Abstract
RPA is known to stimulate the helicase activity of Werner syndrome protein (WRN), but the exact stimulation mechanism is not understood. We use single-molecule FRET and magnetic tweezers to investigate the helicase activity of WRN and its stimulation by RPA. We show that WRN alone is a weak helicase which repetitively unwind just a few tens of base pairs, but that binding of multiple RPAs to the enzyme converts WRN into a superhelicase that unidirectionally unwinds double-stranded DNA more than 1 kb. Our study provides a good case in which the activity and biological functions of the enzyme may be fundamentally altered by the binding of cofactors.
INTRODUCTION
Werner syndrome protein (WRN) is a human helicase which plays important roles in maintaining the genome integrity by participating in a variety of cellular processes such as DNA replication, repair, telomere maintenance, and homologous recombination (1–3). A defect in the wrn gene causes a autosomal recessive disease called Werner syndrome (WS), which is characterized by premature aging and predisposition to cancer (4,5). WRN belongs to the RecQ helicase superfamily, which is well conserved from Escherichia coli to human (3). WRN possesses not only 3′-5′ unwinding activity but also 3′-5′ exonuclease and single-stranded DNA (ssDNA) annealing activities (6–8). Like many other RecQ helicases, WRN can resolve a variety of secondary structures like forked duplex, displacement loops, bubbles, Holliday junctions, G-quadruplexes and triplexes (9,10). Structure specificity and versatile catalytic activities makes WRN indispensable for DNA metabolism including preservation of genome such as branch migration, regression of stalled fork, and the resolution of telomeric secondary structures (1).
WRN interacts with many proteins in DNA metabolism (1,11–13). These include replication protein A (RPA), the ssDNA binding protein in eukaryote which is essential in replication, repair and recombination (14). This is one of WRN’s strongest interactors and greatly stimulates the unwinding activity of WRN (15,16). While WRN alone can only unwind a short stretch of base pairs, the addition of RPA enables the enzyme to unwind much longer stretches (16). In-vivo, RPA has been found to co-localize with WRN at foci of DNA damage and replication arrest (17–19), suggesting that the two proteins collaborate in DNA repair. We can imagine two different ways by which RPA stimulates the unwinding activity of WRN. First, RPA can inhibit the rewinding of the unwound duplex by coating the unwound single-stranded DNA. Second, RPA can enhance the unwinding activity of WRN through protein–protein interaction. Biochemical studies showed that RPA and WRN physically interact, and that the interaction is critical for the enhanced helicase activity of WRN in the presence of RPA (16,20,21). A high affinity RPA-interaction site in WRN was mapped to the two strongly acidic amino acid repeats located between the exonuclease domain and the helicase core domain (21). Using truncated RPA mutants it was shown that the interaction with WRN is mediated by the residues of the RPA70 subunit, which overlaps with the ssDNA-binding domains (20). Based on these observations, one can imagine that direct binding of RPA to WRN might induce a conformational change which can affect robust unwinding. However, the mechanisms by which WRN unwinds DNA and how RPA stimulates its the helicase activity are not understood.
Here, we report single molecule FRET and magnetic tweezers assays to investigate the helicase activity of WRN alone and its stimulation by RPA. To study solely the effects of protein–protein interaction, all critical experiments were performed in the absence of free proteins. Like other RecQ helicases, WRN alone exhibited repetitive unwinding on a forked DNA substrate. When WRN was incubated with low concentration of RPA (1 nM), the repetitive unwinding was maintained, but the number of unwound base pairs slightly increased. When WRN was incubated with higher concentration of RPA (20 nM), WRN became a superhelicase that unidirectionally unwounds the duplex up to 1 kb. Therefore, our study clearly reveals that WRN’s activity is differently modulated by RPA in a concentration dependent manner, and that full stimulation of the helicase activity of WRN requires the binding of multiple RPAs.
MATERIALS AND METHODS
Purification of human WRN protein
Human WRN and its mutant were expressed and purified using a baculovirus/Sf9 insect cell system as previously described (22). Human RPA was expressed from plasmid vector p11d-tRPA (gift from Dr Marc S. Wold) (23) in E. coli BL21 (Novagen) and purified as previously described (24). Purified WRN protein is shown in Supplementary Figure S1.
DNA fork substrate for single-molecule FRET assay
DNA strands were purchased from Integrated DNA Technologies (Coralville, IA). Oligonucleotide sequences are described in Supplementary Table S1. Dyes were labeled at the amine-modified thymine bases (/iAmMC6T/). DNA fork were prepared by mixing two strands, and slowly cooling down from 95°C to 4°C with a cooling rate of −1°C per minute.
DNA hairpin constructs for magnetic tweezers assay
DNA hairpin constructs with stem of 50, 588 and 1038 bp were used in the experiment and preparation procedure was explained in Supplementary Methods in detail. The constructs consist of three part; hairpin, insert, and digoxigenine handle. The hairpin contains biotins at its 5′ end for attachment to streptavidin coated magnetic beads and 30 bases single stranded region for helicase loading. The digoxigenine handle is for tethering to glass surface coated with anti-digoxigenine (Roche Diagnostics). The dsDNA of 800 bp was inserted as a spacer between the hairpin and the digoxigenine handle to prevent nonspecific attachment of magnetic beads to the glass surface. Hairpin part of the 50 bp construct was made by annealing two oligonucleotides; One biotinylated and the other with a sequence for ligation with insert (Integrated DNA Technologies, Coralville, IA). For longer hairpin constructs, long dsDNA serving as a stem was ligated with a loop oligonucleotide and a partial duplex which contain biotins and the sequence for subsequent ligation on each end. The insert, the stem, the digoxigenine handle were prepared by digesting PCR products with restriction or nicking endonucleases to have proper the appropriate sticky end. Digoxigenine labels of the handle were introduced by PCR carried out in the presence of digoxigenine-dUTP (Roche Diagnostics). The entire constructs was completed by ligating the three. To increase the yield of ligation between hairpin and insert, we used nonpalindromic 10 base overhang generated with nicking endonuclease as the sticky end as described previously (25).
