Abstract
Background
Single-gene mutation syndromes account for some familial glioma (FG); however, they make up only a small fraction of glioma families. Gliomas can be classified into 3 major molecular subtypes based on isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion. We hypothesized that the prevalence of molecular subtypes might differ in familial versus sporadic gliomas and that tumors in the same family should have the same molecular subtype.
Methods
Participants in the FG study (Gliogene) provided samples for germline DNA analysis. Formalin-fixed, paraffin-embedded tumors were obtained from a subset of FG cases, and DNA was extracted. We analyzed tissue from 75 families, including 10 families containing a second affected family member. Copy number variation data were obtained using a first-generation Affymetrix molecular inversion probe (MIP) array.
Results
Samples from 62 of 75 (83%) FG cases could be classified into the 3 subtypes. The prevalence of the molecular subtypes was: 30 (48%) IDH-wildtype, 21 (34%) IDH-mutant non-codeleted, and 11 (19%) IDH-mutant and 1p/19q codeleted. This distribution of molecular subtypes was not statistically different from that of sporadic gliomas (P = 0.54). Of 10 paired FG samples, molecular subtypes were concordant for 7 (κ = 0.59): 3 IDH-mutant non-codeleted, 2 IDH-wildtype, and 2 IDH-mutant and 1p/19q codeleted gliomas.
Conclusions
Our data suggest that within individual families, patients develop gliomas of the same molecular subtype. However, we did not observe differences in the prevalence of the molecular subtypes in FG compared with sporadic gliomas. These observations provide further insight into the distribution of molecular subtypes in FG.
Keywords: familial glioma (FG), IDH-mutant and 1p/19q-codeleted, IDH-mutant non-codeleted, IDH-wild type, molecular inversion probe (MIP)
Importance of the study
The etiology for patients with FG, with the exception of rare familial syndromes, is still unclear. The development of molecular subtypes to classify sporadic gliomas has shown prognostic value by improving genetic insight and clinical decision making. The use of molecular subtyping in nonsyndromic FG will be critical in defining the molecular characteristics of these gliomas. Here we provide preliminary evidence that FG patients display similar molecular signatures to the sporadic glioma population. We also provide evidence that within families containing 2 or more gliomas, individuals tend to develop gliomas of the same molecular subtype. This suggests that nonsyndromic FG and sporadic glioma develop through similar molecular pathways.
Gliomas are the most common primary central nervous system tumor in adults. They account for 29% of all primary brain tumors and 82% of malignant brain tumors, leading to approximately 17000 deaths in the US each year.1,2 Gliomas are traditionally classified as either high-grade glioblastoma (GBM) or low-grade astrocytomas and oligodendrogliomas, which differ significantly in their prognosis and treatment.3 Typically, a person with a diagnosis of GBM has a poor prognosis despite treatment, and the median survival is approximately 14 months.2 In comparison, a person with a diagnosis of lower-grade glioma tends to have a better prognosis, but the disease still has a significant impact on the patient’s quality of life, with an average survival of approximately 7 years.4
Familial glioma (FG) accounts for approximately 5%–10% of all glioma cases, similar to the percent of familial cases seen in other, more common cancers, such as breast and colorectal cancer.5,6 The development of glioma within families is not well understood, but some single-gene Mendelian syndromes (eg, neurofibromatosis 1 or 2, tumor protein 53 [TP53]) are known to have a significantly higher risk for FG. Collectively, these syndromes are rare, and known genes explain only a small fraction of familial risk.7–9 Our research has identified deleterious mutations in known tumor suppressors (eg, TP53), and we were the first to suggest that mutations in protection of telomeres 1 (POT1) are causative in FG.10 Case-control studies have consistently reported a 2-fold increased risk of developing gliomas in first-degree relatives of affected glioma patients.5,6,11 While evidence suggests a potential genetic link between family members, more research needs to be performed to identify mutations that aggregate in pedigrees which do not carry mutations in some of these known familial syndrome genes.
