Abstract
CARM1 is a protein arginine methyltransferase (PRMT) that has been firmly implicated in transcriptional regulation. However, the molecular mechanisms by which CARM1 orchestrates transcriptional regulation are not fully understood, especially in a tissue-specific context. We found that Carm1 is highly expressed in the mouse testis and localizes to the nucleus in spermatids, suggesting an important role for Carm1 in spermiogenesis. Using a germline-specific conditional Carm1 knockout mouse model, we found that it is essential for the late stages of haploid germ cell development. Loss of Carm1 led to a low sperm count and deformed sperm heads that can be attributed to defective elongation of round spermatids. RNA-seq analysis of Carm1-null spermatids revealed that the deregulated genes fell into similar categories as those impacted by p300-loss, thus providing a link between Carm1 and p300. Importantly, p300 has long been known to be a major Carm1 substrate. We found that CREMτ, a key testis-specific transcription factor, associates with p300 through its activator, ACT, and that this interaction is negatively regulated by the methylation of p300 by Carm1. Thus, high nuclear Carm1 levels negatively impact the p300•ACT•CREMτ axis during late stages of spermiogenesis.
INTRODUCTION
Arginine methylation is a widespread post-translational modification (PTM) that has been linked to the regulation of a broad swath of biological processes (1). Quantitative high-resolution mass-spectrometry analysis has revealed that ∼7% of all arginine residues in ∼3300 human proteins are methylated in the HEK293 cells, which is comparable to global serine phosphorylation and lysine ubiquitination levels (2). Methyl marks on arginine residues are capable of providing docking sites for ‘reader’ proteins, and competitively masking the deposition of neighboring PTMs in downstream signaling cascades (3). Arginine methylation is catalyzed by nine protein arginine methyltransferases (PRMT1-9), which are classed into three enzyme types. CARM1 (along with PRMT1, 2, 3, 6 and 8) is a Type I enzyme, which catalyzes the asymmetrical di-methylation of arginines (ADMA). Type II enzymes (PRMT5 and 9) deposit symmetrical di-methylarginines (SDMA) marks, and there is a single Type III (PRMT7), which can only monomethylate substrates (1,4–6). Both histones and non-histone proteins can serve as substrates for PRMTs, and in the context of transcriptional regulation, these enzymes function as both transcriptional activators and repressors (5,7).
CARM1, also referred to as PRMT4, was the first member of this family to be identified as a transcriptional regulator, through its ability to be recruited by nuclear receptors, via the p160 coactivator family, to chromatin (8). The recruitment of CARM1 to transcriptional promoters results in the methylation of the p160 coactivator family (SRC-1, SRC-2/GRIP1 and SRC-3/NCOA3/AIB1), the histone acetyltransferases (p300/CBP), and histone H3 (8–10). These methylation events generally enhance gene activation (11). Therefore, CARM1 is considered a secondary coactivator for nuclear receptor-mediated transcription. Moreover, H3R17me2a ChIP studies showed elevated levels at a number of gene promoters (12–19), indicating that CARM1 functions as a rather general coregulator for a large number of transcription factors including p53, YY1, NF-κB, PPARγ, RUNX1 and E2F1 (20). However, there are also scenarios (with CREB and RUNX) in which CARM1 functions as a transcriptional repressor (21,22). Gene ablation studies in mice revealed that CARM1 is vital for survival after birth (23). Enzyme-dead CARM1 knock-in mice phenocopy the null mice, indicating that CARM1’s enzymatic activity is required for most of its in vivo functions (24). CARM1 knockout embryos are smaller in size, and display a number of cell differentiation defects such as a partial block in T-cell development (25) and improper differentiation of lung alveolar cells (17) and adipocytes (19). The functional importance of CARM1 in germline development has yet to be investigated, and it is the focus of this study.
Our interest in studying the role of CARM1 in germline development was piqued for three reasons: First, Carm1 is highly expressed in the mouse testis. Furthermore, Carm1 is found in the cytoplasm and excluded from the nucleus in many cell types (26), but during spermatogenesis its localization moves from cytoplasmic to nuclear during the spermatocyte to spermatid transition, suggesting an important role for Carm1 in the late stage of sperm development. Second, methylarginine marks are ‘read’ by Tudor domain (TDRD)-containing proteins (27), and a large number of these TDRD proteins are preferentially expressed in the male germline. These testis-specific TDRD proteins recognize the methylated arginine motifs at the N-terminal of PIWI family proteins (MILI, MIWI and MIWI2), which bind a specific population of small RNAs (piRNAs) (28,29). Mouse knockout studies demonstrated these TDRD family proteins (TDRD1, 2, 7, 9 and 12) are essential for retro-transposon repression and germ cell development in the mouse testis by regulating the PIWI-piRNAs pathway (30–33), thus strongly implicating the PRMTs in these processes. Third, using pan SDMA and ADMA antibodies, it has been reported that arginine-methylated proteins are abundant in the testis (27,34–37). Genetic studies revealed that the conditional loss of Prmt5 (the enzyme responsible for over 90% of the SDMA production) in germ cells led to the depression of retro-transposons due to the failure of H4R3 methylation, and the induction of apoptosis (38) and Prmt5 deletion in the postnatal testis resulted in an early developmental arrest in the spermatocytes owing to a defective PIWI-piRNA pathway (39). Thus, SDMA deposition is important for germline development, but the biological implication of ADMA deposition is still unclear.
In this study, we discovered that CARM1 is abundantly expressed in the testis, as compared with other organs, with especially strong expression at the late stages of haploid spermatids. We generated a mouse model with a conditional germline loss of Carm1 using Stra8-Cre, and found that this PRMT is not required for spermatocyte development. It is however essential for the late stages of haploid spermatids development. We show that the normal transcriptional regulatory functions of Carm1 in the late stages of spermiogenesis are orchestrated, at least in part, through the methylation of GRIP1 binding domain (GBD) domain of p300, which abolishes the interaction between p300 and ACT coactivators, thus repressing the expression of a cohort of CREMτ-bound target genes. Therefore, CARM1 counteracts the positive transcriptional activity driven by the p300•ACT•CREMτ axis, uncovering a unique ‘co-repressor’ role for this PRMT in germline development that is distinct from its traditional ‘co-activator’ role in nuclear receptor signaling.
MATERIALS AND METHODS
Mice
Stra8-Cre in the C57BL/6J background was purchased from the Jackson Laboratory. Floxed Carm1 mice (Carm1fl/fl) were generated as described previously in the mixed C57BL6J/129 background. Male Stra8-Cre mice were first crossed with female Carm1fl/fl mice to generate the Stra8-Cre/Carm1+/fl males. Stra8-Cre/Carm1+/fl male mice were then bred with female Carm1fl/fl to get the Stra8-Cre/Carm1fl/Δ (designated as cKO) males. All animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center.
Antibodies
The antibodies used in this study were as follows (listed in the format of name, supplier, catalogue, dilution): Rabbit α-CARM1 (Bethyl lab, A300-421A, IP/5μg, WB/1:1000); Rabbit α-CARM1 (gift from McGill University, Dr Stephane Richard, IF/1:500; WB/1:1000); Rabbit α-H2B (Active Motif, No. 39125, 1:500 for WB); Rabbit α-H2A (Active Motif, No. 39209, 1:500 for WB); Mouse α-p300 (Santa Cruz, sc-48343, IP/10μg, WB/1:200); Mouse α-FHL5/ACT (Santa Cruz, sc-101045, IP/10μg, WB/1:500); Rabbit α-p300 (Elabscience, E-AB-32456, WB/1:1000); Rabbit α-p300R2142me2a (gift from University of South California, Dr Michael R. Stallcup, WB/1:200); Rabbit α-D4H5 (In-house generated, α-ADMA, WB/1:1000); Rabbit α-BL8242 (In-house generated, α-SDMA, WB/1:1000); Mouse α-Actin (Sigma, 1:10 000 for WB); HRP conjugated Sheep α-mouse IgG (GElifescience, NA931-1ML, 1:10 000 for WB); HRP conjugated Donkey α-rabbit IgG (GElifescience, NA934-1ML, 1:10,000 for WB).
Plasmids
GST-tagged plasmids for ACT fusion series were constructed in the pEGFP-C1 vector (Clontech). Total testicular RNAs were purified from the adult mouse testis using the Trizol reagent (Invitrogen). First cDNA synthesis was performed by using a Superscript III first strand synthesis kit (Invitrogen). Different deletion mutants of FHL5 were amplified by corresponding PCR primer pairs carrying two restriction enzyme cut site sequences (BamHI and XhoI) using The Advantage 2 PCR Kit (Clontech, Cat No: 639206). Following double enzyme digestion, the deletion fragments were ligated to the pEGFP-C1 vector plasmid using the T4 ligase (Invitrogen). The inserts were verified by colony-PCR and Sanger sequencing. Full length of WT Carm1 and Enzyme-dead Carm1 plasmid (CARM1d) were generated as described before. Full-length His-tagged p300 plasmid (pcDNA3.1-p300) was a gift from Dr Xiaobing Shi (MDACC). GST-tagged fusion p300 plasmids were generated using the pGEX-6p-2 vector (GE lifescience) from Dr Michael Hottiger (University of Zurich) as follows: GST-p300 (1–672), GST-p300 (672–1193), GST- p300 (1069–1459), GST- p300 (1459–1892), GST-p300 (1893–2414). Flag-tagged p300 plasmids were a gift from Dr Christine Neuveut (Institut Pasteur). Plasmids for Flag-tagged CREMτ, HA-tagged GCNF, CRE/NR-PRL-Luc were generated as previously described (40). Dual binding site sequence (5′-ggtatcctttgtgaggtcaacaatga-3′) for CREMτ and GCNF transcription factors was inserted upstream of the minimal rat prolactin (PRL) promoter in the pGL3-basic vector (Promega).
