Abstract
Retinoid X receptor (RXR) is an obligate heterodimeric partner of several nuclear receptors (NRs), and as such a central component of NR signaling regulating the immune and metabolic phenotype of macrophages. Importantly, the binding motifs of RXR heterodimers are enriched in the tissue-selective open chromatin regions of resident macrophages, suggesting roles in subtype specification. Recent genome-wide studies revealed that RXR binds to thousands of sites in the genome, but the mechanistic details how the cistrome is established and serves ligand-induced transcriptional activity remained elusive. Here we show that IL-4-mediated macrophage plasticity results in a greatly extended RXR cistrome via both direct and indirect actions of the transcription factor STAT6. Activation of STAT6 leads to chromatin remodeling and RXR recruitment to de novo enhancers. In addition, STAT6 triggers a secondary transcription factor wave, including PPARγ. PPARγ appears to be indispensable for the development of RXR-bound de novo enhancers, whose activities can be modulated by the ligands of the PPARγ:RXR heterodimer conferring ligand selective cellular responses. Collectively, these data reveal the mechanisms leading to the dynamic extension of the RXR cistrome and identify the lipid-sensing enhancer sets responsible for the appearance of ligand-preferred gene signatures in alternatively polarized macrophages.
INTRODUCTION
Retinoid X receptor (RXR) is a unique and enigmatic member of the nuclear receptor superfamily due to its heterodimerization capacity with several different nuclear receptors (1). This widespread dimerization capacity of RXR puts this receptor on the crossroads of nuclear receptor-mediated transcriptional regulation, but at the same time it also harbors independent regulatory roles (1–3). RXR has indispensable roles during prenatal development (4,5). Furthermore, drugs targeting RXR are in use for cancer therapy and others are in preclinical trials to tackle insulin resistance and atherosclerosis (6,7). Recently, there have been emerging pieces of evidence pointing to the significance of RXR in modulating the immunological state of macrophages (8–10). To date, several studies report about the multifaceted roles of macrophage RXR in controlling autoimmune disease, the phagocytic capacity of macrophages, the clearance of amyloid-β by brain microglia in an Alzheimer's disease model and leukocyte migration (8,10). According to these studies, there seems to be a consensus that RXRs are important regulators of macrophage function. In addition, open chromatin landscapes of tissue-resident macrophages revealed the enrichment of RXR heterodimer-binding motifs at the accessible chromatin regions of the cells in a tissue-selective manner (11,12). There are only a few established examples of causative relationships between NRs and macrophage specification. It has been shown that in the absence of LXRα (13) and PPARγ (14), the size of spleen- and lung-resident macrophage populations is greatly diminished, respectively. Interestingly, these studies indicate that the appearance of specific RXR heterodimers are defining features of tissue-resident macrophage subtypes, but the molecular triggers and mechanisms mediating the development of these are not known. RXR is part of the heterodimeric NR family and their behavior is different than that of homodimeric, steroid NRs such as ER and GR. The cistromes of steroid receptors are principally driven by ligand binding, which triggers their rapid translocation to the nucleus leading to tens of thousands of de novo, receptor-bound sites in the genome. Counter to this mechanism, RXR heterodimers are considered to be genome-bound molecular switches, repressing transcription in the absence of their ligands, which case can be reversed to an active state once the activating ligand is present, known as the ‘molecular switch model’ (45). This model implies that most if not all chromatin bound receptors act as switches flipped by ligand.
Recent studies reported that the enhancer repertoire of macrophages are dynamically rearranged by the two main signal-dependent transcription factors (SDTFs) (interleukin 4 (IL-4)-activated STAT6 or Kdo(2)-lipid A (KLA)-activated NF-κB) controlling classical and alternative macrophage polarization (15). The fact that these can take place in terminally differentiated cells indicates that cell type-specific gene expression patterns can be rearranged (16,17). Therefore, we hypothesized that upon macrophage polarization, RXR-bound regulatory regions are reprogrammed in order to support the changed signaling requirements.
Recently, it has been shown that RXR heterodimers are able to act on the pre-determined enhancer landscape of macrophages occupied by LDTFs (3,18,19). However, signals initiating the establishment, rearrangement and maintenance of NR cistromes remain elusive in the context of RXR heterodimers. Our recent study in non-polarized macrophages show that ligand stimulation does not significantly affect the genomic distribution of RXR, suggesting that RXR heterodimers behave in a rather static way if compared to steroid receptors, which rapidly occupy thousands of sites in the genome upon ligand binding (3). These observations raise the questions how such RXRs function and what type of mechanistic pathways may play roles in the deposition and redistribution of the receptor in the macrophage genome.
In this study, we have investigated the behavior of the RXR cistrome upon alternative macrophage polarization. Genome-wide characterization of RXR binding revealed a substantially extended RXR cistrome. We show that IL-4-activated STAT6 drives the extension of the RXR cistrome in a PPARγ-dependent manner in the vast majority of the cases. Systematic assessment of open chromatin structure and the binding sites of STAT6, PU.1, RXR, PPARγ, P300 and RAD21 revealed the kinetics and the epigenomic determinants of de novo RXR-bound regulatory sites. Furthermore, PPARγ:RXR heterodimers are required for the proper development of the polarization-specific open chromatin landscape. Genome-wide mapping of RNAPII-pS2 in the presence of the specific ligands, rosiglitazone (RSG) and LG100268 (LG268), allowed us to functionally characterize the PPARγ:RXR-bound enhancer-gene network and pinpoint the dominant/selective effects of these ligands.
MATERIALS AND METHODS
Materials
Ligands: LG268 (Sigma), RSG (Sigma), mIL-4 (Peprotech).
Mouse strains
All strains are on C57BL/6 genetic background. The RXR-deficient macrophage-specific knockout mice were gifts from Pierre Chambon's laboratory. We crossed Rxra fl/fl Rxrb +/− lysozyme-Cre (LysCre)+ males with Rxra fl/fl Rxrb −/− LysCre+ females and used the Rxra fl/fl Rxrb −/− LysCre+ 3 months old male mice.
Mice carrying null or floxed alleles of Pparg were created as described previously (20). These mice were backcrossed to the C57BL/6J strain for eight generations. Mice were bred with LysCre transgene animals to create the following genotypes: Pparg +/+ LysCre+, Pparg fl/fl LysCre+, Pparg +/− LysCre+ and Pparg fl/- LysCre+.
Stat6 KO is a full body knockout, and it was obtained from Jackson laboratories. In this case we maintained them homozygous for Stat6 ablation and we mated knockout male with female mice. For all of our experiments using knockout cells, we used C57BL/6 wild type male mice.
Differentiation of bone marrow-derived macrophages
Isolation and differentiation were completed as described earlier (3). Isolated bone marrow-derived cells were differentiated for 6 days in the presence of L929 supernatant. Cells were either exposed to IL-4 (5 ng/ml) during the whole differentiation process or polarized on the sixth day of the differentiation with IL-4 (20 ng/ml) for the indicated period of time.
Immortalization of mouse bone marrow-derived macrophages
Bone marrow-derived cells were immortalized using the J2 cell line continuously producing the J2 virus encoding v-raf and v-myc oncogenes (21). J2 cells were grown in DMEM containing 20% FBS. Bone marrow cells were seeded in immortalization media I (90% J2 supernatant, 5% HyClone FBS, 10 ug/ml Polybrene 0.1%, L929 supernatant 5%) and incubated overnight. On the second day, supernatant was collected and spun down to pellet floating cells. Adherent cells were scraped and re-plated into a new petri dish using immortalization media II (20% J2 supernatant, 10% HyClone FBS, 10ug/ml Polybrene 0.1%, L929 supernatant 10%, 60% DMEM) and incubated for 6 days. After the immortalization, cells were kept in regular macrophage differentiation media (20% FBS, 30% L929 supernatant and 50% DMEM containing 1% antibiotics).
Treatment conditions
Primary macrophages were treated with IL-4 (5ng/ml) for 6 days during long-term exposure. Upon short-term exposure macrophages were treated with IL-4 (20 ng/ml) for 1, 6 and 24 h. These treatment conditions were used for ChIP-seq. RNAPII-pS2 ChIP-seq and gene expression measurements were performed in macrophages differentiated with M-CSF, on the 6th day cells were exposed to IL-4 (20 ng/ul) for 24 h. After 24 h of polarization, LG268 (100 nM) and RSG (1 uM) were added to the cells for an additional 1 h. ATAC-seq experiments were carried out in the short-term and long-term exposure system using the IL-4 concentrations described above. GRO-seq experiments were carried out after 24 h of IL-4 (20 ng/ul) treatment followed by 1 h of ligand exposure (LG268 100 nM, RSG 1 uM).
