Abstract
Supercoiling can alter the form and base pairing of the double helix and directly impact protein binding. More indirectly, changes in protein binding and the stress of supercoiling also influence the thermodynamic stability of regulatory, protein-mediated loops and shift the equilibria of fundamental DNA/chromatin transactions. For example, supercoiling affects the hierarchical organization and function of chromatin in topologically associating domains (TADs) in both eukaryotes and bacteria. On the other hand, a protein-mediated loop in DNA can constrain supercoiling within a plectonemic structure. To characterize the extent of constrained supercoiling, 400 bp, lac repressor-secured loops were formed in extensively over- or under-wound DNA under gentle tension in a magnetic tweezer. The protein-mediated loops constrained variable amounts of supercoiling that often exceeded the maximum writhe expected for a 400 bp plectoneme. Loops with such high levels of supercoiling appear to be entangled with flanking domains. Thus, loop-mediating proteins operating on supercoiled substrates can establish topological domains that may coordinate gene regulation and other DNA transactions across spans in the genome that are larger than the separation between the binding sites.
INTRODUCTION
Supercoiling is an inherent and dynamic feature of DNA that sensitively regulates genome-based reactions (1–4), altering base-pairing or the form of the helix (5,6) to modify protein binding. It affects the thermodynamic stability of regulatory, protein-mediated loops (7,8), and may become trapped within such loops (7,9,10). In mouse liver cells, topologically associating domains (TADs) with loops catalyzed by CTCF (11) are flanked by topoisomerase (TOP2B), an enzyme that modifies supercoiling (12). This suggests that the interplay between supercoiling and looping is fundamental to eukaryotic gene regulation. Indeed, psoralen incorporation has shown that transcriptionally active TADs are negatively supercoiled (13). The presence of TOP2B at the border between TADs may decouple supercoiling levels in adjacent domains such that one domain can be highly supercoiled and looped, independently of the status of adjacent domains. This would enable discrete loop/supercoiling-mediated gene regulation in each TAD. Supercoiled TADs could be a natural consequence of the supercoiling generated by transcription. In models of divergent transcription operating in 600 kbp regions, supercoiled and torsionally relaxed domains spontaneously became interspersed, and the three-dimensional proximity of DNA segments exhibited a pattern like that observed experimentally in Schizosaccharomyces pombe using chromosome conformation capture assays (14). Supercoiling also affects the hierarchical organization and function of chromatin in prokaryotes like Streptococcus pneumonia which has genes organized in topology-sensitive clusters (15).
This evidence suggests that supercoiling plays a role in establishing domains in chromatin. It is straightforward to imagine the conformational differences between a linkage between two sites in a gently curved DNA molecule that forms a simple loop versus a linkage between juxtaposed sites in a solenoidally or plectonemically coiled molecule that traps supercoiling and may even produce knots. The entanglement that accompanies supercoiling creates topology that, when stabilized by a protein-mediated junction, may have regulatory consequences due to the compaction of the DNA, the difference in supercoiling within and outside the loop, and the partitioning that isolates segments inside a loop from those outside the loop (16).
Although the supercoiling and topology of DNA segments are dynamic and so complex in vivo that measurements would be difficult, single molecule manipulation in vitro allows arbitrary twisting of DNA while monitoring the overall extension of the molecule to detect loop formation. Therefore, to investigate the interplay between supercoiling and looping, superparamagnetic beads were tethered to the surface of a glass microchamber by DNA molecules containing two lac repressor (LacI) binding sites separated by 400 bases, and LacI was added to the solution. LacI protein is a paradigmatic DNA looping protein (17) with two DNA-binding domains connected by a hinge, and can bind simultaneously two, non-adjacent binding sites, producing a loop in the intervening DNA. We used magnetic tweezers (MT) (18) to impart controlled amounts of supercoiling to the DNA tethers, and monitored intermittent shortening of the molecules due to LacI-mediated looping and entanglement, as well as the amount of writhe (19) trapped by loops. LacI-mediated loops often sequestered supercoiling and topologically constrained domains larger than the loop. Thus, protein-mediated loops in supercoiled DNA affect not only the circumscribed DNA but also topologically extend to flanking sequences. This newly discovered way of creating extended topological domains may enable the coordinated regulation of serially arranged elements of the genome.
MATERIALS AND METHODS
Preparation of DNA tethers
DNA tethers were 2121 bp (0.7 μm) in length and contained the O1 and O2 lac repressor operators separated by 400 bp at approximately the center of the tether (Figure S1, supplementary information). These constructs were produced by PCR using plasmid pO1O2_401 (9) as a template for 5′-TGCCCGGGACCCGGAAAGACATGC and 5′-CTGGGCCCGGTGAATCCGTTAGCGA primers, digesting the amplicon with ApaI and XmaI (New England BioLabs, Ipswich, MA, USA), and subsequently purifying with silica-membrane-column kits (Qiagen, Germantown, MD, USA). Biotin- or digoxigenin-labeled DNA end fragments were generated using PCR with pBluKSP+ as plasmid template for 5′-TGGGTGAGCAAAAACAGGAAGGCA and 5′-GCGTAATCTGCTGCTTGCAA primers with dATP, dCTP, dGTP, dTTP (Fermentas-Thermo Fisher Scientific Inc., Pittsburgh, PA) and digoxigenin-11-dUTP (Roche Life Science, Indianapolis, IN, USA) or biotin-11-dUTP (Invitrogen, Life Technologies, Grand Island, NY, USA) in a molar ratio of 1:1:1:0.9:0.1. These primers amplify a 2 kb segment with the multi-cloning site of pBluKSP+ at the center. Cleavage of these bio- or dig-labeled segments with ApaI or XmaI generated ∼1 kb, labeled fragments with 4-bp overhangs for ligation. These labeled DNA fragments were ligated (T4 DNA ligase, New England BioLabs), to opposite ends of the central fragment to produce DNA for tethering streptavidin-labeled beads to an anti-digoxigenin-coated coverglass. The maps of the plasmids and sequences of the primers used are shown in Supplementary Figure S1.
