Abstract
Specific manipulation of RNA is necessary for the research in biotechnology and medicine. The RNA-binding domains of Pumilio/fem-3 mRNA binding factors (PUF domains) are programmable RNA binding scaffolds used to engineer artificial proteins that specifically modulate RNAs. However, the native PUF domains generally recognize 8-nt RNAs, limiting their applications. Here, we modify the PUF domain of human Pumilio1 to engineer PUFs that recognize RNA targets of different length. The engineered PUFs bind to their RNA targets specifically and PUFs with more repeats have higher binding affinity than the canonical eight-repeat domains; however, the binding affinity reaches the peak at those with 9 and 10 repeats. Structural analysis on PUF with nine repeats reveals a higher degree of curvature, and the RNA binding unexpectedly and dramatically opens the curved structure. Investigation of the residues positioned in between two RNA bases demonstrates that tyrosine and arginine have favored stacking interactions. Further tests on the availability of the engineered PUFs in vitro and in splicing function assays indicate that our engineered PUFs bind RNA targets with high affinity in a programmable way.
INTRODUCTION
The post-transcriptional processing and modification of RNAs are essential steps in gene expression, which in turn control many biological processes such as cell differentiation and developmental programs (1,2). Therefore, specific manipulation of RNAs has broad applications in biotechnology and medicine. The RNA-binding proteins play central roles in regulating major steps of RNA processing (1–5), thus manipulation of RNAs with artificially designed RNA-binding proteins presents a unique opportunity to modulate gene expression and cellular function (6–11). A modular design principle for such engineered proteins is combining an RNA-binding scaffold to recognize targets with a functional domain to affect RNA metabolism, as well as a short linker between these two modules (12). The key of this design is to construct RNA-binding scaffolds with programmable sequence specificity, enabling specific recognition of given RNAs with limited off-target effect.
The Drosophila melanogaster Pumilio and Caenorhabditis elegans fem-3 mRNA binding factors (PUF) are sequence-specific RNA binding proteins that recognize mRNA targets with a repetitive scaffold (3–5,13,14). A typical PUF has a C-terminal RNA-binding domain (a.k.a. PUF domain) that comprises eight tandem repeats plus N- and C-terminal flanking regions. The eight PUF repeats arrange along an arch, with each repeat composed of three α-helices (15–19). The RNA binds to the concave surface of the PUF domain in an anti-parallel orientation, with each base recognized by a single repeat (18–22). The second helix of each repeat contains a conserved 5-residue recognition code, designed as ‘12XX5’ (X represents a random residue), where the residue at position 2 stacks with the cognate base and the residues at positions 1 and 5 directly interact with RNA bases through polar interactions (18–20). The stacking residue in each PUF repeat plays a significant role in the binding affinity, whereas the two polar or charged residues determine the specificity of PUF repeats (18–20,23–25).
The unique structure and relative short length of PUF domain make it an ideal RNA-binding scaffold for artificial proteins to modulate RNA processing (6–12,23,25–28). Varying functional domains, several types of artificial PUF factors have been successfully developed with distinct applications (6–11), and the existing and proposed artificial PUF factors will serve as versatile tools in both biomedical research and novel therapies for RNA related diseases (12,26–28). Despite early success, there are several limitations in using customized PUF domain as general programmable RNA-binding scaffolds. One major limitation is that the native PUF proteins contain eight repeats that generally recognize 8-nt RNA sequences. However, in certain cases PUFs recognizing different target length may be required. For example, when recognizing a longer RNA within the entire transcriptome, PUFs with more repeats will minimize the off-target effect. Conversely, a shorter recognition site is more practical for some in vitro applications in order to bind an RNA substrate at multiple sites.
Here we engineered PUFs that target RNAs of different length by modifying the PUF domain of human Pumilio1 (PUF-8R) (17). The RNA-binding affinities of the engineered PUFs with additional repeats are stronger than that of the canonical eight-repeat domain and reach the peak at those with nine or ten repeats. The six-repeat PUF presents weaker binding to its cognate RNA than other engineered PUFs, yet it still has an acceptable affinity. Structural insights of the nine-repeat PUF (PUF-9R) and in complex with its cognate RNA reveal that PUF-9R recognizes the RNA in a modular fashion, whereas PUF-9R presents a more curved architecture than PUF-8R and RNA binding flattens the PUF-9R curvature. Investigation of the residues stacking with RNA bases in PUF-8R shows that tyrosine and arginine have favored stacking interactions. At last, we used these engineered PUFs to study their sequence discrimination and splicing functions. Our results indicate that varying the number of repeats in engineered PUFs will be very useful in manipulating RNA processing with low off-target effect.
MATERIALS AND METHODS
The yeast three-hybrid system
The wild-type PUF domain of human Pumilio1 contains eight tandem repeats (PUF-8R) (17). PUF with 16 repeats (PUF-16R) was obtained from Dr Oliver Rackham from the University of Western Australia (24). Based on PUF-16R, the PUFs with 6, 9, 10, 12 repeats were made by overlapped polymerase chain reaction (PCR) with specific primers (Supplementary Table S1). For yeast expression plasmids, the purified PCR products were digested with BamHI and XhoI sites and inserted into pACT2 plasmids. RNA expression plasmids were made by annealing DNA oligonucleotides and sub-cloning into the pIIIA-MS2-2 plasmids using specific primers with SmaI and SphI sites (Supplementary Table S2).
To analyze PUFs and target RNAs interactions, the yeast three-hybrid assays were performed in YBZ-1 yeast strain as described previously (29,30). Plasmids expressing target RNAs were co-transformed with plasmids expressing PUFs into YBZ1 by standard yeast transformation methods. Transformants were plated on selective synthetic defined (SD) media containing 10 mM 3-aminotriazole and lacking uracil, leucine and histidine. The survived colonies were picked and patched onto fresh plates.
The liquid β-Galactosidase assay
To measure the activity of β-Galactosidase, the yeast colonies were randomly chosen and inoculated into individual wells of a 96-well plate. After overnight incubation at 30°C with shaking to reach mid-log phase, the culture density of each well was determined by reading OD650. A total of 20 μl of yeast culture were transferred into a new plate containing 180 μl of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 1 mM MgCl2, 0.2% (w/v) Sarkosyl, and 0.4 mg/ml O-nitrophenol-β-D-galactopyranoside), followed by 2 h incubation at 37°C. Then 80 μl of 1 mol/l carbonate solution was added into each well to stop the reaction. The A405 was measured to quantify the yellow-colored product (nitrophenol), and the β-galactosidase units was determined as the A405 difference between the sample and the background calibrated by culture densities.
Recombinant protein preparation
PUF proteins with 6, 8, 9, 10 and 12 repeats, as well as mutants of PUF-9R and PUF-8R, were sub-cloned into an engineered pET-Duet-1 vector with a precision protease-cleavable site following the His6 tag. PUF with 16 repeats was sub-cloned into an engineered pGEX-4T-1 vector with a precision protease-cleavable site following the glutathione S-transferase (GST) tag. The DNA sequences were amplified by PCR with specific primers (Supplementary Table S3) using NdeI and XhoI sites. Mutants of PUF-8R and PUF-9R were generated by overlap-PCR using primers listed in Supplementary Table S3.
