Abstract
Gene fusions involving oncogenes have been reported in gliomas and may serve as novel therapeutic targets. Using RNA-sequencing, we interrogated a large cohort of gliomas to assess for the incidence of targetable genetic fusions. Gliomas (n = 390) were profiled using the ArcherDx FusionPlex Assay. Fifty-two gene targets were analyzed and fusions with preserved kinase domains were investigated. Overall, 36 gliomas (9%) harbored a total of 37 potentially targetable fusions, the majority of which were found in astrocytomas (n = 34). Within this lineage 11% (25/235) of glioblastomas, 12% (5/42) of anaplastic astrocytomas, 8% (2/25) of grade II astrocytomas, and 33% (2/6) of pilocytic astrocytoma harbored targetable fusions. Fusions were significantly more frequent in IDH wild-type tumors (12%, n = 31/261) relative to IDH mutants (4%; n = 4/109) (p = 0.011). No fusions were seen in oligodendrogliomas. The most frequently observed therapeutically targetable fusions were in FGFR (n = 12), MET (n = 11), and NTRK (n = 8). Several additional novel fusions that have not been previously described in gliomas were identified including EGFR:VWC2 and FGFR3:NBR1. In summary, targetable gene fusions are enriched in IDH wild-type high-grade astrocytic tumors, which will influence enrollment in and interpretation of clinical trials of glioma patients.
Keywords: FGFR3, Fusion, Genomics, Glioblastoma (GBM), Glioma
INTRODUCTION
In recent years, the field of oncology has undergone impressive advancements due to the development of molecularly targeted treatments. Chromosomal rearrangements (or abnormal transcription) resulting in transcript fusion genes have specifically proved to be therapeutic targets and hence, a focus of intense investigation. Fusion transcripts combine parts of 2 or more genes and are known oncogenic drivers in multiple malignancies (1). Although the occurrence of gene fusion is relatively rare compared to well established oncogenic somatic mutations (e.g. EGFR, KRAS, and TP53), the clinical impact of fusion transcripts has been demonstrated in multiple solid tumors (2–7). A notable example is the EML4:ALK fusion transcript seen in non-small cell lung cancer, which is detected in approximately 7% of patients (8). Screening and subsequent targeting of ALK in selected patients harboring this fusion has improved outcome and subsequently changed the treatment algorithm for this malignancy (9). With such diagnostic and therapeutic potential, the search for novel, targetable fusion proteins has become an integral part of tumor profiling, including in gliomas.
Glioblastoma carries a notoriously poor prognosis, and no targeted therapy has shown substantial survival benefit for it thus far. Specifically, attempts to target molecular aberrations (e.g. EGFR, PDGFR, or PI3K/AKT pathways) have been largely unsuccessful. As such, the need for new therapeutic options and molecularly based diagnostics is dire. Gene fusions have been reported in glioblastoma (GBM), and the functional significance of these fusions is an active area of research (10). The first fusion protein described in GBM was FIG:ROS1 (11), since then, multiple studies and case reports have emerged reporting fusion transcripts, with FGFR family alterations being the most commonly described (12–15). In this study, we report the landscape of fusion transcripts in a large cohort of glioma patients, using RNA sequencing to identify actionable fusion targets (16).
MATERIALS AND METHODS
Patient Population
Three hundred and ninety-three glioma specimens were collected from patients worldwide and were submitted to Caris Life Sciences between 2009 and 2016. Pathological diagnosis was initially determined at the submitting institution and then secondarily independently validated by pathologists at Caris Life Science. In total, 36 cases submitted to Caris were histologically classified as oligodendroglioma; however, IDH status could not be confirmed in 3 cases. Hence, these 3 cases were removed from the analysis. The 390 specimens were then retrospectively analyzed for the presence of genetic fusions. This study is exempt per policy 45 CFR 46.101 (b) and subject information is deidentified.
