrays in Tbx1 LOF embryos, and then selected among genes from known expression patterns and function during mouse embryogenesis that are relevant for cardiac development. We performed qRT-PCR in the same tissue dissection and stage in Tbx1 GOF embryos (Fig. 6). We found that when Tbx1 is constitutively active in the AHF of the distal pharyngeal apparatus, genes were oppositely changed when compared with Tbx1 LOF embryos. Opposite to Tbx1 LOF, we found genes that promote morphogenesis are increased in expression in Tbx1 GOF embryos (Fig. 6A). This includes genes such as Fibroblast growth factor 10 (Fgf10), Cytochrome p450 family 26 subfamily c member 1 (Cyp26c1), Dickkopf WNT signaling pathway inhibitor 1 (Dkk1) and Chordin like 1 [Chrdl1] (Fig. 4A). Furthermore, opposite to Tbx1 LOF, we found a strong decrease of expression of posterior second heart field genes, such as T-box transcription factor 5 (Tbx5) and Wnt family member 2 (Wnt2), as well as cardiac muscle differentiation genes [e.g. Myosin heavy chain 6 (Myh6); Myosin light chain 3 (Myl3); Troponin I3 (Tnni3) Fig. 6B]. Thus, Tbx1 GOF has the opposite findings for some of the same representative genes as Tbx1 LOF embryos (Fig. 6). This expression data provides evidence that opposite changes of Tbx1 expression results in opposite changes in downstream gene expression, but yet it results in a PTA, combined with changes in proliferation, to disrupt proper cardiac OFT formation or septation.
Figure 6.
Gene expression changes in the AHF from Tbx1 GOF versus LOF embryos at E9.5. (A and B) Comparison of representative gene expression changes in the microdissected AHF of Tbx1 GOF and Tbx1 LOF embryos at E9.5. Plotted are differentially expressed genes (P < 0.05 and FC > 1.5) comparing Tbx1 LOF and Tbx1 GOF versus controls by qRT-PCR analysis (FC is fold change).
Discussion
CHD in individuals with 22q11.2DupS
Since the first multi-generation family with 22q11.2DupS was reported (41), many clinical studies on individual subjects or small cohorts have been performed such that a total of 133 have been described in the literature. In this report, we doubled the number of cases with this syndrome to 235 in total. One goal of the study was to further define the frequency and spectrum of cardiac anomalies in individuals with the duplication syndrome. Here we found that CHD occurs in 25% of the total of unrelated individuals with 22q11.2DupS (n = 58 of 235). Among those with CHD, 66% have cardiac OFT defects alone or combined and the rest have other cardiac anomalies.
One major difference is that 22q11.2DupS occurs with reduced penetrance when compared with 22q11.2DS. This is not uncommon for deletion and reciprocal duplication disorders. A genomic duplication tends to be better tolerated in humans than a genomic deletion. For CHD, only 25% of 22q11.2DupS are affected, while over 65% of individuals with 22q11.2DS are affected. Despite the lower frequency of CHD, 22q11.2DupS should be considered as a possible explanation when patients present with cardiac malformations.
Balance of Tbx1 dosage is required for normal cardiac OFT development in mouse models
The cardiac OFT, which is affected in 22q11.2DupS and 22q11.2DS, forms as a result of tissue interactions within the pharyngeal apparatus. There are over 40 genes that map to the 22q11.2 region that is hemizygously deleted in these patients (42). Among the genes on 22q11.2, inactivation of both alleles of Tbx1 and/or Crkl (43,44) result in similar cardiac OFT defects in mice. CRKL maps to the distal half of the 22q11.2 region and could also contribute to CHD in patients with the typical 3 Mb deletion. However, Tbx1 is the only gene mapping to the nested 1.5 Mb deleted region associated with the syndrome. Further, overexpression of Tbx1 can provide genetic rescue of mice with a deletion in the proximal half of the 22q11.2 region (24,25). Tbx1 is expressed very specifically in the pharyngeal epithelia and the mesoderm, including that of the AHF, within during embryogenesis. The stage in mouse embryogenesis in which Tbx1 is highly expressed is equivalent to the first trimester of human pregnancy. TBX1 is not expressed in the blood but it is only expressed in adult skeletal muscle or testes. Therefore, gain or loss of expression in human patients cannot be assessed in the critical tissues during embryogenesis, or in adult proxy tissues such as blood.