Histology, immunohistochemistry and immunofluorescence staining
For Hematoxylin-Eosin (HE) staining, freshly dissected testis and epididymis samples were fixed in Bouin's solution overnight at 4°C, followed by washing in 70% alcohol. All tissues were subjected to standard paraffin-embedding and 5 μm sections were used for HE staining. Periodic acid–Schiff (PAS) staining was performed in the 5μm sections according to the protocol described previously. For immunofluorescence staining, testis samples were fixed in 4% PFA overnight at 4°C. Ten micrometer cryo-sections were generated in the CryoStar™ NX70 Cryostat system (ThermoFisher). Antigen retrieval was performed by submerging the slides in Citrate Buffer (pH 6.0) at boiling temperature for 15min. Following washing in 1XPBS three times, sections were blocked in the 5% BSA buffer for 1 h at room temperature. The primary antibody diluted as appropriate was incubated with the sections overnight at 4°C. The secondary antibody labelled by alexa fluor 488/647 (Invitrogen) was incubated with the primary antibody-labelled sections for 1 h at room temperature. After extensive washing in the 1× PBS buffer for three times, the slides were covered by a coverslip in the ProLong™ Gold Antifade Mountant (ThermoFisher, Cat: P36930) and sealed with nail polish. Imaging was performed using the ZEISS LSM 880 system following manufacturer's instructions. The specificity of the CARM1 antibody was validated using western blot on Carm1−/- MEF cells and immunostaining on Carm1 cKO testis. All immunostaining was performed in three biological replicates.
Western blot
Protein lysates were prepared using the RIPA buffer [50 mM Tris–HCl (1 M, pH 7.5), 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate (DOC) and 1% NP-40, supplemented with protease inhibitor tablet (Roche)]. Protein concentrations were determined by the Bradford protein assay (Bio-rad). 20–40 μg protein samples were resolved in the 11% SDS-PAGE gel and semi-transferred to the PVDF membrane following standard protocols as described previously. Immunoblots were blocked in the 5% milk for 1 h at room temperature and incubated with the primary antibodies overnight at appropriate concentrations as described above. Following three washes in 1× PBS buffer containing 0.05% Tween-20, the blots were further incubated with the HRP-conjugated secondary antibody at room temperature for 1 h. HRP-catalyzed luminescence signals were visualized using the Western Lightning™ Chemiluminescence Reagent Plus kit (PerkinElmer). All experiments were performed in three biological replicates.
Co-immunoprecipitation (Co-IP) assay
HEK293 cells were co-transfected with 2 μg of GFP tagged ACT plasmids (pEGFP-ACT-1 [FL], pEGFP-ACT-2 [1–105], pEGFP-ACT-3 [106–285], pEGFP-ACT-4 [286–468], pEGFP-ACT-5 [106–855], pEGFP-ACT-6[286–855], pEGFP-ACT-7 [469–855], pEGFP-ACT-8 [643–855] vector, together with Flag-tagged p300 plasmid (5 μg), using PEI transfection reagent at 1:5 ratio. Transfection efficiency was monitored by the GFP expression using a fluorescent microscopy. Protein lysates were collected in the RIPA lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1% Triton-100, 0.05% SDS plus the Roche protease inhibitor cocktail) at post-transfection 48 h. Protein concentration was determined by Bradford assay according manufacturer's manual. To determine the interacting domain of ACT with p300, co-IP was performed using the GFP antibody (rabbit) against the GFP-tagged ACT deletion mutants. GFP-ACT/p300 protein complex was pulled down by incubation with protein A/G agarose beads. Following four times of washing, protein complex attached to the beads was dissolved in the SDS sample buffer and subsequently resolved in 7% SDS-PAGE gel. p300 protein was detected through anti-Flag antibody. All experiments were performed in three biological replicates.
Luciferase reporter assay
Luciferase reporter assay was performed in biological triplicates in HEK293 cells (and was repeated in HeLa cells). HEK293 cells were cultured in high-glucose DMEM medium (GIBCO) supplemented with 10% FBS at 37°C, and were split into the 6-well plates one day before transfection. Once the cells grew to 70% confluency the next day, transfection was performed using the PEI transfection reagent at a ratio of 1:2 of total DNA (μg) to PEI (μg) following standard transfection protocol in the presence of complete growth medium with 10% FBS. Renilla luciferase vector was transfected simultaneously as the internal transfection control. At 6 h post-transfection, medium was replaced with fresh complete DMEM medium. Cells were collected for the luciferase assay around post-transfection 48 h using the Dual-Luciferase Reporter Assay kit (Promega, E1960). Luciferase luminescence intensity was recorded in the GloMax 96 Microplate Luminometer (Promega).
For the CARM1 inhibitor (TP-064) treatment, HEK293 cells were cultured in the complete growth medium of the high-glucose DMEM supplemented with 10% FBS TP-064 was added to the complete medium at a concentration of 500 nM. Medium was replenished every day and the cells were cultured for a consecutive 4 days prior to transfection.
Centrifugal elutriation
The testes were collected and dissected free of the tunica albuginea in the 1XKREBS buffer (0.326 g KH2PO4, 13.95 g NaCl, 0.589 g MgSO4, 4 g dextrose, 0.378 g CaCl2, 0.712 g KCl supplemented with 2% BSA). Then the testes tissues were incubated with Collagenase IV at a concentration of 500 μg/ml for 5 min at 37°C. Testes tissues were further broken apart by pipetting up and down briefly using a 1 ml tip-cutted tip and incubated for another 5 min at 37°C. After sedimentation on ice briefly, the interstitial stromal cells in the supernatant were removed, and this procedure was repeated up to five times to make sure all stromal cells were cleared away. Next, 5 ml of 1× HBSS (without calcium and magnesium) solution was added to the tissue pellet supplemented with the Trypsin (700 μg/ml) plus DNase I (30 μg/ml), and incubated at 37°C with continuous shaking for 15 min. The cells were examined under a phase-contrast microscopy to ensure the seminiferous tubules were separated into single cell suspension. Trypsin digestion buffer was removed by centrifugation at 1000 g for 5min. Cell pellets were washed two times using ice-cold 1XKREBS buffer. 10ml KREBS buffer, supplemented with 2% FBS and DNase I (30 μg/ml), was finally added to the single cell pellet and the sample was ready for elutriation. Centrifugal elutriation was performed in the Beckman JE5.0 Elutriator Centrifuge system (Beckman Coulter) following the manufacturer's protocol. The different fractions were collected as follows: 150 ml total volume was collected at rotor speed (3000 rpm) and pump speed (9.7 ml/min) for condensed spermatids (CS); 100 ml total volume was collected at rotor speed (3000 rpm) and pump speed (17.5 ml/min) for elongating and condensing spermatids (ECS); 150 ml total volume was collected at rotor speed (2250 rpm) and pump speed (12.4 ml/min) for round and elongating spermatids (RES); 150 ml total volume was collected at rotor speed (2250 rpm) and pump speed (17.5 ml/min) for round spermatids (RS); 150 ml total volume was collected at rotor speed (2250 rpm) and pump speed (30 ml/min) for spermatocytes. The cell purity for each fraction was confirmed by H&E staining (Supplementary Figure S4) (41).
RNA extraction, RT-qPCR and RNA-seq
Total RNAs were extracted from testis tissue using Trizol reagent following the manufacturer's instructions. For RNA extraction from purified cell types, collected cell pellets were lysed in 500 μl Trizol reagent, followed by a column-based purification protocol using the Direct-zol™ RNA MiniPrep Kit by Zymo Research. Total RNA samples were further treated by DNAase I (Ambion) prior to first strand cDNA synthesis using a Superscript III first strand synthesis kit (Invitrogen). For quantitative reverse transcription-PCR (RT-qPCR) reactions, a PerfeCTa SYBR Green SuperMix kit (Quantabio) was used along with gene-specific primers following manufacturer's instruction. Primers used are described in the Supplementary Table S1.
RNA-seq analysis
RNA-Seq sequencing libraries were generated from total RNA extracted from the two purified spermatid populations (round spermatids [RS] and elongating/condensing spermatids [ECS]) following TruSeq Stranded Total RNA Sample Prep protocol (illumina). The libraries were sequenced using 2 × 75 bases paired end protocol on Illumina HiSeq 2000 instrument. Two biological replicates with each sample purified from 10 testes were prepared for each condition. 22–27 million pairs of reads were generated per sample. Each pair of reads represents a cDNA fragment from the library. The reads were mapped to mouse genome (mm10) by TopHat (version 2.0.10) (42). By reads, the overall mapping rate is 95–97%. 92–95% fragments have both ends mapped to mouse genome. The number of fragments in each known gene from RefSeq database (downloaded from UCSC Genome Browser on 17 July 2015) was enumerated using htseq-count from HTSeq package (version 0.6.0). Genes with less than 10 fragments in all the samples were removed before differential expression analysis. The differential expression between conditions was statistically assessed by R/Bioconductor package DESeq (version 1.16.0). Genes with FDR (false discovery rate) ≤0.05, fold change ≥2 and length >200 bp were called as differentially expressed. Cbp/p300-sensitive genes were determined with non-adjusted P-value <0.05 and fold change >+1.1 or -1.1. The normal expression levels of genes in meiotic (Spc), early (RS) and late (ES) post-meiotic cells were assessed using expression data obtained in the Illumina MouseWG-6 v2.0 expression beadchip array from wild type spermatogenic cells enriched at the corresponding stages from published (GSE46137 and GSE55767) as well as in-house unpublished data. The box plots represent the distributions of quantile normalized expression values of all the genes in each condition. Box plots corresponding to meiotic spermatocytes (Spc), round spermatids (RS) and elongating late spermatids (ES) are respectively colored in red, green and blue.
GST-tag protein expression and purification
GST-tagged plasmids for different deletion mutants of p300 (GST-p300-1 [1–672], GST-p300-2 [672–1193], GST-p300-3 [1069–1459], GST-p300-4 [1459–1892], GST-p300-5 [1893–2414] were generated. GST-tagged plasmids were transformed into BL21 competent E. Coli cells. Protein expression was induced through the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM upon the OD600 of the culture medium reaching 0.6, followed by further shaking in a 37°C incubator for 4 h or at room temperature overnight. Cell pellets were collected in 1 ml 1× PBS and lysed by two rounds of sonication. The supernatant was incubated with the Sepharose 4B agarose overnight in the cold room. After four washes, the bound GST-tagged proteins were eluted with reduced glutathione solution for 4 h at 4°C.
