Abstract
Background
In spite of standard multimodal therapy consisting of surgical resection followed by radiation and concurrent chemotherapy, prognosis for glioblastoma (GBM) patients remains poor. The identification of both differentiated and undifferentiated “stem cell like” populations in the tumor highlights the significance of finding novel targets that affect the heterogeneous tumor cell population. Protein arginine methyltransferase 5 (PRMT5) is one such candidate gene whose nuclear expression correlates with poor survival and has been reported to be required for survival of differentiated GBM cells and self-renewal of undifferentiated GBM cells. In the current study we screened the specificity and efficacy of 4 novel PRMT5 inhibitors in the treatment of GBM.
Methods
Efficacies of these inhibitors were screened using an in vitro GBM neurosphere model and an in vivo intracranial zebrafish model of glioma. Standard molecular biology methods were employed to investigate changes in cell cycle, growth, and senescence.
Results
In vitro and in vivo studies revealed that among the 4 PRMT5 inhibitors, treatment of GBM cells with compound 5 (CMP5) mirrored the effects of PRMT5 knockdown wherein it led to apoptosis of differentiated GBM cells and drove undifferentiated primary patient derived GBM cells into a nonreplicative senescent state.
Conclusion
In vivo antitumor efficacy combined with the specificity of CMP5 underscores the importance of developing it for translation.
Keywords: CMP5, GBM, glioma, PRMT5, senescence
Importance of the study
This is the first study to test specificity and efficacy of PRMT5 inhibitors for GBM therapy. Among the tested PRMT5 inhibitors, CMP5 can effectively and specifically block the PRMT5 activity to cause apoptosis and senescence in the differentiated and primary undifferentiated tumor cell populations, respectively.
Glioblastoma (GBM) is the most common adult malignant form of astrocytoma, accounting for more than 50% of glioma cases.1,2 Standard treatment for GBM involves surgery, radiation, and chemotherapy. The highly invasive nature of GBM makes complete surgical resection challenging; hence, surgery is accompanied by radiation and concurrent chemotherapy. Despite this aggressive therapeutic approach, prognosis for GBM patients remains dismal, with a median survival of around 15 months,1–4 and a significant improvement in patient outcome has yet to be realized. Thus there is an urgent need to discover new targets and develop novel target-specific therapies.
Protein arginine methyltransferase 5 (PRMT5) is a member of the PRMT family of proteins that play a key role in regulation of cellular signaling and gene expression by covalently modifying (methylating) histones as well as nonhistone proteins.5 PRMT5 catalyzes the symmetric dimethylation of guanidine nitrogen atoms within the arginine residues of its substrate. Dimethylation of histone proteins H4 (S2Me-H4R3), H3 (S2Me-H3R8), and H2A directed by it regulates chromatin structure to promote transcriptional repression.6–8
Expression of PRMT5 is increased in high-grade glioma, and its expression negatively correlates with patient survival.9,10 Engineered loss of PRMT5 results in apoptosis or loss of self-renewal for differentiated or undifferentiated GBM cells, respectively.11 Significance of PRMT5 for glioma genesis is further evidenced by the failure of intracranial tumor growth in mice implanted with glioma depleted for PRMT5.9,11 These findings suggest that glioma growth is addicted to PRMT5 expression, which hence might represent a novel druggable target for GBM therapy.
Small-molecule inhibitors for PRMT5 were developed using the crystal structure of rat PRMT1 as a primary template for modeling a human in silico catalytic domain and utilizing the ChemBridge CNS-Set library of 10000 small molecules.12 This is the first account of PRMT5 inhibitors being tested for antitumor efficacy for glioma.12
Here we used in vitro and in vivo zebrafish GBM xenograft models to screen the antitumor activity of these compounds and have identified compound 5 (CMP5) as a novel therapeutic agent for treating GBM.
Materials and Methods
Tissue Culture
As per the guidelines put forth by the institutional review board of The Ohio State University and the University of Texas Health Science Center, de-identified GBM tumor samples were collected from the consenting patients, washed, and dissociated using TrypLE Express (Invitrogen) and the dissociated single cells were grown as GBM neurospheres. Primary patient-derived GBM neurospheres (GBMNS) were cultured in Neurobasal medium (Gibco) with B27 without vitamin A (2%), human epidermal growth factor (50 ng/mL), basic fibroblast growth factor (50 ng/mL), and penicillin/streptomycin (1%) in low-attachment cell culture flasks.13 To obtain differentiated cells, neurospheres were grown in 10% serum for 10 days. To confirm the stemness and differentiation status of the GBMNS and their differentiated cells (GBMDC), respectively, we compared the mRNA expression of stem cell markers such as cluster of differentiation (CD)133, sex determining region Y-box 2, and CD15 and differentiation markers (Tuj1 and glial fibrillary acidic protein) (Supplementary Figure S1).
Mouse Plasma Sample and Brain Sample Collection
All the experiments involving mice were performed as per the guidelines of the Committee on Animal Care at The Ohio State University approved under the protocol Phelps-2009A0196-R1. Thirty-four ICR (imprinting control region) mice were divided into liposome (14 mice), dimethyl sulfoxide (DMSO) (10 mice), and Tween 80 (10 mice) groups. Mice were given 200 μL i.p. injection for 3 formulations. The blood samples and brain tissue were collected at specified timepoints. One hundred milligrams of brain tissue was weighed after quick grind, then homogenized on ice after adding 400 μL 50% MeOH. The mixture was sonicated on ice for 15 min and centrifuged. The supernatant was used as brain extract. Additional pharmacokinetic study methods are provided in the Supplementary methods.
Cell Cycle Analysis
Cells subjected to treatment conditions were washed with phosphate buffered saline and fixed with ethanol (80%). Fixed cells were stained with 50 µg/mL propidium iodide (PI) (Sigma-Aldrich). Flow cytometry analysis was done on stained cells using LSRII (Becton Dickinson).