Plasmids were transformed into BL21 (DE3) strain. The cells were grown in Luria-Bertani (LB) medium and induced with 0.2 mM isopropyl β-D-thiogalactoside (IPTG) for 14 h at 20°C. Cells were harvested by centrifugation and lysed by sonication in buffer containing 50 mM Tris (pH 8.5), 200 mM NaCl, 1 mM phenyl-methane-sulfonyl fluoride (PMSF), 0.5 mg/ml lysozyme, 0.01 mg/ml DNase I and 0.01 M MgCl2. PUF proteins were purified using Ni-NTA columns (Qiagen) or Glutathione-Sepharose 4B columns (GE Healthcare) and eluted with 50 mM Tris (pH 8.5), 250 mM Imidazole or 50 mM Tris (pH8.5), 1.54 g/l reduced glutathione (GSH). Further purification went on with the Source Q column (GE Healthcare) in 50 mM Tris (pH 8.5) and eluted with a gradient of 0–1 M NaCl. Fractions containing the fusion proteins were combined. For PUF-16R, the GST tag was cleaved by precision protease and removed through Glutathione-Sepharose 4B columns. The proteins were concentrated by ultracentrifuge and finally subjected to a Superdex 200 gel filtration column (GE Healthcare) equilibrated with 10 mM Tris (pH 8.0), 150 mM NaCl and 2 mM DL-dithiothreitol (DTT). All proteins were stored at −80°C and the purity was ≥95% as determined by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
The fluorescence polarization assay
The fluorescence polarization (FP) assays were performed as described previously (31), with a few modifications. The 5′-(6-FAM)-labeled RNAs were synthesized by TAKARA and the sequences were listed in Supplementary Table S4. Each reaction sample (total volume of 200 μl) consisted of 5 nM RNA and increasing concentrations of proteins from 0.1 nM to 1 μM in a binding buffer containing 20 mM Tris–HCl, pH 7.5, 50 mM KCl, 0.1 mg/ml bovine serum albumin and 0.5 mM ethylene-diamine-tetraacetic acid (EDTA). The samples were equilibrated at room temperature for 30 min. The FP (mP) values were measured using the FP system on an EnVision Multi-label Plate Reader (Perkin Elmer), referenced against a blank buffer at the beginning of each test. The fraction of RNA bound was calculated by first subtracting the polarization value with no protein added and then dividing by the range of the data for an indicated RNA series. The data were fitted to the quadratic equation by nonlinear least-squares regression using Origin 8 (Origin Lab):
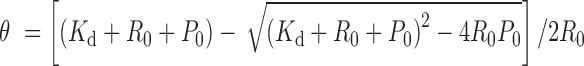 |
where θ is the fraction of RNA bound, R0 is the total concentration of RNA, P0 is the protein concentration and Kd is the dissociation constant.
The alternative splicing assay
The expression constructs of engineered splicing factors (ESFs) were generated as described previously (9). Briefly, all PUFs were sub-cloned into the engineered pGL (pCI-Neo) or pGL-SRSF7 (including residues 123–238 of SRSF7, NP001026854) vectors by PCR with specific primers (Supplementary Table S5) using XbaI and NotI sites. To test the effects of ESFs on exon inclusion, RNA target sequences of indicated PUFs were inserted into the digested reporter pGZ3 by synthesizing and annealing oligonucleotides flanked by XhoI and ApaI sites using specific primers (Supplementary Table S5). The splicing reporters and ESFs were transfected into HEK-293T cells by Lipofectamine 2000 (Invitrogen) following manufacturer’s directions with a ratio of 1:2, unless indicated. Cells were grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and harvested 24 h after transfection.
The purification of total RNA and semi-quantitative RT-PCR were carried out as described previously (9). Briefly, total RNA was purified from transfected HEK-293T cells with TRIzol reagent (Invitrogen) following manufacturer's instruction, followed by 1 h RQ1 RNase-Free DNase (Promega) treatment at 37°C and then heat-inactivation of DNase. Then 2 μg total RNA was reverse transcribed with SuperScript III (Invitrogen) using random primer, and one-tenth of the RT product was used as the template for PCR amplification (25 cycles of amplification, with a trace amount of Cy5-dCTP as label). The resulting gels were scanned with a Typhoon 9400 Imager (GE Healthcare) and quantified with Image Quant 5.2 software (GE Healthcare). The primers used were listed in Supplementary Table S5.
The expression of ESFs was confirmed by western blotting analysis. The total cell pellets were lysed in 1 × SDS-PAGE loading buffer and heated at 95°C for 10 min. Then the mixtures were separated by 4–20% SDS-PAGE Gel (Genscript) and transferred to poly-vinylidene fluoride (PVDF) membrane. The following primary antibodies were used: anti-Flag antibody (F1084, Sigma-Aldrich) and anti-GAPDH antibody (sc-25778, Santa Cruz Biotechnology). Second antibodies were purchased from Santa Cruz Biotechnology (sc-2004, goat anti-rabbit IgG-HRP) and Cell Signaling Technology (#7076, anti-mouse IgG, HRP-linked Antibody). ECL kit (GE Healthcare) was used to visualize the bound second antibodies.
Crystallization, structure determination and refinement
All crystals were grown by vapor diffusion in hanging drops at 20°C. Crystals of the PUF-9R alone (apo-PUF-9R) were grown by mixing the protein (∼6 mg/ml) with an equal volume of reservoir solution containing 20% polyethylene glycol (PEG) 3350 and 0.25 M NaH2PO4. The cognate RNA for PUF-9R (RNA-9nt, 5′-UGUUGUAUA-3′, synthesized by TAKARA) was incubated at 70°C for 5 min followed by 5 min on ice, then the PUF-9R:RNA-9nt complex was prepared by mixing protein and RNA at a molar ratio of 1:1.2 at room temperature for 1 h. Crystals of PUF-9R:RNA-9 nt complex were grown in a reservoir buffer containing 0.1 M BIS-TRIS Propane, pH 8.8, 20% PEG MME 550. All Crystals were equilibrated in a cryoprotectant buffer containing the indicated reservoir solution supplemented with 25% ethylene glycol before flash frozen in liquid nitrogen.
The diffraction data sets were collected at beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) and processed with the HKL2000 (32). The structure of apo-PUF-9R was solved by molecular replacement with the program Phaser_MR from CCP4i suite, using residues from 828–959 and 1032–1146 from the PUF domain of human Pumilio1 (PDB code: 1M8Z) (17) as the search models (33). The PUF-9R:RNA-9nt complex structure was solved by molecular replacement using apo-PUF-9R as the search model. The unmodeled regions were manually built in Coot (34), and the structures were further refined with Coot and Phenix package (35). The data processing and refinement statistics were summarized in Table 1. The structure validation was carried out using Mol-Probity (36). All structural representations were prepared with PyMOL (http://www.pymol.org).