Single-molecule FRET assay
To reduce non-specific binding of proteins and DNA, quartz slides (Finkenbeiner) and cover slips were coated with polyethyleneglycol (mPEG-Succinimidyl Valerate, MW 5000, Laysan) as described (26,27). A fraction of poly-ethyleneglycol has biotin at the end (Biotin-PEG-SVA, MW 5000, Laysan). A narrow channel was made between a quartz slide and a cover slip by using double-sided adhesive tape. For oligonucleotide immobilization, 0.2 mg/ml streptavidin was added to the channel. After 2 min incubation, Fork substrate (100 pM) was added to the channel, and immobilized on a quartz slide surface via biotin-streptavidin interaction. Fork substrate was sequentially incubated with WRN (16 nM, 5 min), and RPA (1 nM or 20 nM, 5 min) in imaging buffer (Tris 50 mM (pH 8.0), BSA 100 μg/ml, DTT 1 mM, 0.4 mg/ml Trolox), and an oxygen scavenger system (0.4% (w/v) glucose, 1 mg/ml glucose oxidase (Sigma) and 0.02 mg/ml catalase (Sigma)). Then unwinding buffer (imaging buffer with ATP–Mg2+) was injected into the channel. DNA forks used are not a good substrate for the nuclease activity of WRN. However, to further inhibit the nuclease activity of WRN, the concentrations of ATP and Mg2+ were always kept the same to reduce free Mg2+ that is required for nuclease activity (28). Single-molecule images were obtained in a wide-field total-internal-reflection fluorescence microscope. Solid-state 532-nm laser (Compass 215M-50, Coherent) and He-Ne 633-nm laser were used to excite the molecules. ALEX (Alternating Laser EXcitation) technique was used (29). Fluorescence signals from single molecules were collected through a water immersion objective (60×, UPlanSApo, Olympus) in an inverted microscope (IX71, Olympus). To block out 532-nm laser scattering, a notch filter (532 ± 8.5 nm, NF03–532E-25, SemRock) was used. To block out 640-nm laser scattering, a notch filter (633 ± 12.5 nm, NF03–633E-25, SemRock) was used. Fluorescence images were detected with an electron multiplying charge coupled device (iXon DV897ECS-BV, Andor Technology), and recorded with a home-made software written in Visual C++. Fluorescence images and single-molecule time traces were extracted with programs written in IDL (ITT Visual Information Solutions) and analyzed with Matlab (MathWorks) and Origin (OriginLab Corporation).
Magnetic tweezers assay
Flow cells were made by sealing two cover glasses with parafilm. The upper glass had two holes for buffer exchange and the bottom glass was functionalized with nitrocellulose (0,1% wt/vol in amyl acetate). Non-magnetic beads (diameter 3.2 μm Thermo Fisher Scientific) which served as reference beads were immobilized to surface by non-specific interaction. The glass surface was further functionalized with anti-digoxigenine solution (100 μg/ml) by incubating overnight and passivated with 10 mg/ml BSA solution (New England Biolabs) for 2 h to prevent nonspecific binding of beads. Streptavidin coated magnetic beads (M270, diameter 2.8 μm, Thermo Fisher Scientific) were mixed with DNA solution in TE tethering buffer (10 mM Tris–HCl (pH 8.0) with 200 mM NaCl) and incubated for 10 min. DNA-beads complex solution was flown into flow cell and after incubation for 1 h, unbound beads were removed by buffer washing. All the magnetic tweezers experiments were carried out in 20 mM Tris–HCl buffer (pH 8.0) supplemented with 40–140 mM KCl, 1 mM MgCl2, 1 mM DTT and 0.02% Triton-X. Higher salt condition than that in FRET assay was used to prevent spontaneous opening of hairpin. For the unwinding experiments with WRN alone, WRN protein (0.5–1 nM) with ATP was flushed in to flow cell to initiate unwinding reaction. To avoid multiple binding events, the concentration of WRN was used as low as possible. When we study RPA effects, RPA (1–20 nM) solution was then added into chamber. Free RPA protein were thoroughly removed after brief incubation (∼5 min) when high concentration of RPA is used. To start the unwinding reaction, buffer containing ATP was flowed in. Flow rate and stretching force during flow were kept as low as possible in order to prevent hairpin opening. High force (>6 pN) was not applied in the experiment involving RPA because RPA alone can melt hairpin molecule at it. Magnetic tweezers assay was performed on the same microscope system as used in FRET experiments.(30) A pair of cube magnets (W-05-N50-G, Supermagnete) mounted on linear and rotational stages (M-126 and C-136, Physik Instrumente) exerted stretching force and torque on beads. Beads were illuminated using a collimated visible LED and imaged to a CCD camera (Pulnix TM-6710Cl, 120 Hz) by the same objective lens used in FRET dye imaging. The x and y positions of the beads were found from defocused image with centroid finding algorithm and z position was determined by matching it with the reference profiles of each beads which was generated at series of positions of objective lens. The uncertainty of z tracking for surface immobilized beads was less than 1.5 nm. Force was determined from x, y fluctuation applying equipartition theorem. However as the fluctuation of beads tethered by short hairpin construct was too fast to capture with 120 Hz camera, instead of measuring force directly, we used calibrated forces as a function of magnet position from those obtained using long dsDNA constructs (21 kb). Blurring and aliasing effect by finite sampling of camera was corrected in force value evaluation (31).
