Abstract
The ratio of the length of the index finger to that of the ring finger (2D:4D) is sexually dimorphic and is commonly used as a non-invasive biomarker of prenatal androgen exposure. Most association studies of 2D:4D ratio with a diverse range of sex-specific traits have typically involved small sample sizes and have been difficult to replicate, raising questions around the utility and precise meaning of the measure. In the largest genome-wide association meta-analysis of 2D:4D ratio to date (N = 15 661, with replication N = 75 821), we identified 11 loci (9 novel) explaining 3.8% of the variance in mean 2D:4D ratio. We also found weak evidence for association (β = 0.06; P = 0.02) between 2D:4D ratio and sensitivity to testosterone [length of the CAG microsatellite repeat in the androgen receptor (AR) gene] in females only. Furthermore, genetic variants associated with (adult) testosterone levels and/or sex hormone-binding globulin were not associated with 2D:4D ratio in our sample. Although we were unable to find strong evidence from our genetic study to support the hypothesis that 2D:4D ratio is a direct biomarker of prenatal exposure to androgens in healthy individuals, our findings do not explicitly exclude this possibility, and pathways involving testosterone may become apparent as the size of the discovery sample increases further. Our findings provide new insight into the underlying biology shaping 2D:4D variation in the general population.
Introduction
It has long been hypothesized that prenatal sex steroids, particularly testosterone, permanently modify the developing nervous system during critical periods of development, which in turn influences behavior in later life (1). Whilst animal models have largely supported this “Organizational hypothesis” (2), evidence from human studies has been much more limited, as accurately measuring prenatal testosterone exposure is extremely difficult. Based on several lines of indirect evidence, it has widely been hypothesized that the ratio of the length of the index finger to the length of the ring finger (2D:4D) is a marker of prenatal androgen exposure (3) and could therefore be used as a retrospective non-invasive biomarker of prenatal testosterone exposure. Males are believed to have greater prenatal testosterone exposure than females, and this is thought to determine the consistently observed lower ratio of the first (index) finger to the third (ring) finger in males (4,5). This sex difference is relatively stable over time (3,6,7), and although there is variation in 2D:4D ratio across ethnic groups (8,9), sexual dimorphism in the digit ratio is consistent across ethnicities (10). Based on the assumption that the digit ratio is a marker of prenatal testosterone exposure, associations have been reported between 2D:4D ratio and a broad range of sex-dependent behaviors and diseases including academic (11) and sporting performance (12,13), social behaviors (14–16), fertility (17,18), Alzheimer’s disease (19), metabolic syndrome indices (20) and autism spectrum disorder (21). Despite the extensive literature regarding 2D:4D ratio (22), most published studies have used small sample sizes (often fewer than 100 individuals) and have been difficult to replicate, raising questions around the utility and precise meaning of the measure (23).
Several twin studies have indicated that 2D:4D ratio is highly heritable (h2: 50–80%) (24–28). Two relatively small genome-wide association studies (GWAS) of 2D:4D ratio have reported two loci influencing variation in the trait (29,30). The minor allele (A) at rs314277 in LIN28B was associated with increased 2D:4D ratio, delayed menarche in females (31) and increased height (32). The minor allele (T) at the second locus, rs4902759 in SMOC1, was associated with decreased 2D:4D ratio. Pedigree studies have shown that mutations in SMOC1 are associated with Waardenburg anophthalmia (OMIM 206920), a syndrome that commonly includes abnormal digits (33–35). Additionally, the protein encoded by SMOC1 has been shown to be up-regulated by androgens (36,37) and down-regulated by estrogen (38), suggesting that SMOC1 could be an intermediate between prenatal sex hormones and digit ratio (29).
Individuals with Complete Androgen Insensitivity Syndrome (CAIS) exhibit more feminine 2D:4D ratios, consistent with an effect of reduced prenatal testosterone exposure on digit ratio (39). These individuals have mutations in the androgen receptor (AR) gene, located on the X chromosome, which codes for a receptor protein that facilitates physiological responses to androgens such as testosterone and dihydrotestosterone (40). In vitro studies suggest that the variable number of CAG repeats in exon 1 of AR is inversely related to the efficiency with which the receptor complex binds to DNA and influences transcription (41). Therefore, an association between the number of CAG repeats at the AR locus and 2D:4D ratio may indicate that sensitivity to androgens is a major driver of the individual differences in 2D:4D ratio seen in the normal population (40).
The aim of this study was to investigate the genetic determinants of 2D:4D ratio by performing the largest GWAS meta-analysis (N = 15 661, with replication N = 75 821) to date. In addition, we aim to leverage genetics to scrutinize the evidence surrounding the hypothesis that 2D:4D ratio reflects prenatal androgen exposure. Specifically, we investigated whether there was association between repeat number in the androgen receptor (XAR) and 2D:4D ratio and we used our GWAS results to examine: (1) whether there was any association between 2D:4D ratio and genetic variants in pathways known to be linked with androgens; and (2) whether there was any relationship between 2D:4D ratio and genetic markers known to be related to (adult) levels of testosterone and/or sex hormone binding globulin (SHBG). We hypothesize that if 2D:4D ratio is truly influenced by levels of prenatal testosterone in utero, then it is reasonable to expect that genetic variants related to androgen sensitivity (XAR) and/or serum levels of testosterone/SHBG might also show association with 2D:4D ratio.
Results
Study population
Table 1 provides a summary of the studies included in the meta-analysis. Full details of the studies in the discovery meta-analysis and replication, including how the 2D:4D ratio was measured and the genotyping methods, are provided in the Supplementary Material. As expected, females had greater 2D:4D ratios than males in all cohorts. The mean and standard deviation of 2D:4D ratios measured on skeletal images (the Generation R Study and the Rotterdam Study) were lower than those of the other cohorts measured on photocopies of the hand, consistent with previous reports using this measure (42).
Table 1.
Descriptive statistics of the discovery and replication cohorts
| Variable | Subset | ALSPAC | Generation R | QIMR | Raine | Rotterdam Study | Twins UK |
|---|---|---|---|---|---|---|---|
| N | All | 5337 | 3059 | 2775 | 1003 | 2091 | 1396 |
| Age (years)a | All | 11.74 (0.23) | 9.80 (0.33) | 15.47 (2.93) | 20.05 (0.43) | 67.84 (7.91) | 54.84 (12.21) |
| Sex (male)b | All | 49% (2615) | 47.9% (1465) | 46.4% (1287) | 50.85% (510) | 42.9% (897) | 9.10% (127) |
| Left 2D:4Da | All | 96.53 (3.25) | 91.15 (2.72) | 97.66 (3.41) | 96.52 (3.45) | 92.40 (2.24) | 96.70 (3.40) |
| Male | 96.05 (3.17) | 90.74 (2.77) | 96.87 (3.38) | 96.16 (3.33) | 91.96 (2.25) | 95.25 (3.24) | |
| Female | 97.00 (3.26) | 91.52 (2.62) | 98.33 (3.29) | 96.93 (3.53) | 92.73 (2.17) | 96.80 (3.42) | |
| Right 2D:4Da | All | 96.37 (3.28) | – | 97.07 (3.43) | 96.99 (3.28) | 92.39 (2.42) | 97.10 (3.50) |
| Male | 95.87 (3.22) | – | 96.12 (3.29) | 96.81 (3.28) | 91.94 (2.39) | 95.58 (3.38) | |
| Female | 96.87 (3.26) | – | 97.88 (3.33) | 97.19 (3.28) | 92.73 (2.39) | 97.24 (3.48) | |
| Mean 2D:4Da | All | 96.45 (2.99) | – | 97.39 (3.10) | 96.76 (3.04) | 92.40 (2.12) | 96.90 (3.10) |
| Male | 95.96 (2.90) | – | 96.51 (3.01) | 96.48 (2.94) | 91.95 (2.13) | 95.42 (2.99) | |
| Female | 96.93 (3.00) | – | 98.14 (2.97) | 97.06 (3.12) | 92.71 (2.05) | 97.02 (3.07) |
Mean (SD).
Percent (number).
Genome-wide complex trait analysis of 2D:4D ratio of the left hand, right hand and mean of both hands
Univariate genetic restricted maximum likelihood (GREML) analysis in ALSPAC revealed that common genome-wide variation explained a substantial proportion of the variance in 2D:4D ratio (left: h2SNP = 0.299, SE = 0.071, P = 4.6 × 10−6; right: h2SNP=0.360, SE = 0.071, P = 6.2 x 10−8; mean: h2SNP = 0.373, SE = 0.071, P = 1.4 × 10−8). The genetic correlation between the left and right hand ratios was not different from 1 (rg = 0.918, SE = 0.074, P = 0.14), indicating that the vast majority of SNPs contributing to variation in the ratio influence both hands. Therefore, in the main text we present the results from the left (typically non-dominant) hand ratio, due to the larger sample size achieved by including the Generation R Study, and include the right hand and mean ratio results in the Supplementary Material.