Table 1. Data collection and refinement statistics.
| apo-PUF-9R | PUF-9R:RNA-9nt | |
|---|---|---|
| Data collection a | ||
| Space group | C2 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 203.05, 32.94, 61.36 | 68.05, 70.32, 98.00 |
| α, β, γ (°) | 90, 105.31, 90 | 90, 90, 90 |
| Resolution (Å) | 50.00–2.45 (2.49–2.45)b | 50–2.25 (2. 29–2.25) |
| R merge (%) | 10.6 (46.1) | 12.0 (50.1) |
| I/σ(I) | 14.9 (8.2) | 17.6 (2.7) |
| Completeness (%) | 98.5 (100) | 99.6 (96.7) |
| Redundancy | 6.9 (7.3) | 9.4 (7.5) |
| Refinement | ||
| Resolution (Å) | 33.62–2.46 (2.55–2.46) | 49.00–2.25 (2.33–2.25) |
| No. of reflections | 14 491 | 22 895 |
| R work/Rfreec | 0.213/0.258 | 0.197/0.236 |
| No. of atoms | ||
| Protein | 3113 | 3114 |
| RNA | 187 | |
| Water | 71 | 144 |
| Average B factor (Å2) | 46.5 | 41.1 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.005 |
| Bond angles (°) | 0.435 | 0.647 |
| Ramachandran Plot | ||
| Favored (%) | 100 | 98.2 |
| Allowed (%) | 0 | 1.8 |
| Disallowed (%) | 0 | 0 |
aEach dataset was collected from a single crystal.
bNumbers in parentheses are for the highest resolution shell.
c R free was calculated by using 4.93 and 5.13% of random data omitted from the refinement of apo-PUF-9R and PUF-9R-RNA, respectively.
RESULTS
Design of PUFs recognizing RNA targets of different length
To generate RNA-binding domains that specifically recognize RNA targets of different length, we engineered several PUFs with 6, 9, 10 and 12 repeats (hereinafter referred to as PUF-6R, PUF-9R and so on) by inserting or deleting different numbers of PUF repeats between the indicated repeats of human Pumilio1 PUF domain, a strategy similar to what was previously used in generating PUF with 16-repeat (24) (Figure 1A and Supplementary Table S1). The engineered PUFs would in principle recognize the cognate RNA targets containing 6, 9, 10, 12 and 16 bases, respectively (hereinafter referred to as RNA-6nt, RNA-9nt and so on) (Figure 1B). The binding of these PUFs to their cognate and off-target RNAs was measured using a yeast three-hybrid system (29,30), and the relative binding activities compared to the wild-type PUF-8R:RNA-8nt pair were plotted (Figure 1C and D). Indeed, all the modified PUFs with different number of repeats specifically bound to their cognate targets as compared to the control RNAs containing unmatched bases (Supplementary Table S2). Unexpectedly, the binding activities of the engineered PUFs peaked at those with nine and ten repeats, while further increase of repeat numbers did not improve the binding activity.
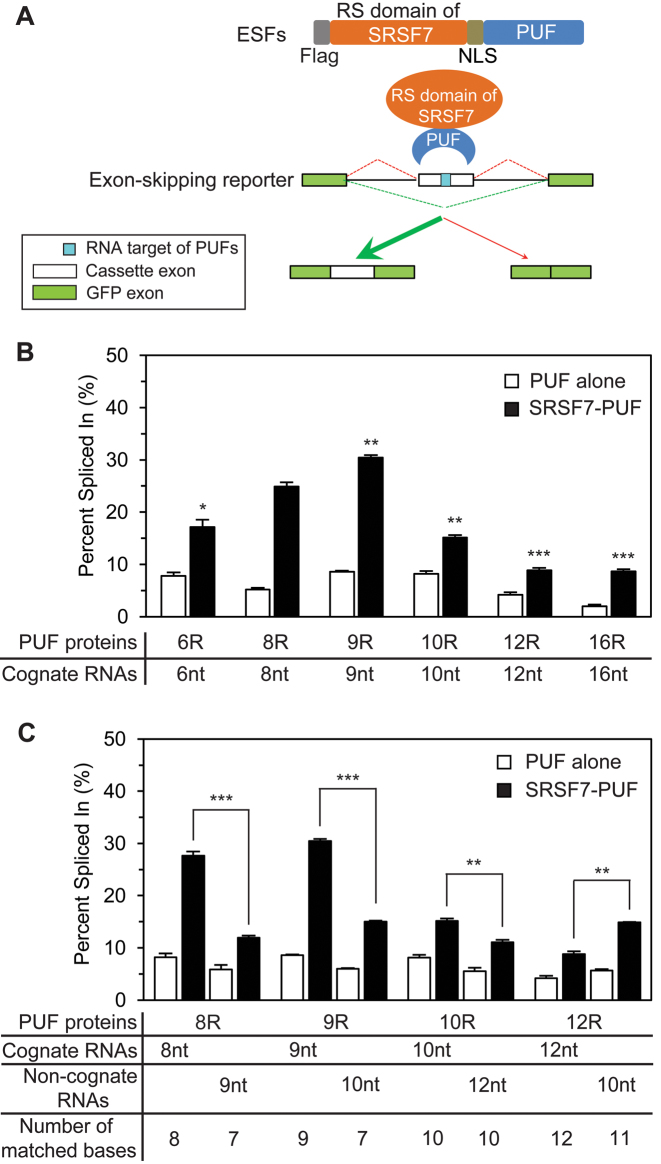
Figure 1.
The engineered PUFs recognize RNA targets of different length. (A) Schematic representation of the engineered PUFs with different number of repeats. The eight repeats in human Pumilio1 PUF domain (PUF-8R) are shown in rainbow color scheme, and numbers in parentheses are the original residue numbers for the native Pumilio1 PUF-8R. The color schemes of the PUF proteins are consistent throughout the schematic representations unless indicated. (B) The cognate RNA sequences of the engineered PUFs. Bases are colored the same as their cognate PUF repeats in panel A. (C) The yeast three-hybrid system used to measure PUF–RNA interactions. This system contains the PUF-Gal4-AD fusion protein, the PUF target RNA with an MS2-binding site, the MS2-LexA fusion protein and the reporter gene LacZ and HIS3. The expression of reporter gene would be triggered by the interaction between PUF and its cognate RNA. Gal4-AD, Gal-4 activation domain. LexA Op, LexA operon. (D) Measurement of binding activities of the engineered PUFs using the yeast-three-hybrid system. The β-gal activities relative to that of the PUF-8R:RNA-8nt pair were plotted (mean ± S.E.M., n > 3). (E and F) Binding affinities of the recombinant PUFs to various RNA targets determined by the FP assay (mean ± S.E.M., n ≥ 3). Statistical analysis was performed by a two-tailed Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., no significance).