RESULTS
Characterization of WRN helicase activity
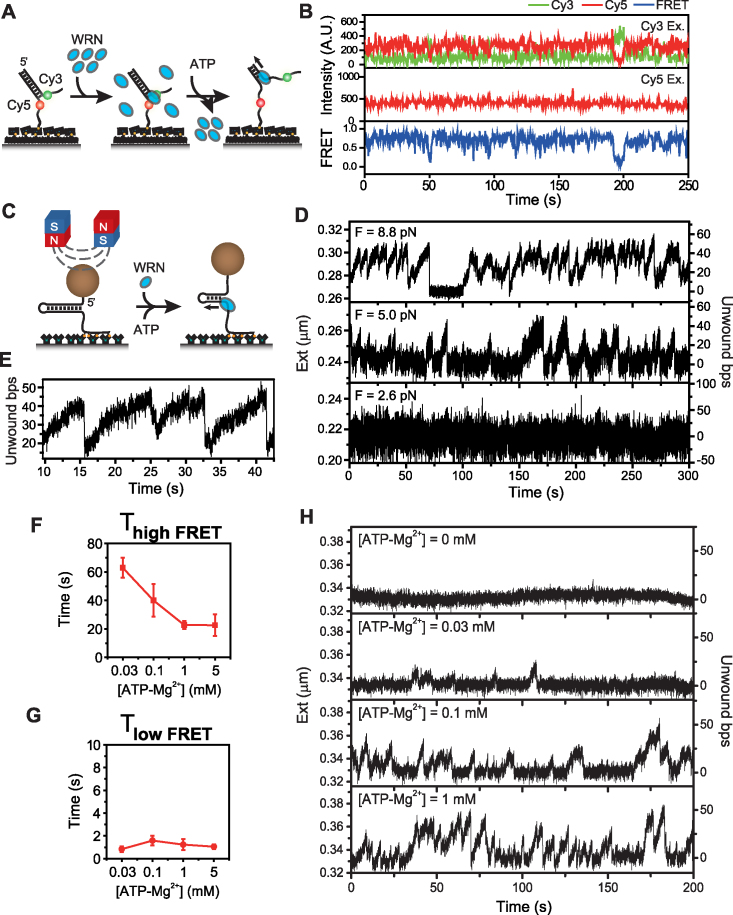
The experimental scheme of single-molecule FRET is depicted in Figure 1A. We prepared a forked DNA substrates with a Cy3 (donor) and a Cy5 (acceptor) in their single-stranded overhangs (Supplementary Table S1) and immobilized them on a polymer-coated quartz surface via streptavidin-biotin interaction within a fluorescence detection chamber. After incubating the immobilized DNAs with WRN (16 nM) for 5 min, 1 mM ATP–Mg2+ was added to the reaction chamber, and changes in fluorescence intensities were measured over time using a total-internal-reflection fluorescence microscope. The buffer exchange system that we used is quite efficient (Supplementary Figure S2), and therefore we can effectively assume that there is no free WRN in solution when unwinding reaction is started.
Figure 1.
Repetitive unwinding of forked DNA by WRN. (A) Forked DNA substrate design and FRET experimental scheme. DNA fork was immobilized on a polymer-coated quartz surface and WRN was added to the chamber to assemble WRN-DNA complex, and then ATP–Mg2+ buffer was added to initiate the unwinding activity of WRN while washing free proteins off. (B) Representative fluorescence intensity time traces for Cy3 (green) and Cy5 (red) with Cy3 excitation (top), for Cy5 with Cy5 excitation (middle), and the corresponding FRET time traces (bottom). The trace shows repetitive unwinding bursts of fork DNA by WRN. The same color convention is used throughout the paper. (C) Simplified drawing of 50 bp DNA hairpin construct and magnetic tweezers experimental scheme. A pair of magnets exert pulling force on the hairpin construct. WRN and ATP–Mg2+ was injected to the chamber at the same time to initiate the unwinding. The concentration of WRN was kept as low as possible to guarantee single protein binding condition. (D) The representative extension time traces of a hairpin at three different forces (F = 8.8, 5.0, and 2.6 pN) plotted with the second y axis showing the number of unwound base pairs calculated from the respective extension by FJC model. (E) The trace enlarged the region between 10 and 45 s of the trace at 8.8 pN (top panel of Figure 1D). ATP concentration dependence of average high FRET (F) and (G) low FRET dwell times. More than 100 molecules were analyzed to get (F) and (G). (H) Representative unwinding traces by WRN measured varying ATP–Mg2+ concentration for 50 bp hairpin at 8.8 pN. Hairpin molecules were incubated with WRN for 5 min and ATP–Mg2+ buffer was flushed in. The traces were recorded sequentially from 0 to 1 mM ATP–Mg2+ for a molecule. Without ATP, no reaction occurred (top panel). As ATP–Mg2+ concentration increased, the unwinding bursts occurred more frequently and unwinding speed became higher (Supplementary Figure S5). Unwinding speed at 0.1 mM was 7.3 ± 1.0 bp/s (S.D. for 13 molecules) while that at 1 mM was 8.7 ± 1.1 bp/s (S.D for 13 molecules). Analysis on those at 0.03 mM was not done because the number of burst was not enough.
Representative fluorescence intensity time traces in Figure 1B suggest that WRN repetitively unwinds the forked DNA in short repetitive bursts with variable length pauses. Since short bursts of DNA unwinding persisted in the absence of free WRN, we speculate that the FRET signal change was not caused by binding/dissociation of WRN to/from the forked DNA substrate, but rather caused by the helicase activity of a single functional unit of WRN. The repetitive unwinding has been reported for many other RecQ helicases (32–35), suggesting that it is a general activity of RecQ helicases.
