Abstract
Introns in mRNA leaders are common in complex eukaryotes, but often overlooked. These introns are spliced out before translation, leaving exon-exon junctions in the mRNA leaders (leader EEJs). Our multi-omic approach shows that the number of leader EEJs inversely correlates with the main protein translation, as does the number of upstream open reading frames (uORFs). Across the five species studied, the lowest levels of translation were observed for mRNAs with both leader EEJs and uORFs (29%). This class of mRNAs also have ribosome footprints on uORFs, with strong triplet periodicity indicating uORF translation. Furthermore, the positions of both leader EEJ and uORF are conserved between human and mouse. Thus, the uORF, in combination with leader EEJ predicts lower expression for nearly one-third of eukaryotic proteins.
INTRODUCTION
The presence of introns is a hallmark of eukaryotic genes. Most eukaryotic genes contain introns, but their numbers vary widely by gene and species (1,2). Higher eukaryotes have an average of 8.8 introns per gene (3). A number of functional roles have been proposed for introns (4–6). However, only some of these roles are well-understood. For example, multi-exon gene structures allow alternative splicing, which produces messenger RNA (mRNA) and protein isoforms with differing roles (7). The diverse functions of introns are reviewed in detail elsewhere (4–6).
Intron position is important for many transcripts. While most introns are located within the CDS (coding sequence of the main open reading frame, mORF), it is well established that some specific introns in the mRNA leader (5′ untranslated regions, 5′UTRs) enhance gene expression in animals and plants (4,8,9). For example, the maize Ubi1 intron enhances expression (10). These 5′UTR introns contain elements that enhance gene expression, at least in part, by promoting transcription and nuclear export (9,10). In addition, some constructs have been engineered to contain a single intron in the mRNA leaders, such as the commercial Promega pGL3 expression vectors. In contrast, introns in the 3′UTRs can reduce expression through nonsense-mediated mRNA decay (NMD) (11–13).
The mRNA leaders of a third to half of eukaryotic mRNAs contain upstream AUGs (uAUGs) and upstream ORFs (uORFs) (14). Some uORFs are well-characterized and known to have a regulatory role and/or encode functional peptides (12,15). A recently developed technique—ribosome profiling (Ribo-seq)—has been used to study uORFs because it provides unprecedented detail of genome-wide translation events (16,17). This technique is based on RNA shotgun sequencing (RNA-seq) that identifies the positions of the ribosomes on an mRNA. It has been used successfully in a range of species (18). Translation of an ORF can be inferred from Ribo-seq data, including uORF translation (19–23).
Several features of the mRNA leader are well-known to affect translation, such as (i) the presence of uORFs and (ii) RNA structures, and (iii) the length of the mRNA leader, and (iv) the sequence context around the translation initiation codon (12,15). However, the relationships between 5′UTR introns (exon-exon or splice junctions at the mRNA leaders, termed leader EEJs after intron removal) and mRNA translation have not been fully investigated. We therefore surveyed the positions of EEJs in human, mouse, zebrafish, fruit fly, and Arabidopsis thaliana and integrated genome-wide datasets from these model organisms to explore new roles of introns.
MATERIALS AND METHODS
Data and accession numbers
The mass spectra and RNA-seq datasets of this study are available on PRIDE (24) (PXD006661) and Gene Expression Omnibus (25) (GSE99697), respectively. Publicly available high-throughput sequencing and shotgun proteomic datasets used in this study are listed in Supplementary Table S1. The processed Ribo-seq and proteomic data are available in Supplementary Tables S2–S4 and S5, respectively.
Reference sequences and gene annotations
The reference sequences and GTF annotation files for human and mouse were retrieved from UCSC Genome Browser (hg19 and mm10) (26) and GENCODE (v19 and vM9) (27,28), unless otherwise mentioned. The reference sequences and annotations for zebrafish, fruit fly, and A. thaliana were retrieved from Ensembl 85 and Ensembl Plant 31 (29).
Ribosomal RNA (rRNA) and transfer RNA (tRNA) sequences for human, mouse, zebrafish, fruit fly were obtained from GtRNAdb (release 30 January 2012) (30). Small nucleolar RNA (snoRNA) sequences for human and mouse were obtained from snOPY (retrieved in March and June 2016, respectively) (31). These sequences were combined with the noncoding RNA sequences obtained from Ensembl.
BED files of mRNA regions were obtained by parsing the GTF files with metagene-maker (32). The BED files were further processed by BEDTools v2.22.0 (33) to obtain the positions of the mRNA leader exons (after removal of 5′UTR introns; Supplementary Table S6).
uORF annotations
Mouse uORF annotation was obtained from previously annotated ORFs of mouse bone marrow-derived dendritic cells (BMDCs) (19). The genomic locations were converted from mm9 to mm10 using Liftover (26) (Supplementary Table S7). Human uORFs were predicted from the Ribo-seq datasets of HeLa and HEK293 cells using RiboTaper v1.3 (20,34,35) (Supplementary Table S7).
Cell culture
All cell culture reagents were obtained from Invitrogen (CA, USA), unless otherwise mentioned. DNA profiling of HepG2 cells was done at DNA Diagnostics Limited (Auckland, New Zealand). Short tandem repeat loci tested are D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818 and FGA, and the gender specific locus amelogenin. The results were matched with the HepG2 cell line specifications at ATCC.
HepG2 cells were grown and maintained in Dulbecco's Modified Eagle Medium (DMEM; without sodium pyruvate) supplemented with 10% fetal bovine serum, and 1% antibiotic-antimycotic cocktail (streptomycin, amphotericin B and penicillin). When the cells had been maintained in an optimal growth rate for 2 weeks, the media was replaced with DMEM for SILAC (stable-isotope labeling by amino acids in cell culture) for three passages to deplete cellular Arg and Lys. This SILAC media was supplemented with 10% dialysed fetal bovine serum, and 0.01 mM of Arg and Lys.
The HepG2 cells acclimated to SILAC media were plated onto a T-25 culture flask at 1 × 105 cells per flask and grown overnight. The cells were then separately grown in the SILAC media supplemented with medium (0.5 mM of l-Arg-13C6, hydrochloride and l-Lys-13C4, hydrochloride) and heavy amino acids (0.5 mM of l-Arg-13C6, 15N4, hydrochloride and l-Lys-13C6, 15N2, hydrochloride). The cells supplemented with 0.5 mM of unlabelled Arg and Lys were used as control.
The HepG2 cells labeled with heavy and medium amino acids were exposed to 100 μM of hemin (Sigma-Aldrich, MO, USA) and 200 μM of deferoxamine (an iron chelator; Sigma-Aldrich, MO, USA). The cells were harvested after 6 h for RNA-seq and mass spectrometry analysis.
Mass spectrometry
The HepG2 cell counts were determined, and equal number of cells were lysed in RIPA buffer (500 μl per 1 × 106 cells) on ice with shaking for 15–20 min. The lysate was centrifuged at 13 000 g for 20 min. Supernatants of medium and heavy protein lysate were mixed at equal volume and electrophoresed in a 10% NuPAGE Bis-Tris gel at 200 V in NuPAGE MOPS SDS Running Buffer (Invitrogen, CA, USA). The gel was stained in Coomassie staining solution.
The entire gel lane was sliced into 10 molecular weight fractions. Each fraction was subjected to in-gel tryptic digestion using a DigestPro liquid handling robotic workstation (Intavis, Germany). The protein digests were dried in a centrifugal concentrator and reconstituted in 5% (v/v) acetonitrile, 0.2% (v/v) formic acid in water. Each sample was then analyzed by nanoflow liquid chromatography coupled to LTQ-Orbitrap XL tandem mass spectrometry (Thermo Fisher Scientific, CA, USA).
The RAW output files were processed through Proteome Discoverer v1.4 (Thermo Fisher Scientific, CA, USA) using default settings. Peak lists were then searched against Human Ensembl 88 protein sequence database using the Sequest HT search engine in Proteome Discoverer. Dynamic modifications of carbamidomethyl (Cys), deamination (Asn), and oxidation (Met) were selected. Post-processing false discovery rate estimation was done using the Percolator algorithm (36). The charge-based Sequest XCorr was adjusted to ensure a false discovery rate of less than 1% on the peptide level. Only peptides assigned at high confidence to master proteins which were identified with two or more peptides were accepted. All peptides that passed the above filters were used for Intensity-Based Absolute Quantification (IBAQ) calculation using pythomics (37).
RNA-seq
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Germany). These samples were submitted to the Otago Genomics and Bioinformatics Facility at the University of Otago (Dunedin, New Zealand) under contract to the New Zealand Genomics Limited for library construction and sequencing. These samples were quality checked using Agilent Bioanalyzer (Agilent, CA, USA), with a threshold of RNA integrity >7. Those samples that passed the quality control were used for library construction. The libraries were prepared using TruSeq stranded mRNA sample preparation kit according to the manufacturer's protocol (Illumina, CA, USA). These libraries were assessed and quantified using Agilent Bioanalyzer (Agilent, CA, USA) and Qubit fluorometer (Invitrogen, CA, USA), respectively. The libraries were paired-end sequenced using HiSeq 2000 (Illumina, CA, USA), generating 2 × 100 nucleotide reads. The gene expression for pairs of replicates had a Spearman correlation of >0.9.
