Abstract
Octopus-toolkit is a stand-alone application for retrieving and processing large sets of next-generation sequencing (NGS) data with a single step. Octopus-toolkit is an automated set-up-and-analysis pipeline utilizing the Aspera, SRA Toolkit, FastQC, Trimmomatic, HISAT2, STAR, Samtools, and HOMER applications. All the applications are installed on the user's computer when the program starts. Upon the installation, it can automatically retrieve original files of various epigenomic and transcriptomic data sets, including ChIP-seq, ATAC-seq, DNase-seq, MeDIP-seq, MNase-seq and RNA-seq, from the gene expression omnibus data repository. The downloaded files can then be sequentially processed to generate BAM and BigWig files, which are used for advanced analyses and visualization. Currently, it can process NGS data from popular model genomes such as, human (Homo sapiens), mouse (Mus musculus), dog (Canis lupus familiaris), plant (Arabidopsis thaliana), zebrafish (Danio rerio), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and budding yeast (Saccharomyces cerevisiae) genomes. With the processed files from Octopus-toolkit, the meta-analysis of various data sets, motif searches for DNA-binding proteins, and the identification of differentially expressed genes and/or protein-binding sites can be easily conducted with few commands by users. Overall, Octopus-toolkit facilitates the systematic and integrative analysis of available epigenomic and transcriptomic NGS big data.
INTRODUCTION
The development of new technologies frequently fosters advances in biology. Next-generation sequencing (NGS) approaches, which enable the rapid and accurate sequencing of short DNA fragments, have changed the ways researchers approach biological problems. The availability of rapidly growing databases containing large-scale genomic information permits the establishment of complex putative molecular maps that serve as hypothesis generators. These hypotheses, in turn, can be tested using more traditional biological experimentation. A variety of NGS-based techniques have been developed. For example, chromatin immunoprecipitation coupled with parallel sequencing (ChIP-seq) is widely used to assess the binding of proteins to the genome (1). RNA sequencing (RNA-seq) can estimate the abundance of whole transcripts and their isoforms (2). Genome-wide nucleosome positioning and open chromatin can be captured by MNase-seq (3) and DNase-seq (4), respectively. As the demand for NGS has increased, several thousand NGS-based data sets have been deposited in public data repositories such as gene expression omnibus (GEO) (5). Notably, novel findings frequently emerge from reanalyzing available NGS-based data sets (6,7). However, there is no easy way to access, download, and process a large set of original (raw) NGS-based data for comparative and integrative analysis, although some web-based applications have been developed to resolve the issue. For example, Galaxy (8) offers users configurable workflows for integrating and analyzing NGS data, but is relatively slow compared with standalone applications. NGS data deposited in the GEO database can be visualized through the genome data viewer function provided by the national center for biotechnology information (NCBI) website. However, basic and advanced analyses cannot be conducted on the website. To fill the gaps between researchers and biological big data deposited in GEO, we have developed an automated epigenomic and transcriptomic NGS-based data analysis workflow called Octopus-toolkit. It is an optimized workflow for retrieving and analyzing hundreds of public epigenomic and transcriptomic NGS-based data sets routinely with a personal computer in a few steps. Various types of epigenomic and transcriptomic NGS-based data, including transcriptomic data, can be processed, visualized, and analyzed in a single step. Advanced analyses, including the identification of differentially expressed genes (DEGs) for RNA-seq and peak calling for ChIP-seq, can be easily conducted with the outputs of Octopus-toolkit. Therefore, The Octopus-toolkit can accelerate the data mining of public epigenomic and transcriptomic NGS data for basic biomedical research.
MATERIALS AND METHODS
Development environment
The Octopus-toolkit was developed using the Java programming language (JDK1.8; Java Development Kit 8) with the Eclipse tool (Neon 1a.service release Java EE IDE, https://www.eclipse.org/), which is an integrated development environment tool for various programming languages. We developed the graphic user interface (GUI) using Swing (version 1.8) and WindowBuilder (version 1.8) in Eclipse. Python (version 2.7.12) was used to connect the analysis steps and the R language (version 3.2.3) was used to create the function generating heatmap and line plot. The Octopus-toolkit has been uploaded to the Github website (https://github.com/kangk1204/Octopus-toolkit2).
