Abstract
Translesion DNA synthesis (TLS) is one mode of DNA damage tolerance that uses specialized DNA polymerases to replicate damaged DNA. DNA polymerase η (Polη) is well known to facilitate TLS across ultraviolet (UV) irradiation and mutations in POLH are implicated in skin carcinogenesis. However, the basis for recruitment of Polη to stalled replication forks is not completely understood. In this study, we used an affinity purification approach to isolate a Polη-containing complex and have identified SART3, a pre-mRNA splicing factor, as a critical regulator to modulate the recruitment of Polη and its partner RAD18 after UV exposure. We show that SART3 interacts with Polη and RAD18 via its C-terminus. Moreover, SART3 can form homodimers to promote the Polη/RAD18 interaction and PCNA monoubiquitination, a key event in TLS. Depletion of SART3 also impairs UV-induced single-stranded DNA (ssDNA) generation and RPA focus formation, resulting in an impaired Polη recruitment and a higher mutation frequency and hypersensitivity after UV treatment. Notably, we found that several SART3 missense mutations in cancer samples lessen its stimulatory effect on PCNA monoubiquitination. Collectively, our findings establish SART3 as a novel Polη/RAD18 association regulator that protects cells from UV-induced DNA damage, which functions in a RNA binding-independent fashion.
INTRODUCTION
Mammalian cells are under endogenous and exogenous attacks every day, causing a variety of genomic lesions. These DNA lesions can often result in replication forks stalling, which if not resolved in time lead to replication fork collapse and even double strand breaks (DSBs), one of the most deleterious DNA damages. In order to cope with this situation, damage tolerance pathways are evolved for the cellular survival (1,2). Translesion DNA synthesis (TLS) is one major mode of DNA damage tolerance, which utilizes specialized polymerases that lack 3′-5′ exonucleolytic proofreading activity and replicate DNA with low fidelity and processivity. The best-characterized TLS polymerases are Y-family polymerases, including Polη, Polι, Polκ and REV1 (3–7). Among them, Polη can correctly bypass ultraviolet (UV)–induced cis–syn thymine–thymine cyclobutane–pyrimidine dimers (CPDs). Loss of functional Polη has been known to cause the variant form of the skin cancer-prone syndrome, Xeroderma Pigmentosum (XPV) (8–10).
It is well known that the TLS pathway can be efficiently triggered by replication stress, which usually generates stretches of single-stranded DNA (ssDNA) through uncoupling of replicative polymerase and helicase activities. The ssDNAs are rapidly coated by replication protein A (RPA), which recruits the ubiquitin E3 ligase RAD18 to stalled replication forks to promote monoubiquitination of Proliferating cell nuclear antigen (PCNA) at Lys164 (11–13). Monoubiquitinated PCNA (PCNA-mUb) then facilitates optimal TLS through its enhanced binding with Y family polymerases (14). Compelling evidence reveals that PCNA-mUb, the key event in TLS, is tightly regulated by several DDR factors, including USP1, MSH2, BRCA1, Polη, REV1 and Parkin (15–20). In contrast to USP1, which can deubiquitinate PCNA-mUb (15,21), MSH2, BRCA1 and Parkin have been shown to facilitate UV-induced PCNA-mUb through promoting ssDNA generation and RPA focus formation (16,19,20). And Polη and REV1, two TLS polymerases, can promote RAD18 recruitment after UV irradiation (17,18). Polη has also been shown to bridge RAD18 and PCNA to promote efficient PCNA-mUb formation after DNA damage. Notably, this Polη scaffolding function is independent of its DNA polymerase activity but relies on the Polη/RAD18 association (17). Interestingly, RAD18 is also required to guide Polη to stalled replication sites through Polη/RAD18 physical interaction (12). Although RAD18 phosphorylation mediated by Cdc7 or JNK enhances the Polη/RAD18 association (22,23), how Polη/RAD18 interaction is regulated remains largely unknown.
SART3 (Squamous Cell Carcinoma Antigen Recognized By T-Cells 3) is a nuclear RNA-binding protein (RBP), which contains half-a-tetracopeptide repeats (HAT) in the N-terminus and two RNA recognition motifs (RRM1 and RRM2) near the C-terminus. Being a U4/U6 recycling factor, SART3 can assist pre-mRNA splicing, thereby regulating gene expression (24). In line with this, SART3 is indispensable for embryonic development, whose deficiency is reported to be embryonic lethal (25,26). Moreover, SART3 also expresses at high levels in the nucleus of malignant tumor cell lines and majority of cancer tissues (27). Here, we identified SART3 to be a novel partner of Polη, whose depletion decreases ssDNA generation, RPA focus formation as well as the chromatin binding of RAD18 in the presence of UVC treatment. Consistently, knockdown SART3 impairs Polη focus formation and CPD lesion bypass after UVC exposure, leading to UV hypersensitivity. Furthermore, we found that SART3 can form homodimers and associate with RAD18. And SART3 can promote the Polη/RAD18 interaction though its coiled-coil domain to facilitate PCNA-mUb formation. Lastly, several missense mutations of SART3 identified in tumor samples fail to augment PCNA-mUb levels and activate TLS pathway. Collectively, we define an RNA binding-independent function for SART3 in TLS by facilitating RAD18 /Polη interaction and RAD18 chromatin accumulation to promote PCNA-mUb formation, providing insights into how SART3 promotes genome integrity and contributes to cancer development.
MATERIALS AND METHODS
Plasmids and reagents
SART3 cDNA was a gift from Dr. Jiahuai Han (Xiamen University). SFB (S-Flag–Streptavidin binding peptide)- and Myc-tagged RAD18 plasmids were gifts from Dr Jun Huang (Zhejiang University). Full-length and truncations of SART3 were PCR amplified and cloned into pEGFP-C3 (Clontech) or pCMV5-Flag to generate GFP- or Flag-tagged fusion proteins. Full-length and truncations of Polη were amplified and cloned into p2xFlag-CMV-14 (Sigma) or pEGFP-C3 vector.
Anti-Flag M2 agarose affinity gel, mouse monoclonal antibody against Flag and BrdU for labeling ssDNA were purchased from Sigma (St. Louis, MO, USA). Antibody against BrdU was from BD science. Antibodies against RAD18 for western blotting, SART3 and RPA32 were from Abcam. Anti-RAD18 antibody for immunofluorescence was purchased from Bethyl Laboratories. Antibodies against CPD was from Cosmo Bio Co (Tokyo, Japan). Monoclonal antibodies against PCNA (PC10) and GFP (FL) were from Santa Cruz Biotechnology. Antibody against USP1 was from Cell Signaling Technology. Alexa Fluor-555-labeled goat anti-mouse-IgG was from Invitrogen.
Cell culture and reagents
Human U2OS and 293T cells were obtained from the American Type Culture Collection (Rockville, MD, USA). SV40-transformed MRC5 (Polη normal) and XP30RO (Polη-deficient) cells were kindly provided by Dr Alan Lehmann. RAD18 knockout 293T cell lines were prepared through TALEN as described (18). All cells were treated with mycoplasma removal agent (MPbio) and cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) at 37°C in the presence of 5% CO2 if not specified. For transient transfection experiments, cells were transfected with indicated constructs using VigoFect (Vigorous Biotechnology Beijing Co., Ltd, China) following the manufacturer's protocol. Forty-eight hours later, transfected cells were collected for further experiments.
For RNAi experiments cells were transfected with siRNAs purchased from GenePharma (Shanghai, China) using RNAiMAX (Invitrogen) according to manufacturer's instruction, and analyzed 72 h later. The gene-specific target sequences were as follows: SART3 (3′ UTR-1) (GGAGACAGGAAATGCCTTA), SART3 (3′ UTR-2) (GATGTGGTGTCCTGAGATA), SART3 (CDS) (GCUGAGAAGAAAGCGUUAA) and POLH (CAGCCAAATGCCCATTCGCAA). The negative control (siNC) sequence (UUCUCCGAACGUGUCACGU) was also obtained from GenePharma. Unless otherwise specified, SART3-3′ UTR-2 was used as the representative siRNA against SART3 in all experiments. Western blots were used to validate knockdown efficiency of theses siRNAs. For focus formation assay, cells were further transfected with GFP-Polη 48 h later after siRNA transfection.
Co-immunoprecipitation and western blotting
HEK293T cells were transfected with Flag-SART3 and GFP-Polη or Flag-Polη and GFP-SART3. Forty-eight hours later, the cells were harvested and lysed with HEPES buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 25 mM NaF, 10 μM ZnCl2). The whole cell lysates were immunoprecipitated with anti-Flag M2 agarose in the presence or absence of RNase A, ethidium bromide (EB), as indicated. For mapping the regions within SART3 responsible for its interaction with Polη and RAD18, GFP-tagged wild-type (WT) and a series of truncated SART3 were co-transfected with Flag-Polη or SFB-RAD18 (SFB: S-Flag-Streptavidin binding peptide) in HEK293T cells for co-immunoprecipitation experiments. The immunoprecipitated products were separated by SDS-PAGE and detected by immunoblotting with indicated antibodies. For isolation of chromatin-fractions, U2OS cells were treated with UVC (15 J/m2), and the triton-insoluble fractions were harvested as previously described (16).
