Abstract
Base excision repair (BER), which is initiated by DNA N-glycosylase proteins, is the frontline for repairing potentially mutagenic DNA base damage. The NTHL1 glycosylase, which excises DNA base damage caused by reactive oxygen species, is thought to be a tumor suppressor. However, in addition to NTHL1 loss-of-function mutations, our analysis of cancer genomic datasets reveals that NTHL1 frequently undergoes amplification or upregulation in some cancers. Whether NTHL1 overexpression could contribute to cancer phenotypes has not yet been explored. To address the functional consequences of NTHL1 overexpression, we employed transient overexpression. Both NTHL1 and a catalytically-dead NTHL1 (CATmut) induce DNA damage and genomic instability in non-transformed human bronchial epithelial cells (HBEC) when overexpressed. Strikingly, overexpression of either NTHL1 or CATmut causes replication stress signaling and a decrease in homologous recombination (HR). HBEC cells that overexpress NTHL1 or CATmut acquire the ability to grow in soft agar and exhibit loss of contact inhibition, suggesting that a mechanism independent of NTHL1 catalytic activity contributes to acquisition of cancer-related cellular phenotypes. We provide evidence that NTHL1 interacts with the multifunctional DNA repair protein XPG suggesting that interference with HR is a possible mechanism that contributes to acquisition of early cellular hallmarks of cancer.
INTRODUCTION
DNA damage can be caused by reactive oxygen species (ROS) produced by both endogenous and exogenous sources (1–4). Faced with ROS-induced DNA damage, cells have evolved the base excision repair (BER) pathway to maintain genome stability. BER is initiated by DNA N-glycosylase proteins, which recognize specific subsets of base damage (1,2). In this initial step, cleavage of the damaged base from the phosphodiester backbone forms an apurinic/apyrimidinic (AP) site (1,5). The bifunctional N-glycosylases also possess associated AP lyase activity and cleave the DNA phosphate backbone, yielding single-strand breaks with ends that require further processing by either apurinic/apyrimidinic endonuclease 1 (APE1) or polynucleotide kinase phosphatase (PNK) (1,5). As BER intermediates are themselves types of DNA damage (6), modulation of BER is crucial to ensure rapid and efficient repair of base damage, while simultaneously ensuring that potentially deleterious BER intermediates do not accumulate.
Cells must achieve a balance between timely initiation of repair and inadvertent DNA damage through tight control of BER glycosylase activity. While a number of studies have assessed loss of function mutants in DNA repair components, few have examined increases in glycosylase protein expression. The best-characterized example of modulating glycosylase expression is analysis of overexpression of the alkylation repair glycosylase, AAG. In budding yeast, overexpression of AAG causes a mutator phenotype as a result of the accumulation of deleterious BER intermediates (6–8). Another defined example is SMUG1 glycosylase where overexpression correlates with poor disease-free survival in gastric cancer patients, although the exact mechanism is currently undetermined (9). Thus, there is evidence to suggest that overexpression of BER proteins could be deleterious.
The bifunctional DNA glycosylase, NTHL1, removes a large subset of oxidative DNA base damage in mammalian cells (10). NTHL1 recognizes oxidative DNA damage and initiates the BER repair pathway (11). Consistent with a critical role for NTHL1 in repairing oxidative damage and protecting the integrity of the genome, a double knockout mouse for the NTHL1 and the NEIL1 N-glycosylases shows spontaneous tumor formation in the lung and liver (12). More recently, studies linked biallelic inactivation of NTHL1 to a novel human colon cancer predisposition syndrome (13,14), presumably through impaired DNA repair. These observations support a tumor suppressive function for NTHL1 consistent with a key role in repair of oxidative base damage. These previous studies have focused primarily on loss of function mutations with NTHL1. Here, we analyze the consequences of increased levels of NTHL1.
An increase the level of cellular NTHL1 could be a protective response that occurs in cancer in an attempt to manage an increased DNA damage burden. Alternatively, an increase in NTHL1 protein levels could directly contribute to tumorigenesis, for example by unbalancing the BER pathway through excessive incision at damaged bases, resulting in accumulation of toxic repair intermediates. Studies have not been performed to assess whether an increase in the level of NTHL1 occurs as a result of tumorigenesis or could contribute to the cause of transformation. To investigate the functional consequences of NTHL1 overexpression, we performed transient overexpression of NTHL1 in the immortalized but non-transformed lung cell line HBEC-3KT. Lung bronchial epithelial cells are a physiologically relevant model system, as lung tissue is repeatedly exposed to various agents that cause oxidative DNA damage (15). Surprisingly, we show that NTHL1 overexpression causes genomic instability and early markers of cellular transformation independent of its catalytic activity.
MATERIALS AND METHODS
Cell lines, cell culture and reagents
All reagents unless otherwise stated, were obtained from Invitrogen (Carlsbad, CA, USA). The human bronchial epithelial cells (HBEC), clone 3T, (16) were a gift from Michael Story (UT Southwestern) and were cultured as previously described (17). Karyotyping of HBEC cells in 2014 verifies the previously published HBEC karyotype (16) and validates cell line identity. The U2OS (osteosarcoma) and A549 (NSCLC adenocarcinoma) cell lines were cultured in DMEM containing 10% fetal bovine serum (FBS). Cell lines derived from HBEC soft agar clones were grown in Keratinocyte Serum-Free Media (SFM) supplemented with bovine pituitary extract and EGF. RPMI-1640 containing 5% FBS in a 1:1 ratio, once isolated from soft agar. Beas2B cells were cultured in MEM containing 5% FBS. H460 and H1299 cell lines were grown in RPMI containing 10% FBS, and were provided by Dr Wei Zhou (Emory University). The Calu-1 cell line was maintained in McCoy's 5a Modified Media and 10% FBS. H1975, H522, H226, H1792 and HCC827 cell lines were maintained in RPMI-1640 media supplemented with 10% FBS, and were obtained from the Winship Cancer Institute (Atlanta, GA, USA). HeLa cells were maintained in DMEM supplemented with 10% FBS. HT29 and HCT116 cells were maintained in McCoy's 5a Modified Media and 10% FBS. Caco-2 and SW480 cells were maintained in DMEM supplemented with 10% FBS, HEPES, l-glutamine and non-essential amino acids. Caco-2 and SW480 cells were a gift from Dr Asma Nusrat (University of Michigan). A description of cell line characteristics and mutational status for lung cell lines (Supplementary Table S1) and colon cancer cell lines (Supplementary Table S2) is provided. All cell lines were supplemented with penicillin and streptomycin and maintained at 37°C in 5% CO2. Cells were routinely screened for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Walkersville, MD, USA).
DR-U2OS cells (18) were provided by Dr M. Jasin (Memorial Sloan-Kettering Cancer Center, USA). Cells were cultured under ambient oxygen levels and 10% CO2 in DMEM supplemented with 10% fetal calf serum and 1% antibiotic/antimycotic.
HEK 293FT cells (Life Technologies Corp.) were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. DsR-7F4 reporter cells were derived from human bladder cancer EJ-30 line as described previously (19,20) and were used for the end-joining assay. The DsR-7F4 cell line contains a transcriptional promoter and a promoterless eGFP gene, separated by an intervening sequence with rare-cutting nuclease I-SceI recognition sites on 5′ and 3′ end. DsR-7F4 cells also contain a promoterless DsRed gene located on a different chromosome with an I-SceI site on the 5′ end. Expression of I-SceI stimulates reciprocal joining of the promoter to the eGFP gene after releasing the intervening fragment, or to the DsRed gene via interchromosomal translocation. Cells were grown in MEM alpha medium (Life Technologies Corp.) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine and 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies Corp.).
cBioPortal data mining
Data was accessed from cBioPortal (www.cBioPortal.org). Non-small cell lung cancer (NSCLC) datasets were searched for the NTHL1 gene amplification and/or mRNA data. Percentages calculated in Figure 1A are the sum of values from each NSCLC dataset. No additional data filtering was performed beyond obtaining numerical values from the oncoprint profile in cBioPortal. Four NSCLC cancer datasets were available in cBioPortal that contained copy number variation data and/or RNA data. The RNA data was set with a Z-score threshold of two. Information about and references for these datasets are provided in Supplementary Table S3.
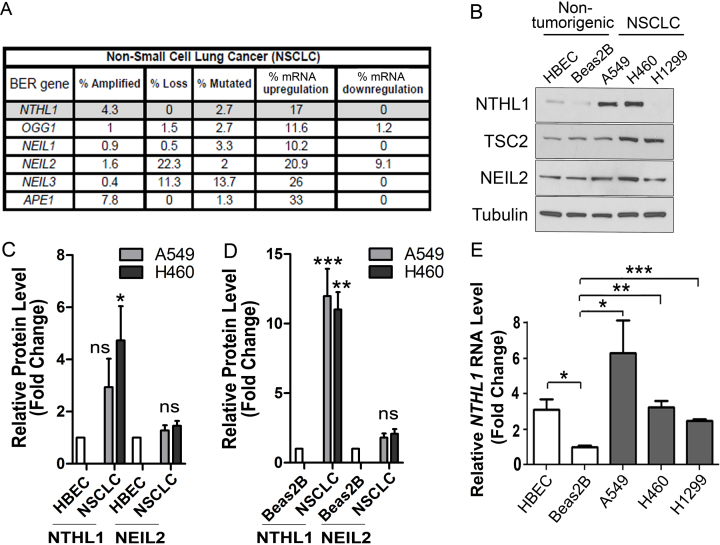
Figure 1.
NTHL1 mRNA and protein levels are increased in non-small cell lung cancer (NSCLC) cell lines. (A) cBioPortal primary tumor data sets across all subtypes of NSCLC demonstrate that multiple base excision repair (BER) genes are amplified in addition to loss or mutation in NSCLC. Messenger RNA (mRNA) dysregulation for each of six BER genes is also observed. (B) Immunoblotting for NTHL1 in a panel of non-transformed, immortalized lung epithelial cell lines (HBEC and Beas2B) and NSCLC cancer cell lines (A549, H460, H1299) reveals variation in NTHL1 protein levels. Levels of another BER glycosylase, NEIL2, do not vary significantly across all cell lines examined. TSC2 protein levels are also comparable in all cell lines examined. (C) Quantification of relative NTHL1 protein levels from A549 and H460 cell lines normalized to the HBEC cell line where the level of NTHL1 was set to 1.0. (D) Quantification of relative NTHL1 protein levels from A549 and H460 cell lines normalized to the Beas2B cell line where the level of NTHL1 was set to 1.0. (E) qRT-PCR analysis of NTHL1 transcript levels reveals that NTHL1 mRNA levels vary between non-transformed and transformed cell lines, but that mRNA levels do not correspond to NTHL1 protein levels. Values were normalized to Beas2B, which was set to 1.0, as this was the lowest value for NTHL1 transcript. NS = not significant; *P≤ 0.05; **P≤ 0.01; ***P≤ 0.001.
