Abstract
The phage Mu DNA transposition system provides a versatile species non-specific tool for molecular biology, genetic engineering and genome modification applications. Mu transposition is catalyzed by MuA transposase, with DNA cleavage and integration reactions ultimately attaching the transposon DNA to target DNA. To improve the activity of the Mu DNA transposition machinery, we mutagenized MuA protein and screened for hyperactivity-causing substitutions using an in vivo assay. The individual activity-enhancing substitutions were mapped onto the MuA–DNA complex structure, containing a tetramer of MuA transposase, two Mu end segments and a target DNA. This analysis, combined with the varying effect of the mutations in different assays, implied that the mutations exert their effects in several ways, including optimizing protein–protein and protein–DNA contacts. Based on these insights, we engineered highly hyperactive versions of MuA, by combining several synergistically acting substitutions located in different subdomains of the protein. Purified hyperactive MuA variants are now ready for use as second-generation tools in a variety of Mu-based DNA transposition applications. These variants will also widen the scope of Mu-based gene transfer technologies toward medical applications such as human gene therapy. Moreover, the work provides a platform for further design of custom transposases.
INTRODUCTION
DNA transposons are genetic elements that are capable of moving within and between genomes, and are widespread both in prokaryotes and eukaryotes (1). They are mobilized by a transposon-encoded transposase protein that excises the transposon from its original DNA context and reintegrates it into a new genomic locus. Profound understanding of DNA transposition mechanisms has enabled the use of transposons as efficient tools in molecular biology and biomedical research, ranging from versatile in vitro genetic engineering and random mutagenesis applications to forward genetic screens and efficient genome manipulation methods in a broad range of organisms (2–5). Importantly, the possibility to introduce new genetic material into the human genome underlies the emerging field of transposition-based gene therapies (6). In contrast to genome engineering tools that are nuclease-activity dependent, such as zinc-finger nucleases, TALENs and the CRISPR/Cas9 system (7), transposons enable the direct insertion of a genetic cargo. This is a desirable feature in applications, where the mutagenic potential of off-targeted nuclease-inflicted DNA double strand breaks would represent a concern (8).
During evolution, intracellularly moving DNA transposons have not been selected for the highest potential activity, because the excessive spread of such elements would be detrimental to the host cell and jeopardize the genome integrity. As a low transposition frequency can complicate the use of transposons in applications, enhancing the transpositional activity has been one of the main targets in DNA transposition technology development. Accordingly, enhanced transposase variants have been reported e.g. for Tn5 (9), Sleeping Beauty (10), PiggyBac (11), Himar1 (12) and Mos1 (13). Conversely, transposons that can escape cells as viruses, such as phage Mu, do not depend on the survival of their host and naturally may encode a highly active transposase. However, to how much further can such transposases be enhanced by mutagenesis is yet to be scrutinized experimentally.
Phage Mu is the first DNA transposition system, for which an in vitro transposition reaction was established (14). The original in vitro system and versions thereof have been instrumental in deciphering the mechanistic details of DNA transposition in general, and have formed a basis for the development of advanced Mu-based genetic tools (15,16). Any DNA sandwiched between Mu transposon ends constitutes a mini-Mu transposon mobilizable by the catalytic action of MuA transposase (17), a member of retroviral integrase superfamily (RISF) proteins, having a common RNase H-like fold with a conserved DDE motif (18). The first step in transposition is the formation of a protein–DNA complex called a transpososome, which contains a tetramer of MuA sequence specifically bound to two transposon ends (Figure 1A). Within this structure, MuA catalyzes two chemical reactions on each transposon end (Supplementary Figure S1): hydrolysis of the transposon–donor DNA junction and subsequent attack of the 3′ end of the transposon on a target DNA, attaching the transposon DNA to target DNA (16).
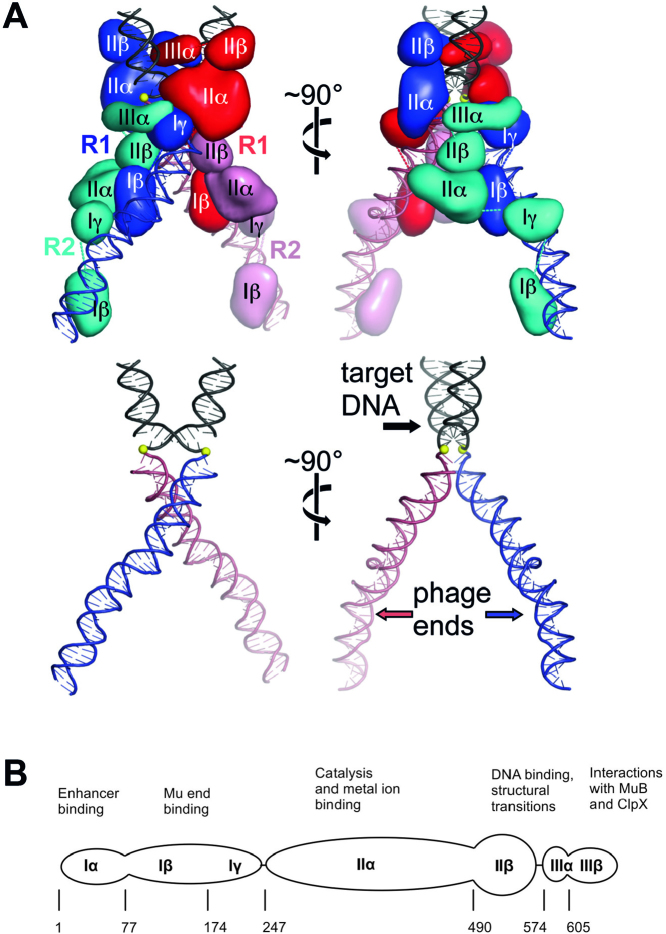
Figure 1.
Mu transpososome structure. (A) Two views of the transpososome with individual protein domains as smoothed surfaces (top). The proteins removed and the scissile phosphates depicted as yellow spheres (bottom). (B) Structural organization of MuA (663 amino acids). The numbers correspond to the amino terminus of each domain. Domain Iα is not required in vitro, but aids transpososome assembly by binding to a transpositional enhancer. Domains Iβ and Iγ recognize bipartite sites within the Mu ends. Domain IIα contains the DDE motif, which coordinates the catalytic metal ions. Domains IIβ and IIIα participate in target DNA binding and transpososome assembly. Domain IIIβ interacts with MuB. This Mu-encoded protein delivers target DNA to the transpososome but is not needed under all conditions. Domain IIIβ also interacts with ClpX, an ATPase responsible for transpososome disassembly.
In its natural context in vivo, the Mu transposition reaction steps also involve the phage-encoded MuB targeting protein, host-encoded DNA architectural proteins (HU and IHF), additional DNA sites (two more MuA binding sites and a transpositional enhancer), as well as the host-encoded ClpX protein, which remodels the product transpososome for disassembly (15,16, Supplementary Figure S1). However, in a minimal in vitro set-up, fully active transpososomes can be assembled efficiently with only MuA and two 50-bp right end segments, each containing two MuA binding sites (termed R1 and R2) (19). The crystal structure of a Mu transpososome at the post-integration stage resembles a pair of scissors where the Mu DNA ends form the handles and the sharply bent target DNA the blades (20). Within the MuA tetramer, the individual domains of the R1- and R2-bound subunits play different roles and make different protein–protein interactions (Figure 1A). This structure provides a useful platform for structure-function studies of DNA transposition and comparisons to similar polynucleotidyl transferase reactions such as HIV integration (21) and V(D)J recombination (22).
The minimal in vitro DNA transposition system of Mu is highly efficient and displays very low target site selectivity (17,23), ideal features for many applications. Mu in vitro transposition technology has been utilized in numerous molecular biology, protein engineering and genomics applications (24–31). In addition, pre-assembled Mu transpososomes can be electroporated into a variety of cell types for efficient gene delivery. This methodology provides a flexible species-non-specific means to modify genomes of bacteria, yeast and even mammalian cells (32–34). Advantageously, it obviates the need to use a plasmid vector for expressing a potentially toxic transposase within cells, thus also eliminating the risk of such plasmids integrating into the genome. Importantly, host factors are not required, broadly potentiating gene delivery among organisms. In addition, the standard conditions with in vitro pre-assembled transpososomes primarily yield single integrations, a desirable outcome not easily obtained with systems utilizing in vivo transposase expression, such as PiggyBac and Sleeping Beauty (10,11). As all Mu-based transposon applications would benefit from the enhanced activity of MuA, mutagenesis of the protein for better performance is warranted.
