Abstract
Background
The purpose of this study was to analyze the potential of radiomics for disease stratification beyond key molecular, clinical, and standard imaging features in patients with glioblastoma.
Methods
Quantitative imaging features (n = 1043) were extracted from the multiparametric MRI of 181 patients with newly diagnosed glioblastoma prior to standard-of-care treatment (allocated to a discovery and a validation set, 2:1 ratio). A subset of 386/1043 features were identified as reproducible (in an independent MRI test-retest cohort) and selected for analysis. A penalized Cox model with 10-fold cross-validation (Coxnet) was fitted on the discovery set to construct a radiomic signature for predicting progression-free and overall survival (PFS and OS). The incremental value of a radiomic signature beyond molecular (O6-methylguanine-DNA methyltransferase [MGMT] promoter methylation, DNA methylation subgroups), clinical (patient’s age, KPS, extent of resection, adjuvant treatment), and standard imaging parameters (tumor volumes) for stratifying PFS and OS was assessed with multivariate Cox models (performance quantified with prediction error curves).
Results
The radiomic signature (constructed from 8/386 features identified through Coxnet) increased the prediction accuracy for PFS and OS (in both discovery and validation sets) beyond the assessed molecular, clinical, and standard imaging parameters (P ≤ 0.01). Prediction errors decreased by 36% for PFS and 37% for OS when adding the radiomic signature (compared with 29% and 27%, respectively, with molecular + clinical features alone). The radiomic signature was—along with MGMT status—the only parameter with independent significance on multivariate analysis (P ≤ 0.01).
Conclusions
Our study stresses the role of integrating radiomics into a multilayer decision framework with key molecular and clinical features to improve disease stratification and to potentially advance personalized treatment of patients with glioblastoma.
Keywords: glioblastoma, MGMT, radiomics
Importance of the study
In this study we present a high-throughput radiomic analysis from MRI data of patients with newly diagnosed glioblastoma prior to standard-of-care treatment. We identify a radiomic signature that improves disease stratification beyond key molecular features (MGMT promoter methylation status, global DNA methylation glioblastoma subgroups), clinical characteristics, and standard imaging parameters. Specifically, prediction errors decreased by 36% for PFS and by 37% for OS when adding the radiomic signature to the model (compared with 29% and 27%, respectively, with molecular + clinical features alone). The radiomic signature was—along with MGMT promoter methylation status—the only parameter with retained independent significance on multivariate analysis. Our study stresses the role of integrating radiomics into a multilayer decision framework with key molecular and clinical features to improve disease stratification and to potentially advance personalized treatment and clinical management of patients with glioblastoma.
Glioblastoma is the most frequent and most aggressive primary brain tumor in adults.1 Standard-of-care treatment consists of maximum safe resection followed by radiotherapy in addition to concomitant and adjuvant chemotherapy with the alkylating agent temozolomide (TMZ).2 Sensitivity to TMZ is primarily mediated by the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT), since epigenetic silencing of the MGMT gene by promoter methylation compromises DNA repair and increases chemosensitivity.3 Methylation status of the MGMT promoter has been shown to be a strong and independent prognostic biomarker in patients with newly diagnosed glioblastoma and a relevant predictive biomarker in the subpopulation of elderly glioblastoma patients.3–6 Assessment of MGMT promoter methylation status has therefore been incorporated into routine clinical management, and clinical trials have started to use it as a patient selection criterion.7–9 In addition, recent large-scale epigenetic profiling studies dissected glioblastoma into meaningful biological subgroups that correlate with distinct molecular-genetic alterations and key clinical parameters.10,11
Parallel to the advancements in the molecular characterization of glioblastoma—with MGMT nowadays considered a pivotal molecular biomarker—comprehensive non-invasive characterization of brain tumors on MRI has recently emerged as a promising field of research.12,13 This approach—termed radiomics—aims to utilize the full potential of medical imaging data and allows non-invasive, three-dimensional, and quantitative characterization of neoplastic tissue and identification of quantitative imaging biomarkers that may complement molecular characterization and thereby improve clinical management of glioblastoma.14–16
In the present study, we analyzed the full potential of radiomics, by automatically extracting and analyzing 1043 quantitative features quantifying tumor image intensity, shape, and texture from MRI in 181 patients with newly diagnosed glioblastoma prior to standard-of-care treatment. Specifically we aimed to investigate whether radiomic profiling allows for superior patient stratification and adds value beyond key molecular, clinical, and standard imaging parameters in patients with glioblastoma.