In the present study we focused on gain of Tbx1 function in the AHF mesoderm marked by the Mef2c-AHF-Cre domain (28), since we recently found LOF in this tissue results in a PTA with complete penetrance (29). We focused on stages E9.5 and E10.5, which are critical stages when the cardiac OFT is elongating. We investigated the basis of the PTA phenotype in the Tbx1 GOF model and compared it with that in the Tbx1 LOF model. When compared with human 22q11.2DupS and 22q11.2DS, the penetrance of CHD in these mouse models is complete and all have a PTA. A PTA is one of the most severe OFT defects in 22q11.2DupS and 22q11.2DS, reflecting the importance of Tbx1 function in the AHF.
We found that CHD occurs in 25% of individuals with 22q11.2DupS, and although most have cardiac OFT defects, few had a PTA. CHD occurs in 60–70% of 22q11.2DS patients and a PTA occurs in ∼3–5% of those with CHD (9,26). In contrast, we found all Tbx1 GOF and LOF embryos had a PTA. This difference between mouse models is because Tbx1 is highly overexpressed in the GOF model and completely inactivated in the LOF model. Nonetheless, consistency of an easily scored phenotype between embryos of the same genotype improves interpretation of molecular experiments.
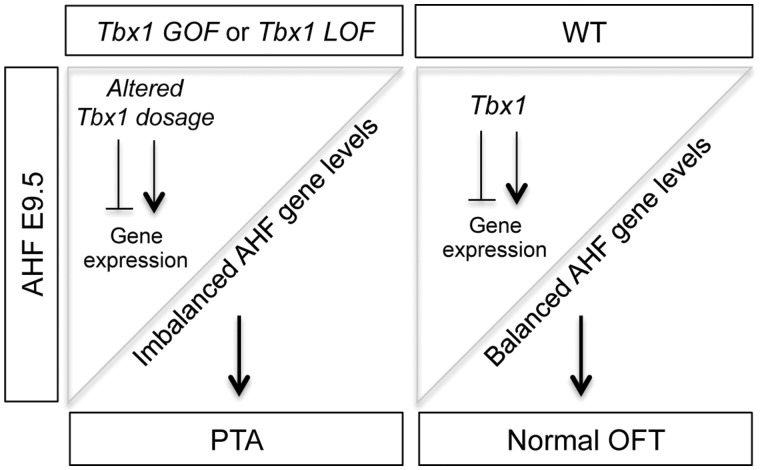
We created a model in Figure 7 that is most consistent with our findings for the AHF specific GOF and LOF mouse models. The cartoon in Figure 7 explains how early perturbations of expression of Tbx1, in either direction, an increase or decrease, can result in the same PTA phenotype in mice. Altered Tbx1 dosage, either by constitutive GOF (Tbx1 GOF) or AHF tissue specific loss of both alleles (Tbx1 LOF), results in imbalance of expression of Tbx1 target genes as shown by our qRT-PCR analyses. Ultimately, this balance is responsible for elongation or later septation of the OFT leading to a PTA. Previous studies of Tbx1 indicate that it promotes proliferation and restricts premature differentiation to cardiac muscle cells in the pharyngeal progenitor cells before they migrate to the cardiac OFT (39,40). The Tbx1 LOF model shares similar gene expression changes as the global null mutant (29). Based upon our gene expression studies of the Tbx1 GOF and Tbx1 LOF model, our data is consistent with opposite effects on expression of downstream genes important for its function in the AHF. The situation with cell proliferation in the Tbx1 GOF and Tbx1 LOF models was somewhat more complicated to understand than the gene expression changes.
Figure 7.
Model for Tbx1 dosage dependent regulation of AHF genes important for OFT septation. In the model, we compare the Tbx1 mutant (left) to control embryos at E9.5. The upper left triangle represents the Tbx1 expression pattern in the AHF that will migrate into the cardiac OFT. Left panel: depicts how altered dosage of Tbx1 results in imbalanced AHF gene expression, which causes a PTA. Right panel: Tbx1 dosage in the AHF of a control embryo. We show that Tbx1 is a key regulator of gene expression for normal cardiac OFT formation resulting in a normal aorta and pulmonary trunk.
