Abstract
ParA and ParB homologs are involved in accurate chromosome segregation in bacteria. ParBs participate in the separation of ori domains by binding to parS palindromes, mainly localized close to oriC. In Pseudomonas aeruginosa neither ParB deficiency nor modification of all 10 parSs is lethal. However, such mutants show not only defects in chromosome segregation but also growth retardation and motility dysfunctions. Moreover, a lack of parB alters expression of over 1000 genes, suggesting that ParB could interact with the chromosome outside its canonical parS targets. Here, we show that indeed ParB binds specifically to hundreds of sites in the genome. ChIP-seq analysis revealed 420 ParB-associated regions in wild-type strain and around 1000 in a ParB-overproducing strain and in various parS mutants. The vast majority of the ParB-enriched loci contained a heptanucleotide motif corresponding to one arm of the parS palindrome. All previously postulated parSs, except parS5, interacted with ParB in vivo. Whereas the ParB binding to the four parS sites closest to oriC, parS1-4, is involved in chromosome segregation, its genome-wide interactions with hundreds of parS half-sites could affect chromosome topology, compaction and gene expression, thus allowing P. aeruginosa ParB to be classified as a nucleoid-associated protein.
INTRODUCTION
The segregation of the bacterial genomes is a precise, complex and still only partially understood process. Most what is known about chromosome segregation is based on studies on low-copy-number plasmids in which partitioning systems have been identified (1,2). Despite their diversity, all these systems contain two protein components: A, an NTPase securing energy supply, and B, a DNA binding protein, and the third vital element, a specific DNA motif designated centromere-like sequence (parC/parS), recognized and bound by the B component (2–4). The discovery of a parA-parB operon located close to origin of replication (oriC) and encoding homologs of plasmid partitioning proteins of class IA in the majority of the bacterial chromosomes sequenced has suggested its potential role in chromosome segregation (5–8).
The chromosomal ParAs are Walker-type adenosine triphosphatases lacking the specific DNA binding domain present in the N-terminal part of the plasmid ParAs (9,10). The chromosomal ParBs, similarly to their plasmidic counterparts, contain an extended helix-turn-helix (HTH) motif, a C-terminal dimerization domain and an N-terminal polymerization domain (4,11–17) and have an ability to spread around the parS sequences (17–20) and silence nearby genes, at least in test plasmids (14). The chromosomal Par homologs, despite their high conservation, exhibit various features in different species. Deletion of parA-parB is lethal in some species, e.g. Caulobacter crescentus and Myxococcus xanthus (21,22), whereas in other species par mutants are viable but show various severity defects of chromosome segregation in the vegetative and/or sporulation phase of growth, e.g. in Bacillus subtilis, Streptomyces coelicolor, Mycobacterium smegmatis, Vibrio cholerae, Streptococcus pneumoniae, Corynebacterium glutamicum and Pseudomonas aeruginosa (23–37).
Chromosomal ParBs recognize multiple (from 2 to 20) parS sequences, highly conserved across the bacterial kingdom and localized mainly in the chromosomal ori domains (defined as 20% around oriC) (8,11,38). They form large nucleoprotein complexes organizing newly replicated ori domains and, with the help of ParAs, separate these domains and hold them in defined locations near the poles until the end of replication and cytokinesis (24,30,39–45). It has been shown that in B. subtilis and S. pneumoniae ParB-bound parS sites act as loading platforms for structural maintenance of chromosomes (SMC) complexes facilitating condensation of ori domains and subsequently of all newly replicated DNA (33,46–50). Two major hypotheses of how the ParBs of Class IA build the large nucleoprotein complexes around the parS sequences have been put forward. Both are based on the data showing that ParBs bind to the palindromic parS sequences as dimers but can also spread laterally along DNA due to dimer–dimer interactions and weak interactions with non-specific DNA (20,51–53). In the looping and bridging model, ParB dimers bound to DNA interact at a distance, which leads to further DNA compaction (20,54). In the nucleation and caging model, ParB bound to parS acts as a nucleation center in which protein–protein and protein–nsDNA interactions spatially entrap additional ParB molecules thus forming a large nucleoprotein complex (52).
Besides their involvement in genome segregation, chromosomal ParAs and ParBs have also been shown to influence bacterial growth and cell cycle by affecting DNA replication and cell division, competence or motility (26,27,32,35,36,39,40,55).
In addition to their main function in plasmid partitioning, plasmidic ParBs of class IA may act as global transcriptional regulators by binding to their operators, spreading and non-discriminately silencing nearby promoters, e.g. in the IncP-1 plasmids (56,57) or IncU plasmids (58). In contrast, oligomers of chromosomal ParBs spread by up to 20 kb from the parSs (18–20,54) under native conditions but seem to affect gene expression very selectively. A comparison of the transcriptomes of B. subtilis wild-type (WT) strain and spo0Jnull mutant (Spo0J is a homolog of ParB) did not reveal significant changes in the expression of genes located around 10 parS sites (19). Transcription of only two out of twenty genes close to oriC and a few outside a cluster of three parS sites was affected in a ΔparB1 mutant of V. cholerae (59). A similar limited regulation was also observed in S. pneumoniae (55) for the competence operon com, one of the operons localized in proximity of a parS cluster.
Pseudomonas aeruginosa is exceptional in this context, since deletion of the parB gene leads to global transcriptional changes affecting more than 1000 genes (60). It remains elusive how ParB of P. aeruginosa affects the expression of so many genes. Ten potential 16-bp palindromic parS sequences have been identified in silico in the genome of PAO1 strain of P. aeruginosa, numbered clockwise, with four of them (parS1–parS4) clustering next to oriC (14). In vitro studies have demonstrated that ParB binds to all ten parSs but with different affinity (61). The highest ParB binding affinity was found toward the perfect palindromic sequences parS2 and parS3, slightly lower toward parS1 and parS4 with one mismatch, and even lower toward the remaining six parSs with two mismatches. parS5 was the poorest binder of ParB in vitro. An analysis of our collection of single and multiple parS mutants has demonstrated that the presence of at least one high affinity site from the parS1–parS4 cluster is necessary and sufficient for proper chromosome segregation (61). The remaining six sites were dispensable for chromosome segregation (61,62).
Here, we used chromatin (nucleoprotein) immunoprecipitation followed by next generation sequencing (ChIP-seq) to analyze the ParB–DNA interactions in P. aeruginosa WT strain, a ParB-overproducing strain and several parS mutants: parSnull with all 10 parS sites modified, parS1-4 mutant with the four parSs of the highest affinity for ParB inactivated and parS2+ with only one high affinity site left intact in the background of the parSnull strain. This allowed us to identify a large number of additional, specific ParB-bound sequences. Consequently, we propose that ParB, having hundreds of defined interaction sites in the genome, represents a new subfamily of nucleoid-associated proteins (NAPs).
MATERIALS AND METHODS
Bacterial strains, growth conditions and plasmid manipulations
Pseudomonas aeruginosa PAO1161 (leu− r−), a derivative of PAO1, was provided by Dr B.M. Holloway (Monash University, Clayton, Victoria, Australia). This strain lacks the inversion between the rrn operons present in the annotated PAO1 genome (63,64). Pseudomonas aeruginosa PAO1161 RifR (WT) strain (35), PAO1161 RifRparBnull (36), PAO1161 RifRparSmut15 with four parS sequences modified (here referred to as parS1-4 mutant), PAO1161 RifRparSmut28 (here referred to as parS2+ mutant) with nine parS sequences modified but with unchanged parS2, and PAO1161 RifRparSnull with all 10 parSs modified (61) were used in the analysis. A ParB-overproducing strain (designated ParB+++) was obtained by transformation of WT strain with pKGB9 (araC-araBADp-parB). Chloramphenicol was added to 75 μg ml−1 (liquid cultures) or 150 μg ml−1 (solid media) to maintain the plasmid and 0.02% arabinose was added to induce the expression of parB (65).
DNA pull-down assay
DNA pull-down analysis was carried out as previously (66) with modifications described in Supplementary Materials and Methods. Biotinylated DNA fragments containing parS1 to parS10 were obtained by polymerase chain reaction (PCR) using appropriate pairs of primers (Supplementary Table S1). ParB binding to the DNA fragments was assessed by detecting ParB in the eluate using western blotting with anti-ParB antibodies (36) and mass spectrometry analysis (Mass Spectrometry Laboratory, IBB PAS).
Nucleoprotein immunoprecipitation (ChIP)
ChIP was performed as described previously (67) with modifications indicated in Supplementary Materials and Methods. Exponentially growing cultures in L broth at 37°C (OD600 ∼0.5) were used. For each biological replicate, two independent 50-ml cultures of each strain were pooled together. The immunoprecipitation was performed with affinity purified rabbit polyclonal anti-ParB antibodies (14).
ChIP-qPCR analysis
The immunoprecipitated DNA was used as a template in quantitative PCR (qPCR) performed with Hot FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne) and primers listed in Supplementary Table S1 in a Roche LightCycler 480. The efficiency of amplification for all primer pairs was between 1.95 and 2.05. The ChIP-qPCR results are presented as ChIP recovery relative to the input DNA used in the procedure, calculated using the formula 100*2∧(CtInput − 9.965 − CtChIP), where Ct are threshold values calculated using the second derivative method with LightCycler 480 software ver 1.5.1.62. Before analysis the input samples were diluted 1000×. Each target sequence was analyzed in samples from at least three biological replicates, each with three technical replicates. Primers amplifying a fragment of proC gene were used to assess non-specific (background) DNA recovery in the ChIP samples.