Micro-chamber preparation
Chambers were prepared as previously reported (16,20,21). In brief, micro-chambers with an approximate volume of ∼30 μl were prepared by laser-cutting a gasket from parafilm and mildly heating to seal it between two coverslips (Fisherbrand, Thermo Fisher Scientific, Waltham, MA, USA). The channel inlet and outlet were narrow to reduce the evaporation of buffer during observation (Supplementary Figure S2). The digoxigenin-labeled ends of DNA molecules were attached to the glass surface coated with polyclonal anti-digoxigenin (Roche Life Science, Indianapolis, IN, USA) and the opposite biotin-labeled ends to 1.0 μm-diameter, streptavidin-coated paramagnetic beads (Dynabead MyOne Streptavidin T1, Invitrogen, Grand Island, NY, USA). The chamber surfaces were passivated with 0.1 mg/ml BSA to prevent non-specific sticking of the DNA and beads to the surface. Prepared chambers were filled with buffer (10 mM Tris–HCl pH = 7.4, 200 mM KCl, 0.5 mg/ml α-casein) and stored in a high humidity box at 4°C up to 24 h or used directly after gently flushing with 200 μl λ buffer (10 mM Tris–HCl pH 7.4, 200 mM KCl, 5% DMSO, 0.1 mM EDTA, 0.2 mM DTT and 0.2 mg/ml α-casein).
Magnetic tweezing
DNA tethers were stretched and twisted using magnetic tweezers, a pair of magnets just above the micro-chamber that can be moved along or rotated around the optical axis of the microscope. The custom instrument is controlled, and data is acquired using MatLab software (Mathworks, Natick, MA, USA). DNA molecules without nicks formed plectonemes that reduced extension upon under- or over-winding at low tension (<0.4 pN) (Supplementary Figure S3) (7,22,23) and were selected for analysis. The supercoiling of a DNA molecule can be quantified using the change in the linking number, ΔLk = Lk – Lk0 in which Lk is the measured linking number, and the linking number of torsionally relaxed DNA, Lk0 equals the number of base-pairs divided by the helical repeat (∼10.4 bp/turn for B DNA). The change in the linking number includes both the excess twist and writhe, ΔLk = ΔTw + ΔWr, compared to torsionally relaxed DNA. DNA molecules were repeatedly wound or unwound, in steps of two turns, to change the linking number by ±10% (±20 turns/(2121 bp/10.4 bp/turn)) at tensions of 0.25 or 0.45 pN, values that are estimated to be relevant for genomic DNA in physiological conditions (24,25). Under 0.25 pN tension, the extension-vs-turns curves without protein were symmetric. At 0.45 pN, extension-vs-turns curves without protein exhibited a longer maximum extension and were slightly asymmetric, due to partial conversion of unwound DNA to left-handed forms (6,22) (Supplementary Figure S3). The extension-versus-turns data was recorded, first without and then after adding 1 nM LacI protein (provided by Kathleen Matthews, Rice University). The protein was present throughout coiling or uncoiling the DNA, and loops that formed in the highly supercoiled tethers were subsequently detected by shifted and/or decreased maximal extensions when the DNA tethers were uncoiled.
Exclusion of artifacts
Non-specific interactions could in principle decrease the length of the DNA tether and might be of three different types: (i) bead to surface, (ii) DNA to surface or bead, (iii) DNA-bound protein to surface or bead. We have ruled out the possibility that any of these could be the cause of the DNA length shortening we observed in this study. First, non-specific sticking of a DNA-tethered bead to the surface of the flow chamber would produce tether length measurements equal to zero. Such tethers were discarded from the analysis. Second, several observations indicated that transient, non-specific interactions between DNA and either the surface of the flow-chamber or the bead did not occur in the analysed data: (i) control measurements in the absence of LacI (the looping protein) showed only one state, and the extension-vs-turns curves recorded in this condition had reproducible maxima; (ii) decreases in the extension of the tethers were greater than or equal to the expected length of the loop segment, Zloop, under the applied tensions. Random, non-specific sticking would have also produced smaller decreases, which were not observed; (iii) the clear correlation between the tether length decrease and number of turns trapped in the LacI-mediated loop would not result from non-specific interactions that caused temporary, sticking of random segments of the DNA to either the surface of the flow-chamber, or the bead. Third, no interaction between LacI and streptavidin, or anti-digoxigenin, has been reported, and observations that allow us to exclude artifacts caused by DNA sticking to the bead or glass surfaces (ii and iii) allow us to exclude protein-mediated DNA sticking as well.
Finally, non-specific binding of LacI to DNA is negligible (if not non-existent) in the buffer condition and LacI concentration used, as seen by atomic force imaging (26,27) and tethered particle microscopy assays where, in the absence of supercoiling, the extension of a DNA tether without operator sequences in the presence of LacI remained constant in time (28–31).