Adult diffuse gliomas can now be classified into 3 subtypes based on 2 molecular alterations: isocitrate dehydrogenase 1 or 2 (IDH1/2) mutation and 1p/19q codeletion. The classification scheme developed by the World Health Organization (WHO)3 corresponds closely with previously established schemes based on telomerase reverse transcriptase (TERT) mutation, IDH mutation, and 1p/19q codeletion.12,13 In addition to the molecular alterations that provide the basis for each subtype, characteristic copy number alterations are found in each subtype as well. Prognosis based on molecular classification of tumors has shown improved accuracy compared with histological subtyping.13 While the importance of classifying sporadic gliomas by molecular subtype has been established, these particular molecular markers have not yet been evaluated in FG.
We hypothesized that if FG can be classified into one of the known molecular subtypes, (i) the prevalence of these subtypes might be different between familial and sporadic glioma cases, and (ii) in families with multiple gliomas, the tumors in affected members should be of similar subtype. To test these hypotheses, we classified FG tumors from the Gliogene Consortium into the 3 molecular subtypes.
Materials and Methods
Gliogene Consortium Families and Study Participants
The Gliogene Consortium sites identified and recruited families that reported at least 2 gliomas through the Internet, social media, foundations, genetic counselors, and clinicians around the world. All participants provided written informed consent under a protocol that was approved by the institutional review boards at participating institutions. Information from 382 families was collected through Gliogene sites. This study focused specifically on adult diffuse gliomas. Excluded were 7 families with multiple ependymomas, juvenile pilocytic astrocytomas, gangliogliomas, dysembryoplastic neuroepithelial tumors, and/or had no adult diffuse glioma, leaving 375 families.
For this study, formalin-fixed, paraffin-embedded (FFPE) tumor tissue and extracted DNA were available from at least 1 individual in 83 of the 375 families. Ten families had tissue available from a second affected family member (first- or second-degree relative); thus, we analyzed a total of 93 FG cases. DNA was extracted from the Gliogene FFPE tissues as previously described.14 The first-generation Affymetrix molecular inversion probe (MIP) copy number array assay was performed using methods previously reported.14 Array data from 8 FG cases were excluded due to excessive noise, leaving 85 FG cases with adequate MIP data.
The familial cases were compared with a cohort of 148 Mayo Clinic sporadic glioma cases. These sporadic cases contained mutation data of known glioma genes and were previously classified into the Eckel-Passow et al13 molecular subtypes through OncoScan array data analysis (D. Brown, unpublished manuscript). These subtypes correspond closely to the WHO subtypes and are based on 1p/19q codeletion, TERT promoter mutation, and IDH1/2 mutation.13 These were analyzed using Affymetrix V1 OncoScan Arrays, the OncoScan Reagent Kit with probe mix 1.0, and Chromosome Analysis Suite version 3.1.0.15. This cohort was used to compare array patterns and the prevalence of the molecular subtypes within sporadic and FG populations.
Copy Number Analysis of the First-Generation Affymetrix MIP Data
Copy number from the first-generation Affymetrix MIP data was estimated by producing whole genome plots of log2 (copy number/2) by chromosome and position of the probes using a 255 probe moving average in Microsoft Excel. A whole genome view of allele difference was generated by plotting (copy number of allele A − copy number of allele B) versus chromosome and position of the probes (Fig. 1B, D, F). Additional plots were generated focusing on specific regions of interest (allele difference for chromosomes 9 and 17, copy number for platelet derived growth factor receptor alpha [PDGFRA]/KIT, epidermal growth factor receptor [EGFR], cyclin-dependent kinase inhibitor 2A or 2B [CDKN2A/B], and GLI1/CDK4/MDM2) for all samples. The plots for all cases were manually analyzed for copy number variations (CNVs) and copy-neutral loss of heterozygosity (cnLOH).
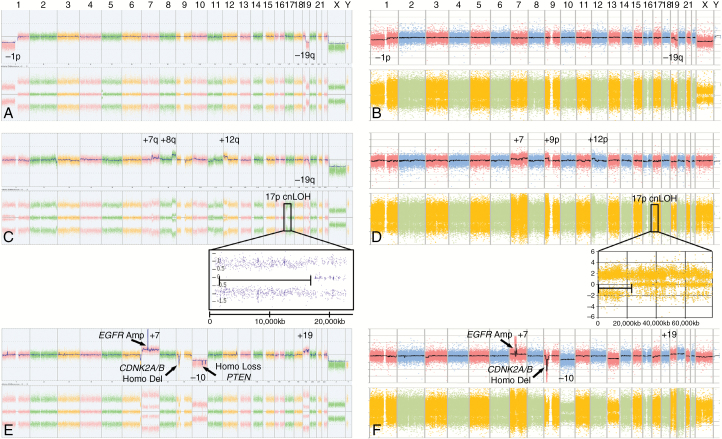
Fig. 1.