ChIP-seq analysis
Library preparation and sequencing on Illumina HiSeq 4000 were performed at Genomed S.A. (Poland). Adapter sequences were removed using Cutadapt (68). Subsequent bioinformatic analyses were performed using the Galaxy platform (http://usegalaxy.org/) (69) and R (https://www.R-project.org/). Reads were mapped to the P. aeruginosa PAO1 (NC_002516.2) genome using Bowtie2 (70). ParB peaks were identified by comparing combined replicates of IP samples (treatment) for each strain with combined negative control (parBnull) samples using the callpeak function of MACS2 ver 2.1.1.20160309 (71) with default options for paired-end binary alignment map (BAM) files, and 0.05 as false discovery rate (FDR) cut off. Visualization of data was performed using DeepTools (72), Integrative Genomics Viewer ver 2.3.91 (73) and Sushi (74). Coverage files, normalized to 1× sequencing depth (RPGC), were generated without binning and smoothing using the bamCoverage tool (72). For simplicity of presentation, the coverage values for individual nucleotides in biological replicates were averaged. ComputeMatrix and plotHeatmap components of deepTools (72) were used to prepare the heatmaps. Identification of the DNA motifs enriched in the ParB peaks was performed using MEME-ChIP (75). Differential binding analysis was performed using Diffbind (76). The ChIP sequencing data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE110158.
RESULTS
ParB binding to parS sequences in vitro and in vivo
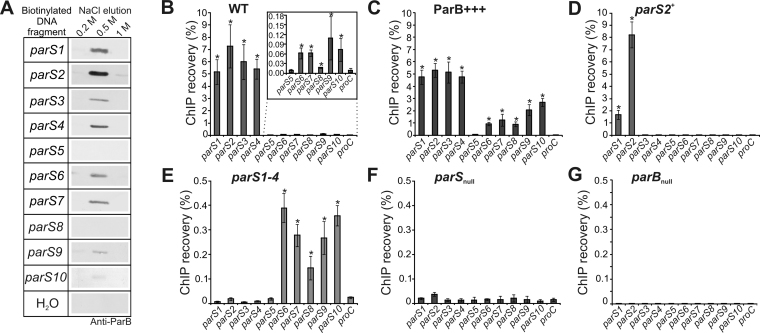
An earlier bioinformatics analysis using the consensus parS sequence of B. subtilis (11) with allowance of two mismatches has led to the identification of ten putative ParB binding sites in the P. aeruginosa PAO1 genome (14). Our previous in vitro studies using electrophoretic mobility shift assay (EMSA) confirmed ParB binding to those sequences and demonstrated the highest affinity of purified His6-ParB to the parS1-parS4 sites, and the lowest to parS5 (61). Since the EMSA was performed with short oligonucleotides and tagged ParB, here we used the DNA pull-down method to analyze the binding of unmodified ParB to different parS sequences in their natural context. Biotinylated DNA fragments (350–450 bp) encompassing the parS sequences were attached to streptavidin-coupled magnetic beads, incubated with PAO1161 WT cell extracts, and washed. Proteins bound to the DNA were eluted and subjected to western blot analysis with anti-ParB antibodies as well as to mass spectrometry analysis. ParB was detected in the elution fractions in all samples containing DNA with any parS sequence with the exception of parS5 and parS8 (Figure 1A). These data indicate that in addition to the parS1–parS4 sequences, required for accurate DNA segregation (61), ParB protein also binds the predicted genomic parS sites: parS6, parS7, parS9 and parS10.
Figure 1.
ParB binding to parS sites in Pseudomonas aeruginosa genome. (A) Western blot analysis of elution fractions from DNA pull-down assays performed with DNA fragments containing parS1–parS10 sequences. Biotinylated DNA was coupled with magnetic beads, incubated with extracts of PAO1161 cells and washed. Proteins bound to the DNA were eluted with increasing concentration of NaCl (0.2–1 M). The negative control contained water instead of biotinylated DNA. Western blotting was performed using polyclonal anti-ParB antibodies. (B–G) qPCR analysis of DNA obtained by nucleoprotein immunoprecipitation using anti-ParB antibodies and extracts from exponentially growing cells of P. aeruginosa strains (B) WT, (C) ParB+++, (D) parS2+, (E) parS1-4, (F) parSnull and (G) parBnull (negative control). qPCR was performed using primers flanking the parS sequences and for the proC gene (background control). Data represent percentage of DNA recovered by ChIP relative to corresponding input samples and are shown as mean ±SD for at least three biological replicates analyzed in three technical replicates. The significance of differences in recovery of parSs relative to the background control (proC) was evaluated by two-sided Student’s t-test assuming equal variance, with P < 0.01 considered as significant (marked by asterix).
To verify the ParB binding to these parS sequences in vivo, ChIP with anti-ParB antibodies followed by quantitative PCR analysis was applied. Since a lack of ParB in P. aeruginosa PAO1161 parBnull mutant produced clear-cut effects under conditions of exponential growth in rich medium at 37°C, but not as severe as during growth in minimal medium (36,77), such milder conditions were used for these experiments. The ChIP procedure was performed with PAO1161 (WT) strain, PAO1161 parBnull mutant (negative control) and PAO1161/pKGB9 (araC-araBADp-parB) strain (ParB+++) with a 5-fold increased ParB level which does not retard bacterial growth (65). Additionally, three parS mutants were included: parSnull with all ten parS sites mutated, parS1-4 mutant with the four high affinity ParB sites inactivated, and parS2+ strain with nine parS sequences modified and only parS2 left intact (61). Despite producing native amounts of ParB, both parS1-4 and parSnull are defective in chromosome segregation to a similar extent as the parBnull mutant (36). The rationale behind using these strains was that it could help to detect ParB binding not only to the known parSs but also to other sites, by either increasing the cellular ParB level or by re-distribution of ParB from the main binding sites involved in chromosome segregation, as shown previously for a parSΔ6 mutant of B. subtilis (19).
DNA recovered after ChIP as well as corresponding input DNA was used in quantitative real-time PCR with primers amplifying fragments encompassing parS1 to parS10 sequences and a fragment of proC gene as a background control (Supplementary Table S1). In WT samples strong ParB binding to parS1 to parS4 was confirmed as manifested by high ChIP recovery (Figure 1B). Additionally, a statistically significant enrichment of sequences encompassing parS6, parS7, parS8, parS9 and parS10, but not parS5, was found (Figure 1B, inset), indicating that under the growth conditions tested ParB did bind in vivo to all the parS sequences with the exception of parS5. Also in the ParB+++ strain ParB was bound to all the parSs but parS5 (Figure 1C). Interestingly, while the regions encompassing parS1 to parS4 were recovered with a similar efficiency as in the WT, the remaining parS sites were significantly more enriched than in WT (Figure 1B and C). As expected, in the parS2+ strain, the region encompassing parS2 showed the highest enrichment (Figure 1D), but also the DNA region around parS1, adjacent to parS2, was significantly enriched, suggesting that ParB bound to parS2 spread to adjacent DNA sequences. In the parS1-4 mutant the fragments containing parS1 to parS4 were not significantly enriched (Figure 1E) but those encompassing parS6 to parS10 were recovered with a higher efficiency than for the WT strain. Finally, no ParB binding to the altered parS sequences was found in the parSnull strain (Figure 1F), and for the parBnull strain (negative IP) the recovery was less than 0.01% for all the sequences analyzed (Figure 1G).
Taken together, these results confirm that the parS1 to parS4 sequences are the major ParB binding sites in P. aeruginosa cells and that they are saturated by ParB in the WT strain. The fact that the lower-affinity sites parS6 to parS10 were also enriched, and that this enrichment was higher in ParB-overexpressing cells and also in cells lacking functional parS1–parS4 sequences, suggests that when a ParB pool not bound to its major targets appears, it may specifically associate with other sites of the bacterial chromosome.
Genome-wide identification of ParB-enriched regions
To gain a global insight into the ParB interactions with the P. aeruginosa chromosome, chromatin immunoprecipitation was combined with next-generation sequencing. The anti-ParB immunoprecipitated DNA from two independent biological samples of each: WT, ParB+++, parS2+, parS1-4, parSnull and parBnull cells was sequenced and the reads were mapped to the P. aeruginosa PAO1 (NC_002516.2) genome. PlotFingerprint analysis indicated the strongest local ChIP enrichment (a large part of the reads mapped to a few genomic bins) for the WT strain and a more dispersed distribution of the ParB-bound DNA regions in strains lacking functional parS1 to parS4 sequences and in the strain overproducing ParB (Supplementary Figure S1A). Some enrichment in specific regions of the genome was observed for the negative control parBnull strain, which was likely due to cross-reactions of the polyclonal antibodies used.
A principal component analysis (PCA) on the mapped reads obtained for the six strains produced three clusters (Figure 2A). WT, ParB+++ and parS2+ formed one group, parS1-4 and parSnull formed a second group, and the parBnull (negative control) strain was clearly separated from those two. Similar grouping was obtained by hierarchical clustering analysis based on correlation coefficients between different samples (Supplementary Figure S1B). Additionally, inspection of the distribution of the sequence reads along the reference genome showed that the reads from the five strains expressing ParB mapped roughly to the same regions which were different from the regions overrepresented in the parBnull strain (Supplementary Figure S1C). Overall, these analyses confirmed that the obtained reads were enriched in sequences specifically bound by ParB.