Measuring the coiling and extent of topological domains
The x, y, z, t coordinates of a tethered and a non-specifically stuck bead were recorded in real-time using a custom MatLab routine to analyze 10 Hz digital video. A set of control traces lasting 2.5 mins were recorded under tensions of 0.25, 0.45 or 0.75 pN and several positive and negative supercoiling levels were measured to determine the nominal DNA extension before adding LacI protein. After adding 1 nM LacI protein, the x, y, z, t data were recorded and analyzed using a ‘change point’ algorithm followed by an expectation-maximization routine (32,33) to identify distinct conformational states and estimate their lifetimes. Note that while looped states were essentially perpetual under 0.25 pN of tension in DNA supercoiled to –2% superhelical density, when the tension was increased to 0.45 or even 0.75 pN, the loop more frequently ruptured and the molecule extended briefly to an unlooped state. A previous report shows that the probability of a looped conformation at a given tension increases with negative supercoiling, and higher levels of supercoiling are necessary to overcome higher tension to maintain equivalent looping probability (7).
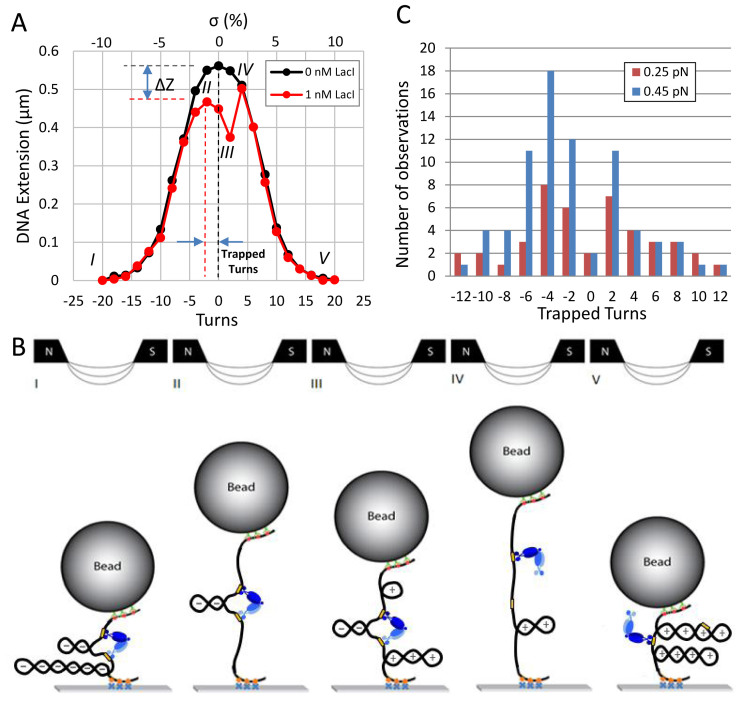
As for measurements of supercoiling trapped in DNA loops mediated by the lambda-CI protein (7), the number of turns trapped by the LacI-mediated DNA loop was indicated by the relative shift of the peak in the extension-vs-turns curves recorded with and without LacI protein (Figure 1A). The difference, ΔZ, between the peak height of the extension-vs-turns curve with and without protein (Figure 1A), was used to estimate the extent of DNA topologically trapped by the LacI-secured loop.
Figure 1.
LacI-mediated loops trap supercoiling and resist torsion. The formation of plectonemes in an O1-400-O2 construct under 0.25 pN of tension without LacI produced a typical extension-vs-turns curve (black). When LacI was added to the buffer, the maxima of subsequent extension-vs-turns curves were often reduced by ΔZ due to loop formation by LacI and shifted according to the number of trapped supercoils. (A) In this example, after unwinding by -20 turns, an extended plectoneme reduced the extension of the tether (state I) with or without LacI. The relative shift of the maxima of the extension-vs-turns curves acquired with (red) or without (black) LacI while winding from –20 to –2 turns indicates that two negative supercoils were trapped within the loop (state II). After further winding to +2 turns to reach state III, the loop ruptured, and the extension became identical to that without LacI (state IV). Further winding to +20 produced extensions equal to those observed without LacI. (B) Schematic representations of supercoiled configurations compatible with the red extension-vs-turns curve in A. At –20 turns a large plectoneme is formed in which LacI can secure a loop between juxtaposed operators (state I). Winding the tether from –20 to –2 turns, relaxed flanking plectonemic gyres but not those within the LacI-mediated loop, which shifted the maximum of the extension-vs-turns curve from 0 to –2 turns (state II). Further winding to +2 turns produced (+) plectonemes in the flanking DNA, while the plectoneme within the loop remained negatively (–) supercoiled (state III). In this state, the LacI junction sustains maximal torsional stress between oppositely supercoiled DNA segments. When the loop spontaneously ruptured, negative supercoiling within immediately relaxed an equal amount of positive supercoiling in the flanking DNA giving a more extended tether with a smaller plectoneme (state IV). Thereafter, further winding extended the plectoneme and the extension-vs-turns curve superimposed with that of an unlooped tether as a large plectoneme with positive supercoils formed (state V). LacI might secure loops between juxtaposed operators in this configuration that could be detected by a shift in the extension-vs-turns maximum during unwinding. (C) Repeatedly winding and unwinding DNA tethers in the presence of LacI and noting the shifts in the extension-vs-turns curves produced distributions of the number of supercoils trapped by the LacI-mediated loops in tethers under 0.25 (red) or 0.45 (blue) pN of tension. While the distribution of trapped supercoils is symmetric about zero for tethers under 0.25 pN of tension for equal numbers of winding and unwinding cycles, negative supercoils are more often trapped in loops formed at 0.45 pN. Note also that few loops formed without trapping any supercoiling.