Ribo-seq and RNA-seq data analysis
Short read alignment was done using STAR 2.5.2b (38). Replicates were combined unless otherwise stated. Only one mismatch was allowed. To remove nonribosomal footprints, reads were first aligned to noncoding RNAs (using parameter –outStd SAM –outReadsUnmapped Fastx –clip3pAdapterSeq {adapter_sequence} –seedSearchLmax 10 –outFilterMultimapScoreRange 0 –outFilterMultimapNmax 255 –outFilterMismatchNmax 1 –outFilterIntronMotifs RemoveNoncanonical > /dev/null). All 3′ end adapter sequences used in the Ribo-seq libraries analyzed are listed in Supplementary Table S1.
Unmapped reads were then aligned to genome (using parameter –clip3pAdapterSeq {adapter_sequence} –seedSearchLmax 10 –outFilterMultimapScoreRange 0 –outFilterMultimapNmax 255 –outFilterMismatchNmax 1 –outFilterIntronMotifs RemoveNoncanonical –outSAMtype BAM SortedByCoordinate). The aligned ribosome footprints were examined for triplet periodicity according to footprint size (read length) using RiboTaper v1.3 (20). Those footprint sizes with good triplet periodicity were further examined to obtain the offset values for adjusting their read start position to ribosomal-A site using RibORF v0.1 (23). This footprint read adjustment step also removed any remaining footprints that did not conform to the triplet periodicity.
In addition, unmapped reads were also aligned to protein coding transcripts (using parameter –clip3pAdapterSeq {adapter_sequence} –seedSearchLmax 10 –outFilterMultimapScoreRange 0 –outFilterMultimapNmax 255 –outFilterMismatchNmax 1 –outFilterIntronMotifs RemoveNoncanonical –outSAMtype BAM Unsorted –outSAMmode NoQS –outSAMattributes NH NM). The aligned RNA-seq reads were used for mRNA isoform quantification using Salmon v0.60 (39). The aligned ribosome footprints were used for isoform level quantification using riboprof (Ribomap v1.2) (40), which was supported by the mRNA isoform abundance data, RNA-seq alignment and the offset values for the footprint read adjustment (above). This step assigned ribosome footprints uniquely to mRNA isoforms by prioritizing the frame 1 of a CDS, followed by the frames 2 and 3, and lastly the UTRs.
RNA folding prediction
For RNA structure prediction, mRNA leader sequences were extracted from genomic sequences using BEDTools v2.22.0 (33). The Minimum Free Energy (MFE) of folding the mRNA leader sequences was predicted using RNAfold (41) and normalized to leader length. For RNA accessibility, the first 100 bases of the 5′ end of mRNA sequences were extracted. Accessibility of these sequences was estimated using Localfold (42) and normalized to the length (100 bases).
Metagene analysis
The distributions of EEJs along the mRNAs were plotted using Guitar v1.11.9 (43). Guitar produces metagene plot based on normalized lengths of the mRNA regions. Metagene ribosome profiles around the translation initiation and termination codons were plotted using RibORF v0.1 (23). For splicing analysis, completed Splicing Index (coSI) was calculated using IPSA (https://github.com/pervouchine/ipsa) (44). For metagene analysis of MLN51, the aligned iCLIP (individual-nucleotide resolution UV crosslinking and immunoprecipitation) reads were ‘hard clipped’ using a custom shell script and converted to BED format using BEDTools v2.22.0 (33). The center position of each processed iCLIP read was used as the RNA protein binding site rather than the read start position (45).
Comparative genomic analysis
Genomic coordinates of the mRNA regions were converted to transcript level coordinates using TransDecoder v3.0.0 (46). The relative positions of the leader EEJ and uAUG were determined. This data was mapped to human and mouse orthologous data downloaded from Ensembl Biomart (accessed in May 2016). To avoid ambiguity, only one-to-one orthologs that contain a single leader EEJ and/or uNUG were analyzed (Supplementary Table S8).
Statistical analysis
Welch two sample t-test, Fisher exact test, one-sided chi-square test and linear regression analysis were done using R v3.4.0 (47). Plots were constructed using ggplot2 (48) unless stated otherwise. The standardized regression coefficients of the linear regression models were plotted using sjPlot (49). A significant threshold of P-value < 0.05 was used unless stated otherwise.
RESULTS AND DISCUSSION
Introns are common in the mRNA leaders of complex organisms
In this study, we focused on multicellular eukaryotes because unicellular eukaryotes can have strikingly varied intron densities. For example, only about 5% of genes in Saccharomyces cerevisiae have introns but other species have many introns (50). The multicellular species chosen were human, mouse, zebrafish, fruit fly and A. thaliana. These five species have high quality gene annotations and the several types of genome-wide datasets required.
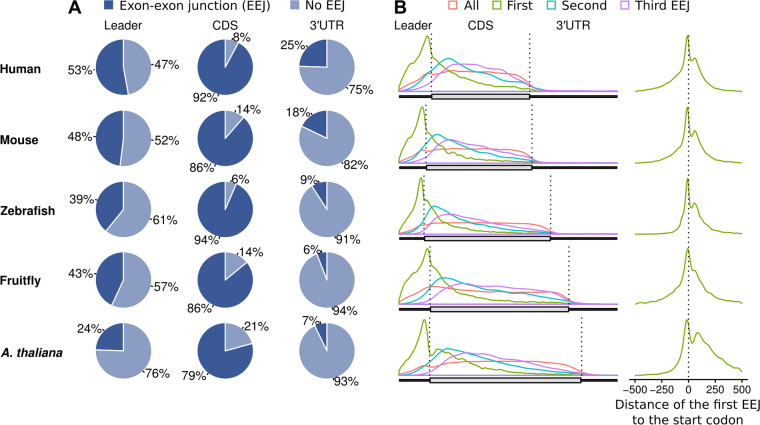
Firstly, the frequency and distribution of EEJs (the junctions between exons after intron removal) in the mRNAs were determined. Notably, EEJs are commonly present in the mRNA leaders (Figure 1A and Supplementary Table S6). In contrast to previous studies, our transcript-based analysis identified more leader EEJs in these species: for example, 24% in A. thaliana compared to 11% that we reported a decade ago (51), and 53% in human compared to 38% (Mammalian Genome Consortium) (52), 35% (RefSeq) (53) and 28% (UTRdb in 2001) (54) in previous studies. This is likely due to different sources of annotations, and improvement of genome assembly and annotation over the years due to high-throughput sequencing.
Figure 1.
Distributions of exon-exon junctions (EEJs) in each mRNA region in the genomes of human, mouse, zebrafish, fruit fly, and A. thaliana. (A) Proportion of EEJs in the mRNA leader (5′UTR), CDS and 3′UTR. (B) Distribution of EEJs along the mRNAs with segment specific scaling (left panel); distribution of the first EEJs centered at the start codon (right panel). CDS, coding sequence of the mORF; EEJ, exon-exon junction; mORF, main open reading frame; UTR, untranslated region.
As expected, most (79% or more) mRNAs of higher eukaryotes contain EEJs in the CDS (Figure 1A). As previously noted (51–54), EEJs are also less common in the 3′UTRs analyzed here (7–25%). These results suggest that EEJs are distributed towards the 5′ end of the mRNAs. This is partly because of strong evolutionary pressure against introns in the 3′UTRs, which may trigger mRNA degradation via NMD pathway (11–13).
To further investigate the distribution of EEJs, metagene plots were constructed to visualize the distributions of the first three EEJs along the mRNAs (Figure 1B, left panel). Remarkably, the most frequent location of the first EEJs is just preceding the main translation initiation codon. These results are consistent with the metagene plots centered at the initiation codon (Figure 1B, right panel).
Strikingly, these EEJ distribution patterns are similar across the five species examined. Based on this and previous studies on specific genes (4,8,9,51,55), we postulated that the juxtaposition of leader EEJ and initiation codon may be linked with the efficiency of translation.
mRNA leaders with EEJs have higher relative ribosome densities
To explore the possible link between the leader EEJs and mRNA translation, we first analyzed publicly available Ribo-seq datasets (and matched RNA-seq) for human (HeLa cells) (35), mouse (liver tissue) (56), zebrafish (48 h embryo) (57), fruit fly (0–2 h embryo) (58), and A. thaliana (Columbia-0, 4 day old etiolated seedlings) (59) (Supplementary Table S1). These particular datasets have good quality metrics (e.g. triplet periodicity), good sequencing depth (at least 20 million transcript mapped footprints), and matched transcriptome (RNA-seq) datasets.