To measure the PUF–RNA interaction in a more quantitative fashion, we purified the recombinant proteins of various engineered PUFs and determined the dissociation constants (Kd) to their cognate targets using the FP assay (31) (Supplementary Figure S1). The association constants Ka (Ka = 1/Kd) were plotted in Figure 1E. Consistent with the in vivo data, PUF-9R and PUF-10R bound to their cognate RNAs more tightly than the wild-type PUF-8R (∼12- and 7-fold, respectively). We also measured the association constants of some engineered PUFs with RNAs other than their cognate targets, with different numbers of the matched pairs between the RNA base and the PUF repeat (Figure 1F). Remarkably, the binding affinities of PUF-9R to different RNA targets dramatically decreased with decreasing matching bases. For PUF-8R, the binding affinity slightly decreased when recognizing the RNA-9nt that contains seven matched base; however, PUF-12R bound 2-fold more tightly to the non-cognate RNA containing 11 matching bases than to the fully matching RNA-12nt. More intriguingly, the binding affinities of PUF-10R decreased when the RNA targets containing the same matching bases but getting longer (RNA-10nt versus RNA-12nt). Furthermore, we constructed a fully experimental matrix of all engineered PUFs and varying RNA targets and determined the relative binding affinities (Supplementary Figure S2). When recognizing the longer RNA targets (≥9 nt), PUF-9R, PUF-10R and PUF-12R showed much more efficient binding compared with PUF-6R, PUF-8R and PUF-16R. The binding affinity reached the peak again at 9–10 matched bases, consistent with our results using yeast three-hybrid system. Notably, for each PUF protein, the binding affinities slightly decreased with longer RNA targets. Taken together, the yeast three-hybrid and in vitro FP data indicated that, in addition to the number of PUF repeats and matching bases, the length of RNA targets might affect the PUF–RNA affinity.
Modulating alternative splicing with engineered PUFs
An important application for PUF domain is to serve as a programmable RNA-binding scaffold in artificial factors to specifically manipulate RNA metabolism (6–12,26–28). Previously we generated ESFs by combining PUF domains with splicing modulation domains (9), and used ESFs to manipulate splicing of various endogenous genes (27,37). To expand the application of our engineered PUF domains, we created ESFs targeting RNAs of different length by fusing these engineered PUF domains with the arginine/serine-rich (RS) domain of SRSF7 splicing factor that promotes exon inclusion (9). We also included a nuclear localization sequence to direct the ESFs to the nucleus and a Flag tag to facilitate the detection of the ESFs (Figure 2A). By co-transfecting HEK-293T cells with plasmids expressing the ESFs and a splicing reporter containing the cognate RNA sequences in an alternatively spliced cassette exon, we tested the activities of the new ESFs. Alternative splicing levels were measured by RT-PCR using the total RNAs purified from the transfected cells as templates and primer pairs (Supplementary Table S5) targeting the GFP exons of the reporter (Supplementary Figure S3A). The expression of splicing factors was confirmed by western blots (Supplementary Figure S3A).
Figure 2.
Effects of ESFs with engineered PUFs on alternative splicing. (A) Schematic diagram of the alternative splicing assay. (B and C) Quantification of the cassette exon inclusion in the presence of indicated PUF and RNA pairs. The percent spliced in values were calculated as the percentage of the cassette exon-included isoform among all isoforms (mean ± S.E.M., n = 3). Statistical analysis was performed by a two-tailed Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
As designed, ESFs containing SRSF7 RS domain and different engineered PUFs (SRSF7-PUF) showed obviously enhanced inclusion of the cassette exon compared to those with the corresponding PUF domain alone, indicating our engineered PUF domains could be utilized to construct artificial factors that efficiently manipulate splicing (Figure 2B). Consistent with the measurement of binding affinity (Figure 1D and E), the ESF containing PUF-9R affected splicing more efficiently than that with the native PUF-8R, while increasing PUF repeat numbers beyond nine did not always enhance the splicing. Furthermore, the efficiency of the engineered ESFs to some non-cognate RNA targets was also consistent with their binding affinities (Figures 1F and 2C), indicating that the length of RNA targets may affect the splicing efficiency as well.
Interestingly, compared to the negative controls with the splicing reporters co-expressed with empty pGL vector (Supplementary Figure S3B and C), expression of PUF domains alone without addition of SRSF7 RS domain have small but diverse influences on inclusion of the alternative exon (Figure 2B). In particular, expression of most PUF domains alone (8R, 9R, 12R) suppressed the splicing of cognate mini-gene reporters compared with empty vector, whereas PUF-10R slightly promoted the splicing of its cognate reporter (comparing Figure 2B to Supplementary Figure S3B). This variable effects of PUF domains alone was observed before, probably because the binding of PUF at exons directly affects spliceosome assembly (9). Thus, we calibrated the splicing regulatory activity of SRSF7-PUF by comparing to the PUFs alone for a more consistent comparison.
We also noticed that while the activities of ESFs are generally correlated with the RNA binding affinities of their PUF domain, the PUF-10R seemed to be an exception because the ESFs containing PUF-10R showed a weaker splicing regulatory activity (Figure 2B) although it binds cognate RNA with a high affinity (Figure 1D and E). We speculate that such inconsistency may due to the relative configuration between the PUF domain and the splicing modulation domain (RS domain SRSF7). Since these two domains have to be in a right conformation to ensure maximal accessibility of PUF domain to RNA and RS domains to spliceosomal components at the same time, the PUF domains with more than 10 repeats to bind to longer RNA targets may reduce the ability of RS domain to interact with spliceosome, thus reducing its activity in regulating splicing.
Overall structure of PUF-9R presents the increased curvature
To unveil the molecular mechanism of the affinity change for PUFs with different number of repeats, we conducted crystallization trials for complexes of all engineered PUFs with their cognate targets and successfully generated diffractable crystals of PUF-9R in isolation and in complex with its cognate RNA-9nt. The structures of the apo-PUF-9R and the PUF-9R:RNA-9nt complex were determined to 2.4 Å and 2.2 Å, respectively (Table 1).
The PUF-9R protein is made up of three fragments of human PUF-8R: residues 828–955, 1028–1142 and 1028–1176 (17), which respectively corresponds to repeats R1′-R3, R4-R6 and R7-R9′ of the engineered PUF-9R (Supplementary Figure S4A). As expected, PUF-9R folds into nine canonical PUF repeats (R1-R9), flanked by two terminal repeats (R1′ and R9′) (Figure 3A). The nine central repeats each comprises three α-helices (α1–α3), while the terminal repeats each have one regular helix and one short 310 helix. The canonical PUF repeats are structurally similar and can be superimposed onto each other with an average root-mean-square deviation of 1.5 Å for 36 Cα atoms (Supplementary Figure S4B). The PUF repeats in this engineered PUF-9R stack together to form a curved right-handed super-helix, similar to the native PUF proteins (15–19,21,22,38–40). The α2 helices of the canonical PUF repeats are located on the inner concave side of the protein, and the α3 helices together with two 310 helices from the terminal repeats form the convex surface (Figure 3A).