Plasmids and site directed mutagenesis
The NTHL1 gene was sub-cloned from the RG214598 plasmid (Origene, Rockville, MD, USA) using the restriction sites SgfI and MluI, and was cloned into the pCMV6-AC-GFP plasmid to create a C-terminally tagged NTHL1-GFP protein used in FACS sorting experiments, micronucleus, and localization studies (Origene). For NTHL1-Flag, NTHL1 was cloned from the pCMV6-AC-NTHL1-GFP plasmid into the pcDNA3.1 (+) vector using the HindIII and BamHI restriction sites (see Supplementary Table S4 for plasmids and primers for Flag tag addition). The pDsRED-Express-N1 plasmid was obtained from Clontech (Mountain View, CA, USA) and used as negative control in the micronucleus experiments.
Site-directed mutagenesis of NTHL1 to create the catalytically dead NTHL1 K220Q mutant was performed on the pcDNA3.1(+) NTHL1-Flag construct (see Supplementary Table S4 for primers), and the Q5 Site-Directed Mutagenesis Kit (New England BioLabs, Ipswich, MA, USA). All plasmids were sequenced to ensure no mutations were inadvertently introduced, and to verify the presence of the NTHL1 K220Q mutant.
Transfection and drug treatments
HBEC cells were plated at a density of 2.3 × 105 cells per well in a six well dish, trypsinized until rounded, then transfected using Fugene HD Transfection Reagent (Promega, Madison, WI, USA) in a 3:1 (Fugene: 1 μg DNA) ratio in OPTIMEM. Cells were incubated for three hours, and transfection media was replaced with fresh HBEC media. U2OS cells were seeded at a density of 1.5 × 105 cells per well of a six well dish and transfected with Lipofectamine 2000 (Invitrogen) for 6 h before fresh media was added. Plasmid concentration was 1 μg per well for all experiments described in six well plate format, and scaled down for 24-well plates based on well area. Replication stress was induced in HBEC and U2OS cells by treatment with 2 mM hydroxurea (Sigma, St. Louis, MO, USA) for 24 h in media. Camptothecin (CPT) (Sigma, St. Louis, MO, USA) treatments were performed in SFM media with 1 μM CPT for 24 h.
RNA isolation and real time PCR
HBEC, Beas2B, A549, H460 and H1299 cell lines were plated the day before at a density of 1.5 × 106 cells per 100 mm dish. Cells were pelleted, resuspended, and divided in half for Immunoblotting and RNA preparation. Trizol RNA isolation was performed as previously described (21). Nucleic acid quantification was carried out using a NanoDrop 2000 system (Thermo Fisher Scientific). Briefly, 1 μg of total RNA isolated from cell pellet was reversed transcribed using M-MLV (Invitrogen) in a reaction volume of 50 μl according to the standard kit protocol. The NTHL1 accession number used for this study is NM_002528.5, and the consensus cDNA sequence is CCDS10457.1 for isoform 1. Primers were designed using the primer-BLAST program from NCBI. All real-time qPCR reactions were performed with 5 ng of cDNA, 0.5 μM of primers (see Supplementary Table S4 for primer sequences), and 10 μl of Quantitect Sybr Green PCR mix (Qiagen) using StepOnePlus system and software (Applied Biosystem). Primers used spanned an exon-exon junction as an internal control to avoid amplification of genomic DNA. PCR cycles were performed as follows: 95°C for 10 min, 95°C for 15 s, 58°C for 30 s, 72°C for 30 s for 45 cycles. Values are reported for three independent experiments containing three technical replicates. No outlier values were discarded. Results were normalized to 18S rRNA subunit. Post real-time analyses were performed using Microsoft Excel and Student's t test.
Immunohistochemistry
Immunohistochemistry (IHC) was performed by the Emory University Pathology Core using a standard IHC protocol on paraffin embedded NSCLC tissue. Novus Biological (Littleton, CO, USA) NTHL1 antibody (NB100–108) was used for IHC staining. NTHL1 antibody was validated, per manufacturer's recommendation, on kidney cancer. Antibody optimization with titrations of 1:100, 1:200 and 1:400 were performed, and an antibody concentration of 1:200 was chosen for NSCLC tissue staining.
Immunoblotting
To prepare cell lysate, cell lines were lysed using RIPA buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), except where indicated and sonicated three times at 3 W and 0.5 amplitude on a Misonix Sonicator 3000 (Newtown, CT, USA). Lysates were spun in a cold room at 13 000 rpm for 15 min to obtain the final cleared lysate. Whole cell lysate protein concentrations were determined using the Bradford assay. The Invitrogen NuPAGE system was used with MOPS or MES buffer based on the molecular weight of the target protein (10% acrylamide gels). The exception was XPG, which was separated using Novex 4–12% Tris–glycine gels (1.0 mm) from Invitrogen. Transfer was performed onto nitrocellulose membranes (0.2 μm pore) in the cold room at 100 V for 70 min, with the exception of overnight transfer at 35 V when blotting for XPG. Blocking was performed in 5% Amersham ECL Prime Blocking Reagent (Pittsburgh, PA, USA) for 1 h at room temperature. Primary antibodies used include NTHL1 1:500 (ab70726, Abcam, Cambridge, MA, USA); NEIL2 1:10,000 (180576, Abcam); Actin 1:1,000 (ab8224, Abcam); RPA 1:750 (Abcam, ab2175, clone 9H8); Tubulin 1:1,000 (F2168, Sigma; Clone DM1A); ATR phosphoThr1989 1:500 (GTX128145, GeneTex, Irvine, CA, USA); ATR 1:500 (sc-1887, Santa Cruz, Santa Cruz, CA USA); kRas 1:1,000 (F234, Santa Cruz); TSC2 1:1,000 (3612, Cell Signaling, Danvers, MA, USA); Flag 1:1000 (2368, Cell Signaling), GFP antibody 1:2000 (#2555, Cell Signaling), and RPA phosphoSer4/Ser8 1:500 (A300–245A, Bethyl Laboratories, Montgomery, TX, USA). XPG was detected using rabbit anti-XPG 97727 antibody R2 at 1:1000 (22). Secondary antibodies used were HRP-conjugated anti-mouse 1:3000 (W4028, Promega) and anti-rabbit 1:3000 (W4018, Promega, Madison, WI, USA), and anti-goat 1:5000 (sc-2020, Santa Cruz). Horseradish peroxidase was detected using the ThermoFisher Scientific pico SuperSignal Chemiluminescent kit (#32106) or (for XPG) the ECL Prime system (Amersham). Immunoblot figures were cropped and made in Photoshop. Blots were quantified using the GE Healthcare ImageQuant program.
To perform semi-quantitative analysis of immunoblots for fold NTHL1 overexpression change, NTHL1 and CATmut lysates were serially diluted three-fold starting at a total protein concentration of 2 μg, for final protein concentrations of 0.5, 1 and 2 μg for NTHL1 and CATmut protein. Empty vector control was serially diluted, starting at 10 μg for final protein concentrations of 2.5, 5 and 10 μg. Ten 10 μg of non-transfected control sample was loaded.
Both DR-U2OS and EJ DsR-7F4 cells, which were also used for the gene conversion assay, and cells used to evaluate replication stress markers were lysed in SDS sample buffer (3% SDS, 10% glycerol, 100 mM Tris–HCl, pH 6.8) and heated at 95°C for 5 min. Protein concentrations were determined by the BCA assay (Pierce). Samples were mixed with SDS sample buffer + 200 mM DTT + 0.5 mM bromophenol blue, prior to resolution on 4–12% Tris–glycine gels (Invitrogen), and were transferred to nitrocellulose membranes and probed with the corresponding antibodies. Antibodies used were mouse anti-HA (clone 16B12, MMS-101P, Covance), mouse anti-tubulin (CP06, Calbiochem), mouse anti-Flag (F3165, Sigma) and sheep anti-mouse (NA931V, GE Healthcare) conjugated to horseradish peroxidase (HRP).
ImageQuant was used to quantitate immunoblots for the replication stress signaling. All protein values were normalized to actin to control for protein loading. The ratio of phosphorylated protein:total protein in each lane was then determined. Values were normalized so that HU control signal was set as 1.0.
Immunoprecipitation
To analyze NTHL1 association with XPG, HBEC cells were grown and transiently transfected with NTHL1-Flag or control vector following manufacturer's protocol (Promega). Forty-eight hours after transfection, cells were subject to either mock or 10 mM H2O2 treatment for 10 min and were washed with PBS. Cells were incubated in complete DMEM medium for 10 min and were collected by trypsinization and lysates were prepared with a lysis buffer containing 50 mM HEPES–KOH (pH 7.5), 250 mM KCl, 0.5% Triton X-100, 2.5 mM MgCl2, 0.5 mM EDTA, 5% glycerol and protease inhibitor cocktail (Roche). The DNA and RNA in the suspensions was digested with 25 U/μl benzonase (Novagene) for 30 min on ice. The suspensions were cleared by centrifugation at 12 000 rpm for 30 min, and the supernatants were collected and used for immunoprecipitation assay with anti-FLAG agarose M2 beads (Sigma). After extensive washing of the M2 beads with the same buffer, NTHL1-Flag protein complexes were eluted in the wash buffer containing 0.4 μg/μl FLAG-peptide (Sigma). The samples were separated by 4–12% acrylamide gel (Novex) and analyzed by immunoblotting including detection with ECL (GE Healthcare) system.
Cytokinesis-block micronucleus assay
HBEC cells were plated at a density of 30 000 cells per well and U2OS cells were plated at a density of 20 000 cells per well in a 24-well plate on glass coverslips. Media in the HBEC wells was supplemented with 5 μl of fetal bovine serum per ml. Cells were treated with 3 μg/ml Cytochalasin B in media and stained as previously described (17). Cells were imaged and scored as described (17) using an Olympus IX81 inverted microscope with SlideBook 5.0 (3i Intelligent Imaging Innovations, Denver, CO, USA).
Immunofluorescence and microscopy
HBEC cells were seeded at a density of 30 000 cells per well on glass coverslips. Cells were transfected as described, and 24 h after transfection, were washed in cytoskeletal (CSK) buffer (0.15% Triton-X 100, 360 mM piperazine diethanesulfonic acid, 25 mM HEPES, 10 mM EGTA, 2 mM magnesium chloride, pH to 6.9) supplemented with protease and phosphatase inhibitors for 5 min on ice, followed by two washes with PBS on ice, in order to retain chromatin bound protein. Cells were fixed in 4% paraformaldehyde. Primary antibodies used were γH2AX 1:500 (Millipore, Billerica, MA, USA) and 53BP1 1:1000 (Novus Biologicals, Littleton, CO, USA). Imaging was performed on a Zeiss LSM510META confocal microscope (Thornwood, NY, USA) using a 20× Plan-Apo objective (NA = 0.75). Three independent experiments were performed, and 50 cells per replicate were counted for quantification.
Comet assay
Comet assays were performed as previously described (23,24). In brief, cells were transfected with each NTHL1 construct and cells were suspended in low melting point agarose 24 h following transfection. Cells were then alkaline lysed, electrophoresed and neutralized, followed by staining with SYBER green. Cells were imaged on an Olympus IX81 inverted microscope with SlideBook 5.0. Fifty cells from each replicate experiment were analyzed using the Comet Score software (TriTek Corporation).