Real-Time Quantitative PCRfor enhancer RNA and mRNA detection (qPCR)
RNA was isolated with Trizol reagent (Ambion). RNA was reverse transcribed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer’s protocol. Transcript quantification was performed by qPCR reactions using SYBR green master mix (BioRad). Transcript levels were normalized to Ppia.
Assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq)
ATAC-seq was carried out as described earlier with minor modification (22). Cells were scraped and counted to achieve 50k/ml in ice-cold PBS. Cell suspension was further diluted to 25k/ml and nuclei were isolated with ATAC-LB (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL). Nuclei from 25k cells were used for tagmentation using Nextera DNA Library Preparation Kit (Illumina) from two biological replicates. After tagmentation DNA was purified with Minelute PCR Purification Kit (Qiagen). Tagmented DNA was amplified with Kapa Hifi Hot Start Kit (Kapa Biosystems) using 9 PCR cycles. Amplified libraries were purified again with Minelute PCR Purification Kit. Fragment distribution of libraries was assessed with Agilent Bioanalyzer and libraries were sequenced on a HiSeq 2500 platform.
ATAC-seq analysis
The primary analysis of ATAC-seq-derived raw sequence reads has been carried out using the updated version of our ChIP-seq analysis command line pipeline (46) including the following steps: Alignment to the mm10 mouse genome assembly was done by the BWA tool (23), and BAM files were created by SAMTools (24). Fragment length was set uniformly to 120 nts by makeTagDirectory (HOMER). Genome coverage (bedgraph) files were generated by makeUCSCfile.pl (HOMER) (18), converted to TDF files by IGVtools, and then visualized with IGV2 (25). Read densities for read distribution heat maps were generated by annotatePeaks.pl (HOMER), median normalized based on a consensus set generated from those peaks detected from at least two samples of all used for the given comparison and visualized by Java TreeView (47). Coverage values of the summits were used for box plots.
ChIP (chromatin immunoprecipitation)
ChIP was performed essentially as previously described (26). Libraries were prepared either with Ovation Ultralow Library Systems (Nugen) or TruSeq ChIP library systems (Illumina) according to the manufacturer's instructions. The following antibodies were used: IgG (Millipore, 12–370), RXR (sc-774), P300 (sc-585), PU.1 (sc-352), RAD21 (ab992), STAT6 (sc-981), PPARG (Perseus #PP-A3409A) and RNAPII-pS2 (ab5095).
ChIP-seq analysis
The primary analysis of ChIP-seq-derived raw sequence reads has been carried out similarly as described for the ATAC-seq analysis. Peaks were predicted by MACS2 (27), and artifacts were removed according to the blacklist of ENCODE (48). Three RXR ChIP-seq replicates derived from those BMDMs differentiated in the presence or absence of IL-4 were analyzed by DiffBind (28) using an input control: consensus peak set was formed from those peaks predicted from at least two of six samples. Peaks showing significant increase upon IL-4 treatment (P>0.05) and those without significant increase were further divided based on their presence in Stat6 KO BMDM cells. The median summit coverage value of the cluster showing no difference between the three conditions (Cl3) was the basis of the normalization of read coverage on both the read distribution heat maps and box plots (in the cases of RXR, PU.1, P300, RAD21 and RNAPII-pS2). The de novo RXR peak set (Cl2) was further divided based on the presence of STAT6 peaks upon 1h of IL-4 treatment. These subdivisions then were further separated and analyzed by the PPARγ-dependent peaks (not present in Pparg KO) and independent RXR peaks (present in Pparg KO).
RNAPII-pS2 abundance on gene bodies (using mm10 RefSeq annotation) was quantified and tested using package Rsubread and edgeR (P ≤ 0.05 and FC ≥ 2.0), respectively. Metagene plots were created using ngs.plot (49) software.
Motif analysis
The middle 100-bp regions of the up to 1000 top peaks were used for motif enrichment analyses, performed by findMotifsGenome.pl (HOMER). P-values were calculated by comparing the target region enrichments with those of an about 50 000-region random (background) set generated by HOMER. The selected motif matrices (Figure S1A) were mapped by annotatePeaks.pl (HOMER). Box plot and bar graph was used to represent motif score distribution. Motif appearances counted in 30-bp windows within 1.5 kb around the chosen regions were determined by using intersectBed (BEDTools) and other command line programs, and then were plotted by java TreeView. Sequence of the top DR1 motif (±16 bp) for each selected RXR/PPARg-bound region (within 200 bp) was recovered by ‘homerTools extract’ and to color sequences, each nucleotides were exchanged to a number (–1, 0 or 1) or ‘x’ (NA). Based on the generated matrix, sequence heat maps were visualized by Java TreeView. Nucleotide frequencies within the annotated peak/DR1 groups were converted to Homer motif matrices with command line tools, and then were visualized by motif2Logo.pl (HOMER).
SubTAD prediction
The insulator motif matrix determined from the top 1000 CTCF peak summits that overlapped with RAD21 peaks (insulator regions) was mapped onto the 200-bp region around CTCF summits, and the top hit for each region was designated as an insulator element. RPKM values for both CTCF and RAD21 ChIP-seq samples (3) were calculated on the 100-bp region around insulators, and those regions showing higher density for both proteins than the 100 000th of the summed density of all regions were used for subTAD prediction. The closest insulators showing convergent direction within a 1-Mb distance but farther than 1 kb were assigned to each other and called subTADs if their coverage showed less than a 2-fold difference for both proteins. In the case of overlapping subTADs, those were kept having the highest insulator coverage. ‘Negative’ subTADs with divergent insulators were defined in the same way as convergent ones.
Global Run-On sequencing (GRO-seq)
GRO and library preparation were performed as described earlier (3) with the following modifications: Libraries were generated from two biological replicates using NEBNext Small RNA Library Prep set for Illumina. Bone marrow-derived macrophages were polarized with IL-4 for 24 h or left untreated, then cells were exposed to RSG, LG268 or Veh (vehicle - DMSO:Ethanol) for one additional hour. Fragment distribution of libraries was assessed with Agilent Bioanalyzer and libraries were sequenced on a HiSeq 2500 platform.
GRO-seq analysis
The primary analysis of GRO-seq-derived raw sequence reads has been carried out similarly as described for the ATAC-seq analysis. The upper decile-normalized RPKM values calculated from the unique reads on protein-coding genes were compared between the three conditions (Veh, RSG and LG268 treatment) examined by both GRO-seq and RNAPII-pS2 ChIP-seq methods. Correlation coefficients were calculated based on the differentially expressed genes between the three conditions.
Enhancer analysis
TSSs of ligand-responsive genes and PPARγ:RXR ‘co-peaks’ within the same subTAD were assigned to each other. Average RNAPII-pS2 densities within the 2-kb region around the annotated peaks were determined by annotatePeaks.pl (HOMER) and visualized as box plots. Fold changes upon ligand treatments were shown on scatter plots.
Western Blot
Whole cell lysates were resolved by electrophoresis in 10% polyacrylamide gel and then transferred to Immobilon-P Transfer Membrane. Membranes were probed with anti-PPARγ (81B8) and anti-RXR (sc-553) antibodies according to manufacturer's recommendations.
Statistical tests
QPCR measurements were presented as means ± SD. We made at least two biological replicates; performed unpaired (two-tailed) t tests and the differences were considered significant at P < 0.05. ChIP-seq densities presented in box plots are analyzed with (two-tailed) paired t-test. Differences were considered significant at P < 0.0001.
Availability of sequencing data sets
Sequencing data have been submitted to GEO under accession number: GSE110465.