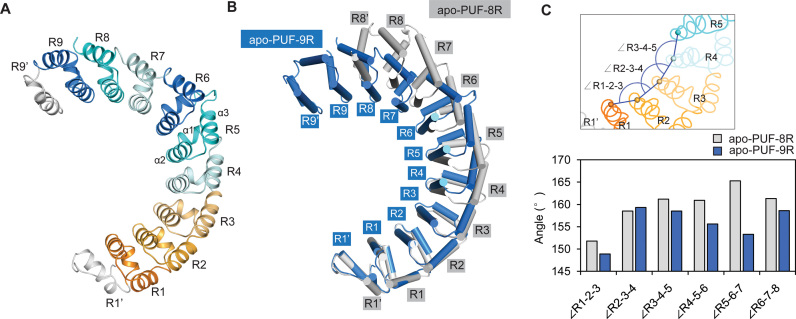
Figure 3.
More curved conformation of PUF-9R. (A) Overall structure of apo-PUF-9R. Repeats are colored as in Figure 1A, and helices α1, α2 and α3 in repeat R5 were indicated. (B) Comparison of apo-PUF-9R (blue) and apo-PUF-8R (gray, PDB code: 1M8Z) upon superposition of repeats R1′-R1. (C) Comparison of the repeat-to-repeat angles of apo-PUF-9R and apo-PUF-8R.
The overall curvature of the engineered apo-PUF-9R is strikingly different from that of the native apo-PUF-8R (PDB code: 1M8Z) (17) (Figure 3B). By aligning the N-terminal repeats R1′-R1 of apo-PUF-9R and apo-PUF-8R, the C-terminus of apo-PUF-9R bends inward significantly, indicating that increasing repeat number leads to a more curved conformation. We quantified the curvature by measuring the angles between the successive equivalent helices along the long axes of the protein (19). Indeed, the repeat-to-repeat angles in the apo-PUF-9R structure are generally smaller than those of the apo-PUF-8R (Figure 3C). Therefore, increasing of repeat number not only extends the linear length of a PUF protein, but also increases its overall curvature.
Flattened conformation of PUF-9R upon RNA binding
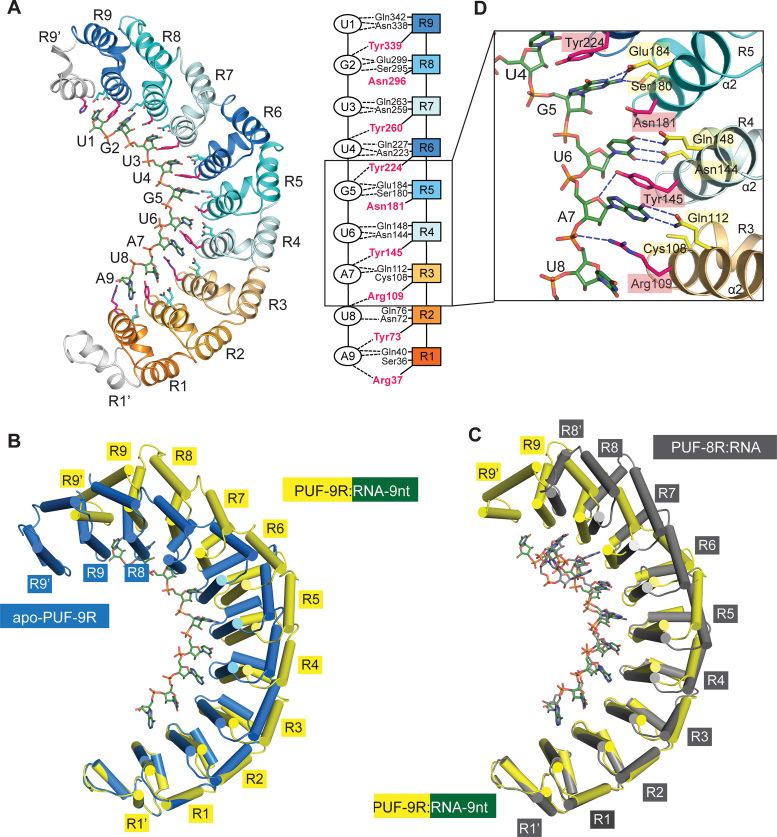
In the PUF-9R:RNA-9nt complex structure, PUF-9R recognizes its cognate RNA-9nt in a modular fashion, with one RNA base bound by one repeat (Figure 4A). All nine matching nucleotides (5′-UGUUGUAUA-3′) were observed in the electron density map (Supplementary Figure S5A). The concave surface of PUF-9R serves as a platform for RNA recognition, and the 3′ end of the RNA binds to the N-terminus of PUF-9R, similarly as that in most native PUF–RNA structures (18–20,22,38,39,41). Unexpectedly, superposition of the apo-PUF-9R and the PUF-9R:RNA-9nt complex revealed a marked difference in their overall curvatures (Figure 4B and Supplementary Figure S5B), and such curvature decrease upon RNA binding is rarely observed among most native PUFs (18–22,38,39,42–45). More intriguingly, the curvature of the RNA-bound PUF-9R is almost the same as that of the PUF-8R–RNA complex (PDB code: 1M8Y) (20) (Figure 4C and Supplementary Figure S5C). The conformational change in the overall curvature of PUF-9R might provide an extended surface for the cognate RNA-9nt to fit in, and thereby is required for tighter binding between PUF-9R and RNA-9nt.
Figure 4.
RNA binding flattens the curvature of PUF-9R. (A) Overall structure of PUF-9R in complex with the 9-nt RNA target (forest). Shown on the right is a schematic diagram of the interactions between PUF-9R and RNA-9nt, with dashed lines representing the polar interactions. (B) Comparison of the PUF-9R conformations in complex with RNA-9nt (yellow) and in isolation (blue) by aligning repeat R1′-R1. (C) Comparison of PUF-9R:RNA-9nt (yellow) and PUF-8R:RNA complex (dark gray, PDB code: 1M8Y) upon superposition of repeats R1′-R1. (D) Close-up view of the interface between PUF repeats R3-R5 and RNA bases G5-A7. Residues stacked between RNA bases (position 2) are highlighted in magenta, and those involved in polar contacts (positions 1 and 5) are colored in yellow.