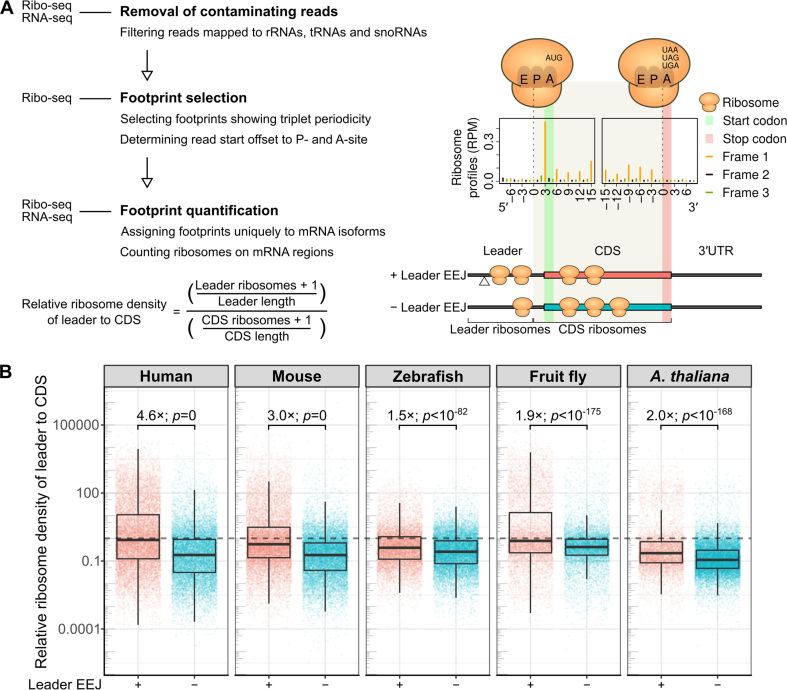
As Ribo-seq provides individual codon resolution of ribosome footprints (16,17), we compared the positions of translating ribosomes on the mRNAs with leader EEJs to those without. Firstly, in order to remove spurious signals, we filtered off the reads that mapped to noncoding RNAs including rRNAs, tRNAs and snoRNAs (Materials and Methods; Figure 2A, left panel).
Figure 2.
Distributions of translating ribosomes on the mRNAs. (A) Schematic representation of Ribo-seq data analysis (left panel, Materials and Methods). The publicly available datasets analyzed are of HeLa cells, mouse liver, zebrafish 48 h embryos, fruitfly 0–2 h embryos, and A. thaliana seedlings (Supplementary Table S1). The read start positions of ribosome footprints were adjusted to ribosomal A-site, as shown in the metagene ribosome profiles of mouse mORFs (right panel). To avoid misassignment, ribosome footprints mapped to three bases upstream of the start codon (gray shading) were counted as CDS ribosomes (Supplementary Table S2). (B) Relative ribosome density of the mRNA leader to CDS (see equation in A). Fold difference of the median and the P-value from t-test are shown. The dotted line marks the ratio of one, when the density ratio of an mRNA is equal. CDS, coding sequence of the mORF; EEJ, exon-exon junction; mORF, main open reading frame; RPM, Reads Per Million mapped reads.
Secondly, we analyzed the distribution of ribosome footprints showing high quality triplet periodicity with appropriate offsets to the ribosomal A-site (Figure 2A, right panel, and Supplementary Figure S10, left panel). The number of ribosome footprints mapped to the mRNA leader and CDS of each mRNA were counted using Ribomap (40) (Supplementary Table S2). Ribomap was chosen because it is the only program that permits quantification of ribosome profiles at the mRNA isoform level (Materials and Methods). In particular, ribosome footprints were uniquely assigned to mRNA isoforms using relative abundance of mRNA isoforms obtained from the matched RNA-seq data. This approach prevents over-representation of rare transcripts. Only the transcripts with mRNA read counts >5, and ribosome footprint counts >5, on either the mRNA leader or CDS were included. We noted a relatively high number of ribosome footprints were mapped to sites immediately preceding the initiation codon, in particular at the +2 positions (see green bars in Figure 2A, right panel, and Supplementary Figure S10, left panel). To prevent misassignment of CDS ribosome footprints to the mRNA leaders, the ribosome footprints mapped to the three bases preceding the main initiation codons were also assigned as CDS ribosomes (Figure 2A, right panel).
Next, the ribosome density of the mRNA leaders relative to CDS was calculated (Figure 2A, equation). The ratios and values in Figure 2B and subsequent figures are presented on log10 scales as these values have wide ranges. Remarkably, this density ratio was significantly higher in the mRNAs with leader EEJs than those without across the five species examined (Figure 2B, Welch two sample t-test, P-values < 10−82). They were over fourfold and threefold higher in human cells and mouse liver, respectively. These results suggest that the leader EEJs may be associated with how ribosomes translate the parts of the mRNAs.
More importantly, these findings motivated us to ask several more questions. (i) What is the relationship between the leader EEJ and translation? (ii) How does the leader EEJ compare to well-known features of mRNAs that affect translation, such as uAUG or RNA structure in the mRNA leader? (iii) How consistent is this observation across other ‘omic’ datasets particularly proteomics?
The frequency of the leader EEJs anti-correlates with translation
To explore the relationships between translation and the leader EEJ and the other mRNA features, we processed a total 15 publicly available Ribo-seq datasets using the isoform level quantification approach described in the previous section and Materials and Methods. These datasets include seven cultured cells (35,60–65) and eight tissues (56,66,67) as detailed in Supplementary Table S1. The processed data is available in Supplementary Table S3. Several of the datasets are of the same or closely related cells or tissues but produced in different labs. Most were based on the protocols of Ingolia and colleagues (68–70), except the human (35) and mouse (56) datasets analyzed in the previous section, which were prepared using a commercial version of the protocol (Epicentre ARTseq Ribosome Profiling Kit). This allowed us to control for the effects of experimental and inter-lab variability.
Translation efficiency of the CDS (CDS TE) was calculated as the number of ribosome footprints mapped to the CDS divided by the number of RNA-seq reads mapped to the corresponding full-length transcript (Supplementary Material, section 1.1). A linear regression model was then fitted to these datasets using CDS TE as the response variable and mRNA features as the explanatory variables (Supplementary Figures S1 and S2).
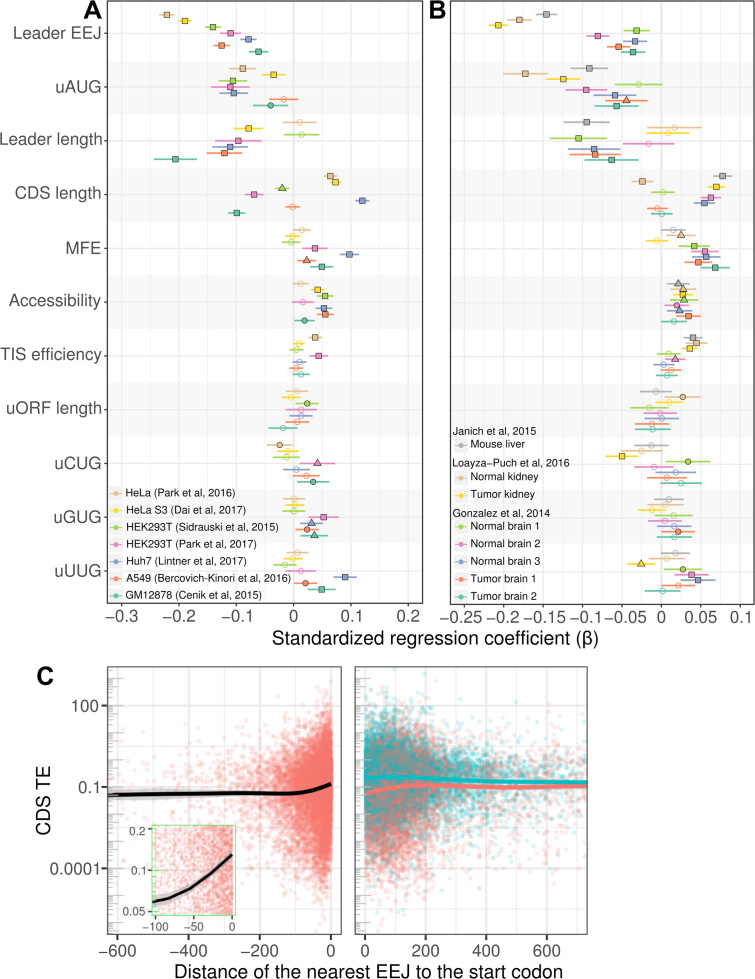
Strikingly, the frequency of the leader EEJs was the only predictor which anti-correlates unambiguously with CDS TE and the strongest predictor in most of the datasets analyzed. Indeed, the leader EEJ correlation had the largest standardized regression coefficients in four out of seven, and three out of eight, cultured cell and tissue datasets, respectively (Figure 3A and B, respectively). These results suggest that the signal from such a strong predictor outweighs noise.
Figure 3.
Relationships between translation and the features of mRNAs. Standardized regression coefficients of the linear regression models of the (A) cultured cell and (B) tissue datasets are shown. The data sources are represented by different colors. Filled squares, triangles and circles denote the P-values of <0.001, <0.01 and <0.05, respectively. Unfilled circles denote not statistically significant. Error bars denote the 95% confidence intervals. (C) Relationships between translation efficiency (CDS TE) and the distance to the nearest EEJ before or after the translation start codon (center of panels). CDS, coding sequence of the mORF; EEJ, exon–exon junction; MFE, minimum free energy of mRNA leader sequence normalized to leader length; mORF, main open reading frame; TIS, translation initiation site; uNUG, upstream cognate or near-cognate triplet; uORF, upstream open reading frame (with AUG initiation codon). The list of sources and processed data are available in Supplementary Tables S1 and S3, respectively. Also see the related Supplementary Figure S5.
We considered possible confounding variables, for example, many mRNAs with leader EEJs or uAUGs have long mRNA leader sequences (Supplementary Figure S3, class i and iii versus class ii and iv). A previous study had shown that the time required for ribosome preinitiation complex to scan for the main initiation codon increases with the mRNA leader length (71), which might affect the translation rate observed here. Therefore, we further re-analyzed data subsets with similar leader lengths. In these matched sets, the leader EEJ remained the strongest predictor except for the brain tissue data (Supplementary Figure S5). Such an anti-correlation between the leader EEJ and translation has not been reported before.
