Abstract
Objectives
Vancomycin-resistant Enterococcus faecium is a leading cause of MDR hospital infection. Two genetically definable populations of E. faecium have been identified: hospital-adapted MDR isolates (clade A) and vancomycin-susceptible commensal strains (clade B). VanN-type vancomycin resistance was identified in two isolates of E. faecium recovered from blood and faeces of an immunocompromised patient. To understand the genomic context in which VanN occurred in the hospitalized patient, the risk it posed for transmission in the hospital and its origins, it was of interest to determine where these strains placed within the E. faecium population structure.
Methods
We obtained the genome sequence of the VanN isolates and performed comparative and functional genomics of the chromosome and plasmid content.
Results
We show that, in these strains, VanN occurs in a genetic background that clusters with clade B E. faecium, which is highly unusual. We characterized the chromosome and the conjugative plasmid that carries VanN resistance in these strains, pUV24. This plasmid exhibits signatures of in-host selection on the vanN operon regulatory system, which are associated with a constitutive expression of vancomycin resistance. VanN resistance in clade B strains may go undetected by current methods.
Conclusions
We report a case of vancomycin resistance in a commensal lineage of E. faecium responsible for an atypical bacteraemia in an immunocompromised patient. A reservoir of transferable glycopeptide resistance in the community could pose a concern for public health.
Introduction
Enterococcus faecium, natively a gastrointestinal (GI) tract commensal organism, emerged as a leading cause of MDR hospital-acquired infection in the 1980s.1 Epidemiological and recent comparative genomic studies reported the existence of distinct subpopulations of E. faecium and the features that distinguished epidemic hospital-adapted isolates [named clade A and including the so-called clonal complex 17 (CC17)] and commensal-type strains (clade B).2–5 The hospital-adapted epidemic strains diverged from the clade B commensal strains by a distance usually associated with speciation and, importantly, clade A strains are frequently resistant to high levels of aminoglycosides, ampicillin and vancomycin, which undoubtedly contributes to their spread among hospitalized patients worldwide.4–7 By contrast, the clade B strains are usually susceptible to most antibiotics, including vancomycin, and rarely cause infection aside from sporadic cases in severely immunocompromised patients.4
In the USA, between 35% and 75% of speciated enterococcal infections are caused by E. faecium, of which a majority are vancomycin resistant.8,9 Glycopeptide antibiotics (namely vancomycin and teicoplanin) are widely used for the treatment of severe infections due to Gram-positive bacteria and act by inhibition of peptidoglycan synthesis.10 Since the early descriptions of vancomycin-resistant enterococci in 1988,11,12 nine variations of gene clusters conferring resistance to glycopeptides have been identified, distinguished by sequence differences in either d-alanyl-d-lactate ligase (VanA, VanB, VanD and VanM) or d-alanyl-d-serine ligase (VanC, VanE, VanG, VanL and VanN).10,13–15 In addition to the ligase, the genes in the resistance cluster encode enzymes involved in the synthesis of modified peptidoglycan precursors, with lower affinity for glycopeptides, as well as enzymes that hydrolyse the competing, natively produced d-Ala-d-Ala precursor.10 The resistance genes are regulated by two-component regulatory systems (VanR and VanS) that, when functional, induce the expression of the resistance in the presence of glycopeptide.10
In addition to the threat posed by VRE, MDR enterococci serve as a reservoir for horizontal gene transfer (HGT) of antibiotic resistance to other pathogens, as illustrated by reports of vanA transfer to Staphylococcus aureus.16 Aside from the VanC-type resistance, which is ordinarily intrinsic to species Enterococcus gallinarum and Enterococcus casseliflavus, van clusters have been acquired through HGT.10 The vanA and vanB gene clusters are generally located on transposons (Tn1546 and Tn1549/Tn5382) often found in conjugative plasmids.10 In addition, both vanE and vanD resistance clusters have been shown to be part of putative conjugative elements, although transferability could not be demonstrated.17,18
The vanN resistance cluster, which we previously showed to be transferable by conjugation of a large plasmid of unknown structure and origin, has been found in two clonal strains isolated from the blood and the faeces (strains UCN71 and UNC72) of a hospitalized patient in France,15 from the faeces of a hospitalized patient in Canada and also in five clonal strains isolated from domestic chicken meat in Japan.19,20 Interestingly, routine molecular typing by MLST indicates that the chromosomes of the three groups of strains are not clonally related, all being outside of the CC17 of E. faecium.15,19,20
To understand the genomic context in which VanN occurred, the risk it posed for transmission in the hospital and its origins, it was of interest to determine where these strains placed within the E. faecium population structure. Here, we show that in UCN71 and UCN72, this VanN type of resistance occurs in strains that cluster with clade B commensal E. faecium, a population in which vancomycin resistance had not been previously reported. Because of the low level of resistance (16 mg/L) conferred by the WT vanN cluster, this phenotype may go undetected.
