Abstract
Background
While recent genome-wide association studies have suggested novel low-grade glioma (LGG) stratification models based on a molecular classification, we explored the potential clinical utility of patient-derived cells. Specifically, we assayed glioma-associated stem cells (GASC) that are patient-derived and representative of the glioma microenvironment.
Methods
By next-generation sequencing, we analyzed the transcriptional profile of GASC derived from patients who underwent anaplastic transformation either within 48 months (GASC-BAD) or ≥7 years (GASC-GOOD) after surgery. Gene set enrichment and pathway enrichment analyses were applied. The prognostic role of a nuclear factor-kappaB (NF-κB) signature derived from GASC-BAD was tested in 530 newly diagnosed diffuse LGG patients comprised within The Cancer Genome Atlas (TCGA) database. The prognostic value of the GASC upstream regulator p65 NF-κB was assessed, by univariate and multivariate Cox analyses, in a single center case study, including 146 grade II LGGs.
Results
The key elements differentiating the transcriptome of GASC isolated from LGG with different prognoses were mostly related to hallmarks of cancer (eg, inflammatory/immune process, NF-κB activation). Consistently, the NF-κB signature extrapolated from the GASC study was prognostic in the dataset of TCGA. Finally, the nuclear expression of the NF-kB-p65 protein, assessed using an inexpensive immunohistochemical method, was an independent predictor of both overall survival and malignant progression-free survival in 146 grade II LGGs.
Conclusion
This study demonstrates for the first time the independent prognostic role of NF-kB activation in LGG and outlines the role of patient-based stem cell models as a tool for precision medicine approaches.
Keywords: glioma-associated stem cells, low-grade glioma prognosis, p65/NF-κB, precision medicine, tumor microenvironment
Importance of the study
Besides assessing the role of NF-κB in low-grade gliomas, this work supports the use of patient-derived cells representative of the tumor microenvironment as a tool for a precision medicine approach. The optimized procedure can be further extended to select novel biomarkers and suggest therapies aimed at targeting the tumor microenvironment.
Gliomas are the most common malignant primary tumors of the central nervous system1 and are classified, according to the World Health Organization (WHO),2 into high grade (grades III–IV) and low grade (grades I–II). WHO grade II glioma is a paradigmatic example of a tumor whose clinical management is challenging3; despite a better prognosis than high-grade glioma (HGG), a complete surgical resection is limited for functional reasons and ≈70% of patients undergo anaplastic transformation within 10 years.4,5 Moreover, the timing of progression is heterogeneous, and definitive criteria for the use of adjuvant chemo- or radiotherapies are still missing. In fact, adjuvant treatments are burdened by important side effects, such as the risk of late cognitive defects following radiotherapy, limiting their employment in patients with long life expectancy.
Precision medicine (PM) aims at tailoring medical treatments, evaluating individual differences in gene, environmental, and lifestyle characteristics.6 Up to now, genomic analyses have been the most used approach, which in the case of low-grade glioma (LGG), has suggested the superior predictive value of molecular classifications over histology.7–9
In addition to genomics, the PM initiative aims at developing new models of human cancer, including human cancer cell lines.10 These latter are instrumental to gain new insights into tumor biology and to better predict the response of patients to treatments.
Therefore, we employed human adult stem cells isolated from gliomas as an approach to PM.11 Specifically, we expanded in vitro a population of glioma-resident stem cells, or glioma-associated stem cells (GASC).11 These, differently from glioma stem cells, are not tumor-initiating cells and are devoid of the genetic alterations characterizing the tumor of origin, but show the ability to support the growth of tumor cells.11 For these reasons, we considered GASC as a model of the activated tumor microenvironment (TME). Indeed, the TME is fundamental in the multistep development of human tumors by modulating the acquisition of the hallmarks of cancer (eg, tumor-promoting inflammation, immune evasion, invasion).12
In this work, we employed the patient-derived in vitro model of the glioma TME for the identification of novel LGG prognostic biomarkers that have been validated at tissue level. Specifically, (i) we compared, by means of next-generation sequencing (NGS), the gene expression profile of GASC isolated from LGG characterized by good prognosis with that of GASC isolated from LGG characterized by a rapid anaplastic transformation; (ii) we showed that a signature from nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) extrapolated from GASC was prognostic in a dataset of 530 newly diagnosed diffuse LGG patients comprised within The Cancer Genome Atlas (TCGA); finally, (iii) we assessed the independent prognostic role of the nuclear expression of the p65/v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA) protein in a large single center case study including 146 grade II LGG tissues (Fig. 1A).
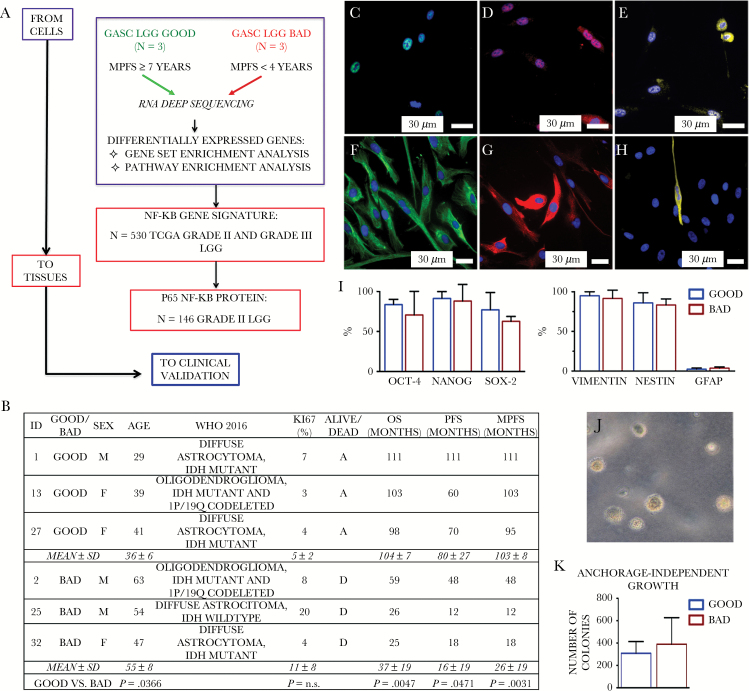
Fig. 1.