Gene conversion and single-strand annealing assays
DR-U2OS cells were seeded, and transfected 24 h later with Lipofectamine2000 (Invitrogen) and 3 μg total DNA plasmid consisting of I-SceI-encoding plasmid (1.5 μg) and pcDNA3.1 plasmid (1.5 μg) with empty vector control, NTHL1-Flag or NTHL1-Flag cat dead (CATmut). Flow cytometry was carried out 72 h later to measure GFP positive cells. GFP conversion was normalized to GFP (%) in the empty vector control for each biological experiment. Statistical significances were determined using the Student's t test.
End joining assay
Lentivirus production for the DsR-7F4 end joining assay was performed as previously described in 10 cm dishes using Lipofectamine 3000 (Life Technologies) (8,19). For each dish, 13.6 μg of control vector, NTHL1, CATmut or I-SceI expressing vector (Plasmid 31484, Addgene), was co-transfected with 3.4 μg pMD2.G (Plasmid 12259, Addgene) which encodes vesicular stomatitis virus (VSV) G coat protein, and 6.8 μg psPAX2 (Plasmid 12260, Addgene) which encodes other lentiviral packaging factors, into HEK 293FT cells. Forty-eight h post-transfection, lentivirus-containing supernatants were harvested and clarified at 800 × g for 20 min. Lentivirus yield was estimated using a Lenti-X GoStix kit (Clontech Laboratories, Inc). Suspensions containing >5 × 105 IFU/ml were used directly or aliquoted and stored at –80°C.
DsR-7F4 reporter cells were infected with empty vector control, NTHL1 or CATmut expressing lentivirus for 14 h at 37°C in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Medium was then exchanged for regular growth medium, and incubated for an additional 10 h. Reporter cells were then infected with I-SceI-expressing lentivirus for 14 h at 37°C in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Following infection, medium was exchanged for regular growth medium. At 48 h post-I-SceI-infection, growth medium containing 25 μM biliverdin (Toronto Research Chemicals) was exchanged to enhance the infrared fluorescent protein (IFP) signal. At 84 h post-I-SceI-infection, cells were harvested by trypsinization followed by centrifugation at 230 × g for 5 min, and resuspended in 0.5 ml PBS (Life Technologies Corp.). FACS Analysis was performed using a FACS LSR II instrument (Becton Dickinson) in Emory University Pediatric Flow Cytometry Core. Data were analyzed using Flowjo VX software (TreeStar, Inc.).
Soft agar colony formation
HBEC cellular transformation was measured by using soft agar colony formation. Transfection was performed as described except that NTHL1 overexpressing cells were plated 72 h following transfection (25). In brief, low melting temperature agarose containing a 1:1 ratio of HBEC media and RPMI-1640 media was diluted to a final concentration of 0.7% and solidified at 4°C in each well of a 6-well plate. After fluorescence activated cell sorting (FACS) of GFP positive cells or following transient transfection, 55 000 cells per well were immediately plated in a top layer of low melting temperature agarose at a final concentration of 0.37%. Cells were cultured at 37°C with 5% CO2 for 3 weeks. Upon colony appearance, the media was changed from HBEC media to a 1:1 ratio of HBEC and RPMI-1640 media. Colonies were visually counted through an Olympus IX81 inverted microscope. Colonies were isolated from soft agar, trypsinized into single cell suspension, and grown into clonal cell lines for loss of contact inhibition assays and persistent genomic instability.
Loss of contact inhibition
Loss of contact inhibition was measured as described (26). Loss of contact inhibition was visualized both macroscopically and on an Olympus IX81 inverted microscope with QCapture (QImage, Surrey, CAN) using a 10× objective.
FACS sorting
Cells were plated at a density of 1.7 × 106 cells per 100 cm2 dish and transfected with NTHL1-GFP plasmid or GFP alone. Twenty-four hours following transfection, cells were trypsinized, spun down, and resuspended in filter-sterilized 1× phosphate buffered saline containing 5 mM EDTA, 25 mM HEPES pH 7.0, 1% BSA and 10 U/ml of DNAse. Cells were sorted on a BD FACSAria machine using a 100 μM diameter nozzle. Cells were binned into three populations based on NTHL1-GFP intensity deemed as high, intermediate, and low based on the observed distribution of GFP positive cells. NTHL1-GFP positive cells from each population were immediately diluted and plated in fresh Keratinocyte serum-free media (SFM) and allowed to recover before each assay or the addition of cytochalasin B.
Cell cycle sorting
Plasmids encoding GFP, NTHL1 or CATmut were transfected into HBEC3kt cells with Fugene. Complete media was restored 6 h after transfection and cells were collected 24 h later. Cells in suspension were fixed for 10 min with 2% paraformaldehyde (PFA) on ice; PFA was quenched with glycine followed by 70% ethanol incubation for 2 h. Cells were washed with PBS and treated with 100 μg RNAse A for 10 min at 37°C. Propidium iodide (PI) was added to a final concentration of 50 μg/ml in PBS with 0.1% triton X100 and incubated for 30 min at room temperature before acquisition in a LSRII Flow Cytometer (BD). Green fluorescent positive cells were gated and PI signal was collected in single cell populations. To calculate the fraction of cells in each phase of the cell cycle, the Watson Pragmatic univariate model was applied following compensation of the samples in FlowJo. Constraints on the means were applied setting a range for G1 and G1–n for G2.
Statistical analysis and data representation
Error bars in figures represent the standard error from the average of at least three independent experiments. Each measurement was performed in duplicate. Two-way ANOVA was performed in Graph Pad for the analysis of HBEC micronucleus formation at 48 and 96 h, with a = 0.05. One-way ANOVA (a = 0.05) was calculated in VasserStats for all other experiments, unless otherwise indicated. Micronuclei frequency distribution and DSB frequency distribution was analyzed using the chi-squared test. P values are as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
RESULTS
NTHL1 protein levels are elevated in non-small cell lung cancer (NSCLC) cell lines
Recent genomic analysis of multiple families links homozygous loss-of-function germline mutations in NTHL1 to a novel colon cancer predisposition syndrome (13,14). Mining of cBioPortal cancer genomic datasets (www.cBioprtal.org) surprisingly revealed that NTHL1 is more frequently amplified than deleted in multiple cancer types and NTHL1 mRNA is upregulated in many instances. Both NTHL1 amplification and mRNA upregulation are found in a variety of cancers including lung, breast, prostate, and pancreas. Either amplification or mRNA upregulation could result in increased NTHL1 protein levels. Examples of gene amplification or mRNA upregulation are known to occur in many cancers (27), prompting us to investigate the potential cellular consequences of NTHL1 overexpression.
Non-small cell lung cancer (NSCLC) shows NTHL1 gene amplification or mRNA upregulation, whereas no loss of NTHL1 is reported (Figure 1A). Collectively, NTHL1 amplification and mRNA upregulation occurs in ∼21% of NSCLC cases (Figure 1A). Expansion of cBioPortal analysis to other BER genes reveals a pattern where mRNA is frequently upregulated in NSCLC (Figure 1A). Therefore, a subpopulation of NSCLC tumors may have increased NTHL1 protein.
To determine whether NTHL1 amplification or mRNA upregulation corresponds to an increase in protein levels, we compared the level of NTHL1 protein in a panel of transformed and non-transformed lung epithelial cell lines. Steady-state NTHL1 protein levels are low in non-tumorigenic HBEC and Beas2B cells, but significantly higher in several NSCLC cell lines examined (Figure 1B, Supplementary Figure S1A and B). While NTHL1 protein levels were not elevated in every instance, the majority of cell lines analyzed display elevated NTHL1 protein (Figure 1B, Supplementary Figure S1A and B). In contrast, protein levels for another BER glycosylase, NEIL2, or protein levels for a gene located adjacent to NTHL1, TSC2, show relatively consistent expression across the cell lines examined (Figure 1B and Supplementary Figure S1C). Quantification of immunoblots reveals that the increase in NTHL1 protein levels in NSCLC cells lines (A549 and H460) is significant when compared to either HBEC or Beas2B cell lines (Figure 1C and D). To determine whether NTHL1 transcript levels correlate with protein levels in these cell lines, qRT-PCR was performed. This analysis reveals that NTHL1 mRNA levels do not necessarily correspond to NTHL1 protein levels in the cell lines examined (Figure 1E) suggesting a complex regulation of NTHL1 at multiple levels. Nonetheless, these results demonstrate that, in general, steady-state NTHL1 protein is increased in NSCLC cell lines compared to control cells.
NTHL1 is primarily localized to the nucleus in both NSCLC and overexpressing HBEC cells
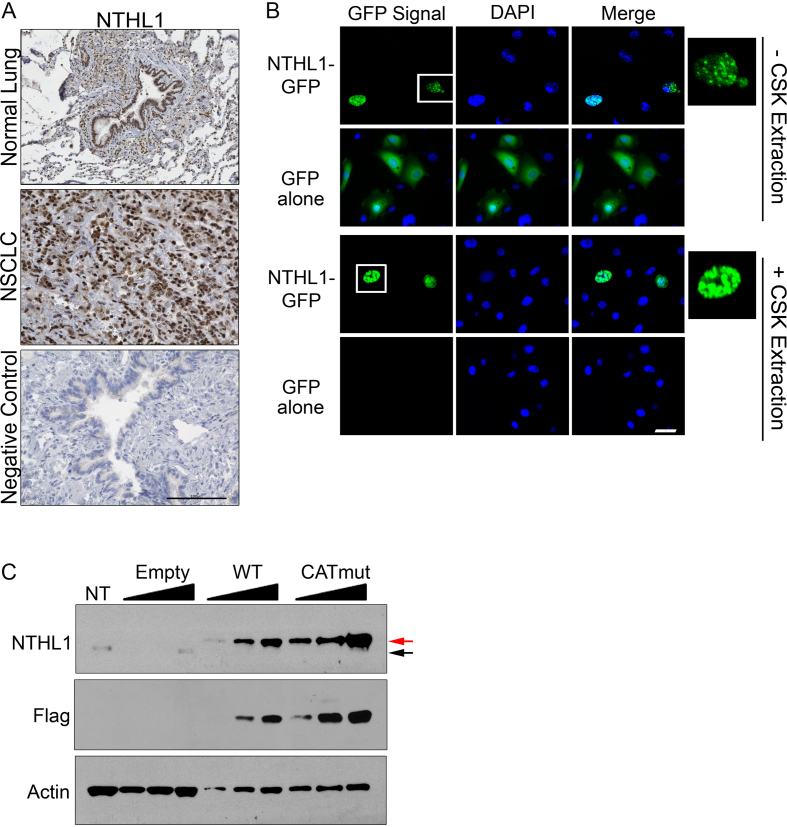
To assess the localization of NTHL1, we performed immunohistochemistry in matched normal tissue and primary NSCLC tumor tissue (Figure 2A). NTHL1 is predominately localized to the nucleus in both normal lung and NSCLC primary tissue. To determine whether overexpressed NTHL1 is also localized to the nucleus, HBEC cells were transfected with a plasmid that transiently overexpresses NTHL1-GFP. As shown in Figure 2B, NTHL1-GFP is localized predominately to the nucleus in these cells. Furthermore, NTHL1 is chromatin-bound, as NTHL1-GFP signal survives pre-extraction of the soluble fraction with cytoskeletal (CSK) buffer (Figure 2B). GFP protein alone is not targeted to the nucleus and is soluble, as GFP is removed by CSK pre-extraction (Figure 2B). Thus, transiently overexpressed NTHL1-GFP localizes to the nucleus and associates with chromatin.