Tbx1 has been previously shown to promote cell proliferation in multipotent cardiac progenitors in cell culture and global loss results in reduced proliferation at E9.5 (39,45). Surprisingly, loss of Tbx1 in the AHF did not significantly alter cell proliferation in the AHF lineage at E9.5 (29). Lack of changes in proliferation may reflect differences in the models used and localized inactivation in the AHF. Another difference in the Tbx1 LOF model, which was not identified before, was at E10.5, there appeared to be an accumulation of GFP+ AHF lineage cells in the posterior second heart field region caudal to the heart. We did not observe this accumulation of cells in global Tbx1 null mutant embryos. This accumulation might be a result of a cell deployment defect apparent at E10.5 in this particular model owing to localized inactivation in the AHF.
Since Tbx1 LOF embryos had a PTA with complete penetrance, despite lack of cell proliferation changes at this critical stage, it is possible that Tbx1 GOF embryos would also not have proliferation changes at E9.5, or possibly an increase in cell proliferation that would be opposite to that of global Tbx1 null mutant embryos (39). In contrast, we found a modest decrease in proliferation in Tbx1 GOF embryos at E9.5 and some reduction of cell numbers at E10.5. One explanation for this unexpected finding is because in this model there is constitutive overexpression of Tbx1 in the AHF thereby impacting these cells affecting the formation of the OFT resulting in a PTA. Further, some of the genes oppositely changed in the Tbx1 GOF model might affect cell proliferation when changed in one direction but not the other.
Tissue autonomous and non-autonomous functions of Tbx1 appear to be important in cardiac OFT development. When compared with what we previously found in Tbx1 LOF embryos, in the Tbx1 GOF embryos at E9.5, we found increased expression of AHF genes that promote morphogenesis of the cardiac OFT (Fgf10 and Chrdl1). Again, opposite to the situation in Tbx1 LOF embryos, for Tbx1 GOF embryos, we found reduced expression of Tbx5, in the second heart field and heart, which is important for promoting cardiomyocyte differentiation (46) and acting upstream of muscle structural protein genes (Myh6, Myl3 and Tnni3), which were also decreased in expression. A PTA results from failed septation of the cardiac OFT to form the separate aorta and pulmonary trunk. Neural crest cells (NCCs), which migrate from the closing neural tube through the pharyngeal apparatus to the cardiac OFT, are required for septation of the OFT to form the aorta and pulmonary trunk (47,48). Based upon our Tbx1 LOF and GOF findings, we suggest that Tbx1 may also have opposite non-autonomous functions to the adjacent NCCs. Genes we found oppositely changed between Tbx1 LOF and GOF that are relevant to NCC migration, survival or function includes genes important in the retinoic acid morphogen pathway, including, Rxrg, Aldh1a2 and Cyp26c1 (49–51). Thus, altered balance of expression of autonomous and non-autonomous genes downstream of Tbx1 is critical for cardiac development, interestingly, leading to the same PTA phenotype.
Conclusions
In conclusion, we found that cardiac OFT defects are a hallmark feature of 22q11.2DupS as they are in 22q11.2DS, with the caveat that they occur with reduced penetrance when compared with 22q11.2DS. There are also more varied types of cardiac anomalies in association with 22q11.2DupS than what occurs in 22q11.2DS, perhaps owing to GOF of additional genes on 22q11.2. To understand the molecular pathogenesis and tissue specificity responsible for cardiac OFT defects we found that gain as well as loss of Tbx1 expression in the AHF is required for normal cardiac OFT development. We suggest that a precise dosage of Tbx1 in the AHF is a key regulator of cardiogenic genes downstream of Tbx1 that if becomes disrupted it leads to cardiac OFT defects. Some of these downstream genes might act as modifiers of phenotypes in the reciprocal duplication and deletion disorders.
Materials and Methods
Human study subjects
A duplication of 22q11.2 in subjects was identified using clinically using fluorescence in situ hybridization, multiplex ligation probe amplification and/or microarray comparative genomic hybridization. A total of 102 subjects were ascertained from the 22q and You Center at the Children’s Hospital of Philadelphia referred secondary to a history of congenital anomalies, developmental delay and/or autism spectrum disorder. Echocardiography and/or cardiology reports were reviewed for each subject. All investigations were conducted under an IRB approved protocol at The Children’s Hospital of Philadelphia (Internal Review Board, 07–005352_CR2 CHOP IRB) and Albert Einstein College of Medicine (Committee on Clinical Investigations-CCI# 1999–201). Informed consent was obtained for all participants.