Data download and format conversion
Next-generation sequencing approaches typically generate FASTQ files of up to several gigabytes. To speed up file transfers, Octopus-toolkit incorporates the Aspera high-speed file transfer protocol (http://asperasoft.com/). It automatically retrieves key information listed on the GEO website, including sample name, organism (reference genome), library strategy (single-end or paired-end), and instrument model. Currently, NGS data sequenced by the Illumina instrument including Hiseq 2500 can only be processed by Octopus-Toolkit. Once the download has been completed, the SRA files are converted to FASTQ files by using SRA Toolkit (http://www.ncbi.nlm.nih.gov/books/NBK158900/).
Quality control
To check the quality of raw sequence data in the FASTQ format, FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), one of most popular quality check programs, was integrated into Octopus-toolkit. After the file conversion, FastQC checks the quality of part of the converted files and provides a quick overview of the sequencing quality.
Quality trimming and alignment
A flexible trimming tool called Trimmomatic (9) removes the unreliable parts (incorrectly called bases) of given reads before aligning reads to a reference genome. Then, the trimmed reads are mapped to the corresponding reference genome using either STAR for RNA-seq (10) or HISAT2 for RNA-seq, ChIP-seq, MeDIP-seq, ATAC-seq, MNase-seq, and DNase-seq (11). The aligned reads are stored in the BAM format (12).
Data processing
Hypergeometric optimization of motif enrichment (HOMER) is a suite of tools for motif discovery and various next-generation sequencing analyses (13). It is a versatile application that can analyze most epigenomic and transcriptomic NGS data types, including RNA-seq, ChIP-seq, MeDIP-seq, ATAC-seq and DNase-seq. Therefore, Octopus-Toolkit primarily utilizes it for advanced analyses. After the alignment, HOMER will convert BAM files into the tag-directory format to generate BigWig files (14) for visualization and to conduct advanced analyses. Directory structures generated by Octopus-toolkit and detailed commands for advanced analyses are described in Supplementary Figure S1.
RESULTS
Octopus-toolkit workflow
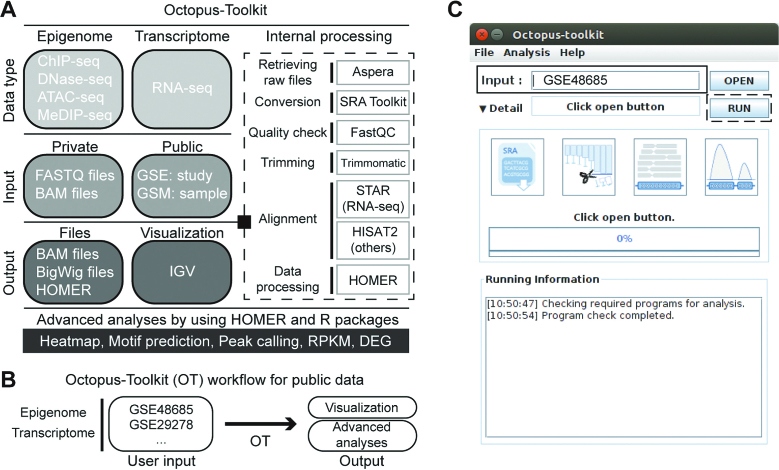
Octopus-toolkit is developed for biologists who lack formal computer science training but are struggling to utilize public NGS-based data sets. Octopus-toolkit operates in two different modes: private and public (Figure 1A). The private mode was designed to process the user's own NGS data (FASTQ files), while the public mode can analyze public NGS data by retrieving raw files (SRA files) from the GEO database. Among various types of NGS data, Octopus-toolkit can process the following two types of epigenomic and transcriptomic data: antibody- or enzyme-mediated experiments (chromatin immunoprecipitation with massively parallel DNA sequencing, ChIP-seq; methylated DNA immunoprecipitation with massively parallel DNA sequencing, MeDIP-seq; assay for transposase-accessible chromatin with high-throughput sequencing, ATAC-seq; sequencing for micrococcal nuclease-sensitive sites, MNase-seq and DNase I-hypersensitive sites sequencing, DNase-seq) and experiments for the quantification of gene expression (whole-transcriptome shotgun sequencing, RNA-seq). Currently, NGS data from human (Homo sapiens), mouse (Mus musculus), dog (Canis lupus familiaris), plant (Arabidopsis thaliana), zebrafish (Danio