(A) Study design. (B) Clinical-pathological features of LGG-bad patients included in the study. Table reports clinical data, histopathological diagnosis, Ki67 expression, and patient prognosis, referred to 3 LGGs with a good prognosis (GOOD) and 3 LGGs with a bad prognosis (BAD). F = female; M = male; D = dead; A = alive; n.s. = not significant. (C–I) GASC phenotype. Representative fluorescence images of the expression of Oct-4 (green, C), Nanog (red, D), Sox2 (yellow, E), vimentin (green, F), nestin (red, G), and glial fibrillary acidic protein (yellow, H) in a GASC line obtained from a patient characterized by a good prognosis. The blue of 4′,6′-diamidino-2-phenylindole identifies nuclei. (I) Quantitative analysis of the fraction of cells expressing the assayed pluripotent state specific transcription factors and intermediate filaments, as assessed by immunofluorescence. Data are presented as mean ± SD. No significant differences between GOOD- and BAD-GASC were detected. (J–K) Anchorage-independent growth of GASC. (J) Representative phase contrast images of colonies acquired 4 weeks after seeding in soft agar. (K) Data are presented as mean ± SD. No significant differences between GOOD- and BAD-GASC were detected.
Materials and Methods
An extended Materials and Methods section is available in the Supplementary materials online.
GASC Isolation and Characterization
The research was approved by the local ethics committee (Consents 102/2011/Sper-196/2014/Em). Written informed consents were obtained from patients. Clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki. GASC were isolated and characterized as previously published.11
GASC RNA Extraction, Library Preparation, and Sequencing
We analyzed GASC lines derived from patients who underwent anaplastic transformation either within 48 months (GASC-BAD; n = 3) or ≥7 years (GASC-GOOD; n = 3) after surgery. RNA was extracted using the Trizol reagent (Invitrogen). Libraries were prepared by TruSeq mRNA Sample Prep kit (Illumina), processed with Illumina cBot for cluster generation on the flowcell, and sequenced on single-end 50 base mode on HiSeq2500 (Illumina) generating 30 M of reads/sample. Cufflinks13 and Cuffdiff14 were used to evaluate gene expression and pairwise differential expression, respectively. Raw data, corresponding to the gene expression profiles, are available in the Gene Expression Omnibus database.
Functional and Gene Set Enrichment Analysis
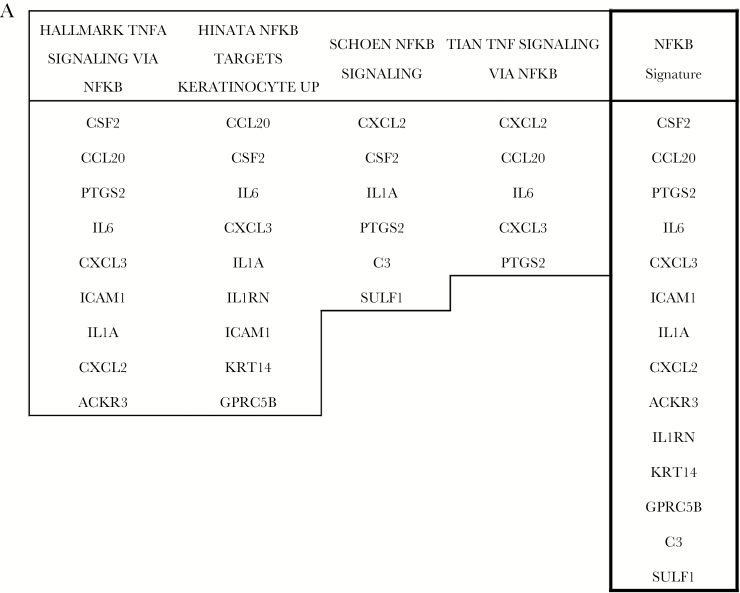
Differentially expressed genes were analyzed using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com). The prediction of the transcription factors and regulative molecules was obtained using the Upstream regulators function (IPA suite). For every upstream regulator an overlap P-value and a z-score were calculated: the P-value indicates the significance based on the overlap between dataset genes and known targets regulated by the molecule, while the z-score is used to infer the possible activation (z-score >2) or inhibition (z-score <−2) of the molecule based on prior knowledge stored in the proprietary Ingenuity Knowledge Base. The enrichment for gene signatures was performed using Gene Set Enrichment Analysis15 interrogating the hallmark gene sets database and the chemical and genetic perturbations database. The NF-κB gene signature was obtained selecting the 14 differentially expressed genes included in 4 gene sets related to NF-κB.
TCGA Data Analysis
Expression data and survival information of 530 LGG samples were obtained using the cgdsr16 package for R/Bioconductor. Tumor subtype and mutational information were obtained from.8 Patients were stratified based on the expression levels of the proposed NF-κB signature using the median value as cutoff. The 2 cohorts of patients were compared by Kaplan–Meier survival plot.
Single Center LGG Tissue Analyses
One hundred and forty-six patients, not previously chemo- and/or radiotreated, who underwent a surgical resection of a newly diagnosed LGG at the Neurosurgery Department of Udine were analyzed. Extensive surgical resection was done at diagnosis, and radio- and/or chemotherapy was administered in case of tumor progression. Histological examination, immunohistochemistry, fluorescence in situ hybridization, and analysis of the genetic status of O6-methylguanine-DNA methyltransferase (MGMT) promoter and isocitrate dehydrogenase 1 and 2 (IDH1/2) genes were performed, with minor modifications.11
Tissue microarray (TMA)
Fourteen TMAs have been constructed incorporating glioma by using the Galileo CK4500 instrument. Three cores of 1 mm were collected from each block. TMAs were stained by immunohistochemistry for the NF-κB-p65 subunit (Abcam) and alpha thalassemia/mental retardation syndrome X-linked protein (ATRX) (Sigma) and digitally scanned with an Aperio CS2 (Leica Biosystems) at a magnification of 40x, keeping constant the acquisition parameters. Total expression of NF-κB-p65 was analyzed by ImageJ v1.44 (National Institutes of Health) evaluating the optical density per unit area of each sample core and calculating, for each patient, the average value. Once set, the threshold for ImageJ analysis was maintained constant for all samples. ImageJ processing and analyses were performed by 2 blinded investigators. Nuclear scoring and cytoplasmic expression of p65 were done by a semiquantitative approach (Histoscore method).17 Briefly, the score values (ranging from 0 to 300) were calculated by multiplying the percentage of labeled cytoplasms or nuclei by the staining intensity (1 = low, 2 = mid, or 3 = high). Cores were scored by 2 experienced researchers in a blind manner. For each tumor, the average value of the respective cores was calculated.