Estimating plectonemic gyre lengths
As DNA is twisted, the polymer absorbs torque until reaching the buckling threshold beyond which additional twist is absorbed as plectonemic writhe, and the end-to-end distance of the tether decreases until the entire tether is plectonemic. The change in the extension due to a plectonemic gyre in moderately supercoiled DNA can be measured from the slope of the linear section of extension-vs-turns curves, for example Supplementary Figure S3, and for 0.25–0.45 pN is about 50 nm (34–38). However, for loops formed in highly supercoiled DNA tethers the relevant size of a plectonemic gyre (ΔZ/turn), Zgyre, was determined by dividing the maximum extension by the maximum number of imposed turns (Ztether/|turnsmax|, Supplementary Table S1). Zgyre was then used to estimate expected values of ΔZ using Equation (1) as described in the Results section.
RESULTS
Loop-securing LacI resists torsional stress in dynamically supercoiled DNA
To investigate supercoiled loops, para-magnetic microspheres were tethered to a cover glass by single 2121 bp DNA molecules that were gently stretched and twisted using magnetic tweezers. Twisting in the absence of LacI produced symmetric extension-vs-turns curves in which progressively larger plectonemes reduced the extension, as the amount of mechanically introduced twist increased (Figure 1A black, S4 black). The extension drops perceptibly with as little as ±2% supercoiling and dropped to <20% of the untwisted length with ±6% supercoiling. Six percent negative supercoiling is commonly measured for plasmids extracted from bacteria (39), and enzymatically measured estimates of the in vivo supercoiling levels not constrained by proteins in bacterial genomes range from 2 to 4% (40). In the presence of LacI, extension-vs-turns curves often exhibited shifted and/or reduced peaks (Figure 1A red and Supplementary Figure S4, red or blue) with respect to controls without LacI. In the representative example depicted in Figure 1A (red), initially the DNA was extensively negatively supercoiled (–20 turns) with an extended plectoneme (Figure 1B, state I). Successive cartoons in Figure 1B represent conformations that the molecule likely visited during winding from –20 to +20 turns (Figure 1A, red curve). Winding progressively relaxed plectonemes in the flanking segments, but plectonemes within the LacI-mediated loop were protected, so that the extension-vs-turns curve displayed a reduced maximum at –2 turns (state II). Further winding produced positive plectonemes in the flanking DNA (+), while the plectoneme within the loop remained negatively (–) supercoiled (state III). This condition, with plectonemes of opposite handedness inside and outside the loop, exerts the maximal possible torsional stress on the LacI tetramer securing the loop. When the loop ruptured, negative supercoiling within the loop immediately relaxed an equal amount of positive supercoiling in the flanking DNA giving a new torsional equilibrium (state IV). Thereafter, the curve was identical to that of an unlooped tether.
LacI-mediated loops preferentially trap negative supercoiling at higher tension
Loops that trapped superhelicity were observed under two levels of tension (Figure 1C). Under 0.25 pN of tension, loops trapped similar distributions of negative and positive supercoils and torsion-free loops rarely formed. Under 0.45 pN of tension, torsion-free loops were rare as well, but loops that trapped supercoiling readily formed and more frequently trapped negative than positive supercoils. At either tension, some of the LacI-mediated loops trapped as many as ±12 turns. Several studies of DNA with moderately sized plectonemes have established the contour length per writhe at ∼50 nm under approximately half a piconewton of tension (34–38). However, twelve plectonemic gyres 50 nm in length do not fit into a 400 bp segment. Note also that at 0.45, but not 0.25 pN, LacI more often trapped negative supercoiling. Since unwinding DNA under 0.45 pN of tension produces a partial transition to L-DNA (5,6), which has a left-handed helical repeat of 15 bp, it would be possible to absorb twelve negative supercoils as L-DNA in a 400 bp loop. However, the phase change to L-DNA does not occur at 0.25 pN for which high numbers of trapped supercoils were also observed. In addition, positive supercoiling under either tension does not induce a helical phase change that might allow +12 turns to become trapped in the loop. All together these observations indicate that reductions of the extension to the maxima observed in the shifted peaks involve a more general mechanism of trapping supercoils.