Single-molecule FRET is a productive way to monitor the unwinding activity of a helicase, but its relatively low imaging speed, and short detecting range of unwound bases are limitations. To gain deeper understanding of the unwinding mechanism of WRN, we performed the unwinding experiments using magnetic tweezers. The scheme of magnetic tweezers experiments are shown in Figure 1C. We prepared a 50 bp DNA hairpin substrate that has the same duplex region sequence as the forked substrate used in FRET assay. The hairpin construct attached at its 5′-end to a magnetic bead via streptavidin-biotin interaction and at another end to a glass surface via digoxigenine and anti-digoxigenine interaction. At the 3′-side of hairpin was 30 nucleotides single stranded region, which served as a helicase loading site. Hairpin molecules showing proper mechanical unzipping behavior, as shown in Supplementary Figure S3, were chosen for further measurements. The extension increase by WRN unwinding could be converted into the number of base pairs unwound using a force versus extension relation for single stranded DNA from Freely Jointed Chain (FJC) model (36). To initiate the reaction, WRN and ATP–Mg2+ was added to hairpin molecules stretched by magnets (Figure 1C). The characteristic trace obtained at 8.8 pN is shown in the upper panel of Figure 1D. Individual events of a slow unwinding followed by a fast re-annealing occurred in a highly repetitive pattern (Figure 1E). This pattern is similar to those reported for BLM (a member of human RecQ) and Arabidopsis thaliana RecQ (AtRecQ2) (32,34). Owing to a faster sampling frequency of magnetic tweezers (120 Hz) than FRET assay (5 Hz), we were able to detect the slopes of DNA extension generated by the translocation of the enzyme during unwinding. The unwinding speed of WRN at 8.8 pN was 6.8 ± 1.7 bp/s (S.D for 13 molecules). Compared with the gradual increase of the unwinding phase, the rewinding events were more abrupt; it rapidly reshuffled to the original position (Figure 1E). A long pause at 70s in the upper panel of Figure 1D is likely due to dissociation of a protein from the hairpin DNA. As the force increased, the average number of unwound base pairs and average unwinding burst frequency increased and the average unwinding speed decreased even though molecule-to-molecule variations were significant (Figure 1D, Supplementary Figure S4).
To learn more about the repetitive unwinding mechanism of WRN, we studied ATP concentration dependence of the unwinding activity of WRN. Kinetic analyses of FRET experiments revealed that high FRET dwell time was ATP concentration dependent and inversely correlated with increasing ATP concentration (Figure 1F). On the other hand, low FRET dwell time was ATP concentration-independent (Figure 1G). In order to detect unwinding and rewinding phases in further detail, we measured the unwinding traces at varying ATP concentrations using magnetic tweezers (Figure 1H) at 8.8 pN. We observed that more base pairs tended to be unwound at the higher ATP concentration (Supplementary Figure S5A). Analysis on the slopes during unwinding and rewinding showed that the average unwinding speed became higher as ATP concentration increased (Supplementary Figure S5B). Specifically, it increased from 7.3 ± 1.0 bp/s (S.D for 13 molecules) at 0.1 mM ATP to 8.7 ± 1.1 bp/s (S.D for 13 molecules) at 1 mM ATP. These observations are consistent with the general consensus on helicases, which consume ATP to separate base pairs. No effect of ATP on the rewinding phase was detected, suggesting that it is the slipping process of the enzyme, not requiring ATP hydrolysis (37). The overall pattern of unwinding burst showed similar trend as in the FRET assay; the higher the ATP concentration, the more frequent bursts occurred (Supplementary Figure S5C).
RPA effects at low concentration
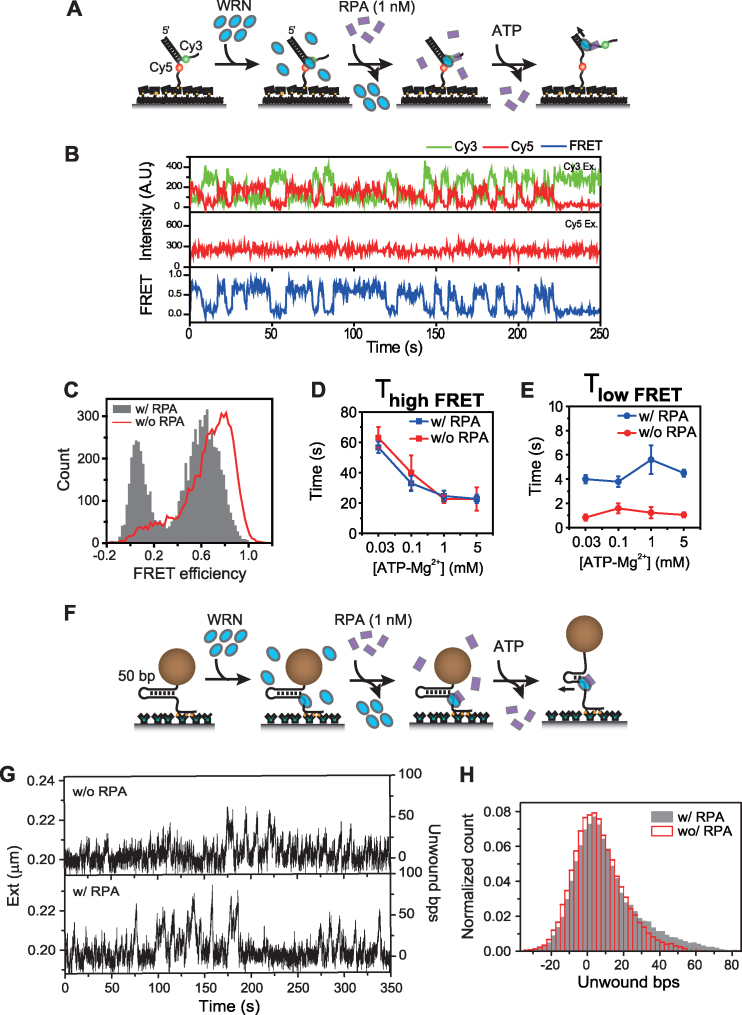
Next, we investigated the effect of RPA at low concentration (1 nM) on WRN unwinding dynamics using FRET. For this experiment, fork substrates were sequentially incubated with WRN and RPA, and then ATP was injected (Figure 2A). We observed clearer repetitive FRET dynamics between two well-defined FRET values after the addition of ATP (Figure 2B and C). The unwinding event was not observed when RPA alone was injected or the helicase dead mutant of WRN, K-WRN, was used instead of wild type WRN (Supplementary Figure S6). Detailed analysis of repetitive unwinding kinetic parameters revealed that RPA had significant effects on the kinetics of the repetitive unwinding of WRN (Figure 2D and E). Whereas the high FRET dwell time (Figure 2D) remained unchanged, the low FRET dwell time was increased (Figure 2E).