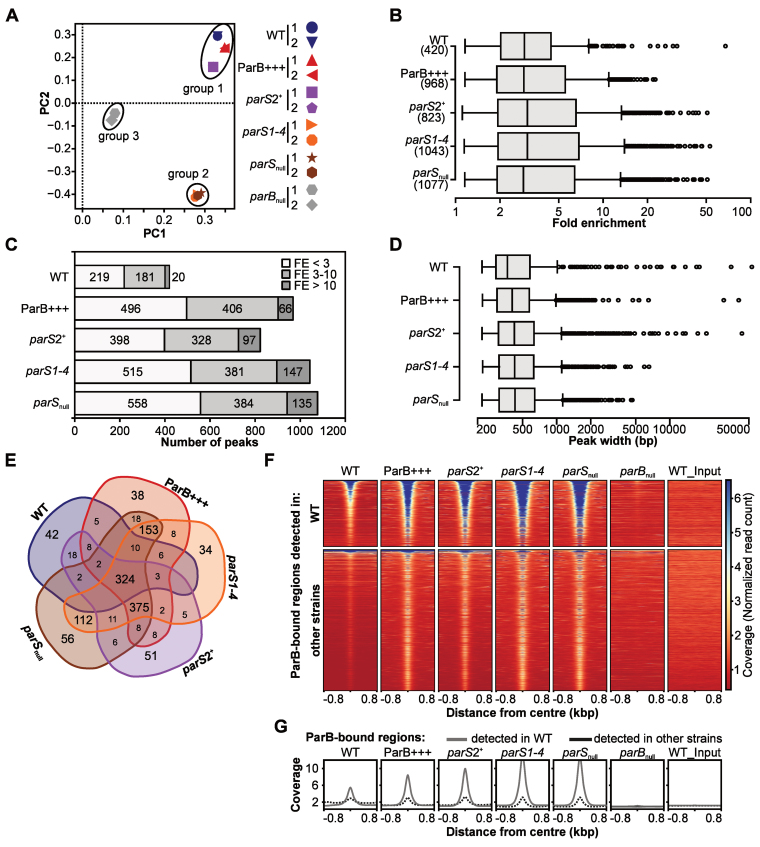
Figure 2.
ChIP-seq analysis of ParB interactions with Pseudomonas aeruginosa chromosome. Reads obtained by ChIP-seq for two biological replicates were mapped to the PAO1 genome using Bowtie2 to generate BAM files, subsequently used in peak calling with MACS2. (A) PCA of BAM files. The identified components represent directions along which the variation in the data (read coverage values) is highest, allowing analysis of variability between the strains and biological replicates of the same strain. (B) Distribution of FE values for detected peaks. (C) Number of ParB ChIP-seq peaks categorized according to their FE. (D) Distribution of width of ParB ChIP-seq peaks. (E) Compilation of ParB-bound regions in five strains. Two peaks in two strains were considered identical if their summits were <200 bp apart. The analysis yielded 1305 unique regions (Supplementary Table S7). (F) Coverage heatmap around centers of ParB-bound regions. The analysis was performed separately for 420 ParB-associated regions detected in WT strain (top) and 885 regions only detected in at least one of the remaining four strains (bottom). Normalized read counts for each nucleotide were averaged for two biological replicates of each strain and colored according to scale. Each line in the heatmap represents ±800 bp around the center of the region. Rows were sorted in descending order of mean coverage value. The same regions analyzed in parBnull and WT input are included as controls. (G) Mean coverage for two groups of analyzed regions (as in F) in different strains.
To identify the sequences corresponding to the ParB binding sites we performed peak calling on merged data for the two biological replicates what was justified by the high correlation of coverage data within each pair for a given strain (Supplementary Figure S1B and C). Using the FDR cut off value of 0.05, 420 specific peaks were found for the WT strain, 968 peaks for ParB+++, 823 for parS2+, 1043 for parS1-4 and 1077 peaks for parSnull (Supplementary Tables S2–6). The peaks identified in the WT displayed fold enrichment (FE) between 1.15 and 67.5, with a median of 2.9 (Figure 2B). A similar median FE (3–3.1) was observed for all the other strains analyzed (Figure 2B). Remarkably, not only the number of peaks but also the number of peaks with FE > 10 was substantially higher in the ParB+++ strain as well as in the strains with the high-affinity parS sites impaired (Figure 2C), indicating feasibility of the approach used for the identification of additional ParB binding sites.
The lengths of the sequences encompassed by the peaks varied greatly between 0.2 kb and about 50 kb in WT, ParB+++ and parS2+ samples (Figure 2D). In parS1-4 and parSnull the broadest peaks reached only 8 and 5 kb, respectively. Nevertheless, the median values of the peak width did not differ substantially among the strains (Figure 2D).
The peaks obtained for the five strains were matched based on the proximity of the peak summits (Supplementary Table S7). Approximately 90% of the corresponding peaks in various strains had their summits <20 bp apart. Overall, the analysis yielded 1305 groups of peaks representing regions with increased ParB occupancy (Supplementary Table S7, hereafter referred to as ParB-bound regions). Of those, 324 were common to all five strains, representing a high-confidence set of ParB-bound regions in the P. aeruginosa genome (Figure 2E and Supplementary Table S7). Three other most numerous pools consisted of: 375 sites identified in four strains (ParB+++ and the three parS mutants), 153 in three strains (ParB+++, parS1-4 and parSnull), and 112 sites common to parS1-4 and parSnull. Less than 10% of the peaks were unique to any single strain.
The ParB-bound regions generally displayed an increased read coverage in their central parts (Figure 2F and G). The 420 regions identified in WT strain (Figure 2F, top) were also occupied by ParB in all the other strains, with the exception of those harboring parS sequences that were inactivated in the respective parS mutants. Importantly, no increased coverage was visible for parBnull or WT_input. A similar inspection of ChIP-seq data for the remaining 885 ParB-bound regions detected only in ParB+++, parS2+, parS1-4 or parSnull revealed that most of these regions also displayed a slight increase of coverage in WT (Figure 2F, bottom and G), suggesting that ParB could also bind to these sites in WT cells. Overall, this data indicated the existence of more than 1000 specific ParB-bound regions in the P. aeruginosa genome. The high reproducibility of these regions in different P. aeruginosa strains implies robust and specific binding of ParB to many more sites beyond the canonical parS sites.
Inspection of ParB ChIP-seq peaks in various strains
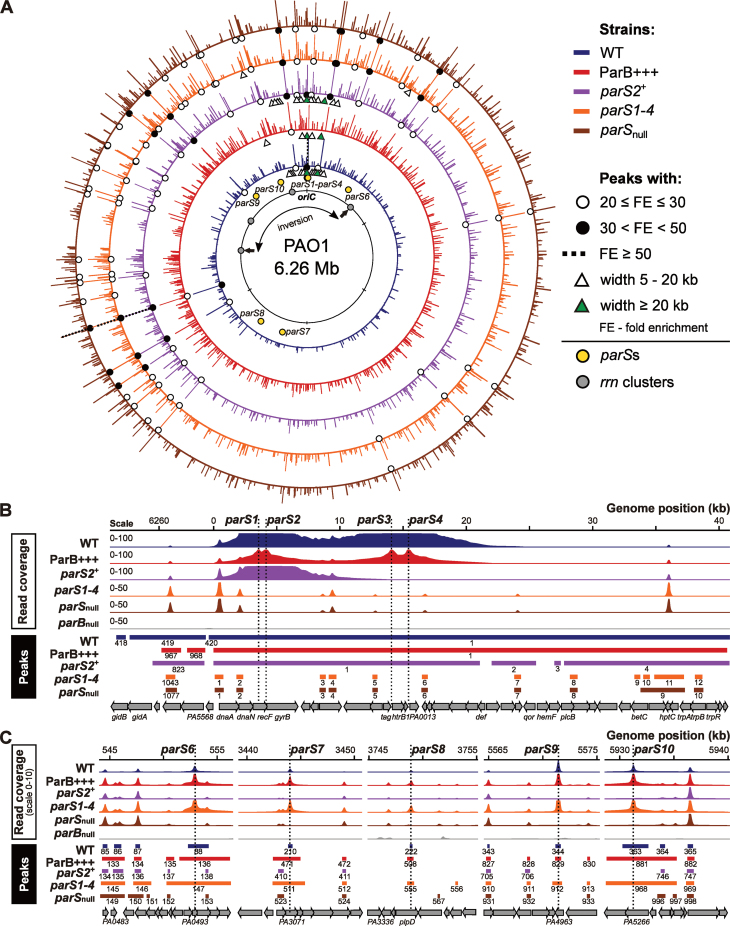
The distribution of the ParB ChIP-seq peaks in various strains was visualized on the P. aeruginosa PAO1 genome (Figure 3A). For clarity, only those with FE > 3 were considered. The ParB peaks were present across the entire genome, but the peaks with the highest FE (marked with dots) were almost exclusively found in the genome half containing oriC. Notably, the broadest peaks (>5 kb), almost exclusively found in only three strains, WT, ParB+++ and parS2+, were tightly clustered in less than ca. 8% of the genome around oriC (triangles in Figure 3A). In the parS1-4 mutant only two peaks broader than 5 kb were detected. Interestingly, these peaks encompass parS6 and parS10.
Figure 3.
Localization and characteristics of ParB ChIP-seq peaks. (A) Distribution of peaks detected in five analyzed strains. Bar height corresponds to peak FE. Only peaks with FE > 3 are shown and all peaks with FE ≥ 30 are shown at the same height. Genomic coordinates are according to the PAO1 genome. The rrnA–rrnD gene clusters are marked with gray dots, arrows indicate rrnA and rrnB loci where inversion occurred in PAO1 (63,64) but not in PAO1161 strain. (B) Coverage of the genome regions encompassing parS1–parS4 cluster and (C) parS6–parS10 sequences in each strain. The histograms show normalized read counts (averaged for two biological replicates) for corresponding positions of the PAO1 genome. Ranges of peaks, their numbers as in Supplementary Tables S2–6 and annotated genomic features are indicated below.