Fusion Detection
For gene fusion detection, anchored multiplex PCR was performed for targeted RNA sequencing using the ArcherDx fusion assay (Archer FusionPlex Solid Tumor panel). The formalin-fixed paraffin-embedded tumor samples were microdissected to enrich the sample to ≥20% tumor nuclei, and mRNA was isolated and reverse transcribed into complementary DNA. Unidirectional gene-specific primers were used to enrich for target regions, followed by Next-Generation sequencing (Illumina MiSeq platform). Targets included 52 genes, and the full list can be found at http://archerdx.com/fusionplex-assays/solid-tumor. Genes selected are known to be associated with various carcinomas. Reads that were matched to a database of known fusions and other oncogenic isoforms (Quiver database, ArcherDx), as well as those novel isoforms or fusions with high reads (>10% of total reads) and high confidence after bioinformatic filtering, were analyzed. Samples with <4,000 unique RNA reads were reported as indeterminate and excluded from analysis, and all the analyzed fusions were in-frame and were predicted to have kinase domains preserved. Fusions among the >11,000 fusions known to be found in normal tissues were excluded (17). The detection sensitivity of the assay allows for detection of a fusion that is present in at least 10% of the cells in the samples tested. The clinical literature was reviewed to locate therapeutic implications of the fusions identified.
Determinations of 1p19q
To determine the codeletion of chromosomes 1p and 19q, FISH (fluorescence in situ hybridization) was performed using the Abbott Molecular probes for 1p36/1q25 and 19p13/19q13. The cut point for declaring both 1p and 19q deletion was when ratios of 1p/1q signals and 19q/19p signals were both <0.80.
Statistical Analysis
The Fisher exact test was used to compare fusion rates between groups (R v3.4.1). p Values of <0.05 were considered significant. Multiple comparison adjustment was not conducted, owing to the exploratory nature of the study.
RESULTS
Patient Characteristics
Gliomas from a total of 390 patients were included and screened for the presence of fusion transcripts (Table 1). The mean age of the entire cohort was 52 years, and the mean ages of patients with and without fusions were 52 years and 54 years, respectively. The 3 most common tumor locations were frontal (n = 113; 36%), temporal (n = 96; 30%), and parietal (n = 57; 18%). There was significant preferential enrichment of fusions based on tumor location (p = 0.0468), as 4% frontal, 9% temporal, 16% parietal, and 8% of other locations harbored targetable fusions. Tumor grade (high vs low) was available for 379 cases and most tumors were high-grade (84%; n = 317). Specific WHO grade was available for 357 cases, of which 66% (n = 237) were grade IV. Data regarding tumor lineage were available for all cases, and astrocytomas were dominant, comprising 81% (n = 316) of cases followed by oligodendrogliomas (8%; n = 33). Forty-one cases were classified as glioma not otherwise specified (NOS) (11%).
TABLE 1.
Patient and Tumor Characteristics
| n | % | ||
|---|---|---|---|
| Age | Adult | 383 | 98.2 |
| Pediatric | 7 | 1.8 | |
| Total | 390 | ||
| Lineage | Astrocytoma | 316 | 81 |
| Oligodendroglioma | 33 | 8.5 | |
| Glioma NOS | 41 | 10.5 | |
| Total | 390 | ||
| High vs Low grade | High-grade | 317 | 83.6 |
| Low-grade | 62 | 16.4 | |
| Total | 379 | ||
| WHO Grade | I | 6 | 1.7 |
| II | 52 | 14.6 | |
| III | 62 | 17.4 | |
| IV | 237 | 66.4 | |
| Total | 357 | ||
| IDH-1 status | Wild-type | 266 | 71.9 |
| Mutant | 104 | 28.1 | |
| Total | 370 | ||
| IDH-2 status | Wild-type | 358 | 98.6 |
| Mutant | 5 | 1.4 | |
| Total | 363 | ||
| Tumor location | Frontal | 113 | 35.5 |
| Temporal | 96 | 30.2 | |
| Parietal | 57 | 17.9 | |
| Occipital | 11 | 3.5 | |
| Posterior fossa | 11 | 3.5 | |
| Other | 19 | 6 | |
| Frontotemporal | 2 | 0.6 | |
| Frontoparietal | 3 | 0.9 | |
| Temporoparietal | 3 | 0.9 | |
| Parieto-occipital | 3 | 0.9 | |
| Total | 318 |
NOS, not otherwise specified.