Here, we used an in vivo screen to identify hyperactive MuA transposase variants within a large library of random mutations. Mapping of the causative substitutions onto the Mu transpososome structure suggests multiple mechanisms inducing hyperactivity. We show that highly active MuA variants can be produced by combining several additively acting substitutions. Our work provides novel MuA variants for multiple applications and offers a platform for further design of transposases.
MATERIALS AND METHODS
Enzymes, reagents, DNA and techniques
Plasmids are described in Supplementary Table S1, commercial proteins and reagents in Supplementary Table S2 and oligonucleotides in Supplementary Table S3. Plasmid DNA was prepared using appropriate kits from QIAGEN. Standard DNA techniques were performed as described (35). Cat-Mu transposon DNA was prepared as described (17). DNA sequencing was done using BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3130 XL sequencer, both from Life Technologies/Applied Biosystems.
Cells and cultures
Escherichia coli DH10B (Invitrogen) was used as a cloning host and DH5α (Invitrogen) for plasmid DNA isolation and papillation analysis. MuA variants were expressed for purification in E. coli BL21(DE3)pLysS (Novagen). Bacteria were cultured in Luria-Bertani (LB) medium or on LB agar plates (35) supplemented with ampicillin (Ap) and/or chloramphenicol (Cm) when required. Bacterial electrocompetent cells and standard competent cells were prepared as described in (32) and (36), respectively.
Mouse AB2.2-Prime embryonic stem cells (Lexicon Genetics) were grown on gelatinized tissue culture dishes in knockout Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% defined fetal calf serum, penicillin–streptomycin, L-glutamine, non-essential amino acids, sodium pyruvate, mercaptoethanol and 500 U/ml LIF (leukemia inhibitory factor) at 37°C in the presence of 5% CO2.
Generation of MuA mutant libraries
MuA (Gene Bank P07636), encoding MuA transposase (663 amino acids), was mutated using error-prone polymerase chain reaction (PCR) as described (37). Altogether five mutant libraries (Supplementary Table S4) were generated with the specifications indicated below.
Taq DNA polymerase was used under three mutagenic PCR conditions with 0, 1 or 2 μl of mutagenic buffer added (37). Each PCR amplification was performed using HSP492/HSP493 primer pair with pTLH2 as a template. PCR products were purified using QIAquick PCR Purification Kit (Qiagen), digested with NcoI and EcoRI, and subjected to preparative electrophoresis using 1.0% SeaPlaque GTG agarose gel in 1 x TAE buffer (40mM Tris-Acetate, 1 mM EDTA; 35). The MuA-encoding 2-kb fragment was isolated using QIAquick MinElute Gel Extraction Kit (Qiagen) and ligated into pTLH1 digested with NcoI and EcoRI to generate plasmids for papillation assay. Ligation products were electroporated into electrocompetent DH10B cells as described (32), and the cells were plated onto LB plates containing Ap (100 μg/ml) and Cm (10 μg/ml). A total of ∼6 × 104 bacterial colonies were pooled for each library, and the bacteria were grown for plasmid isolation at 37°C for 2 h in LB medium containing Ap (100 μg/ml) and Cm (10 μg/ml).
Mutazyme II DNA polymerase was used in two separate PCR reactions, employing 5 or 10 cycles of amplification as described (37). Both amplifications were performed using HSP492/HSP493 primer pair with pALH6 as a template. The MuA-encoding fragment was isolated using preparative agarose gel electrophoresis as above. It was further amplified by PCR using Vent DNA polymerase under non-mutagenic conditions. The reaction (50 μl) contained 10 ng of the fragment as a template, 0.5 μM each of the primers HSP492 and HSP493, 200 μM each dNTPs, and 1 U Vent DNA polymerase in ThermoPol reaction buffer containing 4 mM MgSO4. PCR included 5 min at 95°C; 30 cycles of amplification (45 s at 95°C, 1 min at 59°C, 2.5 min at 72°C); and 5 min at 72°C. The fragment was gel-isolated and ligated into pTLH1 for papillation assay as above. For both library, plasmid DNA was isolated from a pool of ∼6 × 104 bacterial colonies as described above.
Papillation assay
Papillation assay (38) was used to screen MuA mutants. The assay is based on mini-Mu transposon mobilization in vivo and scores transposition events as blue microcolonies (papillae) within whitish E. coli colonies. Briefly, an assay plasmid encoding MuA under the arabinose/glucose controllable E. coli PBAD promoter, and including a reporter transposon, was transformed into standard competent DH5α cells (100 μl), and the cells were plated onto LB agar plates supplemented with Ap (100 μg/ml), Cm (20 μg/ml), lactose (0.05%), Xgal (40 μg/ml) and arabinose (1 × 10−4%). Standard assay included incubation at 30°C for 115 h. To assay highly active MuA variants, the plates were incubated at 25°C for 140 h. When indicated, glucose (0.22%) replaced arabinose to reduce MuA expression. Representative ∼5 mm diameter colonies were photographed using an Olympus (Tokyo, Japan) ColorView II digital camera attached to an Olympus SZX12 stereomicroscope equipped with Zeiss (Oberkochen, Germany) KL1500 LCD cold light source. The papillae were enumerated manually using AnalySIS software (Soft Imaging System, Olympus). Unless otherwise indicated, the mean value and standard deviation (SD) were calculated for each protein variant from six colonies.
Mutant screen
Papillation assay was used for the mutant screen. Bacteria were grown on bioassay trays (240 × 240 × 20 mm, Genetix) with the density of ∼1000 colonies per plate. Increased papillation was evaluated visually, and clonal cultures were produced from each selected colony on LB plates containing Ap (100 μg/ml) and Cm (10 μg/ml). Plasmid DNA was isolated from the clones and re-assayed for papillation to confirm the observed phenotype. The MuA specificity of the phenotype was verified by re-cloning each respective MuA gene into pTLH1 and assaying the ensuing plasmids for papillation. DNA sequencing was used to reveal the mutated residues within the MuA gene of each particular plasmid.
Plasmid constructions
HSP570/HSP571 linker was cloned into BamHI-linearized pET3d expression vector to yield plasmid pTLaH1, in which the linker-borne unique KpnI site substitutes for the original BamHI site. Wild-type (WT) MuA was subsequently cloned from pLHH4 into pTLaH1 using NcoI and KpnI sites, yielding pTLH4. The control plasmid expressing MuA63–663 (pLHH10) was produced by PCR-cloning into pTLH1 a fragment generated with HSP500/HSP351 primer pair and pALH6 template, employing NcoI and XhoI sites. Unique point mutations were generated by initially PCR-amplifying plasmid pTLH4 DNA with mutation-generating specific primer pairs (Supplementary Table S3). Each amplification reaction (50 μl) contained 100 ng pTLH4 as a template, 0.5 μM each primer, 200 μM each dNTPs, and 1U Phusion DNA polymerase in Phusion HF buffer. PCR included an initial 2 min at 98°C. Subsequently, 10 cycles of amplification were performed with 30 s at 98°C, 1 min at a primer-pair-defined temperature (52–66°C) and 7 min at 72°C. The final extension phase employed 5 min at 72°C. Amplified DNA was subjected to preparative electrophoresis on a 0.5% SeaPlaque GTG agarose gel in TAE buffer, purified using QIAquick MinElute Gel Extraction Kit (Qiagen), and phosphorylated using T4 polynucleotide kinase and ATP, after which it was circularized using T4 ligase. Each mutated MuA gene was subcloned into papillation assay plasmid pTLH1 using NcoI and KpnI restriction sites. By repeating the procedure, up to three mutations were generated per MuA gene. Those MuA genes that encoded more than three amino acid substitutions were purchased from GeneCust (Supplementary Table S4). They, or their deletion derivatives made by PCR, were cloned using NcoI and KpnI restriction sites into pTLH1 for papillation analysis and pTLaH1 for protein purification. The authenticity of the constructs was confirmed by sequencing.