GST pull-down
HEK293 cells were transfected with the Flag-tagged full-length FHL5 plasmids in a 60mm dish. Cell lysates were prepared using 500 μl RIPA buffer per dish on ice. Cell lysates were pre-cleared by incubation with Glutathione sepharose beads for 1 h at 4°C. Purified protein (10μg) for the GST-tagged p300 deletion mutants was incubated with 500 μl of lysate for 6 h at 4°C. Beads with the GST-bound protein complex were washed four times using the RIPA lysis buffer. The protein complex was then resolved by SDS-PAGE, and subject to western blot analysis.
RESULTS
CARM1 is highly expressed in haploid germ cells in the testis
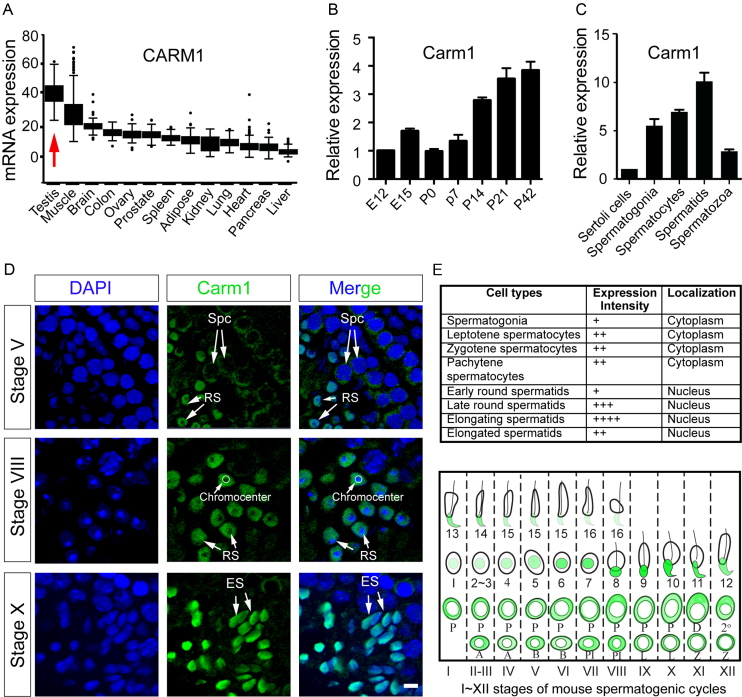
An analysis of PRMT mRNA expression levels in human tissues, using the Genotype-Tissue Expression (GTEx) database (43), revealed that CARM1 is ubiquitously expressed, with the highest expression levels in testis (Figure 1A). Germ cell development is strictly time-defined during embryonic and postnatal development in mammals. Carm1 mRNA is detected in the newly formed embryonic testis at embryonic day 12 (E12), and expression increases through postnatal development into adulthood, suggesting its critical role during late stage of spermatogenesis (Figure 1B). Moreover, since the testis is composed of not only germ cells at different developmental stages, but also Sertoli cells, we investigated which cell types Carm1 expression was predominant. To this end, we purified Sertoli cells and germ cells at different stages, using centrifugal elutriation, followed by qRT-PCR of the purified RNA. We found that Carm1 mRNA is detectable in Sertoli cells, but is more abundantly expressed in the haploid spermatids (Figure 1C). Next, we performed immunofluorescent staining of cryo-sections of adult testis to determine the expression and localization of Carm1 protein. Within the epithelium of seminiferous tubules, the entry of spermatogonia into meiosis, followed by the haploid spermatids elongation, occurs in a cyclic fashion, whereby the differentiating spermatogonia move from the basal compartment towards the lumen. This process is spatially synchronized into 12 (I–XII) classified stages along the length of the tubules, with a single stage being represented in one cross-section of the seminiferous tubules. Each stage is characterized by the distinct combination of spermatogonia, spermatocytes and spermatids at different steps (a total of 16 steps) on basis of acrosome morphology (44). Interestingly, the immunofluorescent staining revealed that Carm1 subcellular localization is highly dynamic (Figure 1D). Carm1 is present in the cytoplasm of the early stages of spermatogenic cells, including spermatogonia and spermatocytes (Figure 1D and E). Following meiosis, Carm1 translocates to the nucleus of haploid round spermatids, with the highest protein levels detected in the nuclei of elongating spermatids (Figure 1D and E). Of note, CARM1 protein is excluded from the heterochromatin structure referred to as Chromocenter, in the round spermatids (Figure 1D).
Figure 1.
Carm1 expression and localization in testis. (A) Carm1 mRNA expression levels in different human tissues as detected by RNA-seq from GTEx database; RPKM, Reads Per Kilobase of transcript per Million; (B) Relative mRNA levels of Carm1 quantified by quantitative real time PCR (qPCR). Testicular samples were collected at embryonic day 12 (E12) and E15, as well as postnatal day 0 (P0), P7, P14, P21 and P42. The mRNA levels at E12 were arbitrarily set as 1; Data were calculated from biological triplicates and shown as mean±SD. (C) Relative mRNA levels of Carm1 quantified by qPCR in purified germ cell populations from the testis; The mRNA levels in Sertoli cells were arbitrarily set as 1. Data were calculated from three biological replicates with 5 mice in each pool, and shown as mean ± SD. (D) Expression and localization of Carm1 protein by immunofluorescent staining (IF) using Carm1-specific antibody. Carm1 protein localization in three representative stages of spermatogenic cycles is presented in seminiferous tubules. The expression intensity and localization of CARM1 were summarized in (E). ‘+’ indicates the relative levels of Carm1 protein (top). Diagrammatic representation of Carm1 subcellular localization is shown in ‘green’ (bottom). Spc, Spermatocytes; RS, Round spermatids; ES, Elongating spermatids. A, A type spermatogonia. B, B type spermatogonia. PL, pre-Leptotene spermatocytes. L, Leptotene. Z, Zygotene. P, Pachytene. D, Diplotene. 2 represents secondary spermatocytes. Roman numerals I–XII represent Stage I–XII in the seminiferous tubules of mouse. Arabic 1–16 numerals illustrate a total of 16 steps of haploid spermatids during elongation. The details of staging seminiferous tubules were described previously (44). Bar, 10 μm.
Carm1 is essential for haploid germ cell development and male fertility
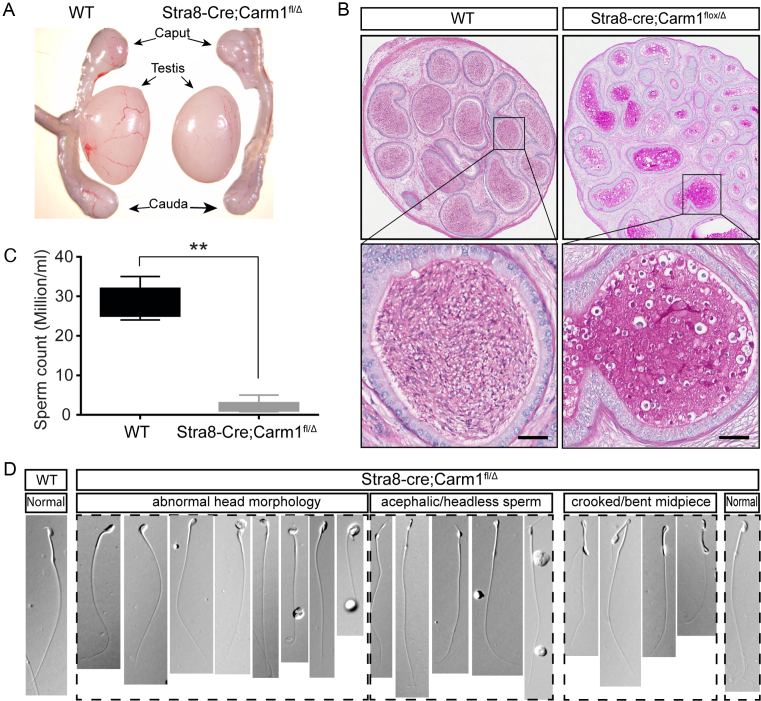
To explore the physiological functions of Carm1 in spermatogenesis, we generated a conditional germline-specific Carm1 knockout mouse line using a well-established Stra8-Cre line. Stra8-Cre is activated in the spermatogonial population starting from postnatal day 3 (P3) (45). Mice harboring a floxed Carm1 allele were generated by flanking the exons 3 and 4 with lox-P sites, and deletion of these two exons causes a frame shift (23,46). Through two steps of breeding, we generated the Stra8-Cre;Carmfl/Δ mice (hereafter referred to as cKO) for all subsequent studies (Supplementary Figure S1A). Mice with germline-specific deletion of Carm1 developed normally without any discernable gross phenotype. Using western blot analysis, we confirmed that CARM1 protein is dramatically decreased in the cKO whole testis (Supplementary Figure S1B) and is totally absent in the Carm1 cKO haploid spermatids at different stages (Supplementary Figure S2A), confirming that Carm1 is tissue-specifically deleted. The testis size and weight of the cKO are comparable to those of WT littermates at 6-weeks of age, with a slight, but significant, decline in the testis weight in 3-month old males (Figure 2A and Supplementary Figure S2B). To test whether Carm1 cKO has an impact on germline development, we performed a fertility test by breeding 2-month old cKO males, as well as WT male littermates, with fertility-proven 3-month old WT females. We found that while the WT male littermates can produce an average of 6.7 pups per litter, half of the cKO males are completely sterile and the other half produce very small litters (an average of 1.7) (Supplementary Figure S2C), indicating that Carm1 is essential for male fertility. Next, we dissected out the epididymis and examined the mature sperm. Not surprisingly, we found that there is a significant drop in the total number of sperm cells in cKO males as compared to WT littermates (Figure 2B). Furthermore, there is an accumulation of immature germ cells and aberrant sperm in the cauda epididymis of cKO mice (Figure 2C). Phase-contrast microscopic analysis revealed numerous anomalies in sperm cells morphology and structure, including abnormal head formations, acephalic/headless sperm and sperm with a bent midpiece (Figure 2D and Supplementary Figure S2D). Moreover, there is a dramatic decrease in the motility of cKO sperm (data not shown). The irregularly shaped sperm heads, declined sperm count and sperm motility, together, account for the infertility/subfertility phenotype observed in Carm1 cKO males.