Overall, the other strong predictors were more consistent, but the weak predictors were more noisy (Figure 3A and B, and Supplementary Figure S5). The second most consistent and strongest predictor was the frequency of uAUGs, followed by the lengths of mRNA leader and CDS. Interestingly, several other mRNA features examined were weaker predictors of translation: (i) the length of the uORF (with AUG initiation codon), (ii) Translation Initiation Site efficiency (TIS efficiency)—the experimentally determined effect of sequence context surrounding the AUG initiation codon (72), (iii) RNA structure in the mRNA leader—MFE estimated by RNAfold (41) normalized to leader length, (iv) RNA accessibility—the accessibility of the first 100 bases of mRNAs predicted by LocalFold (42) normalized to the length (Figure 3A and B, and Supplementary Figure S5). We found that uORF length associates weakly with translation rate, which is in agreement with a previous proteomic study (73). In contrast to previous studies using reporter constructs (15,72,74), the other well-known mRNA features also associate weakly with translation rate at the genome-wide scale.
The relationship between the distance from the EEJ to the main initiation codon and translation (CDS TE) was also examined (Figure 3C, mouse liver data). CDS TE was higher with EEJs located less than 100 bases upstream of the start codon (Figure 3C, left panel, inset, black line and red points). For mRNAs with CDS EEJs (blue points), CDS TE decreased slightly with increasing distance of the CDS EEJ to the start codon (Figure 3C, right panel, blue line). Translation of mRNAs with both leader and CDS EEJs (Figure 3C, right panel, red line) also varied with distance. These findings support the hypothesis that the location of EEJ is associated with translation.
Translation is the lowest for the mRNAs with both leader EEJs and uAUGs
The linear regression models in the previous section showed that translation decreases with increasing number of leader EEJs as well as uAUGs (Figure 3A and B, and Supplementary Figure S5). We therefore divided and defined the mRNAs into four classes: class i, ii, iii and iv (Figure 4 and Table 1). Class i mRNAs harbor both leader EEJs and uAUGs (red). Notably, this is the largest class in the human genome (Table 1, 35% of mRNAs). Class ii have leader EEJs but no uAUGs (green) (14%). Class iii have no leader EEJs but has uAUGs (blue) (20%). Class iv are what could be considered as the ‘typical’ mRNAs—having no leader EEJs and no uAUGs (purple) (30%). In contrast to the conventional view that an mRNA leader would lack both leader EEJs and uORFs, we found that the mRNAs with leader EEJs and uORFs (class i) are the most abundant class of transcripts in the mouse and fruit fly genomes besides human (Table 1).
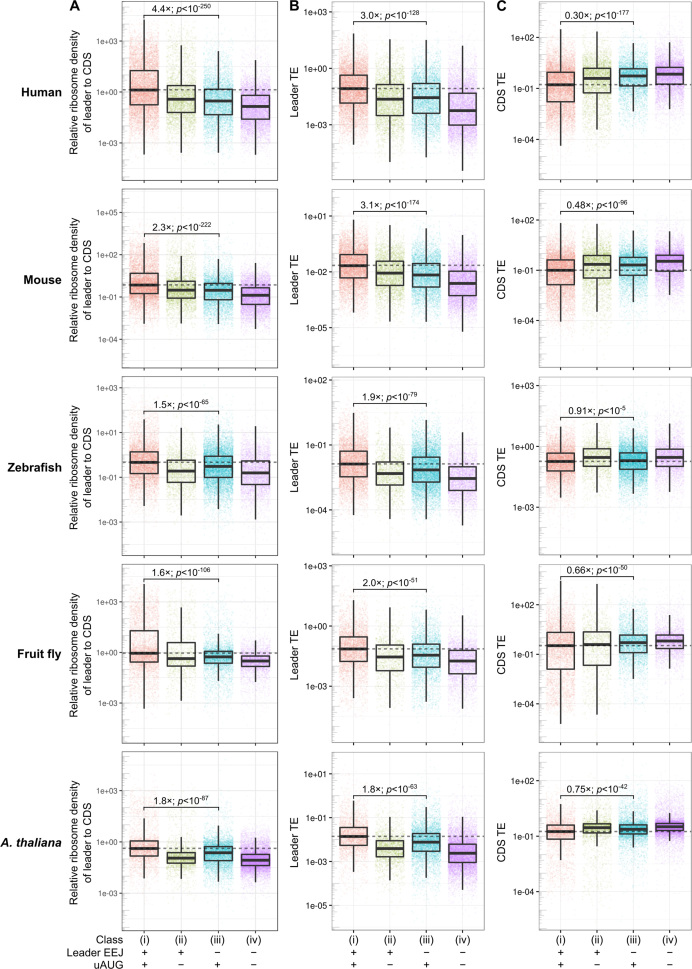
Figure 4.
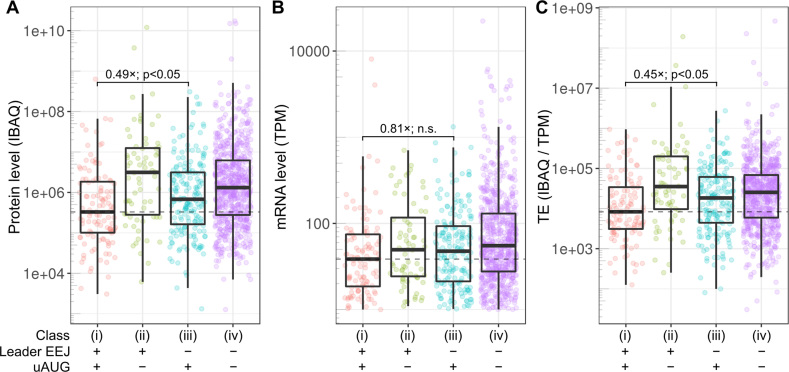
Translation of mRNAs with or without leader EEJs and uAUGs. Statistically significant differences between the mRNA classes are indicated. The scales used include low abundance outliers and enable visual comparison. (A) Relative ribosome density of the mRNA leader to CDS, and TE of (B) the mRNA leader and (C) CDS. mRNAs are grouped into four classes, those that (i) have both leader EEJs and uAUGs (pink), (ii) have leader EEJs but no uAUGs (green), (iii) have no leader EEJs but have uAUGs (blue), (iv) have no leader EEJs and no uAUGs (‘typical’ mRNAs; purple). CDS, coding sequence of the mORF; EEJ, exon-exon junction; mORF, main open reading frame; TE, translation efficiency; uAUG, upstream AUG. The list of sources and processed data are available in Supplementary Tables S1 and S2, respectively. Also see the related Supplementary Figures S7 and S8.
Table 1. Proportion of the mRNA classes in the five model species examined. Only the transcripts detected in both the Ribo-seq and RNA-seq datasets of these species are shown below. The list of sources and processed data are available in Supplementary Tables S1 and S2, respectively.
| mRNA class | Human | Mouse | Zebrafish | Fruit fly | A. thaliana | Total |
|---|---|---|---|---|---|---|
| i | 8900 (35%) | 8392 (33%) | 6965 (31%) | 4626 (34%) | 3155 (14%) | 32038 (29%) |
| ii | 3593 (14%) | 3581 (14%) | 1881 (8%) | 1322 (10%) | 2299 (11%) | 12676 (12%) |
| iii | 5138 (20%) | 5557 (22%) | 9292 (42%) | 4277 (31%) | 5647 (26%) | 29911 (28%) |
| iv | 7547 (30%) | 7956 (31%) | 4208 (19%) | 3554 (26%) | 10773 (49%) | 34038 (31%) |
| Total | 25178 | 25486 | 22346 | 13779 | 21874 |
i, have both leader EEJs and uAUGs; ii, have leader EEJs but no uAUGs; iii, have no leader EEJs but have uAUGs; iv, have no leader EEJs and no uAUGs (‘typical’ mRNAs); EEJ, exon-exon junction; uAUG, upstream AUG. Total numbers are italicized.
Relative ribosome density of the mRNA leader to CDS, leader TE and CDS TE for the four mRNA classes were calculated (Figure 4A, B and C, respectively). The TE of an mRNA region was calculated as the number of ribosome footprints divided by the number of RNA-seq reads mapped to the full-length transcript. Of all the species examined, the density ratio and leader TE of class i mRNAs were consistently the highest and significantly higher than those of class iii (Figure 4A and B, P-values < 10−51). In contrast, the CDS TE of class i mRNAs was consistently the lowest and significantly lower than that of class iii (Figure 4C, P-values < 10−5). These results suggest that the leader EEJ may be associated with a strong expression of the AUG initiated (cognate) uORF but a weak expression of the mORF. Although previous studies have suggested that cognate uORF can reduce protein expression (73,75,76), such a relationship in conjunction with the leader EEJ has not also been reported to date.
We then compared translation of the mRNAs with leader EEJs (class ii) with that of typical mRNAs (class iv). Both mRNA classes lack uAUGs so translation at the mRNA leader must initiate at non-AUG codons. The density ratio and leader TE of class ii was consistently higher than that of typical mRNAs in all species (Figure 4A and B, P-values < 10−51), suggesting that leader EEJ may also be associated with non-canonical uORF (non-uAUG) translation. For the CDS TE, however, not all species had a concomitantly lower CDS TE (Figure 4C). The exception was zebrafish, in which the CDS TE of class ii was 1.2-fold higher than that of class iv (P-value < 10−3). This may be due to the numbers of functional non-canonical uORFs (class ii and iv, non-uAUG) vary at the greater extent than canonical uORFs (class i and iii, uAUG) among the species.