Materials and Methods
Patients
Retrospective evaluation of imaging data was approved by the local ethics committee of the University of Heidelberg and informed consent was waived. All patients were screened who had newly diagnosed brain tumor at the University of Heidelberg Medical Center from August 2009 to May 2016 with available Illumina Infinium HumanMethylation450 array data from tissue specimens of the initial surgery. Specimens were obtained from patients undergoing surgical resection or biopsy at the Department of Neurosurgery at the University of Heidelberg Medical Center and gathered according to the research procedures approved by the institutional review board at the Medical Faculty Heidelberg. Written informed consent was obtained from each patient. We selected patients who met the following criteria: (i) confirmed isocitrate dehydrogenase wildtype glioblastoma based on integrated diagnosis with histology and molecular information according to the 2016 World Health Organization classification of tumors of the central nervous system,17 (ii) availability of a corresponding pretreatment MRI study prior to surgery with an identical sequence protocol including pre- and postcontrast T1-weighted 3D magnetization-prepared rapid acquisition gradient echo (MPRAGE) images, fluid attenuated inversion recovery (FLAIR) images, and T2–turbo spin echo weighted images. Moreover, (iii) only patients receiving adjuvant treatment following surgery consisting of either (a) concomitant radiation plus TMZ followed by adjuvant TMZ or (b) radiation alone or (c) TMZ alone were selected. A total of 181 patients met the outlined inclusion and exclusion criteria and served as the final cohort for the present study.
Baseline epidemiological and clinical characteristics of all patients are shown in Supplementary Table S1. Assessment of treatment response was performed according to the Response Assessment in Neuro-Oncology working group criteria.18 At the time of last assessment (April 2017), 90% of patients (163/181) showed tumor progression and 67% of patients (122/181) had died. Overall survival (OS) was calculated from the time of diagnosis until death or last follow-up. Similarly, progression-free survival (PFS) was calculated from the time of diagnosis until tumor progression or death (whichever occurred first).
DNA Methylation Profiling
The Illumina Infinium HumanMethylation450 array was used to obtain genome-wide assessment of DNA methylation, according to the manufacturer’s instructions at the Genomics and Proteomics Core Facility of the German Cancer Research Center as described previously.10,15 Data were filtered according to the following criteria: removal of probes targeting the X and Y chromosomes (n = 11551), removal of probes containing a single nucleotide polymorphism (dbSNP132 Common) within 5 base pairs of and including the targeted cytosine-phosphate-guanine site (n = 24536), and removal of probes not mapping uniquely to the human reference genome (hg19) allowing for one mismatch (n = 9993). In total, 438370 probes were kept for analysis. The MGMT promoter methylation status (methylated vs unmethylated) was determined from the Illumina Infinium HumanMethylation450 array data as described previously (using the “mgmtstp27” library in R v3.4.0).19,20 Furthermore, a random forest algorithm compared each case with a brain tumor DNA methylation profile reference bank consisting of more than 2800 brain tumor cases to assign each patient to a glioblastoma subgroup based on the individual global DNA methylation pattern (see Supplementary Table S2 and www.molecularneuropathology.org/mnp/classifier/1).10,21,22 The HumanMethylation450 array data of the present study have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE103659.
MR Imaging and Postprocessing
Images were acquired in the routine clinical workup using a 3 Tesla MR system (Magnetom Verio/Trio TIM, Siemens Healthcare) with a 12-channel head-matrix coil. Briefly, the protocol included T1-weighted 3D MPRAGE images both before (T1) and after (cT1) administration of a 0.1 mmol/kg dose of gadoterate meglumine (Dotarem, Guerbet) as well as axial FLAIR and axial T2-weighted images. Details on MRI acquisition parameters are available in Supplementary Table S3.