Figure 2.
Phenotypic analysis of germline-specific conditional Carm1 KO (cKO) mouse model. (A) Gross morphology of the testis and epididymis from WT and cKO mice. (B) Comparison of the average sperm count in the cauda epididymis between the WT and cKO mice at 6-week-old age. Three biological repeats were performed. Data were shown as mean ± SD. (C) Representative Hematoxylin and Eosin (H&E) staining of cauda epididymis and (D) phase-contrast microscopy of cauda sperm from WT and cKO littermates. Note that the majority of sperm exhibit deformed head morphology in the cKO cauda epididymis, as indicated. Data were presented as mean ± SEM.
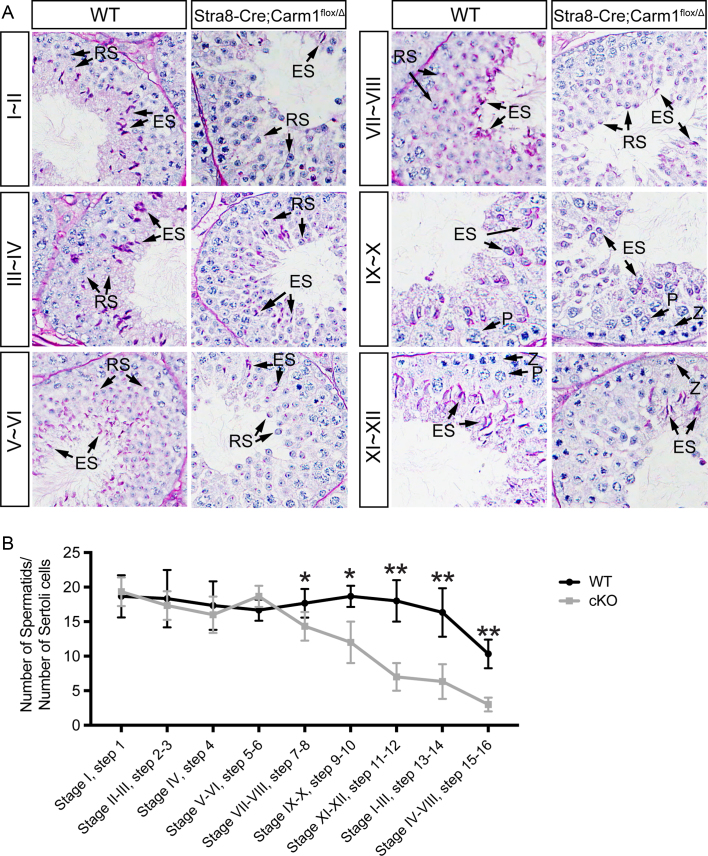
To accurately determine at which step germ cell development is impacted by Carm1 loss, we performed Periodic acid–Schiff (PAS) staining on the cross-sections of paraffin-embedded testis. Germ cell development can be divided into a total of 12 (I–XII) stages, based on the morphology of spermatid acrosome and the combinations of spermatogenic cell types, in a specific cross-section of the seminiferous tubules (47). As shown in Figure 3A, we observed a normal number of spermatogonia and meiotic spermatocytes at different stages in the cKO cross-sections testis. Following meiosis, the total number of early stage round spermatids (Step 1–6) are also unaffected in cKO mice. However, we noticed a significant drop in the number of late stage spermatids, starting from Step 7, in the cKO mice (Figure 3A and B). To further consolidate this morphological data, we performed the qPCR assays using characteristic germline-specific markers (Supplementary Figure S2E–H). As expected, there was a significant reduction at the mRNA levels of Carm1 in the cKO testis (Supplementary Figure S2E). However, MVH, a germline protein that is present in all stages of spermatogenic cells (48), started to decrease from P30, which correlates with late stage haploid spermatid development (elongating spermatids) (Supplementary Figure S2F). The mRNA levels of the spermatid-specific marker H1T2 (49) also decrease in the P30 testis (Supplementary Figure S2G). In contrast, the expression levels of the spermatocyte-specific marker SYCP3 (50) remains constant in the testis of cKO males of different ages (Supplementary Figure S2H). Together, these data suggest that Carm1 is dispensable for meiosis, but critical for haploid spermatids development.
Figure 3.
Defective development of haploid spermatids in Carm1 cKO mice. (A) Periodic acid–Schiff (PAS) staining of the cross-sections of paraffin-embedded testes from WT and cKO mice. Z, Zygotene; P, Pachytene; RS, Round spermatids; ES, Elongating spermatids. (B) Statistic comparison of the average number of spermatids relative to Sertoli cells, calculated from PAS-stained cross-sections. Data were presented as mean ± SEM. *P< 0.05; **P< 0.01 (Student's t test).
Global gene expression changes in the Carm1 KO spermatids
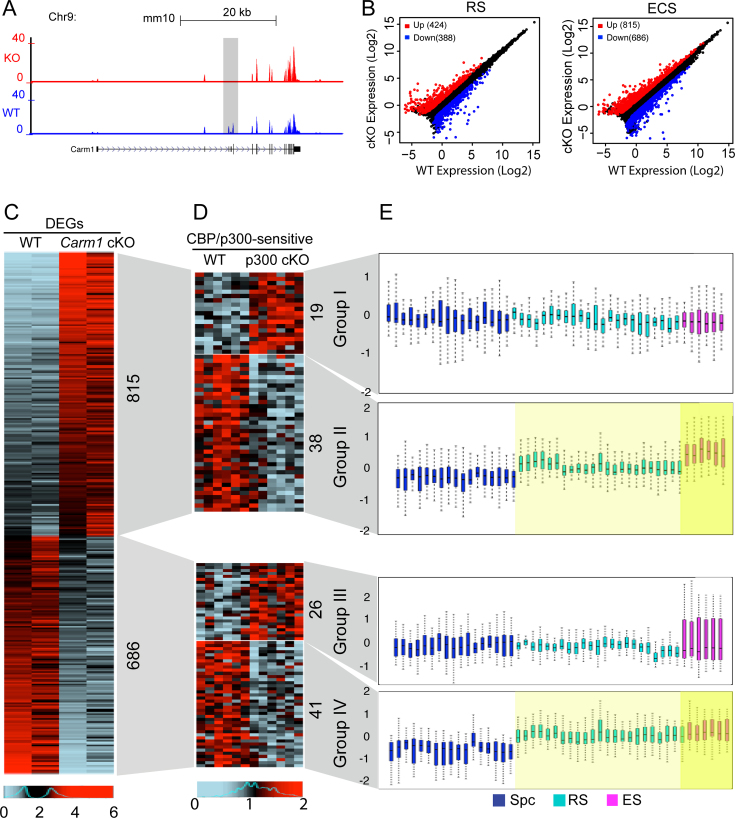
Numerous studies have characterized Carm1 as a transcriptional co-regulator (22,51–53), and we were thus interested in investigating the extent of genome-wide gene expression deregulation in Carm1-null spermatids. To this end, we made cell preparations enriched in spermatid populations at two stages (round spermatids [RS] and elongating/condensing spermatids [ECS]) by centrifugal elutriation from both WT and cKO testes (Supplementary Figure S4), and conducted RNA-seq analyses. A snapshot of the genome browser from the RNA-seq data shows that exon 3 and 4 of Carm1 were exclusively excised in the cKO spermatids, as compared to the WT (Figure 4A), causing a frame shift and thus confirming that Carm1 is specifically abrogated in haploid spermatids. Consistent with its role as a transcriptional regulator, we found 424 up- and 388 down-regulated genes (cutoff: ≥2 fold change, FDR < 0.05) in round spermatids, and 815 up- and 686 down-regulated genes in elongating/condensing spermatids, which represents mostly residual RNA produced from late round and elongating spermatids (Figure 4B, Supplementary Tables S2 and S3). To eliminate the possibility that the DEGs profile results from the disproportional ECS population, we randomly selected a panel of 10 genes that have been demonstrated to exhibit elevated expression trends in the ECS (as compared to RS) in the WT testes (54), and compare their expression levels in the ECS from the WT and cKO testes (Supplementary Figure S5). This analysis reveals that their mRNA expression levels do not vary significantly between WT and cKO spermatids, which strongly supports the fact that the gene expression differences in the ECS between WT and cKO cells are likely not due to gross alterations in the proportions of ECS subpopulations. We expected that a large set of haploid-specific genes would be deregulated in cKO haploid spermatids, since in these cells a large number of germline-restricted proteins are synthesized, which is required for the morphological assembly (e.g. acrosome, flagellum) and nuclear condensation occurring exclusively at this stage. However, a Gene Set Enrichment Analysis (GSEA) suggested that the deregulated gene sets were more particularly involved in specific activities such as metabolic process, oxidative process and glycosylation process (data not shown). Gene Ontology (GO) analyses demonstrated that specific ‘GO terms’, such as cell adhesion, cell proliferation, and immune response were also significantly over-represented in the Carm1 cKO spermatids (data not shown). The gene signature observed here in Carm1-null spermatids is reminiscent of the transcriptional changes observed in CBP/p300 conditional KO spermatids as described in our previous work (54), which suggested that CBP/p300 could play critical roles in specific processes occurring at the late stages of haploid spermatids development, including metabolic reprogramming. Therefore, the present expression profiles of cKO spermatids suggest a mechanistic link between Cbp/p300 and Carm1 in late spermiogenesis, which we investigated next.
Figure 4.
Genome-wide changes of the mRNA transcriptome in the purified round spermatids [RS] and elongating/condensing spermatids [ECS] upon Carm1 cKO. (A) A snapshot of the UCSC genome browser for the Carm1 mRNA expression track in the round spermatids between WT and cKO. Gray rectangle highlights the two floxed exons that were specifically deleted in the cKO haploid spermatids. (B) Scatter plots showing the differentially expressed genes (DEGs) (Cutoff: fold change ≥ 2, FDR < 0.05) in the purified round spermatids and elongating spermatids, respectively, between the WT and cKO testis. Red dots represent up-regulated, while blue dots indicate down-regulated DEGs; (C, D) Heatmaps showing the DEGs in the Carm1 cKO and Cbp/p300-sensitive genes upon Cbp/p300 cKO in elongating spermatids. (C) represents the DEGs in the elongating spermatids between WT and Carm1 cKO. (D) denotes the Cbp/p300-sensitive genes in the elongating spermatids between WT and Cbp/p300 cKO for those up-regulated (top) and down-regulated DEGs (bottom) in the Carm1 cKO spermatids. (E) Box plots indicate the normal expression pattern for the genes clustered into the four groups, respectively, throughout development of spermatocytes, round spermatids to elongating spermatids. Each bar in the box plots represents one biological replicate of the purified germ cell population. Spc, spermatocytes. RS, round spermatids. ECS, elongating/condensing spermatids.