Overall, data regarding IDH status were available for 370 cases, and 109 (28%) mutated cases were identified. The majority of these mutants were IDH1 (n = 104) (Table 1). As expected, the mean age of patients with IDH-mutated tumors (42 years) was significantly lower than those with wild-type tumors (56 years) (p < 0.0001). Most IDH wild-type tumors were of high grade (92%; n = 234/254) and astrocytic lineage (90%; n =235/261). The IDH mutation incidence was distributed in 78% (n = 40/51), 73% (n = 43/59), and 7% (n = 16/221) of grades II, III, and IV tumors, respectively. All grade I lesions (n = 6) were of the IDH wild-type (Table 2). Among GBMs and anaplastic astrocytomas with known IDH status, IDH mutations were observed in 7% (n = 15/219) and 67% (n = 29/43) of cases, respectively. All oligodendrogliomas were IDH mutated. Note, among IDH2-mutated tumors (n = 5), 40% (n = 2), and 60% (n = 3) were from astrocytic and oligodendroglia lineages, respectively. All IDH2 mutant tumors were WHO grade II.
TABLE 2.
IDH Status (IDH1/IDH2) in 370 Glial Tumors
| IDH Status | Status Known (n) | Wild-Type (n) | Mutant (n) |
|---|---|---|---|
| Astrocytoma | 297 | 235 | 62 |
| Oligodendroglioma | 33 | 0 | 33 |
| Glioma NOS | 40 | 26 | 14 |
| Total | 370 | ||
| High-grade | 298 | 234 | 64 |
| Low-grade | 61 | 20 | 41 |
| Total | 359 | ||
| I | 6 | 6 | 0 |
| II | 51 | 11 | 40 |
| III | 59 | 16 | 43 |
| IV | 221 | 205 | 16 |
| Total | 337 | ||
| Grade IV (GBM) | 219 | 204 | 15 |
| Grade III (AA) | 43 | 14 | 29 |
| Grade II (LGG) | 26 | 10 | 16 |
| Grade I (Pilocytic astrocytoma) | 6 | 6 | 0 |
| Grade II Oligodendroglioma | 21 | 0 | 21 |
| Grade III Oligodendroglioma | 11 | 0 | 11 |
NOS, not otherwise specified; GBM, glioblastoma multiforme; AA, anaplastic astrocytoma; LGG, low-grade glioma.
Detectable Fusions in Glioma Specimens
Overall, we observed 37 fusion events in 36 gliomas (9%) (Fig. 1A). A single GBM tumor had 2 fusion transcripts detected (PTPRZ1:MET and EML4:NTRK3). EGFR fusions (EGFR:SEPT14 and EGFR:VWC2) were only observed in GBMs. Of the 36 cases where a fusion was detected, most were in high-grade gliomas (n = 31). Overall 10% (31/317) of high-grade gliomas harbored fusions compared with 6% (4/62) of low-grade gliomas. Additionally, of the 36 fusion cases, 94% (n = 34) were detected in astrocytomas, whereas fusions were detected in 2 glioma NOS cases. Notably, no fusions were detected in oligodendrogliomas (Fig. 1A). Moreover, 1p19q codeletion data were available for 15 tumors showing fusions, and none of them displayed a codeletion. Among the astrocytomas specifically, 11% (25/235) of GBMs, 12% (5/42) of anaplastic astrocytomas (AAs), 9% (2/23) of grade II astrocytomas, and 33% (2/6) of pilocytic astrocytomas carried fusions (Fig. 1B).
FIGURE 1.