Mutation frequency
Mutation frequencies were measured using the supF shuttle vector system as described previously (28), which is used to measure TLS activity in mammalian cells. HEK293T cells were transfected with siNC or SART3 siRNAs twice. Forty-eight hours after the first transfection, cells were transfected with UVC-irradiated (400 J/m2 UVC) pSP189 reporter plasmid. 48 h later, pSP189 plasmid from 293T cells was extracted by using a DNA miniprep kit (Tiangen, China) and the purified plasmid was digested with Dpn1 followed by transformation into the MBM7070 bacterial strain. The transformed MBM7070 cells were cultured on Luria-Bertani plates with 200 μM IPTG, 100 μg/ml X-gal and 100 μg/ml ampicillin. The mutation frequency in the supF coding region was calculated by the ratio of white (mutant) and blue (WT) colonies. The pSP189 plasmid and MBM7070 strain were gifts from Dr M. Seidman (29).
Micronucleus
Micronucleus assay was performed as described (20,30). Briefly, U2OS cells transfected with siNC or siSART3 were irradiated with UVC (3 J/m2) followed by incubating with 6 μg/ml Cytochalasin B in complete medium for 48 h. The cells were trypsinized and washed with PBS once, further treated with 0.075 M KCl for 20 min. Cells were centrifuged to remove most hypotonic buffer and stained with 0.01% acridine orange. Cells with two nuclei were counted to calculate the micronuclei rate.
Immunofluorescence (IF)
IF was performed as described (16,18). Briefly, cells were seeded on cover glasses and irradiated with UVC. The cells were permeabilized with 0.5% Triton X-100 for 5–30 min before being fixed in 4% paraformaldehyde. The samples were then blocked with 5% donkey serum (for RAD18 staining) or 5% FBS and 1% goat serum (for RPA32 staining) for 30 min. The cells were next incubated with indicated antibodies for 45 min followed by incubation with Alexa Fluor 568 goat anti-mouse (Invitrogen, Molecular Probes) for 45 min. The cells were later counterstained with DAPI and images were acquired with a Leica DM5000 (Leica) equipped with HCX PL S-APO 63 × 1.3 oil CS immersion objective (Leica) and processed with Adobe Photoshop 7.0.
For quantitative analysis of UV-induced Polη focus formation, U2OS cells transfected with GFP-Polη were treated with 15 J/m2 UVC and fixed with 4% paraformaldehyde 10 h later as previously described. GFP-Polη-expressing cells with more than 30 Polη foci were counted as GFP-Polη foci positive cells. More than 200 cells were analyzed for each treatment.
Detection of un-replicated CPDs in UV-treated cells
Detection of CPDs in single-stranded DNA templates was performed as previously described (31–33). Briefly, cells cultured on coverslips were treated with 0 or 10 J/m2 UVC. Four hours later, cells were permeabilized with 1% Triton X-100 in PBS for 2 min, followed by fixation with 2% formaldehyde in 0.5% Triton X-100/PBS for 15 min at room temperature. Un-bypassed CPDs in non-denatured DNA were detected by using primary mouse monoclonal antibodies against it (TDM2, CosmoBio). Cells were subsequently incubated with Alexa Flour 568 goat anti-mouse (Invitrogen, Molecular Probes) and stained with DAPI. To check the UV-induced CPDs, cells were further treated with 2 M HCl for 10 min to denature DNA after fixation. Images were obtained using a fluorescence microscope.
Immunofluorescent detection of ssDNA
Cells were seeded on glass coverslips to reach a confluence of about 50% the next day. BrdU (Sigma) (10 μM) was added to the cells and incubated for 48 h. Cells were then irradiated with 20 J/m2 UVC. Two hours later, the cells were permeabilized with solution buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100) for 5 min at room temperature, followed by fixation in 4% paraformaldehyde for 10 min. SsDNAs were identified in non-denatured DNA with primary antibodies against BrdU (BD). To check the BrdU incorporation efficiency, cells were further treated with 2 M HCl for 10 min after fixation. Images were analyzed by fluorescent microscopy.
RESULTS
SART3 interacts with Polη and regulates Polη focus formation after UV radiation
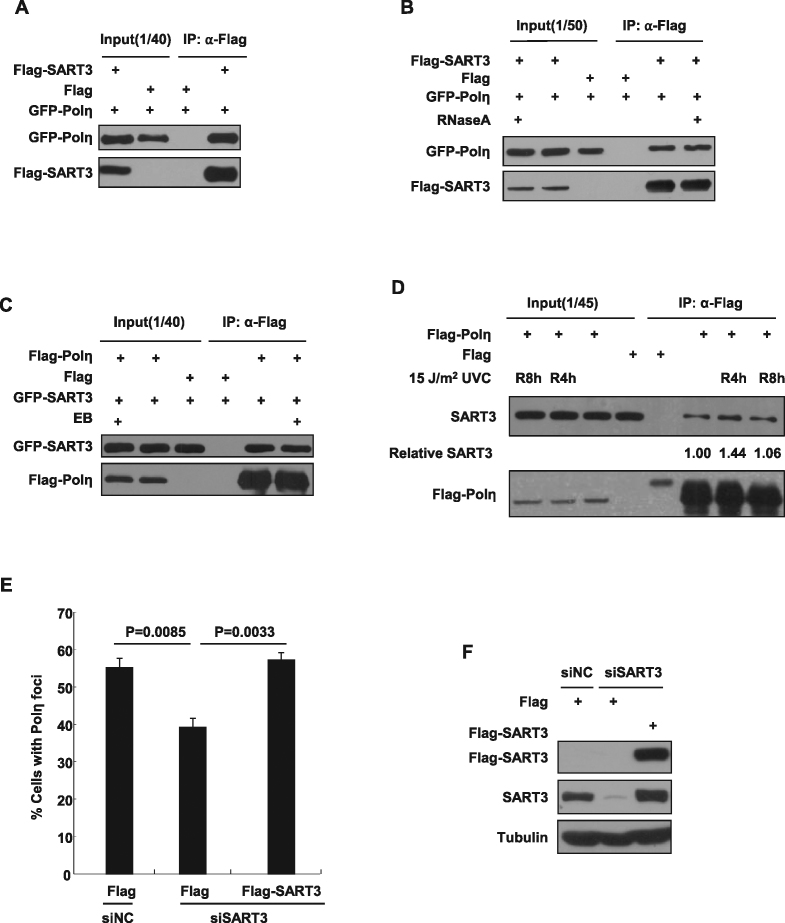
To determine the mechanism of regulation of Polη in response to UV irradiation, we transfected 2xFlag-Polη in HEK293T and performed immunopurification as described (34). The protein samples at the indicated region on a SDS-PAGE gel were digested and analyzed with liquid chromatography-tandem mass spectrometry (Supplementary Figure S1) (34). The analysis identified several potential Polη-interacting proteins, including SART3, which is a factor implicated in pre-mRNA splicing and gene expression. The association of GFP-Polη with Flag-SART3 was confirmed by co-immunoprecipitation (co-IP) (Figure 1A). Given that the C-terminus of SART3 has two RNA-recognition motifs (RRM1 and RRM2), which can recognize and bind RNA, we examined whether RNA is required for the interaction between SART3 and Polη. Cell lysates were treated with RNase A prior to co-IP. The result showed that RNase A treatment had no apparent effect on the association between Flag-SART3 and GFP-Polη (Figure 1B). We further observed that the binding between Flag-Polη and GFP-SART3 was not affected after treatment of cell lysates with ethidium bromide (EtBr), which is known to disrupt protein–DNA interactions (Figure 1C). These results exclude the possibility that the association between SART3 and Polη is mediated via DNA or RNA. Additionally, we found that Flag-Polη could also precipitate endogenous SART3 (Figure 1D). Intriguingly, their association manifested a dynamic change after UV treatment, namely, the level of interaction between the two proteins increased at 4 h and decreased to background at 8 h after UV treatment (Figure 1D).
Figure 1.
SART3 interacts with Polη and regulates Polη focus formation after UV radiation. (A) SART3 interacts with Polη. Flag-SART3 and GFP-Polη were co-transfected into 293T cells. The cell lysates were immunoprecipitated with anti-Flag agarose M2 beads and detected with antibodies against GFP and Flag. The input included 2.5% of the cell lysate used. (B) The interaction between Flag-SART3 and GFP-Polη is independent of RNA. 293T cells were co-transfected with Flag-SART3 and GFP-Polη. The cell lysates were immunoprecipitated using anti-Flag M2 agarose beads in the presence of RNase A (100 ng/μl). (C) The interaction between SART3 and Polη is not mediated by DNA. 293T cells were transfected with Flag-Polη and GFP-SART3. The cell lysates were immunoprecipitated in the presence of EtBr (200 μg/ml) and detected using anti-GFP or anti-Flag antibody. (D) The interaction between Flag-Polη and SART3 after UV irradiation. 293T cells were transfected with Flag-Polη. Forty-eight hours later, the cells were irradiated with 15 J/m2 UVC and further incubated. At the indicated time points, the cells were harvested for immunoprecipitation. Levels of relative SART3 were analyzed by western blot and quantified using Image J. Data represent means from three independent experiments. (E and F) SART3 depletion decreases GFP-Polη focus formation after UV irradiation. U2OS cells transfected with siSART3-3′ UTR or siNC were further co-transfected with Flag-SART3 and GFP-Polη. (E) Cells were treated with 15 J/m2 UVC, and repaired for 10 h. The proportion of GFP-Polη cells with more than 30 foci were measured, and at least 200 cells were counted. Data represent means ± SEM from three independent experiments. Statistical analyses were performed using a two-tailed Student's t-test. (F) The cell pellets were harvested 72 h later, and lysed by RIPA buffer. The expression of Flag-SART3 was detected with anti-Flag antibody. The blot was stripped and re-immunoblotted with a SART3 antibody. Tubulin: loading control.