Figure 2.
NTHL1 is localized to the nucleus in both NSCLC tissue and in HBEC cells. (A) Immunohistochemistry of NTHL1 protein (brown) in normal lung and NSCLC tissue. The negative control was performed in normal lung tissue and represents a normal serum control from mice in the absence of primary NTHL1 antibody. The black scale bar corresponds to 100 μm. (B) HBEC cells expressing NTHL1-GFP reveal that NTHL1 is localized to the nucleus and is chromatin-associated as demonstrated by persistence after pre-extraction with cytoskeletal (CSK) buffer. GFP alone is not targeted to the nucleus or chromatin-associated as demonstrated by CSK pre-extraction. Cells inside the white box have been enlarged and inset on the right. The white bar corresponds to 100 μm. (C) Immunoblot of transiently expressed NTHL1-Flag in HBEC cells. The same protein lysate was serially diluted by half to include final concentrations of 2, 1 and 0.5 μg of total protein for wildtype NTHL1 and CATmut samples. Total protein concentration loaded for assessing endogenous NTHL1 in HBEC cells (non-transfected; NT) was 10 μg. The empty vector control (Empty) was diluted by half to final protein concentrations of 10, 5 and 2.5 μg. The lower limit of detection of endogenous NTHL1 from HBEC cells is 10 μg of total protein lysate. The black arrow indicates endogenous NTHL1, while the red arrow denotes exogenously expressed NTHL1-Flag.
To assess the relative fold overexpression of wildtype NTHL1 (NTHL1) compared to endogenous NTHL1, NTHL1 was Flag-tagged. We also created a Flag-tagged, catalytically dead NTHL1 (CATmut) by replacing lysine 220 with glutamine, which takes advantage of a previously characterized catalytic mutant (28). Because attempts to generate HBEC cell lines stably overexpressing the NTHL1 or CATmut protein were unsuccessful, we used a transient overexpression model for NTHL1 and CATmut. Both NTHL1 and CATmut proteins were highly overexpressed (11–54-fold) compared to endogenous NTHL1 (Figure 2C).
DNA damage accumulates with overexpression of either NTHL1 or CATmut NTHL1
An increase in NTHL1 protein as seen in NSCLC lines could be a protective mechanism in response to increases in cellular reactive oxygen species. However, as BER intermediates are themselves types of DNA damage (6), increases in NTHL1 protein could overwhelm downstream BER processes, resulting in accumulation of BER intermediates. To determine whether NTHL1 overexpression results in DNA damage, we used the alkaline comet assay to analyze the sum of DNA single-strand breaks, double-strand breaks (DSB), and alkaline labile apurinic/apyrimidinic (AP) sites (29,30). Initially, we hypothesized that more DNA damage would occur with wild type NTHL1 versus CATmut, as the enzymatic activity is intact for NTHL1. With NTHL1 N-glycosylase activity and AP lyase activity present, single strand breaks could accumulate upon NTHL1 overexpression. Surprisingly, transient overexpression of either NTHL1 or CATmut resulted in similarly elevated levels of total DNA damage, compared to the control. (Figure 3A and Supplementary Figure S2A). This result suggests that elevated NTHL1 levels increase cellular DNA damage in a manner that does not depend on NTHL1 catalytic activity.
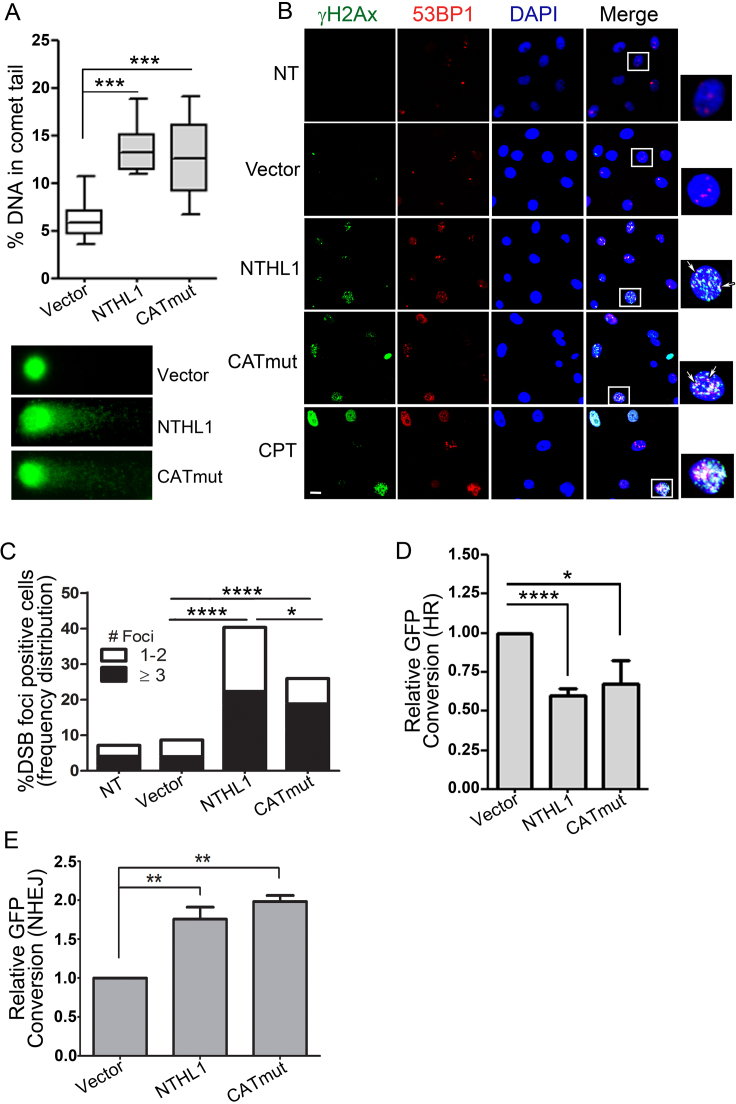
Figure 3.
DNA damage accumulates upon overexpression of wildtype NTHL1 or CATmut. (A) Alkaline comet assay of HBEC cells transiently overexpressing NTHL1 or CATmut NTHL1 24 hours following transfection. DNA damage was measured by %DNA in comet tail. DNA comets were labeled using SYBER green, and representative images of comet tails are shown. (B) Immunofluorescence of double-strand break (DSB) markers following NTHL1 or CATmut transient overexpression in HBEC cells. Twenty-four hours following transfection, cells were stained to assess colocalization of γH2Ax (green) and 53BP1 (red) markers as an indication of DSBs. Foci that show co-localization of γH2Ax and 53BP1 were counted in the quantification (arrows). Camptothecin (CPT) serves as a positive control for DSBs. The white box indicates an enlarged image of a cell with DSBs, and the white bar corresponds to 50 μm. (C) Quantification of the total number of DSB positive nuclei, and DSB foci number per nucleus from experiments in panel B. (D) Transient NTHL1 or CATmut overexpression impairs homologous recombination assayed by a reporter for gene conversion in DR-GFP U2OS cells. The fraction of GFP positive cells was determined and normalized to the empty vector control (Vector) and is presented as Relative GFP Conversion with the Vector set to 1.0. (E) Transient overexpression of NTHL1 or CATmut increases NHEJ as assayed by the DsR-7F4 reporter cell line. The fraction of GFP positive cells was determined and normalized to empty vector control (Vector) and is presented as Relative GFP Conversion with the Vector set to 1.0. P values are as follows: *P≤ 0.05*; **P≤ 0.001; ***P≤ 0.0001.
As the comet assay indicates that total damage induced by overexpression of either NTHL1 or CATmut is approximately equivalent, we examined whether a specific type of DNA damage was more prevalent. CATmut protein lacks enzymatic activity to induce single strand breaks so we tested whether DSBs could be the cause of the DNA damage detected in the comet assay (Figure 3A). Either NTHL1 or CATmut was overexpressed, and co-localization of the DNA double-strand break (DSB) markers γH2Ax and 53BP1 was assessed by immunofluorescence (Figure 3B). As a positive control, camptothecin, a topoisomerase I inhibitor that causes DSBs associated with replication (31), was employed. In the absence of DNA damaging agents, transient overexpression of either NTHL1 or CATmut NTHL1 causes accumulation of DSBs (Figure 3B) as compared to non-transfected (NT) or empty vector (Vector) controls (Figure 3B). Quantification of cells with DSB foci and the number of DSB foci per nucleus (Figure 3C) reveals that either NTHL1 or CATmut overexpression induces a significant increase in the number of cells displaying DSBs (Figure 3C). Overexpression of NTHL1 induces a significantly greater percentage of cells (40%) with DSBs compared to CATmut (25%) (Figure 3C). However, the proportion of cells having large numbers of DSB foci per nucleus (≥3) is equally increased with overexpression of either NTHL1 or CATmut (Figure 3C). Our results support the idea that a novel, catalytically independent function of NTHL1 contributes to the generation of DSBs.
Overexpression of NTHL1 or CATmut could either directly induce DSBs, inhibit DSB repair, or possibly both. To investigate whether NTHL1 overexpression inhibits DSB repair, cells containing a reporter that directly interrogates homologous recombination (HR) or non-homologous end joining (NHEJ) were employed. To measure DSB repair by HR, we assayed gene conversion in the direct repeat GFP (DR-GFP) reporter construct integrated into the DR-U2OS cell line (18,32) (Supplementary Figure S2B). This assay directly interrogates HR activity following an I-SceI-induced DSB in the GFP gene. If HR is active, the cleaved GFP gene can be repaired with a second, transcriptionally inactive GFP gene resulting in recovery of GFP fluorescence. GFP positive cells are detected using flow cytometry. Either NTHL1-Flag or CATmut-Flag protein was transiently overexpressed in the DR-U2OS cells containing this reporter system (Figure 3D and Supplementary Figure S2D). Overexpression of NTHL1 or CATmut causes an apparent ∼50% decrease in HR capacity compared to empty vector control (Vector) (Figure 3D and Supplementary Figure S2C).
To assess whether the decrease in DSB repair is specific to HR or is due to a global inhibition of DSB repair, which could include nonhomologous end joining (NHEJ) processes, we assayed end joining using a distinct reporter in DsR-7F4 cells (Figure 3E). As described in Materials and Methods (19), this system interrogates end joining through the generation of a DSB flanking a puromycin cassette, which separates a transcriptional promoter and a promoterless GFP gene. Induction of an I-SceI DSB yields proximal and distal ends that when re-ligated through end joining processes, results in the restoration of a transcriptionally active GFP gene and thus GFP signal that can be detected by flow cytometry (19,20). GFP expression is reconstituted upon deletion of ∼500 nucleotides. To control for levels of I-SceI, I-SceI is fluorescently tagged with infrared fluorescent protein (IFP). I-SceI expression is controlled following flow cytometry through gating of cells that demonstrate similar IFP expression, and only cells that express similar levels of IFP are included in the analysis. The DsR-7F4 reporter does not allow discrimination between classical and alternative non-homologous end joining (c-NHEJ and alt-NHEJ) and therefore measures changes in total end joining processes. Transient NTHL1 or CATmut overexpression in this reporter cell line results in a modest but significant increase in GFP expressing cells compared to empty vector control (Vector) (Figure 3E and Supplementary Figure S2E). Thus, overexpression of NTHL1 or CATmut results in decreased DSB processing specifically by HR, while simultaneously increasing NHEJ.