New genetic loci associated with 2D:4D ratio
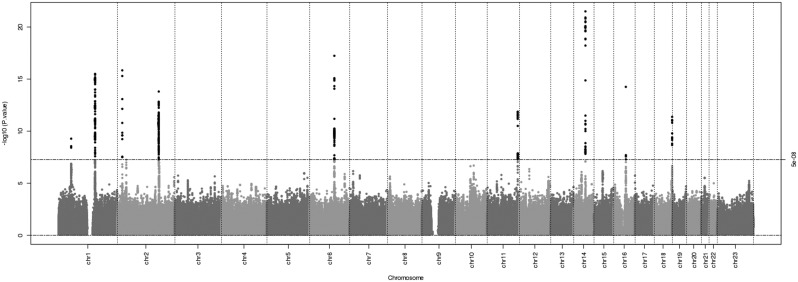
The meta-analysis of approximately 8.4 million 1000 genomes-imputed SNPs, including SNPs on the X chromosome, indicated that the lowest observed P-values for each of the three ratios deviated from the expected null distribution (Supplementary Material, Fig. S1), whereas systematic inflation of the test statistics due to bias was negligible (λleft = 1.020, λright = 1.012, λmean = 1.014). Further, no evidence of heterogeneity was detected between the discovery cohorts (Supplementary Material, Fig. S2). Eleven genomic loci reached genome-wide significance (P < 5 × 10−8) in the discovery meta-analysis of 15 661 individuals for 2D:4D ratio [Fig. 1 for Manhattan plots from the GWAS of the left hand (European only), Supplementary Material, Fig. S3 for the left hand (multiethnic), right hand and average of both hands]. Conditional and joint analysis in the genome-wide complex trait analysis (GCTA) software (43) identified two independent signals in the 16q12.1 locus, totaling 12 independent signals across the three 2D:4D ratio measurements. All 12 signals were replicated in 75 821 (52.8% male) research participants from 23andMe, Inc. (all P < 0.004; Table 2). Of the nine loci reaching genome-wide significance for the first time, six have not previously been described in the context of 2D:4D ratio, including: rs11581730 on chromosome 1q22; rs12474669 on chromosome 2q31.1; rs77640775 on chromosome 7p14.1; rs10790969 on chromosome 11q24.3; rs6499762 and rs1080014 on chromosome 16q12.1; and rs4799176 on chromosome 18q23. Two of the nine novel loci were reported at a suggestive significance level (but not genome-wide) in Medland et al. (30): SNPs in LDAH (previously known as C2orf43) on chromosome 2p24.1 and in GLIS1 on chromosome 1p32.3. The locus on chromosome 2q31.1 is near the HOXD cluster of genes that are hypothesized to be required for growth and patterning of the digits, but this is the first time that convincing evidence for genetic association with 2D:4D ratio has been obtained. The remaining two loci included SNPs in LIN28B, identified previously by Medland et al. (30) and SMOC1, reported by Lawrance-Owen et al. (29). A summary of the meta-analysis results for the lead SNPs at each locus that reached genome-wide significance in the discovery sample are provided in Table 2, with the results from each study presented in Supplementary Material, Table S3 and regional plots in Supplementary Material, Figure S4. The lead SNPs at the 12 replicated signals together explain 3.8% of the variance in 2D:4D ratio; this is equivalent to over half of the variance explained by sex in the Raine Study (5.1%).
Figure 1.
Manhattan plot from the discovery meta-analysis of left hand 2D:4D ratio. The horizontal black line indicates genome-wide significance (P < 5 × 10−8). Black dots indicate those loci that reach genome-wide significance.
Table 2.
Genome-wide-significant loci from the discovery meta-analysis in all individuals for left hand 2D:4D ratio; the most significant SNP from each locus is presented
| Chr | Position [bp (GRCh37/hg19)] | Nearest gene | Effect allele/ other allele | EAFa | Beta | SE | P-value | |
|---|---|---|---|---|---|---|---|---|
| rs4927012 | ||||||||
| Discovery | 1 | 54068016 | GLIS1 | T/C | 0.875 | −0.358 | 0.058 | 5.08 × 10−10 |
| Replication | 1 | 54068016 | GLIS1 | T/C | 0.871 | −0.042 | 0.006 | 3.48 × 10−12 |
| rs11581730 | ||||||||
| Discovery | 1 | 155082158 | EFNA1 | A/T | 0.496 | 0.294 | 0.036 | 3.02 × 10−16 |
| Replication | 1 | 155082158 | EFNA1 | A/T | 0.503 | 0.026 | 0.004 | 4.99 × 10−11 |
| rs340600 | ||||||||
| Discovery | 2 | 20892006 | LDAHb | T/G | 0.199 | −0.379 | 0.046 | 1.38 × 10−16 |
| Replication | 2 | 20892006 | LDAHb | T/G | 0.199 | −0.043 | 0.005 | 1.81 × 10−17 |
| rs12474669 | ||||||||
| Discovery | 2 | 175134232 | OLA1 | A/G | 0.139 | 0.417 | 0.054 | 1.51 × 10−14 |
| Replication | 2 | 175134232 | OLA1 | A/G | 0.143 | 0.043 | 0.006 | 1.92 × 10−13 |
| rs847158 | ||||||||
| Discovery | 2 | 176962102 | HOXD12/HOXD11 | A/G | 0.602 | 0.199 | 0.037 | 1.03 × 10−7 |
| Replication | 2 | 176962102 | HOXD12/HOXD11 | A/G | 0.609 | 0.039 | 0.004 | 3.04 × 10−20 |
| rs314277c | ||||||||
| Discovery | 6 | 105407662 | LIN28B | A/C | 0.155 | 0.428 | 0.050 | 5.55 × 10−18 |
| Replication | 6 | 105407662 | LIN28B | A/C | 0.149 | 0.067 | 0.006 | 1.77 × 10−32 |
| rs77640775d | ||||||||
| Discovery | 7 | 42190714 | GLI3 | A/G | 0.137 | −0.252 | 0.053 | 1.92 × 10−6 |
| Replication | 7 | 42190714 | GLI3 | A/G | 0.146 | −0.033 | 0.006 | 6.64 × 10−9 |
| rs10790969 | ||||||||
| Discovery | 11 | 128529842 | FLI1 | T/C | 0.276 | 0.284 | 0.040 | 1.33 × 10−12 |
| Replication | 11 | 128529842 | FLI1 | T/C | 0.272 | 0.027 | 0.005 | 1.26 × 10−9 |
| rs2332175c | ||||||||
| Discovery | 14 | 70345411 | SMOC1 | A/G | 0.529 | 0.360 | 0.037 | 3.00 × 10−22 |
| Replication | 14 | 70345411 | SMOC1 | A/G | 0.546 | 0.045 | 0.004 | 6.74 × 10−29 |
| rs6499762 | ||||||||
| Discovery | 16 | 51697874 | SALL1 | A/C | 0.125 | 0.441 | 0.056 | 5.33 × 10−15 |
| Replication | 16 | 51697874 | SALL1 | A/C | 0.129 | 0.083 | 0.006 | 2.83 × 10−41 |
| rs1080014 | ||||||||
| Discovery | 16 | 51900171 | TOX3 | C/T | 0.514 | 0.203 | 0.036 | 1.94 × 10−8 |
| Replication | 16 | 51900171 | TOX3 | C/T | 0.501 | 0.012 | 0.004 | 3.35 × 10−3 |
| rs4799176 | ||||||||
| Discovery | 18 | 76378307 | SALL3 | C/T | 0.256 | 0.305 | 0.044 | 4.09 × 10−12 |
| Replication | 18 | 76378307 | SALL3 | C/T | 0.244 | 0.057 | 0.005 | 1.89 × 10−34 |
Replication results are presented from 23andMe where the 2D:4D ratio was reported as a relative measure [i.e. 0 = index finger longer (17.1% of research participants), 1 = index and ring finger the same length (14.0% of research participants), 2 = ring finger longer (68.9% of research participants)].
Average effect allele frequency (EAF) across the cohorts in each of the meta-analyses.
Previously known as C2orf43.
Genetic loci that had previously been associated with 2D:4D ratio in Medland et al. (30).
SNP passed genome-wide significance in the average 2D:4D ratio meta-analysis (see Supplementary Material for results).
We conducted analysis of the X chromosome to investigate whether there was evidence for 2D:4D ratio being partly an X-linked trait. No SNPs on the X chromosome reached genome-wide or suggestive significance (Fig. 1). The HOXA gene cluster, along with the HOXD gene cluster, plays an important role in limb development, and were initially thought to be essential for the development of the 2D:4D ratio. Although the HOXD region was associated with 2D:4D ratio in our meta-analysis, there was no strong evidence for association between variants in the HOXA cluster and 2D:4D ratio (439 SNPs with all P > 0.001, Supplementary Material, Fig. S5).
As a secondary analysis, we also conducted sex-stratified analyses in each of the discovery cohorts and combined the results using a fixed-effects meta-analysis. The majority of the loci reaching genome-wide significance in the male or female only analyses were identified in the combined analysis (Miami plots in Supplementary Material, Fig. S6). One novel locus reached genome-wide significance in the female only analysis of the left hand 2D:4D ratio in the multi-ethnic meta-analysis. The top SNP in this region, rs10105686 [C allele (allele frequency = 0.79) females: β = −0.334, P = 2.42 × 10−9; males: β = −0.024, P = 0.71], is in FGFR1 on chromosome 8. However, this locus only reached genome-wide significance for the left hand 2D:4D ratio in the multi-ethnic meta-analysis (P = 3.4 × 10−7 in the female left hand European analysis), and would not be declared significant after correction for multiple testing given the large numbers of secondary analyses performed [i.e. secondary analyses involved analysis of males, females, left hand (both European and multi-ethnic analyses), right hand and average 2D:4D ratio, plus four sets of genome-wide sex heterogeneity analysis].