Given that Polη plays an important role in UV-induced TLS, to uncover the significance of SART3 binding to Polη in vivo, we examined whether SART3 depletion impairs Polη focus formation after UV irradiation. SART3 in U2OS cells was depleted by siRNA directed to its 3′ UTR. The knockdown cells were transfected with GFP-Polη, followed by UVC (15 J/m2) treatment and incubated for 10 h. We noted that the proportion of GFP-Polη foci-positive cells in the SART3-depleted group (39.3%) was significantly decreased compared to non-targeting siRNA (siNC)-treated control (55.2%) (Figure 1E). To exclude the possibility that the reduced Polη focus formation results from siRNA off-target effect, we also co-transfected Flag-SART3 and GFP-Polη into the SART3-depleted U2OS cells and checked its Polη focus formation. We found that expression of Flag-SART3 (57.3%) could rescue the reduced Polη focus formation in SART3-depleted cells to the siNC-treated control level (55.2%) (Figure 1E and F). This result indicates that SART3 is required for optimal Polη association with stalled replication forks following UV exposure.
SART3 depletion impairs UV-induced PCNA-mUb and RAD18 focus formation
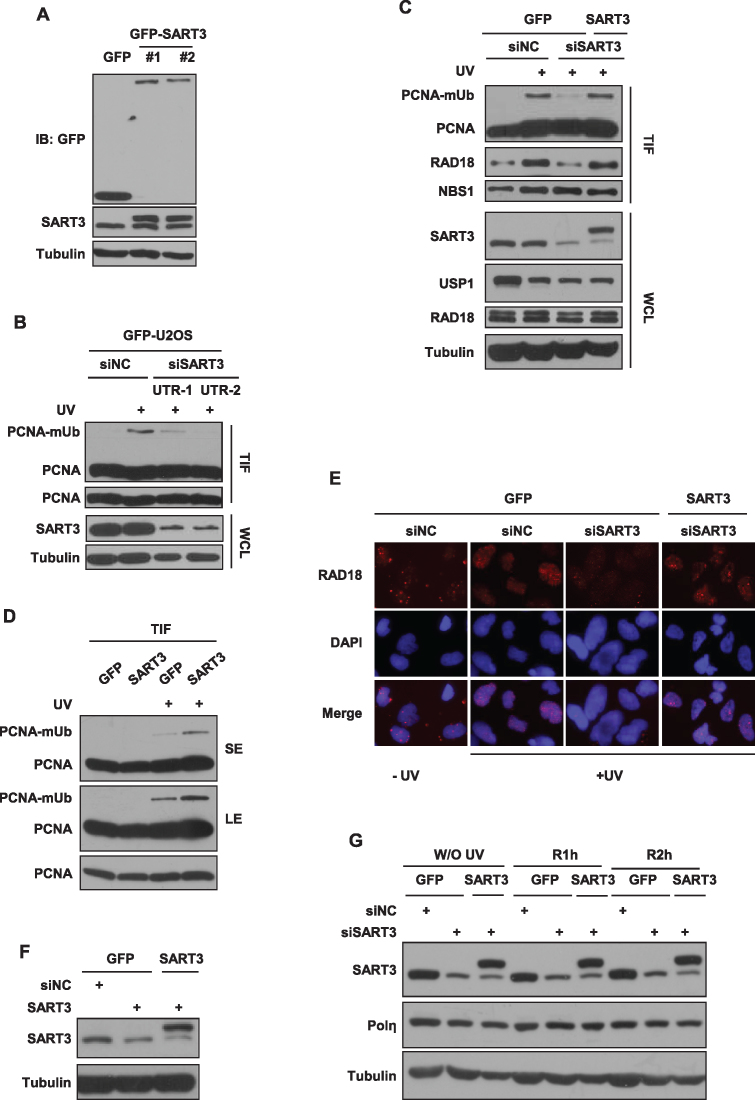
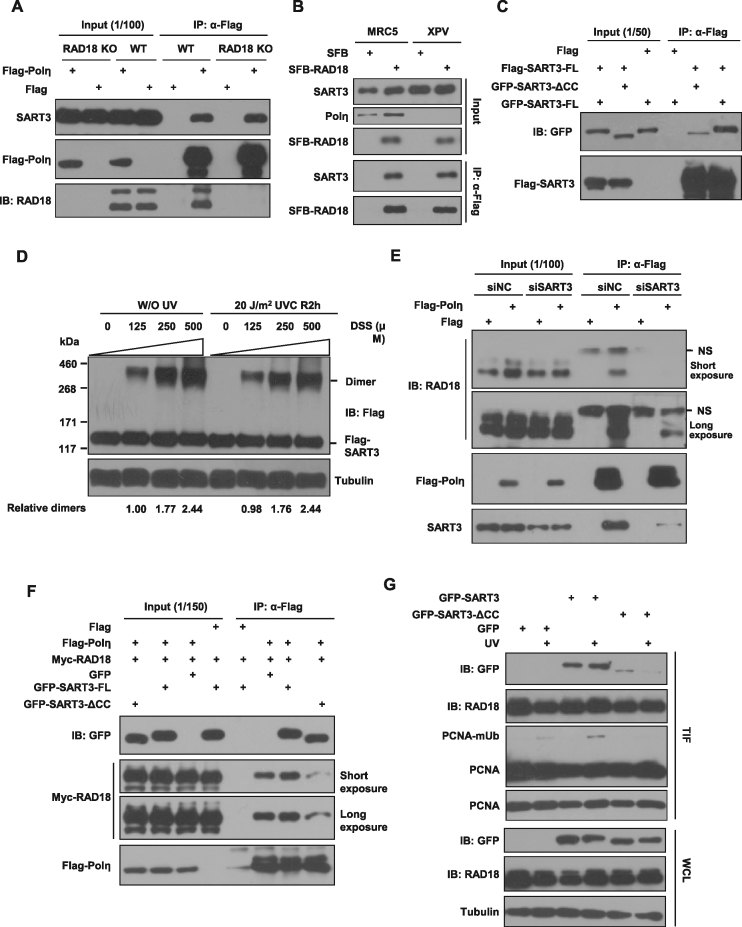
It is well-established that PCNA-mUb plays an important role in the recruitment of Y-family TLS polymerases to stalled replication factories after replication stress. We then determined whether the reduced Polη focus formation caused by SART3 depletion is due to an impaired formation of PCNA-mUb after UV irradiation. To test that, we first tried to establish SART3 knockout cells using the CRISPR/Cas9 method, but were unsuccessful, hinting that SART3 might be important for cell survival. We then generated U2OS stable cells expressing either GFP-SART3 or GFP vector (GFP) by lentivirus infection (Figure 2A). The expression levels of exogenous and endogenous SART3 were comparable in the U2OS stable cells expressing GFP-SART3. The cells were transfected with two independent siSART3 oligos targeting SART3 3′ UTR regions followed by UVC treatment. The triton-insoluble fractions were collected and analyzed by immunoblotting. We noticed that depletion of endogenous SART3 in GFP stably expressing cells led to a dramatically decrease in PCNA-mUb at 4 h after UV irradiation, compared to siNC control (Figure 2B). Similarly, SART3 ablation with two different siRNA oligos targeting its 3′ UTR or CDS regions, attenuated the UV-inducible level of PCNA-mUb (Supplementary Figure S2A). In contrast, depletion of endogenous SART3 in GFP-SART3 stably expressing cells manifested no obvious effect on PCNA-mUb, supporting that the reduced PCNA-mUb observed in SART3-depleted cells results from knockdown of SART3 (Figure 2C). Moreover, U2OS cells stably overexpressing GFP-SART3 showed more efficient formation of PCNA-mUb than GFP-expressing cells (Figure 2D).
Figure 2.