One possible explanation for the apparent decrease in HR and the concurrent increase in NHEJ is that cells overexpressing NTHL1 or CATmut might accumulate in the G1 phase of the cell cycle. Cells in the G1 phase primarily employ NHEJ for DSB repair, while cells in the S and G2 phases primarily employ HR for repair of DSBs. Thus, a change in cell cycle profile could explain these results (Figure 3D and E). Cell cycle profiles in HBEC cells expressing NTHL1-GFP or CATmut-GFP were analyzed by flow cytometry and reveal no significant changes in overall cell cycle profile compared to cells expressing GFP alone (P = 0.27) (Supplementary Figure S2F). A slight but not significant decrease in the proportion of cells in G1 phase was observed, concomitant with a small but not significant increase in the number of S phase cells (Supplementary Figure S2F). Thus, the increase in NHEJ and decrease in HR cannot be attributed to a change in the HBEC cell cycle profile. Our results suggest that increased NTHL1 or CATmut protein inhibits HR-mediated repair of DSBs, providing an explanation for the DSB accumulation in both NTHL1 and CATmut overexpressing cells.
Genomic instability is induced by either NTHL1 or CATmut overexpression
To investigate the cellular consequences of NTHL1 and CATmut-induced DSBs, we performed the cytokinesis-blocked micronucleus assay as a measure of genomic instability (33). To verify that cells displaying genomic instability specifically overexpress NTHL1, we isolated NTHL1-GFP expressing cells by fluorescence activated cell sorting (FACS). We further divided the GFP positive cell population into cells expressing low, intermediate or high levels of NTHL1 based on the intensity of the GFP signal (Supplementary Figure S3A). Forty-eight hours after sorting, binucleated cells were counted and scored for micronucleus formation. A significant increase in micronucleus formation was observed in cells overexpressing NTHL1-GFP but not GFP alone (Figure 4A and Supplementary Figure S3B). Furthermore, micronucleus formation correlated with the level of NTHL1-GFP overexpression and was observed even in the population with low levels of overexpression (Figure 4A). NTHL1 protein levels are displayed (Figure 4B) from each sorted population, verifying that GFP intensity detected by FACS reflects the amount of protein in the cell. U2OS cells overexpressing either NTHL1-GFP or CATmut-Flag also display micronuclei formation (Figure 4C).
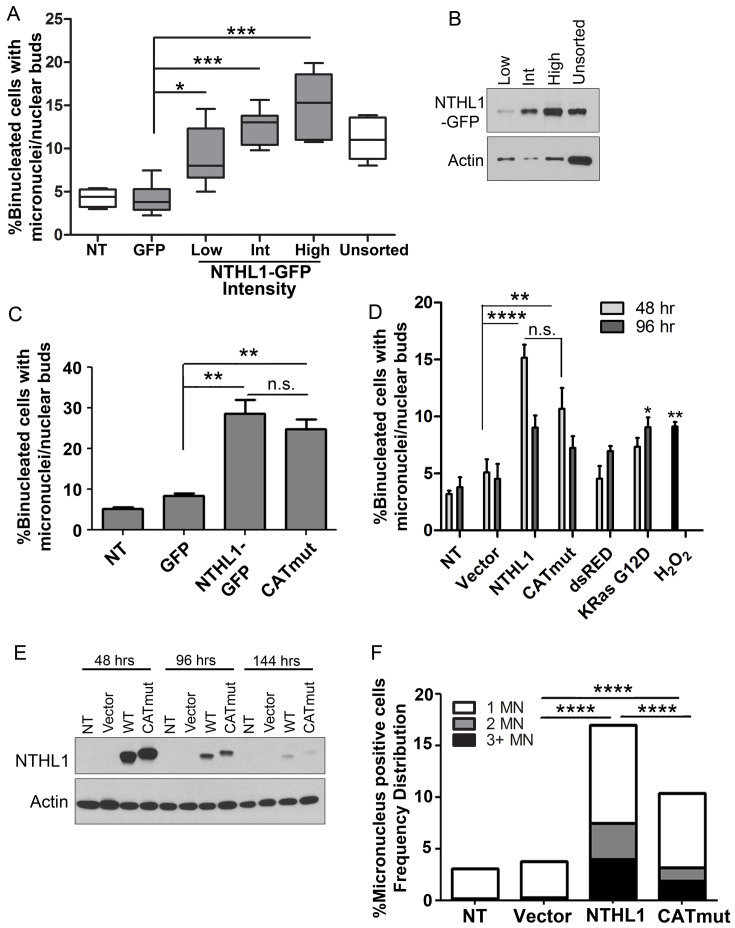
Figure 4.
Genomic instability is induced by wildtype NTHL1 and CATmut overexpression. (A) HBEC cells expressing NTHL1-GFP were FACS sorted based on GFP signal intensity. GFP positive cells were further sorted into three populations based on the intensity of the GFP signal into low, intermediate (Int), and high groups, and micronucleus formation was measured in each population and compared to unsorted NTHL1-GFP cells. Non-transfected (NT) cells and cells expressing GFP alone (GFP), were used as controls. Data from NT, GFP alone, and ‘unsorted’ cells were collected from unsorted cell populations. (B) Immunoblot of NTHL1 from the FACS sorted cell population of low, Int and high GFP intensity signals. (C) U2OS cells transiently overexpressing either NTHL1-GFP or CATmut-Flag were analyzed for micronucleus formation. Like HBEC cells, U2OS cells show an increase in micronucleus formation. (D) Overexpression of NTHL1 or CATmut in HBEC cells causes micronuclei induction at 48 hours following transfection compared to empty vector (Vector) control cells. Expression of red fluorescent protein (dsRED) or of G12D mutant K-Ras and treatment with hydrogen peroxide (H2O2) were all used as controls for micronucleus formation. KRas G12D produced statistically significant micronucleus induction at 96 hours compared to empty vector control. Micronuclei formation in dsRED-expressing cells is not significantly different from vector control at 48 or 96 h. (E) Immunoblots for NTHL1 and CATmut at 48-, 96- and 144-h time points following transient overexpression in HBEC cells show that NTHL1 and CATmut expression is decreased at 96 hours. NTHL1 expression decreases further 144 hours following transfection. (F) The number of micronuclei (MN) per binucleated cell increases in NTHL1 or CATmut overexpressing HBEC cells. Cells were scored as having 1, 2 or 3 or more micronuclei (MN) per binucleated cell. NS = non-statistically significant; *P≤ 0.05; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001.
To assess whether NTHL1 expression correlates with genomic instability over time, NTHL1 and CATmut were overexpressed by transient transfection in HBEC cells. The level of NTHL1 expression as well as micronucleus formation were monitored at 48 and 96 hours following transfection (Figure 4D and E). NTHL1 or CATmut overexpression induces significant micronucleus formation compared to empty vector control at 48 hours following transfection, with no statistically significant difference between NTHL1 and CATmut (Figure 4D). Increased micronuclei formation is still observed at 96 h but is not statistically significant compared to empty vector control. Overexpression of an unrelated protein such as dsRED (Figure 4D and Supplementary Figure S3C) did not induce micronucleus formation, whereas positive controls of treatment with 100 μM H2O2 or overexpression of K-Ras G12D protein both induce micronuclei formation (Figure 4D and Supplementary Figure S3D) (34). Levels of NTHL1 and CATmut expression are high at 48 hours but have significantly declined by 96 h following transfection (Figure 4E). The resulting decline in micronuclei present at 96 h is likely due to the decrease in NTHL1 expression, as NTHL1 levels correlate with genomic instability (Figure 4A). Therefore, the number of micronuclei detected at 48 hours and 96 hours post-transfection differ in a manner that correlates with the level of NTHL1 or CATmut overexpression.
In addition to causing elevated levels of cells with micronuclei, NTHL1 and CATmut overexpression also induce the appearance of multiple micronuclei per binucleated cell (Figure 4F and Supplementary Figure S3E). Control non-treated cells (NT) and cells transfected with empty vector alone (Vector) had <4% of binucleated cells with micronuclei. Cells overexpressing NTHL1 or CATmut display >15% or >10% of binucleated cells with micronuclei formation, respectively (Figure 4F). Remarkably, binucleated cells displaying two (∼3% for NTHL1 or ∼1% for CATmut) or even three or more (∼4% for NTHL1 or ∼2% for CATmut) micronuclei were observed (Figure 4F). We conclude that both NTHL1 and CATmut overexpression are potent inducers of genomic instability.
NTHL1 or CATmut overexpression induces replication stress signaling
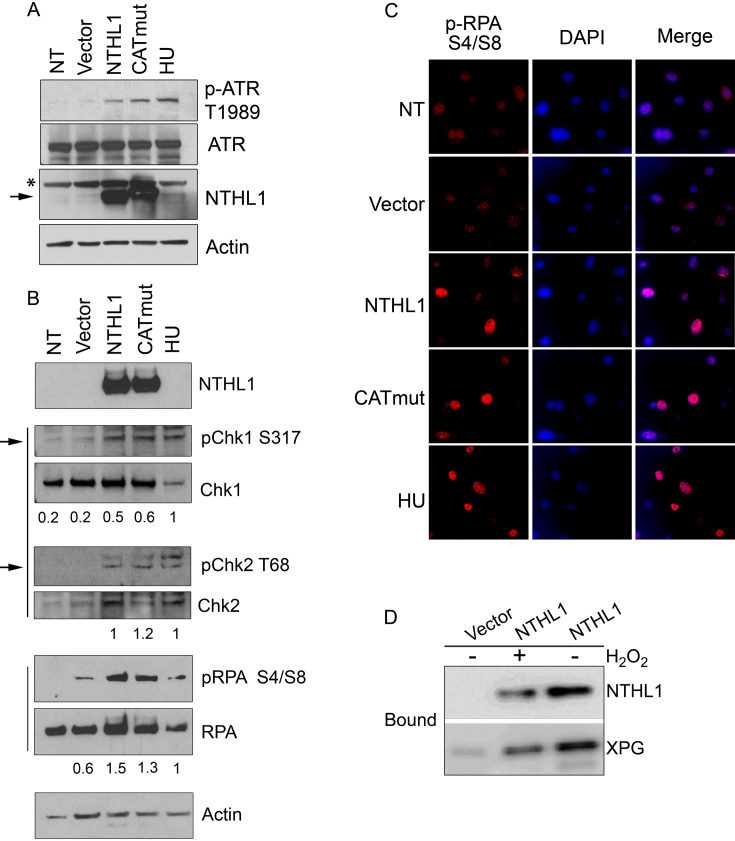
As unrepaired DSBs during S phase can result in genomic instability and micronuclei formation (35–37), we investigated whether replication stress contributes to the observed cellular phenotypes by immunoblotting for known replication stress-signaling proteins. Phosphorylation of the ATR (ataxia telangiectasia and Rad3 related) kinase takes place during the replication stress response (38). As part of this signaling cascade, ATR undergoes autophosphorylation at threonine 1989 (T1989) (38), and then phosphorylates downstream targets such as Chk1 to trigger the intra-S phase checkpoint (39,40). Hydroxyurea (HU) treatment, which induces replication stress and ATR activation (38), was used as a positive control (Figure 5A). Cells overexpressing either NTHL1 or CATmut both activated ATR (pATR T1989) (Figure 5A) and Chk1 (S317) (Figure 5B), consistent with the presence of replication stress (39). DSBs resulting from replication fork collapse activate ATM (40), which phosphorylates downstream effectors such as γH2Ax and Chk2 (40). Phosphorylation of Chk2 on threonine 68 (T68) occurs in response to DNA damage and is essential to mount a proper response to DSBs (40). Cells overexpressing NTHL1 or CATmut both display an increase in pChk2 T68 compared to negative controls (Figure 5B). The presence of γH2Ax and 53BP1 foci (Figure 3), together with pChk2, indicate that overexpression of NTHL1 or CATmut induces DSBs during replication stress through a mechanism that does not require NTHL1 catalytic activity and is consistent with the demonstrated reduction in HR.