Gene by sex interaction
GREML analysis in ALSPAC showed no significant indication of gene by sex interaction (left: vgxe = 0.000, SE = 0.138, P = 0.5; right: vgxe = 0.117, SE = 0.144, P = 0.21; mean: vgxe = 0.000, SE = 0.140, P = 0.5). Consistent with this, only one locus on chromosome 9 reached genome-wide significance for difference in the magnitude of the regression coefficients between males and females for the average of both hands 2D:4D ratio (top SNP, rs16929125, A allele frequency = 0.91, heterogeneity P = 1.17 × 10−8; Supplementary Material, Fig. S7 for Manhattan plots and Supplementary Material, Fig. S8 for QQ plots). However, this locus only reached genome-wide significance for average 2D:4D ratio, and would not be declared significant after correction for multiple testing given the large number of secondary analyses performed.
Gene prioritization, pathway and tissue analysis
We used Data-driven Expression-Prioritized Integration for Complex Traits (DEPICT) (44) to identify the most likely causal gene at each locus and to investigate enriched pathways. DEPICT identified the nearest gene to the top associated signal to be the most likely causal gene in 10 of our 12 signals (Supplementary Material, Table S4); GLIS1, LDAH (previously known as C2orf43), OLA1, LIN28B, GLI3, FLI1, SMOC1, SALL1, TOX3 and SALL3. At the 2q31.1 locus, DEPICT prioritized two genes, HODX11 and HOXD12, whilst at the 1q22 locus, five genes were prioritized: EFNA1, DPM3, EFNA3, KRTCAP2 and SLC50A1. We will subsequently refer to this locus as EFNA1, which is the nearest gene to the top association signal.
When using the meta-analysis results from the average 2D:4D ratio of both hands, one gene set reached a false discovery rate (FDR) P < 0.01, which mapped to the MSX1 PPI sub-network (Supplementary Material, Table S5). The tissue enrichment analysis did not identify any tissues with a FDR P < 0.01 (Supplementary Material, Table S6). Based on the expression data of 53 tissue types from the Genotype-Tissue Expression (GTEx) Consortium, four of the nearest genes to our lead SNPs showed high tissue expression in the testis or adrenal gland (LDAH, LIN28B, SMOC1 and C16orf97; Supplementary Material, Fig. S9) relative to the other available tissues. Three of these four also showed expression in the brain (LDAH, LIN28B and SMOC1), in addition to two nearest genes, which showed high expression in the brain (OLA1 and SALL3).
Association between 2D:4D ratio and testosterone sensitivity
We examined the association of 2D:4D ratio variation with the length of an established CAG repeat polymorphism in the AR gene, a proxy of testosterone sensitivity, in a meta-analysis of the ALSPAC and QIMR cohorts. We found nominal evidence for a weak positive association between the number of CAG repeats in the AR gene on the X chromosome and mean 2D:4D ratio in females (mean of repeats: β = 0.056, P = 0.02; lower length repeat: β = 0.047, P = 0.03), but not in males (cf. Table 3, see Supplementary Material, Table S7 for left and right 2D:4D ratio). The 91 SNPs in AR in the GWAS showed little evidence for association with 2D:4D ratio (minimum P = 0.03; Supplementary Material, Fig. S10).
Table 3.
Association between the number of CAG repeats in the AR gene and the mean of the left and right hand 2D:4D ratios
| ALSPAC | QIMR | Meta-analysis | |
|---|---|---|---|
| All individuals | |||
| N = 5328 | N = 498 | N = 5826 | |
| Mean | 0.014 (0.016), P = 0.19 | 0.040 (0.052), P = 0.22 | 0.016 (0.015), P = 0.14 |
| High | 0.012 (0.015), P = 0.21 | 0.014 (0.046), P = 0.38 | 0.012 (0.014), P = 0.20 |
| Low | 0.013 (0.016), P = 0.21 | 0.041 (0.057), P = 0.24 | 0.015 (0.015), P = 0.16 |
| Male | |||
| N = 2615 | N = 231 | N = 2846 | |
| Mean | −0.002 (0.020), P = 0.54 | −0.099 (0.083), P = 0.88 | −0.007 (0.019), P = 0.65 |
| Female | |||
| N = 2713 | N = 287 | N = 3000 | |
| Mean | 0.046 (0.028), P = 0.05 | 0.125 (0.072), P = 0.04 | 0.056 (0.026), P = 0.02 |
| High | 0.030 (0.025), P = 0.12 | 0.068 (0.058), P = 0.12 | 0.036 (0.023), P = 0.06 |
| Low | 0.040 (0.026), P = 0.06 | 0.112 (0.082), P = 0.09 | 0.047 (0.025), P = 0.03 |
Displayed are beta (SE) and P-values in each of the cohorts and the combined estimates from the fixed effects, inverse-variance weighted meta-analysis. ‘Mean’ refers to analyses involving the average CAG repeat length, ‘High’ refers to analyses involving the highest length repeat and ‘Low’ refers to analyses involving the lower length repeat. One-tailed P-values testing for a positive association between CAG repeat length and 2D:4D ratio are presented.
2D:4D associated variants and other traits
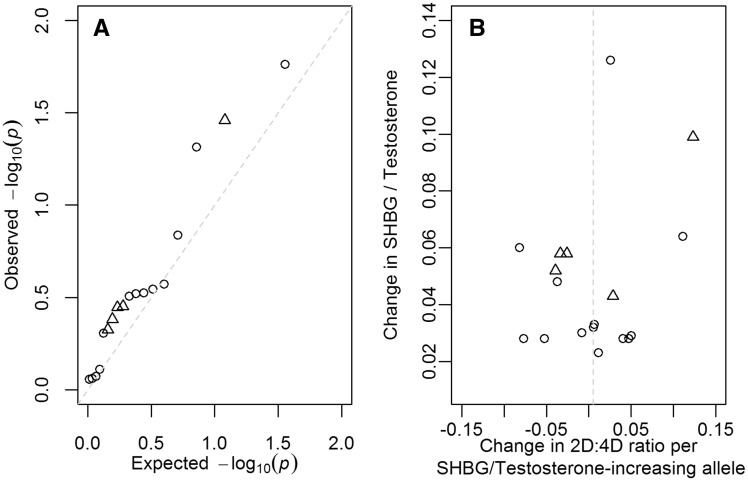
Given the putative relationship between prenatal testosterone levels and 2D:4D ratio, we also examined the association between five published SNPs (rs12150660, rs5934505, rs10822186, rs10822184 and rs72829446) shown to influence testosterone levels (45,46) using our 2D:4D ratio meta-analysis of the discovery cohorts; one other reported SNP was not tested, rs6258, as it was excluded from our meta-analysis as the minor allele frequency was <1%. Due to the high correlation between testosterone and its principal binding protein, SHBG, we also tested the association between the 13 published loci for SHBG (47, 48) and 2D:4D ratio. We observed three associations with left hand 2D:4D ratio at P < 0.05: the C allele at rs1641537 (allele frequency = 0.87) was associated with increased 2D:4D ratio (β = 0.111, P = 0.05) and increased SHBG, the T allele at rs1573036 (allele frequency = 0.39) was associated with decreased 2D:4D ratio (β = −0.077, P = 0.02) and increased SHBG and the T allele at rs72829446 (allele frequency = 0.11) was associated with increased 2D:4D ratio (β = 0.123, P = 0.04) and increased testosterone (Fig. 2) (i.e. two out of the three associations were in the opposite direction to expected). We also failed to detect enrichment for association with 2D:4D ratio over all 18 testosterone and SHBG SNPs (Fisher’s combined probability test P = 0.10). Additionally, there was no difference in the male and female effect sizes from the sex-stratified analyses across the 18 SNPs (Fisher’s combined probability test for the heterogeneity P = 0.23). Only one SNP, rs3779195, showed heterogeneity between males and females with the SHGB increasing T allele negatively associated with 2D:4D ratio in males and positively associated with 2D:4D ratio in females (βmale = −0.183, Pmale = 0.01; βfemale = 0.053, Pfemale = 0.39; Phet = 0.01).
Figure 2.
Plots highlighting the relationship of 13 SHBG and five testosterone associated SNPs (45–48) with the left hand 2D:4D ratio discovery meta-analysis. (A) Q-Q plot of the meta-analysis P-values for each of the SNPs. (B) Plot of the β coefficient for the left hand 2D:4D ratio meta-analysis against the β coefficient previously reported for SHBG or testosterone (the 13 SHBG associated SNPs are aligned to the SHBG increasing allele and the five testosterone associated SNPs are aligned to the testosterone increasing allele). Triangles indicate SNPs associated with testosterone, circles indicate SNPs associated with SHBG.
Given the previous observational epidemiological associations between 2D:4D ratio and sex-dependent behaviors, we used publicly available GWAS summary results for a variety of traits to estimate genetic correlations with 2D:4D ratio using linkage-disequilibrium (LD) score regression (49). We present the genetic correlation results in Supplementary Material, Table S8, but advise caution in their interpretation, considering the recommendation by Bulik-Sullivan et al. (49) regarding conducting genetic correlation analysis for traits with heritability z-scores below 4, as the estimates tend to be noisy and less reliable (the z-scores for the left hand, right hand and average of both hands were 2.9, 3.0 and 3.3, respectively).