SART3 depletion impairs UV-induced PCNA-mUb and RAD18 focus formation. (A) The comparable expression levels of exogenous and endogenous SART3 in GFP-SART3 stably expressing cells. U2OS stable cells expressing either GFP or GFP-SART3 were harvested and analyzed with antibodies against GFP, SART3 and Tubulin. (B) SART3 deficiency decreases PCNA-mUb in GFP stably expressing cells. SiNC or two different siSART3 oligos were transfected in GFP stably expressing cells. 72 h later, cells were treated with 15 J/m2 UVC and further incubated for 4 h. The triton-insoluble fractions (TIF) were harvested and analyzed with anti-PCNA antibody. (C) The decreased level of PCNA-mUb was perfectly rescued in GFP-SART3 stably expressing cells depleted of endogenous SART3. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3, and irradiated with 15 J/m2 UVC, repaired for 4 h. The triton-insoluble fractions (TIF) and whole-cell lysates (WCL) were harvested and analyzed with indicated antibodies. (D) SART3 promotes PCNA-mUb formation in GFP-SART3 stably expressing cells after UV irradiation. U2OS cells stably expressing either GFP or GFP-SART3 were irradiated with 15 J/m2 UVC and further incubated. The triton-insoluble fractions (TIF) were harvested and analyzed with anti-PCNA antibody. (E and F) SART3 depletion impairs UV-induced RAD18 focus formation. U2OS stable cells expressing either GFP or GFP-SART3 were transfected with siNC or siSART3 and incubated for 72 h. Then the cells were irradiated with 15 J/m2 UVC, further cultured for 4 h. (E) Representative images of cells stained with DAPI or antibody against RAD18 after UV irradiation. The cells were permeabilized with 0.5% Triton X-100 for 5min and fixed, then immunostained with anti-RAD18 antibody and mounted with DAPI. (F) Western blots showed the knockdown efficiency of siRNA in cells. (G) SART3 depletion exhibits no appreciable reduction in Polη levels. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3. Seventy-two hours later, the cells were irradiated and further incubated. Then the cell pellets were lysed by RIPA buffer and analyzed with indicated antibodies.
Given that the level of PCNA-mUb is negatively regulated by the USP1 deubiquitinase (15), we compared the levels of USP1 in SART3-depleted and control U2OS cells under the presence and absence of UV radiation. SART3 depletion did not cause appreciable alterations in USP1 expression (Figure 2C), excluding the possibility that the compromised PCNA-mUb formation in SART3-depleted cells is caused by upregulation of USP1.
The level of PCNA-mUb is also positively regulated by the RAD18-RAD6 ubiquitin ligase complex. We first compared the level of RAD18 in the control and SART3-depleted cells and found that SART3 depletion had no obvious effect on RAD18 expression (Figure 2C). Considering that PCNA-mUb occurs in a chromatin context, we further analyzed whether SART3 regulates the chromatin association of RAD18. Interestingly, the result showed that depletion of SART3 reduced the level of chromatin-associated RAD18 (Figure 2C), while expressing siRNA-resistant GFP-SART3 could rescue this reduction. We also wanted to know whether SART3 is required for RAD18 focus formation after UV irradiation. IF results showed that depletion of SART3 in GFP but not GFP-SART3 expressing cells led to a dramatic decrease in the RAD18 focus formation at 4 h after UV treatment (Figure 2E–F). These results indicated that the reduced formation of PCNA-mUb observed in SART3-depleted cells after UV treatment results from impaired RAD18 accumulation at stalled replication forks.
To rule out the possibility that SART3 depletion reduces Polη focus formation through downregulation of Polη, we knocked down the endogenous SART3 in U2OS cells stably expressing GFP-SART3 or GFP and examined the protein level of Polη with or without UVC treatment (Figure 2G). No apparent reduction in Polη level was noted after SART3 depletion. In addition, SART3 ablation attenuated UV-inducible PCNA-mUb in MRC5 cells as well as in XP30RO (Polη-deficient) cells (Supplementary Figure S2B), suggesting that SART3 regulates formation of PCNA-mUb in a Polη-independent pathway.
Collectively, these results indicate that SART3 regulates UV-induced Polη focus formation through RAD18 and PCNA-mUb-dependent fashion.
SART3 regulates UV-induced RPA focus formation and single-stranded DNA generation
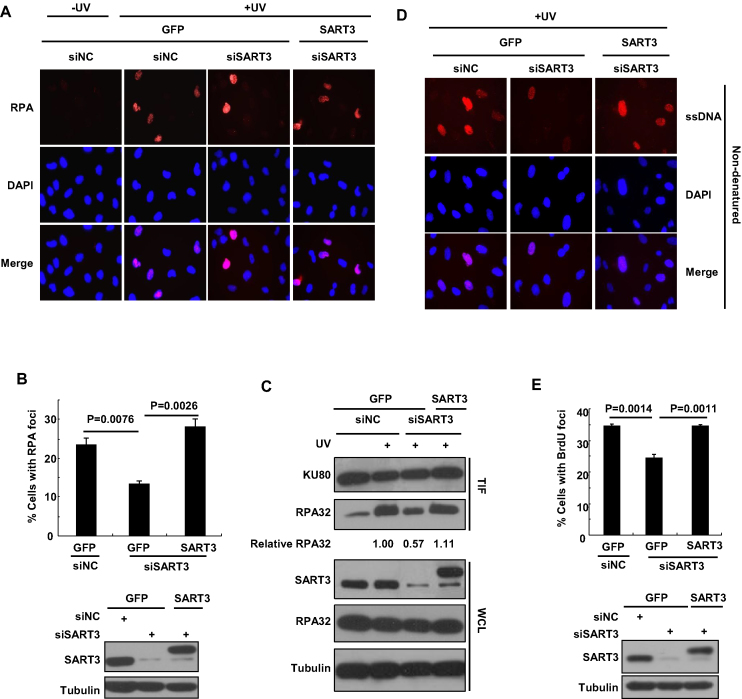
As RAD18 is recruited to chromatin through its direct interaction with RPA-coated ssDNA (35), we then tested whether SART3 depletion impairs the chromatin association of RPA. Endogenous SART3 in GFP or GFP-SART3 expressing cells was depleted and RPA focus formation as well as the extents of RPA binding to chromatin after UV exposure were compared. IF results showed that UV-induced RPA32 focus formation was dramatically decreased in SART3-depleted cells (13.3%) relative to siNC control (23.3%) (Figure 3A and B). Expression of siRNA-resistant GFP-SART3 was able to rescue the impaired RPA focus formation (28.0%). Consistently, we found that depletion of SART3 in GFP but not GFP-SART3 expressing cells reduced the level of RPA32 on chromatin (Figure 3C), while it had no effect on RPA32 expression (Figure 3C). We also pulsed labeled SART3-depleted U2OS cells with EdU (10 μM) for 1 h followed by reaction with Alexa488-azide. After checking the proportion of EdU positive cells in siNC and siSART3, we found that SART3 depletion also impaired DNA replication (Supplementary Figure S3A).
Figure 3.
SART3 regulates UV-induced RPA focus formation and single-stranded DNA generation. (A and B) SART3 ablation attenuates RPA focus formation after UV irradiation. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3 and incubated for 72 h. Then the cells were irradiated with 15 J/m2 UVC and further cultured. (A) Representative images of cells stained with DAPI or antibody against RPA32 after UV irradiation. The cells were permeabilized with 0.5% Triton X-100 for 30 min and fixed, immunostained with anti-RPA32 antibody and mounted with DAPI. (B) Quantification of the percentage of cells with RPA32 foci. The proportion of cells with more than 10 RPA32 foci were measured, at least 200 cells were counted. Data represent means ± SEM from three independent experiments. Western blots showed the knockdown efficiency in cells. (C) Depletion of SART3 reduces the level of RPA32 on chromatin. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3, and irradiated with 15 J/m2 UVC, repaired for 4 h. The triton-insoluble fractions (TIF) and whole-cell lysates (WCL) were harvested and analyzed with the indicated antibodies. (D and E) SART3 deficiency displays an obvious reduction in ssDNA formation. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3 and incubated overnight, then the cells were labeled with 10 μM BrdU for 48 h, followed by 20 J/m2 UVC irradiation, further incubated for 2 h. The cells were permeabilized with 0.5% Triton X-100 for 5 min and fixed, immunostained with anti-BrdU antibody. Representative images are shown (D). (E) The percentage of cells with more than 10 BrdU foci was quantified (at least 200 cells were counted). Data represent means ± SEM from three independent experiments. Western blots showed the knockdown efficiency of siRNA in cells.
RPA32 is known to be recruited to stalled replication forks through its avid affinity with ssDNA. We then checked whether the compromised RPA32 chromatin loading in SART3 knockdown cells results from impaired ssDNA generation after UV exposure. The cells were labeled with a thymidine analogue 5-bromo-2′-deoxyuridine (BrdU) for 48 h prior to UVC exposure. BrdU corresponding to ssDNA but not dsDNA was monitored by immunofluorescence with an anti-BrdU antibody without denaturation of DNA. The result revealed that, at 2 h post-UV, SART3-depleted cells exhibited a significant reduction in UV-induced ssDNA formation (Figure 3D and E). The percentage of BrdU-positive cells in SART3-depleted cells (24.3%) was remarkably reduced, compared with that in siNC controls (34.5%). Moreover, exogenous expression of SART3 could rescue the ssDNA formation defect (34.5%) (Figure 3D and E). We also performed a denatured immunofluorescence to show that the entire nuclear DNA was more or less evenly labeled with BrdU (Supplementary Figure S3B). These results indicate that SART3 facilitates efficient ssDNA generation after exposure to UV.