PUF-9R mainly interacts with the bases of RNA-9nt, with the ribose and phosphate groups facing the solvent (Figure 4A). Each PUF repeat recognizes one RNA nucleotide in a modular fashion, as reported in wild-type PUF–RNA structures (18–20,22,38,39,43). The 5-residue recognition codes ‘12XX5’ on each α2 helix protrude from the inner surface of the protein, and are responsible for the specific interaction with the corresponding RNA base (Figure 4A). Two polar or charged side chains of residues at positions 1 and 5 form multiple hydrogen bonds with the edge of each base which ensure the specificity of each repeat, while residues at position 2 are stacked between two adjacent RNA bases which mainly contributes to the binding affinity. In PUF-9R, three kinds of stacking residues are presented: tyrosine on repeats R2, R4, R6, R7 and R9; arginine on repeats R1 and R3; asparagine on repeats R5 and R8 (Figure 4A and Supplementary Figure S4A). The arginine residue contains three hydrophobic methylene groups and one positively charged guanidine group, which make Van der Waals contacts, as well as the cation–π interactions, with the RNA bases (46–48). The basic guanidino group of arginine also participates in hydrogen bonding to the polar RNA backbone (Figure 4D). The aromatic side chain of tyrosine forms π–π interactions with two adjacent purine and/or pyrimidine rings, and the presence of the hydroxyl group allow the formation of direct or water-mediated hydrogen bonds with RNA backbone as well. By contrast, the smaller, uncharged side chain of asparagine barely contacts the RNA bases. Thus, a PUF repeat with tyrosine or arginine as the ‘stacking’ residue would bind more tightly to RNA bases (Figure 4D).
Enhanced binding affinity for PUF repeat with tyrosine or arginine as stacking residue
In addition to residues Arg, Tyr and Asn presented in PUF-9R, a forth residue, histidine, is also found to be the ‘stacking’ residue in native PUF proteins (18–20,22,38,39,42,45). For instance, PUF-8R contains a histidine on helix α2 of repeat R4 (Figure 5A) (17,20), however, the imidazole ring of histidine makes relatively weak stacking interactions with adjacent bases when compared to the bulky side chains of tyrosine and arginine. Therefore, we proposed that tyrosine or arginine would serve as the ‘stacking’ residue much better than histone or asparagine.
Figure 5.
Tyrosine and arginine serve as better ‘stacking’ residues. (A) Stacking interactions between PUF-8R repeats R4-R7 and its cognate RNA (PDB code: 1M8X). Residues stacked between RNA bases are highlighted in magenta. (B) Binding affinities of various PUF-8R stacking mutants to the native RNA-8nt determined by the FP assay (mean ± S.E.M., n = 3). (C) Quantification of the cassette exon inclusion for ESFs containing indicated PUF-8R mutant (mean ± S.E.M., n = 3). The splicing reporters and ESFs were transfected into HEK-293T cells with a ratio of 1:4. Statistical analysis was performed by a two-tailed Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., no significance).
To evaluate this hypothesis, we generated a series of point mutations on the wild-type PUF-8R, which contains all four aforementioned kinds of ‘stacking’ residues and the association constant Ka of each mutant to the native target RNA-8nt was determined through FP assay (Figure 5B and Supplementary Figure S6A). When the small, polar Asn1080 on helix α2 of repeat R7 was replaced by Arg or Tyr, the RNA-binding affinities of PUF-8R mutants N1080R and N1080Y increased 2- to 3-fold compared with the wild-type protein. Mutation of His972 on repeat R4 to Tyr (H972Y) similarly enhanced the RNA binding efficiency, whereas replacement of His972 with an Asn (H972N) markedly reduced its affinity. In addition, substitution of Arg1008 in repeat R5 to Tyr (R1008Y) had little effect, while mutating it to Asn (R1008N) resulted in a dramatic decrease in the binding affinity. Significantly, the Ka value of PUF-8R (N1080Y) was more than 8-fold to that of PUF-8R (R1008N) (Figure 5B). These results demonstrated that the bulky side chains of Tyr and Arg rendered them the better ‘stacking’ residues while a single Asn substitution of an 8-repeat PUF protein could markedly reduce the RNA binding efficiency.
We further confirmed these results using the alternative splicing system (9). We constructed the ESFs with the above PUF-8R mutants as described previously, and co-transfected them in HEK-293T cells with the plasmids expressing the splicing reporter containing the cognate 8-nt RNA target. Inclusion of the cassette exon levels were measured with RT-PCR and the expression of splicing factors was confirmed by western blots (Supplementary Figure S6B). The splicing efficiency of the indicated PUF-8R mutants was generally consistent with the above FP assay data (Figure 5C). Particularly, arginine mutation (N1080R) of the ‘stacking’ residue on repeat R7 had >3-fold higher splicing efficiency than the asparagine mutation (R1008N) on repeat R5. Effects of these PUF mutants on alternative splicing suggested that PUF modules could be optimized as more efficient tools in RNA manipulation by modifying the stacking residues to increase their binding affinities with the RNA targets.
Modular binding of PUF-9R to RNA targets ensures high-sequence discrimination
To extend the application, the engineered PUFs are expected to recognize targets of any sequence precisely and tightly. The native PUF-8R only recognizes adenine, uracil and guanine and the 5-residue recognition code for cytosine has been demonstrated as SYXXR (24,44). Meanwhile, our engineered PUF-9R had the highest binding affinity to its cognate RNA-9nt (Figure 1D and E) and the ESF containing PUF-9R showed the most efficient splicing (Figure 2B, black bars). In addition, it was reported that the mutations in the C-terminal three repeats (R6–R8) of native PUF proteins and their cognate bases they recognized affected the PUF–RNA binding more profoundly than those elsewhere (43,49).
Based on these considerations, we used PUF-9R as a representative and mutated the ‘12XX5’ recognition codes on N-terminal repeats R1, R2 and R3 into SYXXR, individually or in combination (Figure 6A). We also mutated the C-terminal repeat R9 to have a test. The binding affinities of these modified PUFs to various RNA targets were measured by FP assay and the relative binding affinities were represented in a form of heat map (Figure 6B and Supplementary Table S6). Indeed, all modified PUF-9R recognized their cognate RNA targets with higher affinities compared with other RNAs, which clearly indicated that the engineered PUFs could be modified to recognize the designed targets specifically in a programmable way. Notably, though PUF-9R (R9C) and PUF-9R (R1,9C) showed relatively low affinities compared to other modified PUF-9R proteins, they still had acceptable specificity to their cognate targets. The low affinities of the two R9 mutants again confirmed the low mutability of the C-terminal repeats of PUF domains, consistent with the conserved ‘UGU’ triplet in almost all RNA targets recognized by natural PUF proteins (49).
Figure 6.
Recognition of PUF-9R mutants to RNA targets containing cytosine. (A) Schematic diagram of PUF-9R mutants and their cognate RNA sequences. Repeats bearing the C code mutation are colored in orange. (B) Heat map of the relative binding affinities of PUF-9R mutants to different RNA targets. The relative binding affinity of each PUF:RNA pair (500 nM protein, 5 nM RNA) to the PUF-9R:RNA-9nt pair was listed in Supplementary Table S6 (mean ± S.E.M., n = 3).