Discussion
The 2D:4D ratio, a sexually dimorphic trait, has been extensively used in adults as a biomarker for prenatal androgen exposure. In the largest genetic association study of 2D:4D ratio to date, we identified nine novel loci for 2D:4D ratio, in addition to replicating two previously identified loci, LIN28B and SMOC1. These 11 loci explained 3.8% of the variance in mean 2D:4D ratio. After assessing association between 2D:4D ratio and a range of testosterone-related traits, we found no conclusive evidence of the 2D:4D ratio constituting a marker of prenatal androgen exposure, although it is possible that pathways involving testosterone may become apparent as the size of our GWAS increases in the future. Yet, associations at distinct novel loci provide additional insight into the underlying biology shaping 2D:4D ratio variation.
The association signal on 1p32.3 spans GLIS1, which is expressed across several organs in the reproductive system, including the prostate, vagina, testis and cervix. Glis1, the protein encoded by GLIS1, is a Kruppel-like zinc finger protein that appears to have a critical role in controlling gene expression during specific stages of embryogenesis (50).
At 1q21-q22, the closest gene to the associated variants is EFNA1, which encodes a member of the ephrin family and has been implicated in mediating developmental events, notably in the nervous system. SNPs in the region of EFNA1, DPM3 and KRTCAP3 have previously been associated with prostate cancer risk (51) and γ-glutamyl transferase (GGT), an indicator of liver disease (52). Based on the results from GTEx, EFNA1 is also mainly expressed in the liver, which is the most active site of lipid metabolism. A proxy for our lead 2D:4D ratio associated SNP at this locus, rs11264329, was associated with total and LDL cholesterol levels (53). Additionally, the top SNP identified at 2p24.1 is in LDAH (previously known as C2orf43); SNPs in LDAH have also been associated with prostate cancer risk (54,55). The protein encoded by LDAH is involved in cholesterol mobilization (56). Testosterone, which is linked to prostate cancer risk, is created when luteinizing hormone (LH) triggers the testicular Leydig cells to convert cholesterol to testosterone. Therefore, these two loci could implicate cholesterol metabolism in steroidogenesis as a link to testosterone exerting a role on 2D:4D ratio variation, albeit requiring further investigation into the functional implications.
OLA1, which maps to the 2q31.1 locus, plays multiple roles in the regulation of cell proliferation and cell survival. Ding et al. show that mouse embryos lacking OLA1 have delayed development leading to immature organs and stunted growth, which were frequently lethal prenatally (57). Their data suggests that there is a defect in cell proliferation due to a delay in cell cycle progression meaning that the mutant embryos appeared to undergo fewer proliferation cycles resulting in growth restriction.
SNPs in the 7p14.1 region map within GLI3. The gene encodes a zinc finger transcription factor that functions in the hedgehog signal transduction pathway. SNPs in this region have also been associated with facial morphology, namely nose wing breadth (58), and implicated in several Mendelian disorders which are characterized by craniofacial and limb abnormalities. Specifically, there are several disorders and conditions where polydactyly is a feature (59), including Greig cephalopolysyndactyly syndrome (GCPS), and Pallister-Hall syndrome (PHS). There is some evidence for brachydactyly (shortened digits) in a mouse null Gli3 mutant developed by Sheth et al. (60), and in patients with PHS (59).
The association signal arising from the 11q24.1-q24.3 locus is intergenic between FLI1 and ETS1. Through GWAS, SNPs in the FLI1 gene have been shown to be associated with height, with similar effects in both males and females (61). In addition, SNPs in ETS1 have been shown to be associated with rheumatoid arthritis (62) and celiac disease (63) in European populations and with systemic lupus erythematosus (64–66) in Chinese populations; all of these diseases have a higher prevalence in females. This gene encodes the protein Ets1, which is expressed in a variety of tissues throughout the development of an embryo and plays a role in pituitary hormone secretion (67). In mice, the ETS factor family defines Shh spatial expression in limb buds and alterations define pathogenetic mechanism leading to preaxial polydactyly (68).
The GWAS signal on chromosome 16q12.1 maps in the vicinity of the SALL1, TOX3 and C16orf97 genes. Not much is known about the function of C16orf97. However, SALL1 was identified by DEPICT as being the most likely causal gene for the rs6499762 association; mutations in SALL1 cause Townes-Brocks syndrome (69), a condition characterized by hand malformations, abnormally shaped ears and anal atresia, among other genital malformations (70). Additionally, SALL3 on chromosome 18q23, is also part of the human spalt-like gene family, which is associated with syndromic forms presenting with skeletal abnormalities. Kohlhase et al. (71) characterized this gene and implicated it in the 18q deletion syndrome, which results in mental and growth retardation, developmental delay, hearing loss, and facial and limb abnormalities including tapered fingers (72). Altogether, several links between 2D:4D ratio and testosterone metabolism can be derived from these associations, involving hormonal pathways and the process of sexual differentiation during early development. Further, SNPs in the SALL3 region have also been associated with prostate cancer (51), which may suggest additional links between 2D:4D ratio and testosterone.
One gene set was identified as being associated with 2D:4D ratio, the MSX1 PPI subnetwork. An Msx1-interacting network of transcription factors has been shown to operate during early tooth development (73). Therefore, the identification of this gene set may be highlighting a network that is involved in several areas of development.
Interestingly, we didn’t find strong evidence of association between variants within the HOXA gene cluster and 2D:4D ratio. This lack of association does not preclude variation in distal enhancers acting through effects on the expression of HOXA cluster genes nor that variants of smaller effect act from within the cluster itself [with an alpha of 5 × 10−8 we had 80% power to detect a genetic variant that explained approximately 0.28% of the variance in the left hand 2D:4D ratio in Europeans only (N = 14 382)]. We did, however, find an association involving a variant in HOXD12 (the most strongly associated SNP was rs847158, P = 9.58 × 10−11, which replicated in the 23andMe dataset). The exact role of HOXD12 has not yet been determined; however, the homeobox family of genes plays an important role in morphogenesis and is particularly relevant in the development of the limbs and genitals (74).
2D:4D ratio as a marker of testosterone exposure
2D:4D ratio has been used extensively in adults as a biomarker for prenatal androgen exposure. However, whether the digit ratio reliably reflects prenatal androgen exposure has not been convincingly demonstrated. Most of the data linking 2D:4D ratio with prenatal androgen exposure is based on preclinical or indirect evidence, including studies that indicate that 2D:4D ratio is fixed early in gestation and is associated with adult levels of circulating testosterone (3,75). The most direct test of this hypothesis to date was performed by Lutchmaya et al. who showed that testosterone levels in amniotic fluid from the second trimester of pregnancy were not associated with 2D:4D ratio at 2 years of age (76). However, the authors did find that an increased ratio of testosterone to estradiol was associated with a lower (or more male like) 2D:4D ratio, suggesting that the relationship between digit ratio and prenatal hormones may be more complicated and not only reflect testosterone levels (76,77).
In the present study we attempted to use genetic evidence to find support for the testosterone biomarker hypothesis. Our rationale was that if prenatal testosterone affects 2D:4D ratio, then it is logical that polymorphisms in genes related to androgen sensitivity (e.g. in XAR) and/or SNPs robustly associated with androgen levels/levels of SHBG, should also be related to 2D:4D ratio. Whilst we did detect some evidence of a positive association between the number of CAG repeats in AR and 2D:4D ratio in females, we note that the small effect size would not be significant after adjusting for the multiple statistical tests we performed. Power calculations suggest that our combined sample of N = 5826 individuals in the XAR meta-analysis was well powered (∼78%) to detect a locus responsible for 0.001% of the phenotypic variance in 2D:4D ratio (one-tailed α = 0.05). In comparison, all of our genome-wide significant SNPs explained >0.001% of the variance in 2D:4D ratio (most explained much more variance than this). This suggests that if genetic variation in XAR does contribute to variation in 2D:4D ratio through, for example, sensitivity to testosterone, its effect is likely to be small relative to other sources of genetic variation. Two recent smaller meta-analyses also failed to find an association between length of the repeat in XAR and 2D:4D ratio (78,79).
Likewise, using SNPs that are associated with testosterone and SHBG, we were unable to detect any enrichment for association with 2D:4D ratio. We were also unable to identify any genetic correlation between 2D:4D ratio and a range of traits and diseases previously implicated with 2D:4D ratio variation. This indicates that the previously identified observational associations may not be driven by known genetic loci that are shared between the traits, although we acknowledge the power of analysis was low and confidence intervals around our estimates were large.