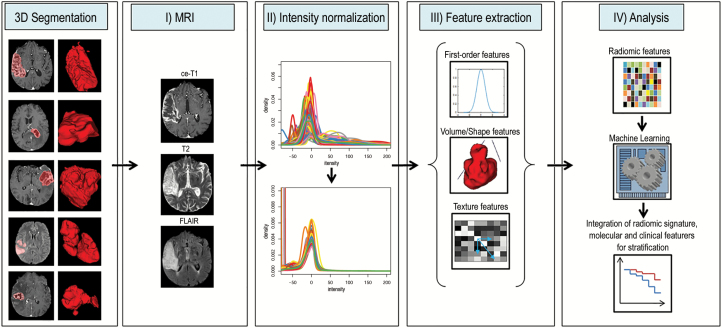
Postprocessing was performed as described previously (Fig. 1). First, image intensity normalization (using the ANTsR23 and WhiteStripe24 packages implemented in R v3.4.025) was performed to transform arbitrary MR signal intensities into standardized intensity ranges for each imaging modality across all subjects, to generate well-defined inputs for quantitative radiomic feature extraction (details are shown in Supplementary Table S4).26 The Functional Magnetic Resonance Imaging of the Brain (FMRIB) software library (FSL; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL) was used for image registration. First, brain voxels were isolated by generating a binary brain mask from the T1 volume using the brain extraction tool27 and transferred to all other imaging volumes (cT1, FLAIR, T2) for each patient. These image volumes were then registered to the brain-extracted T1 volume using the linear image registration tool28,29 with a mutual information algorithm and a 6 degree of freedom transformation. T1 subtraction volumes (subT1) were generated by voxel-wise subtraction of the T1 from the cT1 volume. Tumor segmentation was then performed semi-automatically (by P.K., a radiology resident with 4 years of experience, and subsequently checked by D.B., a board-certified radiologist and neuroradiologist with 15 years of experience in image processing; discrepancies were resolved through a consensus discussion) to select the contrast-enhancing (CE) portion of the whole tumor (on the subT1 images), as well as the non-enhancing FLAIR hyperintense (NE) portion (defined as FLAIR hyperintense abnormality excluding the CE and necrotic [NEC] tumor portions, that is, including both FLAIR hyperintense tumor and potentially vasogenic edema) and the NEC portion of the tumor (on the cT1 images) using a region-growing segmentation algorithm implemented in ITK-SNAP (www.itksnap.org30), as described previously.31,32 Radiomic feature extraction was performed either (i) from the CE segmentation mask if CE but not NEC tumor was present, (ii) from a combined CE+NEC segmentation mask if both CE and NEC tumor were present, or (iii) from the NE segmentation mask if only NE but not CE and NEC tumor was present. A holefilling algorithm (using the imdilate function in Matlab) was applied to slightly dilate the segmentation mask and thus include single-voxel holes within the segmentation mask.
Fig. 1.
Radiomics image postprocessing workflow. Left: different tumors have different shapes and intensities, as shown on representative slices on the left (tumor segmentations in red), with the volume-rendered 3D segmentations on the right. Right: workflow from tumor segmentation to analysis. (I) Different MRI sequences are skull-stripped and coregistered to each other. (II) Image intensities are normalized into a common parameter space that allows referencing across different subjects. (III) Multiple radiomic features are automatically calculated from intensity-normalized images using 3D segmentations, including first-order, volume/shape, and texture features. (IV) The large number of radiomic feature parameters are then subjected to machine learning–based classification algorithms to identify radiomic-based imaging signatures that are related to an outcome of interest. Finally, integrative assessment of radiomic signatures with molecular and clinical characteristics aims to improve stratification of patients.
Radiomic features were calculated automatically using in-house software implemented within the medical imaging interaction toolkit (www.mitk.org)33 and included (i) 21 first-order features (FO), (ii) 17 volume and shape features (VSF), and (iii) 321 texture features (TF). The final set consisted of 342 FO and TF for each of the intensity normalized modalities (T1, cT1, FLAIR), and 17 VSF, resulting in 1043 radiomic features for each patient. Definitions of radiomic features are given in Supplementary Table S5.
Statistical Analysis
Subsequent analysis was performed using R v3.4.0. Patients were randomly allocated to a discovery and validation set (2:1 ratio with n = 120 patients in the discovery set and n = 61 patients in the validation set) with the distribution of MGMT promoter methylation kept balanced between both sets (stratified random split). Distribution of epidemiological, clinical, and molecular characteristics between the discovery and validation sets was compared with the chi-square test for categorical parameters and the Wilcoxon test for continuous parameters.
A total of 386 out of the 1043 extracted radiomic features (37.0%) were identified as stable and reproducible based on a separate prospective test-retest study and selected for further analysis (methodology and results of this preceding analysis are outlined in Supplementary Table S6).
A Cox regression model via penalized maximum likelihood (lasso) was fitted on the discovery set to identify a subset of radiomic features and construct a radiomic signature from the high-dimensional radiomic dataset associated with outcome (as measured by OS; using the glmnet package34,35). The tuning parameter λ, which is the global regularization parameter, was identified via 10-fold cross-validation. The performance of the identified radiomic signature for stratifying PFS and OS in the discovery and validation sets was assessed by comparing models that included (i) molecular features alone (MGMT promoter methylation status and global DNA methylation subgroups), (ii) clinical features alone (including patient’s age, KPS at diagnosis, extent of resection [EOR; gross total resection (GTR) vs subtotal resection (STR) or biopsy] and adjuvant treatment [radiotherapy plus concomitant and adjuvant TMZ (RT+TMZ) vs RT or TMZ only]), (iii) standard imaging features alone (tumor volumes from contrast enhancement, necrosis, and edema), (iv) radiomic signature alone, and (v) different combinations of the above stated models to assess the incremental value of combining parameters from different layers (ie, molecular, clinical, standard imaging, radiomics).