Volumetric analysis
Pre- and postoperative tumor volume, extent of resection (EOR), and values of the preoperative difference between T2- and T1-weighted MR images (ΔVT2T1) were calculated as previously described.5
Statistical Analysis
Study population was described using standard methods. Overall survival (OS), progression-free survival (PFS), and malignant PFS (MPFS) were defined as the time between initial surgery and, respectively, death (OS), demonstration of increase in tumor size on follow-up imaging, malignant progression, and/or death (PFS), and demonstration of gadolinium enhancement on follow-up imaging and/or higher-grade tumor on subsequent biopsy or death (MPFS). OS, PFS, and MPFS were described using the Kaplan–Meier approach. Survival was analyzed by Cox proportional hazards models. Covariates with P < 0.1 at univariable analysis were selected for multivariable stepwise analysis. Analyses were conducted with Stata/SE 14.1 for Mac software and an R software environment for statistical calculation.
Results
Frozen GASC Obtained from LGG Patients Retain Their Properties
Fig. 1B summarizes the clinicopathological features of the 6 LGG patients whose GASC underwent transcriptional analysis. Noteworthy, the “bad prognosis” group did not include only IDH1/2 wild-type gliomas, known to be characterized by a poor prognosis.7–9
Since selected GASC were stored in liquid nitrogen for ≈10 years, we first verified if they retained key GASC properties. As previously shown, all cell lines displayed a rather homogeneous mesenchymal stem cell immunophenotype when analyzed by flow cytometry, being mainly positive for cluster of differentiation (CD)13, CD29, CD44, CD49a, CD49d, CD59, CD73, CD90, CD105, and N-cadherin, while hematopoietic markers, such as CD45, CD34, CD38, CD14, and human leukocyte antigen‒D related were expressed in a small fraction of cells, usually less than 1%; similarly, the glioma stem cell marker CD133 was present in less than 0.5% of the cells (Supplementary Figure S1). All cell lines expressed pluripotent state specific transcription factors (ie, octamer-binding transcription factor 4 [Oct-4], Nanog, and sex determining region Y–box 2 [Sox2]) and early intermediate filaments (ie, nestin and vimentin), while glial fibrillary acidic protein was scarcely represented (Fig. 1C–I). All cells were multipotent, being able to differentiate into glial, oligodendroglial, and neuron-like cells (Supplementary Figure S2), and all cells grew in an anchorage-independent way (Fig. 1J–K).
As a whole, GASC were confirmed to possess an undifferentiated phenotype and aberrant growth properties.
A Signature Comprising Different Hallmarks of Cancer Characterizes GASC Isolated from LGG with a Bad Prognosis
The gene expression profile of GASC was evaluated by NGS. Supplementary Table S1 displays the 82 genes differently expressed between GASC from LGG with different prognoses. Functionally, GASC-BAD were characterized by an upregulation of genes involved in the inflammatory response and in the communication between innate and adaptive immune cells, and a deregulation of pathways involved in metabolism (Supplementary Table S2), features known to be related to the hallmarks of cancer.12 Accordingly, the upstream regulators analysis identified, as molecules potentially responsible for the transcriptional program observed in GASC-BAD, several members of the interleukin (IL) family (IL-1A, IL-1B, IL-4, IL-6, IL-17, IL-4, IL-13), tumor necrosis factor (TNF), interferon-γ (IFNG), as well as elements of NF-κB signaling (eg, RELA, NF-κB1, inhibitor of nuclear factor kappa B kinase subunit beta [IKBKB]) (Table 1 and Supplementary Table S3). Interestingly, among the list of molecules potentially able to interfere with the transcriptional program characterizing GASC-BAD could be found anti-inflammatory drugs (eg, cyclosporine A, glucocorticoid, dexamethasone, salicylic acid, tacrolimus, bortezomib, simvastatin), food supplements (eg, curcumin, resveratrol), and NF-κB inhibitors (Sn50 peptide and pyrrolidine dithiocarbamate) (Table 1).
Table 1.
Upstream regulators analysis*
| Upstream Regulator | Molecule Type | Activation z-Score | P-value of Overlap |
|---|---|---|---|
| IL-1A | Cytokine | 3.902 | 1.44E-17 |
| IL-1B | Cytokine | 4.863 | 1.48E-17 |
| TNF | Cytokine | 4.845 | 6.42E-16 |
| RELA | Transcription regulator | 2.922 | 4.82E-14 |
| IFNG | Cytokine | 3.713 | 1.54E-13 |
| TLR5 | Transmembrane receptor | 2.767 | 4.02E-13 |
| NF-κB (complex) | Complex | 3.509 | 5.47E-13 |
| TLR3 | Transmembrane receptor | 3.141 | 1.46E-12 |
| IL-4 | Cytokine | 2.346 | 2.94E-12 |
| IL-17F | Cytokine | 2.395 | 4.11E-12 |
| NFKB1 | Transcription regulator | 1.847 | 1.04E-11 |
| IL-13 | Cytokine | 1.316 | 2.48E-11 |
| FOXL2 | Transcription regulator | 2.804 | 2.50E-11 |
| IL-17A | Cytokine | 3.023 | 3.20E-11 |
| IKBKB | Kinase | 3.197 | 7.23E-10 |
| IL-6R | Transmembrane receptor | 2.179 | 3.02E-09 |
| IL-6 | Cytokine | 2.338 | 2.79E-06 |
| Cyclosporine A | Biologic drug | −2.358 | 3.21E-03 |
| Etanercept | Biologic drug | −2.2 | 5.01E-08 |
| PD98059 | Chemical, kinase inhibitor | −3.361 | 2.23E-07 |
| SB203580 | Chemical, kinase inhibitor | −3.344 | 1.59E-09 |
| U0126 | Chemical, kinase inhibitor | −3.006 | 4.34E-07 |
| SP600125 | Chemical, kinase inhibitor | −2.683 | 3.99E-07 |
| LY294002 | Chemical, kinase inhibitor | −2.604 | 4.76E-06 |
| Bay 11–7082 | Chemical, kinase inhibitor | −2.424 | 2.39E-08 |
| Glucocorticoid | Chemical drug | −2.609 | 1.52E-08 |
| Curcumin | Chemical drug, food supplement | −2.591 | 1.58E-04 |
| N-acetyl-L-cysteine | Chemical drug | −2.346 | 4.90E-05 |
| Dexamethasone | Chemical drug | −2.266 | 1.12E-09 |
| Resveratrol | Chemical drug, food supplement | −2.118 | 2.49E-03 |
| Genistein | Chemical drug, food supplement | −2.031 | 5.24E-05 |
| Salicylic acid | Chemical drug | −1.983 | 2.61E-05 |
| Tacrolimus | Chemical drug | −1.969 | 4.77E-05 |
| Simvastatin | Chemical drug | −1.607 | 5.16E-06 |
| Aspirin | Chemical drug | −1.527 | 1.39E-05 |
| Bortezomib | Chemical drug | −1.45 | 3.19E-04 |
| Indomethacin | Chemical drug | −1.394 | 8.48E-03 |
| Troglitazone | Chemical drug | −1.371 | 3.91E-05 |
| Sn50 peptide | Chemical toxicant, NF-κB inhibitor | −2.406 | 4.16E-08 |
| Pyrrolidine dithiocarbamate | Chemical reagent, NF-κB inhibitor | −2.233 | 2.79E-05 |
| MiR-155-5p (miRNAs w/seed UAAUGCU) | Mature microRNA | −2.395 | 1.29E-05 |
Included in Ingenuity Pathway Analysis. The table shows a list of selected molecules possibly able to activate (positive z-score) or revert (negative z-score) the transcriptional program observed in GASC with BAD prognosis. For each molecule, the prediction z-score and the P-value of the overlap are shown.