Whilst we were unable to find any convincing evidence that sensitivity to/levels of androgens is a major driver of the individual differences in 2D:4D ratio seen in the normal population, there are several key assumptions underlying the use of genetic variation to investigate the link between prenatal androgen exposure and 2D:4D ratio. First, investigating the association between the number of CAG repeats in AR and 2D:4D ratio relies on the assumption that CAG length reflects androgen sensitivity. There is fairly good evidence for this, at least in vitro as derived from the Chamberlain et al. functional study showing a linear relationship between increased CAG length and decreased transactivation function (41). Second, we assume that the genotyping of the CAG repeat is accurate. Although there was some discordance in the replicate genotyping in ALSPAC, the majority of discrepancies were only one CAG repeat different between the replicates. Our simulations presented in the Supplementary Material indicate that this degree of measurement error had little influence on the power of our association analysis. Third, we assume that the SNPs associated with adult levels of testosterone/SHBG also reflect testosterone/SHBG levels prenatally and that the effect of these SNPs are similar in males and females. The testosterone-associated SNPs were identified in GWAS of adult men only and one of the two GWAS for SHBG, identifying only one novel locus, was in post-menopausal women only. However, Coviello et al. conducted sex-stratified analyses and identified only one locus with significant heterogeneity on SHBG between males and females (48). They also showed that the SHBG SNPs identified explained ∼15.6% of the variation in SHBG in men and ∼8.4% of the variation in women. This indicates that although the SNPs have a greater overall effect in males, they are still likely to be associated with androgens in females. It is too difficult to measure androgen levels in the fetus so we are unable to confirm that these SNPs are also associated with androgen levels prenatally. Finally, although none of our novel loci showed direct evidence of being related to pathways involving testosterone, it does not preclude the very real possibility that testosterone influences 2D:4D ratio by down-regulating or up-regulating the expression of genes involved in its determination. Indeed, in GWAS of height (80) and BMI (81), the role of expected hormone pathways only appeared when the studies were sufficiently powered to find far greater number of genome-wide significant hits than in the present study.
In conclusion, we have conducted the largest GWAS of 2D:4D ratio to date and identified nine novel loci robustly associated with 2D:4D ratio in Europeans, bringing the total number of robustly associated loci to 11. We were unable to find strong evidence from our genetic study to support the hypothesis that 2D:4D ratio is a direct biomarker of prenatal exposure to androgens in healthy individuals, although our findings do not explicitly exclude this possibility, and pathways involving testosterone may become apparent as the size of the discovery sample increases further. Our findings provide new insight into the underlying biology shaping 2D:4D variation in the general population.
Materials and Methods
Participants
We drew on data from six cohorts for the discovery genome-wide meta-analysis and association with the CAG repeat in AR including the Avon Longitudinal Study of Parents and Children (ALSPAC), the Generation R Study, the Rotterdam Study, the Western Australian Pregnancy Cohort (Raine) Study, TwinsUK and the Queensland Institute of Medical Research (QIMR) sample, which was drawn from the Brisbane Adolescent Twin Study [BATS; also known as the Brisbane Longitudinal Twin Study (BLTS)]. The 23andMe cohort was used for replication of the genome-wide significant findings. Details of each of these studies, including how the 2D:4D ratio was measured and the genotyping methods, are provided in the Supplementary Material.
Ethics statement
All cohorts in the discovery meta-analysis or replication obtained ethical approval from their local ethics review boards; ALSAPC from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees, The Generation R Study from the Medical Ethics Committee of the Erasmus Medical Center in Rotterdam, QIMR from the QIMR Human Research Ethics Committee, the Raine study from the King Edward Memorial Hospital and Princess Margaret Hospital for Children Human Research Ethics Committees, the Rotterdam Study from the Medical Ethics Committee of the Erasmus Medical Center in Rotterdam, TwinsUK from the Guy’s and St Thomas’ (GSTT) Ethics Committee and research participants from 23andMe provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review).
Statistical analysis
The 2D:4D ratio was calculated as the length of the second digit divided by the length of the fourth digit, multiplied by 100 so as to avoid computational difficulties due to the low variance of the trait. In all studies, the measure was normally distributed, so no further transformation was applied.
Genome-wide complex trait analysis of 2D:4D ratio
To estimate the proportion of additive genetic variance in 2D:4D ratio explained by directly genotyped SNPs, we conducted a univariate GREML analysis using the GCTA software (82) in >4900 individuals from ALSPAC. Sex was included as a fixed effect in the model. Bivariate GREML analysis (83) was used to estimate the genetic correlation between the left and right hand 2D:4D ratio, which will indicate whether the same genetic variants contribute to variation in the ratio of each hand. Additionally, a gene by sex analysis was conducted using the gene by environment test, to indicate whether the SNPs associated with 2D:4D ratio differed between males and females.
Genome-wide association analysis: discovery
Genome-wide association analysis using imputed dosages to account for uncertainty in the imputation was performed using linear regression in each cohort, adjusting for sex. In addition, the QIMR and TwinsUK cohorts accounted for zygosity and relatedness. The Generation R Study (European subset), the Rotterdam Study and Raine adjusted for four principal components for population stratification. A sensitivity analysis to maximize power was conducted by including all individuals of the Generation R Study with adjustment for 20 principal components to account for the multi-ethnic sample as performed earlier (84–88). Results presented in the main text are derived from the meta-analysis including the European subset only, with the multi-ethnic analysis presented in the Supplementary Material. SNPs were tested for association with left 2D:4D ratio, right 2D:4D ratio (all cohorts excluding the Generation R Study) and the mean of the left and right hand 2D:4D ratios (all cohorts excluding the Generation R Study). Results were combined using fixed-effects inverse-variance weighted meta-analysis in METAL (89), adjusting for genomic control. Within each study, SNPs with a MAF < 1%, an INFO score < 0.4 or R2 for imputation quality < 0.3 were excluded from the meta-analysis and SNPs that were reported in less than 50% of the total sample size were excluded from further follow-up.
We also tested whether the regression coefficients differed between males and females. We carried out genome-wide association analysis in males and females separately in each of the discovery cohorts and, as with the main analysis, we excluded variants with a MAF <1% and poor imputation quality (INFO score <0.4 or R2 for imputation quality < 0.3). We performed a fixed-effects inverse-variance weighted meta-analysis of each sex in METAL (89), adjusting for genomic control. Finally, we excluded variants that were reported less than 50% of the male and female sample sizes, and performed a chi-square test of heterogeneity between the meta-analysed male and female effects in METAL (89) to test for the difference between the effect sizes in males and females and produce an overall level of significance.
Conditional and joint association analysis
We performed approximate conditional and joint SNP association analysis using the GCTA software (43), which utilizes meta-analysis summary statistics and LD structure from a reference sample. We used this approach to identify additional signals in regions of association, using a subset of 15 000 UK Biobank (90) individuals as the reference sample to approximate LD patterns. The selected subset of the UK Biobank individuals were of European descent and unrelated to anyone else in the subset.
Genome-wide association analysis: replication
SNPs that reached genome-wide significance (P < 5 × 10−8 for the left, right or average 2D:4D ratio) in the discovery meta-analysis were replicated in the 23andMe dataset. If the imputed SNP passed quality control or the genotyped SNP was unavailable, then the imputed SNP was used for analysis, otherwise the genotyped SNP was used. Analysis of each SNP was performed using linear regression, adjusting for sex, age, the first five principal components for population stratification and genotyping platform. Results were adjusted for a genomic control inflation factor of λ = 1.074.
Variance explained
The variance explained by each SNP was calculated using the effect size from the discovery meta-analysis (beta, β), the minor and major allele frequencies (p and q, respectively) and the variance of the 2D:4D ratio (Var(Y)) using the following formula:
We used the median standard deviation in 2D:4D ratio across the cohorts, which was 3.40 for the left hand, 3.35 for the right hand and 3.06 for the average of both hands to calculate the phenotypic variance in this formula. Under the assumption that all SNPs independently contribute to 2D:4D ratio, we computed the total variance explained by the lead SNPs at the genome-wide significant loci as the sum of the single-SNP explained variances.
Gene prioritization, gene set and tissue/cell type enrichment analysis
We conducted three analyses implemented in DEPICT (44) to establish the functional connections with our lead signals. First, we prioritized genes which are most likely to be causal for 2D:4D ratio by correlating the reconstituted gene set membership of each gene nearby the associated signal to genes from other associated loci and adjusting for potential sources of bias such as gene size. Second, we performed a gene set enrichment analysis, which tests if the genes in the associated loci are enriched in the reconstituted gene sets. Third, we analysed expression enrichment across particular tissues or cell types, by testing whether genes associated with 2D:4D ratio loci were seen highly expressed in any of the 209 Medical Subject Heading (MeSH) annotations using data from 37 427 expression arrays. In all three analyses, we used FDR to adjust for multiple testing, with an FDR P < 0.01 defined as significant.
The DEPICT analyses were based on independent lead SNPs (r2 < 0.1, European populations 1000 genomes reference panel) with P-values below the genome-wide significant threshold (P < 5 × 10−8). Gene-set enrichment was further grouped into ‘meta gene sets’ by similarity clustering, as described previously (44).
Additionally, we investigated the functionality of the genes closest to the lead SNPs identified in these analyses using expression data on 53 tissue types from the GTEx Consortium (91).
Analysis of the CAG repeat polymorphism in AR
Information on the CAG repeat polymorphism in AR was available in the ALSPAC and QIMR cohorts (see Supplementary Material for genotyping information). In ALSPAC, we performed linear regression of 2D:4D ratio (left, right and mean of the left and right) on length of CAG repeat (in females either average repeat length, the highest length repeat or the lower length repeat). The QIMR analyses were conducted using full information maximum likelihood structural equation models in openMx (92) which explicitly accounted for relatedness and zygosity while estimating the linear effect of the CAG repeat on 2D:4D ratio. We performed analyses including all participants (with sex as a covariate), females separately and males separately. A fixed-effects inverse-variance weighted meta-analysis was used to combine the results from the two cohorts using the rmeta package in R (version 3.0.0) (93). A one-tailed hypothesis was used to test whether there was a positive association between the number of CAG repeats and 2D:4D ratio.