Figure 2.
The effect of low concentration RPA on the repetitive unwinding of WRN. (A) Scheme of the experiment to observe the effect of RPA at low concentration (1 nM) on WRN unwinding dynamics. Fork substrate was sequentially incubated with WRN (16 nM, 5 min), and RPA (1 nM, 5 min), and then imaging buffer containing 1 mM ATP–Mg2+ was injected. No free WRN or RPA was present during the measurements. (B) Representative fluorescence intensity time traces for Cy3 (green) and Cy5 (red) with Cy3 excitation (top), for Cy5 with Cy5 excitation (middle), and the corresponding FRET time traces (bottom). The trace shows repetitive unwinding of WRN with RPA. (C) Comparison of unwinding FRET histograms of WRN incubated with 1 nM RPA (gray bars) and WRN alone (red line). The histograms were made from time windows showing repetitive unwinding behavior. ATP concentration dependence of average high FRET (D) and (E) low FRET dwell times after incubation with 1 nM RPA (blue) and without RPA (red). More than 100 molecules were analyzed to get (D) and (E). (F) Magnetic tweezers experimental scheme with RPA. 50 bp DNA fork was sequentially incubated with WRN and RPA (1 nM, 5 min). Free protein was removed after each incubation steps. Unwinding was initiated by adding ATP–Mg2+ buffer. (G) Representative extension time traces of repetitive unwinding of WRN with or without RPA measured at F = 3.1 pN. Unwinding trace was first recorded after the injection of WRN and 1 mM ATP–Mg2+ (without RPA, upper panel), and then the trace of the same molecule was recorded after subsequently adding RPA (1 nM) and 1 mM ATP–Mg2+ (lower panel). The traces were moving-averaged. (H) Comparison of histograms of the number of unwound basepairs with and without RPA (gray bars and red lines, respectively) for the traces shown in (G).
To find the exact cause of the alteration of the low FRET dwell time, we examined the effect of RPA on the unwinding phase of WRN using magnetic tweezers (Figure 2F). In this experiment, we reduced the force to 3.1 pN because unwinding became very active by force itself at a higher force and it was hard to clearly distinguish the sole effect of RPA. The upper panel in Figure 2G is the unwinding trace by WRN alone and the lower is the unwinding trace of the same DNA molecule after WRN was incubated with 1 nM RPA. Because of the thermal noise in the traces at low forces, we could not analyze the initiation frequency and the unwinding speed. However, it is clear that WRN unwound more base pairs in the presence of RPA (Figure 2H). Figure 2H shows that WRN can unwind 50 bp-hairpin almost completely in the presence of bound RPA. With this range of unwinding, the distance between Cy3-Cy5 FRET pair is well beyond the Förster distance, and no appreciable FRET change is expected. Therefore, the disappearance of a middle FRET population in Figure 2B and longer dwell time in low FRET state upon the addition of RPA in Figure 2E is quite consistent with the magnetic tweezers data in Figure 2H.
RPA effects at high concentration
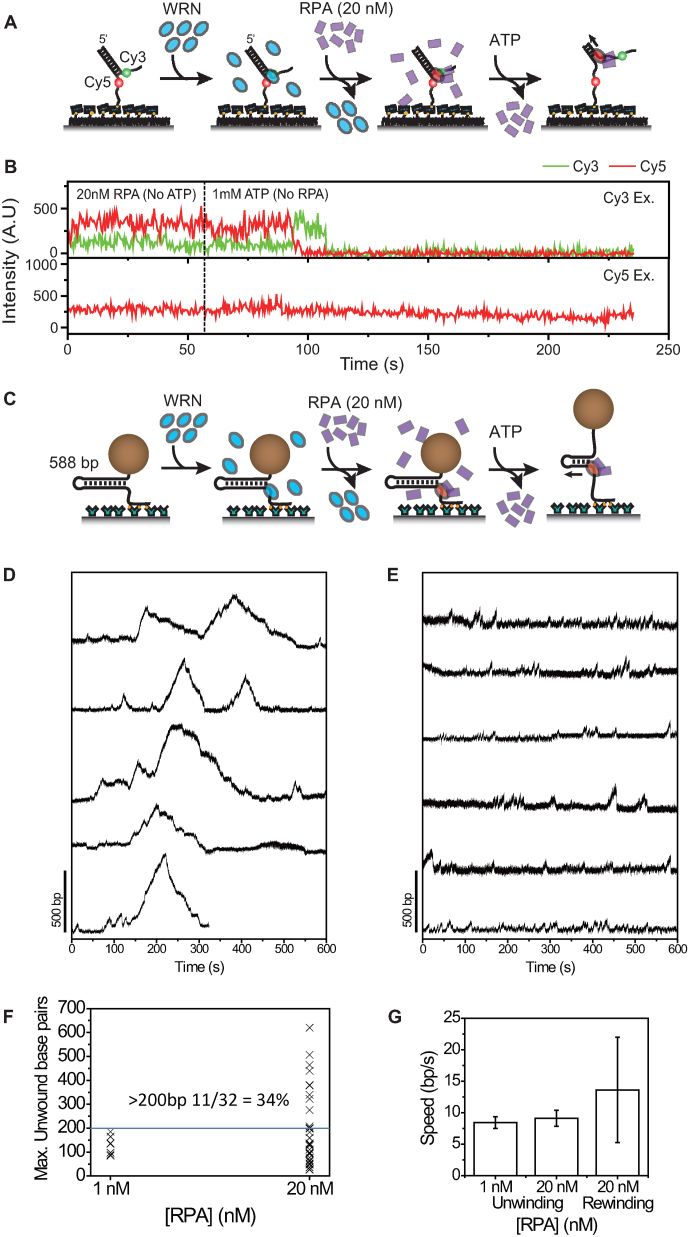
It has been reported that WRN can unwind several hundred base pairs in the presence of RPA (16). Our results above did not show any dramatic increase of WRN processivity when it is incubated with a few nanomolar RPA (Figure 2B). To study how the processivity of WRN is affected when it is incubated with higher concentrations of RPA, we sequentially incubated forked DNA substrates with WRN and 20 nM RPA, and started the unwinding reaction by adding 1 mM ATP (Figure 3A). As FRET time traces in Figure 3B show, about 40% (23 out of 57) of DNA substrates was completely unwounded once unwinding occurred. The disappearance of Cy3 signal is hardly due to the photobleaching of the fluorophore because the average unwinding time was measured as 69 s whereas photobleaching time in the experimental condition was 862 s.