SART3 is required for optimal cellular response after UV treatment
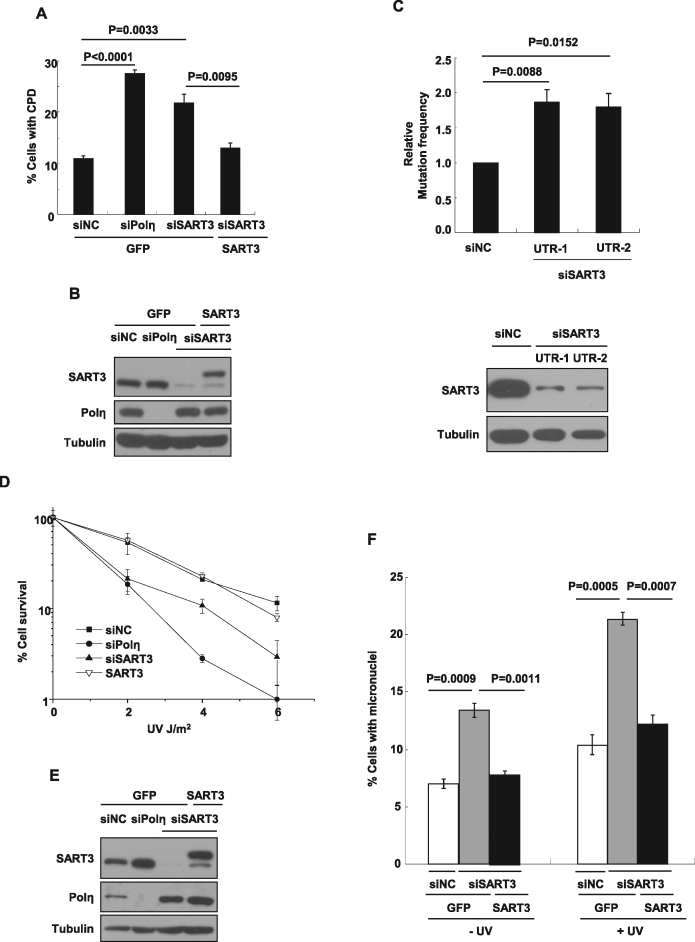
To further define the role of SART3 in UVC-induced TLS pathway, we determined the ability of SART3-depleted cells to bypass UV-induced CPD lesions as previously reported. Analogous to Polη-depleted cells (27.4%), SART3 knockdown cells (21.9%) exhibited an obviously increased CPD signal in ssDNA compared to siNC-treated control cells (10.9%) (Figure 4A and B). In line with the result that SART3 is required for optimal Polη focus formation after UV treatment, expressing siSART3-resistant GFP-SART3 in SART3 knockdown cells reversed the CPD signal in ssDNA (13.1%) close to siNC level (Figure 4A and B and Supplementary Figure S3C). Notably, equivalent amounts of CPDs were detected in denatured DNA in these cells (Supplementary Figure S3C). These results reveal that SART3 is required for gap filling opposite genomic CPD lesions.
Figure 4.
SART3 is required for optimal cellular response after UV treatment. (A and B) SART3 deficiency exhibits significantly increased levels of CPD lesions. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3, siPolη. 72 h later, the cells were irradiated with 10 J/m2 UVC, further incubated for 2 h. Then the cells were permeabilized and fixed, immunostained with anti-CPD antibody and mounted with DAPI. (A) Data represent means ± SEM from three independent experiments. More than 200 cells were counted. (B) Knockdown efficiency was checked by immunoblotting. (C) SART3 ablation increases UV-induced mutation frequency. 293T cells were transfected with siNC or siRNAs of SART3 twice. Forty-eight hours after the first transfection, cells were transfected with UVC-irradiated (400 J/m2) pSP189 reporter plasmids. 48 h later, pSP189 plasmids were extracted from 293T cells and digested with Dpn1, followed by transformed into the MBM7070 bacterial strain for mutation frequency analysis. Data represent means ± SEM from three independent experiments. Statistical analyses were performed using a two-tailed Student's t-test. Knockdown efficiency was confirmed by immunoblotting. (D and E) The depletion of SART3 exhibits hypersensitivity to UV irradiation. Cell survival assays in SART3-depleted or Polη-depleted cells after UV exposure. SiNC, siSART3 or siPolη transfected cells were irradiated with indicated dosage of UVC, which were permitted to grow for 14 days before being counted. (D) Experiments were performed in triplicate. Error bars represent standard error. (E) Knockdown efficiency was confirmed by immunoblotting. (F) Percentage of cells containing one or more micronuclei in SART3 knockdown cells significantly increases under both basal and UV irradiation conditions. U2OS cells stably expressing either GFP or GFP-SART3 were transfected with siNC or siSART3, followed by 3 J/m2 UVC exposure or not, then further incubated with 6 μg/ml CB for 48 h. Cells with micronuclei were counted. Data represent means ± SEM from three independent experiments.
Previous studies have shown that cells depleted of Polη exhibit an elevated mutation frequency (3,36). Given that depletion of SART3 abrogates Polη recruitment after UVC exposure, we speculated that SART3 depletion may also increase UV-induced genome mutagenesis. To test that, a mutagenesis assay based on a UV–irradiated shuttle vector pSP189 that carries a mutant supF suppressor tRNA33 was performed (29). As expected, the normalized mutation frequencies in SART3 depleted cells were elevated (1.87-fold increase for siUTR-1, 1.79-fold increase for siUTR-2, as compared to siNC control) (Figure 4C). We also noticed that depletion of SART3 in U2OS cells sensitized the cells to UV killing, although the extent was less than that of depletion of Polη in GFP-expressed U2OS cells (Figure 4D and E). Moreover, the SART3 depletion-derived UV hypersensitivity could be rescued by exogenously expressed siRNA-resistant SART3 (Figure 4D). To further uncover the biological significance of SART3 in the cellular response to UV irradiation, we also performed a micronucleus (MN) assay. We found that knockdown of SART3 significantly increased the proportion of cells with unperturbed MN (13.4% vs 7.03%) or MN after UV exposure (21.4% versus 10.4%), which was completely rescued by exogenously expressed siRNA-resistant SART3 (Figure 4F). These data suggest that SART3 plays an important role in preventing genomic instability in response to UV irradiation.
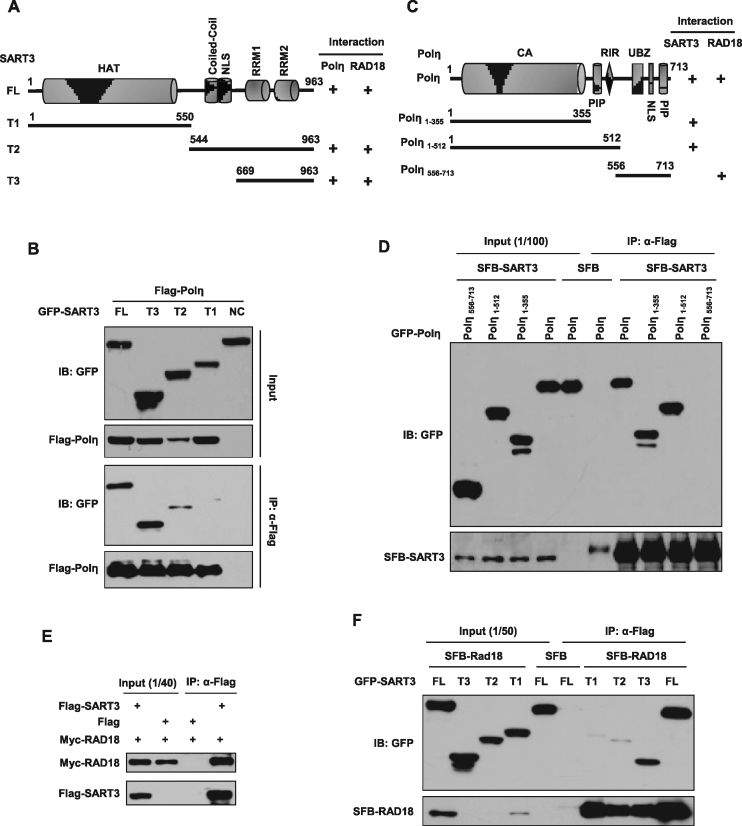
SART3 interacts with RAD18 and Polη through its RRMs
To identify the domains in SART3 responsible for its interaction with Polη, Flag-Polη and a series of GFP-SART3 truncations (Figure 5A) were transiently co-transfected into 293T cells followed by co-IP. We found that peptides containing RRMs (RNA recognition motifs) of SART3 associated with Polη (Figure 5B), indicating that the HAT domain does not appear to be the major interactor with Poln. We also examined the interactions between SFB-SART3 and GFP-Polη truncations (Figure 5C). We found that SART3 bound to the N terminus of Polη, which is distinct from the Polη-RAD18 binding region (Figure 5D). Similar result was obtained through a GST-SART3 pull down assay (Supplementary Figure S4A).