Previous studies have found that the use of cycloheximide in Ribo-seq experiment can produce spurious reads mapped to the mRNA leaders, in particular in the Saccharomyces cerevisiae data (16,77). To investigate this possibility, we analyzed publicly available matched Ribo-seq datasets of human cells with and without cycloheximide treatment (HeLa cells) (78). Our observations on these mRNA classes, in particular class i versus iii, were consistent regardless of cycloheximide treatment (Supplementary Figures S6 and S7).
A limitation of the prior analysis is the use of a single reference annotation, thus all the biological material analyzed were assumed to have the same mRNAs, in particular the transcription start sites (TSS). However, it is known that some mRNA isoforms have identical CDS but different promoters and mRNA leader sequences, which is challenging problem for assigning ribosome footprint at the mRNA isoform level (17,40). The TSS (79) and splicing (80) may be cell type- and tissue-specific, and changed dynamically in different physiological states (81). To address this issue, we first assembled the transcriptomes of each cell type using the matched RNA-seq datasets from Ribo-seq experiments using Cufflinks ab initio (81) (detailed in Supplementary Material, section 2.2). The TSS were precisely adjusted using the relevant ENCODE/RIKEN CAGE (Cap Analysis Gene Expression) or RAMPAGE (RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression) datasets (79,82,83). To avoid ambiguity, the TSS nearest to the main initiation codon were chosen and only the genes with a single mRNA leader variant were analyzed (Supplementary Table S4). These allow unambiguous grouping of genes into classes. Our observations on these classes, in particular class i versus iii, were consistent with the prior analysis (Supplementary Figure S8).
Protein expression is the lowest for the genes with both leader EEJs and uAUGs
To examine if protein expression is also the lowest in class i mRNAs, we did shotgun proteomics in parallel with RNA-seq using a different human cell line (HepG2; hepatocellular carcinoma). This is a pulsed SILAC experiment in testing cellular response to iron (manuscript in preparation, Materials and Methods). We then estimated the abundance of pre-existing proteins (unlabeled) at the gene level using IBAQ approach. Next, we estimated the abundance of mRNAs at the gene level from the control RNA-seq datasets using Salmon (39). Only the proteins supported by at least two or more high confidence peptides and a gene expression unit of 10 TPM (Transcripts Per kilobase Million mapped reads) were used for further analysis. In addition, we analyzed previously published, post-processed proteomic and matched RNA-seq datasets on human (HeLa cells) (84), mouse (NIH3T3 cells) (85,86), and zebrafish (24 h embryo) (87) as listed in Supplementary Table S1. The processed data is available in Supplementary Table S5.
Indeed, as predicted from the results in Figures 3 and 4, the class i genes (with both leader EEJs and uAUGs) have the lowest protein level and TE, which were significantly lower than that of class iii (Figure 5A and C, and Supplementary Figure S9A and C, P-values < 0.05). The class i mRNA level was also the lowest, but not significantly different to that of class iii (Figure 5B and Supplementary Figure S9B). It was notable that both the translation and steady state protein level for class i were consistent lower than class iii in all datasets analyzed, regardless of species, technologies, methodologies, and other sources of variation.
Figure 5.
Protein expression of HepG2 cells. (A) Protein, (B) mRNA and (C) TE levels were quantified at the gene level. Classes are as in Figure 4. EEJ, exon-exon junction; IBAQ, Intensity-Based Absolute Quantification; n.s., not significant (P-value > 0.05); TE, translation efficiency (ratio of protein to mRNA); TPM, Transcripts Per kilobase Million mapped reads; uAUG, upstream AUG. The processed data is available in Supplementary Table S5. Also see the related Supplementary Figure S9.
From the genome-wide, multi-omic data presented here, the 5′UTR introns of class ii genes correlate inconsistently with the translation rate and protein level (Figures 4 and 5, and Supplementary Figures S8 and S9). However, previous studies have found that adding a single 5′UTR intron to specific constructs enhances gene expression (4,8,9,51,55). It is likely this effect is mainly due to transcriptional enhancement, but at least in part, it is due to translation (6). This discrepancy in the genome wide analyses may be because most genes have multiple introns and complex regulation, in contrast to the constructs commonly tested with a single intron.
Previous studies have shown that uORF translation could trigger NMD of the transcripts (12,13,15). The low level of class i mRNAs may be due to NMD, and therefore resulted in a low protein level. We analyzed a list of mRNAs targeted by UPF1 (a key regulator of NMD pathway) in HeLa cells determined recently (88). Notably, the NMD targets were distributed proportionally across the mRNA classes (Supplementary Table S9). For example, there were 33% and 18% of class i and iii mRNAs subjected to NMD, respectively, which were comparable to their respective abundance (Table 1, 35% and 20%, respectively). The results suggest that uORF containing mRNAs (class i and iii) are not enriched as NMD targets.
Ribosome footprints on the uORFs of the mRNAs with leader EEJs show strong triplet periodicity
A known issue in some Ribo-seq data is contamination with RNA-seq fragments (16). To exclude this possibility, we took advantage of the observation that genuine ribosome footprint profiles show triplet periodicity (19,20), rather than being random pieces of mRNA. This triplet pattern of translating ribosomes should be clear in both uORFs and mORFs.
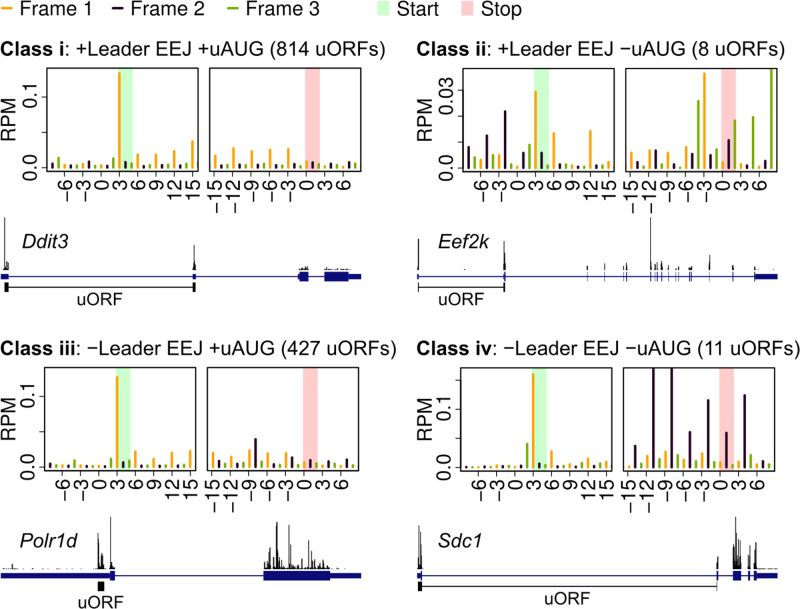
The uORF annotations (Materials and Methods) were used to examine the triplet periodicity of ribosome footprints on uORFs. Notably, nearly two-thirds of previously identified mouse uORFs (19) were class i uORFs (Figure 6 and Supplementary Table S7, uAUG with leader EEJ). In the mouse liver data, the metagene ribosome profiles of class i uORFs showed a strong triplet periodicity, comparable to that of mORFs (Figure 6 versus Figure 2A, right panel). This was also observed in the human HeLa ribosome footprints using different sources of uORF annotation (Supplementary Figure S10 and Table S7). Further examination of the periodicity of ribosome footprints showed that uORFs are also being translated in the other mRNA classes. For example, the uORFs of Eef2k, Polr1d and Sdc1 indeed have clear triplet periodicity (representing class ii, iii, and iv, respectively, in Figure 6). However, the translated uORFs were predominantly class i uORFs.
Figure 6.
Metagene ribosome profiles of the mouse liver uORFs. Distribution of ribosomes on the cognate (AUG; left panel), near-cognate (CUG, GUG and UUG; right panel) uORFs and their respective examples. EEJ, exon-exon junction; RPM, Reads Per Million mapped reads; uAUG, upstream AUG; uORF, upstream open reading frame. The uORF annotation is available in Supplementary Table S7. Also see the related Supplementary Figure S10.
An example of class i uORF is shown in Figure 6: the well-studied uORF of Ddit3 (89,90) (DNA damage-inducible transcript 3 protein) that was actively being translated rather than the mORF. Ddit3, or better known as CHOP, is a multifunctional transcription factor that is transiently expressed in response to endoplasmic reticulum stress (91,92). Other genes in this class such as Atf4 and Eif5, are also well-characterized as having regulatory uORFs (15,62,93–95). As both human and mouse cells have widespread translation of class i uORFs, we postulated that many uAUGs and leader EEJs may be juxtaposed for a regulatory function.