Chromosome Analysis Suite (ChAS) Affymetrix OncoScan arrays and first-generation Affymetrix MIP arrays from gliomas in 3 molecular subtypes show comparable characteristic alterations. OncoScan arrays from sporadic gliomas (left) and MIP arrays from familial gliomas (right). Characteristic CNVs are labeled for (A, B) IDH-mutant and 1p/19q codeleted glioma, (C, D) IDH-mutant non-codeleted glioma, and (E, F) IDH-wildtype glioma. The insets for IDH mutant non-codeleted glioma arrays (C, D) illustrate regions of chromosome 17p cnLOH, a characteristic in most IDH-mutant non-codeleted gliomas. Since ChAS 3.1.0.15 software could not be used to view first-generation MIP array data (B, D, F), these data were plotted using Microsoft Excel (see Materials and Methods).
DNA Sequencing
If blood was available, subjects were also sequenced for germline mutations in the POT1 and TP53 genes using previously reported methods.10
Classification of Glioma Molecular Subtype
Subjects were classified into the 3 most prevalent molecular subtypes—IDH-wildtype, IDH-mutant and 1p/19q codeleted, as well as IDH-mutant non-codeleted—in the WHO 2016 classification system, based on characteristic copy number alterations.3
Affymetrix first-generation MIP array data collected on the Gliogene families were characterized for CNVs and cnLOH. Each tumor was classified into one of the molecular subtypes based on abnormalities in array pattern and compared with a Mayo Clinic cohort of 148 sporadic gliomas (D. Brown, unpublished manuscript). For the FG cohort, cases classified as IDH-mutant 1p/19q codeleted were distinguished by their characteristic translocation between 1p/19q codeleted arms (Fig. 1B). FG cases classified as IDH-mutant non-codeleted always contained 17p cnLOH and other acquired abnormalities, such as duplication of 7q, duplication of 8q, and/or deletion of 19q (Fig. 1D). FG cases classified as IDH-wildtype contained gain of chromosome 7, loss of chromosome 10, gain of chromosome 19, gain of chromosome 20, amplification of EGFR, and homozygous deletion of CDKN2A/B and/or phosphatase and tensin homolog (PTEN) (Fig. 1F). Mutation analysis of the known glioma genes was precluded due to lack of sufficient DNA.
To ensure that the FG cases with tumor MIP data were an accurate representation of all the probands in the Gliogene study, sex, age at glioma diagnosis, glioma histology, and glioma grade of the FG cases with (n = 75) and without (n = 300) MIP array data were compared.
Statistical Analysis
Differences between molecular subtypes were evaluated using Fisher’s exact test for categorical values and t-tests for continuous variables. The kappa statistic was used to assess concordance of molecular subtypes within families.
Results
Study Participant Demographics
The distribution of demographics from FG cases in this study with MIP array data available (n = 75) is shown in Supplementary Table S1. The mean age at diagnosis was 49, and the FG cases were 56% male (n = 42). Histological information was available for 97% of 75 FG cases (n = 73); the majority were glioblastomas (GBMs) (n = 44, 59%), followed by astrocytomas (n = 12, 16%), oligodendrogliomas (n = 9, 12%), and oligoastrocytomas (n = 8, 11%). Grade was available for 74 patients; grade IV was the most prevalent (n = 44, 59%), followed by grade III (n = 18, 24%) and grade II (n = 12, 16%). One individual had no grade specified (1%).
The subset of FG cases with tumor MIP data was similar to that of the whole population of FG (Supplementary Table S1). No statistically significant difference in age at diagnosis, sex, histological subtype, or grade (P = 0.87, 0.75, 0.54, and 0.34, respectively) was observed when the FG cases with and without MIP array data were compared.
The distribution of demographic factors in the FG cases was compared with a cohort of 148 Mayo Clinic sporadic adult diffuse gliomas, shown in Supplementary Table S2. The mean age was 49, and 57% were male (n = 84). The distribution of histology was GBM (n = 53, 36%), oligoastrocytoma (n = 40, 27%), astrocytoma (n = 34, 23%), and oligodendroglioma (n = 21, 14%). There was a statistical difference in histological subtype designation and grade (P = 0.001 and 0.001, respectively) between the FG cases in this study compared with the sporadic cohort. There were more GBMs and fewer oligoastrocytomas among the FG cases.