For each model, we assessed the overall performance with prediction error curves (PECs) over time and the integrated Brier score (IBS) (using the pec function of the pec library36,37). The IBS can range from 0 for a perfect model to 0.25 for a non-informative model with a 50% incidence of the outcome. Specifically, the discovery set was supplied to the traindata argument of the pec function, whereas the validation set was used for estimating the PECs and IBS (data argument of the pec function). Furthermore, ANOVA was used to determine whether additional predictors significantly increase the model fit (ie, reduction in the log-likelihood). Multivariate Cox regression models were used to assess the independent significance of the different predictors (ie, radiomic signature and molecular and clinical characteristics).
The linear predictor (a weighted sum of the covariates in the Cox regression model, where the weights are the regression coefficients) was used to generate a conditional version of a kernel smoothed Kaplan–Meier estimator using nearest neighborhoods (with the prodlim library38). For illustration, we show the estimated survival curves for low, medium, and high levels of the linear predictor, determined as the survival curve estimate for the neighborhood of the smallest (<25th percentile), medium (25th–75th percentile), and largest values (>75th percentile) of the signature, respectively. P-values <0.05 were considered significant.
Results
Distribution of epidemiological (patient’s age [P = 1.00]), clinical (pretreatment KPS [P = 0.50], EOR [P = 0.65], adjuvant treatment following surgery [P = 0.29]), and molecular characteristics (MGMT promoter methylation status [P = 1.0], global DNA methylation glioblastoma subgroups [P = 0.61]) was balanced between the discovery and validation sets (2:1 ratio with n = 120 in the discovery and n = 61 in the validation set) (Supplementary Figure S1).
External selection of stable and reproducible radiomic features was performed in an independent prospective test-retest study prior to radiomic signature discovery. A subset of 386 of 1043 radiomic features (37.0%) were identified as reproducible (based on a concordance correlation coefficient >0.8) and selected for further analysis (see Supplementary Table S6 for detailed results). The lasso-penalized Cox model (Coxnet algorithm) was fitted on the reproducible radiomic features in the discovery set and identified 8 features that were most important for predicting treatment outcome using a 10-fold cross-validated threshold. These features were derived from all imaging sequences (cT1, FLAIR, and T2) and included texture features from gray-level co-occurrence and run-length matrices (6/8 features on FLAIR and T2) as well as volumetric features (2/8 features on cT1) (Supplementary Table S7). The volumetric feature tumor asphericity was the parameter with the highest positive Coxnet coefficient, thereby suggesting that greater asphericity (shape irregularity) of the tumor was associated with worse outcome.
The identified radiomic signature allowed stratification of PFS with hazard ratios (HRs) of 1.46 (P < 0.01) in the discovery set and 2.23 (P < 0.01) in the validation set. Similarly, stratification of OS based on the radiomic signature was achieved with an HR of 2.72 in both discovery and validation sets (P < 0.01 each). Adding the radiomic signature as an additional explanatory variable to a model that already included key molecular (ie, MGMT promoter methylation status and global DNA methylation subgroups) and clinical features (ie, patient’s age, KPS at diagnosis, EOR, and type of adjuvant treatment) significantly increased the model fit for stratifying PFS and OS in both the discovery and the validation set (P ≤ 0.01 each) (Table 1a). In contrast to the radiomic signature, less sophisticated standard imaging parameters such as tumor volumes (including those from contrast enhancement, necrosis, and edema) did not exhibit additional explanatory value for stratifying PFS or OS beyond key molecular or clinical characteristics (P > 0.05 each) (Table 1a).
Table 1.