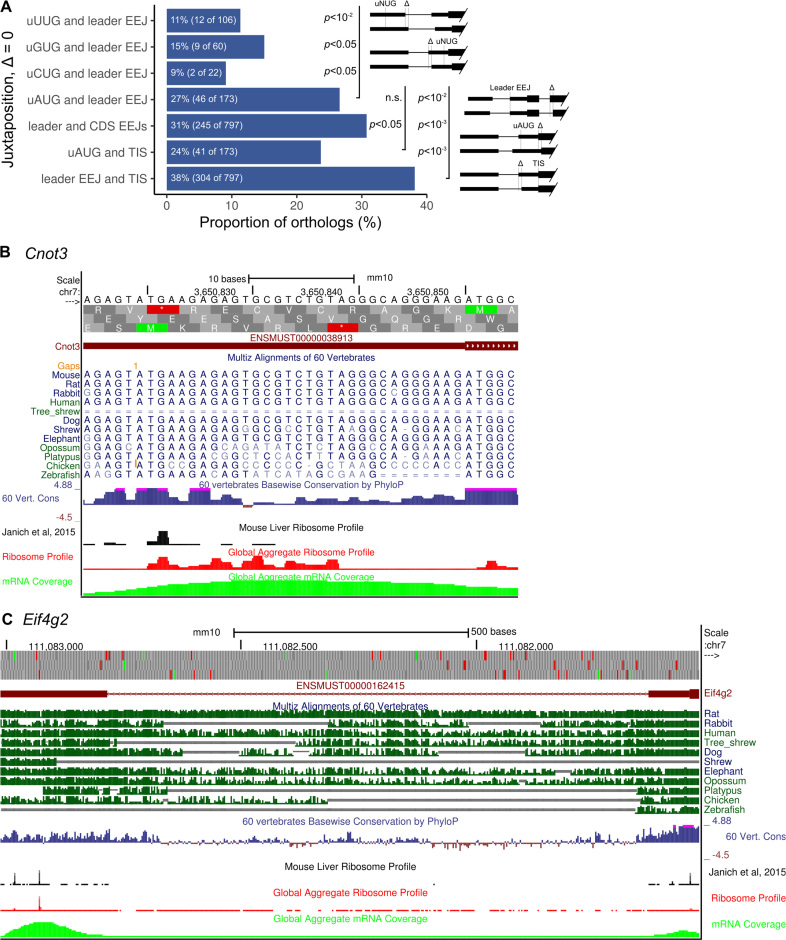
Juxtaposition of the uAUG and leader EEJ is evolutionarily conserved
To investigate relative positions of the uAUGs and leader EEJs, human and mouse orthologs were compared. To avoid ambiguity, only one-to-one orthologs with a single leader EEJ and/or uNUG were used (Supplementary Table S8). Firstly, the positions of the leader EEJs are conserved in 38% of the orthologs (Figure 7A). Secondly, the juxtapositions of the uAUGs and leader EEJs are as conserved as that of the uAUGs and main initiation codons (27% and 24% conserved, respectively). Thirdly, the juxtapositions of the uAUGs and leader EEJs are significantly more conserved than that of the near-cognate triplets and leader EEJs (both Fisher exact and one-sided chi-square tests, P-values < 0.05). Some examples of these are Cnot3 and Eif4g2 orthologs (Figure 7B and C, respectively), with strong ribosome peaks at their uAUGs. Overall, these results suggest that these uORFs in combination with leader EEJs are evolutionarily conserved features and likely to have inhibitory functions.
Figure 7.
Conservation of the positions of leader EEJs and uAUGs in human and mouse orthologs. The orthologs that contain a single leader EEJ and/or uNUG were compared. (A) Proportion of the conserved distance of between two positions. These positions are considered to be conserved if their relative distance did not change between species. Only the largest P-values, either from Fisher exact or one-sided chi-square test are shown. Specific examples of genes: the leader EEJ positions in the (B) Cnot3 and (C) Eif4g2 orthologs are conserved. The uAUGs of Cnot3 and Eif4g2 are located at the downstream and upstream of the leader EEJ, respectively, as shown in the UCSC Genome Browser with the conservation tracks, mouse liver ribosome profiles, and GWIPS-Viz global aggregate tracks. CDS, coding sequence of the mORF; EEJ, exon-exon junction; mORF, main open reading frame; n.s., not significant (P-value > 0.05); TIS, translation initiation site of the mORF; uNUG, upstream cognate or near-cognate triplet. The ortholog data is available in Supplementary Table S8.
Splicing of the leader exons is complete
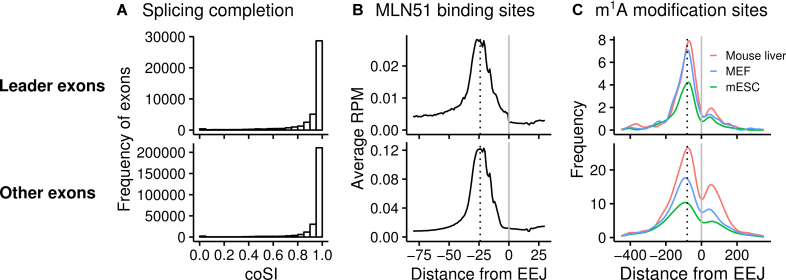
Incomplete splicing of the mRNA leaders (intron retention) could also confound these results, although this was controlled by using matched RNA-Seq data (above). To address splicing, we calculated the completeness of slicing using the completed Splicing Index (coSI) in an independent HeLa RNA-seq dataset (96) (Figure 8A). This dataset also has relevant matched iCLIP datasets (Figure 8B). We compared the coSI of the leader exons with all the other exons. Both types of exons showed a similar proportion of completed splicing (Figure 8A, 40% at a coSI of one).
Figure 8.
Splicing and dynamic marks on the mRNAs. (A) Completeness of splicing at the mRNA leader and CDS. (B) Metagene profiles of MLN51 binding sites and (C) m1A modification sites centered at the EEJs (gray lines). The leader exons and all the other exons are shown at the top and bottom panels, respectively. CDS, coding sequence of the mORF; coSI, completed Splicing Index; EEJ, exon-exon junction; MEF, mouse embryonic fibroblasts; mESC, mouse embryonic stem cells; mORF, main open reading frame; RPM, Reads Per Million mapped reads.
As a result of splicing, the exon junction complex (EJC) is deposited 20–24 bases preceding the EEJ (11,13). It was previously shown that the EJCs and their positions affect translation (11–13). We therefore examined the presence of MLN51 (a key EJC core protein) surrounding the leader EEJs and all the other EEJs using the iCLIP dataset from the same study (96). Canonical EJC deposition sites were observed in both types of EEJs (Figure 8B), which are consistent with the coSI results (Figure 8A).
The relative position of junctions and uORFs may be important (Figure 7A). If the leader EEJ is located upstream of the uAUG or not far downstream, a preinitiation complex may first contact the EJC before initiating. To explore these possibilities, we analyzed the class i uORF translation using the same Ribo-seq datasets used for linear regression analysis (Supplementary Material, section 3). To avoid ambiguity, only the class i mRNAs with a single leader EEJ and uAUG were analysed. uORF translation was expressed as normalized ribosome profile: sum of the ratio of ribosome profile to mRNA profile at the single nucleotide level, and divided by uORF length (Supplementary Material, equation in section 3). The class i uORFs chosen did not overlap with the mORFs.
For all the datasets analyzed, uORF translation increased in close proximity to the leader EEJ (∼200 bases) and peaked around the co-location point of the EJC and uAUG (Supplementary Figure S11A, red dotted line, and Figure S11B, blue lines). These results suggest that the uAUGs located around the EJCs may initiate and translated better, as the nearby EJCs may interact dynamically with the ribosome preinitiation complexes to promote initiation (97). A similar relationship was also observed for the mORFs (Figure 3C, right panel).
In addition to the EJC, a dynamic mark on the mRNAs that could modulate translation is m1A (N1-methyladenosine) RNA modification, which is commonly present at the 5′ end of mRNAs (98). Therefore, previously reported m1A modification sites were analyzed (98). Strong signals of m1A modification were found at −88 position of the leader EEJs (Figure 8C), similar to that of all the other EEJs, further supporting some possible regulatory roles of the leader EEJs.
Concluding remarks
Overall, our cross species and multi-omic analysis has shown that the position of EEJ in the mRNA leader is associated with a site of translation initiation. When translation starts at cognate (AUG) uORFs, in particular for those mRNAs with leader EEJs (class i), protein expression is significantly lower than those without leader EEJs (class iii). Based on these findings and previous work, here we present two possible models.
The first model is motivated by an evolutionary perspective. Introns are distributed unevenly along genes as a result of purifying selection (Figure 1). In particular, 5′UTR introns and some flanking sequences are maintained because they may contain regulatory sequences controlling gene expression (51,53,99). A common regulatory element is a uORF. Therefore, the leader EEJs and uORFs are frequently juxtaposed (Figure 7A). The frequency of leader EEJs is a strong predictor of lower mORF translation (Figure 3A and B, and Supplementary Figure S5), which may be due to the co-occurrence of nearby regulatory elements such as uORFs and RNA structures. The leader EEJs in conjunction with uORFs predict the lowest translation rate of the mORFs (Figures 4 and 5, and Supplementary Figures S8 and S9), which may be because these uORFs are likely more inhibitory than the other classes of uORFs.