DISCUSSION
Most of the native PUF proteins consist of eight repeats and bind their RNA targets in a modular fashion, which is selected during evolution with near optimal specificity and affinity. Previously the PUF with 16 repeats (PUF-16R) has been constructed by inserting R1–R8 between R5 and R6 of the original wild-type PUF-8R (24). Here, we further constructed artificial PUFs with different numbers of repeats as a tool to specifically target RNAs. According to the previous work and our results (Supplementary Figure S2), the PUF-16R showed higher binding affinity than PUF-8R when recognizing RNA-16nt, and bound stronger to RNA-16nt compared with RNA-8nt. In both cases, there are more binding sites for PUF-16R to RNA-16nt, which will result in their increased binding affinities. In addition, PUF-8R presented more efficient binding than PUF-16R when recognizing RNA-8nt, which is consist with our results (Supplementary Figure S2) and the conclusion that the number of PUF repeats will affect the binding affinity. However, the binding affinity of PUF-16R:RNA-16nt is comparable to that of PUF-8R:RNA-8nt as judged by yeast-three-hybrid and FP assay, suggesting a modest improvement of PUF-16R as a more specific RNA-binding scaffold (Figure 1D and E). Instead, the binding affinity of the engineered PUF-9R and PUF-10R are more than 5-fold higher than that of PUF-8R. Therefore, PUFs with more repeats have more efficient binding, but further increasing repeats will not always improve binding.
PUFs with additional repeats would have more binding sites for nucleotides, thus they are expected to bind more tightly to their cognate targets. However, our data showed that the binding affinities of the engineered PUF-12R and PUF-16R were similar to that of the native PUF-8R, less than that of the PUF-9R and PUF-10R (Figure 1D and E). Structural comparison of the native PUF-8R and the engineered apo-PUF-9R suggests that additional number of PUF repeats increase the overall curvature (Figure 3B). Similarly, the structure of Nop9, a PUF-like protein with 11 repeats, also reveals a dramatic curved, C-shaped conformation (40) and even more curved than apo-PUF-9R (Supplementary Figure S7). These results suggest that the curvature of PUFs would increase with the addition of PUF repeats. Interestingly, a remarkable decrease of the overall curvature is found in PUF-9R upon the binding of its cognate RNA-9nt. The similar case occurs in Nop9 as the 11-nt RNA binding also extends the protein curvature (41). Moreover, a slight conformational change is also presented between the unbound PUF-8R and its complex with the cognate RNA (17,20). We therefore believe that the change of overall curvature is required for the formation of stable PUF–RNA complex, and PUFs with more repeats would undergo more dramatic structural changes to open the more curved conformation upon RNA binding. Considering such conformational change is an energy-consuming process, the engineered PUFs with more repeats would spend more energy for binding their cognate RNAs and thereby display weak affinities.
In addition, PUFs with more repeats may bind to their cognate RNA targets in imperfect modes. Some native PUF proteins could bind RNA targets more than 8nt with a binding efficiency much lower than that of PUF-8R from human Pumilio1 (18,19,39). Such a lower affinity may be owing to their different RNA-binding modes. For example, Puf5p from Saccharomyces cerevisiae presents the most extended architecture among the native PUFs and binds to RNA sequences from 8 to 12 nt, but shows divergent modes for RNA binding when targets are more than 8 nt, with only part of the bases being recognized (39). Structures of C. elegans FBF-2 and S. cerevisiae Puf4p in complex with their cognate 9-nt RNA targets show that one or two bases flipped away from the binding surface (18,19). Furthermore, structures of PUF-8R in complex with some non-cognate 9-nt RNAs also reveal similar binding mode with one base not bound by the protein (21). Recently reported 11-repeat Nop9, with the most curved conformation of the PUF structures determined up to now, binds 18S rRNA with only eight bases being recognized (41). Thus, in addition to opening the curvature, PUFs with more repeats may adopt altered patterns to bind RNA targets, of which only part of the bases could be recognized. In both the energy-consuming conformational changes upon RNA binding and the partial/incomplete recognition modes, binding affinities of PUFs with more repeats would be interfered. Therefore, combining the results of engineered PUFs to different RNA targets (Figures 1F, 2C and Supplementary Figure S2), the binding affinities of the engineered PUFs are affected by the number of matched bases, the structure of engineered PUFs and the length of RNA targets.
Among the known PUF proteins with canonical 8-repeat architectures (18–20,22,38,39,42), there are five different residues that stack with RNA bases. In addition to the aforementioned four ‘stacking’ residues, another uncommon residue, cysteine, appears in S. cerevisiae Puf4p and Puf5p (18,39). Compared to tyrosine and arginine, the stacking effect of cysteine would be weak, similar to that of asparagine. Previously, it was reported that mutating the stacking residue asparagine on repeat 7 of C.elegans FBF-2 broadened the specificity of the recognized base at +3 position, but not +1 position (49). R288Y mutation of FBF-2 decreased the specificity for recognizing the base at position +7 but had little effect on +8 base (45). Furthermore, H454Y mutation on repeat 7 of FBF-2 would increase the specificity for both of the stacked bases, but H454R mutation could only increase the specificity for +2 position base (45). Here, our results on the examination of the four common types of stacking residues in human PUF-8R show that tyrosine and arginine present preferred stacking interactions with RNA bases. Although tyrosine and arginine showed interchangeable contributions to RNA-binding affinity in our assays, these two residues are not completely equivalent in their side chain properties. Unlike tyrosine, arginine is positively charged, which directly form ionic bonds with the adjacent nucleotides. In addition, arginine has a relatively longer and more flexible side chain, which is more advantageous than tyrosine when the adjacent bases are pyrimidine or the stacked bases shift from the stacking axes. These differences between tyrosine and arginine may affect the selection of the preferred bases they stack. Therefore, modifying the stacking residues should be considered for designing PUFs with desired affinity and specificity as well.
The engineered PUF-9R and PUF-10R are over 10 times more efficient compared to the canonical eight-repeat PUF domains in binding RNAs, while still being small enough to be packed into most gene therapy vectors, indicating that they will be very useful in engineering artificial PUF factors recognizing and manipulating various RNA targets inside cells (such as pre-miRNA, disease-related mRNA, etc.). The PUF domains with reduced numbers of repeats may be useful for other biotechnical applications, such as generating small fragments of long RNAs during the construction of high-throughput sequencing libraries. Recently, CRISPR-Cas13a system has been engineered as a specific and programmable RNA recognition module to manipulate RNA degradation (50), or RNA editing (51). The targeting mechanisms of this system depend on RNA–RNA pairing without manipulation of proteins, making it more specific than PUF-based RNA recognition system. However, this system requires co-folding of Cas13a with guide RNA, which may not be very efficient. In addition, compare to human PUF proteins, Cas13a is a bacterial protein that may cause immune response in human. Finally, the size of the CRIPSR-Cas13a will limit its application in some cases. On the other hand, PUF is a single-component programmable RNA binding protein and has proven to be efficient with many distinct applications (6–12,23,26–28). Meanwhile, PUF is relatively easy to manipulate, with small size and low off-target effects (20,23,25,49). Further studies using such engineered PUF scaffolds may focus on investigating the binding and specificity of each repeat, since the conformation will be more curved for PUFs with much more repeats. This will enable the engineering of better RNA binding factors with designed specificity, which can be fused to various functional modules to manipulate different steps of RNA metabolism.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 5YKH for the apo-PUF-9R and 5YKI for the PUF-9R:RNA-9nt structures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF) and Dr S. Fan at Tsinghua Center for Structural Biology for assistance in data collection, and the China National Center for Protein Sciences Beijing for providing facility support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2016YFA0502004 to J.-W.W.]; National Natural Science Foundation of China [31621063 to J.-W.W., 31570823 to Z.W. and 31400726 to W.-J.Z.]; National Institutes of Health [R21AR061640 to Z.W.]. Funding for open access charge: National Natural Science Foundation of China [31621063].