The functions of Carm1 and Cbp/p300 are entwined in a number of different ways: (i) Cbp/p300 are well-characterized substrates for CARM1 (10,22), (ii) Cbp/p300 acetylation of the histone H3 tail primes Carm1 for methylation (52,55,56) and (iii) recent structural studies show that p300 complexes with Carm1, and that this interaction impacts the enzymatic activity of both these enzymes (11). We thus further explored the overlap of gene-sets that are Cbp/p300 and Carm1 regulated in vivo. An analysis of the deregulated gene expression profiles in Carm1 cKO elongating spermatids reveals that the loss of Carm1 induces similar numbers of genes to be up-regulated (815) and down-regulated (686) (Figure 4C). We next investigated which of these genes were also regulated by Cbp/p300 by integrating previously generated microarray-based transcriptome data (GSE55767) (54) with our RNA-seq data, and again identified both up- and down-regulated genes (Figure 4D). Importantly, the RNA-seq analysis designed to look for Cbp/p300 regulated genes was performed on Cbp/p300 depleted (not null) spermatids, leading to the identification of the deregulated transcriptional programs likely to be the most sensitive to fluctuating Cbp/p300 activity (54). Independently, we re-analyzed illumina-based transcriptome data from multiple biological repeats of purified germ cell populations (spermatocytes, round spermatids and elongating spermatids) from WT testes which we had previously used as controls in our investigations of various mouse models (54,57). By comparing these expression data, we explored the normal expression pattern of the four differentially expressed gene clusters (Groups I-IV) whose expression is affected by Carm1 cKO, and stratified them by the effect of Cbp/p300 on their regulation (Figure 4E). We were particularly interested in gene expression patterns which are different between spermatocytes and round spermatids (when CARM1 translocates into the nucleus—Figure 1D and E), and between round and elongating spermatids (when spermatid numbers drop in the cKO—Figure 3B). Genes in Group II (Carm1 acts as a repressor; Cbp/p300 acts as an activator) and Genes in Group IV (Carm1 acts as an activator; Cbp/p300 acts as an activator) exhibit a rising expression trend during post-meiotic development, and may thus be targets for Carm1 regulation as their expression changes correlate with the nuclear translocation of Carm1 and subsequent elevated levels. Carm1 and p300 have long been known to have synergistic coactivator activity (58,59), which likely accounts for the Group IV genes. What is unexpected here is the relatively large proportion of genes in Group II, repressed by Carm1 and activated by p300. We next investigated the possibility that Carm1 could function as a repressor by directly down-regulating the coactivator activity of p300.
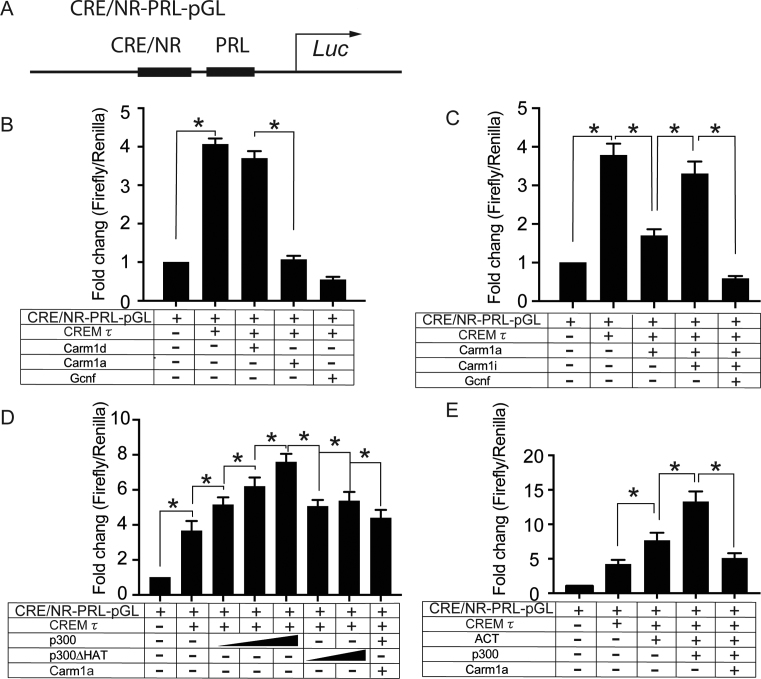
CARM1 represses transcriptional activity of testis-specific CREMτ transcription factor
Carm1 has been best characterized as a repressor through its ability to block the activation of the cAMP response element-binding protein (CREB) (22). CREB is closely related to CREMτ (cAMP response element modulator tau) (60), which is a master transcription factor that is exclusively expressed in haploid germ cells, and CREMτ KO resulted in the early haploid germ cell developmental arrest (61). Thus, we reasoned that CARM1 may have an impact on the transcriptional activity of CREMτ. To investigate this possibility, we utilized a luciferase reporter in which the DNA binding consensus sequences for CREMτ and GCNF, an orphan male germ cell nuclear factor, were placed upstream of a minimal rat prolactin (PRL) promoter (designated as CRE/NR-PRL-Luc) (40) (Figure 5A). We performed dual luciferase assays through the co-transfection of CARM1 and CREMτ into HEK293 cells. While CREMτ transfection alone can significantly induce the luciferase expression, transfection together with full-length WT CARM1 (Carm1a) significantly dampened the transcriptional activity of CREMτ (Figure 5B). This repressor activity of Carm1 is dependent on a functional methyltransferase domain (Carm1d). Also, co-transfection of GCNF (a factor known to counteract CREMτ activity) (40), significantly reduces luciferase activity. We also performed the dual luciferase assay in the presence of the CARM1-specific small-molecule inhibitor (CARM1i) TP064 and, as shown in Figure 5C, the transcriptional repressor activity of Carm1 was abolished by CARM1i treatment. Together, these data (Figure 5B andC) support a role for Carm1 and its activity as a repressor of CREMτ-regulated transcription.
Figure 5.
Luciferase reporter assay showing that CARM1 represses transcriptional activity of testis-specific CREMτ, ACT and CBP/p300. (A) Schematic diagram showing the structure of CREMτ-responsive luciferase reporter CRE/NR-PRL-pGL. The consensus sequence (cAMP-responsive element, CRE, and nuclear receptor, NR) for the DNA binding of CREMτ and GCNF transcription factors was inserted upstream of a minimal rat Prolactin promoter (PRL) in the pGL3.0 luciferase vector (Promega). (B–E) Relative fold changes of luciferase expression calculated from the transfection with different combinations of plasmids as indicated. Plasmids were generated as described in methods and materials. Dual-luciferase activity of three independent biological replicates were recorded for each assay. Renilla luciferase activity was used as the internal control. Data were presented as mean ± SEM. *P< 0.05; **P< 0.01 (Student's t test).
In somatic cells, CREM and CREB directly bind the CBP/p300 coactivators in a phospho-dependent manner to enhance its transcriptional activity (62). Using p300, we next examined how Carm1 affects p300 activity in the context of CREMτ-responsive luciferase reporter. Co-transfection of p300 significantly enhanced the expression of CREMτ reporter activity in a dose-dependent manner, while a histone acetyltransferase (HAT) mutant form of p300 did not augment CREMτ-regulated transcriptional activation (Figure 5D). Importantly, the transcriptional activation of CREMτ by p300 was significantly abrogated in the presence of CARM1 (Figure 5D). In haploid spermatids, unlike somatic cells, CREMτ is not phosphorylated at the key residue (S117) required for Cbp/p300 docking (63). Thus, in the context of germ cells, CREMτ and CBP/p300 likely interact through some other mechanism. Indeed, a yeast two-hybrid screen with CREMτ as bait identified Activator of CREMτ in the Testis (ACT), encoded by the Fhl5 gene, which is specifically expressed in haploid spermatids (63). Consistent with previous studies (63), co-transfection of ACT significantly augmented the CREMτ-mediated transcriptional activation, and we found that the additional transfection of p300 further enhanced luciferase activity (Figure 5E). This data suggests that ACT may be a linker between CREMτ and p300, a hypothesis that we test below. Importantly, the co-activator activity of ACT and p300 on the CREMτ reporter was largely abolished upon the addition of CARM1 (Figure 5E).
CBP/p300 interacts with ACT
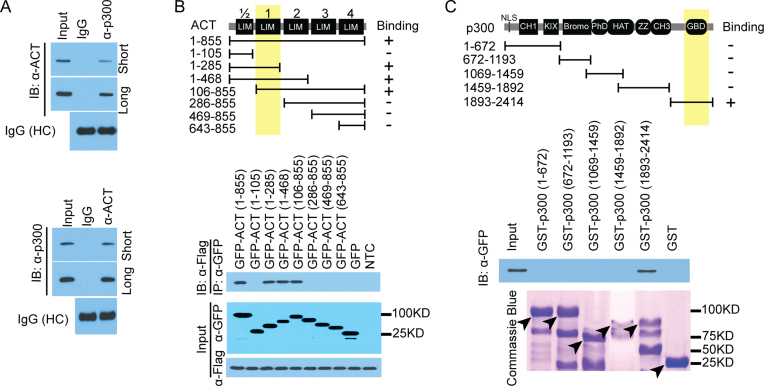
ACT is likely an adaptor molecule as it is composed almost entirely of LIM domains, which are involved in protein-protein interaction (64). It has been proposed that ACT functions by recruiting other coactivators, to facilitate CREMτ-driven transcription programs (65). Intriguingly, Carm1 cKO mice exhibit similar developmental defects in the haploid spermatids as observed in the KO mice of CREMτ, Cbp/p300 and ACT (54,61,66,67), supporting a mechanistic link between these four factors. Additionally, luciferase assays demonstrated the functional interplay among CREMτ, p300, ACT and CARM1 (Figure 5). We therefore tested whether ACT might associate with p300 and recruit these coactivators to CREMτ, and reciprocal endogenous co-immunoprecipitation (co-IP) experiments in the WT haploid spermatids using p300- and ACT-specific antibodies demonstrated an interaction between these two proteins (Figure 6A). Next, we mapped the interaction domain on ACT and p300. ACT has four and a half LIM domains in the full-length protein (Figure 6B). The third LIM domain of ACT interacts with the P-box of CREMτ (63). Co-IP experiments with deletion mutants of GFP-ACT mapped the p300 interaction to the first LIM domain (Figure 6B). Reciprocal mapping using GST fusions of the different domains of p300 and a GST pull-down approach (68), revealed an interaction between the C-terminal GBD domain and full-length GFP-ACT (Figure 6C). These results demonstrated a direct interaction, in cells and in vitro, between the first LIM domain of ACT and the GBD domain of p300.