Materials and methods
Bacterial strains
The strains used in this study are listed in Table S1 (available as Supplementary data at JAC Online). Comparative genomic analysis was performed on VanN-type strains UCN71 and UCN72; a VanN transconjugant, UCN73;15 and comparator E. faecium strains (Table S1), representing the breadth of the E. faecium population and previously used to distinguish and characterize clade A and clade B lineages.4 Specifically, these strains are from 51 distinct STs carefully chosen to maximize the diversity and representation of MLST types and clusters.4
Genome sequencing
For Illumina sequencing, total DNA was purified (DNeasy DNA extraction; Qiagen, Valencia, CA, USA), quantified (Qubit dsDNA High-Sensitivity; Invitrogen, Carlsbad, CA, USA) and sequencing libraries were prepared from 50 ng of DNA per sample (Nextera DNA Sample Preparation; Illumina, San Diego, CA, USA). Libraries were normalized to 2 nM, multiplexed and subjected to 200 bp paired-end sequencing on the MiSeq platform at the Massachusetts Eye and Ear Infirmary Ocular Genomics Institute (Boston, MA, USA). On average, 5 million high-quality paired-end reads were collected for each strain, representing >250-fold coverage of the ∼3 Mb genomes. For 454 sequencing, a DNA fragment library was prepared using the GS DNA Library Preparation Kit (Roche, Indianapolis, IN, USA) followed by emulsion-based PCR amplification. Sequencing was performed using a 454 Life Sciences (Roche) GS-FLX system. For UCN72 pyrosequencing, 164 965 reads (read length between 250 and 300 nt) were collected. Draft genomes and raw reads of the strains UCN71, UCN72, UCN73 and BM4107 are accessible in GenBank: Bioproject number PRJNA245745.
Assemblies, PCR for plasmid closure, annotation and orthogrouping
Sequence reads were assembled de novo (CLC Genomics Workbench; Cambridge, MA, USA). Gene prediction, annotation and orthoclustering were conducted as previously described.4 For strain UCN72, Illumina and 454 reads were pooled and assembled de novo to produce a high-quality hybrid assembly of 12 supercontigs, ranging in length from 8063 to 1 094 650 bp (N50 = 3 contigs, mean coverage >250-fold). Contig #8 (154 330 bp) from the UCN72 genome assembly contains the vancomycin resistance cluster and was putatively identified as a plasmid. In order to determine its entire sequence, we designed specific primers pUV24F (5′-GCACTAGGGCTATACGCCTGT-3′) and pUV24R (5′-TCTCAACTTCTGGTGCTGGATCTG-3′) reading outward from the ends, sequenced the PCR product and filled the 6 nt gap to obtain the circularized plasmid, named pUV24 (154 336 bp).
Phylogeny, SNP identification, whole-genome alignment and mobilome identification
A phylogenetic tree (1344 single-copy core orthogroups, mid-point rooted) was determined using PhyML with 1000 bootstrap replicates.21 The UCN72 high-quality assembly was used as the reference for mapping of E. faecium UCN71 sequence reads. Quality-based variant detection, based on the Neighborhood Quality Standard algorithm,22 was used to identify polymorphisms. High-confidence variants, using a combination of quality filters and thresholds for coverage (>100×) and frequency (>95%), were identified. Contigs of UCN72, UCN73 and BM4107 were ordered and oriented to the genome of reference strain E. faecium Aus0004 using progressiveMauve,23 and plotted using GenoPlotR.24 The prevalence of glycopeptide resistance in strains of a given ST was obtained by analysing all strains (n = 3077) from the MLST database as well as all strains (n = 764) for which the genome was available in the GenBank WGS database (last accessed October 2017). Plasmids and phage-related genes were predicted using PlasmidFinder (http://cge.cbs.dtu.dk/services/PlasmidFinder/) and PHAST (http://phast.wishartlab.com/).