As a further confirmation, we compared the genes differentially expressed in GASC with 2 major collections of gene sets present in the Molecular Signatures Database (MSigDB).18 Interrogating the 50 “hallmark” gene sets, we observed that the differentially expressed GASC genes comprise elements of 11 “hallmark” gene sets, including the gene set regulated by NF-κB in response to TNF and several gene sets involved in the inflammatory and immune process, as well as a gene set involved in the epithelial to mesenchymal transition (Supplementary Table S4). Again, analyzing the curated gene sets of MSigDB, we confirmed the presence of 3 NF-κB–related gene sets (Supplementary Table S5). Finally, 14 of the 82 genes differentially expressed between GASC with different prognoses were attributable to NF-κB–related pathways and we decided to include them in an NF-κB GASC signature (Fig. 2A).
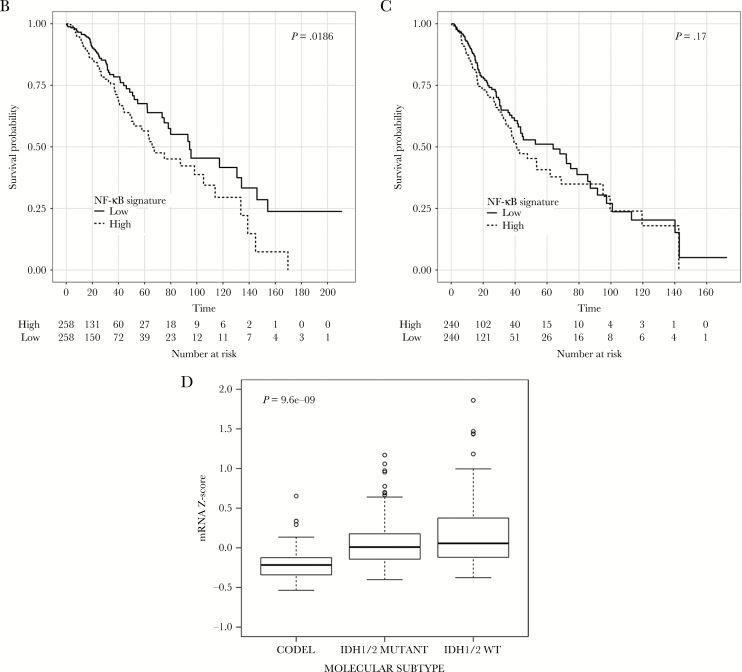
Fig. 2.
Prognostic role of the NF-κB signature in the 530 TCGA lower-grade gliomas. (A) The NF-κB signature has been obtained combining the genes of 3 NF-κB–related signatures enriched in genes differentially expressed in GASC with different prognoses. Kaplan-Meier curves showing OS (B), and PFS (C) in LGG patients stratified according to the NF-κB signature. (D) Level of expression of the NF-κB signature in the 3 molecular classes. Data are presented as box and whiskers. CODEL = gliomas IDH1/2 mutant and 1p/19q codeleted; IDH1/2 MUTANT = gliomas IDH1/2 mutant but not 1p/19q codeleted; IDH1/2 WT = gliomas without IDH1/2 gene mutation.
The NF-κB GASC Signature Is Prognostic in a Large Dataset and Correlates with the LGG Molecular Class
To investigate the prognostic significance of the NF-κB GASC signature, we interrogated the dataset of TCGA Brain LGG comprising 530 tumors.8 Survival analysis showed that tumors with higher expression of the signature had shorter OS (Fig. 2B) and a trend toward a shorter PFS (Fig. 2C). The prognostic value of the signature was possibly related to its level of expression across tumor subtypes. In particular, tumors presenting IDH mutation and 1p/19q codeletion expressed the signature at significantly lower levels with respect to tumor with IDH wild-type (P = 1.33e-10) or with IDH gene mutation only (P = 7.85e-13) (Fig. 2D). Interestingly, the signature had lower expression in gliomas harboring mutations in capicua transcriptional repressor and in the telomerase reverse transcriptase promoter (Supplementary Figure S3A, B), which are both associated with favorable clinical outcomes.8 Last, members of the NF-κB complex positively correlated with the signature (Pearson’s correlations, NFKB1 = 0.59, NFKB2 = 0.56, RELB = 0.53, REL = 0.46, P < 2.2e-16), indicating that in gliomas the signature is possibly activated via NF-κB (Supplementary Figure S3C).
Nuclear NF-κB-p65 Expression at Tissue Level Is an Independent Predictor of Bad Prognosis in LGG
Next, we decided to assess the presence, at tissue level, of the expression of RELA, also known as NF-κB transcription factor p65 (NF-κB-p65), which we previously identified as one of the molecules (upstream regulators) possibly responsible for the enhanced pro-tumorigenic environment characterizing LGG rapidly evolving to anaplasia.
The expression of NF-κB-p65 was evaluated in TMAs including 146 grade II glioma samples (Table 2). According to the WHO 2016 classification, 55.5%, 39.4%, and 5.5% of the gliomas were diffuse astrocytomas IDH mutant, oligodendrogliomas IDH mutant and 1p/19q codeleted, and diffuse astrocytoma IDH wild type, respectively. The preoperative volume of the tumors was a median 47 cm3 (range, 6–260 cm3) and 54.1% of the tumors presented, at MRI, an infiltrative pattern (ΔVT2T1 >15 cm3).5 The EOR was on average 86% (range, 49%–100%).
Table 2.