Prevalence of Molecular Subtypes in FG Cases
The newer-generation Affymetrix OncoScan V1 array data from the sporadic cases (Fig. 1A, C, E) were compared with the first-generation Affymetrix MIP data from the Gliogene families (Fig. 1B, D, F).
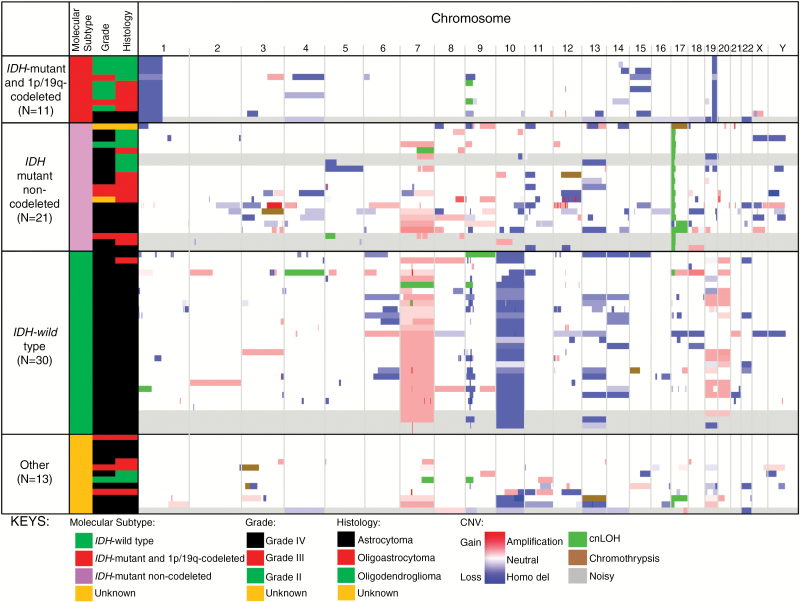
Out of 75 FG cases, 62 (83%) could be classified into one of the molecular subtypes: 30 (48%) IDH-wildtype, 21 (34%) IDH-mutant non-codeleted, and 11 (18%) IDH-mutant and 1p/19q codeleted gliomas. In the absence of additional mutation data, the remaining 13 (21%) cases could not be classified into molecular subtypes (Fig. 2 and Supplementary Tables S2 and S3).
Fig. 2.
The prevalence of glioma molecular subtypes in FG cases is similar to the Mayo Clinic sporadic control population. Integrated Genomic Viewer was used to illustrate the copy number alterations, chromothrypsis, and cnLOH observed in these gliomas. The cases are sorted based on molecular subtypes (horizontal columns) and further categorized vertically by grade (II, III, or IV) and histology (astrocytoma, oligoastrocytoma, oligodendroglioma). On the right, intensity of red or blue corresponds to degree of gain or loss, respectively, in copy number. Green indicates cnLOH, and brown indicates chromothrypsis. Samples labeled gray contained arrays that had exceptional background noise or were difficult to interpret. Vertical columns represent chromosome number (listed at the top). Size of color bands corresponds to size of chromosome alteration.
Out of 148 sporadic glioma patients, 64 (43%) were IDH-wildtype, 40 (27%) were IDH-mutant non-codeleted, and 34 (23%) were IDH-mutant and 1p/19q codeleted. Ten (7%) were classified into the smaller subtypes (TERT and IDH-mutant and triple-negative) defined by Eckel-Passow et al,13 or were unclassifiable (Supplementary Table S2).
The molecular distribution of the FG cases was not statistically different than the sporadic population (P = 0.54) (Supplementary Table S2).