rerio), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans) and budding yeast (Saccharomyces cerevisiae) species can be automatically processed through Octopus-toolkit. The only difference between the private and public modes is the initial process, for which the public mode requires original files to be downloaded, while the private mode uses users’ own FASTQ files. Therefore, we focused on the public mode to explain how a given data set can be processed via Octopus-toolkit (Figure 1A). Briefly, a high-speed file transfer application called Aspera (http://asperasoft.com/) is used to download original files in the SRA format. When original FASTQ files are submitted to the GEO repository, the files are stored in the SRA format to reduce file size. Therefore, SRA files have to be converted to FASTQ files using the SRA toolkit application (http://www.ncbi.nlm.nih.gov/Traces/sra/). After the conversion, the quality of sequenced reads in the FASTQ files is determined by using the FastQC program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Subsequently, low-quality portions of reads are removed with Trimmomatic (9). There are two types of data: genome-based or transcriptome-based (splicing events frequently occur) data. For genome-based data including ChIP-seq, MeDIP-seq, ATAC-seq, MNase-seq, and DNase-seq, trimmed reads are aligned to corresponding reference genomes using the HISAT2 aligner (11). On the other hand, trimmed reads in RNA-seq data are mapped to a reference genome using the STAR aligner (10) for fast processing. Both aligners generate mapped reads in the BAM format. Lastly, the BAM files are processed using a versatile NGS-data analysis tool called HOMER (13) for visualization and advanced analyses. For the visualization, BigWig files (14) can be used to examine any loci (or genes) of interest via the integrative genomics viewer (IGV) (15). When it comes to RNA-seq data, the abundances of gene expression levels are estimated by means of the reads per kilobase per million mapped reads (RPKM) method (2) and are stored as text files in a designated directory (Supplementary Figure S1). All of the required programs are automatically installed on the user's Linux- or Mac-based computer at the first execution of Octopus-toolkit. In addition, all the steps mentioned above are internally processed; therefore, the daunting task for novice users can be shortened. In sum, the input of Octopus-toolkit is a list of GEO accession numbers, such as GSE (study) and GSM (sample) (Figure 1B and C), and the output is processed files for visualization and advanced analyses (Supplementary Figure S1). In the following sections, we have demonstrated the usefulness of Octopus-toolkit by reanalyzing available mouse and budding yeast NGS data.
Figure 1.
Octopus-Toolkit workflow. (A) Detailed information on data types, input, and output for Octopus-Toolkit is shown. Programs associated with Octopus-Toolkit and their purposes are described (dashed line box). (B) An example of its use is depicted. (C) Graphical user interface of Octopus-Toolkit is shown. The only input required for Octopus-Toolkit is an accession number for epigenomic and transcriptomic NGS data sets (GSE accession number) or a single piece of NGS data (GSM accession number) (black box). Multiple NGS data sets can be sequentially processed by providing a list of GSE (or GSM) accession numbers as a text file. Octopus-Toolkit runs all the steps after the Run icon (dotted line box) is clicked on.