The 20 peaks with FE > 10 identified in the WT (Figure 2C) were analyzed in detail in the four other strains (Table 1). The cluster of four high affinity parS sites (parS1–parS4) in the WT strain produced an extended peak of 53 kb with the maximum FE of 67 (Figure 3B). The corresponding 50-kb peak in ParB+++ strain was enriched only 20-fold. In the parS2+ mutant a peak with the summit at parS2 encompassed 21 kb and had an FE of 33. No such broad peaks were detected in the parS1-4 or parSnull strains but the absence of ParB binding to the parS1–parS4 cluster in these mutants unmasked an additional 15 or 16 peaks, respectively, one of them upstream of dnaA with an FE of ca. 19. The role of the ParB binding in the expression of dnaA operon requires further studies.
Table 1. Selected ParB-bound regions in P. aeruginosa.
| Coordinates in PAO1 genome | Peak fold enrichment (FE) | Locus tag | Additional | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak | Peak | closest | information/ | ||||||||
| Number (WT) | Start | End | Summit | width | WT | ParB+++ | parS2+ | parS1-4 | parS null | to summit | name |
| 1 | 14 | 53 134 | 14 136 | 53121 | 67.5 | 19.7 | 33.5 | nd | nd | PA0011 (ig) | parS1-4 |
| 12 | 123 814 | 158 647 | 147 478 | 34834 | 29.8 | 19.1 | 40.7 | 42.7 | 41.6 | PA0129 (ig) | d |
| 69 | 325 608 | 326 332 | 326 024 | 725 | 13.8 | 18.0 | 28.1 | 33.2 | 31.9 | PA0290 | |
| 94 | 620 609 | 621 160 | 620 843 | 552 | 12.5 | 19.5 | 29.7 | 35.8 | 35.0 | PA0564 | *1 |
| 103 | 713 611 | 714 166 | 713 888 | 556 | 10.4 | 14.3 | 22.7 | 26.5 | 26.6 | PA0660 | |
| 133 | 1 366 827 | 1 367 323 | 1 367 079 | 497 | 10.0 | 13.1 | 21.7 | 26.4 | 24.3 | PA1259 | |
| 166 | 2 354 358 | 2 354 890 | 2 354 602 | 533 | 10.7 | 11.9 | 22.3 | 26.6 | 24.0 | PA2138 | h |
| 225 | 3 839 047 | 3 839 623 | 3 839 369 | 577 | 12.9 | 13.9 | 24.3 | 28.7 | 28.0 | PA3430 | *3 |
| 232 | 3 987 269 | 3 987 755 | 3 987 519 | 487 | 11.0 | 15.5 | 24.2 | 29.7 | 28.0 | PA3559 | |
| 238 | 4 099 347 | 4 099 809 | 4 099 572 | 463 | 11.1 | 15.0 | 27.5 | 32.2 | 31.1 | PA3659 | *2 |
| 245 | 4 170 680 | 4171 338 | 4 171 009 | 659 | 26.4 | 19.8 | 44.4 | 46.4 | 45.4 | PA3725 | g |
| 262 | 4 381 238 | 4 381 931 | 4 381 579 | 694 | 31.7 | 21.8 | 50.8 | 52.9 | 50.8 | PA3911 | f |
| 279 | 4 586 068 | 4 587 049 | 4 586 456 | 982 | 15.7 | 13.4 | 27.3 | 29.1 | 28.6 | PA4102 | e |
| 310 | 5 098 227 | 5 098 844 | 5 098 560 | 618 | 13.4 | 13.0 | 24.4 | 26.4 | 26.5 | PA4550 | a |
| 324 | 5 378 372 | 5 378 982 | 5 378 701 | 611 | 12.7 | 16.2 | 26.9 | 31.8 | 30.8 | PA4790 | |
| 337 | 5 496 504 | 5 497 129 | 5 496 819 | 626 | 12.5 | 17.0 | nd# | nd# | nd# | PA4899 | |
| 338 | 5 498 127 | 5 499 905 | 5 498 743 | 1779 | 20.9 | 19.2 | 38.0 | 41.5 | 40.7 | PA4901 | b |
| 359 | 5 775 771 | 5 776 395 | 5 776 084 | 625 | 11.7 | 15.4 | 23.1 | 27.6 | 27.9 | PA5126 | |
| 398 | 6 140 926 | 6 143 944 | 6 142 506 | 3019 | 23.5 | 22.6 | 42.4 | 46.8 | 46.6 | PA5452 | c |
| 412 | 6 203 326 | 6 217 050 | 6 215 975 | 13725 | 11.6 | 16.1 | 23.8 | 29.0 | 27.6 | PA5523 | |
| 88 | 552 324 | 554 217 | 552 921 | 1894 | 5.8 | 11.1 | nd | 15.5 | nd | PA0493 | parS6 |
| 210 | 3 443 861 | 3 444 275 | 3 444 056 | 415 | 3.6 | 7.5 | nd | 11.1 | nd | PA3071 | parS7 |
| 222 | 3 748 815 | 3 749 078 | 3 748 950 | 264 | 2.2 | 4.6 | nd | 6.2 | nd | PA3339 | parS8 |
| 344 | 5 571 227 | 5 571 683 | 5 571 466 | 457 | 7.8 | 10.9 | nd | 20.9 | nd | PA4963 | parS9 |
| 363 | 5 930 304 | 5 932 676 | 5 931 318 | 2373 | 6.0 | 11.0 | nd | 15.1 | nd | PA5266 | parS10 |
nd: not detected
nd#: not detected as a separate peak but included in the region around PA4901 locus
Shown are parameters of ParB ChIP-seq peaks with FE higher than 10 as determined by MACS2 analysis of ChIP-seq data for P. aeruginosa PAO1161 (WT) strain using parBnull strain as a control. Corresponding peaks in other strains were identified by peak matching (Supplementary Table S7). Data for peaks containing parS6–parS10 sequences are also included. Last column provides names of parS sequences and designations of the ParB binding sites identified before (62).
The remaining 19 highest peaks detected in WT strain were also present in the four other strains and had the FE > 10. Interestingly, in the three parS mutants the accumulation of ParB at these 19 loci was much higher than in WT or ParB+++ (Table 1). Notably, none of these 19 ParB-associated peaks corresponded to known parS sequences. Nevertheless, the parS6 to parS10 sites (but not parS5) were ParB-bound in three strains, with 3- to 4-fold higher ParB occupancy in ParB+++ and parS1-4 strains compared to WT (Table 1 and Figure 3C).
The width of the parS-encompassing peaks varied for different parSs and also depended on the relative availability of ParB. The parS6- and parS10-containing peaks encompassed ca. 2 kb each in WT, but were ca. 6 kb wide when an excess of ParB was available, i.e. in ParB+++ and parS1-4. ParB bound to parS7, parS8 and parS9 occupied ∼400 bp in WT and under conditions of ParB overabundance the ParB-bound region extended significantly (∼3 kb) only for parS7. These data suggest that ParB bound to a parS site can spread for variable distances depending on the individual parS sequence and possibly its context, and the availability of ParB.
Overall, the ChIP-seq analysis demonstrates that in vivo ParB binds not only to the parS sequences identified previously, with the exception of parS5 (Figure 3B and C; Table 1), but also to additional regions across the entire genome creating a specific, highly reproducible pattern of distribution (Figure 3A).
DNA motif other than palindromic parS is bound by ParB in P. aeruginosa genome
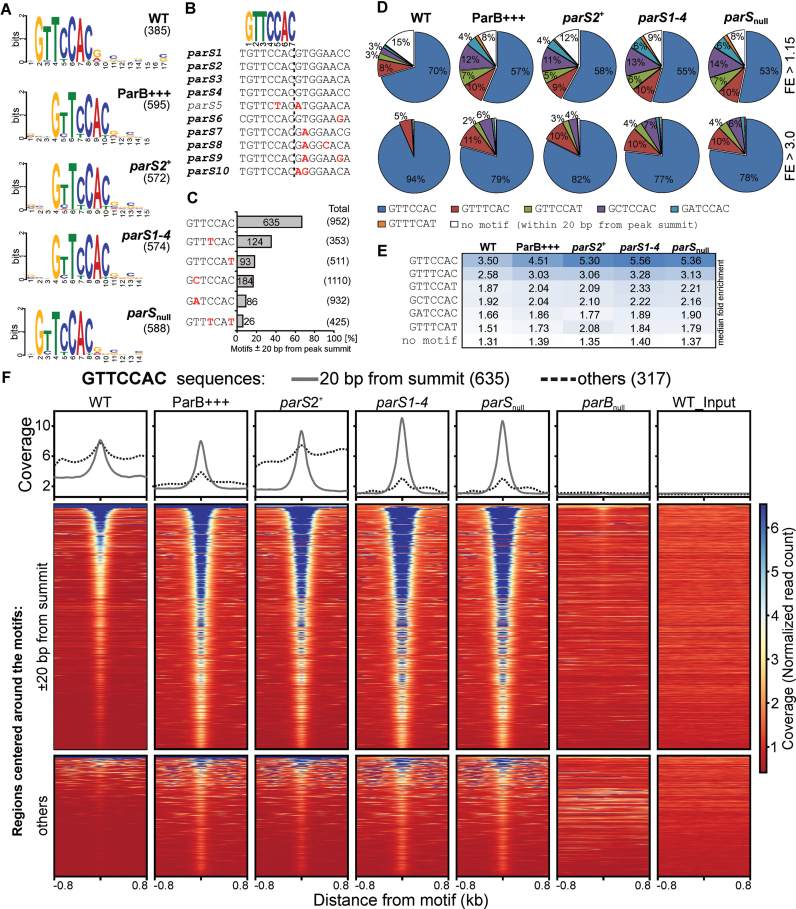
To check if the genome-wide ParB binding involved a defined DNA motif, we performed a search of DNA sequences flanking the summits (±75 bp) of the peaks using MEME ChIP (75). Remarkably, a similar 7-bp DNA sequence logo was identified in the data from all five strains (Figure 4A). The GtTcCAc motif showed strict conservation of positions 1, 3, 5 and 6 whereas at the three other positions some freedom (T→C/A at position 2, C→T at positions 4 and 7) was allowed (Figure 4A). Notably, this motif resembles the GTTCCAC sequence, which is a conserved part of the 8-bp inverted arms of the four major parSs critical for chromosome segregation, parS1–parS4 (Figure 4B), and is also present in one arm of the remaining parS sequences, with the exception of parS5, the only parS to which ParB binding was not found (Figure 1). In parS6–parS10 the other arm carries substitutions at positions 1, 3 and/or 6, the positions strictly conserved in the deduced motif (Figure 4B), suggesting that these parts of parS6–parS10 might not be involved in initial ParB binding.