LacI-mediated loops dynamically trap large topological domains in supercoiled DNA
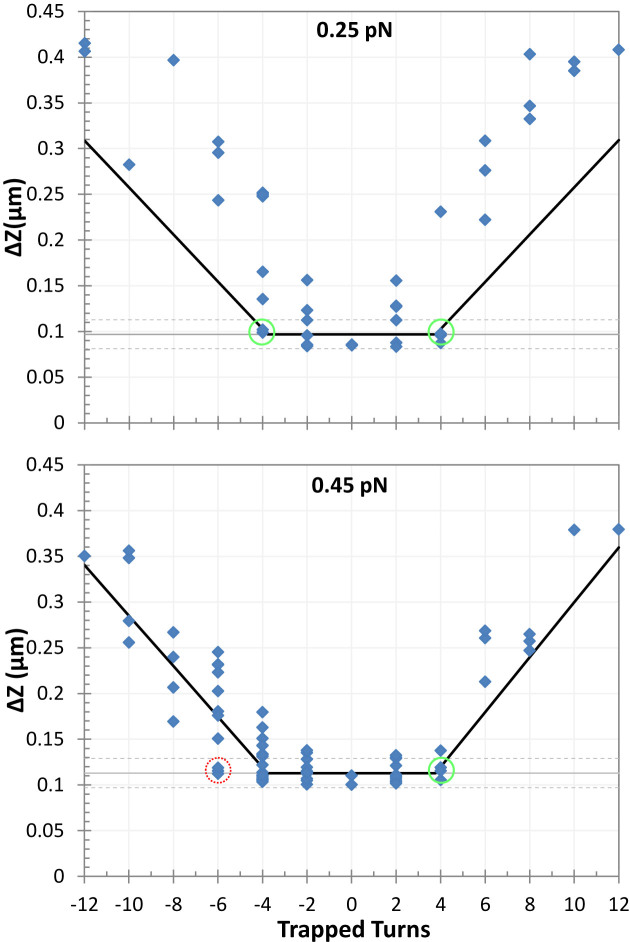
A naïve expectation is that the boundaries of topological domains induced by looping proteins should correspond to the contour length between the delimiting protein binding sites. In this study, such a topological domain would reduce the maxima of extension-vs-turns curves by a value, Zloop, proportional to the length of the loop, Lloop. Figure 2 reports the reduction of the extension-vs-turns maxima for LacI-mediated loops relative to unlooped DNA tethers, ΔZ, as a function of the number of turns trapped by the loop. Expected Zloop values were estimated using the proportionality constants described in Supplementary Figure S3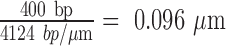 or
or 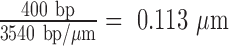 for 0.25 pN or 0.45 pN, respectively. Zloop values were expected to lie along the grey, horizontal lines. The largest ΔZ observed at 0.25 pN, 0.41 μm, is 4.3-fold larger than Zloop. Under 0.45 pN of tension, ΔZ values as large as 0.38 μm, 3.4 times Zloop, were observed. Thus, LacI-mediated loops in supercoiled DNA frequently established topological domains much larger than the loop length, and the size of the increase was inversely correlated with tension. In Supplementary Figure S5, a cumulative histogram of ΔZ values for DNA tethers shows that at 0.45 pN, 50% of the topological domains were greater than Zloop by as much 3.4-fold. At 0.25 pN 65 percent of ΔZ values exceeded Zloop reaching a maximum of 4.3-fold. This difference may be due to tension which opposes coiling as well as the juxtaposition of coils that might become entangled when a loop forms. It is remarkable that these topological domains constrained 64 to 80% of the tether while the loop segment was only 19% of the total length.
for 0.25 pN or 0.45 pN, respectively. Zloop values were expected to lie along the grey, horizontal lines. The largest ΔZ observed at 0.25 pN, 0.41 μm, is 4.3-fold larger than Zloop. Under 0.45 pN of tension, ΔZ values as large as 0.38 μm, 3.4 times Zloop, were observed. Thus, LacI-mediated loops in supercoiled DNA frequently established topological domains much larger than the loop length, and the size of the increase was inversely correlated with tension. In Supplementary Figure S5, a cumulative histogram of ΔZ values for DNA tethers shows that at 0.45 pN, 50% of the topological domains were greater than Zloop by as much 3.4-fold. At 0.25 pN 65 percent of ΔZ values exceeded Zloop reaching a maximum of 4.3-fold. This difference may be due to tension which opposes coiling as well as the juxtaposition of coils that might become entangled when a loop forms. It is remarkable that these topological domains constrained 64 to 80% of the tether while the loop segment was only 19% of the total length.
Figure 2.
Loops that trapped high levels of supercoiling greatly reduced the extension of tethers under tensions of 0.25 (upper) and 0.45 (lower) pN. Thin gray horizontal lines in both panels show ΔZ values corresponding to the loop size (400 bp). The gray dashed lines represent ±2 standard error of ΔZ measurements for the unlooped tether (Supplementary Table S1). The black lines represent estimates of Zloop values derived from Equation (1) using the average Zgyre values reported in Supplementary Table S1. They are minima, because the loop was assumed to have absorbed its full capacity of trapped supercoiling before any additional supercoiling was attributed to flanking plectonemes entangled with the loop. Note that extensions equal to Zlooponly occurred between ntrapped values of –4 to +4 at 0.25 pN (green circles). At 0.45 pN, extensions equal to Zloop were observed at between ntrapped values of –6 (red circle) and +4 (green circle).
ΔZ values equal to Zloop only occur when all trapped supercoiling, ntrapped, is entirely within the loop (Figure 2, grey horizontal line). To determine the amount of supercoiling trapped within or peripheral to the loop, one can estimate the maximum number of supercoils that might be contained in the looped segment, nloop, by dividing Zloop by the extension corresponding to a plectonemic gyre, Zgyre. Using the Zgyre estimates in Supplementary Table S1 indicates that 3.8 gyres could fit within a loop segment. In Figure 2, it is noteworthy that values equal to Zloop only occurred below |ntrapped| values of ∼4 at 0.25 pN or +4 at 0.45 pN (green circles). At 0.45 pN, ntrapped values as low as –6 (red circle) were observed. This may occur, because negative supercoiling DNA under 0.45 pN of tension likely partitions a fraction of the DNA tether into left-handed DNA. With respect to B-DNA, left-handed helicity constrains a linking number change of (–2) into 4.8 nm of contour length (41). Small segments of left-handed DNA easily fit into a loop and likely extend the range of negative supercoiling that can become trapped.