Clinical pathological features of the 146 patients included in the tissue microarray
| Clinicopathologic Features | Patients, no. | % | Median (range) |
|---|---|---|---|
| Sex | |||
| Male | 86 | 58.9 | – |
| Female | 60 | 41.1 | – |
| Age at surgery, y | – | – | 39 (18–70) |
| KPS | 100 (80–100) | ||
| Preoperative volume (cc) | – | – | 47 (6–260) |
| ΔVT2T1 category (cc) | 15 (0–84) | ||
| Proliferative tumoral pattern, ΔVT2T1 <15 cm3, n (%) | 67 | 45.9 | |
| Infiltrative tumoral pattern, ΔVT2T1 ≥ 15 cm3, n (%) | 79 | 54.1 | |
| Postoperative volume (cc) | – | – | 8 (0–125) |
| EOR (%) | – | – | 86 (49–100) |
| Histology (WHO 2007) | |||
| Astrocytoma | 80 | 54.8 | – |
| Oligodendroglioma | 19 | 13.0 | – |
| Oligoastrocytoma | 47 | 32.2 | – |
| WHO 2016 classification | |||
| Diffuse astrocytoma IDH mutant | 81 | 55.5 | – |
| Diffuse astrocytoma IDH wild type | 8 | 5.5 | – |
|
Oligodendroglioma IDH mutant and 1p/19q
codeleted |
57 | 39.0 | – |
| Molecular class | |||
| IDH wild type | 8 | 5.5 | – |
| IDH1/2 mutation only | 81 | 55.5 | – |
| IDH1/2 mutation and 1p/19q codeletion | 57 | 39.4 | – |
| Ki67 expression (%) | 5 (1–20) | ||
| ≤4 | 71 | 48.6 | – |
| >4 | 75 | 51.4 | – |
| Number of mitoses / 10 high power fields | – | – | 1 (0–10) |
| P53 expression (%) (n = 144) | 40 (0–100) | ||
| ATRX downregulation (n = 141) | 73 | 51.8 | |
| IDH1 mutation | 134 | 91.8 | – |
| IDH2 mutation | 4 | 2.7 | – |
| IDH1 or IDH2 mutation | 138 | 94.5 | – |
| Chromosome 1p deletion | 58 | 39.7 | – |
| Chromosome 19q deletion | 69 | 47.3 | – |
| Chromosome 1p and 19q codeletion | 57 | 39.0 | – |
| MGMT promoter methylation | 135 | 93.8 | – |
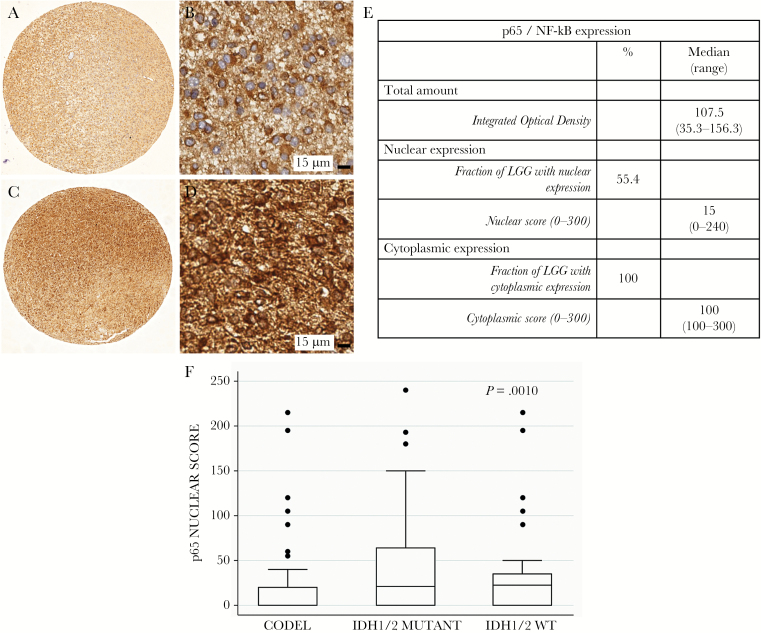
Considering NF-κB-p65, this latter was detected both in the nucleus and in the cytoplasm (Fig. 3A–D) of glioma tissues. However, while NF-κB-p65 was localized in the cytoplasm of all gliomas, its presence in the nucleus, where it can act as a master transcriptional regulator, was detected in about half of the analyzed samples. Indeed, the total amount of protein, evaluated as integrated optical density per unit area, was quite similar in all samples, while the nuclear score was quite wide, ranging from 0 to 240, thus indicating a possible different activation of NF-κB in the different tissues (Fig. 3E). Therefore, we evaluated the ability of NF-κB-p65 to predict OS, MPFS, and PFS.
Fig. 3.
Expression of NF-κB-p65 protein in TMA comprising 146 grade II LGGs. (A–D) Representative pictures of the immunohistochemical detection of NF-κB-p65 in the 1-mm cores of LGG presenting the protein only in the cytoplasm (A–B) or in both the cytoplasm and the nucleus (C–D). (E) Table summarizing the quantification of total amount, nuclear expression, and cytoplasmic expression of p65 NF-κB-p65. (F) Nuclear expression of NF-κB-p65 (expressed as nuclear score) in the 3 molecular classes. Data are presented as box and whiskers. CODEL = gliomas IDH1/2 mutant and 1p/19q codeleted; IDH1/2 MUTANT = gliomas IDH1/2 mutant but not 1p/19q codeleted; IDH1/2 WT = gliomas without IDH1/2 gene mutation
Overall survival
Overall, there were 83 deaths (57%) and the median follow-up in the surviving patients was 86 months (range, 43–193 mo). The estimated 5- and 10-year OS were 74% and 34%, respectively.
As summarized in Table 3, the prognostic factors positively associated with OS at univariate analysis (P < 0.05) were the Karnofsky performance status (KPS), the EOR, and the presence of mutated IDH1 or IDH2 genes, while OS was significantly poorer in older patients, as well as in patients showing high expression of Ki67 and, at MRI, an infiltrative pattern. With respect to IDH1/2 wild-type glioma, glioma characterized by either IDH1/2 mutation or IDH1/2 mutation and 1p/19q chromosome codeletion presented a better OS. Finally, the multivariate Cox analysis showed as independent predictors of OS the nuclear expression of p65 (hazard ratio [HR] 1.005, 95% CI: 1.001–1.009, P = 0.028), together with the well-established prognostic markers Ki67 (HR 1.766, 95% CI: 1.104–2.825, P = 0.018), age (HR 1.021, 95% CI: 1.002–1.040, P = 0.030), EOR (HR 0.948, 95% CI: 0.930–0.967, P < 0.0001), high infiltrative pattern index (HR 1.020, 95% CI: 1.003–1.036, P = 0.021), as well as presence of gliomas characterized by IDH1/2 mutation (HR 0.151, 95% CI: 0.059–0.385, P < 0.0001) or IDH1/2 mutation and 1p/19q codeletion (HR 0.113, 95% CI: 0.044–0.293, P < 0.0001), considering the molecular class IDH1/2 wild-type as a reference.