Mouse mutant alleles
The following mouse mutant alleles used in this study have been previously described: Tbx1f/+ (flox = f), Tbx1-GFP, Mef2c-AHF-Cre/+ and ROSA26-GFPf/+ (RCE: loxP). Tbx1 GOF embryos (Tbx1 GOF: Mef2c-AHF-Cre/+; Tbx1-GFPf/+) were generated by crossing male Mef2c-AHF-Cre/+ mice with Tbx1-GFPf/f females. To generate Mef2c-AHF-Cre/+; Tbx1f/- mutant embryos (Tbx1 LOF), Mef2c-AHF-Cre/+ transgenic male mice were crossed to Tbx1+/- mice to obtain male Mef2c-AHF-Cre/+; Tbx1+/- mice, and these were then crossed with Tbx1f/f females. Wild-type, Tbx1-GFPf/+ and Me2fc-AHF-Cre/+; Tbx1f/+ littermates were used as controls for the experiments.
The Mef2c-AHF-Cre/+; Tbx1+/- and the Mef2c-AHF-Cre/+ mice were backcrossed 10 generations to a Swiss Webster background from a mixed C57Bl/6, Swiss Webster background. The PCR strategies for mouse genotyping have been described in the original reports and are available upon request. All experiments including mice were carried out according to regulatory standards defined by the NIH and the Institute for Animal Studies, Albert Einstein College of Medicine (https://www.einstein.yu.edu/administration/animal-studies/), IACUC protocol # 2016-0507.
Mouse embryo heart histology and phenotypic analysis
Mouse embryos were isolated in phosphate-buffered saline (PBS) and fixed in 10% neutral buffered formalin (Sigma Corp.) overnight. Following fixation, the embryos were dehydrated through a graded ethanol series, embedded in paraffin and sectioned at 5 µm. All histological sections were stained with hematoxylin and eosin using standard protocols. Staining was performed in the Einstein Histopathology Core Facility (http://www.einstein.yu.edu/histopathology/page.aspx). A total of 24 hearts, including controls, at E14.5 were obtained from more than 10 to 15 independent crosses and analyzed morphologically using light microscopy.
Mef2c-AHF-cre cell lineage tracing using fluorescence imaging
Images were generated from GFP expressing embryos by direct fluorescence using a Zeiss Discovery-V12 Stereo microscope immediately following dissection. For tissue sections, embryos were fixed for 2 h with embryos stage ≤ E10.5 (30–32 somite pairs). Fixation was carried out in 4% PFA in PBS at 4°C. After fixation, tissue was washed in PBS and then cryoprotected in 30% sucrose in PBS overnight at 4°C. Embryos were embedded in optimal cutting temperature and cryosectioned at 10 µm. Images were then captured using a Zeiss Axio Observer microscope.
Cell number quantification, proliferation and apoptosis assays on tissue sections
To count cell number, we obtained 10-micron serial sagittal sections of Tbx1 GOF embryos, which were collected and stained with an antibody for GFP (ab6673). To ensure that the GFP positive cell quantification was accurate, we counted GFP positive cells in the pharyngeal apparatus in every third section throughout each embryo. We did not count the cells in the OFT or right ventricle. When counting cells, we matched the embryos by stage using somite counts, and we matched the sections by position within the embryo. We also ensured that for each pair of control and mutant embryos we counted the same number of sections (10–12 per embryo). While calculating the cell frequency or density could be useful, since the morphology of the embryo changed throughout the sections, there was some variability between how many GFP positive cells were present per section. We graphed the median number of GFP positive cells per section instead of the mean to account for this variability. Proliferation and apoptosis assays were performed as following: after fixation, frozen sections were obtained as described and then permeabilized in 0.5% Triton X-100 for 5 min. Blocking was performed with 5% serum (goat or donkey) in PBS/0.1% Triton X-100 (PBT) for 1 h. Primary antibody was diluted in blocking solution (1:500) and incubated for 1 h. Proliferation of cells was assessed by immunofluorescence using the primary antibody anti-phospho Histone H3 (Ser10), a mitosis marker (06-570 Millipore). Sections were washed in PBT and incubated with a secondary antibody for 1 h. Secondary antibody was Alexa Fluor 568 goat a-rabbit IgG (A11011 Invitrogen) at 1:500. Slides were mounted in hard-set mounting medium with DAPI (Vector Labs H-1500). Images were captured using a Zeiss Axio Observer microscope. We counted all proliferating cells in each section and calculated the ratio of proliferating cells within the Mef2c-AHF-Cre lineage. Then, we estimated the mean and standard error of the average ratios of proliferating cells to GFP+ cells for controls and Tbx1 GOF embryos and compared them using the t-test. Apoptosis was assessed on 10 μm thick frozen sections by using TMR Red In situ Cell Death kit (2156792 Roche) following the manufacturer’s instructions. Natural GFP from the reporter or an antibody for GFP (Abcam 6290) was used to distinguish the AHF cells in both assays described above. Representations of the complete AHF region from at least 4 embryos per genotype from at least 3 independent litters were used in each assay.