RESULTS
IL-4-activated STAT6 is required for the extension of the RXR cistrome in alternatively polarized macrophages
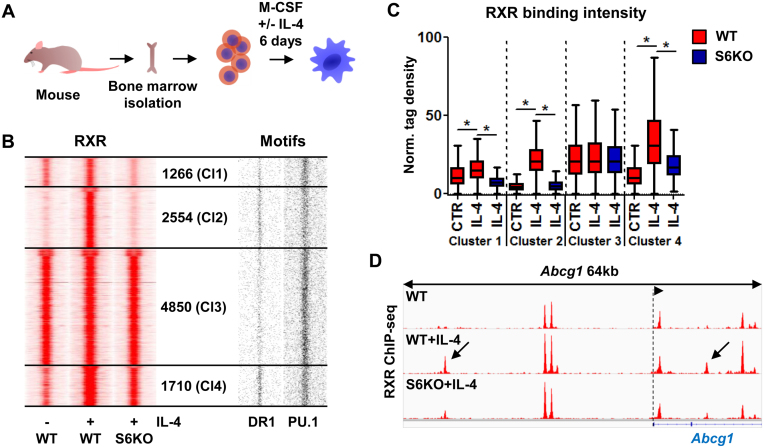
In order to investigate whether and how alternative macrophage polarization impacts the RXR cistrome, we utilized mouse bone marrow-derived macrophages (BMDMs) differentiated in the presence of IL-4 (Figure 1A). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) for RXR revealed an extensively remodeled and expanded RXR-bound set of genomic regions upon IL-4 stimulation. The magnitude of the expansion prompted us to address the contribution of STAT6 to this process. In order to dissect this aspect we used STAT6-deficient macrophages from Stat6 knockout (S6KO) animals. Using ChIP-seq experiments we determined the RXR cistrome in S6KO macrophages and clustered the RXR-bound regions based on their IL-4 and STAT6 dependence (Figure 1B). This analysis yielded four major RXR clusters: Cluster 1 (Cl1) contains those regions having slight induction in the presence of IL-4 and exhibit strong STAT6-dependence; Cluster 2 (Cl2) has de novo RXR-bound regulatory regions and complete IL-4/STAT6-dependence; Cluster 3 (Cl3) includes regions unaffected by IL-4/STAT6; and Cluster 4 (Cl4) is similar to Cl2 with regards to the IL-4-induced RXR deposition, but its component regions exhibited a lower level of STAT6-dependence (Figure 1B–D). De novo motif enrichment analysis of the four clusters reported the presence of the expected macrophage-specific transcription factor motifs, for instance PU.1, C/EBP, TRE (AP-1) and RUNX (Supplementary Figure S1A). Importantly, we observed marked differences between Cl2 and Cl3, specifically that Cl2 displayed robust enrichment for the direct repeat 1 (DR1) motif indicating that likely PPARγ is the predominant partner of RXR at these sites, while in Cl3, nuclear receptor (NR) half sites dominate with some enrichment for DR4 (specific for e.g. LXR:RXR heterodimers). Cl1 showed similar results to those of Cl3, and the results of Cl4 were similar to those of Cl2 (Supplementary Figure S1A). Importantly, we observed the enrichment of the EGR2 motif in Cl2 and Cl4, which has been described as a transcription factor induced upon alternative macrophage polarization (29,30). Mapping the matrices of the enriched motifs provided a more detailed picture about motif distribution (Figure 1B, Supplementary Figure S1B). NR half sites were enriched near all RXR-bound regions, and the putative STAT6 elements showed a low enrichment in the IL-4/STAT6-dependent Cl2. DR1 showed the expected high frequency in both clusters 2 and 4, but there were DR1 sites scattered also in the other two clusters. TREs showed similar distribution as DR1s, but EGR2 motifs had a higher frequency in the de novo cluster. In contrast, a high abundance of putative PU.1 elements could be observed in clusters 1 and 3, and fewer hits were found in Cl2 (Figure 1B and Supplementary Figure S1B), indicating that beside the LDTF PU.1 and the SDTF STAT6, other transcription factors also have major roles in Cl2, including PPARγ.
Figure 1.
Alternatively polarized macrophages greatly extend their RXR cistrome in an IL-4/STAT6-dependent manner. (A) Model system used to achieve alternative macrophage polarization. (B) Read distribution plots of RXR ChIP-seq signal intensities in wild type (WT) and Stat6 knockout (S6KO) macrophages either left untreated (–) or treated with IL-4 (+) (left). Clusters were discriminated based on IL-4 responsiveness and STAT6 dependence, and the number of genomic regions within them is indicated (middle). Distribution of DR1 and PU.1 motifs around RXR peaks (right). 1.5-kb regions are represented in 30-bp bins. (C) Box plot depicting the normalized tag density for each RXR clusters as shown on panel B. Significant differences are indicated with asterisks, determined by two-tailed paired t test, P< 0.0001. (D) Genome browser view about the RXR-bound genomic regions on the Abcg1 locus under the indicated conditions. Arrows indicate de novo RXR-bound regulatory regions, exhibiting STAT6 dependence.
Altogether, these results show that IL-4-polarized macrophages have an extended RXR cistrome where PPARγ may be the dominant partner, and STAT6 is indispensable in the development of this state.
Dynamic establishment of de novo RXR binding is initiated by STAT6 and leads to PU.1, P300, RAD21 recruitment and chromatin opening
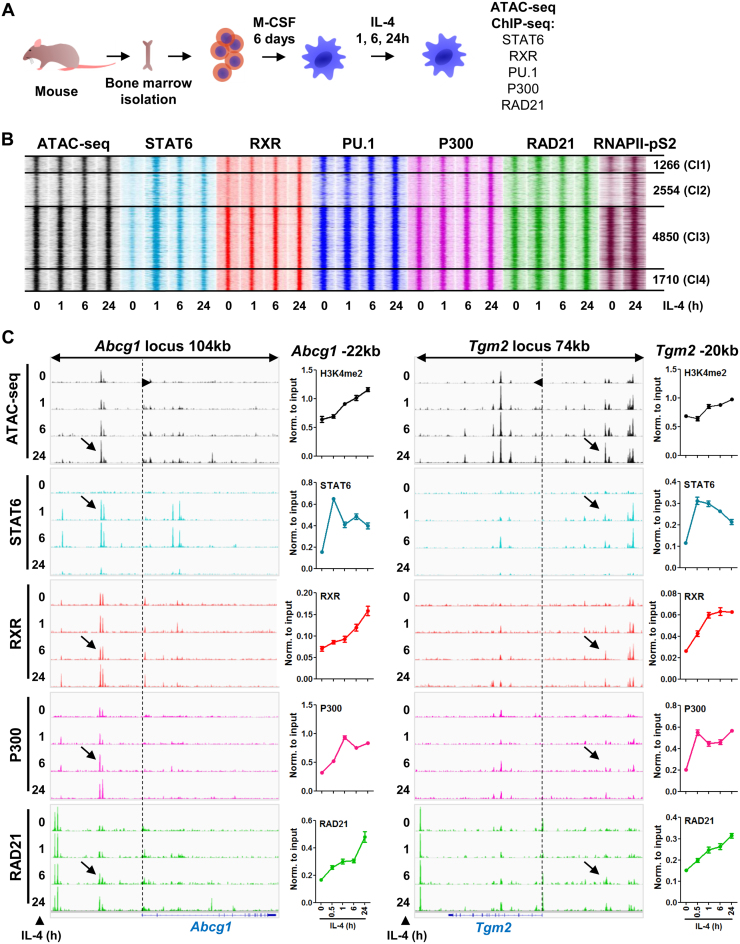
Regarding the extensive effect of IL-4 on the RXR cistrome during macrophage differentiation, we were wondering about the dynamics and epigenomic features of this rearrangement. To test these, we applied time-course experiments upon the IL-4 treatment of differentiated macrophages and determined the chromatin accessibility (ATAC-seq) and the binding of STAT6, RXR, PU.1, P300 (universal co-activator, active enhancer mark) and RAD21 (genome architectural protein) by ChIP-seq at the 0, 1, 6 and 24-h time points (Figure 2A). Overlaying the RXR clusters (as shown on Figure 1) with the time-course data sets revealed robust changes in Cl2, while the other clusters did not exhibit major differences, except for the binding of STAT6, which got elevated upon IL-4 stimulation in all clusters (Figure 2B, Supplementary Figure S2A). Therefore we focused our attention to the most dynamically changing, de novo RXR cluster (Cl2).
Figure 2.