(a) Analysis of deviance for different Cox regression models (ANOVA) was used to determine whether the radiomic signature or the tumor volume increased the model fit beyond key molecular and clinical characteristics. (b) Performance metrics of the different Cox regression models based on prediction error curves over time with the integrated Brier score (lower values indicate better performance)
| (a) Analysis of Deviance for Different Cox Regression Models (ANOVA) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Discovery Set | Validation Set | ||||||
| OS | PFS | OS | PFS | ||||||
| P | chi2 | P | chi2 | P | chi2 | P | chi2 | ||
| Molecular1 + Clinical2 | Molecular1 + Clinical2 + Radiomic signature | <0.01 | 34.3 | 0.01 | 6.2 | <0.01 | 10.4 | <0.01 | 8.0 |
| Molecular1 + Clinical2 | Molecular1 + Clinical2 + Tumor volumes3 | 0.79 | 1.0 | 0.19 | 4.7 | 0.21 | 4.6 | 0.14 | 5.4 |
| (b) Performance Metrics of the Different Cox Regression Models | |||||||||
| Model |
Integrated Brier Score (IBS) (percent reduction of IBS compared with the null model4) |
||||||||
| OS | PFS | ||||||||
| Single layer | Molecular1 | 0.149 | −9% | 0.121 | −12% | ||||
| Clinical2 | 0.133 | −18% | 0.126 | −9% | |||||
| Tumor volumes3 | 0.160 | −2% | 0.135 | −2% | |||||
| Radiomic signature | 0.137 | −16% | 0.125 | −9% | |||||
| Two layers | Molecular1 + Clinical2 | 0.119 | −27% | 0.098 | −29% | ||||
| Clinical2 + Radiomic signature | 0.116 | −29% | 0.117 | −15% | |||||
| Molecular1 + Radiomic signature | 0.122 | −25% | 0.109 | −21% | |||||
| Three layers | Molecular1 + Clinical2 + Radiomic signature | 0.103 | −37% | 0.089 | −36% | ||||
Annotation: 1 = including MGMT promoter methylation status and global DNA methylation glioblastoma subtypes; 2 = including patient’s age, KPS, EOR, and adjuvant treatment; 3 = including tumor volumes from contrast enhancement, necrosis, and edema; 4 = IBSs for the null (reference) models were 0.163 for OS and 0.138 for PFS.
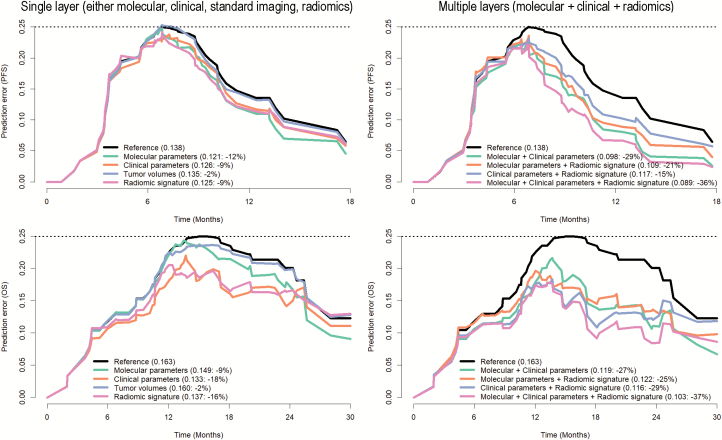
The PECs and IBS not only confirmed the incremental value of the radiomic signature but also highlighted the importance of combining information from multiple layers (ie, clinical and molecular characteristics and radiomic signatures) to improve disease stratification. Specifically, predicting PFS and OS with information from a single layer (either molecular, clinical, standard imaging, or radiomic characteristics) allowed reduction of the prediction error (compared with a non-informative model) by 12% for PFS and by 9% for OS with molecular characteristics alone, by 9% for PFS and 18% for OS with clinical characteristics alone, and by 9% for PFS and 16% for OS with the radiomic signature alone (Fig. 2, left). The preoperative tumor volumes (including those from contrast enhancement, necrosis, and edema) as less sophisticated standard imaging parameters did not exhibit relevant explanatory value (reduction of prediction error by only 2% for both OS and PFS) and were therefore excluded from further analysis.
Fig. 2.
Prediction error curves for stratifying PFS (upper row) and OS (lower row) based on a single layer (left column)—ie, either molecular (including MGMT promoter methylation status + global DNA methylation glioblastoma subtypes) or clinical (patient’s age + KPS, EOR, adjuvant treatment) information or standard imaging parameters (tumor volumes from contrast enhancement, necrosis, and edema) or the radiomic signature—or (right column) combining the information from multiple layers. Prediction error rates are given in brackets (including the percentage reduction compared with the null model with no explanatory value). Combining the information from multiple layers (right column) allowed reduction of the prediction error beyond every single layer model (left column). The identified radiomic signature reduced the prediction error beyond molecular and clinical features and combining molecular + clinical information and the radiomic signature yielded the highest accuracy, with a reduction of the prediction error by 36% for PFS and 37% for OS (compared with 29% and 27% for a model without the radiomic signature that includes only molecular and clinical information).