β-Galactosidase Staining
GBMNS were trypsinized every 3 days and treated with fresh media and CMP5 each time for 20 days. At the end of the treatment, cells were trypsinized into single cells and subjected to β-galactosidase (β-gal) assay with the help of the Senescence Cells Histochemical Staining Kit, per manufacturer’s instructions (Sigma-Aldrich).
Zebrafish Xenograft Model
Transparent casper mutant zebrafish (roy;nacre)14 procured from Children’s Hospital Boston (Dr Leonard Zon laboratory) were maintained according to the approved Institutional Animal Care and Use Committee protocol (2009A0141-R2, principal investigator Christine Beattie) at The Ohio State University. When a clear midbrain boundary was visible, fish were recruited for the xenograft transplantation. Xenotransplants were carried out at 36 hours post fertilization.
Green fluorescent protein (GFP)–positive GBMNS-30 neurospheres (about 1 mm3) were dissociated using TrypLE (Gibco). Fish were immobilized by adding 4.2 mL of 25× stock solution of tricaine (Sigma-Aldrich) to 100 mL fresh fish water, and ~50 cells per animal were injected into the midbrain-hindbrain boundary with a pulled borosilicate glass pipette (Sutter Instruments) as described.15 Transplanted animals were allowed to recover in 24-well plates (Corning) containing fish water and penicillin/streptomycin (Invitrogen) and maintained on a warming plate at 32°C for the duration of the study. All fish were screened for tumor take 24 hours posttransplant.
Five days posttransplant (dpt) of tumor cells, tumor establishment was confirmed and animals were placed in 1 mL fish water in 24-well plates and treated with either 1% DMSO (Thermo-Fisher Scientific), HLCL66 (0.1 µM), HLCL65 (20 µM), CMP5 (20 µM), or CMP12 (20 µM) for 5 days at 28°C in 24-well plates. Fish water and drugs were replaced every day during the 5-day treatment period (5–10 dpt).
Animals were followed for survival up to 20 days post tumor implantation. SPSS was used to create Kaplan–Meier survival curves.
Statistical Analysis
Results are presented as mean values ± standard deviation of the mean. Statistical analysis was carried out by unpaired Student’s t-test using GraphPad Prism software. For survival data, the Kaplan–Meier method was used to estimate survival fractions and the log-rank test was used to evaluate difference in survival between different groups. P-values were adjusted for multiple comparisons by Holm’s procedure. P-values <0.05 were considered statistically significant.
Results
In Vitro Screening of PRMT5 Inhibitors
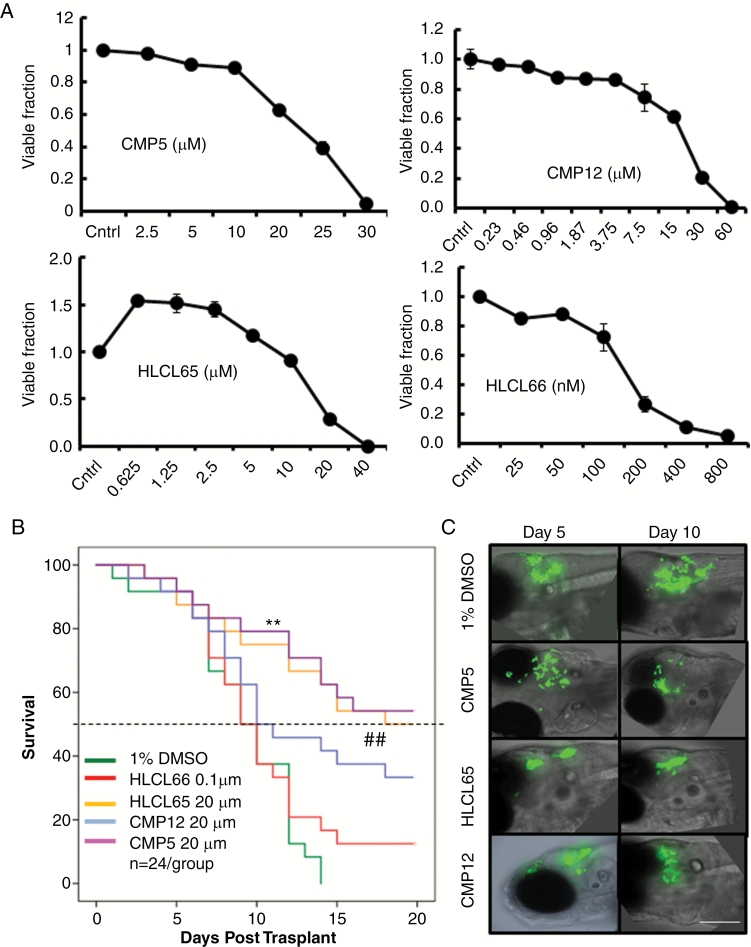
Four second-generation PRMT5 inhibitors: CMP5, CMP12, compound 65 (HLCL65), and compound 66 (HLCL66) were selected to test the utility of PRMT5 inhibition as a druggable target for GBM. Supplementary Figure S2 shows the structures as well as the modeled binding within the catalytic site of PRMT5 for each of these 4 compounds. To test the antiglioma utility of these inhibitors, we compared the dose-dependent cytotoxicity of these agents against the primary patient-derived neurospheres (GBMNS-30) 3 days after treatment by a standard MTT assay (Fig. 1A). All 4 of the PRMT5 inhibitors had a dose-dependent cytotoxicity against GBMNS-30 with lethal dose concentrations to kill 50% of test animals (LD50) of 22.5 µM, 25 µM, 150 nM, and 15 µM for CMP5, CMP12, HLCL66, and HLCL65, respectively.
Fig. 1.