Table 3.
Univariate analysis of clinical, histological, and molecular parameters with OS, PFS, and MPFS in 146 patients with low-grade-gliomas
| OS | PFS | MPFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Age (modeled as continuous variable) | 1.021 | 1.003–1.040 | 0.021 | 1.008 | 0.993–1.024 | 0.287 | 1.014 | 0.997–1.030 | 0.104 |
| Sex | |||||||||
| Male | 1 | 1 | 1 | ||||||
| Female | 0.706 | 0.446–1.120 | 0.139 | 0.906 | 0.627–1.309 | 0.600 | 0.718 | 0.480–1.073 | 0.106 |
| KPS (modeled as continuous variable) | 0.963 | 0.931–0.996 | 0.027 | 0.987 | 0.957–1.018 | 0.410 | 0.970 | 0.940–1.000 | 0.052 |
| Preoperative volume (T2_pre) (modeled as continuous variable) | 1.002 | 0.998–1.006 | 0.357 | 1.003 | 1.000–1.007 | 0.071 | 1.006 | 1.002–1.009 | 0.002 |
| Infiltrative growth index (T2_ T1) (modeled as continuous variable) | 1.028 | 1.014–1.043 | <0.0001 | 1.028 | 1.015–1.042 | <0.0001 | 1.038 | 1.024–1.053 | <0.0001 |
| Postoperative (T2_post) (modeled as continuous variable) | 1.007 | 0.998–1.015 | 0.112 | 1.007 | 1.001–1.015 | 0.030 | 1.017 | 1.010–1.024 | <0.0001 |
| % EOR (modeled as continuous variable) | 0.942 | 0.926–0.959 | <0.0001 | 0.955 | 0.941–0.970 | <0.0001 | 0.951 | 0.936–0.966 | <0.0001 |
| Histotype (WHO 2007) | |||||||||
| Astrocytoma | 1 | 1 | 1 | ||||||
| Oligodendroglioma | 1.15 | 0.622–2.137 | 0.651 | 0.983 | 0.576–1.676 | 0.950 | 0.922 | 0.513–1.657 | 0.786 |
| Oligoastrocytoma | 0.830 | 0.506–1.360 | 0.460 | 0.825 | 0.549–1.241 | 0.355 | 0.855 | 0.555–1.317 | 0.477 |
| % Ki67 | |||||||||
| ≤4 | 1 | 1 | 1 | ||||||
| >4 | 1.952 | 1.247–3.057 | 0.003 | 1.222 | 0.851–1.755 | 0.278 | 1.635 | 1.102–2.425 | 0.015 |
| Number of mitoses 10 HPF (modeled as continuous variable) | 1.052 | 0.924–1.198 | 0.442 | 1.042 | 0.927–1.171 | 0.496 | 1.037 | 0.922–1.166 | 0.541 |
| IDH1 or IDH2 mutation | |||||||||
| No | 1 | 1 | 1 | ||||||
| Yes | 0.157 | 0.070–0.355 | <0.0001 | 0.348 | 0.160–0.756 | 0.008 | 0.235 | 0.106–0.518 | <0.0001 |
| Chromosome 1p/19q codeletion | |||||||||
| No | 1 | 1 | 1 | ||||||
| Yes | 0.710 | 0.453–1.112 | 0.134 | 0.671 | 0.462–0.973 | 0.035 | 0.579 | 0.385–0.869 | 0.008 |
| Molecular class | |||||||||
| IDH1/2 wild type | 1 | 1 | 1 | ||||||
| IDH1/2 mutant | 0.172 | 0.075–0.397 | <0.0001 | 0.399 | 0.181–0.878 | 0.022 | 0.283 | 0.127–0.631 | 0.002 |
| IDH1/2 mutant and 1p/19q codeletion | 0.136 | 0.057–0.324 | <0.0001 | 0.282 | 0.125–0.636 | 0.002 | 0.175 | 0.076–0.405 | <0.0001 |
| P53 expression (modeled as continuous variable) | 1.477 | 0.797–2.735 | 0.215 | 1.619 | 0.970–2.701 | 0.065 | 1.696 | 0.990–2.908 | 0.055 |
| ATRX downregulation | |||||||||
| Yes | 1 | 1 | 1 | ||||||
| No | 1.185 | 0.764–1.838 | 0.448 | 0.873 | 0.608–1.255 | 0.464 | 0.876 | 0.592–1.296 | 0.507 |
| MGMT promoter methylation | |||||||||
| No | 1 | 1 | 1 | ||||||
| Yes | 0.525 | 0.227–1.213 | 0.132 | 0.858 | 0.399–1.846 | 0.695 | 0.771 | 0.337–1.763 | 0.537 |
| Total NF-κB/p65 (modeled as continuous variable) | 0.998 | 0.990–1.006 | 0.655 | 0.996 | 0.989–1.003 | 0.259 | 0.999 | 0.991–1.006 | 0.766 |
| Nuclear NF-κB/p65 (modeled as continuous variable) | 1.003 | 0.999–1.007 | 0.100 | 1.002 | 0.999–1.006 | 0.220 | 1.004 | 1.001–1.008 | 0.015 |
Boldface represents statistical significance values from 2-sided tests (Cox regression) statistically significant when <0.05.
Tumor progression
Tumor progression was identified in 123 (84%) cases and the estimated 5- and 10-year PFS rates were 43% and 4%, respectively. At the univariate analysis (Table 3), the EOR, the presence of mutated IDH1/2 gene, and 1p/19q chromosome codeletion were associated with a significant improvement in PFS. Instead, a large postoperative volume and an infiltrative pattern were associated with a worse PFS. Finally, the multivariate Cox analysis showed that independent predictors of PFS were EOR (HR 0.958, 95% CI: 0.943–0.973, P < 0.0001), an infiltrative pattern index (HR 1.018, 95% CI: 1.003–1.033, P = 0.020), and gliomas characterized by either IDH1/2 mutation (HR 0.335, 95% CI: 0.150–0.748, P = 0.008) or IDH1/2 mutation and 1p/19q codeletion (HR 0.222, 95% CI: 0.096–0.514, P < 0.0001), considering the molecular class IDH1/2 wild-type as a reference.