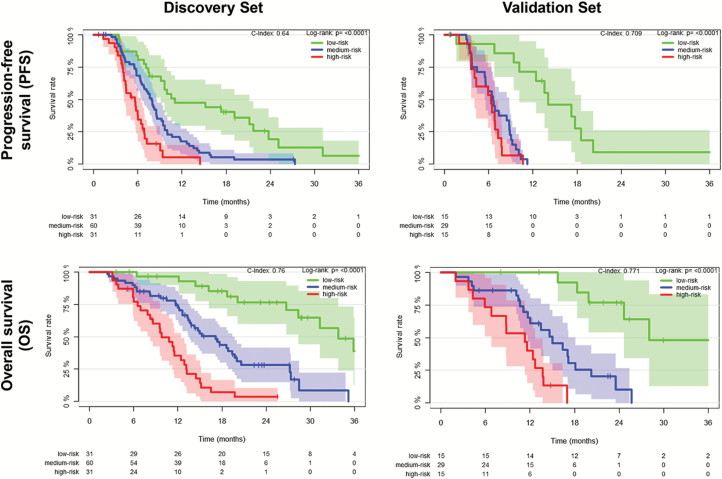
Combining the information from multiple layers allowed reduction of the prediction error beyond every single-layer model. The highest accuracy was achieved for a model that included information from all layers (ie, clinical + molecular characteristics + radiomic signature) with a reduction of the prediction error by 36% for PFS and 37% for OS (compared with 29% and 27% for a model without the radiomic signature that includes only molecular and clinical characteristics) (Fig. 2, right). Fig. 3 illustrates the integrative performance of a model that leverages the information from all layers (ie, clinical + molecular characteristics + radiomic signature) for predicting PFS and OS in both discovery and validation sets (for illustration purposes, estimates are shown for low, medium, and high levels of the linear predictor).
Fig. 3.
Progression-free and overall survival in the discovery and validation sets stratified based on key molecular characteristics (MGMT promoter methylation status + global DNA methylation glioblastoma subtypes), clinical characteristics (patient’s age + KPS, EOR, adjuvant treatment), and the radiomic signature. Survival curves were derived computing nearest-neighbor estimate of bivariate distribution of survival and linear predictor levels. For illustration purposes, we show estimates for low, medium, and high levels of predictors.
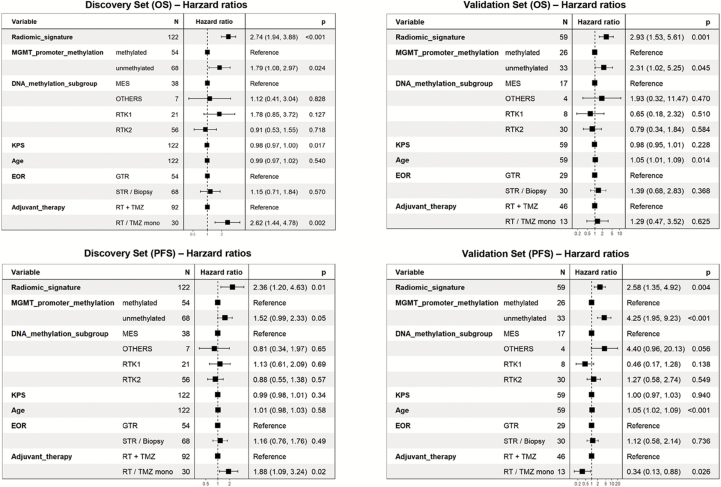
The radiomic signature was, along with MGMT promoter methylation status, the only parameter within a multivariate Cox regression model (including all parameters from all layers, that is, molecular and clinical features and radiomic signature) that consistently showed independent significance for predicting PFS and OS in both discovery and validation sets (Fig. 4). Specifically the radiomic signature and MGMT promoter methylation status were associated with HRs of 2.36 (P = 0.01) and 1.52 (P = 0.05) for stratifying PFS and HRs of 2.74 (P < 0.01) and 1.79 (P = 0.02) for stratifying OS in the discovery set. Similarly, in the validation set the radiomic signature and MGMT promoter methylation status were associated with HRs of 2.58 (P = 0.01) and 4.25 (P < 0.01) for stratifying PFS and HRs of 2.93 (P < 0.01) and 2.31 (P = 0.05) for stratifying OS. In contrast, neither the global DNA methylation glioblastoma subgroups (ie, receptor tyrosine kinase [RTK] II, mesenchymal, and RTK I, which resemble the classical, mesenchymal, and proneural subgroups according to The Cancer Genome Atlas classification [Supplementary Figure S2] 10,11) nor any of the clinical parameters demonstrated consistent and independent association for predicting PFS and OS in both the discovery and the validation set.