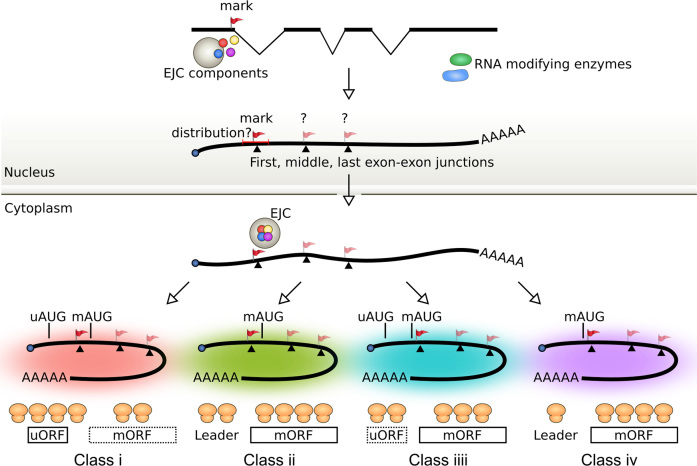
In an additional model, we postulated that a mark(s) is retained on mRNAs after splicing and nuclear export, which modulates such translation events in the cytoplasm (Figure 9). mRNAs can be marked in several ways in the nucleus which subsequently affects cytoplasmic events (11,13,98,100–104). These marks may be covalently or noncovalently bound, be persistent, or dynamic. Well-established persistent covalent modifications are the 5′-m7G-cap (5′-7-methylguanosine cap) and poly(A) tail—marking the two ends of most mRNAs. These two persistent marks dictate the fate of mRNAs, in particular in nuclear export, translation and stability. Notably, recent groundbreaking high-throughput studies have mapped dynamic epitranscriptomic marks, in particular methylated adenosine, on some mRNAs (98,100,101). One recent study showed that m1A modification occurs predominantly at the 5′ end of mRNAs, in particular the downstream of first EEJ (98). Our analysis also showed that the peak of this modification in the mRNA leaders is prior to the leader EEJs (Figure 8C). This modification correlates with an increase in mORF translation and is consistent with the uORF translation data presented here.
Figure 9.
Model of mRNA translation regulated by the putative marks around the (leader) EEJs. Introns are removed from the pre-mRNAs in the nucleus. This splicing leaves a mark(s) in the mRNA (red flag). One possibility for this mark(s) is the exon junction complex (EJC). The EJCs remain bound to the mRNAs from nuclear export to translation, serving as a ‘memory’ of splice junctions. Other dynamic modifications and potential marks would be RNA modifications. Both EJCs and methyladenosine have been shown to mark the mRNAs dynamically and are related to translation. The positions of these marks may influence the translation of either uORFs and/or mORFs. EEJ, exon-exon junction; EJC, exon junction complex; mAUG, main AUG; mORF, main open reading frame; uAUG, upstream AUG; uORF, upstream open reading frame (with AUG initiation codon).
A more well-established possibility for the mark at the leader EEJ is the exon–junction complex (EJC). The EJC is deposited 20–24 bases precedes almost every EEJ after splicing (11,13,102,103). The EJC contains four core proteins (MLN51, Y14, Magoh and eIF4A4) and accessory proteins. The EJCs remain bound to the mRNAs after nuclear export. Thus, the mRNAs in the cytoplasm retains a ‘memory’ of the splice junctions (11,105,106). Several EJC components have been previously shown to enhance translation in the cytoplasm, in particular MLN51, a translation activator that binds to eIF3 (11,107–112). Our analysis of MLN51 binding site data showed that these sites were bound before the leader EEJs (Figure 8B). Although the EJCs are removed during the pioneer round of translation (113,114), it is possible that the interaction between MLN51 and eIF3 persists for the subsequent round of translation (11).
There are at least two proteins that link the EJC to translation initiation, which may in part explain why the translation rate peaks around the co-location point the EJC and initiation codon: PYM (Partner of Y14 and Magoh) and SKAR [S6 kinase 1 (S6K1) ALYREF-like] (11,97). During the pioneer round of translation, a ribosome preinitiation complex may interact with PYM, which promotes translation initiation and disassembly of EJC (112,114). Whereas SKAR interacts with the EJC core protein eIF4A3 to promote the pioneer round of translation initiation for a subset of mRNAs, which are regulated by the mammalian target of the rapamycin complex 1 (mTORC1)-S6K1 signalling pathway (115). In general, this mTORC1 pathway is promoted by growth factors but inhibited by stress (116). In future, it would be interesting to compare the translation events of the CBC-bound and eIF4E-bound mRNAs using Ribo-seq, in order to understand the effects of EJCs on the pioneer round and steady-state of translation, respectively.
DATA AVAILABILITY
The mass spectra and RNA-seq datasets of this study are available on PRIDE (24) (PXD006661) and Gene Expression Omnibus (25) (GSE99697), respectively. Publicly available high-throughput sequencing and shotgun proteomic datasets used in this study are listed in Supplementary Table S1. The processed Ribo-seq and proteomic data are available in Supplementary Tables S2–S4 and S5, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
C.S.L. is a recipient of a Dr Sulaiman Daud 125th Jubilee Postgraduate Scholarship. We are grateful to Mr Andrew Gray for fruitful discussions of the linear regression analysis. We thank Dr Daniel Garama for his helpful guidance throughout the proteomics experiments, and Dr Monika Zavodna of Otago Genomics and Bioinformatics Facility for valuable discussions on the RNA-seq experiments.
Author Contributions: All authors designed the experiments. C.S.L. did the bioinformatics analysis. S.J.T.W. and T.K. did the proteomics experiments. All authors wrote and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Otago Research Grants (to C.M.B. and T.K.). Funding for open access charge: University of Otago.
Conflict of interest statement. None declared.
REFERENCES
- 1. Koonin E.V., Csuros M., Rogozin I.B.. Whence genes in pieces: reconstruction of the exon–intron gene structures of the last eukaryotic common ancestor and other ancestral eukaryotes. Wiley Interdiscip. Rev. RNA. 2013; 4:93–105. [DOI] [PubMed] [Google Scholar]
- 2. Csuros M., Rogozin I.B., Koonin E.V.. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput. Biol. 2011; 7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koonin E.V. Intron-dominated genomes of early ancestors of eukaryotes. J. Hered. 2009; 100:618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chorev M., Carmel L.. The function of introns. Front. Genet. 2012; 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jo B.-S., Choi S.S.. Introns: the functional benefits of introns in genomes. Genomics Inform. 2015; 13:112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaul O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017; 91:145–155. [DOI] [PubMed] [Google Scholar]
- 7. Chen M., Manley J.L.. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009; 10:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laxa M. Intron-Mediated Enhancement: A tool for heterologous gene expression in plants. Front. Plant Sci. 2016; 7:1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gallegos J.E., Rose A.B.. The enduring mystery of intron-mediated enhancement. Plant Sci. 2015; 237:8–15. [DOI] [PubMed] [Google Scholar]
- 10. Vain P., Finer K.R., Engler D.E., Pratt R.C., Finer J.J.. Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep. 1996; 15:489–494. [DOI] [PubMed] [Google Scholar]
- 11. Le Hir H., Saulière J., Wang Z.. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016; 17:41–54. [DOI] [PubMed] [Google Scholar]
- 12. Hellens R.P., Brown C.M., Chisnall M.A.W., Waterhouse P.M., Macknight R.C.. The emerging world of small ORFs. Trends Plant Sci. 2016; 21:317–328. [DOI] [PubMed] [Google Scholar]
- 13. Shoemaker C.J., Green R.. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andrews S.J., Rothnagel J.A.. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014; 15:193–204. [DOI] [PubMed] [Google Scholar]
- 15. Hinnebusch A.G., Ivanov I.P., Sonenberg N.. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016; 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brar G.A., Weissman J.S.. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015; 16:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014; 15:205–213. [DOI] [PubMed] [Google Scholar]
- 18. Michel A.M., Fox G., M Kiran A., De Bo C., O’Connor P.B.F., Heaphy S.M., Mullan J.P.A., Donohue C.A., Higgins D.G., Baranov P.V.. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014; 42:D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T. et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015; 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U.. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016; 13:165–170. [DOI] [PubMed] [Google Scholar]
- 21. Raj A., Wang S.H., Shim H., Harpak A., Li Y.I., Engelmann B., Stephens M., Gilad Y., Pritchard J.K.. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife. 2016; 5:e13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malone B., Atanassov I., Aeschimann F., Li X., Großhans H., Dieterich C.. Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res. 2017; 45:2960–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyner C., Barber G.P., Casper J., Clawson H., Diekhans M., Eisenhart C., Fischer C.M., Gibson D., Gonzalez J.N., Guruvadoo L. et al. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 2017; 45:D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mudge J.M., Harrow J.. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015; 26:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshihama M., Nakao A., Kenmochi N.. snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 2013; 6:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flynn R.A., Do B.T., Rubin A.J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E.T., Wysocka J. et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 2016; 23:231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X., Wan J., Liu B., Ma M., Shen B., Qian S.-B.. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015; 12:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park J.-E., Yi H., Kim Y., Chang H., Kim V.N.. Regulation of Poly(A) tail and translation during the somatic cell cycle. Mol. Cell. 2016; 62:462–471. [DOI] [PubMed] [Google Scholar]
- 36. Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J.. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007; 4:923–925. [DOI] [PubMed] [Google Scholar]
- 37. Cutler J.A., Tahir R., Sreenivasamurthy S.K., Mitchell C., Renuse S., Nirujogi R.S., Patil A.H., Heydarian M., Wong X., Wu X. et al. Differential signaling through p190 and p210 BCR-ABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia. 2017; 31:1513–1524. [DOI] [PubMed] [Google Scholar]
- 38. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H., McManus J., Kingsford C.. Isoform-level ribosome occupancy estimation guided by transcript abundance with Ribomap. Bioinformatics. 2016; 32:1880–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lange S.J., Maticzka D., Möhl M., Gagnon J.N., Brown C.M., Backofen R.. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012; 40:5215–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cui X., Wei Z., Zhang L., Liu H., Sun L., Zhang S.-W., Huang Y., Meng J.. Guitar: an R/bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. Biomed Res. Int. 2016; 2016:8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pervouchine D.D., Knowles D.G., Guigó R.. Intron-centric estimation of alternative splicing from RNA-seq data. Bioinformatics. 2013; 29:273–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hauer C., Curk T., Anders S., Schwarzl T., Alleaume A.-M., Sieber J., Hollerer I., Bhuvanagiri M., Huber W., Hentze M.W. et al. Improved binding site assignment by high-resolution mapping of RNA-protein interactions using iCLIP. Nat. Commun. 2015; 6:7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013; 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2017; Vienna: http://www.R-project.org/. [Google Scholar]
- 48. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016; 2nd ednNY: Springer [Google Scholar]
- 49. Lüdecke D. sjPlot: Data Visualization for Statistics in Social Science. 2018; R package version 2.4.1 https://CRAN.R-project.org/package=sjPlot.