Figure 6.
Mapping the interaction between ACT and p300. (A) Reciprocal endogenous Co-IP showing that ACT physiologically associates with p300 in vivo in the round spermatids. (B) ACT interacts with p300 through its first LIM domain. GFP tagged ACT full-length and deletion mutants harboring various number of LIM domains were constructed as indicated. Co-IP conducted using Flag-tagged p300, followed by western blot using GFP antibody against GFP-tagged ACT fragments. (C) p300 interacts with ACT through its GBD domain at the C-terminal. GST-pulldown was performed using GST-tagged p300 deletion mutants, followed by Western blot using the GFP antibody against the full-length ACT. Five deletion mutants of p300 constructs were constructed as described previously. Black arrowheads point to the expected protein size as predicted.
CARM1 methylation of p300 restricts its binding to ACT
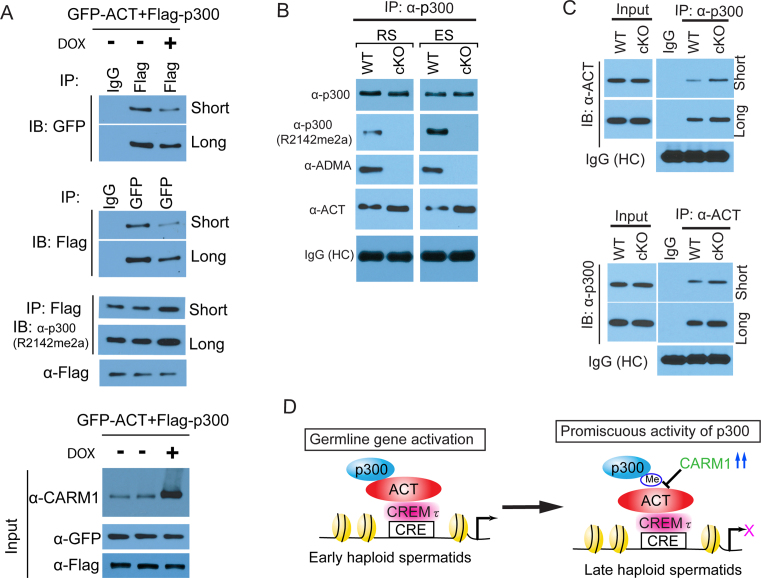
The ability of Carm1 to methylate p300 has been well-documented, and these methylation motifs are located in both the N-terminal end encompassing the KIX domain and the GBD domain at the C-terminal (10,22,69). Among the three potential methylated arginine residues of GBD, R2142 has been validated to be the bona fide CARM1-mediated methylation site in somatic cells (10). R2142 is highly conserved across the mammal species. Since we found that p300 specifically interacts with ACT through its GBD domain (Figure 6C), we wondered how methylation of p300 affects its ability to interact with ACT. To investigate this possibility, we first utilized a previously generated Carm1 flip-in HEK293 cell line (70), in which Carm1 protein levels can be dramatically induced. We confirmed the induction of Carm1 with doxycycline (Supplementary Figure S2A), and that elevated CARM1 levels boost the methylation of p300 using a pan ADMA antibody (Supplementary Figure S2B). Next, we performed co-IP assays using lysates from cells that were co-transfected with the plasmids of p300 and ACT, with and without doxycycline-inducible CARM1 overexpression. Using a methyl-specific p300 antibody (R2142me2a) that was previously generated (10), we again saw that methylation of p300 is elevated after Carm1 induction (Figure 7A). In reciprocal co-IP experiments, we observed that CARM1 overexpression weakened the interaction between p300 and ACT (Figure 7A), suggesting CARM1-deposited methylation negatively regulates this protein-protein interaction.
Figure 7.
p300 methylation at the GBD domain attenuated the interaction between ACT and p300 in vitro and in vivo. (A) The effect of overexpression of CARM1 on the interaction between ACT and p300. CARM1 was induced for overexpression by doxycycline in the inducible CARM1 Flip-in HEK293 cell line. Reciprocal co-IP was performed as indicated using Flag- and GFP-tag antibodies, followed by immunoblotting with the indicated antibodies. (B) p300 is endogenously methylated at R2142 in the haploid spermatids. Endogenous p300 protein was immuno-precipitated with specific p300 antibody, followed by immunoblotting with various antibodies as indicated. Note that Carm1 cKO abolished the ADMA mark on the p300 protein, and more bound ACT protein was detected upon Carm1 cKO in the p300 immunoprecipitates. The heavy chain (HC) of IgG served as an antibody loading control. (C) Endogenous co-IP was carried out in the round spermatids purified from the WT and cKO testes. Note that elevated binding affinity was observed between ACT and p300 in the cKO spermatids. (D) A working model for the role of Carm1-mediated methylation in haploid spermatids development. In the early stage of haploid spermatids, ACT recruits p300 co-activators to activate germline-specific target gene expression. In the late stage of haploid spermatids, a cohort of CREMτ/ACT-bound target genes must be temporally inhibited through the disassembly of the p300•ACT•CREMτ axis by the methylation of p300 by Carm1, for the last wave of metabolic reprogramming in the elongating spermatids.
To examine the potential in vivo relevance of this methylation-regulated protein-protein interaction, we next asked whether the R2142 of p300 is methylated in haploid spermatids, and how Carm1-loss impacted the interaction. First, we immunoprecipitated endogenous p300 and performed Western analysis using antibodies against ADMA and the R2142me2a mark, and found that p300 protein is indeed methylated in vivo in both round spermatids and elongating spermatids purified from wild type testes, but not in Carm1-null spermatids (Figure 7B). Moreover, p300 is more heavily methylated at the R2142 site in elongating spermatids as compared to round spermatids (Figure 7B), consistent with the fact that CARM1 is more abundant in the elongating spermatids (Figure 1D and E). Importantly, with Carm1-loss, we observed a stronger interaction between endogenous ACT and p300 in both round and elongating spermatids (Figure 7B). Finally, we performed reciprocal co-IP using lysates from purified elongating spermatids. Again we see that in the absence of Carm1, there is a stronger interaction between endogenous ACT and p300 (Figure 7C). These results suggest that the methyl mark in the GBD domain of p300 attenuates its interaction with ACT in vivo, and that CARM1 negatively regulates p300•ACT•CREMτ mediated transcription in haploid spermatids.
DISCUSSION
Arginine methylation and spermiogenesis
PRMT5, 6 and 7 have been studied in male germ cells to varying degrees. The functional significance of arginine methylation in germline development was first recognized with the genetic deletion of Dart5 (the fly ortholog of mammalian Prmt5) (71), and subsequently the conditional deletion of Prmt5 in mice (38). The primary targets for PRMT5 methylation in germ cells are the PIWI proteins, which are preferentially expressed in the gonads in both vertebrates and invertebrates (28) PIWI bind to a distinct population of small non-coding RNAs, termed PIWI-interacting RNAs (piRNAs), ranging from 26- to 31-nt in length, which are also primarily expressed in the gonads (28,29). Methylated PIWI proteins are recognized by members of the TDRD protein family (29), and genetic manipulation of components of the TDRD/PIWI/piRNAs pathway induces de-repression of the transposable elements, thus leading to the spermatogenic arrest and male infertility (28,30,32). Genome-wide association studies (GWAS) linked the Prmt6 variants to the infertility in the non-obstructive azoospermia (NOA) patients (72). Furthermore, PRMT6 is localized to the nucleus of spermatogonia and spermatocytes, with low levels of expression in Leydig and Sertoli cells (73). Knockdown studies in GC-1 and GC-2 germ cell lines, suggest that PRMT6 is involved in cellular processes including cell migration and apoptosis, although Prmt6−/− males are fertile and no mechanism of action was elucidated. Finally, PRMT7 has been implicated in male germline genomic imprinting through its ability to interact with CTCFL (also known as BORIS), methylate histone H4, and promote the recruitment of DNA methyltransferases (74). It is important to note that CARM1 does not methylate the same type of substrates as PRMT6, and unlike PRMT5 and 7, it deposits an ADMA mark. It is thus very likely that CARM1 exhibits distinct functions during germline development, which are not redundant with other PRMTs.
The pleiotropic roles of CARM1 in spermiogenesis
Compared with other somatic organs, Carm1 is highly expressed in the testis, and CARM1 protein exhibits dynamic expression levels and distinct subcellular localization in the germ cells at different stages (Figure 1). We thus initially hypothesized that CARM1-mediated deposition of arginine methylation marks may be essential for the functional integrity of TDRD/PIWI/piRNAs pathway, and therefore anticipated that CARM1 would be involved in the development of early spermatocytes. Unexpectedly, we found that Carm1-null germ cells progressed normally through meiosis to the haploid round spermatids during the first wave of spermatogenesis. Nonetheless, transcriptome analysis of Carm1-null germ cells revealed significant perturbations in the expression profiles of round spermatids (Figure 4), leading to the defective development of the elongating spermatids (Figures 2 and 3). This transcriptome profiling provided us with gene sets that were both up- and down-regulated, and we attempted to perform Carm1 ChIP-seq experiments in round and elongating spermatids to help establish which of these gene sets were potentially direct targets of Carm1 regulation, but these experiments were unsuccessful. Importantly, the spectrum of Carm1-regulated gene sets was implicated in metabolic processes and not, as we expected, signatures related to haploid-specific gene sets. The similarity of the Carm1 and p300 deregulated gene signatures thus prompted us to investigate this pathway.