Fig. 4.
Multivariate Cox regression model for PFS and OS with key molecular parameters (MGMT promoter methylation status, molecular glioblastoma subtypes), clinical features, and the radiomic signature as explanatory variables. Independent significance for both PFS and OS in both discovery and validation sets was only retained for the radiomic signature and MGMT promoter methylation status. MES = mesenchymal; RT = radiation therapy.
Association of the radiomic signature with global DNA methylation glioblastoma subgroups showed that radiomic signature levels were evenly distributed throughout the different subgroups (Supplementary Table S8).10,11
Discussion
Radiomics applies advanced computational methods to convert medical images into a large number of quantitative descriptors of oncologic tissues.12,39 In the present study, we used a high-throughput approach to automatically extract 1043 quantitative radiomic features from the multiparametric MRI to comprehensively characterize the imaging phenotype in 181 patients with newly diagnosed glioblastoma. Lasso-penalized Cox regression modeling (on a subset of 396 radiomic features that were identified as stable and reproducible in an independent test-retest dataset) identified an 8-feature-based radiomic signature that allowed non-invasive prediction and stratification of PFS and OS with additive performance beyond key molecular and clinical features, but also compared with less sophisticated standard imaging parameters such as preoperative tumor volumes. Integrating the radiomic signature into a model with key molecular and clinical features significantly increased the fit and accuracy of the model. Moreover, the radiomic signature was—along with MGMT promoter methylation status—the only parameter with retained independent significance on multivariate analysis. Our study therefore suggests that radiomic subtyping of glioblastoma may complement disease stratification of patients with glioblastoma beyond key molecular characteristics such as MGMT promoter methylation status and global DNA methylation subgroups, but also beyond clinical and standard imaging parameters and thereby potentially improve clinical decision making for patients with glioblastoma.
Our approach is based on comprehensive quantitative information derived from 3 different MRI sequences that allow a multiparametric three-dimensional characterization of the entire tumor. Stability and reproducibility of the present analysis were maximized by (i) performing methodically robust feature selection in an independent test-retest dataset, (ii) applying a biologically motivated normalization technique for multisequence MRI (white stripe normalization) that sets the ground for reliable quantitative analysis of MRI data, and (iii) performing internal validation of the radiomic signature through 10-fold cross-validation as well as external validation in an independent dataset.
Several recent studies already highlighted the potential of radiomics to identify prognostic and potentially predictive imaging biomarkers and to predict molecular subgroups and pathway activities. 14–16,40–43 The present study adds important information to the existing literature, since it is the first study that performed (i) independent test-retest analysis to identify stable radiomic features in neuro-oncology and (ii) comparison of radiomic imaging signatures with already established hallmark molecular, clinical and imaging parameters as a benchmark to prove incremental benefit of radiomics for clinical decision making.
Imaging-related limitations may result from the limited through-plane resolution of the T2 and FLAIR data compared with the higher-resolution T1 data. As a result, assessment of fine structural detail in one of the 3 spatial dimensions on the FLAIR data was affected by some degree of blurring. Furthermore, despite the high degree of automation of the current postprocessing workflow, supervision by the radiologist for semi-automated outlining of the tumor for radiomic feature extraction is still required. However, deep learning based tumor segmentation approaches may allow further automation of the workflow and promote seamless integration of this technology into clinical practice.44 Moreover, the sophisticated postprocessing workflow involves many steps and currently requires about 60 minutes of computation time per patient. With the use of customized high-performance and parallel computing, postprocessing time could, however, be shortened significantly, thus meeting the requirements of translating this technology into clinical practice. Moreover, translation of the results from our hypothesis-generating study to a broader multicenter scale and reaching consensus on a standardized set of radiomic features and postprocessing workflow are crucial for homogenization and comparability of different study findings and to allow successful adoption and integration of radiomics in clinical decision making.
In conclusion, our study stresses the role of integrating radiomics into a multilayer decision framework with key molecular and clinical features to improve disease stratification and to advance personalized treatment and clinical management of patients with glioblastoma.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
R.T.S. is funded partially by the National Institutes of Health under award numbers R01NS085211 and U24CA189523. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency.
Supplementary Material
Acknowledgments
P.K. is supported by the Medical Faculty Heidelberg Postdoc-Program and the Else-Kröner Memorial Scholarship of the Else Kröner-Fresenius Foundation. DNA methylation analysis was performed as part of the National Center for Tumor Diseases (NCT) Precision Oncology Program of the Heidelberg Center for Personalized Oncology (German Cancer Research Center–HIPO). We would further like to thank the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) for excellent technical assistance.