d,d-Peptidase activity
Cultures were grown in the absence or presence (8 mg/L) of vancomycin, cells were pelleted and lysed, and the cytoplasmic fraction was assessed for d,d-peptidase activity as described previously.15
Results
VanN is a rare occurrence of vancomycin resistance in clade B E. faecium
VRE E. faecium UCN71 and UCN72 (16 mg/L) were isolated from the same patient, in France.15 The patient was diagnosed with Burkitt’s lymphoma (the primary GI non-Hodgkin’s lymphoma) and intravenous vancomycin therapy was started on day 4 and ended on day 29 (Figure S1A). UCN71 was isolated from the blood on day 33 and UCN72 was isolated from the stool on day 55. Notably, the patient developed VRE bacteraemia on day 33, 1 day after a negative screen for VRE in the faeces.
Comparison of nucleotide polymorphisms in the 1344 core genes of UCN71 and UCN72 to a diverse set of E. faecium genomes places the VanN-type isolates within clade B (Figure 1a). When all E. faecium strains within the MLST database were searched for the occurrence of glycopeptide resistance, it was found only in the clade A hospital-adapted strains, making VanN unique in its occurrence within clade B E. faecium (Figure 1a). In addition, when 764 publically available E. faecium genomes were searched and assigned to a clade (Figure S1B), 64% (n = 486 vancomycin-resistant E. faecium) were found to harbour a vancomycin resistance cluster (Figure 1b). Of 486 vancomycin-resistant E. faecium, >99% belong to the MDR hospital-adapted clade A, leaving only three clade B vancomycin-resistant E. faecium (Figure 1b), including UCN71 and UCN72 VanN isolates. The only other occurrence of vancomycin resistance outside of clade A was found in EnGen0263 (accession number NZ_AJAD00000000), an isolate that possesses a vanA cluster on a rep7 family conjugative plasmid (data not shown).
Figure 1.
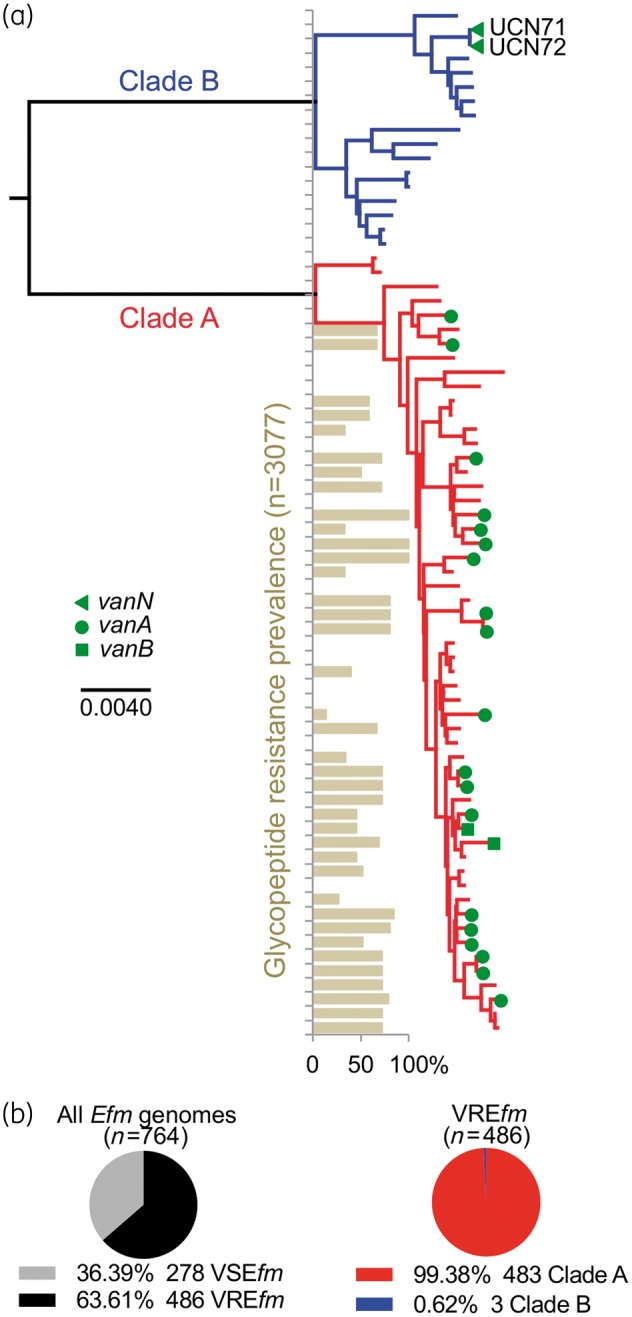