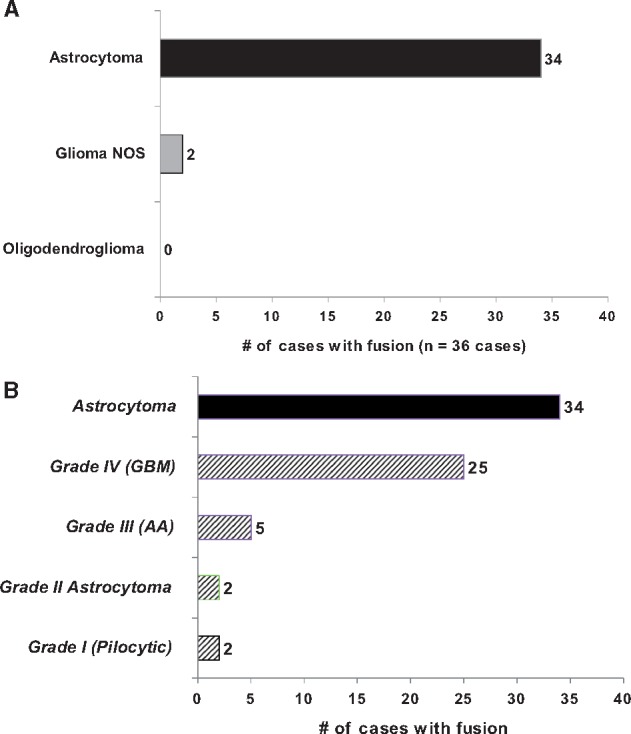
(A) Bar chart showing the distribution of cases with a fusion detected. Fusions were detected in 34 astrocytoma cases and 2 glioma NOS cases; no oligodendrogliomas harbored a fusion. (B) Bar chart showing distribution the 34 astrocytoma cases harboring fusion stratified by WHO grade.
Among the therapeutically-targetable fusions detected (37 fusion events), FGFR-family alterations were the most common (n = 12) (Fig. 2). Within FGFR alterations, the most common fusion was with TACC3 (FGFR3:TACC3) (83%; n = 10 cases) (Table 3). The 2 remaining cases were FGFR3:BRAP (GBM) and FGFR3:NBR1 (AA). The second most commonly encountered fusion type involved MET alterations, making up 30% (11/37) of the fusion events. The MET fusions were exclusive to high-grade gliomas (10 GBMs and 1 AA) (Fig. 2). The most common fusion was PTPRZ1:MET (4 GBM, 1 AA), followed by ST7:MET (3 GBMs) and CAPZA2:MET (2 GBMs) (Table 3). Of note, only 2 IDH1-mutant tumors (1 GBM, 1 AA) carried a MET fusion, and both were PTPRZ1:MET. The distribution of NTRK fusions (n = 8) among glioma grades was more diverse (Fig. 2), and there was no predominant NTRK fusion. The remaining types of fusion were categorized as “other,” and among these, 50% involved EGFR fusions, followed by BRAF, and PDGFRA (Table 3).
FIGURE 2.
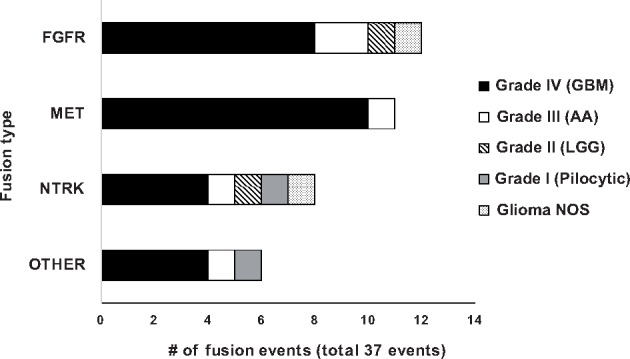
Bar chart showing the distribution of specific gene fusion types. A total of 37 fusion events (in 36 cases) were detected.
TABLE 3.