Concordance of Molecular Subtypes within Paired Families
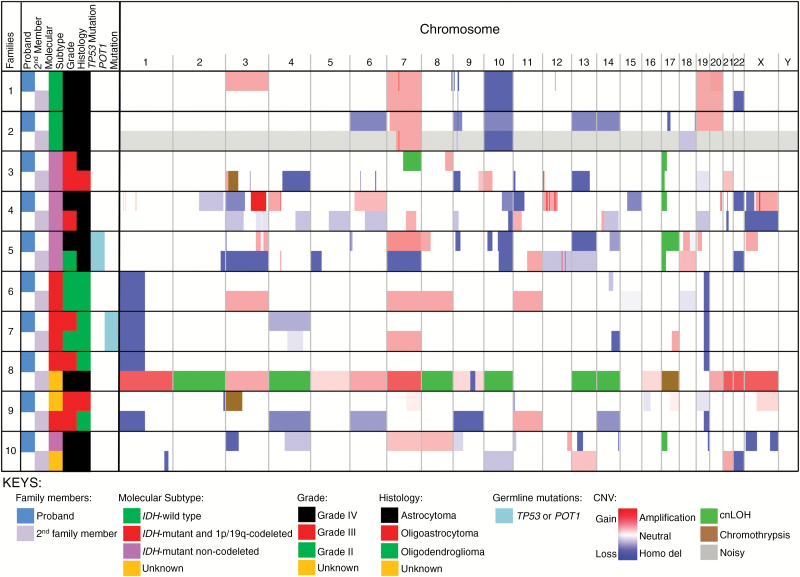
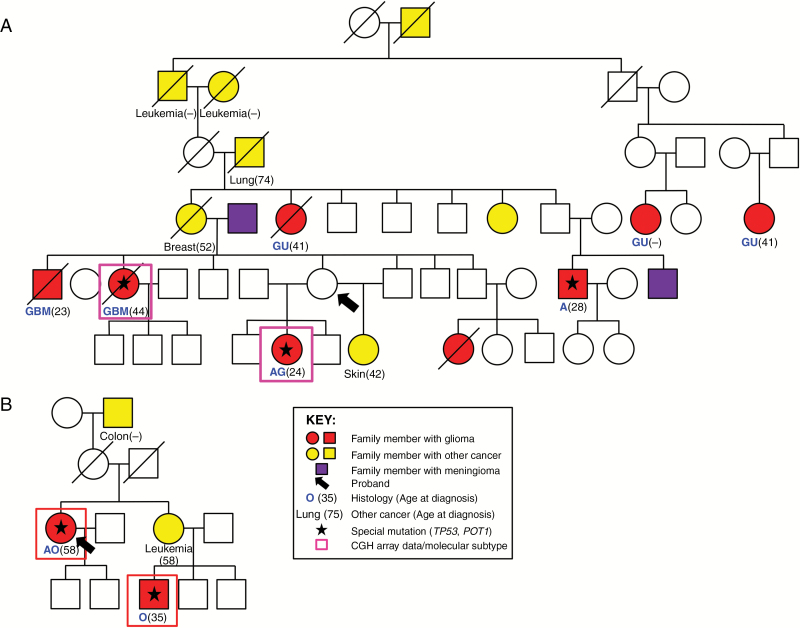
Within the dataset there were 10 families with 2 glioma cases; 7 (70%) were concordant based on molecular subtype: 3 were IDH-mutant non-codeleted, 2 IDH-wildtype, and 2 IDH-mutant and 1p/19q codeleted (Fig. 3 and Supplementary Table S4). The remaining 3 families were discordant for molecular subtypes. Seven of the 10 families were also concordant for histologic subtype. In family 5, two individuals had TP53 germline mutations, and both developed IDH-mutant non-codeleted tumors (Fig. 4A). In family 7, two individuals with IDH-mutant and 1p/19q codeleted tumors also had POT1 germline mutations (Fig. 4B). The kappa statistic of these families was 0.59.
Fig. 3.
Seven of the 10 paired gliomas from families with 2 available tumors show concordance for molecular subtype. Left: family attributes, including designation as proband or second family member, molecular subtype, grade, histology, or the presence or absence of germline mutations (see keys). Right: Integrated Genomic Viewer figure illustrating CNVs (with gray indicating samples that were noisy and/or difficult to interpret). Every 2 horizontal rows represent one family, with the top being the proband and the bottom being the second family member. Intensity of red or blue color corresponds to degree of gain or loss, respectively, of copy number, while green depicts cnLOH, and brown depicts chromothrypsis. Vertical columns represent chromosome number (listed at the top). Size of color bands corresponds to size of chromosome alteration.
Fig. 4.