Non-specific looping is unlikely
ΔZ values larger than Zloop result from topological domains larger than the loop. One possibility is that superhelicity promotes non-specific binding giving rise to a variety of loop sizes. Although Kramer et al. (42) concluded that negative supercoiling enhanced LacI binding to operator-bearing minicircles, Sasmor et al. (43) found that LacI bound with higher affinity to torsionally relaxed DNA and 100-fold higher affinity to a torsionally relaxed O1 operator compared to a non-operator sequence. In more recent experiments, LacI-mediated looping of supercoiled plasmids produced looped domains commensurate with specific binding according to atomic force microscopy measurements (9). In addition, just recently, we reported that supercoiling facilitates the formation of a LacI-mediated loop of the expected size in DNA under tension (Yan, Y., Leng, F., Finzi, L. and Dunlap, D. (2018) Protein-mediated looping of DNA under tension requires supercoiling. Nucleic Acids Research, ePub ahead of print). Given the low likelihood of high non-specific affinity and the recent observations of loop sizes in supercoiled DNA corresponding to expectations from the known sequences, it is unlikely that non-specific loops of various sizes produced the wide range of extensions observed.
A general mechanism for trapping high levels of supercoiling
Changes in ΔZ larger than the loop might also result if not only the loop segment, but also flanking sequences become plectonemically entangled. Since high values of writhe must be partitioned between the loop and flanking segments, ntrapped – nloop represents the topologically constrained writhe in the flanking DNA. The length of this topologically constrained flanking DNA will be Zgyre (ntrapped – nloop). Thus, a minimum estimate for ΔZ would include Zloop plus flanking plectonemic entanglements:
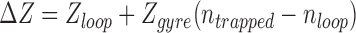 |
(1) |
Estimation of the minimum ΔZ was obtained by substituting 3.8 for nloop in Equation (1) (Figure 2, black lines). The data roughly fall along or above these lines suggesting that simple models might explain topologies that trap minimum amounts of supercoiling.
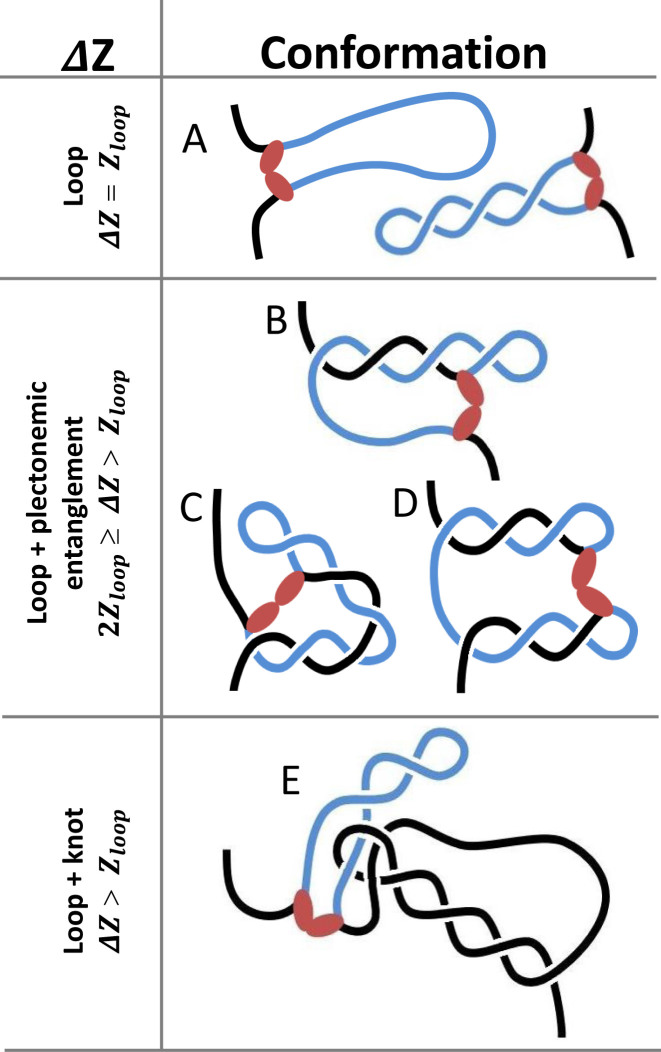
For the nanoscale system described here, Figure 3 depicts open or plectonemic loops (Figure 3A) that would produce ΔZ values equal to Zloop. These are only two examples from a set in which the loop segment does not become entangled with flanking DNA. Such a plectoneme that is completely contained within the 400 bp (136 nm) loop can accommodate at most about four gyres of 36 nm/gyre. If flanking DNA becomes entwined, the loop segment may entangle a flanking segment of lesser or equal length. Thus, whereas approximately four plectonemic gyres can be constrained within a loop, the loop segment entwined with flanking DNA can constrain up to eight plectonemic gyres. Four or six are diagrammed in Figure 3B or C and D respectively. Such topological configurations with flanking entanglements would be expected to exhibit ΔZ values in the range 2Zloop ≥ ΔZ > Zloop. Knotting in which LacI binds to a site threaded through a loop of flanking DNA might also create topological entanglements that restrict extension of the flanking DNA (Figure 3E). in this case ΔZ values, may range from slightly greater than the loop size to very long contours that include almost all of the available DNA.
Figure 3.
Three distinct modes of entanglement trap different levels of supercoiling and topologically constrain different lengths of DNA. LacI might secure a loop without entangling any flanking DNA and shorten the tether by a value proportional to the loop size, Zloop (A). Alternatively, a blue looped segment may entangle with flanking black segments to further reduce the extension. For example, LacI may connect operators in a plectoneme and flanking DNA (B, C), in two different plectonemes (D), or in knotted DNA (E). Entanglements between the loop and flanking segments can reduce the extension by up to twice Zloop (B–D). Knotting between the loop and flanking segments can reduce the extension by more than Zloop (E).