Distribution of Fusion Types in 390 Gliomas (37 Fusion Events in 36 Cases)
| Fusion | GBM | AA | LGG | Pilocytic Astro. | GliomaNOS | |
|---|---|---|---|---|---|---|
| FGFR (n = 12) | FGFR3:TACC3 | 7 | 1 | 1 | 1 | |
| FGFR3:BRAP | 1 | |||||
| FGFR3:NBR1 | 1 | |||||
| MET (n = 11) | PTPRZ1:MET | 4 | 1 | |||
| CAPZA2:MET | 2 | |||||
| ST7:MET | 3 | |||||
| TPR:MET | 1 | |||||
| NTRK (n = 8) | BCAN:NTRK1 | 1 | ||||
| GKAP:NTRK2 | 1 | |||||
| KCTD8:NTRK2 | 1 | |||||
| NOS1AP:NTRK2 | 1 | |||||
| SQSTM1:NTRK2 | 1 | |||||
| TBC1D2:NTRK2 | 1 | |||||
| VCAN:NTRK2 | 1 | |||||
| EML4:NTRK3 | 1 | |||||
| Other (n = 6) | EGFR:SEPT14 | 2 | ||||
| EGFR:VWC2 | 1 | |||||
| LOC1000093631:BRAF | 1 | |||||
| RAB3IP:PDGFRA | 1 | |||||
| ZSCAN23:BRAF | 1 |
IDH Status Impacts Frequency of Fusion
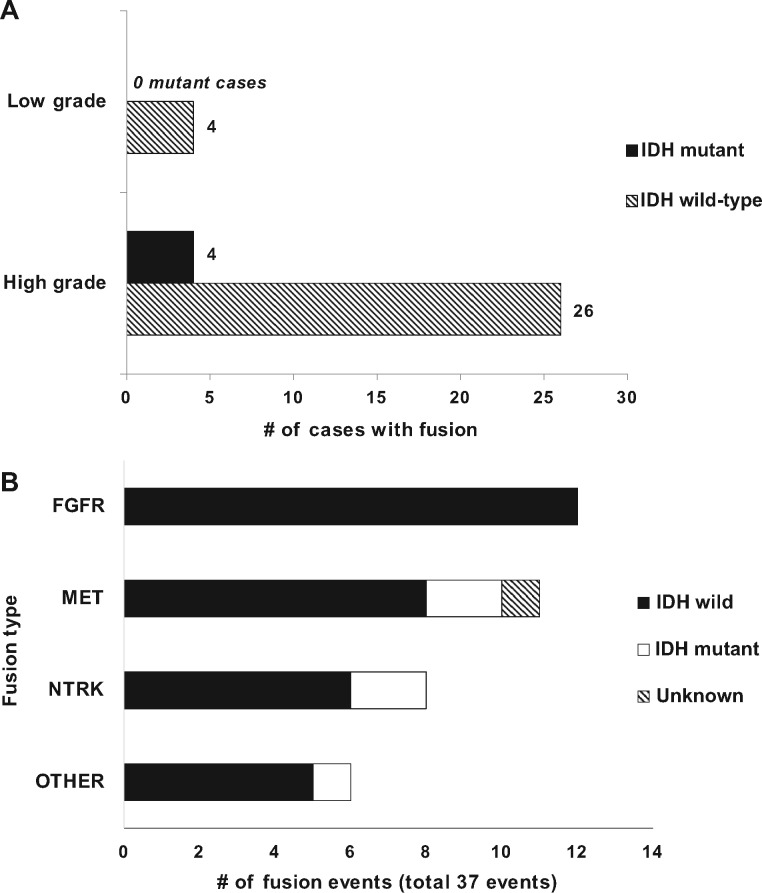
Various potential clinical-pathological associations with fusion were examined including age, tumor location, and IDH status. IDH status was found to be associated with the presence of fusion. Specifically, fusion proteins were significantly more frequent in IDH wild-type tumors (31/261; 12%) relative to IDH mutants (4/109; 4%) (p = 0.011) (Fig. 3A). There was a single fusion case of unknown IDH status. Among high-grade gliomas, 11% (n = 26/234) of IDH wild-type gliomas displayed enriched fusions relative to 6% (n = 4/64) of high-grade IDH mutants (p = 0.3505) (Fig. 3A). This may partly stem from the fact that FGFR fusion proteins were observed almost exclusively in IDH wild-type tumors (Fig. 3B). Low-grade IDH wild-type tumors had a significantly higher frequency of fusions (4/20; 20%) than their IDH-mutant counterparts (0/41; 0%) (p = 0.0093) (Fig. 3A).
FIGURE 3.
(A) Bar graph displaying the distribution of cases with detectable fusion in high- and low-grade gliomas, stratified by IDH status. Overall, fusion proteins were more frequent in IDH wild-type tumors relative to IDH mutants. Low-grade IDH wild-type tumors in particular had a significantly higher frequency of fusions (4 out of 20 cases; 20%) compared to low-grade IDH mutants (0 cases) (p = 0.0268). (B) Bar chart showing the distribution of specific gene fusion types stratified by IDH status.