PRMT5 inhibitors affect the viability and tumor growth in vitro and in vivo. (A) Viability of GBMNS-30 treated with increasing doses of the indicated drugs; 72 h posttreatment, cells were subjected to MTT assay to measure the viability. (B) Kaplan–Meier survival curves of GBMNS-30 bearing fish treated with the indicated drugs (treated day 5 post-implantation) for 5 days. Animals were followed for survival post tumor implantation (n = 24/group); P-values were adjusted for multiple comparisons by Holm’s procedure (** and ## P ≤ 0.05). (C) Spinning disk confocal fluorescent imaging of relative tumor growth on day 5 (day of treatment initiation) and day 10 post tumor cell implantation in one representative fish from each group from the survival study in (B) (bar: 50 µm). MTT = 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
PRMT5 Inhibitors Are Effective in an In Vivo Zebrafish GBM Model
We utilized a zebrafish GBM model to screen these drugs for potential anti-GBM efficacy.15 Briefly zebrafish implanted with GFP expressing GBMNS-30 were treated with DMSO or the indicated compounds for 5 days delivered in the fish water in a 24-well plate, and the animals were observed for survival (n = 24/group) (Supplementary Figure S3A). Fig. 1B shows the Kaplan–Meier survival curves of tumor-bearing fish treated with the indicated drugs at close to LD50 concentrations of each agent (Fig. 1A). While control DMSO-treated animals died with a median survival of 8 days, animals treated with HLCL65 or CMP12 showed significant improvement in survival, with the median survival advantage of >1.5-fold compared with control animals. Interestingly, animals treated with HLCL66, the drug that showed the highest potency in vitro, did not show a survival benefit after treatment. More significantly, more than 50% of animals treated with CMP5 were long-term survivors, indicating the potential antitumor efficacy of CMP5 in vivo. Consistent with the survival of tumor-bearing animals, fluorescent imaging of a representative animal from each group for GFP-positive tumor progression over time revealed obvious tumor shrinkage in fish treated with CMP5 and HLCL65 (Fig. 1C and Supplementary Figure S3B). To test the potential toxicity of these compounds, we evaluated the survival of non-tumor-bearing zebrafish, treated with the indicated compounds in water (n = 24/group) (Supplementary Figure S3C). At the doses used in the efficacy study, HLCL66 was toxic to zebrafish, and treated animals died with a median survival of 13 days (P < 0.001) (Supplementary Figure S3C). None of the other compounds showed any significant toxicity in non-tumor-bearing zebrafish (P ≥ 1 between DMSO and CMP5, CMP12, and HLCL65).
CMP5 Decreases the Proliferation of GBMNS
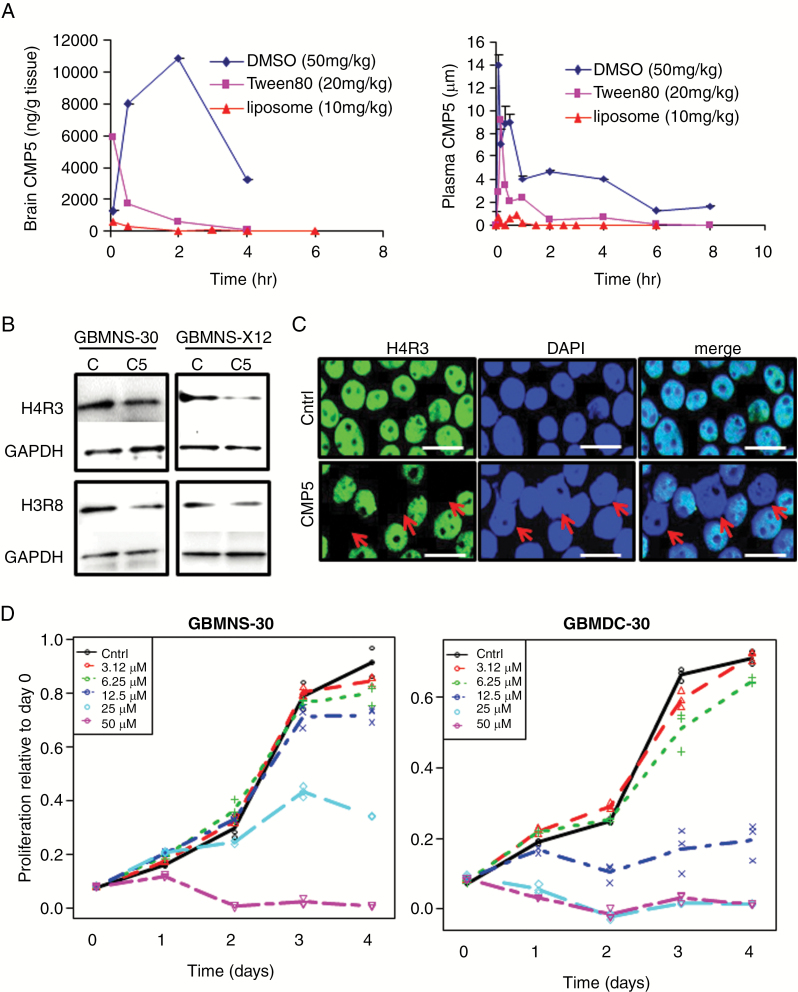
The above results indicated that both CMP5 and HLCL65 had antiglioma effects as well as a favorable safety profile. Our initial pharmacokinetic study with 3 different formulations of CMP5 (Fig. 2A, Supplementary Table S2) revealed CMP5 to have brain tissue accumulation after i.p. injection without causing toxicity in mice. Thus we selected CMP5 as a potential drug candidate for further development. We evaluated the impact of CMP5 on PRMT5 function by comparing the methylation status of histone H4 on arginine 3 and histone H3 on arginine 8 [S(Me2)-H4R3 and S(Me2)-H3R8] by western blot after treatment of GBMNS-30 and GBMNS-X12 cells grown as neurospheres (Fig. 2B). CMP5 reduced the methylation of both PRMT5 target histones. Immunofluorescence for H4R3 further confirmed the reduction in PRMT5 activity in GBMNS-30 treated with CMP5 (note the absence of green H4R3 staining in cells indicated by red arrows after treatment; Fig. 2C). Interestingly CMP5 treatment completely abolished the presence of histone methylation markers in only a fraction of glioma cells. Since blocking PRMT5 function inhibits H4R3 and H3R8 methylation but does not demethylate histones that have already been methylated by PRMT5 prior to its inhibition, this likely reflects the long half-life of H4R3 methylation in different cells. Methylated histones have been previously described to have a long half-life.16,17 Thus while reduced methylation indicates PRMT5 inhibition, complete loss of this marker is not observed in all the cells. It is important to note that 100% of cells are positive for H4R3 methylation prior to treatment and that there is complete absence of H4R3 in a third of the cells after a single treatment with CMP5. These results indicate that CMP5 effectively blocks PRMT5 activity.