These diagrams illustrate the three distinct modes of entanglement that determine the relationship between trapped supercoils and the contour length of the topological domains. For DNA tethers under sub-picoNewton levels of tension, LacI-mediated loops frequently formed plectonemic entanglements and knots that topologically constrained up to 80% of the DNA tether. This implies that loops forming in supercoiled DNA in macromolecularly crowded spaces are likely to produce highly condensed DNA topologies. It is obvious that transient dissociation of the loop-securing protein from a binding site might permit rapid slithering that changes the extent of entanglement and the size of the topological domain (Figures 2 and 3, Supplementary Figure S4). Indeed, records of the extension of supercoiled DNA with and without LacI, clearly showed rapid, LacI-mediated switching between multiple states of different length (Supplementary Figure S6).
DISCUSSION
These data show that LacI can: (i) separate domains of oppositely supercoiled DNA withstanding considerable torsional stress, (ii) mediate loops in DNA under higher tension (0.45 pN), that likely contain L-DNA and preferentially trap negative supercoiling (Figures 1C and 2), (iii), topologically constrain segments larger than the distance between LacI operators (Figure 2) by entwining, or knotting, flanking and loop segments of DNA (Figure 3) and (iv) catalyze variations of the size of these topological domains (Supplementary Figure S6). The latter agrees with the recent suggestion, from microarray assays and Monte Carlo simulations, that the sizes of topological domains in the chromosome change dynamically (44). Such dynamics might moderate extensive genomic restructuring or compaction due to the cooperative binding of architectural proteins, such as HU or HNS in prokaryotes, or histone octamers in eukaryotes.
Topological domains of different sizes have also been observed with λ CI repressor-mediated loops in extensively supercoiled DNA (Supplementary Figure S7). This suggests that constraining topological domains larger than the distance between binding sites in supercoiled genomes may be a general feature of protein-mediated DNA looping. These findings have intriguing implications for DNA/chromatin packaging, especially for coordinated gene expression. For example, it is straightforward to envision that a supercoiled loop might encompass nearby genes and influence the transcription from a nearby promoter, but coarse-grained models show that the interaction of two distant sites along contiguous DNA is most sensitive to supercoiling (45). Plectonemes in distant DNA segments topologically pinned by protein-mediated loops may explain this observation.
There might be significant advantages to topologically entangling more DNA than just the segment between the binding sites of a loop-mediating protein. First, a larger topological domain protects larger segments of DNA from damage following single-stranded nicks (Supplementary Figure S8). Second, larger topological domains may catalyze DNA damage repair by keeping the two ends to be re-joined closer to each other, since they must slither to untangle from each other instead of being free to separate along multiple three-dimensional trajectories (45). Such confinement would be particularly important when double-strand breaks must be repaired through non-homologous end joining. Third, larger domains will produce greater compaction of the chromosome. Fourth, these larger topological domains exhibit different configurational isomers of DNA, which may increase the opportunities for regulation. Fifth, larger topological entanglements facilitate bridging by proteins that maintain structural integrity of the chromosome, such as HNS in prokaryotes (46). Lastly, protein-mediated knotting involving a distant plectoneme radically alters topology and perhaps DNA transactions quite distant from the loop (Figure 3E).
This work suggests that large, dynamic domains, which characterize genomes across biological kingdoms, may result from protein-mediated loops that topologically isolate segments of supercoiled DNA larger than the actual contour length between the protein binding sites. Although relatively small DNA loops, in DNA molecules only five times longer, were examined in this study, the possibility of the formation of extensive plectonemic structures between large loops and flanking DNA is predicted to significantly impact chromosome structure, dynamics and function, including coordinated gene expression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kathleen Matthews at Rice University for lac repressor protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM084070 to L.F., 1R15GM109254-01A1 to F.L.]. Funding for open access charge: National Institutes of Health [R01GM084070].
Conflict of interest statement. None declared.
REFERENCES
- 1. Higgins N.P. Species-specific supercoil dynamics of the bacterial nucleoid. Biophys. Rev. 2016; 8:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mogil L.S., Becker N.A., Maher L.J. III. Supercoiling Effects on Short-Range DNA Looping in E. coli. PLoS ONE. 2016; 11:e0165306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Badrinarayanan A., Le T.B., Laub M.T.. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015; 31:171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sobetzko P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016; 44:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Efremov A.K., Winardhi R.S., Yan J.. Transfer-matrix calculations of DNA polymer micromechanics under tension and torque constraints. Phys. Rev. E. 2016; 94:032404. [DOI] [PubMed] [Google Scholar]
- 6. Marko J.F., Neukirch S.. Global force-torque phase diagram for the DNA double helix: structural transitions, triple points, and collapsed plectonemes. Phys. Rev. E. 2013; 88:062722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding Y., Manzo C., Fulcrand G., Leng F., Dunlap D., Finzi L.. DNA supercoiling: a regulatory signal for the λ repressor. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitson P.A., Hsieh W.T., Wells R.D., Matthews K.S.. Supercoiling facilitates lac operator-repressor-pseudooperator interactions. J. Biol. Chem. 1987; 262:4943–4946. [PubMed] [Google Scholar]