Genetic correlation with associated traits
We used LD score regression, which has been described in detail elsewhere (49), to calculate the genetic correlation between 2D:4D ratio and a range of traits and diseases it has been associated with in observational studies. Note that, we conducted this analysis using the European only meta-analysis results as LD score regression cannot accommodate LD variation between diverse populations. Briefly, the LD score is a measure of how much genetic variation each SNP tags; so if a SNP has a high LD score then it is in high LD with many nearby SNPs. SNPs with high LD scores are more likely to contain more true signals and hence provide more chance for overlap with genuine signals between GWAS. The method uses summary statistics from the GWAS meta-analyses of 2D:4D ratio and the traits of interest, calculates the cross-product of test statistics at each SNP, and then regresses the cross-product on the LD score. The slope of the regression is a function of the genetic correlation between traits. If there is overlap between the samples used in each of the meta-analyses (or cryptic relatedness between samples) it will only affect the intercept of the regression, and will not bias the estimate of the genetic covariance.
Summary statistics from the GWAS meta-analysis for traits and diseases of interest were downloaded from the relevant consortium website (see Supplementary Material, Table S8 for references). The summary statistics files were reformatted for LD score regression analysis using the munge_sumstats.py python script provided on the developer’s website (https://github.com/bulik/ldsc; date last accessed August 10, 2017); we filtered the summary statistics to the subset of HapMap3 SNPs, as advised by the developers, to ensure that no bias was introduced due to poor imputation quality. Where the sample size for each SNP was included in the results file was flagged using –N-col; if no sample size was available then the maximum sample size reported in the reference for the GWAS meta-analysis was used (i.e. the summary statistics for each SNP was assumed to have been estimated using the same sample size). SNPs were excluded if the minor allele frequency was <0.01, the strand was ambiguous, the rs number was duplicated or they had a sample size less than 60% of the total sample size available. Once all the files were reformatted, we used the ldsc.py python script, also on the developer’s website, to calculate the genetic correlation between 2D:4D ratio and each of the traits and diseases. The European LD Score files that were calculated from the 1000 Genomes reference panel and provided by the developers were used for the analysis.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
Generation R Study: The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond [STAR-MDC], Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The generation and management of GWAS genotype data for the Generation R Study was done at the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We would like to thank Karol Estrada, Dr. Tobias A. Knoch, Anis Abuseiris, Luc V. de Zeeuw, and Rob de Graaf, for their help in creating GRIMP, BigGRID, MediGRID, and Services@MediGRID/D-Grid, [funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G] for access to their grid computing resources. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Manoushka Ganesh, Lizbeth Herrera and Marjolein Peters for their help in creating, managing and QC of the GWAS database.
QIMR: We thank the Brisbane twins and siblings for their participation; Marlene Grace, Ann Eldridge and Natalie Garden for sample collection; Kerrie McAloney for study co-ordination; Harry Beeby, Daniel Park, and David Smyth for IT support, Anjali Henders and the Molecular Genetics Laboratory for DNA sample preparation, and Scott Gordon for genotyping QC.
The Rotterdam Study: The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Karol Estrada, PhD, Yurii Aulchenko, PhD, and Carolina Medina-Gomez, PhD, for the creation and analysis of imputed data. We would like to thank Dr. Karol Estrada, Dr. Fernando Rivadeneira, Dr. Tobias A. Knoch, Marijn Verkerk, Anis Abuseiris, Dr. Linda Boer and Rob de Graaf (Erasmus MC Rotterdam, The Netherlands), for their help in creating and maintaining GRIMP. The authors are very grateful to the study participants, the staff from the Rotterdam Study (particularly L. Buist and J.H. van den Boogert) and the participating general practitioners and pharmacists.
Raine: The authors are grateful to the Raine Study participants, their families, and to the Raine Study research staff for cohort coordination and data collection. The authors gratefully acknowledge the assistance of the Western Australian DNA Bank (National Health and Medical Research Council of Australia National Enabling Facility).
23andMe: We thank the 23andMe research participants and employees for their contributions to this study. We thank the following members of the 23andMe Research Team: Michelle Agee, Babak Alipanahi, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, David A. Hinds, Bethann S. Hromatka, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Matthew H. McIntyre, Joanna L. Mountain, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, J. Fah Sathirapongsasuti, Olga V. Sazonova, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Vladimir Vacic, and Catherine H. Wilson.
Conflict of Interest statement: Y.J., A.A., and J.Y.T. are employees of 23andMe, Inc. and hold stock or stock options in 23andMe.
Funding
N.M.W was supported by a National Health and Medical Research Council Early Career Fellowship (APP1104818). E.S. was supported by the European Commission within the framework of the Erasmus-Western Balkans (ERAWEB). M.H was supported by a National Health and Medical Research Council Practitioner Fellowship (APP1058935). F.R. was supported by the Netherlands Scientific Organization (NWO) and ZonMW Project number: NWO/ZONMW-VIDI-016-136-367. S.E.M was supported by a National Health and Medical Research Council Senior Research Fellowship (APP1103623). D.M.E was supported by an Australian Research Council Future Fellowship (FT130101709) and an MRC programme grant (MC_UU_12013/4).
ALSPAC: GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. The UK Medical Research Council and the Wellcome Trust (grant reference: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This work is supported by a Medical Research Council program grant (grant reference: MC_UU_12013/4 to D.M.E). The androgen receptor CAG repeat data was generated with funding from the Medical Research Council (grant reference: G0500953 to Barbara Maughan). This publication is the work of the authors, and D.M.E will serve as guarantor for the contents of this paper.
Generation R Study: The general design of Generation R Study was made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Netherlands Organisation for Scientific Research (NWO), the Ministry of Health, Welfare and Sport and the Ministry of Youth and Families. The musculoskeletal research of the Generation R Study was partly supported by the European Commission grant HEALTH-F2-2008-201865-GEFOS. Additionally, the Netherlands Organization for Health Research and Development supported authors of this manuscript (ZonMw 907.00303, ZonMw 916.10159, ZonMw VIDI 016.136.361 to V.W.J. and ZonMw VIDI 016.136.367 to F.R.). This project also received funding from the European Union’s Horizon 2020 research and innovation programme under the following grant agreements: No. 633595 (DynaHEALTH) and No. 733206 (LIFECYCLE), and from the European Research Council (ERC Consolidator Grant, ERC-2014-CoG-648916 to V.W.J.).
QIMR: The QIMR studies were supported by funding from the Australian National Health and Medical Research Council (grant numbers: 241944, 339462, 389927, 389875, 389891, 389892, 389938, 443036, 442915, 442981, 496739, 552485 and 552498, and most recently 1049894) and the Australian Research Council (grant numbers: A7960034, A79906588, A79801419, DP0212016 and DP0343921).
The Rotterdam Study: The GWAS datasets were supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project no. 050-060-810. Dr Fernando Rivadeneira received an additional grant from the Netherlands Organization for Health Research and Development ZonMw VIDI 016.136.367. The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam.
Twins UK: The study was funded by the Wellcome Trust (Ref: 105022/Z/14/Z); European Community’s Seventh Framework Programme (FP7/2007–2013). The study also receives support from the National Institute for Health Research (NIHR)—funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR.
Raine: The following Institutions provide funding for Core Management of the Raine Study: The University of Western Australia (UWA), Raine Medical Research Foundation, UWA Faculty of Medicine, Dentistry and Health Sciences, The Telethon Institute for Child Health Research, Curtin University, Edith Cowan University and Women and Infants Research Foundation. This study was supported by project grants from the National Health and Medical Research Council of Australia (grant numbers: 403981, 003209 and 1021105; http://www.nhmrc.gov.au; date last accessed April 13, 2018) and the Canadian Institutes of Health Research (grant number: MOP-82893; http://www.cihr-irsc.gc.ca/e/193.html; date last accessed April 13, 2018). This work was also supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Funding to pay the Open Access publication charges for this article was provided by the University of Bristol.