Figure 4.
Recurrent sequence motifs in ParB-bound regions of Pseudomonas aeruginosa genome. (A) Sequence logos of DNA motifs enriched in genome sequences ±75 bp from summits of ParB ChIP-seq peaks identified in indicated strains. Motif search was performed using MEME-ChIP. The number on the right indicates the total amount of sequences used to build the logo. (B) Alignment of parS sequences from the P. aeruginosa PAO1 genome. Logo represents fully conserved sequence in both arms of parS1–parS4 palindromes and in one arm of the remaining parS sequences, with the exception of parS5. Mismatches relative to GTTCCAC are indicated in red. (C) GTTCCAC sequence variants enriched around summits of ParB ChIP-seq peaks. The percentage of the sequences found within ±20 bp of any peak summit relative to the total number of copies in the PAO1 genome is shown. Only motifs enriched more than four times are presented. Data for remaining variants are included in Supplementary Table S8. Mismatches relative to GTTCCAC are indicated in red. (D) Percentage of peaks containing the indicated heptanucleotide motifs as closest to the summit. The analysis was performed for all peaks (upper panel) and for peaks with FE > 3 (bottom panel). Category ‘no motif’ includes peaks with none of the analyzed variants present ±20 bp from summit. (E) Median FE of ParB ChIP-seq peaks carrying indicated variant of the heptanucleotide motif as closest to the summit in different strains. (F) Read coverage around GTTCCAC motifs in the PAO1 genome in ParB ChIP samples from different strains. Each line in the heatmap represents normalized read counts for each nucleotide of region ±800 bp around a GTTCCAC motif sorted in descending order of mean coverage value, averaged for two biological replicates and colored according to scale. Upper part corresponds to fragments with GTTCCAC motif identified within 20 bp from ParB peaks summit, bottom part to fragments with remaining GTTCCAC motifs. The same regions analyzed in parBnull and WT input are included as controls. Plots above the heatmaps indicate mean coverage score separately for the two groups of GTTCCAC motifs.
To identify the sequence variants which could be recognized by ParB, a search of DNA motifs in the PAO1 genome was performed using the GTTCCAC sequence with the allowance of two mismatches. The total pool of such variants was then limited to the 183 ones detected at least once within 20 bp from any peak summit (Supplementary Table S8). Overall, the sequences in proximity of peak summits analyzed here represent 1.3% of the genome. Remarkably, six variants: GTTCCAC, GTTTCAC, GTTCCAT, GCTCCAC, GATCCAC and GTTTCAT showed at least a 4-fold enrichment in sequences ±20 bp of peak summits relative to the total genome (Figure 4C and Supplementary Table S8). Among them, GTTCCAC was the most overrepresented, since 66.7% of the 952 sequences present in the PAO1 genome were found close to peak summits. The second most enriched motif in proximity of summits was GTTTCAC (35.1%).
An analysis of the peaks in all five strains revealed that ≥85% contained at least one GTTCCAC, GTTTCAC, GTTCCAT, GCTCCAC, GATCCAC or GTTTCAT motif within 20 bp from the summit (Figure 4D), with approximately 70% of peaks in each strain containing the motif <5 bp from the summit (Supplementary Figure S2). GTTCCAC was the most frequently identified variant localized closest to the summit (Figure 4D, upper panel), followed by GTTTCAC. Remarkably, when the analysis was limited to the peaks with FE > 3 these two variants were identified in 94 and 5% of WT peaks, respectively (Figure 4D, bottom panel). A similar trend was observed for other strains. Strikingly, peaks without any of the six analyzed motifs ±20 bp from summit represented <1% of the peaks with FE > 3 (Figure 4D, bottom panel).
A complementary analysis of the FE of the peaks containing indicated variant as the closest one to the summit gave the highest median FE for peaks with GTTCCAC, followed by those with GTTTCAC (Figure 4E). The median FE for the peaks without motif ±20 bp from summit was <1.4 suggesting that these peaks might represent low affinity binding sites (Figure 4E).
To further confirm that the genome fragments carrying the six indicated sequences were enriched by precipitation with anti-ParB antibodies we analyzed normalized read coverage around each motif in the genome, separately for those found ±20 bp from the summits and for the remaining ones. As expected, the mean normalized read count around GTTCCAC present ±20 bp from a peak summit revealed a strong central increase in all ParB-expressing strains, but not for parBnull (negative control) or WT_input DNA (Figure 4F, upper part of the heatmap). A similar analysis for the remaining GTTCCAC motifs also revealed an increase in read coverage for sequences in proximity of most of these motifs in all five strains (Figure 4F, bottom part of the heatmap). The observations were confirmed by quantification of the coverage data for both groups (Figure 4F, plots). Similar results were obtained for GTTTCAC, GCTCCAC, GTTCCAT, GATCCAC and GTTTCAT motifs (Supplementary Figure S3). Overall, the high enrichment of six sequence variants related to one arm of parS in proximity of ChIP-seq peak summits strongly suggests that these are true targets for ParB binding.
To check whether ParB binding to the 7-bp motif is followed by ParB spreading (as observed after binding to palindromic parSs) we analyzed the width of peaks containing the motifs. Positive correlation between the peak width and the number of motifs encompassed by the peak (Supplementary Tables S2–6), but not between FE and the number of motifs was observed, revealing that appearance of broad peaks results from the presence of multiple closely located ParB binding sites (Supplementary Figure S4A and B). Concominantly, analysis of the widths of peaks containing a single parS half-site revealed that the vast majority of such peaks spanned <600 bp (Supplementary Figure S4C), twice the average size of the DNA fragments used for ChIP, indicating no obvious ParB spreading around the parS half-sites.
ParB bound to parS1-4 cluster provides a platform for extended ParB–DNA interactions
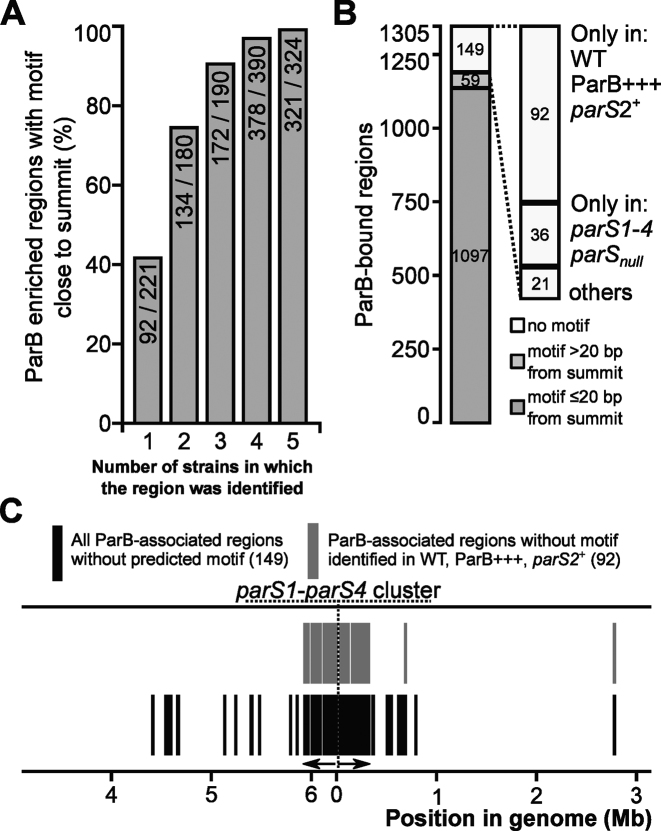
Most ChIP-seq peaks contained one of the six variants of the GTTCCAC motif <20 bp from the summits (Supplementary Figure S2), but a fraction of the peaks lacked such motifs. We asked therefore how consistent was presence of motif-less peaks among different strains, how strongly enriched regions they represented and whether they occurred in specific locations. Combined ChIP-seq data from the five analyzed strains led to the identification of 1305 ParB-bound regions that were then grouped according to the number of strains they were found in (Figure 2E and Supplementary Table S7). The ParB-bound 324 regions detected in all five strains were considered to be the most reliable ones, whereas those found in the ChIP-seq data of any single strain (221 in total) were considered the least reliable. Presence of any of six variants of the 7-bp motif showed positive correlation with the number of strains in which the region was identified (Figure 5A). Out of the 324 ParB-associated regions common to all strains, 321 (99.1%) contained the motif close to the summit in comparison with only 41% (92 out of 221) found in any single strain. Overall, the motif was identified ±20 bp from the peak summit in 1097 of the 1305 ParB-bound regions (Figure 5B), and in 59 ParB-bound regions the motifs were also found but more than 20 bp away from the summit (Figure 5B). Only 149 regions lacked the analyzed motifs. Of those, 70% were detected in a single strain only (Supplementary Table S7) and showed low enrichment (median FE 1.26), suggesting that they may represent non-specific ParB binding. Notably, most of the motif-less peaks were detected in the WT, ParB+++ and/or parS2+ strains, i.e. the strains carrying at least one parS from the parS1-4 cluster (Figure 5B) in which a large nucleoprotein complex formed at oriC (Figure 3B). Indeed, almost all these motif-less peaks were in the vicinity of oriC (Figure 5C) suggesting that the ParB binding to a high affinity site(s) resulted in DNA regions fairly distant from the parS1-4 cluster and lacking the 7-bp motif being recovered in the anti-ParB immunoprecipitate. Hence the large complex formation at oriC involves specific ParB binding to high affinity parS1–parS4 sites and multiple lower affinity parS half-sites (e.g. 39 such motifs are present in the oriC encompassing peak in WT strain) as well as non-specific DNA interactions.