Pedigrees of families 5 and 7 with concordant molecular subtypes and germline mutations of individuals with comparative genomic hybridization (CGH) array data. (A) Glioma family 5 with 2 individuals concordant for IDH-mutant non-codeleted gliomas and germline TP53 mutations. (B) Glioma family 7 with concordant IDH-mutant and 1p/19q codeleted gliomas and germline POT1 mutations. Individuals with glioma are filled in red, with meningioma filled in purple, and with other cancers filled in yellow (cancer and age of diagnosis in parentheses if available). Deceased individuals are indicated with a slash. Age at diagnosis is listed in parentheses. Red or pink boxes indicate individuals with CGH array data for analysis in this study. Lastly, glioma type is indicated in blue text with age of diagnosis in parentheses. Histology: AA, anaplastic astrocytoma; AG, astrocytoma, gemistocytic; AO, anaplastic oligodendroglioma; AU, astrocytoma unclassified; GG, ganglioglioma; GBM, glioblastoma multiforme; GNOS, glioma not otherwise specified; MG, malignant glioma; MOA, mixed oligoastrocytoma; O, oligodendroglioma.
Discussion
Gliomas are the most common primary CNS tumors in adults. Approximately 5%–10% of gliomas are aggregated in families for which there is still no clear etiology.5,6,15 Of that 5%–10%, a small percentage carry mutations in known syndromic genes and are more rigidly defined due to the high penetrance of the underlying gene mutations.8,9 The biology for the remaining families is still not well understood. The Gliogene Consortium enrolled patients and pedigrees that had a family history of gliomas (2 or more gliomas in the family). Some of these patients were in syndromic families, such as TP53 and POT1 families described here and elsewhere. However, most pedigrees showed no evidence of having a mutation in a known familial gene.10,16,17 It is unknown if the remaining families developed multiple gliomas because of random tumor co-occurrence or because of multifactorial inheritance.
Characteristic molecular alterations have been identified in glioma. These alterations, particularly 1p/19q codeletion, TERT promoter mutation, and IDH1/2 mutation, served as the basis for a molecular classification system developed by Eckel-Passow et al, which has better predicted age of onset and survival compared with histological subtyping in sporadic gliomas.12,13 The TERT-mutant only, IDH-mutant only, and TERT-mutant–IDH-mutant codeleted (triple-positive) subtypes correspond closely with the WHO IDH-wildtype, IDH-mutant non-codeleted, and IDH-mutant codeleted subtypes, respectively. Due to the strong concordance between the 2 classification methods, the new WHO nomenclature was used for this paper.
Single nucleotide polymorphisms (SNPs) have been identified to play a role in predisposition to glioma. Because of the significant odds ratio (OR) and allele frequencies of SNPs identified through a glioma genome-wide association study,18 we hypothesized that the prevalence of one (or more) molecular subtypes might be greater in FGs, since a genetic predisposition should increase the prevalence of specific gliomas. Therefore, we sought to investigate whether the prevalence of the molecular subtypes differed between sporadic and familial populations. However, when analyzing probands from sporadic and familial gliomas, there was no statistically significant difference in the prevalence of the molecular subtypes (Fig. 2). This lack of difference may be due to a sample size that was not large enough to achieve significance (eg, small differences in prevalence will be challenging to detect due to constraints in collecting sufficient probands). Alternatively, there are at least 2 other possible explanations for this observation.
First, it may be that all of the gliomas included in this study were not familial. The Gliogene Consortium defined glioma families as those with 2 or more members with glioma. Approximately 83% of the families included in Gliogene were observed to have only 2 gliomas per family.15 Thus, some of the families in Gliogene may represent co-occurrence of sporadic gliomas. Previous linkage studies performed by Gliogene and others also attest to this, as some families were found to be linked, but the majority showed no evidence of linkage.16,19,20 This would indicate that some families may not be truly representative of nonsyndromic familial heritability and rather suggest that these gliomas were aggregated in a family by chance.