Conflict of interest statement. Z.W. has co-founded a company, Enzerna Biosciences, Inc., to commercialize the artificial RNA binding proteins using PUF scaffold. The authors declare that a patent application involving the new enzymes has been filed (application number PCT/US2011/040933).
REFERENCES
- 1. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hudson W.H., Ortlund E.A.. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014; 15:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker D.D., Wang C., Moore J., Dickinson L.K., Lehmann R.. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992; 6:2312–2326. [DOI] [PubMed] [Google Scholar]
- 4. Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P.. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997; 390:477–484. [DOI] [PubMed] [Google Scholar]
- 5. Gerber A.P., Herschlag D., Brown P.O.. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004; 2:E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ozawa T., Natori Y., Sato M., Umezawa Y.. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods. 2007; 4:413–419. [DOI] [PubMed] [Google Scholar]
- 7. Tilsner J., Linnik O., Christensen N.M., Bell K., Roberts I.M., Lacomme C., Oparka K.J.. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 2009; 57:758–770. [DOI] [PubMed] [Google Scholar]
- 8. Furman J.L., Badran A.H., Ajulo O., Porter J.R., Stains C.I., Segal D.J., Ghosh I.. Toward a general approach for RNA-templated hierarchical assembly of split-proteins. J. Am. Chem. Soc. 2010; 132:11692–11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Cheong C.G., Hall T.M., Wang Z.. Engineering splicing factors with designed specificities. Nat. Methods. 2009; 6:825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooke A., Prigge A., Opperman L., Wickens M.. Targeted translational regulation using the PUF protein family scaffold. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choudhury R., Tsai Y.S., Dominguez D., Wang Y., Wang Z.. Engineering RNA endonucleases with customized sequence specificities. Nat. Commun. 2012; 3:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Wang Z., Tanaka Hall T.M.. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 2013; 280:3755–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zamore P.D., Williamson J.R., Lehmann R.. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997; 3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 14. Macdonald P.M. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992; 114:221–232. [DOI] [PubMed] [Google Scholar]
- 15. Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K.. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001; 105:281–289. [DOI] [PubMed] [Google Scholar]
- 16. Jenkins H.T., Baker-Wilding R., Edwards T.A.. Structure and RNA binding of the mouse Pumilio-2 Puf domain. J. Struct. Biol. 2009; 167:271–276. [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Zamore P.D., Hall T.M.. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001; 7:855–865. [DOI] [PubMed] [Google Scholar]
- 18. Miller M.T., Higgin J.J., Hall T.M.. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008; 15:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Opperman L., Wickens M., Hall T.M.. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20186–20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., McLachlan J., Zamore P.D., Hall T.M.. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 21. Gupta Y.K., Nair D.T., Wharton R.P., Aggarwal A.K.. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008; 16:549–557. [DOI] [PubMed] [Google Scholar]
- 22. Lu G., Hall T.M.. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011; 19:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adamala K.P., Martin-Alarcon D.A., Boyden E.S.. Programmable RNA-binding protein composed of repeats of a single modular unit. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2579–E2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Filipovska A., Razif M.F., Nygard K.K., Rackham O.. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 2011; 7:425–427. [DOI] [PubMed] [Google Scholar]
- 25. Cheong C.G., Hall T.M.. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang W., Wang Y., Dong S., Choudhury R., Jin Y., Wang Z.. Treatment of Type 1 myotonic dystrophy by engineering site-specific RNA endonucleases that target (CUG)n repeats. Mol. Ther. 2014; 22:312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y., Chen D., Qian H., Tsai Y.S., Shao S., Liu Q., Dominguez D., Wang Z.. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014; 26:374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wei H., Wang Z.. Engineering RNA-binding proteins with diverse activities. Wiley Interdiscip. Rev. RNA. 2015; 6:597–613. [DOI] [PubMed] [Google Scholar]
- 29. SenGupta D.J., Zhang B., Kraemer B., Pochart P., Fields S., Wickens M.. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hook B., Bernstein D., Zhang B., Wickens M.. RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA. 2005; 11:227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lea W.A., Simeonov A.. Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discov. 2011; 6:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 33. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 35. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. 3rd, Snoeyink J., Richardson J.S. et al. . MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007; 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Ma M., Xiao X., Wang Z.. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 2012; 19:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu D., Stumpf C.R., Krahn J.M., Wickens M., Hall T.M.. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20192–20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilinski D., Qiu C., Lapointe C.P., Nevil M., Campbell Z.T., Tanaka Hall T.M., Wickens M.. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 2015; 6:8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J., McCann K.L., Qiu C., Gonzalez L.E., Baserga S.J., Hall T.M.. Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 2016; 7:13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang B., Ye K.. Nop9 binds the central pseudoknot region of 18S rRNA. Nucleic Acids Res. 2017; 45:3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiu C., Kershner A., Wang Y., Holley C.P., Wilinski D., Keles S., Kimble J., Wickens M., Hall T.M.. Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket. J. Biol. Chem. 2012; 287:6949–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valley C.T., Porter D.F., Qiu C., Campbell Z.T., Hall T.M., Wickens M.. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dong S., Wang Y., Cassidy-Amstutz C., Lu G., Bigler R., Jezyk M.R., Li C., Hall T.M., Wang Z.. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 2011; 286:26732–26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koh Y.Y., Wang Y., Qiu C., Opperman L., Gross L., Tanaka Hall T.M., Wickens M.. Stacking interactions in PUF-RNA complexes. RNA. 2011; 17:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilson K.A., Kellie J.L., Wetmore S.D.. DNA-protein pi-interactions in nature: abundance, structure, composition and strength of contacts between aromatic amino acids and DNA nucleobases or deoxyribose sugar. Nucleic Acids Res. 2014; 42:6726–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gallivan J.P., Dougherty D.A.. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang H., Li C., Yang F., Su J., Tan J., Zhang X., Wang C.. Cation-pi interactions at non-redundant protein–RNA interfaces. Biochemistry (Mosc.). 2014; 79:643–652. [DOI] [PubMed] [Google Scholar]
- 49. Campbell Z.T., Valley C.T., Wickens M.. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 2014; 21:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A. et al. . RNA targeting with CRISPR-Cas13. Nature. 2017; 550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F.. RNA editing with CRISPR-Cas13. Science. 2017; 358:1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 5YKH for the apo-PUF-9R and 5YKI for the PUF-9R:RNA-9nt structures.