Figure 5.
SART3 interacts with RAD18 and Polη through its RRMs. (A) Schematic representation of full-length and truncations of SART3 in the following experiments. HAT: half-a-tetracopeptide repeats; NLS: nuclear localization sequences; RRM1: RNA recognition motif 1; RRM2: RNA recognition motif 2. (B) RRMs of SART3 are responsible for Polη binding. 293T cells were co-transfected with a series of GFP-SART3 truncations and Flag-Polη. Co-IP assay was performed with flag M2 beads, and detected with antibodies against GFP and Flag. (C) Schematic representation of full-length and truncations of Polη in the following experiments. CA: catalytic core; PIP: PCNA interaction peptide; RIR: Rev1 interaction region; UBZ: Ub-binding zinc finger; NLS: nuclear localization signal. (D) The N terminal of Polη mediates its interaction with SART3. 293T cells were co-transfected with a series of GFP-Polη truncations and SFB-SART3. Cell lysates were immunoprecipitated with Flag M2 beads, and detected using anti-GFP or anti-Flag antibody. (E) SART3 associates with RAD18. 293T cells were co-transfected with Flag-SART3 and Myc-RAD18, co-IP was performed using Flag M2 beads, and checked with anti-Myc or anti-Flag antibody. (F) The RRMs of SART3 are required for its binding to RAD18. 293T cells were co-transfected with a series of GFP-SART3 truncations and SFB-RAD18. Co-IP assay was performed with Flag M2 beads, and checked using antibodies against anti-GFP and anti-Flag.
Interestingly, we found that SART3 also associated with RAD18 through a co-IP assay (Figure 5E), which was further confirmed by showing that GST-SART3 bound to endogenous RAD18 but not GST protein alone (Supplementary Figure S4B). To map the domain in SART3 required for its interaction with RAD18, we co-transfected GFP-SART3 truncations and SFB-RAD18 into 293T followed by co-IP. Intriguingly, the RRMs in SART3 also bound with SFB-RAD18 (Figure 5F), suggesting that SART3 binds both RAD18 and Polη through the same domain.
SART3 promotes RAD18/Polη association through its homodimerization
Since RAD18 can associate with Polη (12), it was necessary to rule out the possibility that the interaction between SART3 and Polη is mediated by RAD18. To do this, we transfected Flag-Polη into WT or RAD18 knockout 293T cells (18) followed by co-IP experiments (Figure 6A). The result showed that the association between Flag-Polη and endogenous SART3 were comparable in these two cell lines, indicating that RAD18 is not necessary for the interaction. Meanwhile, Flag-Polη precipitated endogenous RAD18 as well (Figure 6A). We then determined whether Polη mediates the interaction between RAD18 and SART3 by transfecting SFB-RAD18 into MRC5 and XP30RO (Polη-deficient) cells followed by a co-IP assay. We found that SFB-RAD18 still associated with SART3 in the absence of Polη (Figure 6B). To further support this result, we generated a GFP-RAD18 mutant lacking the binding domain for Polη (GFP-RAD18-ΔPID). We found that although GFP-RAD18-ΔPID failed to interact with Flag-Polη (Supplementary Figure S4C), it still bound to Flag-SART3 (Supplementary Figure S4D, lane 5). These results suggest that SART3 interacts with both RAD18 and Polη independently.
Figure 6.
SART3 promotes RAD18/Polη association through its homodimerization. (A) The binding of SART3 to Polη is independent of RAD18. WT or RAD18 knockout 293T cells were transfected with Flag-Polη. Co-IP was performed using Flag M2 beads, and checked using indicated antibodies. (B) Polη is dispensable for the association between endogenous SART3 with SFB-RAD18. MRC5 and XP30RO cells were transfected with SFB-RAD18 (SFB: S-Flag-Streptavidin binding peptide) followed by an immunoprecipitation with Flag M2 beads. The immunoprecipitates were checked with the indicated antibodies. (C) The self-assembly of SART3 depends on its coiled-coil domain. 293T cells were co-transfected with GFP-SART3 or GFP-SART3-ΔCC (deletion of coiled-coil domain, residues 559–620) and Flag-SART3, co-IP was performed using Flag M2 beads. Western blot analysis was performed as indicated. (D) Flag-SART3 forms homodimers in vivo. 293T cells were transfected with Flag-SART3, and left untreated or treated with 20 J/m2 UVC, repaired for 2 h. Then the cells were crosslinked with the indicated concentrations of DSS at room temperature for 20 min. The crosslinking reaction was quenched by the addition of 1M Tris (pH 8.0) to 100 mM final concentration for 20 min and sonicated at 4 °C in a water bath sonicator. The products were separated by 3–8% Tris-Acetate Protein Gels along with the HiMark Pre-Stained Protein Standard molecular mass marker followed by immunoblotting with an anti-Flag antibody. Tubulin: loading control. (E) SART3 is essential for the binding of RAD18 to Polη. 293T cells were transfected with siNC or siSART3, followed by transfection with Flag-Polη. 48 h after transfection, co-IP was performed using Flag M2 beads, and detected with indicated antibodies. NS: non-specific band. (F) SART3 homo-dimerization enhances RAD18/Polη association. The indicated plasmids were transfected into 293T cells, co-IP was performed using Flag M2 beads and analyzed with indicated antibodies. (G) SART3 homodimerization elevates the level of PCNA-mUb. U2OS cells were transfected with the indicated plasmids, irradiated with 15 J/m2 UVC, further cultured. The triton-insoluble fractions (TIF) and whole-cell lysates (WCL) were harvested and checked with indicated antibodies.
Given that SART3 interacts with Polη and RAD18 through the same RRMs and the coiled-coil domain is one of the principal subunit oligomerization motifs in proteins (37), we wondered whether SART3 can interact with itself through its coiled-coil domain, then promote Polη and RAD18 association through forming a complex. To test this hypothesis, we first co-transfected 293T cells with GFP-SART3 and Flag-SART3 followed by immunoprecipitation with anti-Flag antibodies. The result showed that GFP-SART3 but not GFP-SART3-ΔCC (deletion of the coiled-coil domain) could be efficiently co-precipitated (Figure 6C). To further confirm that, 293T cells transfected with Flag-SART3 were treated with different concentrations of a crosslinking reagent. We found that Flag-SART3 could form homodimers in vivo, which was not affected by UVC exposure (Figure 6D). These data indicate that SART3 can form homodimer mediated via its coiled-coil domain.
We then determined whether SART3 enhances the association between Polη and RAD18. We found that depletion of SART3 in 293T cells significantly impaired the interaction between Flag-Polη and endogenous RAD18 (Figure 6E). In addition, overexpression of GFP-SART3, which could form homodimers, enhanced the interaction between Flag-Polη and RAD18 (Figure 6F). In contrast, overexpression of GFP-SART3-ΔCC showed no stimulatory effect (Figure 6F), although it still localized in the nucleus (Supplementary Figure S4E). Considering that Polη/RAD18 interaction plays a key role in targeting RAD18 to PCNA and promoting formation of PCNA-mUb at stalled replication forks (17), we isolated the chromatin fractions to examine whether SART3 homodimerization is vital for PCNA-mUb as well. As shown in Figure 6G, the overexpression of GFP-SART3, instead of GFP or GFP-SART3-ΔCC, promotes formation of PCNA-mUb. Notably, GFP-SART3-ΔCC manifested a dramatically reduced chromatin association (Figure 6G).