Characterization of the epigenomic components of RXR-bound regulatory regions upon macrophage polarization. (A) Scheme of the model system used to study the dynamic features of the RXR cistrome extension and other transcription factors’ binding kinetics using a time-course upon IL-4 exposure. (B) Read distribution plot of ATAC-seq signals (chromatin accessibility) and ChIP-seq intensities for STAT6, RXR, PU.1, P300, RAD21 and RNAPII-pS2 at the indicated time-course of IL-4 stimulation in wild type macrophages (h, hours). Clusters established on Figure 1 panel B are represented for each experiment, and the number of genomic regions within each cluster is indicated. 1.5-kb regions are represented in 30-bp bins. (C) Genome browser views of ATAC-seq and ChIP-seq read enrichments for the presented factors in the presence of IL-4 for the indicated period of time on the Abcg1 and Tgm2 loci. Arrows indicate the enhancers of Abcg1 and Tgm2 located –22 kb and –20 kb from the transcription start site of the genes, respectively. Dashed lines indicate the transcription start sites, and the arrowhead on the lines indicates the direction of the genes. ChIP-qPCR experiments performed on the two enhancers for the indicated factors are also presented next to the genome browser images using a time-course of IL-4 stimulation.
Assessment of chromatin accessibility shed light on the progressively opening nature of these genomic regions in time. This observation could be explained by our ChIP-seq data sets, showing that all the investigated factors were recruited to these sites upon IL-4 stimulation, although with slightly different binding kinetics (Figure 2B). More precisely, STAT6 recruitment was the first indicator of enhancer activation, which was followed by RXR, PU.1, P300 and RAD21 binding. Investigation of the enrichment of the elongation-specific RNA polymerase II phosphorylated on serine 2 (RNAPII-pS2) revealed that the majority of these sites should be considered active enhancers. In addition, we selected five de novo RXR-bound genomic regions and measured enhancer RNA levels. Three out of the five showed significantly induced enhancer transcription in the presence of IL-4 (Supplementary Figure S2B).
After providing a genome-wide and temporal view of this process, we went on to validate these findings on two individual enhancers on the Abcg1 and Tgm2-associated genomic regions (Abcg1 -22 kb, Tgm2 -20 kb). Using H3K4me2 as a general regulatory element mark we observed a similar, continuous signal increment as with ATAC-seq, and the binding patterns of STAT6, RXR, P300, RAD21 and PolII revealed similar recruitment modalities as observed in the ChIP-seq data sets (Figure 2C). Based on these results, we noted a slight difference in the kinetics of RXR and P300 recruitment at the two measured genomic regions, which may suggest that there are different mechanisms for the reprogramming of the RXR cistrome. An additional observation, which further supports this view, is the weaker occupancy of STAT6 in Cl2 if compared to the other clusters (Supplementary Figure S2A), suggesting that the STAT6-mediated direct and indirect means control the remodeling of the RXR-bound regulatory regions.
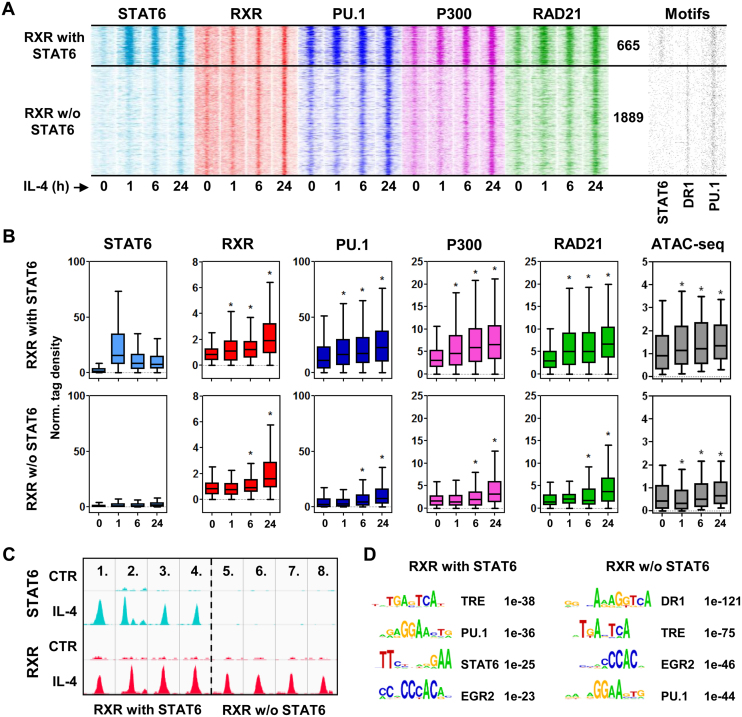
The formation of de novo RXR-bound regulatory elements are STAT6-dependent, but only partially occupied by STAT6
Our previous results indicated that there are likely to be direct and also indirect STAT6 effects that are contributing to the development of the de novo RXR-bound genomic regions (Cl2). Therefore, we decided to investigate STAT6 binding in Cl2, which revealed that out of the 2554 sites, only 26% (655) were bound by STAT6 (Figure 3A–C). Overlaying the binding kinetics of RXR, PU.1, P300 and RAD21 with the STAT6 positive and negative RXR clusters from the time-course experiments revealed that these factors followed the recruitment of STAT6 as early as 1 h after stimulation unlike STAT6 negative RXR sites, which appeared to recruit the receptor and the additional factors after 6 h of stimulation (Figure 3B). A similar phenomenon was observed in chromatin openness, namely that STAT6-bound genomic regions are more accessible at the basal state than those showing a prolonged opening dynamics, and IL-4/STAT6 is able to further open these sites and contribute to the maintenance of this chromatin status probably by secondary effects (Supplementary Figure S3A). In addition, STAT6 positive sites readily-bound by PU.1, P300 and RAD21 at the basal state, further increase the presence of these factors upon IL-4 stimulation. Sorting the motif distribution plots accordingly, we observed that STAT6 binds exclusively to its specific elements, and PU.1 has higher motif enrichment in the STAT6-bound regions (Figure 3A, Supplementary Figure S3B). This is probably also due to the sequence similarity between the two motifs as the ETS core motif (GGAA) might serve as a half site of STAT6. DR1, in contrast, shows enrichment in the STAT6 negative RXR sites, which, in light of the induced PPARγ protein level upon IL-4 treatment (Supplementary Figure S3C), may explain the shift in the dynamics of protein binding events (Figure 3A and B). A de novo motif enrichment analysis also resulted in the notion that there is a partial exclusion between STAT6 and DR1 elements, and there are additional transcription factors, which contribute to the activation of de novo RXR-bound enhancers (Figure 3D).
Figure 3.
Extension of the RXR cistrome is carried out by the direct and indirect actions of STAT6. (A) Read distribution plots of the indicated ChIP-seq experiments in the de novo, IL-4/STAT6-dependent RXR cluster (Cluster2 defined on Figure 1 panel B) separated based on STAT6 binding (h, hours). The number of genomic regions within the STAT6 positive and negative RXR sub-clusters is indicated. Distribution of STAT6, DR1 and PU.1 motifs around RXR peaks (right). 1.5-kb regions are represented in 30-bp bins. (B) Box plots depicting the normalized tag density for the indicated factors in the STAT6 positive and negative RXR clusters. Significant differences were determined by two-tailed paired t test at P< 0.0001 and indicated by asterisks. (C) Genome browser view of examples for STAT6 positive (1–4.) and negative RXR sites (5–8.): 1. chr18:88820090–88820456, 2. chr8:46868575–46869011, 3. chr11:70114345–70114844, 4. chr11:70114345–70114864, 5. chr15:10869138–10869551, 6. chr2:134730555–134731000, 7. chr15:10869138–10869551, 8. chr15:10758849–10759232. (D) De novo motif enrichments in the STAT6 positive and negative RXR sub-clusters. Motif logos, names and the P-values are indicated for each motifs enriched during the analysis.
Collectively, these results indicate that the remodeling of the RXR binding landscape is accomplished by STAT6-initiated and mediated direct and indirect effects, and PPARγ, among other factors (AP-1, EGR2), likely to have critical roles in this process.