Purification of MuA transposase variants
MuA transposase proteins were expressed in BL21(DE3)pLysS using pET3c- or pET3d-derived plasmids and purified as described (39) with following modifications. Cells were grown in LB medium (120 ml) containing Ap (100 μg/ml) and Cm (35 μg/ml) at 37°C to an OD600 of ∼0.9, collected by centrifugation, transferred into LB medium (1200 ml) containing Ap (100 μg/ml), and grown at 28°C to an OD600 of ∼0.5. Protein expression was induced by the addition of 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were collected 2 h post-induction by centrifugation at 4°C and resuspended in an equal weight of 50 mM Tris–HCl (pH 8.0), 10% sucrose, 1 mM dithiothreitol (DTT). The cell suspension was frozen under liquid nitrogen and stored at −80°C until used for protein purification. Cell lysis and ammonium sulfate precipitation (146 mg/ml, ∼25% saturation) were performed as described (39), except that the ammonium sulfate precipitate was resuspended in HEDGK (25 mM HEPES, pH 7.6, 0.1 mM ethylenediaminetetraacetic acid (EDTA) 1 mM DTT, 10% (w/v) glycerol, 0.5 M KCl) with the volume equal to that of the original lysis extract. Phosphocellulose and hydroxylapatite columns were used as described (39) with the following modifications. HEDG (25mM HEPES, pH 7.6, 0.1 mM EDTA, 1 mM DTT, 10% glycerol) was added to the protein solution in order to adjust the conductivity to that of 0.3 M KCl (in HEDG). Subsequently, the solution was loaded onto a 1.4 ml phosphocellulose (P11, Whatman) column (Poly-Prep, Bio-Rad). Proteins were eluted by gravity using a step gradient (0.8 ml increments, 0.1 M intervals) from 0.3 M to 1.5 M KCl (in HEDG). Peak fractions were pooled, and HDG (25 mM HEPES, pH 7.6, 1 mM DTT, 10% glycerol) was added for the adjustment of the conductivity to that of 10 mM potassium phosphate (in HDG supplemented with 0.5 M KCl). The solution was loaded onto a 0.45 ml hydroxyapatite (Macro-Prep Ceramic Hydroxyapatite Type I, Bio-Rad) HR 5/2 column (GE Healthcare Life Sciences). Proteins were eluted using a 10 column volume gradient from 10 mM to 1 M potassium phosphate (in HDG supplemented with 0.5 M KCl). Peak fractions were pooled and dialyzed against HEDG buffer supplemented with 0.3 M NaCl, after which the preparation was frozen under liquid nitrogen and stored at −80°C. Protein concentration was determined spectrophotometrically using the value A280 of 1.58 = 1 mg/ml (40). For all preparations, upon excessive protein loading only one major protein species was detected in a Coomassie blue stained sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (data not shown). Furthermore, only a negligible level of nuclease activity was detected with supercoiled plasmid DNA upon prolonged incubation under in vitro integration reaction conditions (see below), confirming a high purity level of the preparations (data not shown).
In vitro integration
Reactions (25 μl) contained 0.5 pmol Cat-Mu transposon DNA, 500 ng pUC19 DNA as a target, 2.7 pmol (0.22 μg) MuA, 25 mM Tric–HCl, pH 8.0, 100 μg/ml bovine serum albumin (BSA), 15% (w/v) glycerol, 0.05% (w/v) Triton X-100, 126 mM NaCl and 10 mM MgCl2. Reactions were carried out for 1 h at 30°C, unless otherwise indicated, and stopped by freezing in liquid nitrogen. For the qualitative assessment of transposition products, 1.5 μl of loading dye (0.1% bromophenol blue, 2.5% SDS, 50 mM EDTA, 25% Ficoll 400) was added to a 5 μl aliquot of each reaction mixture and the samples were analyzed by electrophoresis on a 0.8% SeaKem LE agarose gel in TAE buffer. Biological selection was used to score transposition products quantitatively. Each reaction mixture was thawed and an aliquot of it (5 μl) was transformed into competent MC1061 cells (200 μl). Alternatively, an aliquot (1 μl) was electrotransformed into MC1061 electrocompetent cells (25 μl). Bacteria were plated onto LB plates containing Ap (100 μg/ml) and Cm (10 μg/ml) to score integration products.
In vivo integration
Cat-Mu transpososomes were assembled at 30°C for 2 h as described (32), after which the preparations were frozen under liquid nitrogen and stored at −80°C. An aliquot of each preparation was thawed and analyzed qualitatively for assembled transpososomes by the use of agarose/BSA/heparin gels as described (32). For the quantitative analysis of genomic integration, each transpososome preparation was thawed and diluted with water (1:8). Subsequently, an aliquot (1 μl) was electroporated into electrocompetent MC1061 cells (25 μl) as described (32). Genomic integrations were scored by plating the bacteria onto LB plates containing Cm (10 μg/ml).
Electroporation of mammalian cells
AB2.2 cells were harvested with trypsin–EDTA, pH 7.4 and washed three times with 1 × phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4). Standard electroporation mixtures contained ∼5 × 106 cells in 800 μl of 1 × PBS and 0.5 μg of transposon DNA assembled in vitro with MuA as described next. The standard in vitro transpososome assembly reaction mixture (40 μl) contained 2.2 pmol of Kan/Neo-Mu transposon DNA (34), 9.8 pmol of MuA, 150 mM of Tris–HCl (pH 6.0), 50% (v/v) glycerol, 0.025% (w/v) Triton X-100, 150 mM NaCl and 0.1 mM EDTA. The assembly reaction was carried out at 30°C for 2 h. The cells with transpososomes were subjected, using a BTX ECM 630 gene pulser (Harvard Apparatus), to a single voltage pulse at room temperature with the following settings: capacitance 500 μF, voltage 250 V (0.4-cm electrode spacing cuvettes, VWR). Following electroporation, the cells were allowed to rest in the cuvette for 10 min, after which 9.2 ml of warm medium (supplemented knockout DMEM) was added, and different volume aliquots were transferred onto gelatinized tissue culture dishes. Selection with 150 μg/ml G418 was initiated 2 days after electroporation and continued for ∼10 days. For the enumeration of colonies, cells were fixed with cold methanol and stained with 0.2% methylene blue.
Structural analyses
The structural and functional consequences of substitutions were assessed by investigating the variants within the secondary and tertiary structures of MuA protein and Mu transpososome structure. The NMR and X-ray structures for isolated MuA protein domains and transpososome crystal structure were from PDB IDs: 1TNS, 2EZK, 2EZH, 1BCO and 4FCY. For consistent secondary structural element identification we used DSSP. UCSF Chimera and PyMOL (The PyMOL Molecular Graphics System Version 1.3) were used for visualizations.
Statistical analyses
To evaluate statistical significance, two-tailed one-sample t-test was performed.
RESULTS
Mutant generation and screening for hyperactive MuA variants
Five different mutagenesis protocols (Supplementary Table S4) were used to generate a broad spectrum of substitutions within all MuA domains (Figure 1B), and ∼3 × 105 independently generated MuA-encoding mutant plasmid clones were produced. Using an in vivo assay (papillation assay, Supplementary Figures S1 and S2), we screened 64 000 clones with 1–3% of them producing an increased level of papillae.
Plasmids were isolated from 222 clones producing the highest numbers of papillae, and these plasmids were re-assayed for papillation. The results verified the plasmid-borne origin of the observed phenotypic change with all of the clones. We then re-cloned the MuA gene from 89 selected plasmids into the original papillation plasmid (pTLH1). The reconstructed plasmids produced papillation phenotypes identical to their respective original counterparts, demonstrating that the observed phenotypic changes were caused by mutations in the MuA gene. The encoded changes were determined by DNA sequencing.
We identified 71 unique MuA sequences, most of which included several mutations (Supplementary Table S5). In papillation analysis, these variants portrayed a broad spectrum of hyperactivity (Supplementary Table S5). Although many changes were identified only once (Supplementary Figure S3), a number of clear hot spots could be discerned (e.g. residues 97, 160, 233, 345, 374, 447, 483, 495) and certain substitutions were identified frequently (e.g. D97G, E233K, E233V, E483G).