Fig. 2.
CMP5 mediated inhibition of PRMT5 catalytic activity in mice and tumor cell proliferation. (A) The brain tissue (0–6 h) and plasma (0–8 h) concentration-time profile of CMP5 for 3 formulations. (B) Western blot analysis of GBMNS-30 and GBMNS-X12 treated with DMSO (Ctrl) or CMP5 (25 µM) for 72 hours. Posttreatment cells were lysed and probed for methylated histone H4R3 and H3R8. Glyceraldehyde 3-phosphate dehydrogenase was used as the internal control. (C) Immunocytofluorescent imaging for H4R3 in GBMNS-30 cells after treatment with CMP5 for 72 hours (bar: 15 µm). (D) Relative growth of GBM30 cells grown as neurospheres (GBMNS) or differentiated cells (GBMDC) after treatment with increasing doses of CMP5 over time. A linear mixed model was used to account for the covariance structure due to repeat measures at different timepoints.
We have previously shown that PRMT5 knockdown differentially impacts GBMNS and differentiated GBM cells. To evaluate the effect of CMP5 on the proliferation of undifferentiated GBMNS and their differentiated counterpart (GBMDC) grown in serum (Supplementary Figure S1), GBMNS and GBMDC were treated with the indicated doses of CMP5 (Fig. 2D). CMP5 treatment decreased the proliferation of both GBMNS and GBMDC in a dose-dependent manner. Interestingly GBMDC were significantly more sensitive to CMP5 compared with the undifferentiated primary GBMNS.
CMP5 Decreases the Self-Renewal Capacity of GBMNS and Increases Apoptosis in GBMDC
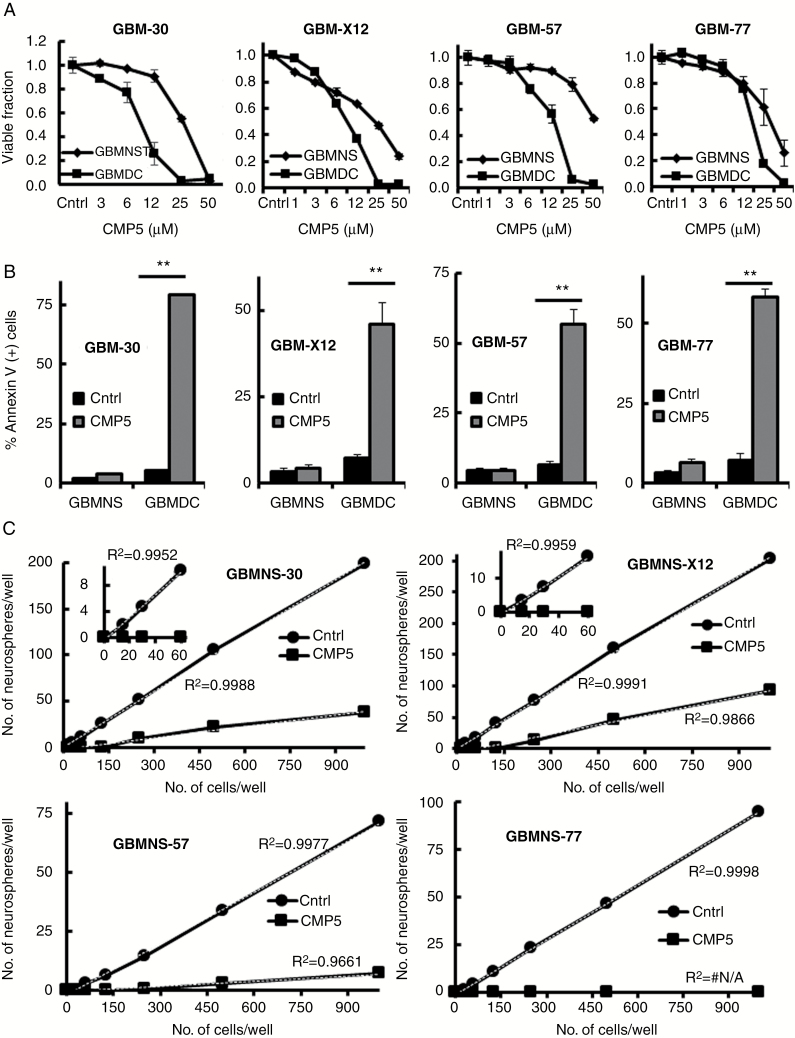
To assess the effect of CMP5 on the viability of GBMNS and GBMDC, we treated 4 different primary patient-derived GBM cells (GBMNS-30, GBMNS-X12, GBMNS-57, and GBMNS-77) grown as neurospheres or differentiated GBM cells (GBMDC-30, GBMDC-X12, GBMDC-57, and GBMDC-77) with different doses of CMP5 and followed their viability by MTT assay (Fig. 3A). Differentiation status of GBMDC was validated by quantitative real-time PCR analysis of genes indicative of differentiation (Supplementary Figure S1). While CMP5 treatment decreased the viability of both GBMNS and GBMDC in a dose-dependent manner, GBMDC were more susceptible to CMP5 treatment.