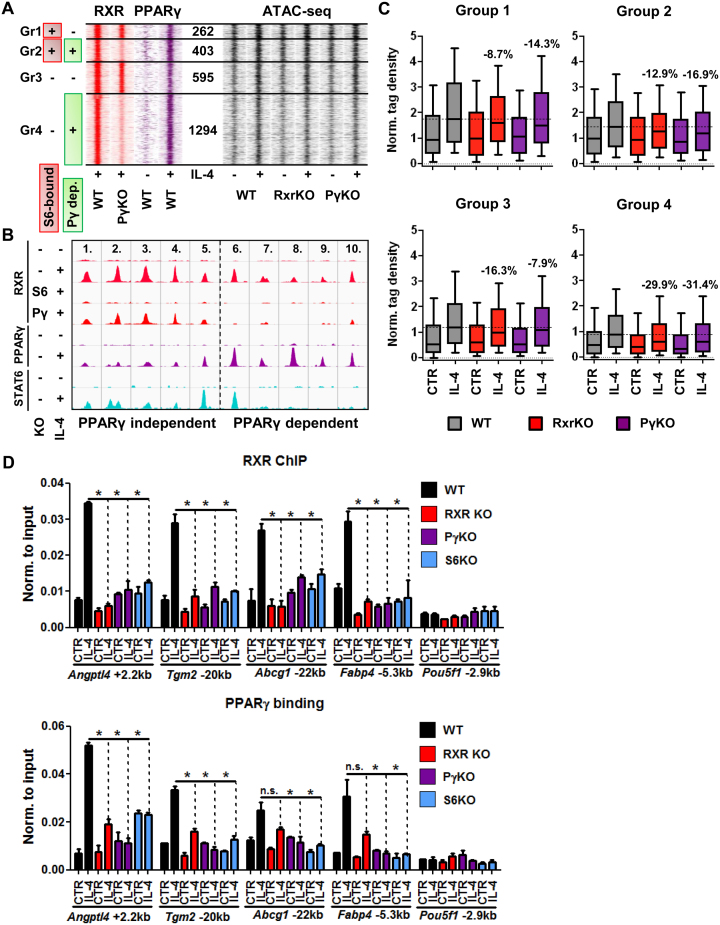
STAT6-induced PPARγ is required for the extension of the RXR cistrome and the heterodimer is needed for chromatin remodeling
Along the lines of our observations made above, we intended to study how PPARγ contributes to the extension of the RXR cistrome. This is important in order to understand the development of the cistrome, because RXR protein levels are not changing whilst, as it has been described and we confirm here, PPARγ levels showed robust up-regulation upon alternative macrophage polarization (31–33), underscoring its possible contribution to RXR redistribution (Supplementary Figure S3C). Using ChIP-seq for RXR in Pparg KO (PγKO) macrophages revealed that a significant part (1697 sites, 66%, Groups 2 and 4) of the de novo RXR sites cannot be established by IL-4 in the absence of PPARγ (Figure 4A and B). Using ATAC-seq in RxrKO and PγKO macrophages we found that both nuclear receptors contribute to chromatin opening. More specifically, sites that showed PPARγ-dependent RXR binding but no STAT6 recruitment (Group 4) exhibited the highest necessity of the presence of PPARγ:RXR to become fully opened (Figure 4A and C; see control set on Supplementary Figure S4A). Group 3 was less dependent on PPARγ, and groups 1 and 2 were less dependent on RXR. Regarding the overall openness, STAT6 binding sites without later PPARγ binding (Group 1) are the most opened, and the originally closed regions with later PPARγ binding (Group 4) are the least opened independent of IL-4 treatment. Motif distributions indicate that only the PU.1 motif frequency follows this order (Group 1 to 4), TRE is more or less equally distributed throughout cluster 2, and interestingly, EGR2 motifs show a negative correlation with PPARγ dependency (Supplementary Figure S4B). Although DR1 is not highly enriched in Group 2, we still see a robust effect of PPARγ on RXR binding and this result is in agreement with the ATAC-seq data in some ways (Figure 4A and C). As both KOs have major effects on the genomic regions in Group 4, we can assume that this is due to the high frequency of DR1 elements and the direct DNA-binding by the receptors. In contrast, in Group 2 we can probably see indirect binding events to a greater extent.
Figure 4.
The appearance of PPARγ is a major component in the genome-wide redistribution of RXR. (A) Read distribution plots of RXR and PPARγ signals from wild type (WT) and Pparg knockout (PγKO) macrophages in the presence (+) or absence (–) of IL-4 as determined by ChIP-seq (left). RXR sub-clusters shown on Figure 3 panel A were further divided based on the dependence of RXR binding on the presence of PPARγ (Pγ-dep.) and STAT6 binding (S6-bound). The number of genomic regions within the four groups (Groups 1–4) is indicated (middle). Read distribution plots of ATAC-seq signals from wild type (WT), Rxr (RxrKO) and Pparg knockout (PγKO) macrophages in the presence (+) or absence (–) of IL-4 (right). 1.5-kb regions are represented in 30-bp bins. (B) Genome browser view of examples for PPARγ-independent (1–5.) and PPARγ-dependent RXR sites (6–10.): 1. chr18:88820090–88820456, 2. chr8:46868575–46869011, 3. chr11:70114345–70114844, 4. chr11:70114345–70114864, 5. chr17:35809550–35809982, 6. chr9:72485489–72485854, 7. chr3:9556170–9556746, 8. chr13:113138467–113138843, 9. chr16:93732883–93733288, 10. chr2:134730555–134731000. (C) Box plots depicting the normalized tag density of ATAC-seq data for each group in the de novo RXR cluster as shown on panel A. Percentages represent the contribution of the knocked-out nuclear receptor to chromatin openness, calculated based on the median values for the IL-4-treated samples. (D) ChIP-qPCR-based determination of the binding modalities of RXR (upper panel) and PPARγ (bottom panel) in Rxra/b (RXR KO), Pparg (PγKO) and Stat6 knockout (S6KO) macrophages in the presence or absence of IL-4. Asterisk represents significant difference determined by two-tailed unpaired t test at P< 0.05 (n = 2).
These results prompted us to carry out ChIP-qPCR experiments to validate and study the relationship between RXR, PPARγ and STAT6 at individual enhancers using Rxra/b, Pparg and Stat6 KO immortalized bone marrow-derived macrophages (iBMDMs). ChIP-qPCR experiments on a select set of enhancers (Angptl4 +2.2 kb, Tgm2 –20 kb, Abcg1 –22 kb, Fabp4 –5.3 kb and Pou5f1 –2.9 kb) revealed that the IL-4-mediated recruitment of PPARγ and RXR is completely dependent on STAT6. The receptors’ recruitment also exhibited mutual dependence on each other's presence, but PPARγ binding was less sensitive to the absence of RXR (Figure 4D). Finally, investigation of the total RXRα protein levels in Pparg KO macrophages revealed that the loss of Pparg does not affect RXRα expression at the protein level (Supplementary Figure S4D).
Altogether, these results establish a hierarchy between these factors and show that PPARγ is a major component in the extension of the RXR cistrome, though other mechanisms triggered by STAT6 may also play significant roles.
Loss of Rxra/b does not affect alternative polarization on a select set of genes
Since our ATAC-seq results indicated that RXRα/β proteins affect chromatin structure, we made an attempt to elucidate whether RXR has a prominent role in the IL-4/STAT6-mediated induction of a select set of M2 polarization marker genes based on recent reports of macrophage polarization (29,30). We polarized primary BMDMs from WT and Rxra/b KO bone marrows and exposed the cells to IL-4 for 24 h. We did not find significant differences in the IL-4-induced expression of Chil3, Irf4, Klf4, Mrc1, Egr2 and Batf3, but we observed significantly lower Retnla mRNA expression in the absence of Rxra/b (Supplementary Figure S4E).
Altogether, we concluded that RXRα/β is not required for the proper induction of this M2 polarization-related gene panel, but further, unbiased analyses are required to clarify the role of the receptor in polarization in its entirety.