DISCUSSION
Our study revealed that approximately 9% of gliomas and 11% of GBMs harbor potentially therapeutically targetable gene fusions. Furthermore, this analysis highlights an association between gene fusions and IDH wild-type astrocytic tumors. This observation is also consistent with prior large-scale profiling initiatives, although it may not have been directly addressed (18, 19). This is not to say that fusions absolutely do not occur in oligodendrogliomas or 1p/19q-deleted gliomas. For example, 1 study demonstrated a BRAF fusion in a pediatric oligodendroglioma, but it is notable that this tumor failed to show 1p/19q codeletion or expression of the IDH1 mutation, which calls into question whether this should have been characterized as an oligodendroglioma (20). A second oligodendroglioma report described an association with a BRAF fusion, but the clinical course was highly atypical, with leptomeningeal spread (21). Finally, a larger prior study of 185 adult diffuse gliomas demonstrated an incidence of 9.4% (17/180) of KIAA1549:BRAF fusions, which predominated in 88% (15/17) of oligodendroglial neoplasms. However, a review of the integrated low-grade glioma and GBM TCGA database revealed that of the approximately 380 cases that were IDH mutant (which is ubiquitous in oligodendrogliomas), there appeared to be only 1 case with BRAF mutation. Without copy number analysis to show the tandem duplications, this latter study is likely to have a high false positive rate due to the methodology or alternatively, that KIAA1549:BRAF fusions may be a feature of a specific subtype of gliomas (i.e. diffusely infiltrating gliomas). Finally, the association of gene fusions with both IDH1 wild-type and astrocytic tumors is also consistent with the reciprocal association of IDH1 mutations and oligodendrogliomas (22, 23).
A notable observation from our study was the identification of heterogeneous types of NTRK fusions (NTRK1, NTRK2, and NKTR3) in which the majority were NTRK2. Although a prior study observed NTRK1 fusions at an incidence of 1% in GBMs (12), we only detected a single NTRK1 fusion—in a pilocytic astrocytoma case. A variety of NTRK fusion types (NTRK1, NTRK2, and NKTR3) have also been described in pediatric high-grade gliomas (24). The NTRK fusion has been previously targeted with Entrectinib (TrkA inhibitor), with a therapeutic effect (25). Alternatively, these patients could be considered for treatment with RXDX-101 in the STARTRK-1 global phase I/II clinical trial NCT02097810 or NCT02568267. The most common fusion detected in our study (at 3%) was in the FGFR family—specifically FGFR3:TACC3, which is the predominant fusion described in the literature for multiple cancers (6, 7, 19). FGFR gene fusions in GBM have been previously documented with a frequency of 2–3% (15, 18, 26). We also observed 2 novel FGFR3 fusions involving BRAP and NBR1, which have not been previously reported in gliomas. Preclinical data supports the therapeutic efficacy of FGFR3:TACC3 inhibition in a GBM xenograft model (14, 15, 26). A phase I study of the FGFR inhibitor JNJ-42756493 reported a partial response in 3 of 4 patients carrying a TACC3 fusion,1 of which was a GBM patient. Overall, we observed MET fusions in ∼3% of gliomas, which is lower than in a prior analysis of 272 gliomas wherein this fusion was detected in 7.7% of anaplastic astrocytomas and 15% of secondary GBMs (27). This fusion has also been targeted successfully with crizotinib (a MET inhibitor) (28).
As these cases were acquired from a worldwide commercial dataset, one of the limitations of this study is that we cannot discern the impact of conventional treatment on the expression of fusion proteins or the impact of these fusions on patient survival. Secondly, we cannot differentiate the fusion expression profiles of secondary vs primary GBM, which would be informative from a mechanistic perspective. Further studies will be needed to uncover the functional significance of gene fusions in GBM and whether directed inhibitors can be combined with conventional treatment. Additionally, the interaction between these fusion proteins and the host immune system is unclear. A recent study indicated that the FGFR3 pathway was activated in non-T cell-inflamed bladder tumors (i.e. those with limited T-cell infiltration), impacting susceptibility to immunotherapy (29). This finding may indicate a connection between fusion transcripts such as the ones involving the FGFR family and the immune microenvironment, which would warrant further investigation.