Figure 5.
Wildtype NTHL1 or CATmut overexpression results in replication stress. (A, B) Immunoblot of HBEC cells transiently overexpressing NTHL1 or catalytically dead NTHL1 (CATmut) 24 h following transfection. The band corresponding to NTHL1 is indicated by the arrow. The asterisk denotes a non-specific band in the immunoblot. Hydroxyurea (HU) treatment serves as a positive control for replication stress. Controls also include non-transfected (NT) cells and cells transfected with Vector. (A) Immunoblot to detect autophosphorylation of ATR at threonine 1989 (p-ATR T1898), which is a replication stress marker. (B) Immunoblots showing that cells overexpressing NTHL1 or CATmut display replication stress response signaling as indicated by phosphorylation of Chk1 (pChk1 S317) and Chk2 (pChk2 T68). The arrows indicate the bands at the correct size for each pChk1 S317 and pChk2 T68, while the upper bands are non-specific binding of the antibody. Total RPA and RPA phosphorylation at serines 4 and 8 (pRPA S4/S8) are shown as a marker of ssDNA generated by HR resection. Numbers below each lane correspond to the ratio of phosphorylated protein to total protein content. (C) Immunofluorescence of the HR marker, pRPA (pRPA S4/S8), in HBEC cells transiently overexpressing NTHL1 or CATmut. The white bar corresponds to 50 μm. (D). Co-purification analysis of overexpressed NTHL1-Flag and endogenous XPG in HBEC cells. The Bound fractions are shown for purification of NTHL1-Flag from HBEC cells that transiently overexpress NTHL1-Flag or control cells transfected with Vector. Cells were either treated with 10 mM H2O2 to assess whether the interaction depends on oxidative DNA damage.
DSBs generated from replication stress are primarily repaired by HR (41), which is initiated by DNA end resection to create single-stranded DNA (ssDNA) (41,42). Replication protein A (RPA) coats the ssDNA and is phosphorylated at serine 4 and 8 (pRPA S4/S8) (43). NTHL1 and CATmut overexpression both induce increased pRPA S4/S8 signal compared to non-transfected (NT) and empty vector controls (Figure 5B and C). Each induces at least a two-fold change in the amount of phosphorylated protein for pChk1 and pRPA compared to empty vector control.
Taken together, our data show a novel NTHL1 enzymatically independent mechanism causes genomic instability through inhibition of DSB repair. There is no evidence suggesting any direct role for NTHL1 in HR. We considered that one way by which overexpression of NTHL1 or CATmut might generate these phenotypes is through a common protein interactor that functions in the HR pathway. A potential candidate is the NTHL1 protein binding partner XPG, a multi-functional protein from the nucleotide excision repair pathway that interacts with and stimulates NTHL1 activity (10). XPG has plays a key role in HR through direct interactions with BRCA1 and BRCA2 (32). Importantly, cells deficient for XPG display elevated levels of micronuclei, accumulated DSBs, replication stress, and HR impairment similar to the phenotypes observed upon overexpression of NTHL1 (32). Direct interaction between XPG and NTHL1 has primarily been investigated with purified proteins (10). We therefore tested whether NTHL1 interacts with XPG in the cellular context. In HBEC cells over-expressing NTHL1-Flag, a substantial fraction of XPG co-purifies with NTHL1-Flag on anti-Flag resin with only minimal background binding of XPG to the resin in control cells (Figure 5D). No significant change in the co-purification is observed in cells treated with H2O2 so the cellular interaction does not depend on oxidative DNA damage. This result is consistent with the possibility that overexpression of NTHL1 could hinder HR repair of DSBs by sequestering XPG.
Early cellular hallmarks of cancer are conferred by NTHL1 and CATmut overexpression
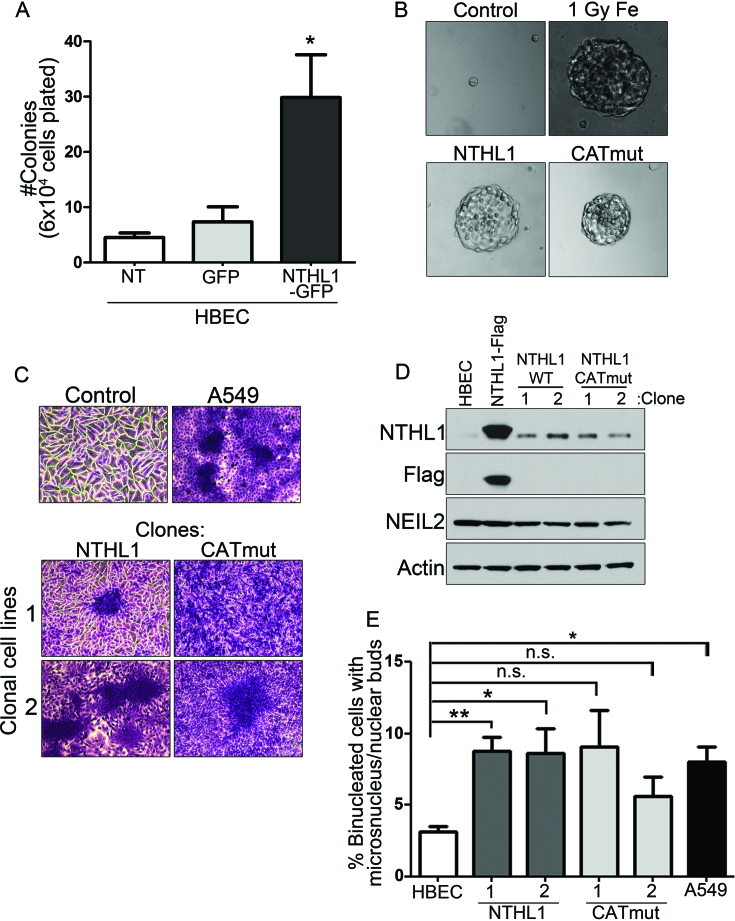
As replication stress and genomic instability are inducers of early cellular markers of transformation (41,44,45), we investigated whether NTHL1 overexpression could induce these phenotypes. The ability to form colonies in soft agar is an established criterion for assessing one of the earliest cancer hallmarks, anchorage independent growth (46). HBEC cells expressing NTHL1-GFP were FACS sorted and GFP-positive cells were plated for the soft agar assay. NTHL1-GFP expressing cells form colonies in soft agar, while cells expressing GFP alone display minimal colony formation (Figure 6A). HBEC cells overexpressing CATmut-Flag also form colonies in soft agar (Figure 6B).
Figure 6.
Cellular hallmarks of cancer are conferred by wildtype NTHL1 or CATmut overexpression. (A) HBEC cells were FACS sorted for NTHL1-GFP expression. Cells were assayed for colony formation in soft agar. (B) HBEC cells overexpressing NTHL1-Flag or CATmut-Flag were grown in soft agar. Parental HBEC cells (control) did not form soft agar colonies, while HBEC cells transformed with 1 Gy of Iron ion irradiation (1 Gy Fe) did. (C) Cell lines derived from HBEC soft agar colonies transformed by NTHL1 or CATmut overexpression were crystal violet stained to display loss of contact inhibition, in contrast to the (control) monolayer growth of HBEC parental cells and comparable to the A549 NSCLC line. Two independent cell lines were created from soft agar colonies for NTHL1 and CATmut. (D) Immunoblot of NTHL1 in each of the isolated clonal cell lines. NTHL1-Flag overexpression in lane two is a transient overexpression that serves as a control. Another BER glycosylase, NEIL2, shows no change in any of the samples analyzed. The Flag epitope is not detected in cells clonally derived from soft agar. (E) Clonal cell lines derived from soft agar demonstrate persistent genomic instability measured by the micronucleus assay compared to non-transfected (NT) HBEC cells and the A549 line as a positive control. NS = non-statistically significant; *P≤ 0.05; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001.
Colonies were isolated from soft agar, and two surviving clones for each NTHL1 variant were expanded into cell lines to test for loss of contact inhibition. Non-transformed HBEC cells form an organized, epithelial monolayer in striking contrast to the disorganized growth of the NSCLC cell line A549 (Figure 6C, top panels). Two different clonal lines of cells expressing either NTHL1 or CATmut show loss of contact inhibition with foci formation similar to that of A549 (Figure 6C, bottom panels). Thus, loss of contact inhibition develops in overexpressing cells that acquired the capability to grow in soft agar.
We assessed NTHL1 protein expression in each clonal cell line and determined that NTHL1 levels were only moderately elevated over control HBEC cells, in contrast to the high levels seen in transiently transfected HBEC (Figure 6D). Transient transfection was employed for the soft agar assay. Therefore, the length of time to complete the soft agar assay and grow the clonal cell lines makes it unlikely that the transiently expressed NTHL1-Flag is still present, unless the NTHL1-Flag construct became integrated into the genome. However, the Flag epitope is not detected in the immunoblot of cells clonally derived from the soft agar assay (Figure 6D), suggesting that the NTHL1 detected is endogenous protein. Quantification showed that NTHL1 expression levels were not statistically different from levels in non-transformed HBEC cells (Supplementary Figure S4A). To investigate whether pre-existing high endogenous levels of NTHL1 pre-disposed for ability to form colonies in soft agar, cells from the parental HBEC cell line were clonally expanded to analyze the variation in expression of endogenous NTHL1. Levels of endogenous NTHL1 in individual parental cells do not reach comparable levels to NTHL1 detected in the soft agar clones (Supplementary Figure S4B). Thus, growth in soft agar is most likely not due to pre-existing high levels of endogenous NTHL1 found in a subset of parental HBEC cells (Supplementary Figure S4B). We conclude that transient overexpression of NTHL1 is sufficient to induce cellular transformation.
Because we show that transient NTHL1 and CATmut overexpression leads to genomic instability, we evaluated whether the clonal cell lines derived from soft agar colonies demonstrated persistent genomic instability as another cancer hallmark. These clones display an elevated level of micronucleus formation that is comparable to the A549 lung cancer cell line (Figure 6E). NTHL1 overexpression therefore contributes to the generation of genomic instability that results in transformation of HBEC cells, and, persistent genomic instability is a permanent characteristic of these transformed cells.