Second, it is possible that a large proportion of apparently sporadic gliomas actually have a polygenic, low-penetrance, nonsyndromic genetic predisposition. If this is true, it would be predicted that the prevalence of molecular subtypes might be similar between familial and sporadic glioma. Similar observations have been made in other cancers. For example, a study on non-BRCA1/2 breast cancer families revealed that the prevalence of molecular groups was similar to sporadic breast cancer.21
The breast cancer studies suggest that the distribution of nonsyndromic familial and sporadic breast cancer is likely caused by co-occurrence of multiple low-penetrance alleles.21 The same is likely true for glioma. The Glioma International Case-Control Consortium (GICC) recently observed that 26 different glioma loci collectively account for inherited risk in approximately 27% of GBM and 34% of non-GBM gliomas, respectively.22 Furthermore, previous FG studies also suggest the likelihood of multifactorial genetic heritability.6,23–25 Taken together, the data suggest a polygenic contribution to the development of a large proportion of gliomas, and those glioma patients who aggregate in families may carry certain genetic factors that are more penetrant than others. For example, ~40% of individuals with IDH-mutant gliomas carry at least one copy of the 8q24 SNP, rs55705857.13,18 Therefore, assessing polygenic risk in glioma family members in the future could further elucidate the contributions that low-penetrance factors play in familial heritability.
For the paired specimen analysis, 7 of the 10 families demonstrated concordance of molecular subtypes (Fig. 3 and Supplementary Table S4). The 3 families that were discordant contained an individual with an unknown molecular subtype. In families 8 and 9, the unknown samples lack 1p/19q codeletion, so the families must be discordant. Despite these 3 discordant families, the kappa statistic revealed moderate to strong concordance of molecular subtype in families rather than simple occurrence by chance. This provides more insight into the similar underlying genetic predisposition of FG and the significance of subtyping tumors according to these molecular features.
Two of the paired families with concordant tumors were likely syndromic in that they carried germline mutations in TP53 and POT1,17,26,27 and concordant tumors might be expected for such families (Fig. 4). Importantly, the molecular subtype was concordant in most of the remaining families. This provides evidence for the heritability of genetic factors that correlate to a specific molecular subtype.
There were some limitations to this study. TERT and IDH mutation status could not be measured in these tumors. Despite this, characteristic CNVs were observed and compared with previously classified cohorts with mutation data (D. Brown et al, unpublished manuscript). Since TERT, IDH, and 1p/19q codeletion are mutations that occur early in tumor development,28 and subsequent copy number alterations are characteristic of the presence or absence of their initial driver mutations, subtypes can be inferred with some accuracy without initial mutation data. However, it should be noted that subtyping by CNVs alone, in the absence of mutation data, is less accurate than mutation data combined with copy number data. Also, there was limited array data from most of the multiplex families, preventing additional comparisons of molecular subtypes within these families. Since tumor collection was not a part of the Gliogene study, the samples included in this study were not systematically collected and not available for all participants; many were from deceased family members as opposed to the proband. Future studies will determine mutation status in critical genes such as TERT, IDH, and POT1 and will genotype individuals and families for critical SNPs in glioma risk loci.
Overall, this study provides evidence that the prevalence of glioma molecular subtypes is similar in FG and sporadic glioma. In addition, gliomas of similar molecular subtype co-occur in families with more than one glioma. Whether these observations are due to stochastic co-occurrence of gliomas in relatives or are evidence for a low-penetrance heritable predisposition of specific molecular glioma subtypes should be a subject of further research. Future studies should attempt to uncover additional genetic alterations and variants that contribute to the predisposition to glioma, and should provide further insight into the biological mechanisms underlying sporadic and familial gliomas.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by grants from the US National Institutes of Health, R25GM075148 (V.Y.R), R01CA139020 (M.L.B. and B.S.M.), R01CA52689 (M.L.B.), P50CA108961 (R.B.J.), and RC1NS068222Z (R.B.J.); the Bernie and Edith Waterman Foundation (R.B.J.); the Ting Tsung and Wei Fong Chao Family Foundation (R.B.J.); the National Brain Tumor Society (M.L.B.); and the American Brain Tumor Association (M.L.B.).
Supplementary Material
Acknowledgments
We acknowledge the contributions of the overall brain tumor research programs that support the Gliogene effort. The list of contributors can be found at: www.gliogene.org.
We would like to thank the patients for participating in this research.
Conflict of interest statement. None declared.