Case one: Integrative analysis of two independent mouse STAT5 studies (GSE48685 and GSE 31578)
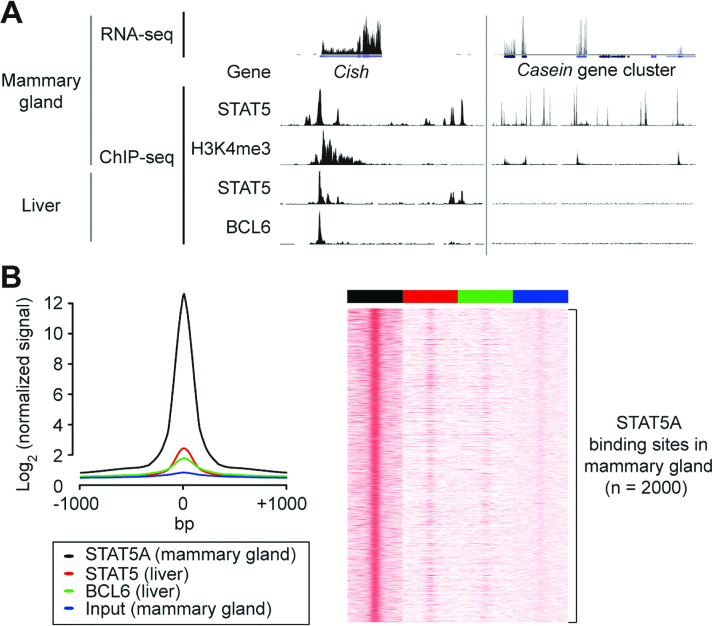
Our previous study (7) reveals that STAT5 acts at an early stage of mouse mammary gland development to establish transcription complexes on mammary-specific genes. STAT5A and H3K4me3 ChIP-seq as well as RNA-seq data were used as the primary approach. To demonstrate the usefulness of Octopus-Toolkit, we reanalyzed the data (GSE48685) (7) along with the mouse liver STAT5 ChIP-seq set (GSE31578) (16). By inputting two GSE accession numbers into Octopus-toolkit, all steps necessary for processing the NGS data were conducted automatically. Briefly, a total of one RNA-seq and 29 ChIP-seq samples (20GB of SRA files) were transferred within 10 min (65 megabytes per second, depending on the network environment) and converted to FASTQ files (∼195GB). All of the files were sequentially processed on a desktop computer (Intel i5–2550K 4-Core 3.40GHz; 32GB memory) with a single step. Approximately, it took 11 h to complete the process. Some of the BigWig files were uploaded to the UCSC genome browser for visualization (Figure 2). Mammary-specific STAT5 binding in the mammary gland but not liver tissues was observed within the Casein gene cluster (milk protein genes), while Cish seemed to be a generic target of STAT5 regardless of cell types as previously described (7). Accordingly, there is strong STAT5 binding and H3K4me3 (active histone mark) enrichment near the Casein genes, while no binding of STAT5 and BCL6 was observed in liver tissue, suggesting mammary-specific regulation of the locus (Figure 2). At this point, users can explore any loci or genes of interest through the genome browser, since NGS analysis generates molecular maps of the genome. For example, the top ten most highly expressed genes at day 1 of lactation (RNA-seq data)—Csn2, Csn1s2a, Wap, Glycam1, Csn1s1, Csn3, Spp1, Trf, Csn1s2b, and Lao1 (7)—were easily validated with the expression table generated by Octopus-toolkit. In sum, Octopus-toolkit provides users with an easy way to generate and explore various molecular maps deposited in the GEO database, which can fill the gap between users and public NGS data.
Figure 2.
UCSC browser snapshot of RNA-seq and STAT5 ChIP-seq performed in mouse mammary gland (GSE48685) and liver (GSE31578) tissues. (A) Each track indicates either an RNA-seq or ChIP-seq sample. Peaks on the ChIP-seq tracks represent binding (or enrichment) of proteins, while peaks on the RNA-seq track indicate relative expression levels of genes. (B) Line plot and heatmap generated by Octopus-toolkit clearly show tissue-specific binding of STAT5 between mammary gland and liver tissues.