Quantitative RT-PCR
Embryos at E9.5 (19–21 somite pairs) were used for quantitative gene expression studies of the distal pharyngeal apparatus, containing the AHF, microdissected from each of the following genotypes: Tbx1 LOF and its control (Tbx1f/+) and Tbx1 GOF were pooled in groups according to genotype. Tbx1f/+ was used as control. Gene expression assays were performed as previously described (29). Statistical significance of the difference in gene expression was estimated using ANOVA and the two-tailed t-test independently when type of comparison allowed it.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank Dr Tao Wang in the Department of Epidemiology & Population Health and Genetics for consultation on the statistical analyses of cell number, proliferation and apoptosis assays.
Conflict of Interest statement. None declared.
Funding
We want to thank the Histopathology Facility at Einstein. Foundation Leducq supported this work, NIH grants, P01HD070454 (BM) and an AHA grant 12POST9100003 (SER).
References
- 1. Gross S.J., Stosic M., McDonald-McGinn D.M., Bassett A.S., Norvez A., Dhamankar R., Kobara K., Kirkizlar E., Zimmermann B., Wayham N.. et al. (2016) Clinical experience with single-nucleotide polymorphism-based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet Gynecol., 47, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McDermid H.E., Morrow B.E. (2002) Genomic disorders on 22q11. Am. J. Hum. Genet., 70, 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edelmann L., Pandita R.K., Morrow B.E. (1999) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet., 64, 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E. (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet., 8, 1157–1167. [DOI] [PubMed] [Google Scholar]
- 5. Shaikh T.H., Kurahashi H., Emanuel B.S. (2001) Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: an update and literature review. Genet. Med., 3, 6–13. [DOI] [PubMed] [Google Scholar]
- 6. Goldmuntz E., Driscoll D., Budarf M.L., Zackai E.H., McDonald-McGinn D.M., Biegel J.A., Emanuel B.S. (1993) Microdeletions of chromosomal region 22q11 in patients with congenital conotruncal cardiac defects. J. Med. Genet., 30, 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keyte A.L., Alonzo-Johnsen M., Hutson M.R. (2014) Evolutionary and developmental origins of the cardiac neural crest: building a divided outflow tract. Birth Defects Res. C Embryo Today, 102, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lewin M.B., Lindsay E.A., Jurecic V., Goytia V., Towbin J.A., Baldini A. (1997) A genetic etiology for interruption of the aortic arch type B. Am. J. Cardiol., 80, 493–497. [DOI] [PubMed] [Google Scholar]
- 9. Peyvandi S., Lupo P.J., Garbarini J., Woyciechowski S., Edman S., Emanuel B.S., Mitchell L.E., Goldmuntz E. (2013) 22q11.2 deletions in patients with conotruncal defects: data from 1, 610 consecutive cases. Pediatr. Cardiol., 34, 1687–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shaikh T.H., Kurahashi H., Saitta S.C., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L.. et al. (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet., 9, 489–501. [DOI] [PubMed] [Google Scholar]
- 11. Saitta S.C., Harris S.E., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H.. et al. (2004) Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet., 13, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McDonald-McGinn D.M., Sullivan K.E., Marino B., Philip N., Swillen A., Vorstman J.A., Zackai E.H., Emanuel B.S., Vermeesch J.R., Morrow B.E.. et al. (2015) 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers, 1, 15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ou Z., Berg J.S., Yonath H., Enciso V.B., Miller D.T., Picker J., Lenzi T., Keegan C.E., Sutton V.R., Belmont J.. et al. (2008) Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet. Med., 10, 267–277. [DOI] [PubMed] [Google Scholar]
- 14. Turner D.J., Miretti M., Rajan D., Fiegler H., Carter N.P., Blayney M.L., Beck S., Hurles M.E. (2008) Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet., 40, 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wentzel C., Fernstrom M., Ohrner Y., Anneren G., Thuresson A.C. (2008) Clinical variability of the 22q11.2 duplication syndrome. Eur. J. Med. Genet., 51, 501–510. [DOI] [PubMed] [Google Scholar]