Unpublished material: D. Brown et al., unpublished manuscript
References
- 1. Ostrom Q, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015; 17(Suppl 4): iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bondy ML, Scheurer ME, Malmer B, et al. ; Brain Tumor Epidemiology Consortium Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113(7 Suppl):1953–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Ohgaki H, Wiester OO, et al.(eds) WHO Classification of Tumors of the Central Nervous System. 2016; Lyon: IARC. [Google Scholar]
- 4. Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38(1):E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wrensch M, Lee M, Miike R, et al. Familial and personal medical history of cancer and nervous system conditions among adults with glioma and controls. Am J Epidemiol. 1997;145(7):581–593. [DOI] [PubMed] [Google Scholar]
- 6. Malmer B, Grönberg H, Bergenheim AT, Lenner P, Henriksson R. Familial aggregation of astrocytoma in northern Sweden: an epidemiological cohort study. Int J Cancer. 1999;81(3):366–370. [DOI] [PubMed] [Google Scholar]
- 7. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malmer B, Adatto P, Armstrong G, et al. GLIOGENE: an international consortium to understand familial glioma. Cancer Epidemiol Biomarkers Prev. 2007;16(9):1730–1734. [DOI] [PubMed] [Google Scholar]
- 9. Malmer B, Iselius L, Holmberg E, Collins A, Henriksson R, Grönberg H. Genetic epidemiology of glioma. Br J Cancer. 2001;84(3):429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bainbridge MN, Armstrong GN, Gramatges MM, et al. ; Gliogene Consortium Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107(1):384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hemminki K, Tretli S, Sundquist J, Johannesen TB, Granström C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009;10(5):481–488. [DOI] [PubMed] [Google Scholar]
- 12. Brat DJ, Verhaak RG, Aldape KD, et al. ; Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. NEJM. 2015; 372(26): 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson PA, Brewster AM, Kim-Anh D, et al. Selective genomic copy number imbalances and probability of recurrence in early-stage breast cancer. PLoS One. 2011;6(8):e23543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sadetzki S, Bruchim R, Oberman B, et al. ; Gliogene Consortium Description of selected characteristics of familial glioma patients—results from the Gliogene Consortium. Eur J Cancer. 2013;49(6):1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shete S, Lau CC, Houlston RS, et al. ; Gliogene Consortium Genome-wide high-density SNP linkage search for glioma susceptibility loci: results from the Gliogene Consortium. Cancer Res. 2011;71(24):7568–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robertson LB, Armstrong GN, Olver BD, et al. Survey of familial glioma and role of germline p16INK4A/p14ARF and p53 mutation. Fam Cancer. 2010;9(3):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44(10):1122–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malmer B, Haraldsson S, Einarsdottir E, Lindgren P, Holmberg D. Homozygosity mapping of familial glioma in Northern Sweden. Acta Oncol. 2005;44(2):114–119. [DOI] [PubMed] [Google Scholar]
- 20. Paunu N, Lahermo P, Onkamo P, et al. A novel low-penetrance locus for familial glioma at 15q23-q26.3. Cancer Res. 2002;62(13):3798–3802. [PubMed] [Google Scholar]
- 21. Larsen MJ, Thomassen M, Tan Q, et al. RNA profiling reveals familial aggregation of molecular subtypes in non-BRCA1/2 breast cancer families. BMC Med Genomics. 2014; 9(7): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; GliomaScan Consortium Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu J, Burnett MG, Shpak M. A comparative study of the molecular characteristics of familial gliomas and other cancers. Cancer Genomics Proteomics. 2016;13(6):467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Neill BP, Blondal H, Yang P, et al. Risk of cancer among relatives of patients with glioma. Cancer Epidemiol Biomarkers Prev. 2002;11(9):921–924. [PubMed] [Google Scholar]
- 25. de Andrade M, Barnholtz JS, Amos CI, Adatto P, Spencer C, Bondy ML. Segregation analysis of cancer in families of glioma patients. Genet Epidemiol. 2001;20(2):258–270. [DOI] [PubMed] [Google Scholar]
- 26. Paunu N, Syrjäkoski K, Sankila R, et al. Analysis of p53 tumor suppressor gene in families with multiple glioma patients. J Neurooncol. 2001;55(3):159–165. [DOI] [PubMed] [Google Scholar]
- 27. Malmer B, Grönberg H, Andersson U, Jonsson BA, Henriksson R. Microsatellite instability, PTEN and p53 germline mutations in glioma families. Acta Oncol. 2001;40(5):633–637. [DOI] [PubMed] [Google Scholar]
- 28. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.