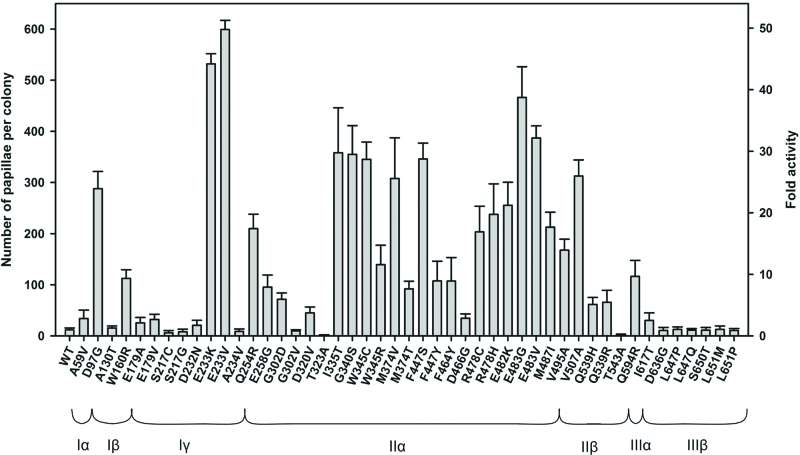
To pinpoint causative mutations, 47 single-substitution MuA variants were generated and analyzed for papillation. Activating substitutions were observed in all seven MuA domains (Figure 2), with the most strongly activating mutations mapping to the central domains, from Iβ to IIIα.
Figure 2.
Activities of MuA single-amino-acid substitution variants in the papillation assay. Left Y-axis: papillae were enumerated from six colonies for each data point. The error bars indicate SD above the average value for each variant. Right Y-axis: fold activity for each protein variant relative to WT MuA. Below the column bars are indicated as the amino-acid substitutions and the MuA subdomain structure. Under the assay conditions used, MuA77–663 produces approximately two times more papillae than WT MuA (38).
In total, 34 substitutions (in 26 specific residues) enhanced the protein activity by at least 2-fold, 27 substitutions by >5-fold, and one by >50-fold. Supplementary Figure S4, Figure 3 and Supplementary Movies 1 and 2 show the mutation positions mapped onto the structures of individual MuA domains and the transpososome. While some mutations cluster near the catalytic site, many others (including the frequently-mutated positions 97, 233 and 483) cluster at the interface between the R1- and R2-bound subunits: between domains Iβ and Iγ (R1) as well as IIα and IIIα (R2). See ‘Discussion’ section for more details.
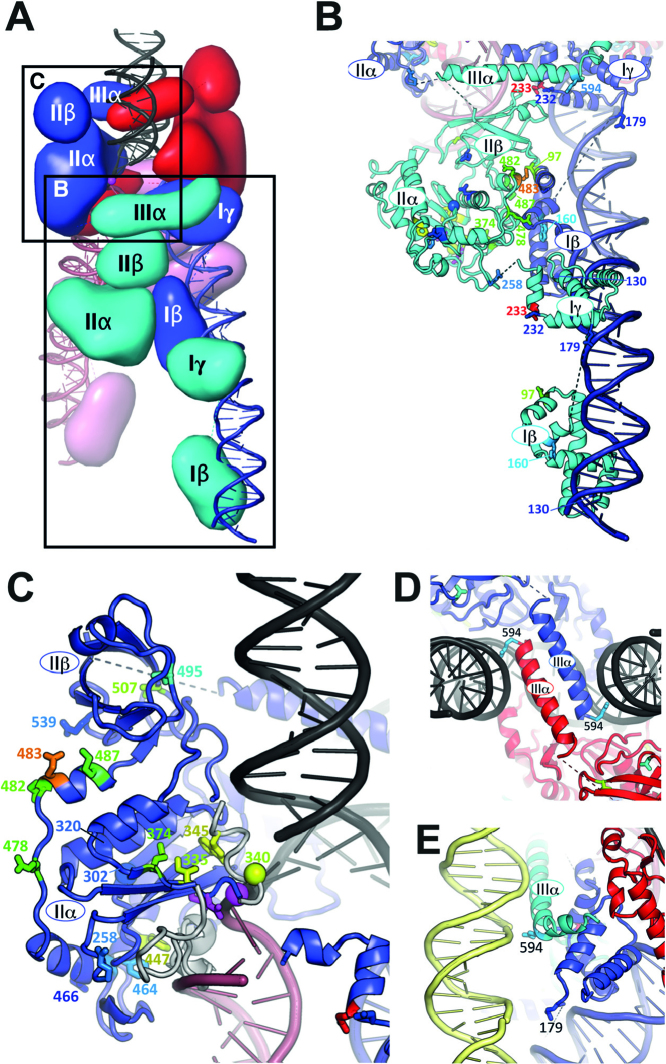
Figure 3.
Activity-enhancing substitutions mapped onto the Mu transpososome structure. (A) The structure represented as in Figure 1A, but from a similar viewpoint as in parts B and C of this figure. (B) The R2-bound subunit, showing its interactions with DNA and other subunits. (C) Closeup of the R1-bound subunit’s domain II, with other proteins removed for clarity. (D) View looking down on the complex as shown in part A, showing how domains IIIα of the R1-bound subunits aid in target capture. (E) Proposed interactions of the R2-bound subunit’s domain IIIα with flanking DNA (yellow), viewed from the right side of the complex as shown in panel A. Side chains whose substitution enhanced activity by at least 2-fold in the papillation assay are shown, colored according to activity from deep blue (lowest) to red (highest). Spheres indicate glycines. The active site DDE and scissile phosphate groups are highlighted in magenta. See also Supplementary Movies 1 and 2.
Synergistic effects of critical substitutions
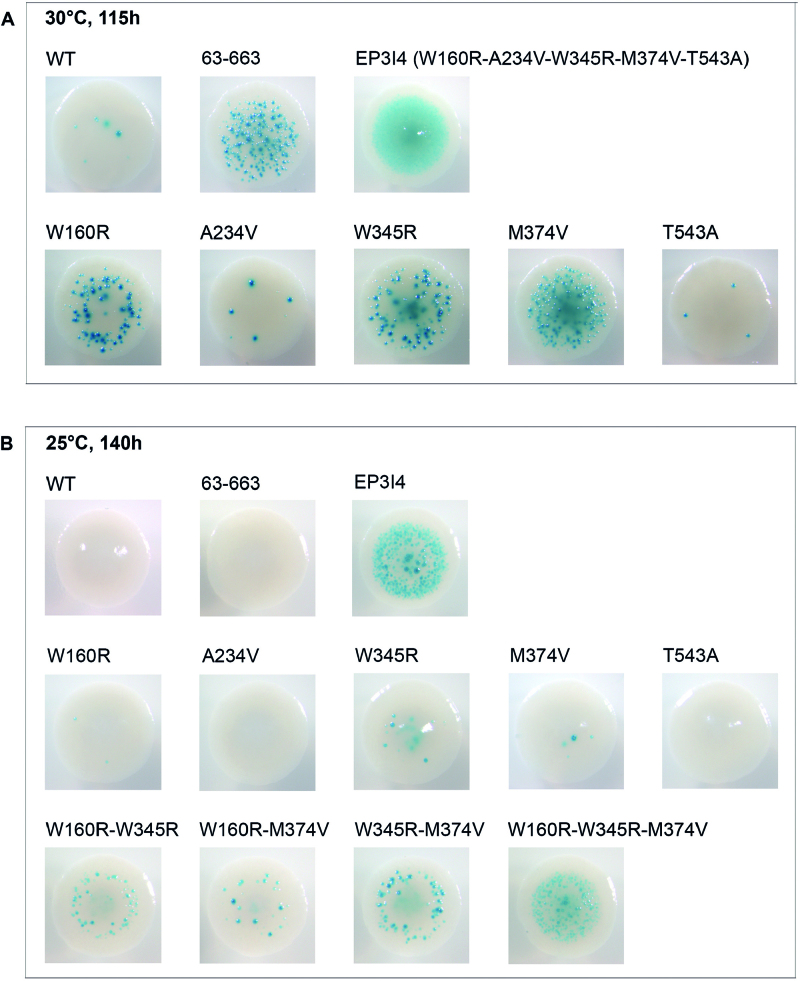
The most active MuA variant identified in our screen (clone EP3I4) contained five substitutions: W160R, A234V, W345R, M374V and T543A (Supplementary Table S5). To discover the causative mutations, we analyzed these substitutions individually and in combinations (Table 1 and Figure 4). Three of the substitutions, W160R, W345R and M374V, clearly enhanced the activity. Combining the substitutions enhanced the activity further. The triple mutant (W160R-W345R-M374V) was the most active and reached the level of the original variant EP3I4. To explore the additivity of substitutions in different domains, we replaced the domain IIα substitution M374V with the domain Iγ substitution E233K, yielding the variant W160R-E233K-W345R, which carries substitutions in three different domains. This variant was the most active yet, producing over 500 times more papillae than WT MuA (Table 1). The success of this engineering emphasizes the independent role of functional domains in generating hyperactive protein variants.