- 16. Portnoi M.F. (2009) Microduplication 22q11.2: a new chromosomal syndrome. Eur. J. Med. Genet., 52, 88–93. [DOI] [PubMed] [Google Scholar]
- 17. Hassed S.J., Hopcus-Niccum D., Zhang L., Li S., Mulvihill J.J. (2004) A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin. Genet., 65, 400–404. [DOI] [PubMed] [Google Scholar]
- 18. Brunet A., Gabau E., Perich R.M., Valdesoiro L., Brun C., Caballin M.R., Guitart M. (2006) Microdeletion and microduplication 22q11.2 screening in 295 patients with clinical features of DiGeorge/Velocardiofacial syndrome. Am. J. Med. Genet. A, 140A, 2426–2432. [DOI] [PubMed] [Google Scholar]
- 19. Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S.. et al. (2003) Role of TBX1 in human del22q11.2 syndrome. Lancet, 362, 1366–1373. [DOI] [PubMed] [Google Scholar]
- 20. Zweier C., Sticht H., Aydin-Yaylagul I., Campbell C.E., Rauch A. (2007) Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet., 80, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rauch R., Hofbeck M., Zweier C., Koch A., Zink S., Trautmann U., Hoyer J., Kaulitz R., Singer H., Rauch A. (2010) Comprehensive genotype–phenotype analysis in 230 patients with tetralogy of Fallot. J. Med. Genet., 47, 321–331. [DOI] [PubMed] [Google Scholar]
- 22. Pan Y., Wang Z.G., Liu X.Y., Zhao H., Zhou N., Zheng G.F., Qiu X.B., Li R.G., Yuan F., Shi H.Y.. et al. (2015) A novel TBX1 loss-of-function mutation associated with congenital heart disease. Pediatr. Cardiol., 36, 1400–1410. [DOI] [PubMed] [Google Scholar]
- 23. Jerome L.A., Papaioannou V.E. (2001) DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet., 27, 286–291. [DOI] [PubMed] [Google Scholar]
- 24. Lindsay E.A., Vitelli F., Su H., Morishima M., Huynh T., Pramparo T., Jurecic V., Ogunrinu G., Sutherland H.F., Scambler P.J.. et al. (2001) Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature, 410, 97–101. [DOI] [PubMed] [Google Scholar]
- 25. Merscher S., Funke B., Epstein J.A., Heyer J., Puech A., Lu M.M., Xavier R.J., Demay M.B., Russell R.G., Factor S.. et al. (2001) TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell, 104, 619–629. [DOI] [PubMed] [Google Scholar]
- 26. Ryan A.K., Goodship J.A., Wilson D.I., Philip N., Levy A., Seidel H., Schuffenhauer S., Oechsler H., Belohradsky B., Prieur M.. et al. (1997) Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J. Med. Genet., 34, 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Z., Huynh T., Baldini A. (2006) Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development, 133, 3587–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verzi M.P., McCulley D.J., De Val S., Dodou E., Black B.L. (2005) The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol., 287, 134–145. [DOI] [PubMed] [Google Scholar]
- 29. Racedo S.E., Hasten E., Lin M., Devakanmalai G.S., Guo T., Ozbudak E.M., Cai C.L., Zheng D., Morrow B.E. (2017) Reduced dosage of beta-catenin provides significant rescue of cardiac outflow tract anomalies in a Tbx1 conditional null mouse model of 22q11.2 deletion syndrome. PLoS Genet., 13, e1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao J., Kochilas L., Nowotschin S., Arnold J.S., Aggarwal V.S., Epstein J.A., Brown M.C., Adams J., Morrow B.E. (2004) Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum. Mol. Genet., 13, 1577–1585. [DOI] [PubMed] [Google Scholar]
- 31. Vitelli F., Huynh T., Baldini A. (2009) Gain of function of Tbx1 affects pharyngeal and heart development in the mouse. Genesis, 47, 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freyer L., Nowotschin S., Pirity M.K., Baldini A., Morrow B.E. (2013) Conditional and constitutive expression of a Tbx1-GFP fusion protein in mice. BMC Dev. Biol., 13, 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Z., Cerrato F., Xu H., Vitelli F., Morishima M., Vincentz J., Furuta Y., Ma L., Martin J.F., Baldini A.. et al. (2005) Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development, 132, 5307–5315. [DOI] [PubMed] [Google Scholar]