Malignant transformation
Anaplastic transformation was observed in 107 (72%) cases. The estimated 5- and 10-year MPFS rates were 58% and 21%, respectively. The prognostic factors positively associated with the MPFS at univariate analysis were the EOR and the presence of mutated IDH1 or IDH2 genes and of a 1p/19q codeletion. Conversely, the MPFS was significantly poorer in patients showing high Ki67 and nuclear NF-κB/p65 and, at MRI, larger preoperative and postoperative volumes and an infiltrative pattern (Table 3). Finally, the multivariate Cox analysis showed as independent predictors of MPFS the nuclear expression of NF-κB-p65 (HR 1.005, 95% CI: 1.001–1.008, P = 0.020) together with the well-established prognostic markers age (HR 1.019, 95% CI: 1.002–1.037, P = 0.029), Ki67 >4% (HR 1.538, 95% CI: 1.021–2.319, P = 0.040), EOR (HR 0.957, 95% CI: 0.941–0.974, P < 0.0001), an infiltrative pattern at MRI (HR 1.033, 95% CI: 1.018–1.048, P < 0.0001), as well as having gliomas characterized by either IDH1/2 mutation (HR 0.228, 95% CI: 0.095–0.548, P = 0.001) or IDH1/2 mutation and 1p/19q codeletion (HR 0.131, 95% CI: 0.053–0.327, P < 0.0001), considering the molecular class IDH1/2 wild-type as a reference.
Altogether, these results indicate that the nuclear expression of NF-κB-p65 was an independent predictor of both OS and MPFS, even when compared with the state-of-the-art clinical, neuroradiological, and molecular LGG prognostic markers.
Interestingly, when we evaluated the NF-κB-p65 nuclear score in the 3 molecular classes, its expression was significantly different in the 3 groups, similarly to that shown for the NF-κB gene signature in the dataset of TCGA LGG (Fig. 3F).
Discussion
The ambition of PM in oncology is to identify subgroups of patients who are homogeneous in terms of biological aggressiveness and risk of progression, as well as response/susceptibility to specific treatments,6,19 and to tailor, for each subgroup, the best therapeutic choices and follow-up strategies.19
The rapid development of PM derives from the progress in high-throughput techniques combined with bioinformatics tools able to process and model the “big data” generated.19
Besides genomics, information is expected to derive also from other sources, including the development of new patient-derived cell models20–22 and the study of the TME, a factor crucially involved in the development of intratumor heterogeneity, which is responsible for tumor invasiveness and drug resistance.12,23–26 Regarding cell lines and PM, most of the literature is focused on the use of patient-derived xenograft models that, being able to recapitulate patients’ tumor histology and heterogeneity, can become a powerful tool to predict drug sensitivity.24 Similarly, there is an increasing interest in demonstrating that the use of patient-derived tumor cell cultures, including induced pluripotent stem cells, is feasible, thus providing investigators and clinicians with models whose genetic setting is similar to that of the tumor of origin, to either suggest predictive biomarkers, perform drug screening assays, or develop innovative genome-directed targeted therapies.20–22 However, xenograft models and patient-derived tumor cells cannot be efficiently obtained from every neoplastic patient and are substantially missing for adult LGG.27 Conversely, we showed that from the TME of LGG it is possible to isolate a population of stem cells, named GASC, that sustain tumor growth.11 Since patient-derived TME cellular models have never been used to infer prognostic factors to be detected at tissue level, we have optimized and clinically validated an experimental procedure that, analyzing GASC with state-of-the-art sequencing techniques and bioinformatics tools, defined biomarkers relevant for the prognostic stratification of LGG patients.
Transcriptomic analyses of GASC-GOOD and GASC-BAD showed that these latter were enriched in elements related to the hallmarks of cancer, suggesting that the TME of LGG with different prognoses differed in the ability to support tumor growth and infiltration, favor immune escape, and activate programs in response to drugs and radiation.12,18 Interestingly, Ceccarelli et al, by studying adult WHO grade II–IV gliomas, showed that progression of LGG to glioblastoma is characterized by changes in the microenvironmental transcriptional program.9 Specifically, with respect to LGG, glioblastomas were enriched in gene sets involved in the inflammatory and immune response as well as in the inhibitor of κB/NF-κB kinase cascade.9
Ingenuity Pathway Analysis identified several soluble factors (eg, chemokines, interleukins, interferons, tumor necrosis factors) as upstream regulators, thus supporting the importance of the secretome in contributing to the hallmarks of cancer.28 NF-κB complex, including the transcriptional regulators RELA (NF-κB-p65) and NF-κB1, were represented as activated upstream regulators. Accordingly, by interrogating both the “hallmark” gene set and the curated gene set collections of the MSigDB, NF-κB–related gene sets emerged as differentially represented in BAD versus GOOD GASC. From these gene sets, we identified an NF-κB signature composed of 14 genes that was able to predict OS when tested in the dataset of TCGA, including 530 newly diagnosed LGG tissues.8 Therefore, the NF-κB–related transcriptional program, inferred by the TME in vitro model, is actually present in glioma tissues and is associated with patient outcome.
In unstimulated cells, NF-κB is retained in the cytoplasm, while, following different stimuli (eg, cytokines, growth factors, DNA damage, oncogenic stress), NF-κB is translocated to the nucleus, regulating, as a transcriptional factor, key cellular functions, including cell survival, inflammation, and immunity.29,30 In glioblastoma, NF-κB is constitutively activated, thus promoting cell growth and survival.31 Moreover, in proneural glioma stem cells, NF-κB induces the transition into a mesenchymal phenotype, associated with radioresistance and poor prognosis.32
To confirm the putative prognostic role of NF-κB activation in LGG, we analyzed the nuclear expression of NF-κB-p65 in 146 newly diagnosed grade II LGGs within the new molecular subgroups as defined by the new WHO classification.2,8 In this study the molecular stratification of glioma into 3 classes (IDH wild-type, IDH mutant only, and IDH mutant and 1p/19q codeleted) was endowed with prognostic significance for OS, PFS, and MPFS. More interestingly, the nuclear expression of NF-κB-p65 resulted to be, together with well-established LGG prognosticators, an independent predictor of both OS and MPFS.5,11
In glioblastoma, there are 2 major mechanisms of NF-κB activation: aberrant epidermal growth factor receptor signaling and deletion of the NF-κBIA gene that encodes inhibitor of κBα.33 Moving from GASC data, we can speculate that in LGG, a major role is played by cytokine stimulation related pathways, such as TNFα and IL-6/signal transducer and activator of transcription 3 activation, as confirmed by Gene Set Enrichment Analysis. However, future investigations on the link between reduced expression of NF-κB and both IDH mutation and 1p/19q codeletion (demonstrated both in the dataset of TCGA and in the TMA case study) will help to explain the mechanism of NF-κB activation in LGG.