Fig. 3.
CMP5 causes apoptosis in GBMDC and decreases self-renewal capacity in GBMNS. (A) Viability of the indicated primary GBM cells grown as neurospheres or differentiated cells treated with increasing doses of CMP5 for 72 hours was measured by MTT assay. (B) GBMNS and GBMDC treated with Ctrl (DMSO) or CMP5 (25 µM) for 72 hours were analyzed for percentage of cells undergoing apoptosis using annexin V and PI staining. Data shown are percent of annexin-positive cells (±SD) for the indicated GBM cells (**P ≤ 0.001). (C) Increasing number of indicated GBMNS ranging from 10 to 1000 cells per well were seeded and treated with CMP5 (25 µM) or Ctrl (DMSO). Seven days posttreatment, number of neurospheres formed in each well were counted and plotted against the number of cells seeded. The thin black line represents the linear trend. All the experiments were conducted in biological triplicates.
We further evaluated the effect of CMP5 on the apoptosis of GBMNS and GBMDC. GBMNS and differentiated GBMDC were treated with CMP5 for 72 h and percent of apoptotic cells was evaluated by annexin PI staining (Fig. 3B, Supplementary Figure S4A). Consistent with PRMT5 knockdown (Supplementary Figure S4B), we observed a significant increase in apoptotic cells in GBMDC with CMP5 treatment (Fig. 3B).11 Interestingly while GBMNS were resistant to CMP5-mediated apoptosis, there was significant reduction in their self-renewal as assessed by the frequency of neurosphere formation in GBMNS-30, GBMNS-X12, GBMNS-57, and GBMNS-77, analyzed by linear regression (Fig. 3C and Supplementary Figure S5). CMP5 treatment resulted in reduction of both number as well as size of the neurospheres, suggesting the effectiveness of CMP5 in countering the self-renewal capacity of GBMNS. Overall these results show that CMP5 treatment decreases the self-renewal capacity of GBMNS without affecting viability, and results in apoptotic death of differentiated GBM cells.
CMP5 Blocks Cell Cycle Progression in GBMNS
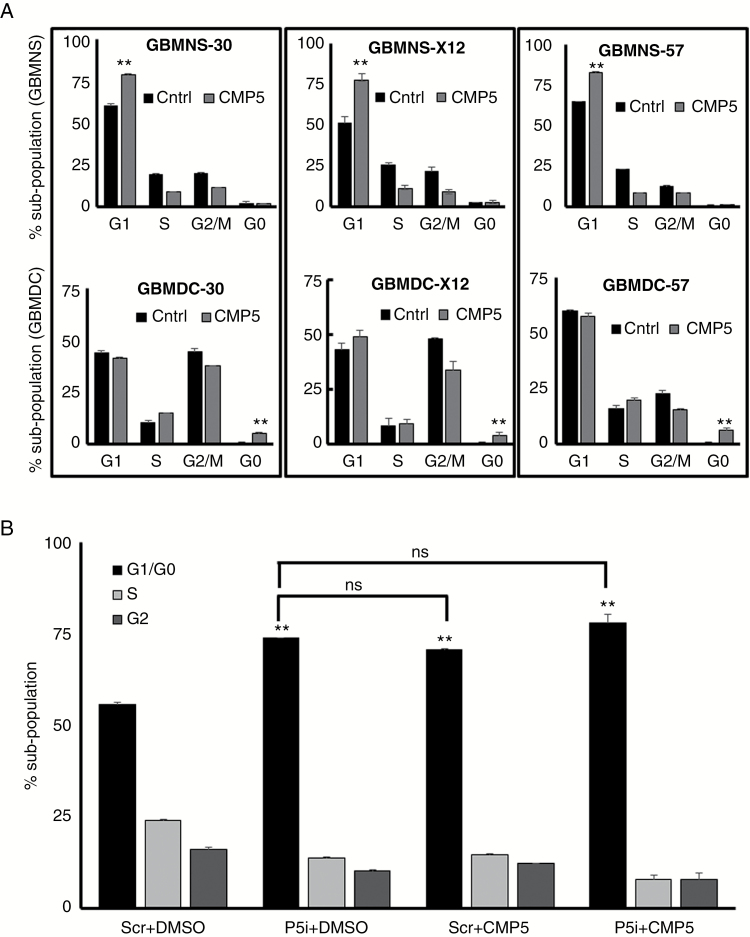
Next we tested the impact of CMP5 treatment on cell cycle progression (Fig. 4A, B). Briefly GBMNS and GBMDC were treated with CMP5 (25 µM) for 24 hours and then cell cycle analysis was done using PI staining to evaluate the effect of CMP5 on the progression of these cells (Fig. 4A). CMP5 treatment increased the G1 subpopulation by at least 20%, 25%, and 15% in GBMNS-30, GBMNS-X12, and GBMNS-57, respectively (upper panel), whereas CMP5 treatment of GBMDC did not affect cell cycle progression but revealed an increase in G0 subpopulation indicative of apoptosis. These results suggest that the decreased proliferation and self-renewal of GBMNS is, in part, a result of G1 cell cycle arrest.
Fig. 4.