Identification of the ligand-activated gene network by the PPARγ:RXR heterodimer in alternatively polarized macrophages
After providing mechanistic details of RXR redistribution throughout the genome, we aimed to identify the target genes of this extended regulatory network. Mapping RNAPII-pS2 by ChIP-seq in the presence of LG268 (RXR agonist) or RSG (PPARγ agonist) revealed the affected gene bodies. We grouped the significantly changing gene bodies based on their ligand sensitivity, and created four gene groups based on a fold change cut-off of two-fold relative to the control cells. Genes exhibiting an at least two-fold induction in the presence of the RXR or PPARγ ligand were considered preferentially responsive to LG268 (‘RXR ligand-preferred’, n = 31) or RSG (‘PPARγ ligand-preferred’, n = 41). If both ligands were able to drive the gene body enrichment of RNAPII by at least two fold, then those were termed ‘Permissive’ genes (n = 29) (Figure 5A, B). We also noted the presence of repressive effects by the ligands, but these were negligible in number compared to the activated ones (‘Repressed’; n = 16) (Supplementary Figure S5A). Gene body enrichment analysis of RNAPII-pS2 further corroborated our findings (Figure 5C and Supplementary Figure S5B) along with our Global Run-On sequencing (GRO-seq) results representing the nascent RNA level in the same treatment paradigm, which positively correlates with the RNAPII-pS2 data on the regulated genes (Figure Supplementary Figure S5C). Furthermore, Ingenuity Pathway Analysis (IPA) revealed the potential functional consequences of the activated and repressed gene programs mediated by the various ligands (Supplementary Figure S5D–G). Interestingly, permissive genes appeared to be responsible for lipid uptake and concentration inside the cells, while preferentially RXR ligand-mediated genes showed enrichment for the regulation of cholesterol flux (Supplementary Figure S5D and E). In addition, preferentially PPARγ ligand-regulated genes seem to be responsible for beta-oxidation and the accumulation of very long-chain fatty acids (Supplementary Figure S5F). In contrast, genes repressed by LG268 appear to be regulating the migration and inflammatory responsiveness of the cells (Supplementary Figure S5G). For validation purposes we measured the expression of the following genes from the ligand-activated gene groups in the presence of both LG268 and RSG at the mRNA level: Permissive—Angptl4, Cd36, Plin2, Cpt1a; RXR ligand-preferred—Abca1, Scd2, Cxcl14, Abcg1; PPARγ ligand-preferred—Fabp4, Sort1, Cfa2t3. These analyses confirmed the ligand preference of the gene groups we identified based on RNAPII-pS2 enrichments and also confirmed that permissive genes are indeed exhibit sensitivity to both nuclear receptor ligands. Of note, we observed antagonism between the two ligands in the PPARγ ligand-preferred group (Fabp4 and Cbfa2t3), while RXR ligand-preferred genes do not show any signs of permissiveness (Supplementary Figure S5D–F).
Figure 5.
Characterization of the genome-wide effects of ligand-responsive PPARγ:RXR heterodimers in IL-4-polarized macrophages. (A) Heat maps representing the significantly changed, ligand-sensitive genes. Ligand dominance was calculated based on a 2-fold cut-off threshold. Permissive genes show an at least 2-fold induction to both ligands, while the RXR and PPARγ ligand-preferred genes were induced by at least 2-fold only in the presence of either ligand. (B) Genome browser views of permissive (Angptl4), RXR ligand-dominated (Tgm2) and PPARγ ligand-preferred (Fabp4) genes. ChIP-seq experiments for RXR, PPARγ and the elongation-specific form of RNAPII (RNAPII-pS2) under the indicated conditions are shown. (C) Metagene plot representation of RNAPII-pS2 signals on the gene bodies of permissive, RXR ligand-preferred and PPARγ ligand-preferred gene groups defined on panel A. Coverage is defined as read count per million mapped reads. Transcription start (TSS) and termination (TTS) sites are indicated.
Our analyses identified the target gene bodies/networks of ligand-mediated PPARγ:RXR heterodimers and point to the presence of preferential ligand effects by the receptors, which appear to represent functionally distinct cellular programs.
Mapping RNAPII-pS2 on PPARγ:RXR-bound genomic elements allows the identification of ligand-responsive heterodimers
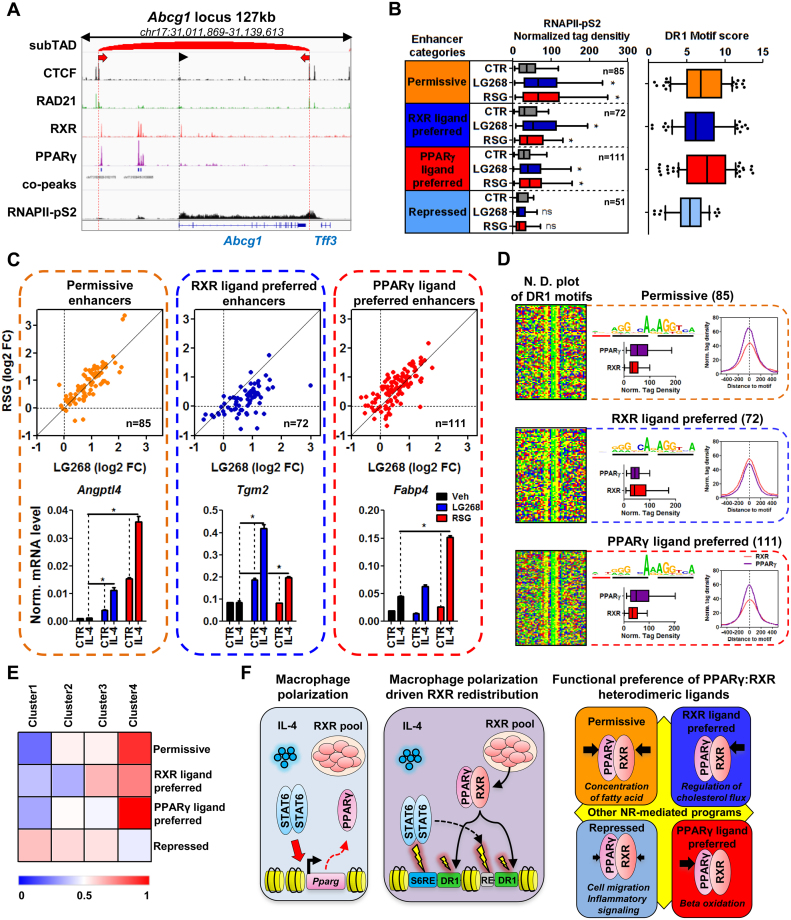
Finally, we decided to annotate PPARγ:RXR heterodimer-bound regions to ligand-responsive genes. Using ChIP-seq experiments for two main architectural proteins, CTCF and RAD21, we predicted the sub-(topologically associating domain) (subTAD) structure of macrophages. We applied an algorithm that utilizes both the insulator coverage and direction, which have been shown recently as indicators of topological domain borders (3,34,35). Next we assigned all PPARγ:RXR co-bound regulatory regions to those genes falling into the same subTAD (Figure 6A). As a result, we annotated 85 permissive, 72 RXR ligand-preferred, 111 PPARγ ligand-preferred and 52 repressed PPARγ:RXR-bound regulatory regions. Plotting the density of RNAPII-pS2 at these genomic loci almost fully recapitulated the changes we observed on the target gene bodies except for enhancers preferring PPARγ ligand-activation, which exhibited an LG268-mediated induction as well, but as expected these sites were still more responsive to RSG (Figure 6B left, C). The repressive effects of the ligands were not significant on the regulatory regions near repressed genes, but they showed a trend towards repressed RNAPII-pS2 binding (Figure 6B and Supplementary Figure S6A). The distribution and plotting of DR1 motif scores reported that the strongest motifs are located within the highly permissive and PPARγ ligand-preferred sites, followed by the RXR ligand-preferred regions, and the repressed regions possessed the weakest motifs (Figure 6B, Supplementary Figure S6B). Validation of the consequences of altered RNAPII-pS2 enrichment on the various ligand-responsive enhancer categories at the mRNA level of target genes (Angptl4—permissive; Vegfa—RXR ligand-preferred; Fabp4—PPARγ ligand-preferred) showed that the changing level of RNAPII-pS2 on the enhancers is a reliable indicator of altered gene expression, moreover at these genes we observed an enhanced ligand responsiveness in IL-4-stimulated cells, as a result of the reorganized PPARγ:RXR-bound regulatory regions (Figure 6C).
Figure 6.