Table 1. Additive and synergistic effects of substitutions.
| MuA transposase | # Lac+ papillaea |
|---|---|
| 30°C, 115 h | |
| WT | 9 ± 2 |
| W160R | 98 ± 6 |
| A234V | 5 ± 2 |
| E233K | 549 ± 9 |
| W345R | 110 ± 5 |
| M374V | 248 ± 23 |
| T543A | 3 ± 1 |
| 25°C, 140 h | |
| WT | 0 ± 0 |
| W160R | 3 ± 0 |
| A234V | 0 ± 0 |
| W345R | 4 ± 0 |
| M374V | 17 ± 2 |
| T543A | 0 ± 0 |
| W160R,W345R | 144 ± 17 |
| W160R,M374V | 70 ± 7 |
| W345R,M374V | 127 ± 4 |
| W160R,W345R,M374V | 264 ± 57 |
| EP3I4 | 295 ± 23 |
| W160R, E233K, W345R | 519 ± 36 |
a In vivo transposition activity was measured by papillation assay. MuA variants were constructed according to amino acid changes detected in the most active mutant screened (EP3I4). It included five amino acid substitutions (W160R, A234V, W345R, M374V, T543A). In addition, a triple mutant carrying substitutions in three separate subdomains was constructed (W160R, E233K, W345R). Effect of single amino acid changes and their various combinations are shown in two papillation assay conditions (30°C, 115 h and 25°C, 140 h) on standard papillation medium (LB, Ap 100 μg/ml, Cm 20 μg/ml, Xgal 40 μg/ml, lactose 0.05%, arabinose 1 × 10−4%). The given data represent averages from triplicate measurements with SDs shown.
Figure 4.
Colonies from the papillation assay showing synergy. The most active mutant (EP3I4) detected in the papillation assay carried five amino acid substitutions. Effect of those substitutions separately and in various combinations are shown in two papillation assay growth conditions. (A) Papillation analysis was performed by incubating transformants at 30°C for 115 h and (B) at 25°C for 140 h on standard papillation medium (LB, Ap 100 μg/ml, Cm 20 μg/ml, Xgal 40 μg/ml, lactose 0.05%, arabinose 1 × 10−4%). WT MuA and deletion variant MuA63–663 were used as controls for initial and enhanced activity, respectively.
Activities of purified proteins
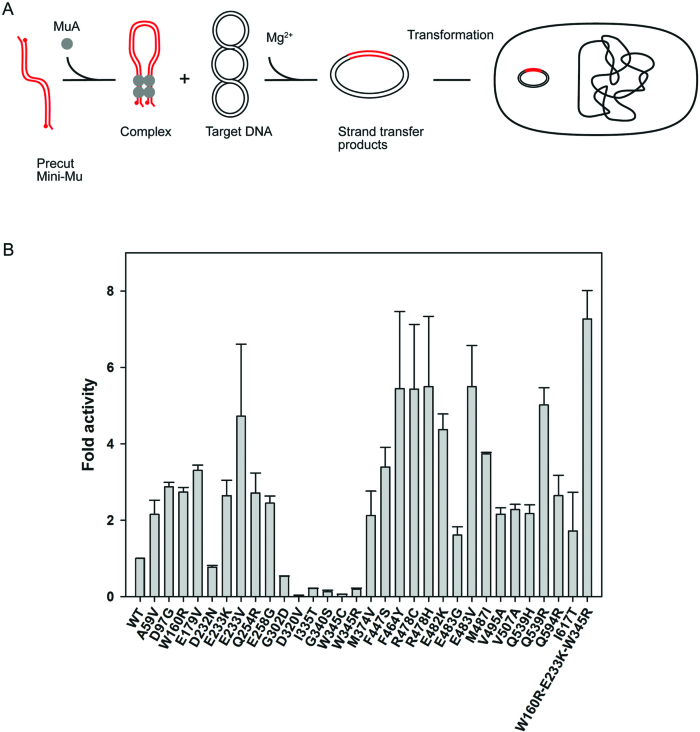
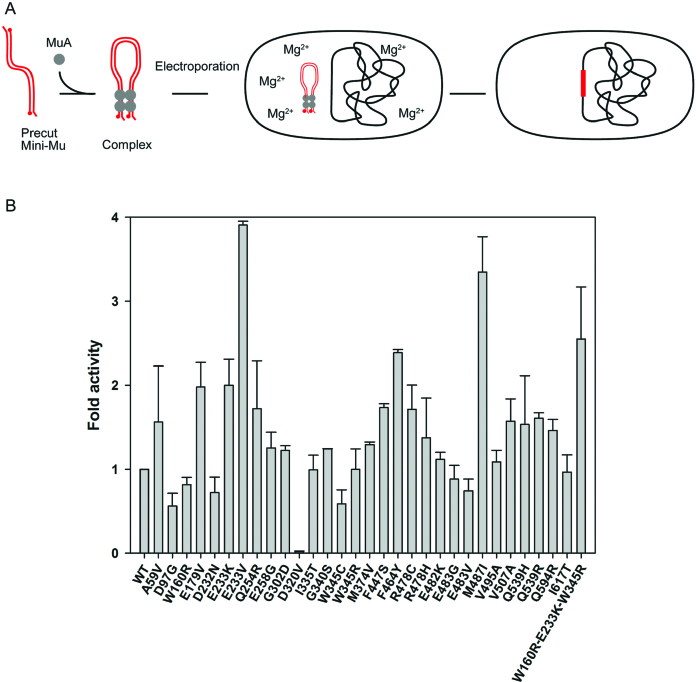
We next purified 30 single-substitution MuA variants along with the WT and triple-mutant (W160R-E233K-W345R) proteins and tested them in two assays relevant for MuA-based applications: (i) in vitro integration into a target plasmid (17) and (ii) genomic integration via electroporation of assembled transpososomes (32).
MuA was incubated with cat-Mu transposon DNA in the presence of pUC19 plasmid DNA as a target. Reaction products were analyzed qualitatively by agarose gel electrophoresis (Supplementary Figure S5) and scored quantitatively (Figure 5) using biological selection (17). The presence of visible reaction products in the gel assay generally correlated well with the number of integrant plasmids scored. Most variants were more active than WT MuA with up to 6-fold enhancement. Activity levels in this assay largely compared well with the papillation assay results (Figure 2). However, five substitutions, all residing between residues 320–345, decreased the plasmid integration activity.
Mu transpososomes were assembled in vitro in the absence of divalent cations, and transpososome formation was analyzed by native agarose gel electrophoresis (Supplementary Figure S6). Transpososomes were then electroporated into E. coli cells, and subsequent integration of the transposon DNA was detected through the resulting chloramphenicol resistance of the host cells (Figure 6). All MuA variants except D320V were proficient in transpososome assembly and in genomic integration. Roughly two-thirds showed enhanced integration activity, E233V being the most active with 4-fold enhancement.
Figure 5.
Scheme for in vitro transposition integration and activities of MuA variants. Schematics of the in vitro transposition assay reaction is shown on top. A tetramer of MuA transposase and mini-Mu transposon ends assemble into a stable transpososome. Under reaction conditions with Mg2+ and supercoiled plasmid target DNA, the transpososome executes transposon integration into the target DNA. Data from the biological selection of integrants are shown on the bottom. The integration events into the target plasmid were analyzed by transforming products from the in vitro transposition reactions into competent Escherichia coli cells and scoring for colonies resistant to both chloramphenicol and ampicillin. The efficiency of each variant is represented as a fold activity relative to the activity of WT MuA transposase. The data are shown as a mean of two experiments. The error bars indicate SD/2 above the average value for each variant.