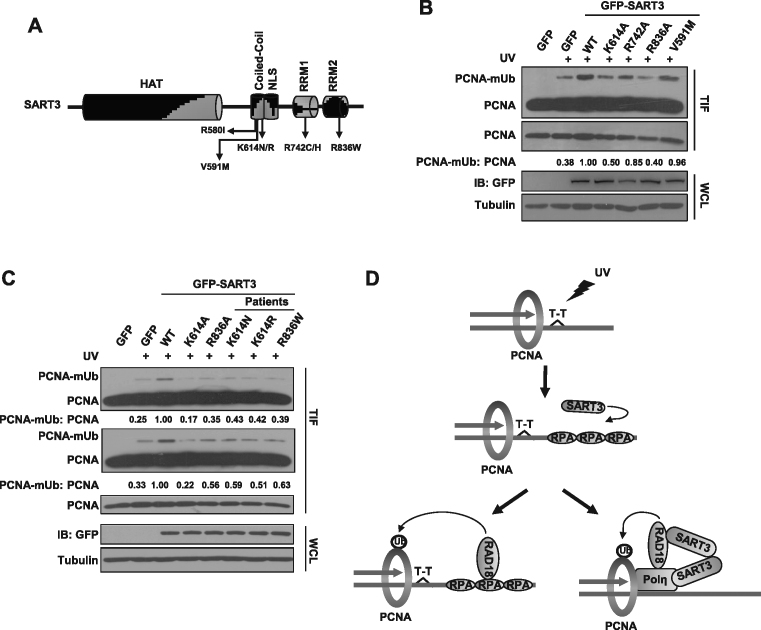
Cancer-associated SART3 mutants affect TLS
Given that TLS is important for genome stability and cancer progression, we wondered whether SART3 mutants identified in patients affect the TLS process. SART3-V591M (Valine591 is mutated to Methionine) is a SART3 missense mutation found in a Chinese pedigree with disseminated superficial actinic porokeratosis (DSAP) (38). When complemented into SART3-depleted cells, both WT and SART3-V591M were able to restore Polη focus formation and PCNA-mUb formation in SART3 depleted cells (Supplementary Figure S5A-C), suggesting that SART3-V591M does not lose its ability to activate TLS. Through exploring the cBioPortal database (http://www.cbioportal.org/index.do) (39,40), several missense mutations with repeated incidence in SART3 coiled-coil and RRM domains were identified in multiple cancers, including melanoma, cervical squamous cell carcinoma and breast cancer (Figure 7A, Table 1). We first constructed several GFP-SART3 mutants in which the target amino acids were mutated into alanine (K614A, R742A and R836A) and transfected them into U2OS cells. In contrast to WT and SART3-V591M, K614A and R836A mutants were less efficient in promoting PCNA-mUb formation (Figure 7B), while R742A mutation did not obviously compromise SART3 ability to stimulate PCNA-mUb formation (Figure 7B). Then we generated several mutants as are the cases in patients, including K614N, K614R and R836W. We found that they displayed lower efficiency in promoting PCNA-mUb formation than that of WT (Figure 7C). We found that the mutations at K614 (K614N, K614R and K614A) did not impair SART3 homodimerization (Supplementary Figure S6A). However, K614N, K614R and R836W mutations significantly compromised SART3 ability to stimulate RAD18/Polη association (Supplementary Figure S6B). We further found that K614N, K614R and R836W mutations also impaired the ability of SART3 binding to chromatin after UV irradiation (Supplementary Figure S6C), indicating that SART3 chromatin binding is likely a prerequisite for its effect on RAD18/Polη association. We also noticed that mutation of R580I did not impair SART3 homodimerization as well as its chromatin association (Supplementary Figures S6D and S6E). Taken together, the ability to promote PCNA-mUb formation after UV was impaired in several SART3 missense mutants of coiled-coil and RRMs domains, hinting that the dysfunction of SART3 mutants in promoting RAD18/Polη association and activating TLS may contribute to genome instability and cancer progression. Notably, taking all the SART3 coding mutations into comparison, we found that the mutation burden of cancer patients with SART3 mutants was higher than those with WT SART3 (Supplementary Figure S7).
Figure 7.
Cancer-associated SART3 mutants affect TLS. (A) Schematic representation of SART3 mutants with repeated incidence found in tumor samples. (B) Two SART3 mutants exhibit lower efficiency in promoting PCNA-mUb. U2OS cells were transfected with a series of GFP-SART3 mutants (targeted amino acids were mutated into alanine), then the cells were treated with 15 J/m2 UVC and incubated for 4 h. The triton-insoluble fractions (TIF) and whole-cell lysates (WCL) were harvested and checked with antibodies against PCNA, GFP and Tubulin. Tubulin: loading control. Levels of PCNA and PCNA-mUb were quantified using ImageJ. (C) Several SART3 mutants with repeated incidence from cancer patients manifest impaired PCNA-mUb formation as compared to that of WT. U2OS cells were transfected with a series of GFP-SART3 mutants (found in patients), then treated with 15 J/m2 UVC and further cultured. The triton-insoluble fractions (TIF) and whole-cell lysates (WCL) were harvested and separated by SDS-PAGE. Levels of PCNA and PCNA-mUb were quantified using Image J. (D) The working model of how SART3 regulates TLS after UV irradiation. SART3 functions in TLS at dual mode. On the one hand, SART3 facilitates ssDNA generation followed by promoting RPA and RAD18 recruitment. On the other hand, SART3 enhances RAD18/Polη interaction via its homodimerization, which synergistically promotes PCNA-mUb formation, modulating TLS.
Table 1. SART3 missense mutations recurrently identified in cancer patients.
| Sample ID | Cancer Study | AA change | Chr | Position | Ref | Var | Location |
|---|---|---|---|---|---|---|---|
| TCGA-JW-A5VL-01 | Cervical | K614N | chr12 | 1.09E+08 | C | G | CC |
| TCGA-HU-A4HB-01 | Stomach | K614R | chr12 | 1.09E+08 | T | C | CC |
| TCGA-AN-A046-01 | Breast | R580I | chr12 | 1.09E+08 | C | A | CC |
| TCGA-AX-A0J0-01 | Uterine | R580I | chr12 | 1.09E+08 | C | A | CC |
| coadread_dfci_2016_3523 | Colorectal | R742C | chr12 | 1.09E+08 | G | A | RRM1 |
| TCGA-AA-A010-01 | Colorectal | R742H | chr12 | 1.09E+08 | C | T | RRM1 |
| DLBCL-Ls1899 | DLBCL | R836W | chr12 | 1.09E+08 | G | A | RRM2 |
| TCGA-FW-A3R5-06 | Melanoma | R836W | chr12 | 1.09E+08 | G | A | RRM2 |
DISCUSSION
Polη/RAD18 physical interaction plays very important roles in TLS regulation. On one hand, RAD18 can guide Polη to stalled replication sites (12), and on the other hand, Polη has been shown to bridge RAD18 and PCNA to promote efficient PCNA-mUb formation after DNA damage (17). Therefore, it seems that RAD18 and Polη play mutually dependent roles in TLS pathway activation through their association. Given their interaction confers cellular resistance to UV killing and regulates genome mutagenesis (17,41). It is necessary to explore how Polη/RAD18 interaction is regulated in vivo.
SART3 was previously regarded as a pre-mRNA splicing factor, which can promote the formation of the U4/U6 di-snRNP to regulate pre-mRNA splicing and gene expression (42–44). In this study, we have provided several lines of evidence to show that SART3 is a critical regulator of Polη/RAD18 interaction and PCNA-mUb formation in response to UV damage. First, SART3 physically interacts with Polη and RAD18. Second, depletion of SART3 significantly impairs UV-induced Polη and RAD18 focus formation. Third, SART3 depletion remarkably decreases PCNA-mUb and RPA focus formation after UV irradiation. Fourth, cells depleted of SART3 displays a defective TLS efficiency, an increased genome instability and hypersensitivity in response UV treatment. Finally, SART3 promotes Polη/RAD18 association through its dimerization. Notably, the novel role of SART3 in TLS is independent of its RNA binding activity. Hence, our data suggest that SART3 not only promotes the generation of ssDNAs at stalled replication forks to facilitate RPA focus formation, but also promotes Polη/RAD18 association, which synergistically enhances PCNA-mUb and Polη focus formation after UV irradiation, modulating TLS process (Figure 7D).
The TLS pathway is known to be efficiently triggered by replication stress, which leads to uncoupling of replicative polymerase and helicase activities, and generating stretches of ssDNA (45). However, how ssDNA generation is regulated remains largely unclear. Here, we found that SART3 regulates ssDNA generation after UV irradiation. Considering that SART3 can function as a histone chaperone to associate with USP15 for the H2B deubiquitination (46), while the absence of H2BK123 ubiquitination (H2Bub1) in yeast leads to an ‘open’ chromatin structure (47), it is plausible that SART3 might modulate ssDNA generation by regulating chromatin dynamics. In support of it, H2Bub1-deficiency cells accumulate unrepaired DNA lesions and/or replication intermediates enriched with RPA foci indicative of ssDNA gaps (48).
We found that SART3 could form homodimers in vivo, which is consistent with a recent report (49). In addition, we noticed that SART3 interacts with both RAD18 and Polη. Interestingly, we found that depletion of SART3 attenuates the association of RAD18 with Polη, indicating that, through dimerization, SART3 is required for enhanced RAD18/Polη interaction. Moreover, based on the fact that K614N and K614R mutations compromised SART3 abilities in chromatin binding and in promoting RAD18/Polη association but not in SART3 dimerization, it is likely that SART3 chromatin binding is a prerequisite for its stimulatory effect on RAD18/Polη association. This mode of regulation of SART3 on RAD18/Polη interaction is distinct from that of Cdc7, which was reported to function via phosphorylation of RAD18 (23). Moreover, unlike SART3, Cdc7 depletion has no effect on PCNA-mUb formation.
SART3 has been implicated as a candidate gene in DSAP, which is an uncommon autosomal dominant chronic disorder of keratinization and develops on sun-exposed areas of skin (38). We found that V591M mutation does not affect UV-induced PCNA-mUb and Polη focus formation, suggesting that the potential DSAP-causing mutation does not function through regulation of TLS to contribute to this disease. We also found that several SART3 missense mutations in SART3 coiled-coil and RRMs with repeated incidence detected in cancer patients impair their ability to promote UV-induced PCNA-mUb formation. Notably, the mutation burden of cancer patients harboring SART3 coding mutations was higher than patients with WT SART3, hinting that other Polη-independent error-prone DDR pathways might be responsible for it.