(a) VanN-type vancomycin resistance occurs in clade B E. faecium. Phylogenomic tree of a collection of E. faecium strains, including the VanN-type UCN71 and UCN72 isolates. The clade structure of the population is shown: clade B commensal strains (blue); and clade A hospital-adapted MDR strains (red). The vancomycin resistance genotype of each strain is shown. The prevalence of glycopeptide resistance of E. faecium strains from the MLST database for each particular ST is shown. Additional strain information is available in Table S1. (b) Prevalence of E. faecium strains (n = 764 publicly available genomes) harbouring (vancomycin-resistant E. faecium) or not (vancomycin-susceptible E. faecium) a vancomycin resistance cluster and distribution of vancomycin-resistant E. faecium within clades A and B. Efm, E. faecium; VREfm, vancomycin-resistant E. faecium; VSEfm, vancomycin-susceptible E. faecium.
VanN strains lack the accessory genome enriched in hospital-adapted E. faecium
In addition to distance between the clades stemming from genetic drift, MDR epidemic clade A and clade B commensal E. faecium strains also differ in gene content.4 We thus compared the genome of commensal strain UCN72 (as representative of both clonal strains) with the complete 3.0 Mb genome of E. faecium Aus0004, as representative of the hospital-adapted epidemic clade (Figure S2). Aus0004, a bacteraemia isolate, contains various mobile genetic elements including four prophages, two genomic islands, as well as three circular plasmids.25 Genome alignment revealed that none of the chromosomally integrated mobile genetic elements identified in E. faecium Aus0004 is present in UCN72 (Figure S2), which otherwise contain only a single chromosomally integrated element (a putative prophage predicted as incomplete by PHAST) (Figure S2). It has been postulated that the paucity of mobile genetic elements in commensal enterococci correlates with the presence of clustered, regularly interspaced, short palindromic repeat (CRISPR) loci, which provide a type of acquired immunity.26 Unlike Aus0004, UCN72 contains both CRISPR1 and CRISPR2 loci and functional CRISPR-associated genes (cas). The genome of strain UCN72 was assembled into 12 scaffolds; 4 of these were putatively identified as plasmid fragments and named pUV24, pUV25, pUV26 and pUV27 (Figure S2).