- 50. Neuvéglise C., Marck C., Gaillardin C.. The intronome of budding yeasts. C. R. Biol. 2011; 334:662–670. [DOI] [PubMed] [Google Scholar]
- 51. Chung B.Y.W., Simons C., Firth A.E., Brown C.M., Hellens R.P.. Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics. 2006; 7:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hong X., Scofield D.G., Lynch M.. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006; 23:2392–2404. [DOI] [PubMed] [Google Scholar]
- 53. Cenik C., Derti A., Mellor J.C., Berriz G.F., Roth F.P.. Genome-wide functional analysis of human 5′ untranslated region introns. Genome Biol. 2010; 11:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pesole G., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S.. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001; 276:73–81. [DOI] [PubMed] [Google Scholar]
- 55. Shalev A., Blair P.J., Hoffmann S.C., Hirshberg B., Peculis B.A., Harlan D.M.. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002; 143:2541–2547. [DOI] [PubMed] [Google Scholar]
- 56. Janich P., Arpat A.B., Castelo-Szekely V., Lopes M., Gatfield D.. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015; 25:1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C. et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014; 33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu M.-J., Wu S.-H., Wu J.-F., Lin W.-D., Wu Y.-C., Tsai T.-Y., Tsai H.-L., Wu S.-H.. Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 2013; 25:3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dai A., Cao S., Dhungel P., Luan Y., Liu Y., Xie Z., Yang Z.. Ribosome profiling reveals translational upregulation of cellular oxidative phosphorylation mRNAs during vaccinia virus-induced host shutoff. J. Virol. 2017; 91:e01858-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sidrauski C., McGeachy A.M., Ingolia N.T., Walter P.. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015; 4:e05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park Y., Reyna-Neyra A., Philippe L., Thoreen C.C.. mTORC1 balances cellular amino acid supply with demand for protein synthesis through Post-transcriptional control of ATF4. Cell Rep. 2017; 19:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lintner N.G., McClure K.F., Petersen D., Londregan A.T., Piotrowski D.W., Wei L., Xiao J., Bolt M., Loria P.M., Maguire B. et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 2017; 15:e2001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bercovich-Kinori A., Tai J., Gelbart I.A., Shitrit A., Ben-Moshe S., Drori Y., Itzkovitz S., Mandelboim M., Stern-Ginossar N.. A systematic view on influenza induced host shutoff. Elife. 2016; 5:e18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cenik C., Cenik E.S., Byeon G.W., Grubert F., Candille S.I., Spacek D., Alsallakh B., Tilgner H., Araya C.L., Tang H. et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015; 25:1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Loayza-Puch F., Rooijers K., Buil L.C.M., Zijlstra J., Oude Vrielink J.F., Lopes R., Ugalde A.P., van Breugel P., Hofland I., Wesseling J. et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016; 530:490–494. [DOI] [PubMed] [Google Scholar]
- 67. Gonzalez C., Sims J.S., Hornstein N., Mela A., Garcia F., Lei L., Gass D.A., Amendolara B., Bruce J.N., Canoll P. et al. Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J. Neurosci. 2014; 34:10924–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. Genome-wide annotation and quantitation of translation by ribosome profiling. Current Protocols in Molecular Biology. 2013; 103:John Wiley & Sons, Inc; doi:10.1002/0471142727.mb0418s103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vassilenko K.S., Alekhina O.M., Dmitriev S.E., Shatsky I.N., Spirin A.S.. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011; 39:5555–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Johnstone T.G., Bazzini A.A., Giraldez A.J.. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016; 35:706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ye Y., Liang Y., Yu Q., Hu L., Li H., Zhang Z., Xu X.. Analysis of human upstream open reading frames and impact on gene expression. Hum. Genet. 2015; 134:605–612. [DOI] [PubMed] [Google Scholar]
- 77. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stadler M., Fire A.. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011; 17:2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R.. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013; 23:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Batut P., Gingeras T.R.. RAMPAGE: promoter activity profiling by paired-end sequencing of 5′-complete cDNAs. Current Protocols in Molecular Biology. 2013; 104:John Wiley & Sons, Inc; doi:10.1002/0471142727.mb25b11s104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M.. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011; 7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 86. Li J.J., Bickel P.J., Biggin M.D.. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ. 2014; 2:e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Alli Shaik A., Wee S., Li R.H.X., Li Z., Carney T.J., Mathavan S., Gunaratne J.. Functional mapping of the zebrafish early embryo proteome and transcriptome. J. Proteome Res. 2014; 13:5536–5550. [DOI] [PubMed] [Google Scholar]
- 88. Imamachi N., Salam K.A., Suzuki Y., Akimitsu N.. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2016; 27:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D., Fafournoux P.. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 2001; 29:4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Palam L.R., Baird T.D., Wek R.C.. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011; 286:10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nishitoh H. CHOP is a multifunctional transcription factor in the ER stress response. J. Biochem. 2012; 151:217–219. [DOI] [PubMed] [Google Scholar]
- 92. Oyadomari S., Mori M.. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2003; 11:381–389. [DOI] [PubMed] [Google Scholar]
- 93. Vattem K.M., Wek R.C.. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., Walter P.. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016; 351:aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P.. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012; 40:2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hauer C., Sieber J., Schwarzl T., Hollerer I., Curk T., Alleaume A.-M., Hentze M.W., Kulozik A.E.. Exon junction complexes show a distributional bias toward alternatively spliced mRNAs and against mRNAs coding for ribosomal proteins. Cell Rep. 2016; 16:1588–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Maquat L.E., Tarn W.-Y., Isken O.. The pioneer round of translation: features and functions. Cell. 2010; 142:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016; 530:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bicknell A.A., Cenik C., Chua H.N., Roth F.P., Moore M.J.. Introns in UTRs: why we should stop ignoring them. Bioessays. 2012; 34:1025–1034. [DOI] [PubMed] [Google Scholar]
- 100. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wan Y., Tang K., Zhang D., Xie S., Zhu X., Wang Z., Lang Z.. Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015; 16:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bono F., Gehring N.H.. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011; 8:24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Le Hir H., Séraphin B.. EJCs at the heart of translational control. Cell. 2008; 133:213–216. [DOI] [PubMed] [Google Scholar]
- 104. Singh G., Pratt G., Yeo G.W., Moore M.J.. The clothes make the mRNA: Past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015; 84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mufarrege E.F., Gonzalez D.H., Curi G.C.. Functional interconnections of Arabidopsis exon junction complex proteins and genes at multiple steps of gene expression. J. Exp. Bot. 2011; 62:5025–5036. [DOI] [PubMed] [Google Scholar]
- 106. Nyikó T., Kerényi F., Szabadkai L., Benkovics A.H., Major P., Sonkoly B., Mérai Z., Barta E., Niemiec E., Kufel J. et al. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 2013; 41:6715–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nott A., Le Hir H., Moore M.J.. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004; 18:210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kamo K., Kim A.-Y., Park S.H., Joung Y.H.. The 5′UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in Gladiolus and Arabidopsis. BMC Plant Biol. 2012; 12:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Matsumoto K., Wassarman K.M., Wolffe A.P.. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998; 17:2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wiegand H.L., Lu S., Cullen B.R.. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11327–11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chazal P.-E., Daguenet E., Wendling C., Ulryck N., Tomasetto C., Sargueil B., Le Hir H.. EJC core component MLN51 interacts with eIF3 and activates translation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5903–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Diem M.D., Chan C.C., Younis I., Dreyfuss G.. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 2007; 14:1173–1179. [DOI] [PubMed] [Google Scholar]
- 113. Sato H., Maquat L.E.. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009; 23:2537–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gehring N.H., Lamprinaki S., Kulozik A.E., Hentze M.W.. Disassembly of exon junction complexes by PYM. Cell. 2009; 137:536–548. [DOI] [PubMed] [Google Scholar]
- 115. Ma X.M., Yoon S.-O., Richardson C.J., Jülich K., Blenis J.. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008; 133:303–313. [DOI] [PubMed] [Google Scholar]
- 116. Ma X.M., Blenis J.. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009; 10:307–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectra and RNA-seq datasets of this study are available on PRIDE (24) (PXD006661) and Gene Expression Omnibus (25) (GSE99697), respectively. Publicly available high-throughput sequencing and shotgun proteomic datasets used in this study are listed in Supplementary Table S1. The processed Ribo-seq and proteomic data are available in Supplementary Tables S2–S4 and S5, respectively.