DISCUSSION
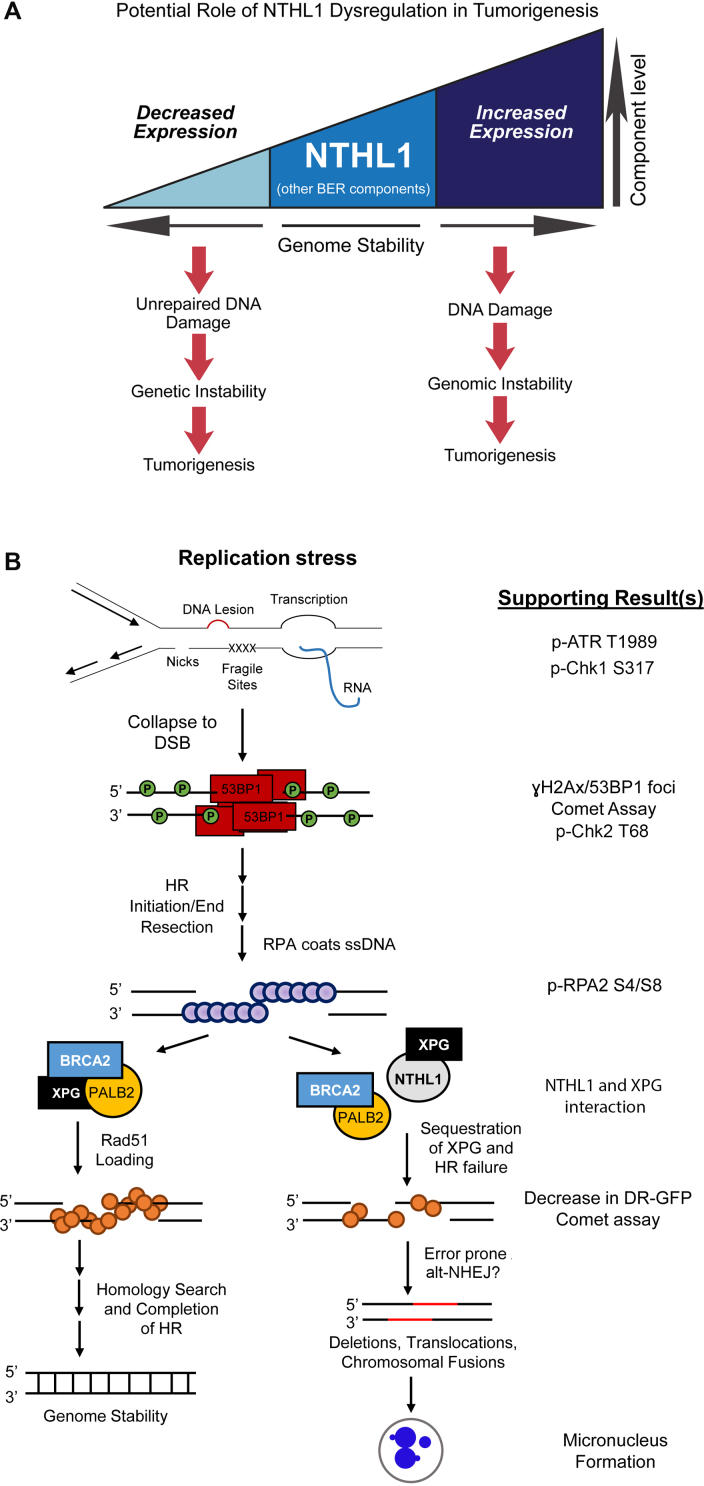
Our results demonstrate that overexpression of NTHL1 can cause genomic instability and cellular transformation independent of catalytic activity. Previous studies identified a contribution of loss of NTHL1 to a colon cancer predisposition syndrome (13,14), presumably through the accumulation of mutations and transformation over time (Figure 7A, left side). However, as we demonstrate, overexpression of NTHL1 can also contribute to transformation (Figure 7A, right side). Therefore, as illustrated in Figure 7A, a proper balance of BER protein components is needed to protect genome stability, as either loss or overexpression of the initiating DNA glycosylase can negatively impact the genome.
Figure 7.
Proposed models for NTHL1 regulation/dysregulation. (A) Loss or decreased expression of NTHL1 is linked to a novel cancer predisposition syndrome (13,14), presumably through lack of DNA repair for NTHL1 substrates, yielding mutations, genetic instability, and ultimately tumorigenesis. Alternatively, increased expression of NTHL1 protein levels can lead to DNA damage that drives genomic instability and contributes to tumorigenesis. Therefore, optimal protein levels of BER components are needed to balance the cellular need for DNA damage repair without initiating genomic instability. (B) A model for events leading to genomic instability. Results supporting the model that is presented in this work are listed on the right. Replication stress that arises during S phase may occur through multiple means, including unrepaired DNA lesions, accumulated DNA repair intermediates, nicks in the phosphodiester backbone, fragile sites, and encounters with transcription, among other causes. A collapsed replication fork results in a DSB that is repaired by homologous recombination (HR). Phosphorylation of γH2Ax and binding of 53BP1 signals the presence of a DSB. BRCA1 displaces 53BP1 in S phase to promote end resection to initiate HR. Single-stranded DNA is then coated by RPA, and RPA phosphorylation on serine 4 and serine 8 (pRPA S4/S8) signal for RAD51 loading. Initiation of HR requires formation of the RAD51 presynaptic filament through displacement of RPA by BRCA2 with PALB2 and DSS1, and efficient RAD51 loading is also dependent on XPG. Completed HR results in genome stability, as a homologous template is used to repair the DSB. Upon NTHL1 overexpression, XPG could be sequestered through protein interaction, thus inhibiting functions required for proper RAD51 loading. This could result in chronic replication stress signaling, as evidenced by pATR T1989. Inefficient RAD51 loading onto DNA results in HR failure, and competing end joining increases. This end joining may include alt-NHEJ to salvage DSB repair, as DNA resection has already occurred. Alt-NHEJ is error prone and can lead to genome instability, as assessed by micronucleus formation.
In our system, transient overexpression of NTHL1 was employed to investigate the consequences of BER dysregulation. A range of levels of NTHL1 overexpression was observed; however, the induction of DNA damage (Figure 3) and genomic instability (Figure 4) did not greatly vary between experimental replicates, suggesting that a minimum threshold for NTHL1 overexpression exists to trigger the observed phenotypes. Interestingly, the observed genomic instability and cellular transformation did not require sustained high levels of NTHL1 overexpression (Figure 6), suggesting that transient high levels of NTHL1 are sufficient to initiate processes that can lead to transformation. In support of this concept, variants of the BER protein DNA polymerase β require only transient overexpression to generate loss of contact inhibition and growth in soft agar in non-transformed mouse mammary tissue (47), a result which parallels the transient NTHL1 overexpression analyzed here.
Our data reveal a novel mechanism of inducing genomic instability and cellular transformation that does not depend on the catalytic activity of NTHL1 (Figures 3, 4 and 6). This surprising result suggests that a protein-protein interaction may underlie some consequences of NTHL1 overexpression (Figure 7B). One candidate interacting partner for NTHL1 that could affect HR activity is XPG (10,48). XPG is an endonuclease that functions in NER (49) and interacts with NTHL1 to enhance NTHL1 binding to and excision of DNA damage substrates (10). XPG is also critical for HR activity and for recovery from replication fork collapse (32). Cells deficient for XPG show increased DSBs as marked by γH2Ax and 53BP1 foci, elevated micronucleus formation, and decreased HR activity (32), consequences that are mirrored by NTHL1 overexpression in this study. The decrease in HR activity upon NTHL1 overexpression (Figure 3) is similar to the decrease in HR activity detected in cells deficient for XPG when analyzed using the same DR-GFP reporter assay (32). Furthermore, NTHL1 or CATmut overexpression increases NHEJ (Supplementary Figure S2F). Importantly, there is no significant change in the cell cycle profile of HBEC cells overexpressing either NTHL1 or CATmut compared to controls (Supplementary Figure S2F), demonstrating that the associated increase in NHEJ is not due to an increase in the proportion of cells in the G1 phase of the cell cycle.
Further supporting disruption of HR repair by NTHL1 overexpression, we show that pRPA S4/S8 signaling is increased (Figure 5B and C), as it is upon loss of XPG (32). Given the similarity in phenotypes, we suggest that independent of catalytic activity, excess NTHL1 could bind to and sequester XPG, thereby impairing the loading of RAD51 to form the presynaptic filament (Figure 7B). This BER/HR pathway crosstalk would render cells vulnerable in S phase when HR is tasked with repairing DNA damage that might give rise to genomic instability (32,41,44). Indeed, our results (Figure 5D) show that overexpressed NTHL1 interacts with XPG in HBEC cells.
Upon HR pathway disruption, DSBs can also be repaired by competing end joining processes, including c-NHEJ and alt-NHEJ (50,51). While c-NHEJ preferentially functions during the G1 phase of the cell cycle (52), alt-NHEJ is most active in S phase and is a highly error prone process (41,51,53). Upon disruption of HR, an increase in alt-NHEJ activity may occur, and this HR/alt-NHEJ interplay is independent of c-NHEJ (42). As end resection has already occurred in this scenario (42,51), error prone alt-NHEJ may be utilized to complete DSB repair. In support of this idea, our results demonstrate an impairment of HR repair (Figure 3), an increase in replication stress (Figure 5), and an increase in NHEJ activity (Figure 3). We propose a model in which cells that overexpress NTHL1 sequester the XPG protein and thus are deficient in the correct repair by HR of DSBs that routinely arise in each cell cycle through collapse of stalled replication forks. These cells may then utilize alt-NHEJ to complete DSB repair, resulting in genomic instability (Figure 7B). Further studies will be needed to determine whether alt-NHEJ plays a role in causing the cellular phenotypes observed upon NTHL1 overexpression.
A prediction of these results is that cancer cells overexpressing NTHL1 would be vulnerable to clinical agents that mechanistically induce DSBs. In support of this idea, a previous study found that lymphoblastoid cells in tissue culture display increased sensitivity to ionizing radiation upon NTHL1 overexpression (54). This study concluded that the increase in sensitivity was due to processing of closely opposed NTHL1 substrates to generate DSBs. However, our results show that NTHL1 overexpression can interfere with HR-mediated repair of DSBs, which could contribute to radiation sensitivity. This finding is consistent with previous work showing that mutation of genes or loss of proteins in the HR pathway can influence the NHEJ pathway (55).
In further support of our findings, a previous study reported cellular transformation as a result of expressing a distinct, catalytically-inactive germline variant of NTHL1 (D239Y) in the MCF10a breast cell line, in which DSBs, chromatid breaks, and chromosomal fusions were observed (26). However, these phenotypes were attributed to unrepaired cytotoxic lesions. The results thus differ from our findings, in which the resulting cellular transformation can occur independent of catalytic activity. The previous study did not evaluate wildtype NTHL1 or D239Y NTHL1 overexpression levels in comparison to endogenous NTHL1 levels, but the conditions used were stable expression in an immortal MCF10A breast epithelial cell line, in contrast to our high levels of transient overexpression in the HBEC line. Whether the different outcomes relate to differences in overexpression levels, differences in the cell lines used, or different consequences of the inactivating mutation tested is not clear. Nevertheless, both the previous study and our findings dramatically illustrate the importance of properly regulating NTHL1 to maintain genome stability.