IL-4-mediated macrophage polarization allows the remodeling of enhancer activity in a PPARγ:RXR heterodimer ligand-dependent manner. (A) Genome browser view of the Abcg1 locus in order to represent the main features of our method to annotate enhancers to the gene groups defined on Figure 5, panel A. ChIP-seq tracks are shown for CTCF, RAD21, RXR, PPARγ and RNAPII-pS2. The predicted subTAD for this locus is presented, with the direction of the CTCF elements (two red arrows). PPARγ:RXR co-peaks are also shown. Dashed black line with an arrowhead represents the transcription start site of Abcg1. (B) Box plot depicting normalized tag density on those enhancers annotated to the gene groups defined on Figure 5 panel A. The number of enhancers in each category is also indicated. Asterisk represents significant difference determined by two-tailed paired t test at P< 0.0001 (left). Box plot representing DR1 motif scores in each enhancer category (right). (C) Scatter plots depicting RNAPII-pS2 signals on individual enhancers from each enhancer group in the presence of rosiglitazone (RSG) and LG268. Cells were polarized with IL-4 for 24 h followed by 1 h of RXR- (LG268) and PPARγ-ligand (RSG) exposure. Log2 fold change over control is presented. Gene expression measurements of Angptl4, Vegfa and Fabp4 genes in control (CTR) and IL-4 polarized macrophages in the absence (ligand control – Veh) or presence of RSG or LG268. Asterisk represents significant difference determined by two-tailed unpaired t test at P< 0.05 (n = 3). (D) Nucleotide distribution (N.D.) plots from each enhancer category in a 50-bp window around the DR1 motifs. Enriched DR1 motifs are also indicated with the number of regions along with the normalized tag density for PPARγ and RXR in each enhancer category in the presence of IL-4 represented as boxplots and histograms. Normalized tag density on the histograms are represented around the DR1 motif in a ±500 bp window (right). (E) Heat map depicting the enrichment of each enhancer groups in the clusters defined on Figure 1 panel B. (F) Graphical representation about the reorganization of the RXR cistrome and its ligand-sensitive action upon macrophage polarization. IL-4 triggers the activation of STAT6, which upregulate Pparg. There is a significant RXR pool in the cells at the steady-state, which is not modulated by IL-4 (left panel). During the polarization process, STAT6 directly (S6RE – STAT6 response element) initiates the formation of de novo regulatory regions for the PPARγ:RXR heterodimer (DR1 – direct repeat 1). The presence of PPARγ is required for the extension of the RXR cistrome, but there are secondary, unknown factors, which possess major roles binding to their response elements (grey box, RE). Yellow lightnings indicate the chromatin opening capacity of the given factor and the size of the lightning indicates the effect of the factor on chromatin opening (middle panel). The extended PPARγ:RXR heterodimer exhibit different ligand sensitivity towards RSG and LG268, which will result in the activation and repression of functionally distinct gene programs. Typically, de novo PPARγ:RXR sites are not responsive to ligand, suggesting a significant, unknown receptor function, which needs to be explored (right panel).
Motif enrichment analysis focusing on the DR1 motif revealed slight differences between the differentially ligand-sensitive PPARγ:RXR heterodimers. We noted the presence of an extended DR1 motif at PPARγ ligand-dominated enhancers. These sites typically exhibit an ANT extension on the PPARγ side (5′) of the DR1. The RXR side (3′) is more or less similar between the categories, although it is weaker in the repressed group (Figure 6D). Analysis of the binding of PPARγ:RXR heterodimers in the enhancer categories showed that ligand preference is associated with higher occupancy for PPARγ and RXR on the PPARγ and RXR ligand-preferred enhancers, respectively. Interestingly, PPARγ ligand sensitivity and the strength of binding followed the observed, extended DR1 motif, while RXR binding was largely invariant, except for the repressed sites, which exhibited low occupancy for both PPARγ and RXR (Figure 6D). These results suggest that the DNA sequence itself might have a deterministic role in regulating ligand responsiveness, which needs to be further investigated in future studies.
Finally, we were wondering about the localization of these ligand-responsive enhancer categories with regards to the clusters defined on Figure 1B. Surprisingly, ligand-activated enhancers were the most enriched in Cl4 and the least enriched in Cl1, while the repressed sites showed the opposite distribution (Figure 6E). It is important to note that de novo RXR sites in Cl2 do not seem to exhibit strong ligand sensitive action.
These results indicate that ligand-sensitive receptor action might be mediated by the genomic environment (sequence or epigenomic environment) and the redistribution of RXR heterodimers does not necessarily give rise to exogenous ligand-operated genomic switches.
DISCUSSION
The development and signaling operations of heterodimeric nuclear receptors are not well understood for the following reasons (1) they bind chromatin and influence transcription in the absence of ligand, (2) they bind as RXR heterodimers allowing for distinct and complex ligand activation modes. In this study we aimed to investigate the RXR-bound regulatory sites of alternatively polarized macrophages. The biological significance of our model is that IL-4-mediated polarization has a key role in anti-helminth defense (36) and also in certain repair-type macrophages, including those recently identified peritoneal macrophages responding to sterile inflammation in the abdominal cavity, and tissue-specific macrophages such as alveolar macrophages (37,38). At the same time it appears that lipid ligand-regulated transcription factors (NRs) also have key roles in macrophage function and potentially in cell type specification as well (11–14). The biological importance of these NRs has been appreciated due to their critical roles in lipid metabolism, immune responses and also cellular differentiation (39). RXR acts as a promiscuous heterodimerization partner of several NRs and our previous results show that among others it controls lipid metabolism (40) and angiogenesis (3), representing an independent signaling pathway in resting macrophages. An intriguing piece of observation from our previous study is that the number of RXR-bound sites is not changing significantly upon ligand activation (3). This is in stark contrast with how steroid hormone receptors function by occupying the genome rapidly in the presence of their ligands (41–43). Our results bring novel mechanistic insights by showing that the polarization process extensively remodels the RXR cistrome and formally suggests that the RXR cistrome is determined by the cellular state rather than the activating ligand signal.
Molecularly, IL-4-activated STAT6 initiates the formation of the so-called de novo (17) or latent enhancers (16), which serve as molecular beacons for the newly formed RXR heterodimer signaling. Recently, a somewhat similar crosstalk has been identified between the inflammatory signal induced NF-κB and the ligand-dependent estrogen receptor alpha (ERα)-mediated signaling in breast cancer cells, showing that inflammatory stimuli can pave the way for estrogen signaling, establishing latent enhancers (44). However, ligand activation of ER leads to thousands of new binding sites in the genome within minutes (44). In contrast, RXR occupies the genome of macrophages in the steady state and works as a genome-bound lipid sensor. Importantly, exogenous ligand addition is not necessary for the IL-4-mediated extension of the RXR cistrome, although we cannot exclude the role of endogenous ligands here. Nonetheless, it appears to be the heterodimeric partner, which has dominant effects on the positioning of RXR. Upon polarization, STAT6 induces the level of PPARγ, thus attracting RXR to DR1 motifs in the genome. Surprisingly, >60% of the STAT6-dependent de novo RXR sites are established in a PPARγ-dependent manner. The remaining 40% may account for the effect of other transcription factors downstream in the polarization process (secondary wave), most likely EGR2 and AP-1. Importantly, the newly deposited PPARγ:RXR heterodimers contribute to the development of the accessible chromatin profile of polarized macrophages, although these factors are not indispensable for de novo enhancer formation (Figure 6F).
Analysis of the binding kinetics of STAT6, RXR, PU.1, P300 and RAD21 at de novo RXR sites revealed that STAT6 is the first recruited factor as expected, and followed by PU.1, RXR, P300 and RAD21. The formation of de novo RXR-bound enhancers is carried out by the direct and indirect actions of STAT6, including the induction of a second wave of transcription factors like PPARγ. Importantly, RXR is not part of the ‘second wave’, because its protein level is not changing and it is most likely recruited from a non-chromatin-bound pool by STAT6 and its partner PPARγ or potentially by other polarization-specific transcription factors. In addition, STAT6 directly affects the extension of the RXR cistrome at those genomic regions, which are primed by PU.1 and other regulatory factors at the basal state.
As the polarization process is nearing completion, one would expect that the newly established PPARγ:RXR heterodimer cistrome confers vastly reprogrammed lipid responsiveness to the cells, but this is not exactly the case here. It is important to note that we observed enhanced ligand responsiveness on some of the ligand-sensitive genes in alternatively polarized macrophages (Figure 6C) and though there is a significant number of ligand-responsive genes, which are predicted to control lipid metabolism, cholesterol flux and uptake of fatty acids, the vast majority of the PPARγ:RXR-bound regulatory regions may work in a ligand-independent manner and their functional importance is unknown.