CMP5 causes G1 cell cycle arrest in GBMNS. (A) Cell cycle analysis utilizing PI staining of indicated GBMNS and GBMDC treated with DMSO or CMP5 (25 µM) for 24 h posttreatment. Graph represents the percent of cell population in each stage of cell cycle (**P ≤ 0.001). (B) GBMNS-30 transfected with scrambled small interfering (si)RNA (Scr) or PRMT5 siRNA (P5i) were treated with DMSO or CMP5 (25 µM); 72 h posttreatment, cells were subjected to cell cycle analysis and the population of cells in each phase of the cell cycle were quantified. **Statistical significance of P5i + DMSO, Scr + CMP5, and P5i + CMP5 in comparison to control (Scr + DMSO) (**P ≤ 0.001). All the experiments were conducted in 3 biological triplicates.
To reconfirm the specificity of CMP5 to target PRMT5, we treated PRMT5-depleted (P5i) GBMNS-30 with CMP5 and conducted cell cycle analysis (Fig. 4B). While both PRMT5 depletion and CMP5 treatment led to G1 cell cycle arrest of GBMNS, treatment of PRMT5-depleted GBMNS with CMP5 did not further increase G1 cell cycle arrest, indicating that CMP5 treatment–induced G1 cell cycle arrest depended on PRMT5 expression.
Negative Regulation of PTEN by PRMT5 Is Disrupted by CMP5
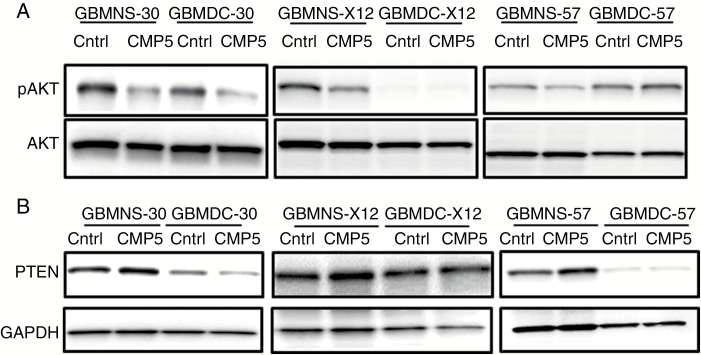
To test if CMP5 affected the PRMT5-phosphatase and tensin homolog (PTEN)-Akt signaling, we treated GBMNS (GBMNS-30, GBMNS-X12, and GBMNS-57) and GBMDC (GBMDC-30, GBMDC-X12, and GBMDC-57) with CMP5 and probed for the expression of both phosphorylated Akt (pAkt) and PTEN expression (Fig. 5). CMP5 treatment decreased the phosphorylation of Akt in GBMNS (Fig. 5A). More importantly, CMP5 treatment resulted in increased expression of PTEN in GBMNS but not in GBMDC (Fig. 5B), indicative of transcriptional repression of PTEN by PRMT5.11 These results confirm that CMP5 derails the negative regulation of PTEN by PRMT5, which in turn decreases the Akt activity in GBMNS.
Fig. 5.
CMP5 affects PTEN-Akt signaling. (A) Western blots of pAkt and total Akt on GBMNS-30, GBMDC-30, GBMNS-X12, GBMDC-X12, GBMNS-57, and GBMDC-57 treated with DMSO or CMP5 (25 µM) for 72 h. (B) Indicated GBMNS and GBMDC were treated with CMP5 (25 µM) for 72 h, lysed, and probed for PTEN expression by western blot. Glyceraldehyde 3-phosphate dehydrogenase served as internal control.
CMP5 Drives GBMNS Toward Senescence
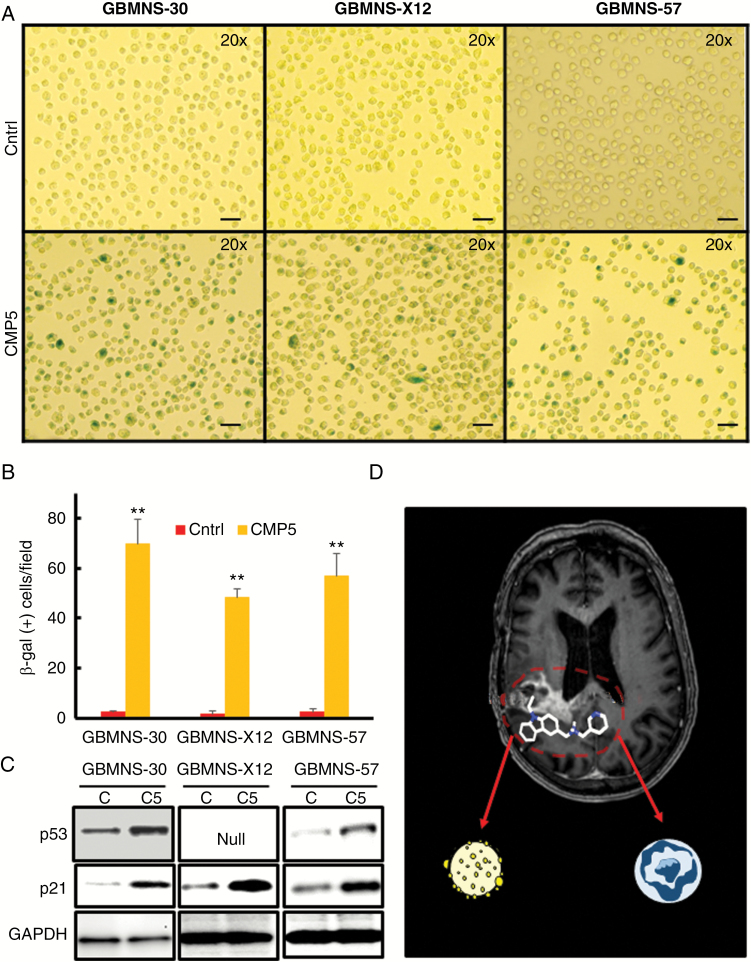
The above results show that CMP5 treatment decreased the proliferation and self-renewal of GBMNS (Figures 2, 3). Since CMP5-treated GBMNS did not undergo apoptotic cell death, we investigated whether CMP5 treatment led to the induction of senescence in these cells after treatment (Fig. 6A, B). Briefly GBMNS (GBMNS-30, GBMNS-X12, and GBMNS-57) were treated with CMP5 (25 µM) for 20 days. Cells were trypsinized every 3 days and fresh media with DMSO or CMP5 were added. Twenty days posttreatment, GBMNS were made into single cells and probed for senescent cells by β-gal staining assay. Bright field images (20x) of GBMNS treated with CMP5 revealed increased cell size along with a significant increase in the number of β-gal positive cells per microscopic view filed (Fig. 6A, B). Irrespective of the signaling pathway involved in inducing senescence, downstream candidates most often involved are p53 and p21.18 Hence, to further confirm the senescent phenotype, GBMNS were treated with CMP5 for 20 days. Posttreatment, we probed these cells for the expression of p53 and p21 protein (Fig. 6C) and mRNA (Supplementary Figure S6). CMP5 treatment increased the expression of both p53 and p21. Together these results suggest that CMP5-treated GBMNS undergo senescence.