References
- 1. Phoenix C.H., Goy R.W., Gerall A.A., Young W.C. (1959) Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology, 65, 369–382. [DOI] [PubMed] [Google Scholar]
- 2. Arnold A.P., Breedlove S.M. (1985) Organizational and activational effects of sex steroids on brain and behavior: a reanalysis. Hormones Behav., 19, 469–498. [DOI] [PubMed] [Google Scholar]
- 3. Manning J.T., Scutt D., Wilson J., Lewis-Jones D.I. (1998) The ratio of 2nd to 4th digit length: a predictor of sperm numbers and concentrations of testosterone, luteinizing hormone and oestrogen. Hum. Reprod., 13, 3000–3004. [DOI] [PubMed] [Google Scholar]
- 4. Ecker A. (1875) Einige Bemerkungen über einen schwankenden Charakter in der Hand des Menschen. Arch. Anthropl., 8, 67–74. [Google Scholar]
- 5. Wilson G.D. (1983) Finger-length as an index of assertiveness in women. Pers. Individ. Diff., 4, 111–112. [Google Scholar]
- 6. McIntyre M.H., Ellison P.T., Lieberman D.E., Demerath E., Towne B. (2005) The development of sex differences in digital formula from infancy in the Fels Longitudinal Study. Proc. Biol. Sci., 272, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trivers R., Manning J., Jacobson A. (2006) A longitudinal study of digit ratio (2D:4D) and other finger ratios in Jamaican children. Hormones Behav., 49, 150–156. [DOI] [PubMed] [Google Scholar]
- 8. Manning J.T., Henzi P., Venkatramana P., Martin S., Singh D. (2003) Second to fourth digit ratio: ethnic differences and family size in English, Indian and South African populations. Ann. Hum. Biol., 30, 579–588. [DOI] [PubMed] [Google Scholar]
- 9. Loehlin J.C., McFadden D., Medland S.E., Martin N.G. (2006) Population differences in finger-length ratios: ethnicity or latitude? Arch. Sex. Behav., 35, 739–742. [DOI] [PubMed] [Google Scholar]
- 10. Manning J.T., Barley L., Walton J., Lewis-Jones D.I., Trivers R.L., Singh D., Thornhill R., Rohde P., Bereczkei T., Henzi P.. et al. (2000) The 2nd: 4th digit ratio, sexual dimorphism, population differences, and reproductive success. Evidence for sexually antagonistic genes? Evol. Hum. Behav., 21, 163–183. [DOI] [PubMed] [Google Scholar]
- 11. Coco M., Perciavalle V., Maci T., Nicoletti F., Di Corrado D., Perciavalle V. (2011) The second-to-fourth digit ratio correlates with the rate of academic performance in medical school students. Mol. Med. Rep., 4, 471–476. [DOI] [PubMed] [Google Scholar]
- 12. Hull M.J., Schranz N.K., Manning J.T., Tomkinson G.R. (2014) Relationships between digit ratio (2D:4D) and female competitive rowing performance. Am. J. Hum. Biol., 27, 157–163. [DOI] [PubMed] [Google Scholar]
- 13. Trivers R., Hopp R., Manning J. (2013) A longitudinal study of digit ratio (2D:4D) and its relationships with adult running speed in Jamaicans. Hum. Biol., 85, 623–626. [DOI] [PubMed] [Google Scholar]
- 14. Kim Y., Kim K., Kim T.H. (2014) Domain specific relationships of 2D:4D digit ratio in risk perception and risk behavior. J. Gen. Psychol., 141, 373–392. [DOI] [PubMed] [Google Scholar]
- 15. Madison G., Aasa U., Wallert J., Woodley M.A. (2014) Feminist activist women are masculinized in terms of digit-ratio and social dominance: a possible explanation for the feminist paradox. Front. Psychol., 5, 1011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam D., Ozorio B. (2014) An exploratory study of the relationship between digit ratio, illusion of control, and risk-taking behavior among chinese college students. J. Gamb. Stud.31, 1377–1385. [DOI] [PubMed] [Google Scholar]
- 17. Oh J.K., Kim K.T., Yoon S.J., Kim S.W., Kim T.B. (2014) Second to fourth digit ratio: a predictor of adult testicular volume. Andrology, 2, 862–867. [DOI] [PubMed] [Google Scholar]
- 18. Klimek M., Galbarczyk A., Nenko I., Alvarado L.C., Jasienska G. (2014) Digit ratio (2D:4D) as an indicator of body size, testosterone concentration and number of children in human males. Ann. Hum. Biol., 41, 518–523. [DOI] [PubMed] [Google Scholar]
- 19. Vladeanu M., Giuffrida O., Bourne V.J. (2014) Prenatal sex hormone exposure and risk of Alzheimer disease: a pilot study using the 2D:4D digit length ratio. Cognit. Behav. Neurol., 27, 102–106. [DOI] [PubMed] [Google Scholar]
- 20. Oyeyemi B.F., Iyiola O.A., Oyeyemi A.W., Oricha K.A., Anifowoshe A.T., Alamukii N.A. (2014) Sexual dimorphism in ratio of second and fourth digits and its relationship with metabolic syndrome indices and cardiovascular risk factors. J. Res. Med. Sci., 19, 234–239. [PMC free article] [PubMed] [Google Scholar]
- 21. Teatero M.L., Netley C. (2013) A critical review of the research on the extreme male brain theory and digit ratio (2D:4D). J. Autism Deve. Disord., 43, 2664–2676. [DOI] [PubMed] [Google Scholar]
- 22. Voracek M., Loibl L.M. (2009) Scientometric analysis and bibliography of digit ratio (2D:4D) research, 1998–2008. Psychol. Rep., 104, 922–956. [DOI] [PubMed] [Google Scholar]
- 23. McIntyre M.H. (2006) The use of digit ratios as markers for perinatal androgen action. Reprod. Biol. Endocrinol., 4, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paul S.N., Kato B.S., Cherkas L.F., Andrew T., Spector T.D. (2006) Heritability of the second to fourth digit ratio (2D:4D): a twin study. Twin Res. Hum. Genet., 9, 215–219. [DOI] [PubMed] [Google Scholar]
- 25. Voracek M., Dressler S.G. (2007) Digit ratio (2D:4D) in twins: heritability estimates and evidence for a masculinized trait expression in women from opposite-sex pairs. Psychol. Rep., 100, 115–126. [DOI] [PubMed] [Google Scholar]
- 26. Gobrogge K.L., Breedlove S.M., Klump K.L. (2008) Genetic and environmental influences on 2D:4D finger length ratios: a study of monozygotic and dizygotic male and female twins. Arch. Sex. Behav., 37, 112–118. [DOI] [PubMed] [Google Scholar]
- 27. Medland S.E., Loehlin J.C. (2008) Multivariate genetic analyses of the 2D:4D ratio: examining the effects of hand and measurement technique in data from 757 twin families. Twin Res. Hum. Genet., 11, 335–341. [DOI] [PubMed] [Google Scholar]
- 28. Voracek M., Dressler S.G. (2009) Brief communication: familial resemblance in digit ratio (2D:4D). Am. J. Phys. Anthropol., 140, 376–380. [DOI] [PubMed] [Google Scholar]
- 29. Lawrance-Owen A.J., Bargary G., Bosten J.M., Goodbourn P.T., Hogg R.E., Mollon J.D. (2013) Genetic association suggests that SMOC1 mediates between prenatal sex hormones and digit ratio. Hum. Genet., 132, 415–421. [DOI] [PubMed] [Google Scholar]
- 30. Medland S.E., Zayats T., Glaser B., Nyholt D.R., Gordon S.D., Wright M.J., Montgomery G.W., Campbell M.J., Henders A.K., Timpson N.J.. et al. (2010) A variant in LIN28B is associated with 2D:4D finger-length ratio, a putative retrospective biomarker of prenatal testosterone exposure. Am. J. Hum. Genet., 86, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He C., Kraft P., Chen C., Buring J.E., Pare G., Hankinson S.E., Chanock S.J., Ridker P.M., Hunter D.J., Chasman D.I. (2009) Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet., 41, 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C.. et al. (2008) Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet., 40, 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abouzeid H., Boisset G., Favez T., Youssef M., Marzouk I., Shakankiry N., Bayoumi N., Descombes P., Agosti C., Munier F.L.. et al. (2011) Mutations in the SPARC-related modular calcium-binding protein 1 gene, SMOC1, cause waardenburg anophthalmia syndrome. Am. J. Hum. Genet., 88, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okada I., Hamanoue H., Terada K., Tohma T., Megarbane A., Chouery E., Abou-Ghoch J., Jalkh N., Cogulu O., Ozkinay F.. et al. (2011) SMOC1 is essential for ocular and limb development in humans and mice. Am. J. Hum. Genet., 88, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rainger J., van Beusekom E., Ramsay J.K., McKie L., Al-Gazali L., Pallotta R., Saponari A., Branney P., Fisher M., Morrison H.. et al. (2011) Loss of the BMP antagonist, SMOC-1, causes Ophthalmo-acromelic (Waardenburg Anophthalmia) syndrome in humans and mice. PLoS Genet., 7, e1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Love H.D., Booton S.E., Boone B.E., Breyer J.P., Koyama T., Revelo M.P., Shappell S.B., Smith J.R., Hayward S.W. (2009) Androgen regulated genes in human prostate xenografts in mice: relation to BPH and prostate cancer. PLoS One, 4, e8384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schaeffer E.M., Marchionni L., Huang Z., Simons B., Blackman A., Yu W., Parmigiani G., Berman D.M. (2008) Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene, 27, 7180–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coleman I.M., Kiefer J.A., Brown L.G., Pitts T.E., Nelson P.S., Brubaker K.D., Vessella R.L., Corey E. (2006) Inhibition of androgen-independent prostate cancer by estrogenic compounds is associated with increased expression of immune-related genes. Neoplasia, 8, 862–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berenbaum S.A., Bryk K.K., Nowak N., Quigley C.A., Moffat S. (2009) Fingers as a marker of prenatal androgen exposure. Endocrinology, 150, 5119–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manning J.T., Bundred P.E., Newton D.J., Flanagan B.F. (2003) The second to fourth digit ratio and variation in the androgen receptor gene. Evol. Hum. Behav., 24, 399–405. [Google Scholar]