Figure 6.
Scheme for genomic integration via electroporation of transpososomes and activities of MuA variants. Schematics of in vivo transposon integration via electroporation of in vitro assembled Mu transpososomes is shown on top. A tetramer of MuA transposase and mini-Mu transposon ends assemble into stable transpososomes under the reaction conditions devoid of divalent cations. Following electroporation, the transpososomes encounter Mg2+ ions in vivo and integrate transposon DNA into the bacterial chromosome. Data for the genomic integration efficiency are shown on the bottom. Cat-Mu transposon DNA was incubated for 2 h with each MuA variant to allow transpososome assembly, after which an aliquot was electroporated into MC1061 electrocompetent cells. Genomic integration events were scored as chloramphenicol-resistant colonies. Efficiencies are represented as a fold activity increase relative to the activity of WT MuA transposase. The data are shown as a mean of two experiments. The error bars indicate SD/2 above the average value for each variant.
Further studies on synergism
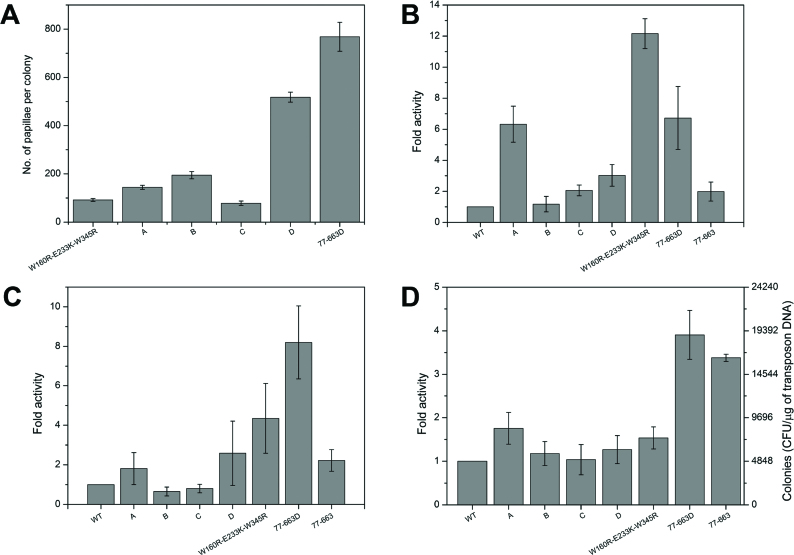
We next studied whether or not MuA’s activity could further be increased by generating novel combinations of the most active individual substitution variants. We made two variants with five (MuAA and MuAC) and two with eight (MuAB and MuAD) substitutions (Supplementary Table S6). As deletion of domain Iα (MuA77–663) had previously been shown to cause a 2-fold activity increase in the papillation assay (38), we also added this deletion to the MuAD variant to generate MuA77–663D. As all of the variants were highly active in an initial papillation assay, they were analyzed under stringent papillation conditions using the highly active W160R-E233K-W345R triple mutant as a control (Figure 7A). MuAD and MuA77–663D performed the best, with 6- and 8-fold activity enhancement over the control, respectively. As the papillation rate for W160R-E233K-W345R is already over 500 times above that of WT MuA (Table 1), we estimate that the rate for MuA77–663D must be at least 4000-fold above the WT level. Thus, on the basis of the single substitution data, we were able to design several MuA variants that were extremely active in vivo.
Figure 7.
Activities of multi-substitution MuA variants. Substitution composition of the variants are shown in Supplementary Table S6. The error bars indicate SD for each variant. (A) Papillation assay. Transformants were incubated for 140 h at 25°C on papillation medium with glucose replacing arabinose (LB, Ap 100 μg/ml, Cm 20 μg/ml, Xgal 40 μg/ml, lactose 0.05%, glucose 0.22%). WT MuA and MuA77–663 do not produce papillae under these conditions, and therefore W160R-E233K-W345R was used as a control. Results are shown as a mean number of papillae from three colonies. (B) In vitro integration assay. The reaction scheme is shown in Figure 5A. Cat-Mu transposon DNA was incubated for 5 min with MuA variants and pUC19 target plasmid. Reaction products were electroporated into MC1061 electrocompetent cells and scored as chloramphenicol and ampicillin double-resistant colonies. Three independent experiments were conducted. P-values (mutants versus WT MuA): A, 0.015; B, 0.603; C, 0.034; D, 0.037; W160R-E233K-W345R, 0.003; 77–663D, 0.039; 77–663, 0.109. (C) Genomic integration assay with Escherichia coli. The reaction scheme is shown in Figure 6A. Cat-Mu transposon DNA was incubated for 1 h with MuA variants to allow transpososome assembly, after which an aliquot was electroporated into MC1061 electrocompetent cells for in vivo integration. Bacterial colonies were scored using chloramphenicol selection. Three independent experiments were conducted. P-values (mutants versus WT MuA): A, 0.225; B, 0.113; C, 0.256; D, 0.123; W160R-E233K-W345R, 0.082; 77–663D, 0.095; 77–663, 0.062. (D) Genomic integration assay with eukaryotic cells. Kan/Neo-Mu transposon DNA was incubated with MuA for 2 h to allow transpososome assembly, after which an aliquot was electroporated into AB2.2 mouse cells for in vivo integration. Genomic integrants were scored using G418 selection. Three independent experiments were conducted. P-values (mutants versus WT MuA): A, 0.07; B, 0.387; C, 0.874; D, 0.286; W160R-E233K-W345R, 0.067; 77–663D, 0.012; 77–663, 0.016.
Next, these MuA variants were purified, and their activity was determined using three assays relevant for transposon-based applications: (i) in vitro integration into plasmid target, (ii) in vivo integration of transposons into the genome of E. coli cells via electroporation of pre-assembled transpososomes, and (iii) in vivo integration of transposons into the genomes of mouse stem cells via electroporation of pre-assembled transpososomes (Figure 7B–D). In general, several variants portrayed higher activities than WT MuA. In the in vitro integration assay (Figure 7B) the most hyperactive mutant was W160R-E233K-W345R with a 12-fold activity increase. In addition, a substantial (6-fold) activity enhancement was gained with variants MuAA and MuA77–663D. In both of the genomic integration assays (Figure 7C and D; Supplementary Figure S7), variant MuA77–663D portrayed the highest activity increase, 8- and 4-fold with bacterial and eukaryotic cells, respectively.
DISCUSSION
An increasing number of transposon systems have recently been adapted for advanced genetic engineering and genome modification applications, spurring efforts to improve their performance. The Mu DNA transposition system constitutes an ideal platform for such applications, and the hyperactive MuA variants described here significantly improve the scope and feasibility of Mu-based DNA transposition technologies.
Like other RISF members, MuA is active only after it has been assembled into a protein–DNA complex containing several subunits. Structures of such molecular machines have shown that despite great diversity, certain architectural principles are conserved (20,41–45). In particular, they are held together by a cooperative network of contacts among multiple protein domains and the two transposon or viral DNA ends. While only two subunits carry out catalysis, some systems, including the Mu transpososome and retroviral intasomes, require additional architectural subunits. For MuA, the R1-bound subunits are catalytic, and the R2-bound ones are architectural. Mapping the hyperactivity data onto the transpososome structure suggests that the selected substitutions affect not just chemical catalysis but also the assembly and stability of the transpososome and its ability to recruit target DNA.
Domains Iα and IIIβ harbored one mildly-enhancing substitution each. These domains are not strictly required for transposition. Domain Iα is inhibitory in the absence of transpositional enhancer sequence (38,46), probably due to weak interactions with other DNA sequences, so it is not surprising that mutating it (A59V) can improve the protein activity. Domain IIIβ interacts with MuB and ClpX (47). Residue I617 of IIIβ is adjacent to the residue R616 previously shown in the context of the Mu transpososome to interact with ClpX (47). Therefore, substitution I617T may introduce a more beneficial interaction at the protein–protein interface. As neither domain Iα nor domain IIIβ is present in the Mu transpososome crystal structure, the effects of the substitutions within these domains currently remain unexplained at the architectural level.