ACKNOWLEDGMENTS
The authors would like to thank Audria Patrick and David M. Wildrick, PhD, for their administrative and editorial support.
REFERENCES
- 1. Parker BC, Zhang W.. Fusion genes in solid tumors: An emerging target for cancer diagnosis and treatment. Chin J Cancer 2013;32:594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bass AJ, Lawrence MS, Brace LE et al. , Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 2011;43:964–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Celestino R, Sigstad E, Lovf M et al. , Survey of 548 oncogenic fusion transcripts in thyroid tumors supports the importance of the already established thyroid fusions genes. Genes Chromosomes Cancer 2012;51:1154–64 [DOI] [PubMed] [Google Scholar]
- 4. Lae M, Freneaux P, Sastre-Garau X et al. , Secretory breast carcinomas with ETV6-NTRK3 fusion gene belong to the basal-like carcinoma spectrum. Mod Pathol 2009;22:291–8 [DOI] [PubMed] [Google Scholar]
- 5. Tomlins SA, Rhodes DR, Perner S et al. , Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–8 [DOI] [PubMed] [Google Scholar]
- 6. Williams SV, Hurst CD, Knowles MA.. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu YM, Su F, Kalyana-Sundaram S et al. , Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soda M, Choi YL, Enomoto M et al. , Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6 [DOI] [PubMed] [Google Scholar]
- 9. Shaw AT, Kim DW, Nakagawa K et al. , Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–94 [DOI] [PubMed] [Google Scholar]
- 10. Frattini V, Trifonov V, Chan JM et al. , The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 2013;45:1141–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charest A, Lane K, McMahon K et al. , Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosom Cancer 2003;37:58–71 [DOI] [PubMed] [Google Scholar]
- 12. Kim J, Lee Y, Cho HJ et al. , NTRK1 fusion in glioblastoma multiforme. PLoS One 2014;9:e91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozawa T, Brennan CW, Wang L et al. , PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev 2010;24:2205–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parker BC, Annala MJ, Cogdell DE et al. , The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 2013;123:855–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh D, Chan JM, Zoppoli P et al. , Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Subramaniam DS, Mehta S, Gatalica Z et al. , RNA-Seq analysis of glioma tumors to reveal targetable gene fusions. J Clin Oncol 2017;35:15_suppl (May 2017) 2019–2019 [Google Scholar]
- 17. Babiceanu M, Qin F, Xie Z et al. , Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res 2016;44:2859–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brennan CW, Verhaak RG, McKenna A et al. , The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ceccarelli M, Barthel FP, Malta TM et al. , Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016;164:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar A, Pathak P, Purkait S et al. , Oncogenic KIAA1549-BRAF fusion with activation of the MAPK/ERK pathway in pediatric oligodendrogliomas. Cancer Genet 2015;208:91–5 [DOI] [PubMed] [Google Scholar]
- 21. Rodriguez FJ, Schniederjan MJ, Nicolaides T et al. , High rate of concurrent BRAF-KIAA1549 gene fusion and 1p deletion in disseminated oligodendroglioma-like leptomeningeal neoplasms (DOLN). Acta Neuropathol 2015;129:609–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H.. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009;174:1149–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan H, Parsons DW, Jin G et al. , IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu G, Diaz AK, Paugh BS et al. , The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cook PJ, Thomas R, Kannan R et al. , Somatic chromosomal engineering identifies BCAN-NTRK1 as a potent glioma driver and therapeutic target. Nat Comms 2017;8:15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Di Stefano AL, Fucci A, Frattini V et al. , Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-type Glioma. Clin Cancer Res 2015;21:3307–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bao ZS, Chen HM, Yang MY et al. , RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 2014;24:1765–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. International Cancer Genome Consortium PedBrain Tumor P. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med 2016;22:1314–20 [DOI] [PubMed] [Google Scholar]
- 29. Sweis RF, Spranger S, Bao R et al. , Molecular Drivers of the Non-T-cell-Inflamed Tumor Microenvironment in Urothelial Bladder Cancer. Cancer Immunol Res 2016;4:563–8 [DOI] [PMC free article] [PubMed] [Google Scholar]