- 9. Fulcrand G., Dages S., Zhi X., Chapagain P., Gerstman B.S., Dunlap D., Leng F.. DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli. Scientific Rep. 2016; 6:19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leng F., Chen B., Dunlap D.D.. Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:19973–19978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dixon J.R., Gorkin D.U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uuskula-Reimand L., Hou H., Samavarchi-Tehrani P., Rudan M.V., Liang M., Medina-Rivera A., Mohammed H., Schmidt D., Schwalie P., Young E.J. et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016; 17:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N.. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013; 20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benedetti F., Racko D., Dorier J., Burnier Y., Stasiak A.. Transcription-induced supercoiling explains formation of self-interacting chromatin domains in S. pombe. Nucleic Acids Res. 2017; 45:9850–9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferrandiz M.J., Martin-Galiano A.J., Schvartzman J.B., de la Campa A.G.. The genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 2010; 38:3570–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Priest D.G., Kumar S., Yan Y., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of interactions between two DNA loops demonstrates loop domain insulation in E. coli cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E4449–E4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adhya S. Multipartite genetic-control elements—communication by DNA loop. Annu. Rev. Genet. 1989; 23:227–250. [DOI] [PubMed] [Google Scholar]
- 18. Finzi L., Dunlap D.D.. Single-molecule approaches to probe the structure, kinetics, and thermodynamics of nucleoprotein complexes that regulate transcription. J. Biol. Chem. 2010; 285:18973–18978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mirkin S.M. Encyclopedia of Life Sciences. 2001; John Wiley & Sons, Ltd [Google Scholar]
- 20. Kumar S., Manzo C., Zurla C., Ucuncuoglu S., Finzi L., Dunlap D.. Enhanced tethered-particle motion analysis reveals viscous effects. Biophys. J. 2014; 106:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovari D.T., Yan Y., Finzi L., Dunlap D.. Tethered particle motion: an easy technique for probing DNA topology and interactions with transcription factors. Methods Mol. Biol. 2018; 1665:317–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao Q., Goyal S., Finzi L., Dunlap D.. Physiological levels of salt and polyamines favor writhe and limit twist in DNA. Macromolecules. 2012; 45:3188–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strick T.R., Allemand J.F., Bensimon D., Croquette V.. Behavior of supercoiled DNA. Biophys. J. 1998; 74:2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blumberg S., Pennington M.W., Meiners J.C.. Do femtonewton forces affect genetic function? A review. J. Biol. Phys. 2006; 32:73–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wuite G.J., Smith S.B., Young M., Keller D., Bustamante C.. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000; 404:103–106. [DOI] [PubMed] [Google Scholar]
- 26. Voros Z., Yan Y., Kovari D.T., Finzi L., Dunlap D.. Proteins mediating DNA loops effectively block transcription. Protein Sci. 2017; 26:1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zurla C., Samuely T., Bertoni G., Valle F., Dietler G., Finzi L., Dunlap D.D.. Integration host factor alters LacI-induced DNA looping. Biophys. Chem. 2007; 128:245–252. [DOI] [PubMed] [Google Scholar]
- 28. Finzi L., Gelles J.. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995; 267:378–380. [DOI] [PubMed] [Google Scholar]
- 29. Johnson S., van de Meent J.W., Phillips R., Wiggins C.H., Linden M.. Multiple LacI-mediated loops revealed by Bayesian statistics and tethered particle motion. Nucleic Acids Res. 2014; 42:10265–10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanzi F., Broggio C., Sacconi L., Pavone F.S.. Lac repressor hinge flexibility and DNA looping: single molecule kinetics by tethered particle motion. Nucleic Acids Res. 2006; 34:3409–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yin H., Landick R., Gelles J.. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys. J. 1994; 67:2468–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manzo C., Finzi L.. Nils GW. Methods In Enzymology. 2010; 475:Academic Press; 199–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watkins L.P., Yang H.. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B. 2005; 109:617–628. [DOI] [PubMed] [Google Scholar]
- 34. Charvin G., Allemand J.F., Strick T.R., Bensimon D., Croquette V.. Twisting DNA: single molecule studies. Contemp. Phys. 2004; 45:383–403. [Google Scholar]
- 35. Forth S., Deufel C., Sheinin M.Y., Daniels B., Sethna J.P., Wang M.D.. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008; 100:148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lipfert J., Skinner G.M., Keegstra J.M., Hensgens T., Jager T., Dulin D., Köber M., Yu Z., Donkers S.P., Chou F.-C. et al. Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15408–15413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neukirch S., Marko J.F.. Analytical description of extension, torque, and supercoiling radius of a stretched twisted DNA. Phys. Rev. Lett. 2011; 106:138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seol Y., Neuman K.C.. Magnetic tweezers for single-molecule manipulation. Methods Mol. Biol. 2011; 783:265–293. [DOI] [PubMed] [Google Scholar]
- 39. Champion K., Higgins N.P.. Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J. Bacteriol. 2007; 189:5839–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rovinskiy N., Agbleke A.A., Chesnokova O., Pang Z., Higgins N.P.. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012; 8:e1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sheinin M.Y., Forth S., Marko J.F., Wang M.D.. Underwound DNA under tension: structure, elasticity, and sequence-dependent behaviors. Phys. Rev. Lett. 2011; 107:108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krämer H., Amouyal M., Nordheim A., Müller-Hill B.. DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988; 7:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sasmor H.M., Betz J.L.. Specific binding of lac repressor to linear versus circular polyoperator molecules. Biochemistry. 1990; 29:9023–9028. [DOI] [PubMed] [Google Scholar]
- 44. Postow L., Hardy C.D., Arsuaga J., Cozzarelli N.R.. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004; 18:1766–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benedetti F., Dorier J., Stasiak A.. Effects of supercoiling on enhancer–promoter contacts. Nucleic Acids Res. 2014; 42:10425–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Muskhelishvili G., Travers A.. The regulatory role of DNA supercoiling in nucleoprotein complex assembly and genetic activity. Biophys. Rev. 2016; 8:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.