The majority of the activating substitutions lie within two protein–protein interfaces between subunits bound to the same Mu end DNA segment. One interface is between domain IIα of the architectural subunit (including residues 478, 482, 483 and 487) and domain Iβ of the adjacent catalytic subunit (including residues 97 and 160). The second interface includes the most highly-activating single substitutions, E233K and E233V. E233 protrudes from the catalytic subunit’s domain Iγ where it packs between the architectural subunit’s domains IIβ and IIIα. The same residues on the opposite subunits (that is, on domain IIα of the catalytic subunit and domains Iβ and γ of the architural subunit) face the solvent. Although the interaction surfaces of domains Iβ and Iγ are highly conserved among MuA-related sequences (20), the Mu-like prophage Hin-Mu contains a lysine in the equivalent position to 233 (48). These observations suggest that this set of substitutions improves the assembly rate, yield and/or stability of transpososomes by changing protein–protein contacts. However, several of the substitutions increase the overall positive charge of the protein, and therefore might also electrostatically enhance DNA binding even though those residues do not directly contact DNA. For example, D232N and E233K yield a highly hyperactive phenotype when combined (Supplementary Table S5).
Several of the activating substitutions lie near the active site and most likely exert their effect via the catalytic subunits. One cluster (G302D, G320V, I335T, G340S, W345C, W345R, M374V and M374T) probably affects the packing of the catalytic core, although D320V could also affect a network of salt bridges between domains IIα and IIβ. Surprisingly, all of these changes except M374V were deleterious in the in vitro transposition assay even though they were activating in the in vivo papillation assay. The most extreme case was D320V, being totally inactive in plasmid targeting and genomic integration assays. Corroborating our data, D320A was previously shown to be inactive for strand transfer in vitro (49). Another cluster of substitutions (Q254R, E258G, F447S, F447Y, F464Y and D466G) packs against the loop containing the DDE-motif residue E392 and/or the adjacent loop. The latter interacts with the non-transferred strand and may undergo conformational changes during transpososome assembly.
The activating substitutions found in domain IIβ are difficult to explain, but like D320V, may affect domains’ IIα and IIβ interaction, which varies slightly between the two types of subunits. Residue Q539 points toward the solvent and toward domain IIα. V495 and V507 flank a cleft that binds a loop extending from domain IIα.
Finally, the E179V (Iγ) and Q594R (IIIα) substitutions most likely improve electrostatic interactions with DNA (Figure 3DE). E179 of both types of subunits is close to Mu end DNA, and on the catalytic subunit it is also close to modeled flanking host DNA. Similarly, on the catalytic subunits Q594R lies near the target DNA, and on the architectural subunits it lies near the modeled flanking host DNA.
Our analysis indicates that the hyperactivity-inducing substitutions exert their effects at several different key steps along the transposition pathway. While many appear to affect protein–protein interactions that are required for tetramer assembly and stability, some may affect catalysis itself, and others (particularly those that increase the net positive charge) may enhance initial binding to Mu end DNA or later binding to target DNA. Mutations at protein–protein interfaces appeared particularly commonly in our screens, perhaps due to the complicated nature of the Mu transpososome architecture, in which different subunits play different roles. Our results show that highly hyperactive variants can be achieved by combining multiple substitutions that act additively or synergistically. This has been noted previously for other transposases (9–13), but may be particularly important for the Mu transpososome because of its remarkably intricate subunit and domain structure. By in vivo analysis, the most hyperactive MuA variant generated in our study contained eight amino acid substitutions.
The effect of each particular hyperactivity-inducing substitution is expected to be manifested primarily via one set of subunits (the R1-bound catalytic versus the R2-bound architectural subunits). However, it is possible that the same mutated residue could exert opposing effects in the two types of subunits. Future work exploiting altered-specificity variants to target specific substitution variants to only the R1 or R2 positions may therefore allow us to design even more highly active variants.
The results of the different assays generally correlated well despite the greatly varying conditions, under which the variants were tested. The papillation assay emulates the entire Mu transposition pathway in vivo, from the search for the transposon ends to the transpososome disassembly, and it also includes MuA protein expression. In contrast, purified protein was used in the other assays (in vitro integration into a plasmid target and in vivo genomic integration via electroporation of pre-assembled transpososome). These assays necessarily differ in the concentrations of MuA and its binding sites, as well as the relative amount of non-specific DNA competing for MuA binding. The latter two assays also utilize pre-cut substrates and therefore circumvent the donor cleavage step, which is mandatory in the papillation assay.
Differences in results among the assays may reflect different relative importances of the many features of the transposase, or in the case of the in vivo papillation assay versus the in vitro integration assay, the involvement of host proteins. Mu transposition proceeds well under in vitro conditions and targets naked DNA. However, in vivo the target DNA would not be naked. As MuA greatly prefers prebent target DNA (50), the multitude of DNA binding proteins found in vivo could have a wide range of effects on the efficiency of target capture by the transpososome. The genomic integration assay also differs from the others in requiring electroporation of functional transpososomes into E. coli. Substitutions that are particularly activating in this assay might therefore primarily affect stability of the transpososomes. In general, our results conform to the common observation that the improvements in an in vivo activity of a protein may manifest only partially under in vitro conditions.
Owing to its lifestyle as a virus that does not depend on the survival of its host, phage Mu has been able to evolve a highly active DNA transposition machinery. Thus, considering that the WT MuA transposase portrays an exceptionally high-intrinsic transpositional activity, it was somewhat surprising that a mutant exhibiting more than 4000-fold activity increase, as measured by in vivo papillation assay, could be engineered. To our knowledge, this is by far the highest increase in the rate of transpositional recombination ever achieved in any experimental system. Clearly, our results show, for the first time, that a transposase of a transposing bacteriophage can be improved considerably for its transpositional activity.
In summary, the mutagenesis approach and structure-function evaluation enabled the creation of highly hyperactive variants of MuA transposase. These variants will improve existing Mu-based techniques and potentiate novel applications, some obvious and others yet to be discovered. We hope that the improvements foster many types of new approaches for genetics and genomics studies, and promote advances in transgenesis as well as developments in medical applications, including those aimed at gene therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pirjo Rahkola, Sari Tynkkynen, Tatjana Saarinen, and Jaakko Heikkilä for technical assistance. Terhi Launiainen is acknowledged for the construction of pTLaH1 and Kiyoshi Mizuuchi for providing plasmids pMK591 and pMK596. We thank Tanja Rämö for the assistance in DNA sequence analysis and Manu Tamminen for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland (SA 251168 to H.S.); Finnish National Technology Agency TEKES (3204/31/35 to H.S.); Finnish Cultural Foundation (to T.S.R.); Emil Aaltonen Foundation (to T.S.R.); Jenny and Antti Wihuri Foundation (to E.P.); University of Turku Graduate School (to E.P.); University of Turku Foundation (to E.P.); Oskar Öflund Foundation (to E.P., T.S.R.); NIH (R01 GM101989 to P.A.R.). Funding for open access charge: University of Turku.
Conflict of interest statement. Hyperactive MuA variants are among the subject-matter covered by US patent No. 9,234,190 and European Patent Application No. 2 855 672 both filed by H. Savilahti.