Case two: Identifying potential DNA-binding proteins recruiting histone-modifying complexes in Saccharomyces cerevisiae (GSE52339)
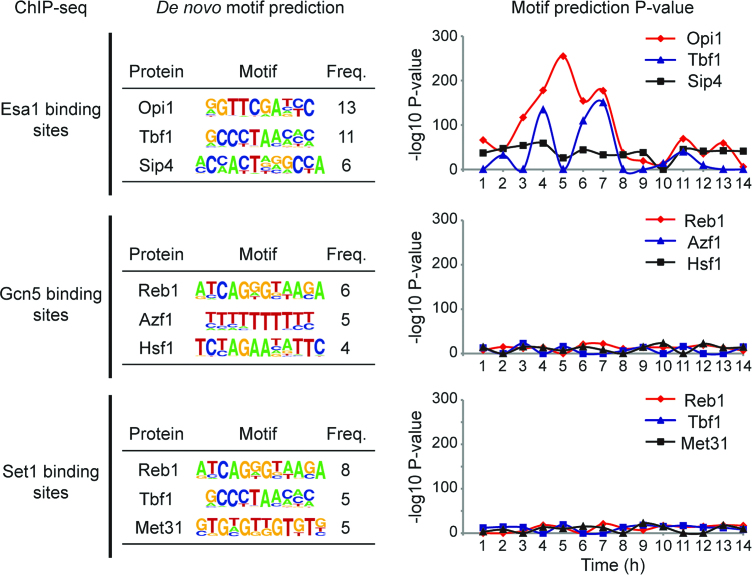
The spatiotemporal recruitment of particular histone-modifying complexes to specific genomic sites is an important mechanism for gene regulation by which a number of histone modifications can be altered dynamically (17). There are four highly conserved core histones—H2A, H2B, H3 and H4—as well as several variants, such as H2A.Z. Each type of histone harbors different sites of post-translational modifications, including acetylation, methylation, phosphorylation, SUMOylation, and ubiquitylation (18). Although the enzymes mediating acetylation on the lysine residues of histone tails have been extensively studied, the molecular mechanism by which histone-modifying enzymes are recruited to specific regions is largely unknown. Recently, Kuang et al. (19) investigated the dynamic changes of histone modifications (H3, H3K4me3, H3K36me3, H3K9ac, H3K56ac, H4K5ac and H4K16ac) and genomic binding of histone-modifying enzymes (ESA1, GCN5 and SET1) at different time points in glucose-limited conditions in S. cerevisiae. To identify potential DNA-binding proteins that might recruit the histone-modifying enzymes, we reanalyzed the ChIP-seq data set (GSE52339) by using the Octopus-Toolkit. Up to 1360 genomic regions were occupied by the histone-modifying enzymes with a false discovery rate (FDR) cutoff of 0.001. We next examined whether any of the DNA sequences (motifs) significantly occurred in these binding sites at each time point by using the motif search function of HOMER (Supplementary Figure S1). Intriguingly, the majority of ESA1-binding sites contained a significant number of OPI1-binding motifs, while the binding regions of GCN5 or SET1 did not include any concordant motifs of known proteins (Figure 3). This result clearly suggested that spatiotemporal recruitment of the ESA1-containing NuA4 histone acetyltransferase complex might be mediated by OPI1. Further experimental validation will reveal the molecular mechanism underlying the recruitment of the NuA4 complex. In support of this, recent studies (20,21) demonstrated the new mechanistic dissection between the NuA4 complex and phospholipid homeostasis, further enforcing the suggested direct link between ESA1 and OPI1.
Figure 3.
De novo motif prediction on ESA1, GCN5 and SET1-binding sites. HOMER was used to predict DNA-binding motifs of proteins significantly associated with each histone-modifying protein. Top three motifs are shown (left panel). Significance of the motifs at each time point is shown (right panel).
DISCUSSION
Owing to its low cost, high throughput, and fast sequencing chemistry, next-generation sequencing has become an effective tool for profiling molecules and their interactions in a variety of fields, such as molecular biology, cancer, immunology, epigenetics, and stem cell research. Although a large amount of NGS data have been accumulated, giving rise to biological big data, they are still in quarantine at least in part. In the era of NGS, research paradigms have shifted from hypothesis-driven approaches to hypothesis-generating science. Therefore, the mining of previous high-throughput data, such as NGS, has become an essential step for setting the research direction. In this study, we have shown the potential of mining epigenomic and transcriptomic big data from GEO using Octopus-toolkit. For example, the reanalysis of ESA1 ChIP-seq data revealed that OPI1 DNA-binding motif significantly occurred in the ESA1 binding regions in S. cerevisiae (Figure 3), which was not highlighted in the previous study (19).