Conflict of interest statement. None
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(suppl 4):iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Wick W, Weller M, van den Bent M, et al. MGMT testing–the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. [DOI] [PubMed] [Google Scholar]
- 4. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 5. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 6. Malmström A, Grønberg BH, Marosi C, et al. ; Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 7. Herrlinger U, Schäfer N, Steinbach JP, et al. Bevacizumab plus irinotecan versus temozolomide in newly diagnosed o6-methylguanine-dna methyltransferase nonmethylated glioblastoma: the randomized GLARIUS trial. J Clin Oncol. 2016;34(14):1611–1619. [DOI] [PubMed] [Google Scholar]
- 8. Wick W, Gorlia T, Bent MJVD, et al. Radiation therapy and concurrent plus adjuvant temsirolimus (CCI-779) versus chemoirradiation with temozolomide in newly diagnosed glioblastoma without methylation of the MGMT gene promoter. J Clin Oncol. 2014;32(suppl 15):2003–2003. [Google Scholar]
- 9. Nabors LB, Fink KL, Mikkelsen T, et al. Two cilengitide regimens in combination with standard treatment for patients with newly diagnosed glioblastoma and unmethylated MGMT gene promoter: results of the open-label, controlled, randomized phase II CORE study. Neuro Oncol. 2015;17(5):708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 11. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grossmann P, Narayan V, Chang K, et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;19(12):1688–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016:160845. [DOI] [PubMed] [Google Scholar]
- 16. Kickingereder P, Gotz M, Muschelli J, et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22(23):5765–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 18. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 19. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. [DOI] [PubMed] [Google Scholar]
- 21. Jones DTW, Capper D, Sill M, et al. Next-generation neuropathology - improving diagnostic accuracy for brain tumors using dna methylation array-based molecular profiling. Neuro-Oncol. 2014;16(suppl 3):iii4. [Google Scholar]
- 22. Capper D, Jones DTW, Sill M, et al. DNA methylation array-based molecular profiling for brain tumor classification. Paper presented at: 60th Annual Meeting of the German Society for Neuropathology and Neuroanatomy (DGNN); August 26, 2015; Berlin. [Google Scholar]
- 23. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shinohara RT, Sweeney EM, Goldsmith J, et al. Normalization techniques for statistical inference from magnetic resonance imaging. UPenn Biostatistics Working Papers. Working Paper 36. http://biostats.bepress.com/upennbiostat/art36.
- 25. R: A Language and Environment for Statistical Computing [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 26. Shinohara RT, Sweeney EM, Goldsmith J, et al. ; Australian Imaging Biomarkers Lifestyle Flagship Study of Ageing; Alzheimer’s Disease Neuroimaging Initiative Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9–19.25379412 [Google Scholar]
- 27. Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 29. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5(2):143–156. [DOI] [PubMed] [Google Scholar]
- 30. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 31. Kickingereder P, Radbruch A, Burth S, et al. MR perfusion-derived hemodynamic parametric response mapping of bevacizumab efficacy in recurrent glioblastoma. Radiology. 2016;279(2): 542–552. [DOI] [PubMed] [Google Scholar]
- 32. Bonekamp D, Mouridsen K, Radbruch A, et al. Assessment of tumor oxygenation and its impact on treatment response in bevacizumab-treated recurrent glioblastoma. J Cereb Blood Flow Metab. 2016;37(2):485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nolden M, Zelzer S, Seitel A, et al. The Medical Imaging Interaction Toolkit: challenges and advances: 10 years of open-source development. Int J Comput Assist Radiol Surg. 2013;8(4):607–620. [DOI] [PubMed] [Google Scholar]
- 34. Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):22. [PMC free article] [PubMed] [Google Scholar]
- 35. Simon N, Friedman JH, Hastie T, et al. Regularization paths for cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011. ;39(5):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mogensen UB, Ishwaran H, Gerds TA. Evaluating random forests for survival analysis using prediction error curves. J Stat Softw. 2012;50(11):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gerds TA. prodlim: Product-Limit Estimation for Censored Event History Analysis. 2017. [Google Scholar]
- 39. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys. 2015;42(11):6725–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui Y, Tha KK, Terasaka S, et al. Prognostic imaging biomarkers in glioblastoma: development and independent validation on the basis of multiregion and quantitative analysis of MR images. Radiology. 2016;278(2):546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Işın A, Direkoğlu C, Şah M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput Sci. 2016;102:317–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.