Figure 3.
Increase of WRN processivity at high RPA concentrations. (A) Scheme of the experiment to observe the effect of RPA at high concentration (20 nM) on WRN unwinding dynamics. Fork substrate was sequentially incubated with WRN (16 nM, 5 min), and RPA (20 nM, 5 min), and then imaging buffer containing 1 mM ATP–Mg2+ was injected. No free WRN or RPA was present during the measurements. (B) Representative intensity time traces showing a full unwinding event when ATP–Mg2+ containing no RPA was injected after forked substrate was sequentially incubated with WRN and 20 nM RPA. Green and red lines are Cy3, and Cy5 signals at Cy3 excitation in the upper panel and red line is Cy5 signal at Cy5 excitation in the lower panel. (C) Magnetic tweezers experimental scheme with RPA. 588 bp DNA fork was sequentially incubated with WRN (3 nM) and RPA. Free protein was removed after each incubation steps. Unwinding was initiated by adding ATP–Mg2+ buffer. Representative extension time traces with 20 nM RPA incubation (D) and 1 nM RPA incubation (E). The experiments were performed at 6.0 pN. (F) Distribution of maximum unwound base pairs for each molecule. In case of 20 nM RPA incubation, 34% of molecules exhibited unwinding more than 200 base pairs whereas none was observed with 1 nM RPA incubation. (G) Comparison of unwinding and rewinding speeds.
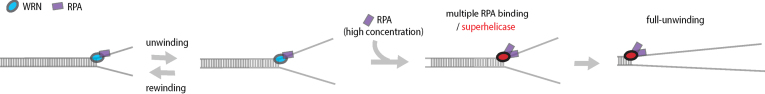
Figure 4.
Model for the RPA effects on WRN helicase activity. In the model, binding of multiple RPAs converts WRN into a superhelicase, but the oligomeric state of active WRN is not fixed. Therefore, multiple RPA binding can occur either to a single WRN, or an oligomeric WRN even though the two possibilities are not mutually exclusive.
To study the unwinding of longer DNA duplexes, we measured unwinding dynamics of hairpins with a long (588 bp) stem (Supplementary Methods) using magnetic tweezers. The incubation and flow scheme were kept the same as in FRET experiments; the hairpin was incubated with WRN and RPA sequentially and the ATP buffer containing no proteins was flushed in (Figure 3C). A burst with very high amplitude corresponding to almost full unwinding of 588 bp stem frequently arose after a couple of repetitive unwinding cycles (Figure 3D). When incubated with 1 nM RPA, however, the DNA substrate with a long stem exhibited only limited unwinding (Figure 3E); 34% of molecules exhibited unwinding more than 200 bp when incubated with 20 nM RPA, but none with 1 nM RPA incubation (Figure 3F). In contrast to the significantly increased unwinding processivity, the speed during a highly processive unwinding mode was not different from the speed during repetitive unwinding bursts (Figure 3F). Another clear difference between highly processive unwinding and repetitive unwinding events is the slope of the rewinding phase. In contrast to one-step rewinding observed from most of the repetitive unwinding events, the rewinding phase of the highly processive unwinding event has clear slopes; after reaching full unwinding, WRN was able to move backward at the speed a little higher than that of the forward direction (Figure 3F). Interestingly, we observed that the highly-processive unwinding events with a long (1 kb) DNA hairpin rarely appears when the low concentration of RPA (1 nM) remained in the flow cell without washing (Supplementary Figure S7). Even though we cannot experimentally justify it, it is probable that long term exposure to low concentration of RPA in solution produced the effect similar to short incubation of high concentration RPA.
DISCUSSION
Using single-molecule FRET and magnetic tweezers assays, we showed that WRN unwinds a limited number of base pairs, regresses back, and reinitiates the unwinding, resulting in repetitive unwinding bursts. This work and other previous single molecule studies (32–35) indicate that this repetitive unwinding is a common characteristic shared by RecQ helicases. In spite of a growing number of single-molecule data on RecQs, there have been debates on the detailed mechanism for how it regresses back after brief unwinding. The single-molecule FRET assay on BLM proposed that it senses the critical length and once it has unwound this length, it switches to the opposite strand and translocases in reverse direction (33). However, according to this work and the previous magnetic tweezers studies on BLM and AtRecQ2 (32,34), the critical length it unwinds at a time was not narrowly distributed as proposed and it was considerably affected by factors such as base composition and applied force. Furthermore, the majority of rewinding events appeared to be instantaneous back sliding. At this point, it is not clear what triggers the termination of continuous unwinding. It would be a mechanical stress which is accumulated while unwound base pairs exceed the critical length as FRET studies suggested (33).