Interestingly, among the molecules that could revert the transcriptional programs characterizing BAD GASC, we identified anti-inflammatory, immunosuppressive drugs and NF-κB inhibitors, corroborating the knowledge that pro-inflammatory programs are active in GASC-BAD and suggesting new therapeutic strategies that target the TME. Consistently, TME-directed therapies are being experimented in clinical trials,34,35 and some results have been obtained for glioblastoma,36–38 where NF-κB inhibitors, used alone or combined with chemo- or radiotherapy, have been tested showing promising, although partial, results.31
In conclusion, there are 2 novelties of this work. First, patient-derived stem cells representative of the TME are a clinically relevant model that can play a role in PM. This is even more useful in tumors with a relatively long survival, such as LGG, since patients can directly benefit from any new discovery obtained by employing their own cells. Second, as in glioblastoma, NF-κB is activated in a subset of LGG, where it acts as an independent prognostic factor and can represent a new target for adjuvant therapies.
Supplementary Material
Supplementary data are available at Neuro-Oncology online.
Funding
This work was supported by FIRB accordi di programma 2011; title: “FIERCE – FInd nEw moleculaR and CEllular targets against cancer.” Pr. RBAP11Z4Z9 (M.E.R., L.M.). 2012–2014; FIRB accordi di programma 2011 pr. RBAP11ETKA_007 “Nanotechnological approaches for tumor theragnostic” (A.P.B.). Programma per la Cooperazione Transfrontaliera Italia-Slovenia 2007–2013. Title: “Identificazione di nuovi marcatori di cellule staminali tumorali a scopo diagnostico e terapeutico,” D35E11000600003 (M.S., C.D.L.); AIRC 5 per mille special program 2011, Pr. 12214. Title: “Application of Advanced Nanotechnology in the Development of Cancer Diagnostics Tools” (M.B.). Project ERC- 7FP SP 2 IDEAS QUIDPROQUO G.A. n. 269051. Title: “Molecular nanotechnology for life science applications: quantitative interactomics for diagnostics, proteomics and quantitative oncology” (D.C., D.M.). Y.C. is supported by the AIRC/FIRC fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement. None declared.
References
- 1. Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Verma V, Mehta MP. Clinical ramifications of “genomic staging” of low-grade gliomas. J Neurooncol. 2016;129(2):195–199. [DOI] [PubMed] [Google Scholar]
- 4. Soffietti R, Baumert BG, Bello L, et al. European Federation of Neurological Societies. Guidelines on management of low-grade gliomas: report of an EFNS-EANO task force. Eur J Neurol. 2010;17(9):1124–1133. [DOI] [PubMed] [Google Scholar]
- 5. Ius T, Isola M, Budai R, et al. Low-grade glioma surgery in eloquent areas: volumetric analysis of extent of resection and its impact on overall survival. A single-institution experience in 190 patients: clinical article. J Neurosurg. 2012;117(6):1039–1052. [DOI] [PubMed] [Google Scholar]
- 6. Committee on a Framework for Development a New Taxonomy of Disease NRC. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 7. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015; 372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ceccarelli M, Barthel FP, Malta TM, et al. TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sankar PL, Parker LS. The Precision Medicine Initiative’s All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med. 2017;19(7):743–750. [DOI] [PubMed] [Google Scholar]
- 11. Bourkoula E, Mangoni D, Ius T, et al. Glioma-associated stem cells: a novel class of tumor-supporting cells able to predict prognosis of human low-grade gliomas. Stem Cells. 2014;32(5):1239–1253. [DOI] [PubMed] [Google Scholar]
- 12. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 13. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobsen A. cgdsr: R-Based API for Accessing the MSKCC Cancer Genomics Data Server (CGDS). R package version 1.2.5 https://cran.r-project.org/package=cgdsr. 2015.
- 17. Tovey S, Dunne B, Witton CJ, Forsyth A, Cooke TG, Bartlett JM. Can molecular markers predict when to implement treatment with aromatase inhibitors in invasive breast cancer?Clin Cancer Res. 2005;11(13):4835–4842. [DOI] [PubMed] [Google Scholar]
- 18. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jameson JL, Longo DL. Precision medicine—personalized, problematic, and promising. N Engl J Med. 2015;372(23):2229–2234. [DOI] [PubMed] [Google Scholar]
- 20. Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim J, Zaret KS. Reprogramming of human cancer cells to pluripotency for models of cancer progression. EMBO J. 2015;34(6):739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quartararo CE, Reznik E, deCarvalho AC, Mikkelsen T, Stockwell BR. High-throughput screening of patient-derived cultures reveals potential for precision medicine in glioblastoma. ACS Med Chem Lett. 2015;6(8):948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seoane J, De Mattos-Arruda L. The challenge of intratumour heterogeneity in precision medicine. J Intern Med. 2014;276(1):41–51. [DOI] [PubMed] [Google Scholar]
- 24. Byrne AT, Alférez DG, Amant F, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(4):254–268. [DOI] [PubMed] [Google Scholar]
- 25. Motaln H, Koren A, Gruden K, Ramšak Ž, Schichor C, Lah TT. Heterogeneous glioblastoma cell cross-talk promotes phenotype alterations and enhanced drug resistance. Oncotarget. 2015;6(38):40998–41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herold-Mende C, Mock A. Microenvironment and brain tumor stem cell maintenance: impact of the niche. Anticancer Agents Med Chem. 2014;14(8):1065–1074. [DOI] [PubMed] [Google Scholar]
- 27. Cesselli D, Beltrami AP, Pucer A, et al. Human low grade glioma cultures. In: Duffau H, ed. Diffuse Low-Grade Gliomas in Adults. London: Springer-Verlag; 2013. [Google Scholar]
- 28. Patel S, Ngounou Wetie AG, Darie CC, Clarkson BD. Cancer secretomes and their place in supplementing other hallmarks of cancer. Adv Exp Med Biol. 2014;806:409–442. [DOI] [PubMed] [Google Scholar]
- 29. Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47(6):921–928. [DOI] [PubMed] [Google Scholar]
- 31. Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci Adv. 2016;2(1):e1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Puliyappadamba VT, Hatanpaa KJ, Chakraborty S, Habib AA. The role of NF-κB in the pathogenesis of glioma. Mol Cell Oncol. 2014;1(3):e963478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. [DOI] [PubMed] [Google Scholar]
- 35. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weathers SP, Gilbert MR. Advances in treating glioblastoma. F1000Prime Rep. 2014;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quail DF, Bowman RL, Akkari L, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016; 352(6288):aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.