Figure 5.
Heptanucleotide motif-independent ParB binding to DNA. (A) Fraction of ParB-associated regions carrying six analyzed variants of heptanucleotide motif ±20 bp from peak summit as a function of number of strains in which the region was detected. Numbers in columns relate to ParB-bound regions with the motif close to the summit versus summed up regions detected in any single strain (221), any two strains (180), any three (190), any four (390) and all five strains (324). (B) Distribution of heptanucleotide motifs (as above) in ParB-bound regions (Supplementary Table S7). The regions are divided into three categories: with a motif ±20 bp from summit, with a motif further than ±20 bp from summit but within the enriched region and with no motif in the enriched region. The motif-less regions were also categorized according to the strains they were detected in. (C) Localization of ParB-bound regions lacking a heptanucleotide motif in the Pseudomonas aeruginosa PAO1 chromosome.
To get an insight into the postulated parS1–parS4 dependent enrichment of certain DNA sequences a differential binding analysis was performed using Diffbind (76). The ChIP-seq data for strains carrying at least one parS of the parS1-parS4 cluster (WT, ParB+++ and parS2+) was compared pairwise with the data for strains lacking such parS site (parS1-4 and parSnull). The analysis was not limited to the ChIP-seq peaks, but instead non-overlapping 200-bp windows covering the whole genome were used. The WT versus parS1-4 comparison revealed that the majority of bins showing statistically significant difference between the strains displayed decreased coverage in WT (Figure 6A). This was in agreement with the previous observation that the ChIP-seq peaks in the parS mutants showed higher FE than the corresponding peaks in the WT (Table 1). Conversely, some bins showed an increased coverage in WT relative to parS1-4 and these were strictly confined to a 400-kb chromosome region encompassing oriC (Figure 6A). The highest differences between the two strains were observed from 10 kb upstream of parS1 to 20 kb downstream of parS4 (Figure 6A) with a clear negative correlation between the magnitude of the difference between the strains and the distance from the parS1–parS4 cluster. This relation was less clear for bins located further away from parS1–parS4. Remarkably, multiple bins in the 115 to 150 kb region showed higher coverage difference between WT and parS1-4 than bins located relatively closer to parS1–parS4 (Figure 6A and B). Similar results were obtained when WT was compared with parSnull as well as when parS2+ or ParB+++ were compared with parS1-4 (Supplementary Figure S5). Significantly increased coverage of sequences from the 115–150 kb region in the WT versus parS1-4 comparison as well as the presence of a 20-kb gap separating this region from the peak encompassing parS1–parS4 were also observed when WT_input was used as a background control in Diffbind analysis (Figure 6B), indicating that the observed distribution is not an artefact caused by an elevated number of reads mapping to this part of the genome in parBnull data, used as a background control in all previous analyses.
Figure 6.
parS1-parS4 dependent ParB distribution in Pseudomonas aeruginosa genome. (A) Comparison of ParB binding to DNA in WT strain versus parS1-4 strain (lacking functional parS1–parS4 cluster). Differential binding analysis was performed in non-overlapping 200-bp windows covering the whole genome using Diffbind. Only bins showing statistically significant difference of coverage (FDR of 0.05) between WT and parS1-4 are shown. On the right, blow up of the oriC region with bins showing higher number of reads in WT than in parS1-4 is shown. (B) Distribution of parS1–parS4 dependent ParB ChIP enriched sequences in proximity of oriC (±200 kb). Diffbind analysis was performed for WT versus parS1-4 and parSnull ChIP-seq data using 200-bp windows. Subsequently the analysis was performed with the 200-bp windows shifted by 100 bp. Data represent windows (or their halves) showing a statistically significantly (FDR ≤ 0.05) higher coverage of reads in WT strain relative to parS1-4 and parSnull for both sets of genomic windows using parBnull or WT_Input data as a background control.
Overall, the data indicate that ParB binding to parS1–parS4 is required for an enrichment of specific 50 kb region around oriC but also at quite distant locations (e.g. the 115–150 kb region) from it. The fact that these regions are discontinuous suggests that the enrichment cannot be a sole consequence of lateral spreading of ParB along the DNA adjacent to the parS1–parS4 cluster but further supports the hypothesis of interactions at a distance involving bridging/looping or caging of DNA (20,52–54,78).
DISCUSSION
Bacterial genomes are highly compact structures whose distribution within cell is tightly controlled spatially and temporally (79,80). Chromosome segregation must be coordinated with DNA replication and cell division to assure delivery of full genome copies to progeny cells. It has been demonstrated that ParA, ParB and parS sequences are engaged in genome partitioning in many bacteria (14,21,24–37). Additionally, they have been shown to be involved, in a species-specific manner, in the regulation of DNA replication initiation by DnaA (26,27), in the formation of a loading platform for SMC on newly replicated DNA to assure its proper condensation (33,46,47,50), in the positioning of oriC domains at defined cell locations (21,24,36,37,43,44,81), coordination of replication and cell division (30,39,82) and also in the regulation of gene expression (55,59,65,82).
Our earlier studies have confirmed the partitioning role of ParA and ParB proteins in P. aeruginosa, identified ten parS sequences bound by ParB with different affinities and showed that any one of the four parS sites closest to oriC, parS1–parS4, is required and sufficient for accurate chromosome segregation (14,61). Remarkably, expression of over 1000 genes was affected by a lack of ParB (60), suggesting that ParB plays a role in global gene regulation. The plausible explanations of that effect were ParB interactions with sites other than parSs and/or an involvement of ParB partners in the regulation. A recent study on ParB in P. aeruginosa (62) claimed that in vivo ParB bound to the parS1–parS4 and eleven other regions but not to other parSs. In contrast, the results of our in vitro (DNA pull-down) and in vivo (ChIP) confirmed ParB binding to parS6–parS10. To solve this apparent discrepancy regarding the ParB–DNA binding in vivo we performed a ChIP-seq analysis using a WT strain, three parS mutants as well as a ParB-overproducing strain, with parBnull serving as a negative control. Unlike in the aftrementioned study (62), untagged ParB and polyclonal anti-ParB antibodies were used rather than a ParB-FLAG chimera. During the data analysis we did not apply a strict and, by its nature, arbitrary FE cut off, but used the criterion of statistical significance alone, which allowed us to identify a high number of ChIP-seq peaks. Importantly, more than 90% of the peaks were identified in at least two different strains (Figure 2E) indicating that, despite their often relatively low FE, these peaks represent bona fide ParB-binding regions.
The ChIP-seq analysis showed unequivocally that in vivo ParB is bound to all the palindromic parS sequences identified previously, except parS5, with the highest degree of ParB association in proximity of oriC coinciding with the parS1–parS4 sites (Figure 3B). Mutations in the parS palindromes blocked the ParB binding which confirmed that the indicated parSs are specifically bound by ParB in vivo. Remarkably, our data indicate that ParB occupies 420 regions in WT strain, covering almost 7% of the genome and ca. 1000 regions under conditions of ParB abundance.
Using the stringent enrichment criterium of FE > 10, 20 high-affinity regions were identified in the WT, including the major one encompassing the parS1–parS4 sites and all 11 ‘secondary ParB binding sites’ detected previously (62) in P. aeruginosa strains grown on minimal medium (Table 1). This partial correspondence of the two sets of results indicates a significant degree of specificity and conservation of ParB binding to the genome. However, the drastically different experimental setups and approaches used in data analysis prevent conclusions about growth conditions-dependent ParB binding to DNA.
When the availability of ParB in the cell is limited, as is the case in the native situation, the protein must discriminate between its binding sites. The occupation of individual sites may be determined by the degree of affinity, interactions with cellular partners (e.g. ParA), local ParB availability, on-going DNA transactions (e.g. transcription and replication), presence of other factors interacting with DNA, DNA conformation, physiological state of the cell or growth conditions. An increased ParB availability in the cells enabled the identification of ParB binding to an additional 885 loci not identified in the WT strain. Importantly, most of these loci do show a read coverage slightly above the background value also in WT ChIP-seq data suggesting low-occupancy ParB binding in WT cells and not only in the mutants used in this study (Figure 2F). Concomitantly, most of the peaks common to WT and mutant cells showed higher FE in the four strains with increased ParB availability. In a similar study in B. subtilis deletion of six out of ten parS sites resulted in re-distribution of Spo0J (ParB) toward the low-affinity parS sites left in the mutant strain (19), whereas in P. aeruginosa the inactivation of the high affinity parS sites led to the re-distribution of ParB not only to the remaining parSs but also to hundreds of additional ParB binding sites.