Fig. 6.
CMP5 causes senescence in GBMNS. (A) β-gal assay on GBMNS-30, GBMNS-X12, and GBMNS-57 that were treated with Ctrl (DMSO) or CMP5 (25 µM) every 3 days for 20 days. Neurospheres were dissociated into single cells for treatment. Twenty days posttreatment they were trypsinized to make into single cells. Cells were then stained for β-gal. Representative microscopic images (20x magnification) demonstrating β-gal positive and negative cells (bar: 40 µm). (B) Quantification of Figure 6A showing the number of β-gal positive cells observed per view field with Ctrl or CMP5 treatment. Experiment was conducted in 3 biological replicates (**P ≤ 0.001). (C) Western blot analysis of the indicated GBMNS treated with DMSO (C) or CMP5 (C5) (25 µM) every 3 days for 20 days. Posttreatment, cells were lysed and probed for the expression of p53 and p21 by western blot. Glyceraldehyde 3-phosphate dehydrogenase is used as an internal control. (D) Schematic representation of the effect of CMP5 on the differentiated and undifferentiated stemlike cells of GBM tumor.
Discussion
Tumor heterogeneity is increasingly recognized as a major hurdle toward the development of anti-GBM therapeutic approaches.19 Therapies targeted to specific driver mutations and signaling events have developed epigenetic shifts in GBM tumor cells, and this is thought to be one of the major contributors for development of resistance to targeted agents.20–23 Recent studies have uncovered the presence of both (i) a small undifferentiated population of cells within GBM that retain plasticity and can adapt to therapy-induced stress, as well as (ii) a large mature differentiated population of tumor cells.19,23 Thus therapies that selectively target only one of these populations result in limited efficacy and eventual disease progression.
Recently we and others have shown that PRMT5 expression is higher in high-grade gliomas and negatively correlates with GBM patient survival.9,10 We utilized patient-derived primary GBM cells (grown in stem cell culture) to uncover the differential role played by PRMT5 in the survival, proliferation, and self-renewal of primary GBMNS, and their differentiated counterpart.11 These studies showed that PRMT5 plays a significant role in both survival and self-renewal of mature and undifferentiated GBM tumor cells, respectively.
Here we tested the potential of PRMT5 as a druggable candidate for GBM therapy. Crystal structure of a highly homologous rat PRMT1 enzyme was utilized to design in silico compounds with the potential to block PRMT5 catalytic site and hence function.12 Treatment of GBM cells with these agents revealed dose-dependent decrease in their proliferation. Of these agents, CMP5 retained specificity for PRMT5 inhibition, and in vivo treatment of zebrafish bearing intracranial tumors revealed that it could effectively curb GBM tumor growth and increase survival.
Conventional therapy for GBM consists mainly of cytotoxic treatment in the form of chemotherapy and radiation. But with time GBM tumor cells develop resistance to the treatment conditions by various mechanisms.24–28 An emerging strategy to overcome this therapy resistance is with cytostatic agents that disable the proliferation and growth of the tumor cells and synergize in combination with cytotoxic drugs.29–31 Cellular senescence is a form of persistent cytostatic response by the cells to the stress, wherein cells undergoing senescence cease to proliferate but can remain metabolically active and viable.32 Therapy-induced senescence is considered an important route to achieving therapeutic sensitivity for tumor cells that are resistant to apoptosis.33,34 More interestingly, senescent cells have also been shown to attract and interact with innate immune cells such as natural killer cells, neutrophils, dendritic cells, and macrophages that augment the clearance of senescent cells from the system.35,36 Further, the identification of molecular targets such as PTEN, p53, S-phase kinase associated protein 2, Aurora kinase A, etc, which are implicated in regulating senescence in cancer,37–43 has led to the development of numerous senescence-inducing small-molecule therapeutic agents. Many of these are currently being tested in GBM and other cancer patients for safety and efficacy (NCT02122770, NCT01164033).44–46
Clinical trials of MLN4924 and RO5045337 drugs in solid and hematological malignancies, respectively,47,48 favor therapeutic senescence. On the contrary, a more recent study by Ouchi et al reported that the mature GBM cells undergoing senescence can be a mechanism by which immature cells can avoid cell death and promote malignant progression of the GBM tumor,49 implying senescence as a way for tumor cells to escape cytotoxic death.
Even though drugs like MLN492447 and Nutlin3a50 have the ability to induce senescence or apoptosis based on cell line type and p53 status, respectively, to the best of our knowledge, as summarized in Fig. 6D, this is the first report of an agent (CMP5) that can induce apoptosis of differentiated cells as well as result in induction of senescence in immature primary tumor cells (Fig. 6D). Thus CMP5 can be considered a class of agents in itself that can differentially regulate differentiated and primary undifferentiated tumor cell populations.