In conclusion, our work identified a novel role of SART3 in TLS regulation. Interestingly, SART3 was found upregulated in cisplatin resistant ovarian cancer cell lines compared with the parental cell lines (P = 0.007 and 0.04, respectively) in two datasets (GSE45553 and GSE43694). Given that Polη can protect tumor cells from cisplatin treatment (50), the function of SART3 in these tumor cells might also contribute to cisplatin chemoresistance. Additional studies of the functions of SART3 in DDR should yield a greater insight into its role in governing genome stability.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Jiahuai Han, Alan Lehmann, Jianguo Ji and Jun Huang for reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Nature Science Foundation of China [81630078]; CAS Strategic Priority Research Program [XDA16010107]; Natural Science Foundation of Beijing [5181001]; National Key Research and Development Program of China [2017YFC1001001]; National Nature Science Foundation of China [91754204, 31570816, 31471331, 31670822, 31701227 and 31470784], CAS Strategic Priority Research Program [XDB14030300]. State Key Laboratory of Membrane Biology and CAS Key Laboratory of Genomic and Precision Medicine. Funding for open access charge: National Nature Science Foundation of China [81630078, 91754204].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ghosal G., Chen J.. DNA damage tolerance: a double-edged sword guarding the genome. Transl. Cancer Res. 2013; 2:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sale J.E., Lehmann A.R., Woodgate R.. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012; 13:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedberg E.C. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005; 6:943–953. [DOI] [PubMed] [Google Scholar]
- 5. Guo C., Kosarek-Stancel J.N., Tang T.-S., Friedberg E.C.. Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 2009; 66:2363–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y., Yuan F., Wu X., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z.. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000; 28:4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z.. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000; 28:4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lehmann A.R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J.F., Kannouche P.L., Green C.M.. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007; 6:891–899. [DOI] [PubMed] [Google Scholar]
- 9. Busuttil R.A., Lin Q., Stambrook P.J., Kucherlapati R., Vijg J.. Mutation frequencies and spectra in DNA polymerase eta-deficient mice. Cancer Res. 2008; 68:2081–2084. [DOI] [PubMed] [Google Scholar]
- 10. Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase eta. Biochemistry. 2014; 53:2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S.. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002; 419:135–141. [DOI] [PubMed] [Google Scholar]
- 12. Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M.. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004; 23:3886–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stelter P., Ulrich H.D.. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003; 425:188–191. [DOI] [PubMed] [Google Scholar]
- 14. Lehmann A.R. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 2011; 585:2772–2779. [DOI] [PubMed] [Google Scholar]
- 15. Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D.. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006; 8:339–347. [DOI] [PubMed] [Google Scholar]
- 16. Lv L., Wang F., Ma X., Yang Y., Wang Z., Liu H., Li X., Liu Z., Zhang T., Huang M. et al. . Mismatch repair protein MSH2 regulates translesion DNA synthesis following exposure of cells to UV radiation. Nucleic Acids Res. 2013; 41:10312–10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durando M., Tateishi S., Vaziri C.. A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013; 41:3079–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z., Huang M., Ma X., Li H., Tang T., Guo C.. REV1 promotes PCNA monoubiquitylation through interacting with ubiquitylated RAD18. J. Cell Sci. 2016; 129:1223–1233. [DOI] [PubMed] [Google Scholar]
- 19. Pathania S., Nguyen J., Hill S.J., Scully R., Adelmant G.O., Marto J.A., Feunteun J., Livingston D.M.. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell. 2011; 44:235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu X., Ma X., Tu Y., Huang M., Liu H., Wang F., Gong J., Wang J., Li X., Chen Q. et al. . Parkin regulates translesion DNA synthesis in response to UV radiation. Oncotarget. 2017; 8:36423–36437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedberg E.C. Reversible monoubiquitination of PCNA: A novel slant on regulating translesion DNA synthesis. Mol. Cell. 2006; 22:150–152. [DOI] [PubMed] [Google Scholar]
- 22. Barkley L.R., Palle K., Durando M., Day T.A., Gurkar A., Kakusho N., Li J., Masai H., Vaziri C.. c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Poleta recruitment to stalled replication forks. Mol Biol Cell. 2012; 23:1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Day T.A., Palle K., Barkley L.R., Kakusho N., Zou Y., Tateishi S., Verreault A., Masai H., Vaziri C.. Phosphorylated Rad18 directs DNA polymerase eta to sites of stalled replication. The J. Cell Biol. 2010; 191:953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whitmill A., Timani K.A., Liu Y., He J.J.. Tip110: Physical properties, primary structure, and biological functions. Life Sci. 2016; 149:79–95. [DOI] [PubMed] [Google Scholar]
- 25. Trede N.S., Medenbach J., Damianov A., Hung L.H., Weber G.J., Paw B.H., Zhou Y., Hersey C., Zapata A., Keefe M. et al. . Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:6608–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Timani K.A., Liu Y., Suvannasankha A., He J.J.. Regulation of ubiquitin-proteasome system-mediated Tip110 protein degradation by USP15. Int. J. Biochem. Cell Biol. 2014; 54:10–19. [DOI] [PubMed] [Google Scholar]
- 27. Yang D., Nakao M., Shichijo S., Sasatomi T., Takasu H., Matsumoto H., Mori K., Hayashi A., Yamana H., Shirouzu K. et al. . Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res. 1999; 59:4056–4063. [PubMed] [Google Scholar]
- 28. Parris C.N., Levy D.D., Jessee J., Seidman M.M.. Proximal and distal effects of sequence context on ultraviolet mutational hotspots in a shuttle vector replicated in xeroderma cells. J. Mol. Biol. 1994; 236:491–502. [DOI] [PubMed] [Google Scholar]
- 29. Parris C.N., Seidman M.M.. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992; 117:1–5. [DOI] [PubMed] [Google Scholar]
- 30. Mansilla S.F., Soria G., Vallerga M.B., Habif M., Martinez-Lopez W., Prives C., Gottifredi V.. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res. 2013; 41:6942–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Temviriyanukul P., van Hees-Stuivenberg S., Delbos F., Jacobs H., de Wind N., Jansen J.G.. Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome. DNA Repair. 2012; 11:550–558. [DOI] [PubMed] [Google Scholar]
- 32. Adam S., Polo S.E., Almouzni G.. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013; 155:94–106. [DOI] [PubMed] [Google Scholar]
- 33. Navaraj A., Mori T., El-Deiry W.S.. Cooperation between BRCA1 and p53 in repair of cyclobutane pyrimidine dimers. Cancer Biol. Ther. 2014; 4:1409–1414. [DOI] [PubMed] [Google Scholar]
- 34. Ma X., Liu H., Li J., Wang Y., Ding Y.H., Shen H., Yang Y., Sun C., Huang M., Tu Y. et al. . Poleta O-GlcNAcylation governs genome integrity during translesion DNA synthesis. Nat. Commun. 2017; 8:1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davies A.A., Huttner D., Daigaku Y., Chen S., Ulrich H.D.. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell. 2008; 29:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maher V.M., Ouellette L.M., Curren R.D., McCormick J.J.. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976; 261:593–595. [DOI] [PubMed] [Google Scholar]
- 37. Burkhard P., Stetefeld J., Strelkov S.V.. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001; 11:82–88. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Z.H., Niu Z.M., Yuan W.T., Zhao J.J., Jiang F.X., Zhang J., Chai B., Cui F., Chen W., Lian C.H. et al. . A mutation in SART3 gene in a Chinese pedigree with disseminated superficial actinic porokeratosis. Br. J. Dermatol. 2005; 152:658–663. [DOI] [PubMed] [Google Scholar]
- 39. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ito W., Yokoi M., Sakayoshi N., Sakurai Y., Akagi J., Mitani H., Hanaoka F.. Stalled Poleta at its cognate substrate initiates an alternative translesion synthesis pathway via interaction with REV1. Genes Cells. 2012; 17:98–108. [DOI] [PubMed] [Google Scholar]
- 42. Harada K., Yamada A., Yang D., Itoh K., Shichijo S.. Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int. J. Cancer. 2001; 93:623–628. [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., Li J., Kim B.O., Pace B.S., He J.J.. HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J. Biol. Chem. 2002; 277:23854–23863. [DOI] [PubMed] [Google Scholar]
- 44. Liu Y., Liu J., Wang Z., He J.J.. Tip110 binding to U6 small nuclear RNA and its participation in pre-mRNA splicing. Cell Biosci. 2015; 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A.. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005; 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Long L., Thelen J.P., Furgason M., Haj-Yahya M., Brik A., Cheng D., Peng J., Yao T.. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 2014; 289:8916–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chandrasekharan M.B., Huang F., Sun Z.W.. Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010; 5:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hung S.H., Wong R.P., Ulrich H.D., Kao C.F.. Monoubiquitylation of histone H2B contributes to the bypass of DNA damage during and after DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2205–E2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park J.K., Das T., Song E.J., Kim E.E.. Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res. 2016; 44:5424–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Srivastava A.K., Han C., Zhao R., Cui T., Dai Y., Mao C., Zhao W., Zhang X., Yu J., Wang Q.E.. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4411–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.