The extent of ParB spreading has been analyzed in diverse chromosomal systems. Most data suggested the spreading for up to 10 kb from a parS site in natural conditions (19,55,59,82). Recently, ParB from C. crescentus has been shown to spread by no more than 2 kb from a single parS site, while the extension of the ParB-associated region in the vicinity of oriC to 10 kb involved ParB binding to a cluster of four parS sequences located in a 5-kb region near oriC (83). The parS1–parS4 cluster located within 15 kb of oriC in P. aeruginosa seems to facilitate even farther ParB–DNA interactions since the most conspicuous region of ParB spreading encompassed more than 40 kb in the WT strain (Figure 6A). A comparison of the distribution of the ParB-associated DNA in oriC region between the WT and parS1-4 (or parSnull) revealed a discontinuous ca. 400 kb stretch around the oriC whose occupation was strictly dependent on the presence of parS1–parS4 (Figure 6). Models explaining how ParB shapes the ori domain presumed binding to the strong affinity parS site(s), spreading due to the non-specific DNA binding activity and forming a large nucleoprotein complex due to 3D interactions between DNA-bound ParB molecules (20,52–54,78). Showing that except parSs, P. aeruginosa ParB has several hundreds of other binding sites in the genome we reveal another important mode of ParB action, which allows it not only to shape the ori domain but also to influence the global chromosome topology.
Notably, we show that the ParB-bound sequences contain the GtTcCAc motif corresponding to seven out of eight nucleotides from a single arm of the canonical parS palindrome (Figure 4A and B). A systematic analysis of the motif variants showed that GTTCCAC, GTTTCAC, GCTCCAC, GTTCCAT, GATCCAC and GTTTCAT motifs were found in proximity of ParB ChIP-seq peak summits significantly more often than expected by chance. In any given strain more than 85% of all peaks and 99% of peaks with FE > 3 carried one of the six sequence variants in proximity of the peak summit. Among these six variants, GTTCCAC was not only the most frequently occurring one but was also found closest to the summits of peaks with the highest FE. In the most consistent peaks, those detected in four or five strains, 97 and 99%, respectively, contained one or more of the six motif variants within 20 bp from the summit (Figure 5A). Remarkably, inspection of the ChIP-seq read coverage around all such motifs in the PAO1 genome showed an increased coverage around them indicating that these motifs were likely targeted by ParB in vivo. Our data also demonstrates that the binding to the parS half-sites (in contrast to the ParB binding to palindromic parSs) does not prompt long distance spreading of ParB.
Recent reports indicate that some ParB proteins do not strictly require a symmetrical parS structure for DNA binding. ParB2 of V. cholerae was shown to bind a non-centromeric site containing a half-parS sequence (82). Additionally, in the multipartite genome bacterium Burkholderia cenocepacia ParBc1, ParBc3 and ParBpBC displayed enhanced affinity for the corresponding half-parS relative to random-sequence DNA (84). This data show that the ability of ParB to bind to one arm of parS is not restricted to P. aeruginosa. However, the biological significance of the ParB binding to parS half-sites in the genome remains elusive.
We speculate that the half-parS sequences scattered across the genome may play a role in concentrating ParB molecules on the nucleoid. The interaction of ParB with multiple sites combined with the interactions between ParB molecules could provide a scaffold for chromosome compaction and affect DNA topology, possibly playing a role in modulation of gene expression. Our earlier transcriptomic analysis of P. aeruginosa parBnull mutant versus WT indicated altered expression of 1166 genes (60). A large fraction of these genes seemed to be involved in the stress response due to disturbances in chromosome segregation (85). A complementary study with a strain showing 5-fold overproduction of ParB (ParB+++), a ParB excess leading to no visible changes in growth rate or genome segregation, revealed altered expression of 211 loci (65). The present ChIP-seq data suggest that the ParB–DNA binding relies on the presence of specific sequence(s). A comparison of the distribution of the analyzed variants of the parS half-site with the transcriptional organization of the genome shows that in PAO1 these sequences are mainly intragenic (GTTCCAC—92%, GTTTCAC—76%, GCTCCAC—89%, GTTCCAT—94%, GATCCAC—93% and GTTTCAT—84%), which suggests their roughly random distribution between coding and non-coding sequences (genes make up 90% of the PAO1 genome). The loci with expression affected by ParB deficiency are putatively expressed from 853 promoters. Only 113 of these promoters contain one of the six indicated sequences. Similarly, the 211 loci affected by ParB overproduction are expressed from 130 promoters only 28 of which contain one of the motifs. Whereas ParB binding to the palindromic parS3 and parS4 is responsible for the repression of PA0011p and PA0013p, respectively (65), the low correlation between the ParB-dependent changes in expression and the presence of half-parS sequences in the promoters of other genes suggests that their ParB-dependent regulation does not rely solely on a direct ParB–promoter interaction. The mechanism linking ParB and gene expression regulation in P. aeruginosa requires further studies.
Summarizing, the most prominent features of the P. aeruginosa partitioning protein ParB are: i) sequence-specific DNA binding to palindromic parS sequences, dedicated primarily to the specific task of oriC separation and chromosome segregation; ii) specific binding to numerous heptanucleotide sites throughout the genome; iii) acting at a distance, probably by a combination of spreading and bridging/looping/or caging; and iv) widespread influence on gene expression. These features allow ParB to be classified as a NAP involved in DNA compaction, genome topology maintenance and modulation of gene expression (79,86,87).
DATA AVAILABILITY
All sequencing data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE110158.
Supplementary Material
ACKNOWLEDGEMENTS
ChIP DNA sequencing was performed by Genomed S.A. (Poland).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre, Poland [2013/11/B/NZ2/02555 granted to G.J.B.]. Funding for open access charge: Institute of Biochemistry and Biophysics.
Conflict of interest statement. None declared.
REFERENCES
- 1. Williams D.R., Thomas C.M.. Active partitioning of bacterial plasmids. Microbiology. 1992; 138:1–16. [DOI] [PubMed] [Google Scholar]
- 2. Gerdes K., Møller-Jensen J., Bugge Jensen R.. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000; 37:455–466. [DOI] [PubMed] [Google Scholar]
- 3. Hayes F. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 2000; 37:528–541. [DOI] [PubMed] [Google Scholar]
- 4. Gerdes K., Howard M., Szardenings F.. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010; 141:927–942. [DOI] [PubMed] [Google Scholar]
- 5. Ogasawara N., Yoshikawa H.. Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 1992; 6:629–634. [DOI] [PubMed] [Google Scholar]
- 6. Gal-Mor O., Borovok I., Av-Gay Y., Cohen G., Aharonowitz Y.. Gene organization in the trxA/B–oriC region of the Streptomyces coelicolor chromosome and comparison with other eubacteria. Gene. 1998; 217:83–90. [DOI] [PubMed] [Google Scholar]
- 7. Yamaichi Y., Niki H.. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:14656–14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Livny J., Yamaichi Y., Waldor M.K.. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 2007; 189:8693–8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Motallebi-Veshareh M., Rouch D.A., Thomas C.M.. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 1990; 4:1455–1463. [DOI] [PubMed] [Google Scholar]
- 10. Davey M.J., Funnell B.E.. The P1 plasmid partition protein ParA. A role for ATP in site-specific DNA binding. J. Biol. Chem. 1994; 269:29908–29913. [PubMed] [Google Scholar]
- 11. Lin D.C., Grossman A.D.. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998; 92:675–685. [DOI] [PubMed] [Google Scholar]
- 12. Autret S., Nair R., Errington J.. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 2001; 41:743–755. [DOI] [PubMed] [Google Scholar]
- 13. Lukaszewicz M., Kostelidou K., Bartosik A.A., Cooke G.D., Thomas C.M., Jagura-Burdzy G.. Functional dissection of the ParB homologue (KorB) from IncP-1 plasmid RK2. Nucleic Acids Res. 2002; 30:1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartosik A.A., Lasocki K., Mierzejewska J., Thomas C.M., Jagura-Burdzy G.. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 2004; 186:6983–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kusiak M., Gapczynska A., Plochocka D., Thomas C.M., Jagura-Burdzy G.. Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J. Bacteriol. 2011; 193:3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Funnell B.E. ParB partition proteins: complex formation and spreading at bacterial and plasmid centromeres. Front. Mol. Biosci. 2016; 3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fisher G.L., Pastrana C.L., Higman V.A., Koh A., Taylor J.A., Butterer A., Craggs T., Sobott F., Murray H., Crump M.P. et al. The structural basis for dynamic DNA binding and bridging interactions which condense the bacterial centromere. Elife. 2017; 6:e28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murray H., Ferreira H., Errington J.. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 2006; 61:1352–1361. [DOI] [PubMed] [Google Scholar]
- 19. Breier A.M., Grossman A.D.. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 2007; 64:703–718. [DOI] [PubMed] [Google Scholar]
- 20. Graham T.G.W., Wang X., Song D., Etson C.M., van Oijen A.M., Rudner D.Z., Loparo J.J.. ParB spreading requires DNA bridging. Genes Dev. 2014; 28:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohl D.A., Gober J.W.. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997; 88:675–684. [DOI] [PubMed] [Google Scholar]
- 22. Iniesta A.A. ParABS system in chromosome partitioning in the bacterium Myxococcus xanthus. PLoS One. 2014; 9:e86897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ireton K., Gunther N.W., Grossman A.D.. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 1994; 176:5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharpe M.E., Errington J.. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol. 1996; 21:501–509. [DOI] [PubMed] [Google Scholar]