Future studies are focused on the drug development of CMP5 to improve its pharmacodynamic and pharmacokinetic properties to use as a therapy for GBM.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by the National Institutes of Health (CA150153 to B.K., CA163205 to B.K., NS045758 to B.K., NS064607 to B.K., NS071346 to R.B., and P30NS045758 to C.B.).
Conflict of interest statement. No conflict of interest.
References
- 1. Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 3. Masui K, Cloughesy TF, Mischel PS. Review: molecular pathology in adult high-grade gliomas: from molecular diagnostics to target therapies. Neuropathol Appl Neurobiol. 2012;38(3):271–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wojton J, Meisen WH, Kaur B. How to train glioma cells to die: molecular challenges in cell death. J Neurooncol. 2016;126(3):377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37(15):4965–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilczek C, Chitta R, Woo E, et al. Protein arginine methyltransferase Prmt5-Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasmin in Xenopus laevis eggs. J Biol Chem. 2011;286(49):42221–42231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan F, Alinari L, Lustberg ME, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74(6):1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han X, Li R, Zhang W, et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J Neurooncol. 2014;118(1):61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banasavadi-Siddegowda YK, Russell L, Frair E, et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene. 2017;36(2):263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alinari L, Mahasenan KV, Yan F, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Otsuki A, Patel A, Kasai K, et al. Histone deacetylase inhibitors augment antitumor efficacy of herpes-based oncolytic viruses. Mol Ther. 2008;16(9):1546–1555. [DOI] [PubMed] [Google Scholar]
- 14. White RM, Sessa A, Burke C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2(2):183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welker AM, Jaros BD, Puduvalli VK, Imitola J, Kaur B, Beattie CE. Standardized orthotopic xenografts in zebrafish reveal glioma cell-line-specific characteristics and tumor cell heterogeneity. Dis Model Mech. 2016;9(2):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148(2):558–567. [DOI] [PubMed] [Google Scholar]
- 17. Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static?Cell. 2002;109(7):801–806. [DOI] [PubMed] [Google Scholar]
- 18. Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196(1):33–39. [DOI] [PubMed] [Google Scholar]
- 19. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karberg S. Switching on epigenetic therapy. Cell. 2009;139(6):1029–1031. [DOI] [PubMed] [Google Scholar]
- 21. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 23. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 24. Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. [DOI] [PubMed] [Google Scholar]
- 25. Brown JM. Tumor microenvironment and the response to anticancer therapy. Cancer Biol Ther. 2002;1(5):453–458. [DOI] [PubMed] [Google Scholar]
- 26. Weller M. Novel diagnostic and therapeutic approaches to malignant glioma. Swiss Med Wkly. 2011;141:w13210. [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Li W, Yang Y, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. [DOI] [PubMed] [Google Scholar]
- 28. Sul J, Fine HA. Malignant gliomas: new translational therapies. Mt Sinai J Med. 2010;77(6):655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Desai AA, Stadler WM. Novel kinase inhibitors in renal cell carcinoma: progressive development of static agents. Curr Urol Rep. 2006;7(1):16–22. [DOI] [PubMed] [Google Scholar]
- 30. Martin L, Schilder RJ. Novel non-cytotoxic therapy in ovarian cancer: current status and future prospects. J Natl Compr Canc Netw. 2006;4(9):955–966. [DOI] [PubMed] [Google Scholar]
- 31. Winquist E, Waldron T, Berry S, Ernst DS, Hotte S, Lukka H. Non-hormonal systemic therapy in men with hormone-refractory prostate cancer and metastases: a systematic review from the Cancer Care Ontario Program in Evidence-based Care’s Genitourinary Cancer Disease Site Group. BMC Cancer. 2006;6:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. [DOI] [PubMed] [Google Scholar]
- 33. Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63(11):2705–2715. [PubMed] [Google Scholar]
- 34. Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104(32):13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sagiv A, Krizhanovsky V. Immunosurveillance of senescent cells: the bright side of the senescence program. Biogerontology. 2013;14(6):617–628. [DOI] [PubMed] [Google Scholar]
- 36. Biran A, Perelmutter M, Gal H, et al. Senescent cells communicate via intercellular protein transfer. Genes Dev. 2015;29(8):791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin HK, Chen Z, Wang G, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464(7287):374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alimonti A, Nardella C, Chen Z, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120(3):681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci U S A. 1997;94(18):9648–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu HJ, Zhou Y, Ji W, et al. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene. 1997;15(21):2589–2596. [DOI] [PubMed] [Google Scholar]
- 41. Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18(18):2789–2797. [DOI] [PubMed] [Google Scholar]
- 42. Lee S, Jeong SY, Lim WC, et al. Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J Biol Chem. 2007;282(31):22977–22983. [DOI] [PubMed] [Google Scholar]
- 43. Kuilman T, Michaloglou C, Vredeveld LC, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. [DOI] [PubMed] [Google Scholar]
- 44. Roberson RS, Kussick SJ, Vallieres E, Chen SY, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res. 2005;65(7):2795–2803. [DOI] [PubMed] [Google Scholar]
- 45. te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62(6):1876–1883. [PubMed] [Google Scholar]
- 46. Coppé JP, Patil CK, Rodier F, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hua W, Li C, Yang Z, et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 2015;17(10):1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yuan Y, Liao YM, Hsueh CT, Mirshahidi HR. Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. J Hematol Oncol. 2011;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ouchi R, Okabe S, Migita T, Nakano I, Seimiya H. Senescence from glioma stem cell differentiation promotes tumor growth. Biochem Biophys Res Commun. 2016;470(2):275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Villalonga-Planells R, Coll-Mulet L, Martínez-Soler F, et al. Activation of p53 by nutlin-3a induces apoptosis and cellular senescence in human glioblastoma multiforme. PLoS One. 2011;6(4):e18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.