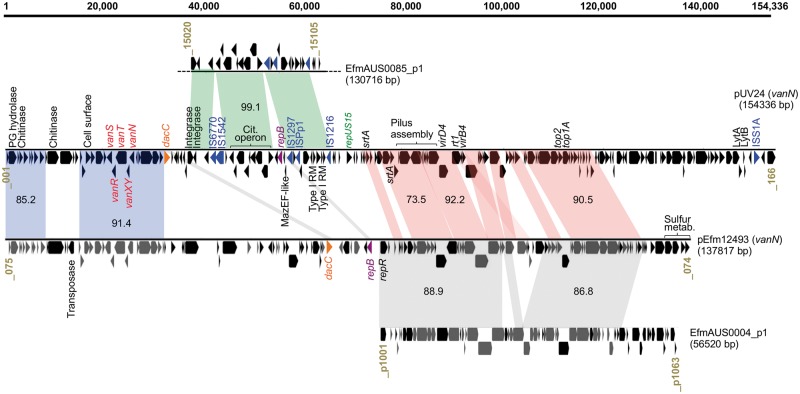
vanN is carried by a conjugative plasmid related to those in MDR epidemic E. faecium
In previous work,15 we obtained vancomycin-resistant transconjugant UCN73 by mating UCN71 with recipient E. faecium strain BM4107. Alignment of UCN73 and BM4107 genomes showed that the gain of vancomycin resistance by UCN73 was associated with the acquisition of conjugative plasmid pUV24 (Figure S2). This large plasmid (154 336 bp, GC content 35.7%) carrying the vanN cluster was closed and circularized using targeted PCR and further compared with the sequence of pEfm12493,19 the only known vanN-carrying plasmid, found in an isolate from the abscess of a patient in Canada (Figure 2). Interestingly, pUV24 (154 kb) is significantly larger than pEfm12493 (138 kb) and is predicted to harbour 166 ORFs. Two main segments (representing ∼80 kb) show an overall synteny and share between 73% and 92% of nucleotide identity between both vanN-carrying plasmids (red and blue syntenic blocks; Figure 2). First, a region of ∼50 kb in pUV24 (red blocks; Figure 2) includes conjugation genes [i.e. a predicted fully functional type IV secretion system (TIVSS)], genes encoding two sortases and genes encoding two topoisomerases. As described previously,19 this region also shares extensive sequence identity (87% overall) and synteny with plasmid Aus0004_p1 from the MDR and epidemic Aus0004 strain (grey blocks; Figure 2). Importantly, pUV24 contains a repA gene [99.9% nucleotide identity with repUS15 (from PlasmidFinder) and 98% identity with repA-pLG1 (accession number ADO66907.1) plasmid families27] instead of the replication gene shared by pEfm12493 and Aus0004_p1. The RepA_pLG1 family groups large plasmids and is highly prevalent in both clades of E. faecium (>78%; 57/72 strains in this study). Other studies have repeatedly associated this family of conjugative plasmid with various resistance genes, including vancomycin and aminoglycosides.27 Besides differences in the rep gene, it is also worth noting the rearrangements between pUV24 and pEfm12493 in the genes coding for the TIVSS. Boyd and co-authors19 noted that, in pEfm12493, the virB4 ATPase gene was truncated and interrupted by the rt1 gene and thus conjugative transfer functions may be compromised. In light of our data, this would explain the conjugative nature of pUV24, which contains a full-length virB4, compared to pEfm12493.
Figure 2.
Schematic diagram of plasmid pUV24 from strain UCN71. Where relevant, genes or groups of genes are indicated by putative function or gene name. Syntenic regions with identity to plasmid pEfm12493 (accession no. KP342511), plasmid p1 of E. faecium AUS0004 (accession no. CP003352) and plasmid p1 of E. faecium AUS0085 (accession no. CP006622) are projected (grey) and average nucleotide identities are indicated. Locus tags are provided at the beginning and end of each linearized plasmid. A scale bar, in bp, is provided. Dotted lines indicate a truncation to avoid representation of non-related material.
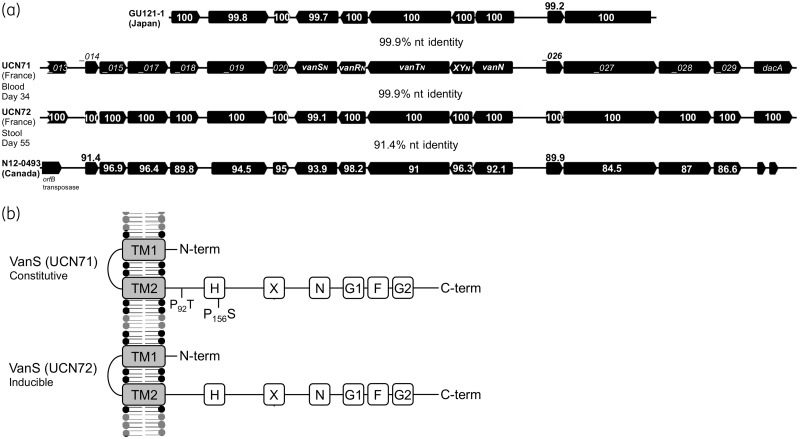
The second region (blue syntenic blocks; Figure 2) shared between pUV24 and pEfm12493 includes the vanN cluster, several cell surface protein genes (including a putative peptidoglycan hydrolase) and a putative chitinase. To investigate in detail the evolution of the vanN resistance cluster, we compared the sequences from the four vanN-VRE reported so far (Figure 3a). Clusters from UCN71, UCN72 (France) and GU121-1 (Japan) share 99.9% nucleotide identity. By contrast the vanN resistance cluster from N12-0493 (Canada) is more distantly related (91% identity). Interestingly, vanN is harboured on a larger segment shared by plasmids pUV24 and pEfm12493 (blue blocks; Figure 2). A predicted transposase sits upstream of this element in pEfm12493 and could be responsible for the movement of the vanN resistance cluster. However, sequence analysis of the boundaries of the shared element did not reveal any obvious signatures of transposition in pUV24 or pEfm12493. Plasmid pUV24 diverges from pEfm12493 by two regions. One shares very high identities (99.1%) with a segment from plasmid EfmAUS0085_p1 in the vancomycin-resistant epidemic E. faecium Aus0085 and contains a complete citrate operon and two putative type I restriction systems (green syntenic blocks; Figure 2). The second region (35 kb at the 3′ end of pUV24) is of unclear origin and BLAST analyses return no significant hits. We identified six insertion sequences (IS6770, IS1542, IS1297, IS1216, ISPp1 and ISS1A) harboured by pUV24 but none seemed directly associated with the movement of VanN. Finally, pUV24 possesses one complete type II toxin–antitoxin (TA) loci (MazEF-like) that may act as plasmid addiction systems to ensure stable maintenance and inheritance. A total of 24 pUV24 ORFs (not shown) encode surface-associated proteins of unknown function or peptidoglycan synthesis-associated proteins, indicating some remodelling of the cell surface in strains carrying pUV24. Among these surface-related proteins are the LytA and LytB N-acetylmuramoyl-l-alanine amidases (Figure 2), which participate in peptidoglycan biosynthesis, with LytB homologues having been described to have autolytic activity and to be involved in cell separation.28 Additionally, a DacC homologue occurs that is thought to possess d-alanyl-d-alanine carboxypeptidase activity, removing the C-terminal d-alanyl residues from sugar-peptide cell wall precursors,29 potentially supplementing the activity of VanXY.
Figure 3.
(a) Comparison of the vanN cluster in strains UCN71 and UCN72 from France, strain GU121-1 from Japan and strain N12-0493 from Canada. Locus tags are provided for the vanN resistance cluster in UCN71. For other resistance clusters, the amino acid identities are indicated for each gene. The overall nucleotide identities of the syntenic regions for each pairwise comparison are provided. (b) Comparison of the VanS protein in strains UCN71 and UCN72. TM1 and TM2 represent the putative transmembrane (TM)-associated sensor domains. The putative catalytic ATP binding domain containing amino acid motifs (H, X, N, G1, F and G2) that are conserved in histidine kinases are indicated, as well as amino acid substitutions. P, proline; S, serine; T, threonine.
In vivo selection affects expression of vancomycin resistance
Since UCN71 and UCN72 were isolated from the same patient 21 days apart, it was of interest to determine whether the strains had diverged. We therefore performed SNP analysis to identify polymorphisms between their genomes. Across the whole genome, a total of three high-confidence SNPs were identified and confirmed by PCR (Table 1). One SNP is synonymous and would not change the amino acid sequence of the corresponding putative ATPase. However, two SNPs surprisingly occurred within the same gene, which encodes VanS, and both are non-synonymous substitutions (Table 1).
Table 1.
Whole-genome SNP comparison between UCN71 and UCN72 serial isolates
| SNP # | Function | Nucleotide (position) | Amino acid (position) |
|---|---|---|---|
| UCN71_2511 | ATPase putative | T→A (321) | R→R (107) |
| UCN71p_021 | VanS histidine kinase | C→A (276) | P→T (92) |
| UCN71p_021 | VanS histidine kinase | C→T (466) | P→S (156) |
We previously hypothesized that amino acid substitutions in VanS of UCN71 were responsible for the constitutive expression of vancomycin resistance in this isolate.15 With the finding of polymorphisms in VanS between UCN71 and UCN72, it was of interest to investigate the resistance expression in strain UCN72. Expression of vancomycin resistance in E. faecium UCN72 was studied by analysing the d,d-dipeptidase activity of VanXYN, with (8 mg/L) or without vancomycin induction (Table 2). Induction of resistance expression in strain UCN72 increased intracellular d,d-dipeptidase activity in cytoplasmic extracts from 8 ± 7 to 37 ± 9 nmol/min/mg of protein. No such increase was noted for strain UCN71 (P < 0.05). Alignment of the VanSN protein sequences of UCN71 (constitutive) and UCN72 (inducible) suggests that either the P → T or P → S substitutions (at positions 92 and 156, respectively) might be responsible for the constitutive expression of the resistance in UCN71 (Figure 3b).