In this study, we showed that WRN unwinding activity is affected by RPA in a concentration dependent manner. When WRN was incubated with low concentration of RPA (1 nM), the repetitive unwinding dynamics was altered and the processivity in a single burst moderately increased. However, when WRN was incubated with a high concentration of RPA (20 nM), double-stranded DNA was fully unwound. Magnetic tweezers experiments with long DNA hairpins revealed that WRN was able to unwind the duplex DNA up to 1 kbp with the aid of RPA. Translocation speed in the rewinding phase was slightly higher than that of the unwinding phase (Figure 3D, F, and Supplementary Figure S7), suggesting that the hindrance effect of RPA on WRN translocation is minor if there is any. This clearly demonstrates that the increased processivity of WRN by RPA is indeed due to the direct protein–protein interaction. Since the significant increase in the processivity of WRN took place only after the exposure to high concentration of RPA, one can suspect that it is a multiple binding effect. The oligomeric state of active WRN for DNA unwinding is not known yet (38), and it is not clear whether multiple RPAs binding to a single WRN or WRN oligomers. Interestingly sgs1, a yeast homolog of human WRN, has been reported to form a complex with multiple RPAs (39), but there is no direct evidence that multiple RPAs bind simultaneously to a single WRN. It has been reported that WRN has two RPA binding domains: the stronger one at acidic repeats in the N-terminal region, and another weaker one at the C-terminal region (21). In case of C. elegans WRN, these two regions interacted with different domains of a single C. elegans RPA molecule (CeRPA73 and CeRPA32 subunits) and both subunits were required for the full stimulation of C. elegans WRN (17). In case of human WRN, however, the mechanism was slightly different in that only hRPA70 subunit is sufficient for the full stimulation of human WRN (17). Considering these together, the multiple RPA binding effect would be due to binding to an oligomeric unit rather than a single WRN molecule, but further studies are required to pin down the question. As the unwinding speed during the highly processive phase was comparable to those during usual repetitive phase, RPA is not likely to affect the translocation speed. This agrees with the previous biochemical study (16) showing that RPA did not increase the ATP hydrolysis rate of WRN.
The repetitive unwinding is an important characteristic defining the RecQ helicases. It may be indispensable for its biological function, for example, to process stalled replication forks, to eliminate potentially deleterious recombination intermediates, or as observed for ssDNA translocases, to strip off other DNA binding proteins. We speculate that the repetitive unwinding is an efficient mechanism to coordinate multifaceted activities for processing higher order DNA structures and to achieve harmony between subunits when it acts as an oligomer. The RPA stimulation of WRN unwinding activity shown in this work implies a biological function working in different mechanism. For decades, RPA has been known to physically interact with WRN (16,17) and to co-localize with WRN on DNA damage foci in a cell (18,19). However, the biological importance of this interaction has been elusive because both RPA and WRN are involved in many DNA processing pathways. Recently it has been reported that RecQ plays an important role in the repair of double-strand break (DSB) by homologous recombination (HR) for various organisms ranging from yeast to human (40–43). The essential step in HR repair is the end resection to form long 3′ single stranded tail for invasion to homologous sequence. There are two pathways for it, Exonuclease 1 (Exo1) dependent and RecQ-Dna2 dependent ones. RPA was known to be essential in the RecQ-Dna2 resection pathway interacting with both RecQ and Dna2 (42–44). Therefore, the highly processive unwinding of WRN stimulated by RPA seems to be required to generate a long ssDNA tail of a few kbps. We found that direct protein–protein interaction alone is enough to induce such stimulation. The failure of the stimulation by heterologous single strand binding proteins reported in a previous study (43) implies that the specific interaction is important to such stimulation. Dna2 is an exonuclease responsible for the degradation of 5′ tailed ssDNA. Interestingly, it can degrade DNA in either direction, but it only digests DNA in 5′ to 3′ direction upon the association with RecQ and RPA (42,45). We suspect that the specific interaction of WRN and RPA may also play important roles to recruit Dna2 in right orientation and to fix its activity to what requires for end resection.
A recent study using single-molecule optical tweezers reported that auxiliary proteins such as an initiation factor greatly increased the processivity of eIF4A DEAD-box helicase (46). Another single-molecule study on PcrA showed that RepD dramatically increased the processivity of PcrA by fixing the conformation of the enzyme in a closed form (47). We speculate that such alteration of enzyme activity by co-factors is widespread, and it is an important mechanism in-vivo in which many co-factors regulate biological processes.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank. Jinwoo Lee for his contribution in the initial stage of the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Creative Research Initiative program [2009-0081562 to S.H., 2015R1A1A1A05001593 to J.K.]; Basic Science Research Program [NRF-2015R1D1A3A01020774 to B.A.] from the National Research Foundation; Intramural program of the National Institutes of Health is appreciated. Funding for open access charge: Creative Research Initiative program [2009-0081562].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rossi M.L., Ghosh A.K., Bohr V.A.. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst.). 2010; 9:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu W.K., Hickson I.D.. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009; 9:644–654. [DOI] [PubMed] [Google Scholar]
- 3. Croteau D.L., Popuri V., Opresko P.L., Bohr V.A.. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014; 83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ozgenc A., Loeb L.A.. Werner Syndrome, aging and cancer. Genome Dyn. 2006; 1:206–217. [DOI] [PubMed] [Google Scholar]
- 5. Orren D.K. Werner syndrome: molecular insights into the relationships between defective DNA metabolism, genomic instability, cancer and aging. Front. Biosci. 2006; 11:2657–2671. [DOI] [PubMed] [Google Scholar]
- 6. Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A.. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997; 17:100–103. [DOI] [PubMed] [Google Scholar]
- 7. Shen J.C., Gray M.D., Oshima J., Kamath-Loeb A.S., Fry M., Loeb L.A.. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem. 1998; 273:34139–34144. [DOI] [PubMed] [Google Scholar]