We should not disregard the potential indirect effects of Carm1 on transcription regulation, which could occur through the methylation of non-histone substrates that may broadly impact splicing, translation and even chromatin structure. For Instance, in late spermiogenesis, there are a number of global chromatin remodeling processes that occur when histones are first replaced by testis-specific transition proteins (TNP1 and 2), and then by protamines (P1 and 2). This remodeling is critical for the formation of highly condensed sperm chromatin. Importantly, both transition proteins and protamines harbor a large percentage of arginine residues (15–50%), which could be targeted for modification by the PRMTs. Indeed, mass spec studies have recently identified a number of arginine-methylated sites on the transition proteins, and CARM1 was shown to methylate TNP2 in vitro (75). The TNP2 protein is exclusively translated in the elongating spermatids (from Step 9 onward). In the Carm1 cKO model, we found defective spermatids occurring from Step 7 of spermatids (Figure 3), which precedes the expression of TNP2. This does not rule out a contribution of TNP2 methylation to normal late stages of spermatid development, but does indicate that perturbations of different molecular pathways may contribute to the defect observed in Carm1-null spermatids. We did not observe altered expression of transition proteins or protamines in our Carm1 cKO model. TP1 and TP2 mRNA were not affected by CARM1 loss, nor was the mRNA encoding H2A.L.2, a histone variant recently shown to be required for the chromatin loading of transition proteins (76). Furthermore, CARM1 is a known regulator of alternative splicing. It is thus likely that Carm1 is regulating normal spermiogenesis at multiple levels, including the structural regulation of chromatin condensation, splicing, and the direct positive and negative regulation of transcription.
p300 is recruited to CREMτ through ACT
One of the key molecular mechanisms that regulates spermatogenesis in a temporal spatially-specific manner is the cAMP-dependent signaling pathway. Gene expression in this pathway is primarily regulated by two members of the bZIP transcription factors–cAMP-response element binding protein (CREB) and cAMP-responsive element modulator (CREM) (60). In somatic cells, in response to external stimuli or metabolic and developmental signals, both transcription factors bind to the consensus palindromic CRE element sequence (TGACGTCA), which are widely distributed in the genome, upon the phosphorylation of a serine residue (S133 in CREB, S117 in CREM) in the activation domain by the cAMP-activated protein kinase. Phosphorylated forms of CREB and CREM enable binding of the CREB binding protein (CBP) and p300, which are well-studied transcriptional co-activators, as well as the recruitment of basal transcriptional machinery. There are multiple variants of CREM present in different tissues owing to the utilization of alternative promoters, polyadenylation signals, and alternative splicing. CREM can function either as a transcriptional repressor or an activator depending on whether it possesses a transactivation domain. Only the shorter CREM variants (lacking the transactivation domain) are detected in the early stage of spermatogenic cells (e.g. spermatogonia). Upon the onset of meiosis, full-length isoform CREM (CREMτ) is expressed, and is an abundant master regulator of transcription in haploid spermatids. In the testis, surprisingly, CREMτ activation bypasses the requirement of serine phosphorylation through binding to a spermatids-specific protein ACT. Here, we found that ACT directly binds p300 through the first LIM domain and the GBD domain, respectively. Thus, the p300•ACT•CREMτ axis represents a powerful transactivation signaling to orchestrate spermiogenesis in the testis.
CARM1 negatively regulates the p300•ACT•CREMτ axis
We have found that Carm1-mediated methylation of the GBD domain of p300 impedes the p300/ACT interaction, and thus Carm1 acts as a repressor of the p300•ACT•CREMτ axis. The enhanced expression of CARM1 in the elongating spermatids reduces the transcriptional activity of CREMτ, particularly of those genes involved in metabolic pathways. By comparing the Carm1 cKO RNA-seq data with the CBP/p300 cKO RNA-seq data, we identified a number of genes that are likely common targets of CARM1 and CBP/p300. With Carm1-loss, 815 genes were significantly up-regulated in elongating spermatids (Carm1 potentially functions as a repressor), and nested within this group were 38 genes that used CBP/p300 as a coactivator. These Group II genes (Figure 4D) are of particular interest to us, because they may be the targets of CREMτ that are repressed by Carm1 in spermatids. Unfortunately, Carm1 ChIP-seq experiments in round and elongating spermatids were unsuccessful. Thus, we have been unable to determine if there are unique Carm1 binding profiles that could act as signatures for Group II genes. An important issue to consider is that while the spermiogenesis phenotypes of CARM1, p300, CREMτ, and ACT knockouts are very similar, CARM1 is acting as a repressor and the other three factors are acting as positive transcriptional regulators. It is possible that these four factors become important at a common point during spermiogenesis, and as a consequence of shared pathway deregulation (positive or negative), similar defective differentiation phenotypes emerge.
CARM1 and p300 were originally identified as the hormone-dependent nuclear receptor co-activators that function synergistically (77). Carm1 can directly methylate p300 at a number of sites, and some of these methylation events promote transcription (78) while others repress transcription (10,22), thereby fine-tuning the activity of certain coactivator complexes. In CREMτ-driven luciferase reporter assays, Carm1 functions as a repressor (Figure 5). We have mapped the interaction of ACT to the GBD domain (Figure 6C), which is the same region that binds GRIP. Importantly, methylation of R2142 by Carm1 (within the GBD domain) results in a block of the GRIP/p300 interaction (10). Similarly, we observed that Carm1 activity prevents the interaction between ACT and the GBD of p300 (Figure 7A–C). Thus, our mechanistic analysis indicates that CARM1 serves as a ‘repressor’ to dampen transcriptional activity of the p300•ACT•CREMτ axis in a developmentally-regulated germline context (Figure 7D).
DATA AVAILABILITY
The GEO accession number for our RNA-Seq data is GSE107984.
Supplementary Material
ACKNOWLEDGEMENTS
Author Contributions: M.T.B. and J.B. conceived the project. J.B. carried out all the cell-based and in vivo experiments. J.S. performed the deep-sequencing. S.R., K.L. and Y.L. performed the bioinformatics analysis of the RNA-Seq data sets. M.T.B. and J.B. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [DK062248 to M.T.B.]; CPRIT [RP120348 to J.S.]; Center for Cancer Epigenetics (CCE) at MDACC provided a fellowship (to J.B.) Funding for open access charge: NIH [DK062248].
Conflict of interest statement. M.T.B. is a cofounder of EpiCypher. Other authors have no competing financial interests.
REFERENCES
- 1. Blanc R.S., Richard S.. Arginine methylation: the coming of age. Mol Cell. 2017; 65:8–24. [DOI] [PubMed] [Google Scholar]
- 2. Larsen S.C., Sylvestersen K.B., Mund A., Lyon D., Mullari M., Madsen M.V., Daniel J.A., Jensen L.J., Nielsen M.L.. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016; 9:rs9. [DOI] [PubMed] [Google Scholar]
- 3. Gayatri S., Bedford M.T.. Readers of histone methylarginine marks. Biochim. Biophys. Acta. 2014; 1839:702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Y., Bedford M.T.. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013; 13:37–50. [DOI] [PubMed] [Google Scholar]
- 5. Iberg A.N., Espejo A., Cheng D., Kim D., Michaud-Levesque J., Richard S., Bedford M.T.. Arginine methylation of the histone h3 tail impedes effector binding. J. Biol. Chem. 2008; 283:3006–3010. [DOI] [PubMed] [Google Scholar]
- 6. Zurita-Lopez C.I., Sandberg T., Kelly R., Clarke S.G.. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J. Biol. Chem. 2012; 287:7859–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alam H., Gu B., Lee M.G.. Histone methylation modifiers in cellular signaling pathways. Cell Mol. Life Sci. 2015; 72:4577–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen D., Ma H., Hong H., Koh S.S., Huang S.M., Schurter B.T., Aswad D.W., Stallcup M.R.. Regulation of transcription by a protein methyltransferase. Science. 1999; 284:2174–2177. [DOI] [PubMed] [Google Scholar]
- 9. Feng Q., Yi P., Wong J., O’Malley B.W.. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 2006; 26:7846–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee Y.H., Coonrod S.A., Kraus W.L., Jelinek M.A., Stallcup M.R.. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:3611–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yi P., Wang Z., Feng Q., Chou C.K., Pintilie G.D., Shen H., Foulds C.E., Fan G., Serysheva I., Ludtke S.J. et al. Structural and functional impacts of ER coactivator sequential recruitment. Mol. Cell. 2017; 67:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bauer U.M., Daujat S., Nielsen S.J., Nightingale K., Kouzarides T.. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002; 3:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carascossa S., Dudek P., Cenni B., Briand P.A., Picard D.. CARM1 mediates the ligand-independent and tamoxifen-resistant activation of the estrogen receptor alpha by cAMP. Genes Dev. 2010; 24:708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. El Messaoudi S., Fabbrizio E., Rodriguez C., Chuchana P., Fauquier L., Cheng D., Theillet C., Vandel L., Bedford M.T., Sardet C.. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13351–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frietze S., Lupien M., Silver P.A., Brown M.. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008; 68:301–306. [DOI] [PubMed] [Google Scholar]
- 16. Kleinschmidt M.A., Streubel G., Samans B., Krause M., Bauer U.M.. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008; 36:3202–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Brien K.B., Alberich-Jorda M., Yadav N., Kocher O., Diruscio A., Ebralidze A., Levantini E., Sng N.J., Bhasin M., Caron T. et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010; 137:2147–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Q., Bruce A.W., Jedrusik A., Ellis P.D., Andrews R.M., Langford C.F., Glover D.M., Zernicka-Goetz M.. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009; 27:2637–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yadav N., Cheng D., Richard S., Morel M., Iyer V.R., Aldaz C.M., Bedford M.T.. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008; 9:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bedford M.T., Clarke S.G.. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009; 33:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vu L.P., Perna F., Wang L., Voza F., Figueroa M.E., Tempst P., Erdjument-Bromage H., Gao R., Chen S., Paietta E. et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 2013; 5:1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu W., Chen H., Du K., Asahara H., Tini M., Emerson B.M., Montminy M., Evans R.M.. A transcriptional switch mediated by cofactor methylation. Science. 2001; 294:2507–2511. [DOI] [PubMed] [Google Scholar]