Recently, we reported an unusual synergy between the activating signals of RXR and STAT6. Interestingly, on a select set of genes ligand activation of RXR alone is not sufficient to trigger gene activation, while STAT6 is capable of doing so. Surprisingly, when both stimuli are present, synergistic gene activation was observed. These results suggest the structural importance of ligand-bound RXR in facilitating another signaling pathway, but insufficient to drive transcription alone (50). The mechanistic details of this phenomenon remained enigmatic and will require additional studies in our model as well.
The integrative approach used here allowed us to functionally characterize the effects of the dominant ligands of PPARγ and RXR. As a result, we observed ligand-selective gene sets from which we predicted ligand-specific cellular functions (Figure 6F), which still attracts attention due to the lack of knowledge about the genome-wide effects of nuclear receptor ligands.
Taken together, here we provide new molecular and mechanistic insights into the polarization process of macrophages and show that the dynamic reorganization of the RXR heterodimer cistrome is a hallmark of this transition, leading to enhancer reprogramming and cistrome extension. The altered set of regulatory regions provides additional options for the cells to extend their functional characteristics and interact with their environment, but there seems to be a large number of heterodimers that are inert to exogenous ligand stimulation, which needs attention and may lead to the reinterpretation of the mechanism of action by nuclear receptors.
DATA AVAILABILITY
Sequencing data have been submitted to GEO under accession number: GSE110465.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the members of the Nagy laboratory for discussions and comments on the manuscript. We thank Peloquin, Cseh, Hathy and Beladi for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hungarian Scientific Research Fund [OTKA K124298, K126885, K116855 to L.N.]; Sanford Burnham Prebys Medical Research Institute; Center of Clinical Genomics and Personalized Medicine of the University of Debrecen; Centre National de Genotypage (CNG) Evry by Steven McGinn, Anne Boland, Doris Lechner and Marie Thérèse Bihoreau and supported by the European Sequencing and Genotyping Infrastructure (funded by the European Commission, FP7/2007–2013) [26205] (ESGI), as part of the ADIPOMACTX transnational access program and also at the Analytical Genomics Core Facility at the Sanford Burnham Prebys Medical Discovery Institute; American Heart Association (AHA) postdoctoral fellowship [17POST33660450 to B.D.]; Hungarian Scientific Research Fund [OTKA PD124843 to N.G.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Evans R.M., Mangelsdorf D.J.. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014; 157:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nunez V., Alameda D., Rico D., Mota R., Gonzalo P., Cedenilla M., Fischer T., Bosca L., Glass C.K., Arroyo A.G. et al. Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daniel B., Nagy G., Hah N., Horvath A., Czimmerer Z., Poliska S., Gyuris T., Keirsse J., Gysemans C., Van Ginderachter J.A. et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014; 28:1562–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sucov H.M., Dyson E., Gumeringer C.L., Price J., Chien K.R., Evans R.M.. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994; 8:1007–1018. [DOI] [PubMed] [Google Scholar]
- 5. Kastner P., Grondona J.M., Mark M., Gansmuller A., LeMeur M., Decimo D., Vonesch J.L., Dolle P., Chambon P.. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994; 78:987–1003. [DOI] [PubMed] [Google Scholar]
- 6. Qu L., Tang X.. Bexarotene: a promising anticancer agent. Cancer Chemother. Pharmacol. 2010; 65:201–205. [DOI] [PubMed] [Google Scholar]
- 7. Dawson M.I., Xia Z.. The retinoid X receptors and their ligands. Biochim. Biophys. Acta. 2012; 1821:21–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roszer T., Menendez-Gutierrez M.P., Cedenilla M., Ricote M.. Retinoid X receptors in macrophage biology. Trends Endocrinol. Metab.: TEM. 2013; 24:460–468. [DOI] [PubMed] [Google Scholar]
- 9. Ma F., Liu S.Y., Razani B., Arora N., Li B., Kagechika H., Tontonoz P., Nunez V., Ricote M., Cheng G.. Retinoid X receptor alpha attenuates host antiviral response by suppressing type I interferon. Nat. Commun. 2014; 5:5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kiss M., Czimmerer Z., Nagy L.. The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: from physiology to pathology. J. Allergy Clin. Immunol. 2013; 132:264–286. [DOI] [PubMed] [Google Scholar]
- 11. Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I.. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014; 159:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014; 159:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. A-Gonzalez N, Guillen J.A., Gallardo G., Diaz M., de la Rosa J.V., Hernandez I.H., Casanova-Acebes M., Lopez F., Tabraue C., Beceiro S. et al. The nuclear receptor LXRalpha controls the functional specialization of splenic macrophages. Nat. Immunol. 2013; 14:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., Kopf M.. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 2014; 15:1026–1037. [DOI] [PubMed] [Google Scholar]
- 15. Glass C.K., Natoli G.. Molecular control of activation and priming in macrophages. Nat. Immunol. 2016; 17:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S., Curina A., Prosperini E., Ghisletti S., Natoli G.. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013; 152:157–171. [DOI] [PubMed] [Google Scholar]
- 17. Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013; 51:310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lefterova M.I., Steger D.J., Zhuo D., Qatanani M., Mullican S.E., Tuteja G., Manduchi E., Grant G.R., Lazar M.A.. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol. 2010; 30:2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R., Koder A., Evans R.M.. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999; 4:585–595. [DOI] [PubMed] [Google Scholar]
- 21. Gandino L., Varesio L.. Immortalization of macrophages from mouse bone marrow and fetal liver. Exp. Cell Res. 1990; 188:192–198. [DOI] [PubMed] [Google Scholar]
- 22. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. bioinformatics. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daniel B., Balint B.L., Nagy Z.S., Nagy L.. Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Methods Mol. Biol. 2014; 1204:15–24. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jablonski K.A., Amici S.A., Webb L.M., Ruiz-Rosado Jde D., Popovich P.G., Partida-Sanchez S., Guerau-de-Arellano M.. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015; 10:e0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roy S., Schmeier S., Arner E., Alam T., Parihar S.P., Ozturk M., Tamgue O., Kawaji H., de Hoon M.J., Itoh M. et al. Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics. Nucleic Acids Res. 2015; 43:6969–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A.W. et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007; 447:1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szanto A., Balint B.L., Nagy Z.S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R.M. et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010; 33:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bouhlel M.A., Derudas B., Rigamonti E., Dievart R., Brozek J., Haulon S., Zawadzki C., Jude B., Torpier G., Marx N. et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007; 6:137–143. [DOI] [PubMed] [Google Scholar]
- 34. Sofueva S., Yaffe E., Chan W.C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S.M., Schroth G.P., Tanay A., Hadjur S.. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013; 32:3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jenkins S.J., Ruckerl D., Cook P.C., Jones L.H., Finkelman F.D., van Rooijen N., MacDonald A.S., Allen J.E.. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011; 332:1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Kubes P.. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell. 2016; 165:668–678. [DOI] [PubMed] [Google Scholar]
- 38. Jones C.V., Williams T.M., Walker K.A., Dickinson H., Sakkal S., Rumballe B.A., Little M.H., Jenkin G., Ricardo S.D.. M2 macrophage polarisation is associated with alveolar formation during postnatal lung development. Respir. Res. 2013; 14:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagy L., Szanto A., Szatmari I., Szeles L.. Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response. Physiol. Rev. 2012; 92:739–789. [DOI] [PubMed] [Google Scholar]
- 40. Szanto A., Narkar V., Shen Q., Uray I.P., Davies P.J., Nagy L.. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004; 11(Suppl. 2):S126–S143. [DOI] [PubMed] [Google Scholar]
- 41. Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F. et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006; 38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 42. Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L.. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011; 145:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011; 474:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franco H.L., Nagari A., Kraus W.L.. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell. 2015; 58:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagy L., Schwabe J.W.. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 2004; 29:317–324. [DOI] [PubMed] [Google Scholar]
- 46. Barta E. Command line analysis of ChIP-seq results. EMBnet.journal. 2011; 17:13–17. [Google Scholar]
- 47. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 48. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Czimmerer Z., Nagy Z.S., Nagy G., Horvath A., Silye-Cseh T., Kriston A., Jonas D., Sauer S., Steiner L., Daniel B. et al. Extensive and functional overlap of the STAT6 and RXR cistromes in the active enhancer repertoire of human CD14+ monocyte derived differentiating macrophages. Mol.Cell. Endocrinol. 2017; doi:10.1016/j.mce.2017.07.034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been submitted to GEO under accession number: GSE110465.