Galaxy (8) and GenePattern (22) are open-source, web-based platforms that consist of various analysis tools for NGS data analysis. Such web-based tools enable users to establish various combinations of analysis pipeline, and avoids tedious manual installation of analysis tools. Moreover, the analyses are processed via servers, eliminating the need for researchers to equip with necessary hardware. However, to make use of the web-based tools, the user needs a substantial pre-knowledge on NGS analysis pipeline (Supplementary Figure S2). Also, web-based tools provide limited space since the space are shared by the users from all over the world. For example, Galaxy provides 250Gb per account with maximum uploading file size of 2Gb. It also takes up a significant time for analyzing large-size data, compared to small-size data, due to the sharing of analysis power. Octopus-toolkit, on the other hand, provides user-friendly interface that users with basic NGS analysis pipeline knowledge can comfortably operate desired analysis; and enables the addition of analysis space if necessary. Octopus-toolkit can deliver the result faster than Galaxy or GenePattern if one is equipped with a computer that is capable of NGS analysis. Nevertheless, Octopus-toolkit has the following limitations that should be improved in the near future. First, although the installation of a Linux or Mac operating system (OS) is much easier than before, the fact that the Octopus-toolkit runs on Linux or Mac can be a difficult barrier for novice users. Second, it cannot process available DNA-seq data including whole-genome sequencing (WGS) or whole-exome sequencing (WES), which are used to detect DNA variants. We hope that this will be implemented in a future version of the Octopus-toolkit. Third, the batch effect, which must be taken into account when conducting meta-analysis using data from different studies, needs further improvements. Although we have provided an example to show how to remove the batch effect for RNA-seq using edgeR (Supplementary Figure S3), users must be aware of the batch effect when it comes to comparing independent studies using the Octopus-toolkit. Fourth, the amount of newly produced NGS data is escalating and there are cases that require the processing of more than 1000 samples. Octopus-toolkit is capable of processing >1000 samples (with no actual limit on the number of samples) but it would take a considerable length of time to process the data on a personal computer. To resolve this issue, a distributing and parallel sample processing system, such as the cloud system or high performance computing (HPC), can be incorporated. We intend to include these features in the Octopus-toolkit for rapid and efficient processing of data in the next version. Despite these shortcomings, the Octopus-toolkit will be useful for users who are willing to mine the many epigenomic and transcriptomic treasures that are publicly hidden as more data becomes available (23).
AVAILABILITY
The Octopus-toolkit software is available at the Github repository (https://github.com/kangk1204/Octopus-toolkit2). Detailed tutorials and manuals are available at the following links: http://octopus-toolkit2.readthedocs.io/en/latest/#.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jinmo Jung for helpful discussion. Lothar Hennighausen is funded by NIDDK/NIH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea [1720100]; Mid-career Researcher Program through the National Research Foundation of Korea (NRF) [2016R1A2B2006354] funded by the Ministry of Science and ICT. Funding for open access charge: National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea [1720100]; Mid-career Researcher Program through the National Research Foundation of Korea (NRF) [2016R1A2B2006354] funded by the Ministry of Science and ICT.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 2. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 3. Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E.. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008; 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang K., Yamaji D., Yoo K.H., Robinson G.W., Hennighausen L.. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol. Cell. Biol. 2014; 34:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005; 15:1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D.. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010; 26:2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Laz E.V., Waxman D.J.. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 2012; 32:880–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Reinberg D.. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001; 15:2343–2360. [DOI] [PubMed] [Google Scholar]
- 18. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuang Z., Cai L., Zhang X., Ji H., Tu B.P., Boeke J.D.. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat. Struct. Mol. Biol. 2014; 21:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salas-Santiago B., Lopes J.M.. Saccharomyces cerevisiae essential genes with an Opi- phenotype. G3 (Bethesda). 2014; 4:761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dacquay L., Flint A., Butcher J., Salem D., Kennedy M., Kaern M., Stintzi A., Baetz K.. NuA4 lysine acetyltransferase complex contributes to phospholipid homeostasis in Saccharomyces cerevisiae. G3 (Bethesda). 2017; 7:1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P.. GenePattern 2.0. Nat. Genet. 2006; 38:500–501. [DOI] [PubMed] [Google Scholar]
- 23. Gewin V. Data sharing: an open mind on open data. Nature. 2016; 529:117–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.