Table 2.
d,d-Dipeptidase enzymatic activities in UCN71 and UCN72 E. faecium strains
| Strain | Vancomycin (mg/L) | Activity (nmol/min/mg of protein) | Resistance |
|---|---|---|---|
| UCN71 | 0 | 49±6 | constitutive |
| UCN71 | 8 | 52±10 | |
| UCN72 | 0 | 8±7 | inducible |
| UCN72 | 8 | 37±9 |
Discussion
The results of our genomic and phylogenetic analyses indicate that the VanN-type strains studied here constitute a very unusual case of vancomycin resistance in the clade B community-associated E. faecium population, contrasting sharply with the clade A hospital-adapted strains that are frequently associated with such resistance (70% prevalence in 694 clade A E. faecium genomes; Figure S1C).
VanN vancomycin resistance was reported in Japan in five E. faecium strains (ST669) isolated from chicken meat.20 Although these strains are not clonally related to UCN71 and UCN72 (ST240), they independently acquired a vanN cluster nearly identical to that in UCN71, suggesting a recent HGT. VanN resistance was also recently reported in a strain (ST955) in Canada isolated in 2008,19 and sequence analyses indicate an independent acquisition. Although these strains were not part of our genome phylogeny, we predict, based on their ST designation (Figure S1D), that strains from Japan are also clade B E. faecium, while the strain from Canada belongs to the hospital-adapted lineage (though does not belong to CC17). This indicates that the vanN vancomycin resistance already has spread within both clade B and clade A MDR E. faecium in different parts of the world. The association with agriculture in Japan highlights the prospect that entry of VanN resistance into the human ecology might have occurred through the food chain. A plasmid of similar length to pUV24 was described to carry the resistance cluster in the Japanese VanN strains.20 By contrast, the plasmid found in the VanN strain from Canada was significantly different from the one in UCN71, again suggesting distinct acquisitions and subsequent rearrangements in both strains. Considerable sequence similarity was noted between fragments of pUV24 and plasmids circulating in MDR epidemic strains (in fact the repA_pGL1 plasmid family is highly prevalent in both clades of E. faecium27), suggesting the absence of a barrier to plasmid transfer between hospital endemic and clade B strains. In fact, vancomycin plasmid transfer has been experimentally observed in vivo in the gastrointestinal tract of mice.30
In the antibiotic era, vancomycin-resistant E. faecium bacteraemia has become a vexing problem because of the lack of effective alternative treatment options.8 Highly hospital-adapted MDR lineages have emerged and account for most of the problem.4,8 The infection model that emerges is that initial administration of antibiotics to a patient results in elimination of key elements of the microbiota that otherwise limit entry of new members into the consortium, leading to large numbers or even predominance of VRE in the gut.31–35 This is then followed by translocation of VRE into the bloodstream and bacteraemia. In addition to antibiotic resistance, hospital-adapted clade A strains acquired new metabolic capabilities which have been shown to enhance their ability to colonize antibiotic-treated mice.4,36 This led to the proposition that clade A MDR strains colonize a microniche in the antibiotic-perturbed GI tract of patients in a manner that is non-competitive with commensal clade B strains.31 This, together with the comparatively low level of vancomycin resistance (16 mg/L) conferred by the WT operon, may account for the inability of the clade B VanN-type E. faecium to grow to large enough numbers in the GI tract to be easily detected in surveillance prior to the onset of bacteraemia. In fact, gut colonization by VanN-type E. faecium appears to have been more insidious (the weakened intestinal barrier in the patient treated for a gastrointestinal lymphoma might have allowed recurrent leakage of the commensal flora into the blood) and only afterward was it found in rectal swab cultures, perhaps influenced by the expectation that the presence of this microbe in the gut usually presages bacteraemia.