- 23. Yadav N., Lee J., Kim J., Shen J., Hu M.C., Aldaz C.M., Bedford M.T.. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6464–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D., Lee J., Cheng D., Li J., Carter C., Richie E., Bedford M.T.. Enzymatic activity is required for the in vivo functions of CARM1. J. Biol. Chem. 2010; 285:1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim J., Lee J., Yadav N., Wu Q., Carter C., Richard S., Richie E., Bedford M.T.. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J. Biol. Chem. 2004; 279:25339–25344. [DOI] [PubMed] [Google Scholar]
- 26. Herrmann F., Pably P., Eckerich C., Bedford M.T., Fackelmayer F.O.. Human protein arginine methyltransferases in vivo–distinct properties of eight canonical members of the PRMT family. J. Cell Sci. 2009; 122:667–677. [DOI] [PubMed] [Google Scholar]
- 27. Chen C., Nott T.J., Jin J., Pawson T.. Deciphering arginine methylation: tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011; 12:629–642. [DOI] [PubMed] [Google Scholar]
- 28. Ku H.Y., Lin H.. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 2014; 1:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vagin V.V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G.J., Aravin A.A.. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009; 23:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saxe J.P., Chen M., Zhao H., Lin H.. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013; 32:1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka T., Hosokawa M., Vagin V.V., Reuter M., Hayashi E., Mochizuki A.L., Kitamura K., Yamanaka H., Kondoh G., Okawa K. et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10579–10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandey R.R., Tokuzawa Y., Yang Z., Hayashi E., Ichisaka T., Kajita S., Asano Y., Kunieda T., Sachidanandam R., Chuma S. et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:16492–16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shoji M., Tanaka T., Hosokawa M., Reuter M., Stark A., Kato Y., Kondoh G., Okawa K., Chujo T., Suzuki T. et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell. 2009; 17:775–787. [DOI] [PubMed] [Google Scholar]
- 34. Kirino Y., Vourekas A., Kim N., de Lima Alves F., Rappsilber J., Klein P.S., Jongens T.A., Mourelatos Z.. Arginine methylation of vasa protein is conserved across phyla. J. Biol. Chem. 2010; 285:8148–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chi M.N., Auriol J., Jegou B., Kontoyiannis D.L., Turner J.M., de Rooij D.G., Morello D.. The RNA-binding protein ELAVL1/HuR is essential for mouse spermatogenesis, acting both at meiotic and postmeiotic stages. Mol. Biol. Cell. 2011; 22:2875–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paronetto M.P., Messina V., Bianchi E., Barchi M., Vogel G., Moretti C., Palombi F., Stefanini M., Geremia R., Richard S. et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009; 185:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen Chi M., Chalmel F., Agius E., Vanzo N., Khabar K.S., Jegou B., Morello D.. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One. 2009; 4:e4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim S., Gunesdogan U., Zylicz J.J., Hackett J.A., Cougot D., Bao S., Lee C., Dietmann S., Allen G.E., Sengupta R. et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell. 2014; 56:564–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Zhu T., Li Q., Liu C., Han F., Chen M., Zhang L., Cui X., Qin Y., Bao S. et al. Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 2015; 5:11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajkovic M., Iwen K.A., Hofmann P.J., Harneit A., Weitzel J.M.. Functional cooperation between CREM and GCNF directs gene expression in haploid male germ cells. Nucleic Acids Res. 2010; 38:2268–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W., Wu J., Kim S.Y., Zhao M., Hearn S.A., Zhang M.Q., Meistrich M.L., Mills A.A.. Chd5 orchestrates chromatin remodelling during sperm development. Nat. Commun. 2014; 5:3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meistrich M.L., Hess R.A.. Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol. Biol. 2013; 927:299–307. [DOI] [PubMed] [Google Scholar]
- 45. Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., Braun R.E.. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kawabe Y., Wang Y.X., McKinnell I.W., Bedford M.T., Rudnicki M.A.. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012; 11:333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahmed E.A., de Rooij D.G.. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 2009; 558:263–277. [DOI] [PubMed] [Google Scholar]
- 48. Tanaka S.S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T.. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000; 14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 49. Tanaka H., Iguchi N., Isotani A., Kitamura K., Toyama Y., Matsuoka Y., Onishi M., Masai K., Maekawa M., Toshimori K. et al. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol. Cell. Biol. 2005; 25:7107–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Hoog C.. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000; 5:73–83. [DOI] [PubMed] [Google Scholar]
- 51. Naeem H., Cheng D., Zhao Q., Underhill C., Tini M., Bedford M.T., Torchia J.. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 2007; 27:120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Daujat S., Bauer U.M., Shah V., Turner B., Berger S., Kouzarides T.. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 2002; 12:2090–2097. [DOI] [PubMed] [Google Scholar]
- 53. Ma H., Baumann C.T., Li H., Strahl B.D., Rice R., Jelinek M.A., Aswad D.W., Allis C.D., Hager G.L., Stallcup M.R.. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 2001; 11:1981–1985. [DOI] [PubMed] [Google Scholar]
- 54. Boussouar F., Goudarzi A., Buchou T., Shiota H., Barral S., Debernardi A., Guardiola P., Brindle P., Martinez G., Arnoult C. et al. A specific CBP/p300-dependent gene expression programme drives the metabolic remodelling in late stages of spermatogenesis. Andrology. 2014; 2:351–359. [DOI] [PubMed] [Google Scholar]
- 55. An W., Kim J., Roeder R.G.. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004; 117:735–748. [DOI] [PubMed] [Google Scholar]
- 56. Yue W.W., Hassler M., Roe S.M., Thompson-Vale V., Pearl L.H.. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007; 26:4402–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Montellier E., Boussouar F., Rousseaux S., Zhang K., Buchou T., Fenaille F., Shiota H., Debernardi A., Hery P., Curtet S. et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013; 27:1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen D., Huang S.M., Stallcup M.R.. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000; 275:40810–40816. [DOI] [PubMed] [Google Scholar]
- 59. Stallcup M.R., Chen D., Koh S.S., Ma H., Lee Y.H., Li H., Schurter B.T., Aswad D.W.. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem. Soc. Trans. 2000; 28:415–418. [PubMed] [Google Scholar]
- 60. Martianov I., Choukrallah M.A., Krebs A., Ye T., Legras S., Rijkers E., Van Ijcken W., Jost B., Sassone-Corsi P., Davidson I.. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics. 2010; 11:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sassone-Corsi P. CREM: a master-switch regulating the balance between differentiation and apoptosis in male germ cells. Mol. Reprod. Dev. 2000; 56:228–229. [DOI] [PubMed] [Google Scholar]
- 62. De Cesare D., Sassone-Corsi P.. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 2000; 64:343–369. [DOI] [PubMed] [Google Scholar]
- 63. Fimia G.M., De Cesare D., Sassone-Corsi P.. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999; 398:165–169. [DOI] [PubMed] [Google Scholar]
- 64. Tran M.K., Kurakula K., Koenis D.S., de Vries C.J.. Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta. 2016; 1863:219–228. [DOI] [PubMed] [Google Scholar]
- 65. Fimia G.M., Morlon A., Macho B., De Cesare D., Sassone-Corsi P.. Transcriptional cascades during spermatogenesis: pivotal role of CREM and ACT. Mol. Cell. Endocrinol. 2001; 179:17–23. [DOI] [PubMed] [Google Scholar]
- 66. Kotaja N., De Cesare D., Macho B., Monaco L., Brancorsini S., Goossens E., Tournaye H., Gansmuller A., Sassone-Corsi P.. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hogeveen K.N., Sassone-Corsi P.. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum. Fertil. (Camb.). 2006; 9:73–79. [DOI] [PubMed] [Google Scholar]
- 68. Hasan S., Stucki M., Hassa P.O., Imhof R., Gehrig P., Hunziker P., Hubscher U., Hottiger M.O.. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 2001; 7:1221–1231. [DOI] [PubMed] [Google Scholar]
- 69. Lee Y.H., Bedford M.T., Stallcup M.R.. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 2011; 25:176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng D., Cote J., Shaaban S., Bedford M.T.. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007; 25:71–83. [DOI] [PubMed] [Google Scholar]
- 71. Gonsalvez G.B., Rajendra T.K., Tian L., Matera A.G.. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006; 16:1077–1089. [DOI] [PubMed] [Google Scholar]
- 72. Hu Z., Xia Y., Guo X., Dai J., Li H., Hu H., Jiang Y., Lu F., Wu Y., Yang X. et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat. Genet. 2011; 44:183–186. [DOI] [PubMed] [Google Scholar]
- 73. Luo M., Li Y., Guo H., Lin S., Chen J., Ma Q., Gu Y., Jiang Z., Gui Y.. Protein arginine Methyltransferase 6 involved in germ cell viability during spermatogenesis and Down-Regulated by the androgen receptor. Int. J. Mol. Sci. 2015; 16:29467–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jelinic P., Stehle J.C., Shaw P.. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006; 4:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gupta N., Madapura M.P., Bhat U.A., Rao M.R.. Mapping of post-translational modifications of transition proteins, TP1 and TP2, and identification of protein arginine methyltransferase 4 and lysine methyltransferase 7 as methyltransferase for TP2. J. Biol. Chem. 2015; 290:12101–12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Barral S., Morozumi Y., Tanaka H., Montellier E., Govin J., de Dieuleveult M., Charbonnier G., Coute Y., Puthier D., Buchou T. et al. Histone Variant H2A.L.2 guides transition Protein-Dependent protamine assembly in male germ cells. Mol. Cell. 2017; 66:89–101. [DOI] [PubMed] [Google Scholar]
- 77. Chen D., Huang S.M., Stallcup M.R.. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000; 275:40810–40816. [DOI] [PubMed] [Google Scholar]
- 78. Chevillard-Briet M., Trouche D., Vandel L.. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002; 21:5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for our RNA-Seq data is GSE107984.