REFERENCES
- 1. Craig N.L., Gellert M., Lambowitz A., Rice P.A., Sandmeyer S.. Mobile DNA III. 2015; Washington, DC: ASM Press [Google Scholar]
- 2. Hickman A.B., Dyda F.. DNA transposition at work. Chem. Rev. 2016; 116:12758–12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ivics Z., Izsvak Z.. The expanding universe of transposon technologies for gene and cell engineering. Mob. DNA. 2010; 1:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kiljunen S., Pajunen M.I., Dilks K., Storf S., Pohlschroder M., Savilahti H.. Generation of comprehensive transposon insertion mutant library for the model archaeon, Haloferax volcanii, and its use for gene discovery. BMC Biol. 2014; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guschinskaya N., Brunel R., Tourte M., Lipscomb G.L., Adams M.W., Oger P., Charpentier X.. Random mutagenesis of the hyperthermophilic archaeon Pyrococcus furiosus using in vitro mariner transposition and natural transformation. Sci. Rep. 2016; 6:36711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vargas J.E., Chicaybam L., Stein R.T., Tanuri A., Delgado-Canedo A., Bonamino M.H.. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 2016; 14:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yin H., Kauffman K.J., Anderson D.G.. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017; 16:387–399. [DOI] [PubMed] [Google Scholar]
- 8. Jamal M., Ullah A., Ahsan M., Tyagi R., Habib Z., Rehman K.. Improving CRISPR-Cas9 on-target specificity. Curr. Issues Mol. Biol. 2017; 26:65–80. [DOI] [PubMed] [Google Scholar]
- 9. Zhou M., Reznikoff W.S.. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 1997; 271:362–373. [DOI] [PubMed] [Google Scholar]
- 10. Voigt F., Wiedemann L., Zuliani C., Querques I., Sebe A., Mates L., Izsvak Z., Ivics Z., Barabas O.. Sleeping beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat. Commun. 2016; 7:11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L.. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lampe D.J., Akerley B.J., Rubin E.J., Mekalanos J.J., Robertson H.M.. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:11428–11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Germon S., Bouchet N., Casteret S., Carpentier G., Adet J., Bigot Y., Auge-Gouillou C.. Mariner Mos1 transposase optimization by rational mutagenesis. Genetica. 2009; 137:265–276. [DOI] [PubMed] [Google Scholar]
- 14. Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983; 35:785–794. [DOI] [PubMed] [Google Scholar]
- 15. Harshey R.M. The Mu story: How a maverick phage moved the field forward. Mob. DNA. 2012; 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harshey R.M. Transposable Phage Mu. Microbiol. Spectr. 2014; 2:doi:10.1128/microbiolspec.MDNA3-0007-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haapa S., Taira S., Heikkinen E., Savilahti H.. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999; 27:2777–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009; 10:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Savilahti H., Rice P.A., Mizuuchi K.. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995; 14:4893–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montaño S.P., Pigli Y.Z., Rice P.A.. The Mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012; 491:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grawenhoff J., Engelman A.N.. Retroviral integrase protein and intasome nucleoprotein complex structures. World J. Biol. Chem. 2017; 8:32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ru H., Chambers M.G., Fu T.M., Tong A.B., Liao M., Wu H.. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell. 2015; 163:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haapa-Paananen S., Rita H., Savilahti H.. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J. Biol. Chem. 2002; 277:2843–2851. [DOI] [PubMed] [Google Scholar]
- 24. Turakainen H., Saarimäki-Vire J., Sinjushina N., Partanen J., Savilahti H.. Transposition-based method for the rapid generation of gene-targeting vectors to produce Cre/Flp-modifiable conditional knock-out mice. PLoS One. 2009; 4:e4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pulkkinen E., Haapa-Paananen S., Turakainen H., Savilahti H.. A set of mini-Mu transposons for versatile cloning of circular DNA and novel dual-transposon strategy for increased efficiency. Plasmid. 2016; 86:46–53. [DOI] [PubMed] [Google Scholar]
- 26. Haapa S., Suomalainen S., Eerikäinen S., Airaksinen M., Paulin L., Savilahti H.. An efficient DNA sequencing strategy based on the bacteriophage Mu in vitro DNA transposition reaction. Genome Res. 1999; 9:308–315. [PMC free article] [PubMed] [Google Scholar]
- 27. Orsini L., Pajunen M., Hanski I., Savilahti H.. SNP discovery by mismatch-targeting of Mu transposition. Nucleic Acids Res. 2007; 35:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poussu E., Vihinen M., Paulin L., Savilahti H.. Probing the α-complementing domain of E. coli β-galactosidase with use of an insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition. Proteins. 2004; 54:681–692. [DOI] [PubMed] [Google Scholar]
- 29. Edwards W.R., Busse K., Allemann R.K., Jones D.D.. Linking the functions of unrelated proteins using a novel directed evolution domain insertion method. Nucleic Acids Res. 2008; 36:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vilen H., Aalto J.-M., Kassinen A., Paulin L., Savilahti H.. A direct transposon insertion tool for modification and functional analysis of viral genomes. J. Virol. 2003; 77:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kekarainen T., Savilahti H., Valkonen J.P.. Functional genomics on Potato virus A: virus genome-wide map of sites essential for virus propagation. Genome Res. 2002; 12:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamberg A., Nieminen S., Qiao M., Savilahti H.. Efficient insertion mutagenesis strategy for bacterial genomes involving electroporation of in vitro-assembled DNA transposition complexes of bacteriophage Mu. Appl. Environ. Microbiol. 2002; 68:705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pajunen M.I., Pulliainen A.T., Finne J., Savilahti H.. Generation of transposon insertion mutant libraries for gram-positive bacteria by electroporation of phage Mu DNA transposition complexes. Microbiology. 2005; 151:1209–1218. [DOI] [PubMed] [Google Scholar]
- 34. Paatero A.O., Turakainen H., Happonen L.J., Olsson C., Palomaki T., Pajunen M.I., Meng X., Otonkoski T., Tuuri T., Berry C. et al. Bacteriophage Mu integration in yeast and mammalian genomes. Nucleic Acids Res. 2008; 36:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambrook J., Russell D.W.. Molecular Cloning: A Laboratory Manual. 2001; NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 36. Pulkkinen E., Haapa-Paananen S., Savilahti H.. An assay to monitor the activity of DNA transposition complexes yields a general quality control measure for transpositional recombination reactions. Mob. Genet. Elements. 2014; 4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rasila T.S., Pajunen M.I., Savilahti H.. Critical evaluation of random mutagenesis by error-prone polymerase chain reaction protocols, Escherichia coli mutator strain, and hydroxylamine treatment. Anal. Biochem. 2009; 388:71–80. [DOI] [PubMed] [Google Scholar]
- 38. Pajunen M.I., Rasila T.S., Happonen L.J., Lamberg A., Haapa-Paananen S., Kiljunen S., Savilahti H.. Universal platform for quantitative analysis of DNA transposition. Mob. DNA. 2010; 1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker T.A., Mizuuchi M., Savilahti H., Mizuuchi K.. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993; 74:723–733. [DOI] [PubMed] [Google Scholar]
- 40. Baker T.A., Mizuuchi M., Mizuuchi K.. MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell. 1991; 65:1003–1013. [DOI] [PubMed] [Google Scholar]
- 41. Richardson J.M., Colloms S.D., Finnegan D.J., Walkinshaw M.D.. Molecular architecture of the Mos1 paired-end complex: The structural basis of DNA transposition in a eukaryote. Cell. 2009; 138:1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hare S., Gupta S.S., Valkov E., Engelman A., Cherepanov P.. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010; 464:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hickman A.B., Ewis H.E., Li X., Knapp J.A., Laver T., Doss A.L., Tolun G., Steven A.C., Grishaev A., Bax A. et al. Structural basis of hAT transposon end recognition by hermes, an octameric DNA transposase from Musca domestica. Cell. 2014; 158:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steiniger-White M., Rayment I., Reznikoff W.S.. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 2004; 14:50–57. [DOI] [PubMed] [Google Scholar]
- 45. Kim M.S., Lapkouski M., Yang W., Gellert M.. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015; 518:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mizuuchi M., Mizuuchi K.. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: Implications for regulation. Cell. 1989; 58:399–408. [DOI] [PubMed] [Google Scholar]
- 47. Abdelhakim A.H., Oakes E.C., Sauer R.T., Baker T.A.. Unique contacts direct high-priority recognition of the tetrameric Mu transposase-DNA complex by the AAA+ unfoldase ClpX. Mol. Cell. 2008; 30:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saariaho A.-H., Lamberg A., Elo S., Savilahti H.. Functional comparison of the transposition core machineries of phage Mu and Haemophilus influenzae Mu-like prophage Hin-Mu reveals interchangeable components. Virology. 2005; 331:6–19. [DOI] [PubMed] [Google Scholar]
- 49. Krementsova E., Giffin M.J., Pincus D., Baker T.A.. Mutational analysis of the Mu transposase. contributions of two distinct regions of domain II to recombination. J. Biol. Chem. 1998; 273:31358–31365. [DOI] [PubMed] [Google Scholar]
- 50. Fuller J.R., Rice P.A.. Target DNA bending by the Mu transpososome promotes careful transposition and prevents its reversal. Elife. 2017; 6:e21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.