While most outcomes in our study showed no statistically significant difference between NTHL1 and CATmut, we detected a difference for the number of DSB foci per cell (Figure 3C) and the number of micronuclei per binucleated cell (Figure 4E). This result is consistent with a potential contribution of NTHL1 catalytic activity to the generation of DNA damage and instability. As we employed transient transfections and obtained a wide range of overexpression, we cannot formally exclude the possibility that differences in NTHL1 protein expression underlie this difference. However, we hypothesize that the catalytic activity of overexpressed NTHL1 may contribute to a portion of our observed genomic instability phenotypes. Further studies that separate the contributions of NTHL1 enzymatic activity and enzymatic-independent activity are needed to further define the mechanisms leading to the observed functional consequences of NTHL1 overexpression.
This study provides evidence suggesting that increases in NTHL1 glycosylase protein levels could contribute to tumorigenesis. Understanding proper regulation of BER components (Figure 7A) will provide further insight into how dysregulation of BER is involved in cancer etiology.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge everyone in the Doetsch, Corbett, Dynan and Cooper labs for their discussions and advice. This study was supported in part by the Emory Flow Cytometry Core (EFCC) and the Emory Integrated Genomics Core (EIGC), part of the Emory Integrated Core Facilities (EICF), and is subsidized by the Emory University School of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [ES011163 to P.W.D. while employed at Emory University, GM058728 to A.H.C., ES019935 to P.K.C.]; NASA [NNX15AD63G to W.S.D.]; National Center for Advancing Translational Sciences of the National Institutes of Health [UL1TR000454]; Winship Cancer Institute of Emory University and NIH/NCI (in part) [P30CA138292]; Maiola Family Fund for lung cancer research; This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (to P.W.D.). The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bauer N.C., Corbett A.H., Doetsch P.W.. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015; 43:10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maynard S., Schurman S.H., Harboe C., de Souza-Pinto N.C., Bohr V.A.. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009; 30:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riley P.A. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 1994; 65:27–33. [DOI] [PubMed] [Google Scholar]
- 4. Marullo R., Werner E., Degtyareva N., Moore B., Altavilla G., Ramalingam S.S., Doetsch P.W.. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS One. 2013; 8:e81162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim Y.J., Wilson D.M. 3rd. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012; 5:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu D., Calvo J.A., Samson L.D.. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012; 12:104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glassner B.J., Rasmussen L.J., Najarian M.T., Posnick L.M., Samson L.D.. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao W., Samson L.. In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdel-Fatah T., Arora A., Gorguc I., Abbotts R., Beebeejaun S., Storr S., Mohan V., Hawkes C., Soomro I., Lobo D.N. et al. Are DNA repair factors promising biomarkers for personalized therapy in gastric cancer. Antioxid. Redox Signal. 2013; 18:2392–2398. [DOI] [PubMed] [Google Scholar]
- 10. Klungland A., Hoss M., Gunz D., Constantinou A., Clarkson S.G., Doetsch P.W., Bolton P.H., Wood R.D., Lindahl T.. Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell. 1999; 3:33–42. [DOI] [PubMed] [Google Scholar]
- 11. David S.S., Williams S.D.. chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998; 98:1221–1262. [DOI] [PubMed] [Google Scholar]
- 12. Chan M.K., Ocampo-Hafalla M.T., Vartanian V., Jaruga P., Kirkali G., Koenig K.L., Brown S., Lloyd R.S., Dizdaroglu M., Teebor G.W.. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst.). 2009; 8:786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rivera B., Castellsague E., Bah I., van Kempen L.C., Foulkes W.D.. Biallelic NTHL1 mutations in a woman with multiple primary tumors. N. Engl. J. Med. 2015; 373:1985–1986. [DOI] [PubMed] [Google Scholar]
- 14. Weren R.D., Ligtenberg M.J., Kets C.M., de Voer R.M., Verwiel E.T., Spruijt L., van Zelst-Stams W.A., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y. et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015; 47:668–671. [DOI] [PubMed] [Google Scholar]
- 15. Mathis C., Poussin C., Weisensee D., Gebel S., Hengstermann A., Sewer A., Belcastro V., Xiang Y., Ansari S., Wagner S. et al. Human bronchial epithelial cells exposed in vitro to cigarette smoke at the air-liquid interface resemble bronchial epithelium from human smokers. Am. J. Physiol. Lung Cell Mol. Physiol. 2013; 304:L489–L503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramirez R.D., Sheridan S., Girard L., Sato M., Kim Y., Pollack J., Peyton M., Zou Y., Kurie J.M., Dimaio J.M. et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004; 64:9027–9034. [DOI] [PubMed] [Google Scholar]
- 17. Werner E., Wang H., Doetsch P.W.. Opposite roles for p38MAPK-driven responses and reactive oxygen species in the persistence and resolution of radiation-induced genomic instability. PLoS One. 2014; 9:e108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z., Hudson F.Z., Wang H., Wang Y., Bian Z., Murnane J.P., Dynan W.S.. Increased mutagenic joining of enzymatically-induced DNA double-strand breaks in high-charge and energy particle irradiated human cells. Radiat. Res. 2013; 180:17–24. [DOI] [PubMed] [Google Scholar]
- 20. Howard S.M., Yanez D.A., Stark J.M.. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015; 11:e1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leung S.W., Apponi L.H., Cornejo O.E., Kitchen C.M., Valentini S.R., Pavlath G.K., Dunham C.M., Corbett A.H.. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene. 2009; 439:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarker A.H., Tsutakawa S.E., Kostek S., Ng C., Shin D.S., Peris M., Campeau E., Tainer J.A., Nogales E., Cooper P.K.. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005; 20:187–198. [DOI] [PubMed] [Google Scholar]
- 23. Morreall J., Limpose K., Sheppard C., Kow Y.W., Werner E., Doetsch P.W.. Inactivation of a common OGG1 variant by TNF-alpha in mammalian cells. DNA Repair (Amst.). 2015; 26:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marullo R., Werner E., Zhang H., Chen G.Z., Shin D.M., Doetsch P.W.. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis. 2015; 36:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J., Farris A.B., Xu K., Wang P., Zhang X., Duong D.M., Yi H., Shu H.K., Sun S.Y., Wang Y.. GPRC5A suppresses protein synthesis at the endoplasmic reticulum to prevent radiation-induced lung tumorigenesis. Nat. Commun. 2016; 7:11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galick H.A., Kathe S., Liu M., Robey-Bond S., Kidane D., Wallace S.S., Sweasy J.B.. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Albertson D.G. Gene amplification in cancer. Trends Genet. 2006; 22:447–455. [DOI] [PubMed] [Google Scholar]
- 28. Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A.H., Seki S., Mitra S.. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 1998; 273:21585–21593. [DOI] [PubMed] [Google Scholar]
- 29. Olive P.L., Banath J.P.. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006; 1:23–29. [DOI] [PubMed] [Google Scholar]
- 30. Shaposhnikov S., Frengen E., Collins A.R.. Increasing the resolution of the comet assay using fluorescent in situ hybridization–a review. Mutagenesis. 2009; 24:383–389. [DOI] [PubMed] [Google Scholar]
- 31. Ryan A.J., Squires S., Strutt H.L., Johnson R.T.. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 1991; 19:3295–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trego K.S., Groesser T., Davalos A.R., Parplys A.C., Zhao W., Nelson M.R., Hlaing A., Shih B., Rydberg B., Pluth J.M. et al. Non-catalytic roles for XPG with BRCA1 and BRCA2 in homologous recombination and genome stability. Mol. Cell. 2016; 61:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fenech M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007; 2:1084–1104. [DOI] [PubMed] [Google Scholar]
- 34. Castagnola P., Giaretti W.. Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim. Biophys. Acta. 2005; 1756:115–125. [DOI] [PubMed] [Google Scholar]
- 35. Fenech M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., Thomas P.. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011; 26:125–132. [DOI] [PubMed] [Google Scholar]
- 36. Xu B., Sun Z., Liu Z., Guo H., Liu Q., Jiang H., Zou Y., Gong Y., Tischfield J.A., Shao C.. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One. 2011; 6:e18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmitelli M., de Campos-Nebel M., Gonzalez-Cid M.. Progression of chromosomal damage induced by etoposide in G2 phase in a DNA-PKcs-deficient context. Chromosome Res. 2015; 23:719–732. [DOI] [PubMed] [Google Scholar]
- 38. Liu S., Shiotani B., Lahiri M., Marechal A., Tse A., Leung C.C., Glover J.N., Yang X.H., Zou L.. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell. 2011; 43:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao H., Piwnica-Worms H.. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001; 21:4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith J., Tho L.M., Xu N., Gillespie D.A.. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010; 108:73–112. [DOI] [PubMed] [Google Scholar]
- 41. Gelot C., Magdalou I., Lopez B.S.. Replication stress in Mammalian cells and its consequences for mitosis. Genes (Basel). 2015; 6:267–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahrabi S., Sarkar S., Pfister S.X., Pirovano G., Higgins G.S., Porter A.C., Humphrey T.C.. A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res. 2016; 44:5743–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shao R.G., Cao C.X., Zhang H., Kohn K.W., Wold M.S., Pommier Y.. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999; 18:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 46. Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C.. Clonogenic assay of cells in vitro. Nat. Protoc. 2006; 1:2315–2319. [DOI] [PubMed] [Google Scholar]
- 47. Sweasy J.B., Lang T., Starcevic D., Sun K.W., Lai C.C., Dimaio D., Dalal S.. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:14350–14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oyama M., Wakasugi M., Hama T., Hashidume H., Iwakami Y., Imai R., Hoshino S., Morioka H., Ishigaki Y., Nikaido O. et al. Human NTH1 physically interacts with p53 and proliferating cell nuclear antigen. Biochem. Biophys. Res. Commun. 2004; 321:183–191. [DOI] [PubMed] [Google Scholar]
- 49. O’Donovan A., Davies A.A., Moggs J.G., West S.C., Wood R.D.. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994; 371:432–435. [DOI] [PubMed] [Google Scholar]
- 50. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the Double-Strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grabarz A., Barascu A., Guirouilh-Barbat J., Lopez B.S.. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am. J. Cancer Res. 2012; 2:249–268. [PMC free article] [PubMed] [Google Scholar]
- 52. Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S.. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998; 17:5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W., Wu X.. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang N., Galick H., Wallace S.S.. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair (Amst.). 2004; 3:1323–1334. [DOI] [PubMed] [Google Scholar]
- 55. Lin Y.F., Nagasawa H., Little J.B., Kato T.A., Shih H.Y., Xie X.J., Wilson P.F. Jr, Brogan J.R., Kurimasa A., Chen D.J. et al. Differential radiosensitivity phenotypes of DNA-PKcs mutations affecting NHEJ and HRR systems following irradiation with gamma-rays or very low fluences of alpha particles. PLoS One. 2014; 9:e93579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.