- 41. Chamberlain N.L., Driver E.D., Miesfeld R.L. (1994) The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res., 22, 3181–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xi H., Li M., Fan Y., Zhao L. (2014) A comparison of measurement methods and sexual dimorphism for digit ratio (2D:4D) in Han ethnicity. Arch. Sex. Behav., 43, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J.. et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. s361–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pers T.H., Karjalainen J.M., Chan Y., Westra H.J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T.. et al. (2015) Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun., 6, 5890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin G., Sun J., Kim S.T., Feng J., Wang Z., Tao S., Chen Z., Purcell L., Smith S., Isaacs W.B.. et al. (2012) Genome-wide association study identifies a new locus JMJD1C at 10q21 that may influence serum androgen levels in men. Hum. Mol. Genet., 21, 5222–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohlsson C., Wallaschofski H., Lunetta K.L., Stolk L., Perry J.R., Koster A., Petersen A.K., Eriksson J., Lehtimaki T., Huhtaniemi I.T.. et al. (2011) Genetic determinants of serum testosterone concentrations in men. PLoS Genet., 7, e1002313.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prescott J., Thompson D.J., Kraft P., Chanock S.J., Audley T., Brown J., Leyland J., Folkerd E., Doody D., Hankinson S.E.. et al. (2012) Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One, 7, e37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coviello A.D., Haring R., Wellons M., Vaidya D., Lehtimaki T., Keildson S., Lunetta K.L., He C., Fornage M., Lagou V.. et al. (2012) A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet., 8, e1002805.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B.. et al. (2015) An atlas of genetic correlations across human diseases and traits. Nat. Genet., 47, 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim Y.S., Lewandoski M., Perantoni A.O., Kurebayashi S., Nakanishi G., Jetten A.M. (2002) Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem., 277, 30901–30913. [DOI] [PubMed] [Google Scholar]
- 51. Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S.. et al. (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet., 45, 385–391. 391e381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chambers J.C., Zhang W., Sehmi J., Li X., Wass M.N., Van der Harst P., Holm H., Sanna S., Kavousi M., Baumeister S.E.. et al. (2011) Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet., 43, 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S.. et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C.. et al. (2011) Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet., 7, e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O.. et al. (2010) Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet., 42, 751–754. [DOI] [PubMed] [Google Scholar]
- 56. Goo Y.H., Son S.H., Kreienberg P.B., Paul A. (2014) Novel lipid droplet-associated serine hydrolase regulates macrophage cholesterol mobilization. Arterioscler. Thromb. Vasc. Biol., 34, 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ding Z., Liu Y., Rubio V., He J., Minze L.J., Shi Z.Z. (2016) OLA1, a translational regulator of p21, maintains optimal cell proliferation necessary for developmental progression. Mol. Cell. Biol., 36, 2568–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adhikari K., Fuentes-Guajardo M., Quinto-Sanchez M., Mendoza-Revilla J., Camilo Chacon-Duque J., Acuna-Alonzo V., Jaramillo C., Arias W., Lozano R.B., Perez G.M.. et al. (2016) A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun., 7, 11616. 11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Al-Qattan M.M., Shamseldin H.E., Salih M.A., Alkuraya F.S. (2017) GLI3-related polydactyly: a review. Clin. Genet., 92, 457–466. [DOI] [PubMed] [Google Scholar]
- 60. Sheth R., Marcon L., Bastida M.F., Junco M., Quintana L., Dahn R., Kmita M., Sharpe J., Ros M.A. (2012) Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science, 338, 1476–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S.. et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature, 467, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S.. et al. (2014) Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature, 506, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adany R., Aromaa A.. et al. (2010) Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet., 42, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang W., Tang H., Zhang Y., Tang X., Zhang J., Sun L., Yang J., Cui Y., Zhang L., Hirankarn N.. et al. (2013) Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet., 92, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang W., Shen N., Ye D.Q., Liu Q., Zhang Y., Qian X.X., Hirankarn N., Ying D., Pan H.F., Mok C.C.. et al. (2010) Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet., 6, e1000841.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Han J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P.. et al. (2009) Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet., 41, 1234–1237. [DOI] [PubMed] [Google Scholar]
- 67. Dittmer J. (2003) The biology of the Ets1 proto-oncogene. Mol. Cancer, 2, 29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lettice L.A., Williamson I., Wiltshire J.H., Peluso S., Devenney P.S., Hill A.E., Essafi A., Hagman J., Mort R., Grimes G.. et al. (2012) Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell, 22, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kohlhase J. (2000) SALL1 mutations in Townes-Brocks syndrome and related disorders. Hum. Mutat., 16, 460–466. [DOI] [PubMed] [Google Scholar]
- 70. Townes P.L., Brocks E.R. (1972) Hereditary syndrome of imperforate anus with hand, foot, and ear anomalies. J. Pediat., 81, 321–326. [DOI] [PubMed] [Google Scholar]
- 71. Kohlhase J., Hausmann S., Stojmenovic G., Dixkens C., Bink K., Schulz-Schaeffer W., Altmann M., Engel W. (1999) SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics, 62, 216–222. [DOI] [PubMed] [Google Scholar]
- 72. Wilson M.G., Towner J.W., Forsman I., Siris E., Summitt R.L. (1979) Syndromes associated with deletion of the long arm of chromosome 18[del(18q)]. Am. J. Med. Genet., 3, 155–174. [DOI] [PubMed] [Google Scholar]
- 73. Zhao M., Gupta V., Raj L., Roussel M., Bei M. (2013) A network of transcription factors operates during early tooth morphogenesis. Mol. Cell. Biol., 33, 3099–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kondo T., Zakany J., Innis J.W., Duboule D. (1997) Of fingers, toes and penises. Nature, 390, 29.. [DOI] [PubMed] [Google Scholar]
- 75. Manning J.T., Wood S., Vang E., Walton J., Bundred P.E., van Heyningen C., Lewis-Jones D.I. (2004) Second to fourth digit ratio (2D:4D) and testosterone in men. Asian J. Androl., 6, 211–215. [PubMed] [Google Scholar]
- 76. Lutchmaya S., Baron-Cohen S., Raggatt P., Knickmeyer R., Manning J.T. (2004) 2nd to 4th digit ratios, fetal testosterone and estradiol. Early Hum. Dev., 77, 23–28. [DOI] [PubMed] [Google Scholar]
- 77. Hickey M., Doherty D.A., Hart R., Norman R.J., Mattes E., Atkinson H.C., Sloboda D.M. (2010) Maternal and umbilical cord androgen concentrations do not predict digit ratio (2D:4D) in girls: a prospective cohort study. Psychoneuroendocrinology, 35, 1235–1244. [DOI] [PubMed] [Google Scholar]
- 78. Voracek M. (2014) No effects of androgen receptor gene CAG and GGC repeat polymorphisms on digit ratio (2D:4D): a comprehensive meta-analysis and critical evaluation of research. Evol. Hum. Behav., 35, 430–437. [Google Scholar]
- 79. Honekopp J. (2013) No evidence that 2D:4D is related to the number of CAG repeats in the androgen receptor gene. Front. Endocrinol., 4, 185.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z.. et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J.. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee S.H., Yang J., Goddard M.E., Visscher P.M., Wray N.R. (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics (Oxford, England), 28, 2540–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kemp J.P., Medina-Gomez C., Estrada K., St Pourcain B., Heppe D.H., Warrington N.M., Oei L., Ring S.M., Kruithof C.J., Timpson N.J.. et al. (2014) Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel Loci in the genetic regulation of bone mass attainment. PLoS Genet., 10, e1004423.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Medina-Gomez C., Felix J.F., Estrada K., Peters M.J., Herrera L., Kruithof C.J., Duijts L., Hofman A., van Duijn C.M., Uitterlinden A.G.. et al. (2015) Challenges in conducting genome-wide association studies in highly admixed multi-ethnic populations: the Generation R Study. Eur. J. Epidemiol., 30, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Medina-Gomez C., Kemp J.P., Dimou N.L., Kreiner E., Chesi A., Zemel B.S., Bonnelykke K., Boer C.G., Ahluwalia T.S., Bisgaard H.. et al. (2017) Bivariate genome-wide association meta-analysis of pediatric musculoskeletal traits reveals pleiotropic effects at the SREBF1/TOM1L2 locus. Nat. Commun., 8, 121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kappen J.H., Medina-Gomez C., van Hagen P.M., Stolk L., Estrada K., Rivadeneira F., Uitterlinden A.G., Stanford M.R., Ben-Chetrit E., Wallace G.R.. et al. (2015) Genome-wide association study in an admixed case series reveals IL12A as a new candidate in Behcet disease. PLoS One, 10, e0119085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. van der Valk R.J., Duijts L., Timpson N.J., Salam M.T., Standl M., Curtin J.A., Genuneit J., Kerhof M., Kreiner-Moller E., Caceres A.. et al. (2014) Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J. Allergy Clin. Immunol., 134, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England), 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Allen N.E., Sudlow C., Peakman T., Collins R. (2014) UK biobank data: come and get it. Sci. Transl. Med., 6, 224ed4. [DOI] [PubMed] [Google Scholar]
- 91. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N.. et al. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Boker S., Neale M., Maes H., Wilde M., Spiegel M., Brick T., Spies J., Estabrook R., Kenny S., Bates T.. et al. (2011) OpenMx: an open source extended structural equation modeling framework. Psychometrika, 76, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ihaka R., Gentleman R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat., 5, 299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.