- 34. Arnold J.S., Werling U., Braunstein E.M., Liao J., Nowotschin S., Edelmann W., Hebert J.M., Morrow B.E. (2006) Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development, 133, 977–987. [DOI] [PubMed] [Google Scholar]
- 35. Jackson A., Kasah S., Mansour S.L., Morrow B., Basson M.A. (2014) Endoderm-specific deletion of Tbx1 reveals an FGF-independent role for Tbx1 in pharyngeal apparatus morphogenesis. Dev. Dyn., 243, 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Baldini A. (2008) In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum. Mol. Genet., 17, 150–157. [DOI] [PubMed] [Google Scholar]
- 37. Chen M., Yang Y.S., Shih J.C., Lin W.H., Lee D.J., Lin Y.S., Chou C.H., Cameron A.D., Ginsberg N.A., Chen C.A.. et al. (2014) Microdeletions/duplications involving TBX1 gene in fetuses with conotruncal heart defects which are negative for 22q11.2 deletion on fluorescence in-situ hybridization. Ultrasound Obstet. Gynecol., 43, 396–403. [DOI] [PubMed] [Google Scholar]
- 38. Tzahor E., Evans S.M. (2011) Pharyngeal mesoderm development during embryogenesis: implications for both heart and head myogenesis. Cardiovasc. Res., 91, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen L., Fulcoli F.G., Tang S., Baldini A. (2009) Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ. Res., 105, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liao J., Aggarwal V.S., Nowotschin S., Bondarev A., Lipner S., Morrow B.E. (2008) Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev. Biol., 316, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edelmann L., Spiteri E., McCain N., Goldberg R., Pandita R.K., Duong S., Fox J., Blumenthal D., Lalani S.R., Shaffer L.G.. et al. (1999) A common breakpoint on 11q23 in carriers of the constitutional t(11; 22) translocation. Am. J. Hum. Genet., 65, 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guna A., Butcher N.J., Bassett A.S. (2015) Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J. Neurodev. Disord., 7, 18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guris D.L., Fantes J., Tara D., Druker B.J., Imamoto A. (2001) Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet., 27, 293–298. [DOI] [PubMed] [Google Scholar]
- 44. Guris D.L., Duester G., Papaioannou V.E., Imamoto A. (2006) Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev. Cell, 10, 81–92. [DOI] [PubMed] [Google Scholar]
- 45. Xu H., Morishima M., Wylie J.N., Schwartz R.J., Bruneau B.G., Lindsay E.A., Baldini A. (2004) Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development, 131, 3217–3227. [DOI] [PubMed] [Google Scholar]
- 46. Brunskill E.W., Witte D.P., Yutzey K.E., Potter S.S. (2001) Novel cell lines promote the discovery of genes involved in early heart development. Dev. Biol., 235, 507–520. [DOI] [PubMed] [Google Scholar]
- 47. Kirby M.L. (1990) Alteration of cardiogenesis after neural crest ablation. Ann. N. Y. Acad. Sci., 588, 289–295. [DOI] [PubMed] [Google Scholar]
- 48. Kirby M.L., Waldo K.L. (1995) Neural crest and cardiovascular patterning. Circ. Res., 77, 211–215. [DOI] [PubMed] [Google Scholar]
- 49. Sucov H.M., Dyson E., Gumeringer C.L., Price J., Chien K.R., Evans R.M. (1994) RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev., 8, 1007–1018. [DOI] [PubMed] [Google Scholar]
- 50. Niederreither K., Vermot J., Messaddeq N., Schuhbaur B., Chambon P., Dolle P. (2001) Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development, 128, 1019–1031. [DOI] [PubMed] [Google Scholar]
- 51. El Robrini N., Etchevers H.C., Ryckebusch L., Faure E., Eudes N., Niederreither K., Zaffran S., Bertrand N. (2016) Cardiac outflow morphogenesis depends on effects of retinoic acid signaling on multiple cell lineages. Dev. Dyn., 245, 388–401. [DOI] [PubMed] [Google Scholar]
- 52. Yu S., Graf W.D., Ramalingam A., Brawner S.J., Joyce J.M., Fiedler S., Zhou X.G., Liu H.Y. (2011) Identification of copy number variants on human chromosome 22 in patients with a variety of clinical findings. Cytogenet Genome Res., 134, 260–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.