- 8. Muftuoglu M., Kulikowicz T., Beck G., Lee J.W., Piotrowski J., Bohr V.A.. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008; 47:10247–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mohaghegh P., Karow J.K., Brosh R.M. Jr, Bohr V.A., Hickson I.D.. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001; 29:2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brosh R.M. Jr, Waheed J., Sommers J.A.. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002; 277:23236–23245. [DOI] [PubMed] [Google Scholar]
- 11. Opresko P.L., Mason P.A., Podell E.R., Lei M., Hickson I.D., Cech T.R., Bohr V.A.. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 2005; 280:32069–32080. [DOI] [PubMed] [Google Scholar]
- 12. Blander G., Kipnis J., Leal J.F., Yu C.E., Schellenberg G.D., Oren M.. Physical and functional interaction between p53 and the Werner's syndrome protein. J. Biol. Chem. 1999; 274:29463–29469. [DOI] [PubMed] [Google Scholar]
- 13. Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A.. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002; 277:41110–41119. [DOI] [PubMed] [Google Scholar]
- 14. Fanning E., Klimovich V., Nager A.R.. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006; 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen J.C., Gray M.D., Oshima J., Loeb L.A.. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998; 26:2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brosh R.M. Jr, Orren D.K., Nehlin J.O., Ravn P.H., Kenny M.K., Machwe A., Bohr V.A.. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999; 274:18341–18350. [DOI] [PubMed] [Google Scholar]
- 17. Hyun M., Park S., Kim E., Kim D.H., Lee S.J., Koo H.S., Seo Y.S., Ahn B.. Physical and functional interactions of Caenorhabditis elegans WRN-1 helicase with RPA-1. Biochemistry. 2012; 51:1336–1345. [DOI] [PubMed] [Google Scholar]
- 18. Sakamoto S., Nishikawa K., Heo S.J., Goto M., Furuichi Y., Shimamoto A.. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001; 6:421–430. [DOI] [PubMed] [Google Scholar]
- 19. Constantinou A., Tarsounas M., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D., West S.C.. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000; 1:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen J.C., Lao Y., Kamath-Loeb A., Wold M.S., Loeb L.A.. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech. Ageing Dev. 2003; 124:921–930. [DOI] [PubMed] [Google Scholar]
- 21. Doherty K.M., Sommers J.A., Gray M.D., Lee J.W., von Kobbe C., Thoma N.H., Kureekattil R.P., Kenny M.K., Brosh R.M. Jr. Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J. Biol. Chem. 2005; 280:29494–29505. [DOI] [PubMed] [Google Scholar]
- 22. Orren D.K., Brosh R.M. Jr, Nehlin J.O., Machwe A., Gray M.D., Bohr V.A.. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999; 27:3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henricksen L.A., Umbricht C.B., Wold M.S.. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994; 269:11121–11132. [PubMed] [Google Scholar]
- 24. Binz S.K., Dickson A.M., Haring S.J., Wold M.S.. Functional assays for replication protein A (RPA). Methods Enzymol. 2006; 409:11–38. [DOI] [PubMed] [Google Scholar]
- 25. Luzzietti N., Brutzer H., Klaue D., Schwarz F.W., Staroske W., Clausing S., Seidel R.. Efficient preparation of internally modified single-molecule constructs using nicking enzymes. Nucleic Acids Res. 2011; 39:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ha T., Rasnik I., Cheng W., Babcock H.P., Gauss G.H., Lohman T.M., Chu S.. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002; 419:638–641. [DOI] [PubMed] [Google Scholar]
- 27. Rasnik I., Myong S., Cheng W., Lohman T.M., Ha T.. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J. Mol. Biol. 2004; 336:395–408. [DOI] [PubMed] [Google Scholar]
- 28. Storer A.C., Cornish-Bowden A.. Concentration of MgATP2-and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem. J. 1976; 159:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michalet X., Lacoste T.D., Weiss S.. Ultrahigh-resolution colocalization of spectrally separable point-like fluorescent probes. Methods. 2001; 25:87–102. [DOI] [PubMed] [Google Scholar]
- 30. Uhm H., Bae S., Lee M., Hohng S.. Single-Molecule FRET combined with magnetic tweezers at low force regime. Bull. Korean Chem. Soc. 2016; 37:408–410. [Google Scholar]
- 31. te Velthuis A.J., Kerssemakers J.W., Lipfert J., Dekker N.H.. Quantitative guidelines for force calibration through spectral analysis of magnetic tweezers data. Biophys. J. 2010; 99:1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klaue D., Kobbe D., Kemmerich F., Kozikowska A., Puchta H., Seidel R.. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 2013; 4:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yodh J.G., Stevens B.C., Kanagaraj R., Janscak P., Ha T.. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009; 28:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang S., Qin W., Li J.H., Lu Y., Lu K.Y., Nong D.G., Dou S.X., Xu C.H., Xi X.G., Li M.. Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res. 2015; 43:3736–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu W.Q., Hou X.M., Zhang B., Fosse P., Rene B., Mauffret O., Li M., Dou S.X., Xi X.G.. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 2017; 7:43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith S.B., Cui Y., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 37. Wu Y. Unwinding and rewinding: double faces of helicase?. J. Nucleic Acids. 2012; 2012:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Compton S.A., Tolun G., Kamath-Loeb A.S., Loeb L.A., Griffith J.D.. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 2008; 283:24478–24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hegnauer A.M., Hustedt N., Shimada K., Pike B.L., Vogel M., Amsler P., Rubin S.M., Van Leeuwen F., Guenole A., Van Attikum H.. An N‐terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012; 31:3768–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan H., Toczylowski T., McCane J., Chen C., Liao S.. Replication protein A promotes 5′→3′ end processing during homology-dependent DNA double-strand break repair. J. Cell Biol. 2011; 192:251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sturzenegger A., Burdova K., Kanagaraj R., Levikova M., Pinto C., Cejka P., Janscak P.. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014; 289:27314–27326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L. et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010; 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen H., Lisby M., Symington L.S.. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell. 2013; 50:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tammaro M., Liao S., McCane J., Yan H.. The N-terminus of RPA large subunit and its spatial position are important for the 5′→3′ resection of DNA double-strand breaks. Nucleic Acids Res. 2015; 43:8790–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garcia-Garcia C., Frieda K.L., Feoktistova K., Fraser C.S., Block S.M.. RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 2015; 348:1486–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arslan S., Khafizov R., Thomas C.D., Chemla Y.R., Ha T.. Protein structure. Engineering of a superhelicase through conformational control. Science. 2015; 348:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.