- 25. Lewis P.J., Errington J.. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol. 1997; 25:945–954. [DOI] [PubMed] [Google Scholar]
- 26. Murray H., Errington J.. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008; 135:74–84. [DOI] [PubMed] [Google Scholar]
- 27. Scholefield G., Whiting R., Errington J., Murray H.. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 2011; 79:1089–1100. [DOI] [PubMed] [Google Scholar]
- 28. Kim H.J., Calcutt M.J., Schmidt F.J., Chater K.F.. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 2000; 182:1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jakimowicz D., Chater K., Zakrzewska-Czerwínska J.. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 2002; 45:1365–1377. [DOI] [PubMed] [Google Scholar]
- 30. Ginda K., Bezulska M., Ziółkiewicz M., Dziadek J., Zakrzewska-Czerwińska J., Jakimowicz D.. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol. Microbiol. 2013; 87:998–1012. [DOI] [PubMed] [Google Scholar]
- 31. Fogel M.A., Waldor M.K.. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006; 20:3269–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kadoya R., Baek J.H., Sarker A., Chattoraj D.K.. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J. Bacteriol. 2011; 193:1504–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minnen A., Attaiech L., Thon M., Gruber S., Veening J.-W.. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 2011; 81:676–688. [DOI] [PubMed] [Google Scholar]
- 34. Donovan C., Schwaiger A., Krämer R., Bramkamp M.. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 2010; 192:3441–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lasocki K., Bartosik A.A., Mierzejewska J., Thomas C.M., Jagura-Burdzy G.. Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 2007; 189:5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartosik A.A., Mierzejewska J., Thomas C.M., Jagura-Burdzy G.. ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology. 2009; 155:1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vallet-Gely I., Boccard F.. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013; 9:e1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bartosik A.A., Jagura-Burdzy G.. Bacterial chromosome segregation. Acta Biochim. Pol. 2005; 52:1–34. [PubMed] [Google Scholar]
- 39. Mohl D.A., Easter J., Gober J.W.. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 2001; 42:741–755. [DOI] [PubMed] [Google Scholar]
- 40. Figge R.M., Easter J., Gober J.W.. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol. Microbiol. 2003; 47:1225–1237. [DOI] [PubMed] [Google Scholar]
- 41. Viollier P.H., Shapiro L.. Spatial complexity of mechanisms controlling a bacterial cell cycle. Curr. Opin. Microbiol. 2004; 7:572–578. [DOI] [PubMed] [Google Scholar]
- 42. Lee P.S., Grossman A.D.. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 2006; 60:853–869. [DOI] [PubMed] [Google Scholar]
- 43. Toro E., Hong S.-H., McAdams H.H., Shapiro L.. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:15435–15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ptacin J.L., Lee S.F., Garner E.C., Toro E., Eckart M., Comolli L.R., Moerner W.E., Shapiro L.. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 2010; 12:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donczew M., Mackiewicz P., Wróbel A., Flärdh K., Zakrzewska-Czerwińska J., Jakimowicz D.. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol. 2016; 6:150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gruber S., Errington J.. Recruitment of condensin to replication origin regions by ParB/Spo0J promotes chromosome segregation in B. subtilis. Cell. 2009; 137:685–696. [DOI] [PubMed] [Google Scholar]
- 47. Sullivan N.L., Marquis K.A., Rudner D.Z.. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009; 137:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X., Le T.B.K., Lajoie B.R., Dekker J., Laub M.T., Rudner D.Z.. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015; 29:1661–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X., Brandão H.B., Le T.B.K., Laub M.T., Rudner D.Z.. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017; 355:524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tran N.T., Laub M.T., Le T.B.K.. SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 2017; 20:2057–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taylor J.A., Pastrana C.L., Butterer A., Pernstich C., Gwynn E.J., Sobott F., Moreno-Herrero F., Dillingham M.S.. Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation. Nucleic Acids Res. 2015; 43:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sanchez A., Cattoni D.I., Walter J.-C., Rech J., Parmeggiani A., Nollmann M., Bouet J.-Y.. Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst. 2015; 1:163–173. [DOI] [PubMed] [Google Scholar]
- 53. Song D., Rodrigues K., Graham T.G.W., Loparo J.J.. A network of cis and trans interactions is required for ParB spreading. Nucleic Acids Res. 2017; 45:7106–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Broedersz C.P., Wang X., Meir Y., Loparo J.J., Rudner D.Z., Wingreen N.S.. Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8809–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Attaiech L., Minnen A., Kjos M., Gruber S., Veening J.-W.. The ParB-parS chromosome segregation system modulates competence development in Streptococcus pneumoniae. Mbio. 2015; 6:e00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jagura-Burdzy G., Macartney D.P., Zatyka M., Cunliffe L., Cooke D., Huggins C., Westblade L., Khanim F., Thomas C.M.. Repression at a distance by the global regulator KorB of promiscuous IncP plasmids. Mol. Microbiol. 1999; 32:519–532. [DOI] [PubMed] [Google Scholar]
- 57. Bignell C.R., Haines A.S., Khare D., Thomas C.M.. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 1999; 34:205–216. [DOI] [PubMed] [Google Scholar]
- 58. Kulinska A., Godziszewska J., Wojciechowska A., Ludwiczak M., Jagura-Burdzy G.. Global transcriptional regulation of backbone genes in broad-host-range plasmid RA3 from the IncU group involves segregation protein KorB (ParB family). Appl. Environ. Microbiol. 2016; 82:2320–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baek J.H., Rajagopala S.V., Chattoraj D.K.. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. Mbio. 2014; 5:e01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bartosik A.A., Glabski K., Jecz P., Mikulska S., Fogtman A., Koblowska M., Jagura-Burdzy G.. Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One. 2014; 9:e87276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jecz P., Bartosik A.A., Glabski K., Jagura-Burdzy G.. A single parS sequence from the cluster of four sites closest to oriC is necessary and sufficient for proper chromosome segregation in Pseudomonas aeruginosa. PLoS One. 2015; 10:e0120867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lagage V., Boccard F., Vallet-Gely I.. Regional control of chromosome segregation in Pseudomonas aeruginosa. PLoS Genet. 2016; 12:e1006428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000; 406:959–964. [DOI] [PubMed] [Google Scholar]
- 64. Klockgether J., Munder A., Neugebauer J., Davenport C.F., Stanke F., Larbig K.D., Heeb S., Schöck U., Pohl T.M., Wiehlmann L. et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 2010; 192:1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kawalek A., Glabski K., Bartosik A.A., Fogtman A., Jagura-Burdzy G.. Increased ParB level affects expression of stress response, adaptation and virulence operons and potentiates repression of promoters adjacent to the high affinity binding sites parS3 and parS4 in Pseudomonas aeruginosa. PLoS One. 2017; 12:e0181726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jutras B.L., Verma A., Stevenson B.. Identification of novel DNA-binding proteins using DNA-affinity chromatography/pull down. Curr. Protoc. Microbiol. 2012; 24:doi:10.1002/9780471729259.mc01f01s24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schulz S., Häussler S.. Chromatin immunoprecipitation for ChIP-chip and ChIP-seq. Methods Mol. Biol. 2014; 1149:591–605. [DOI] [PubMed] [Google Scholar]
- 68. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011; 17:10–12. [Google Scholar]
- 69. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Phanstiel D.H., Boyle A.P., Araya C.L., Snyder M.P.. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics. 2014; 30:2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mierzejewska J., Bartosik A.A., Macioszek M., Płochocka D., Thomas C.M., Jagura-Burdzy G.. Identification of C-terminal hydrophobic residues important for dimerization and all known functions of ParB of Pseudomonas aeruginosa. Microbiology. 2012; 158:1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Song D., Loparo J.J.. Building bridges within the bacterial chromosome. Trends Genet. 2015; 31:164–173. [DOI] [PubMed] [Google Scholar]
- 79. Dame R.T., Tark-Dame M.. Bacterial chromatin: converging views at different scales. Curr. Opin. Cell Biol. 2016; 40:60–65. [DOI] [PubMed] [Google Scholar]
- 80. Badrinarayanan A., Le T.B.K., Laub M.T.. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015; 31:171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lim H.C., Surovtsev I.V., Beltran B.G., Huang F., Bewersdorf J., Jacobs-Wagner C.. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife. 2014; 3:e02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Venkova-Canova T., Baek J.H., Fitzgerald P.C., Blokesch M., Chattoraj D.K.. Evidence for two different regulatory mechanisms linking replication and segregation of Vibrio cholerae chromosome II. PLoS Genet. 2013; 9:e1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tran N.T., Stevenson C.E., Som N.F., Thanapipatsiri A., Jalal A.S.B., Le T.B.K.. Permissive zones for the centromere-binding protein ParB on the Caulobacter crescentus chromosome. Nucleic Acids Res. 2018; 46:1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pillet F., Passot F.M., Pasta F., Anton Leberre V., Bouet J.-Y.. Analysis of ParB-centromere interactions by multiplex SPR imaging reveals specific patterns for binding ParB in six centromeres of Burkholderiales chromosomes and plasmids. PLoS One. 2017; 12:e0177056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schuster M., Hawkins A.C., Harwood C.S., Greenberg E.P.. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 2004; 51:973–985. [DOI] [PubMed] [Google Scholar]
- 86. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 87. Dorman C.J. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 2013; 41:542–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE110158.