Expression of resistance is a transient need for bacterial cells, as they face inhibitory concentrations of drugs only temporarily. Vancomycin resistance is mediated by a sophisticated mechanism that relies on a complex operon and combines synthesis of modified cell wall peptidoglycan precursors and elimination of the normal target precursors.10 Thus, constitutive expression of vancomycin resistance has been shown to have a high fitness cost for the cell.37 However, the same study showed that tight regulation of resistance expression drastically reduces the biological cost associated with vancomycin resistance in Enterococcus. The VanN resistance in UCN71 is one of the few cases, in addition to VanD isolates,38,39 of constitutive vancomycin resistance expression found in clinical settings.15 Interestingly, we found that the clonal UCN72 strain, isolated 21 days after UCN71, showed an inducible vancomycin resistance phenotype, providing strong evidence that it represents the genotype originally acquired in the gut preceding its translocation into the bloodstream. Overall, we found a strong in-host selection that resulted in the accumulation of two (out of three total genome wide) non-synonymous SNPs in the vanS gene that likely affect the phosphatase activity of VanS and thus account for constitutive expression of resistance.
In conclusion, we report a case of vancomycin resistance in a commensal lineage of E. faecium responsible for an atypical bacteraemia in an immunocompromised patient. Because this strain eluded detection prior to its occurrence in the blood, we believe that the prevalence of vancomycin resistance in the community may be underestimated and that such a reservoir of glycopeptide resistance could pose a serious concern for public health.
Supplementary Material
Acknowledgements
We thank Michel Auzou for technical assistance. We are grateful to Pierre Berger and Sabine Camiade for the collection of UCN71 and UCN72 strains, metadata and other clinical relevant information. We thank Paul Godfrey and Jennifer Wortman for their expertise in comparative genomics and computational biology.
Funding
This work was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under NIH/NIAID grants AI072360 and AI083214 (Harvard-wide Program on Antibiotic Resistance). This project has also been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under grant no. U19AI110818 and contract no. HHSN272200900018C to the Broad Institute. K. S. was funded by the Alexander von Humboldt Foundation through a Feodor Lynen Research Fellowship. V. C. was supported by the 7th Framework European Program TROCAR.
Transparency declarations
None to declare.
Supplementary data
Table S1 and Figures S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. Cattoir V, Giard JC.. Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev Anti Infect Ther 2014; 12: 239–48. [DOI] [PubMed] [Google Scholar]
- 2. Willems RJ, Top J, van Santen M. et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 2005; 11: 821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palmer KL, Godfrey P, Griggs A. et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 2012; 3: e00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lebreton F, van Schaik W, Manson McGuire A. et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 2013; 4: e00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raven KE, Reuter S, Reynolds R. et al. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res 2016; 26: 1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leavis HL, Bonten MJ, Willems RJ.. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol 2006; 9: 454–60. [DOI] [PubMed] [Google Scholar]
- 7. Hidron AI, Edwards JR, Patel J. et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 2008; 29: 996–1011. [DOI] [PubMed] [Google Scholar]
- 8. Gilmore MS, Lebreton F, van Schaik W.. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 2013; 16: 10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mikulska M, Del Bono V, Raiola AM. et al. Blood stream infections in allogeneic hematopoietic stem cell transplant recipients: reemergence of Gram-negative rods and increasing antibiotic resistance. Biol Blood Marrow Transplant 2008; 15: 47–53. [DOI] [PubMed] [Google Scholar]
- 10. Depardieu F, Podglajen I, Leclercq R. et al. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 2007; 20: 79–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leclercq R, Derlot E, Duval J. et al. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med 1988; 319: 157–61. [DOI] [PubMed] [Google Scholar]
- 12. Uttley AH, Collins CH, Naidoo J. et al. Vancomycin-resistant enterococci. Lancet 1988; 1: 57–8. [DOI] [PubMed] [Google Scholar]
- 13. Boyd DA, Willey BM, Fawcett D. et al. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel d-Ala-d-Ser gene cluster, vanL. Antimicrob Agents Chemother 2008; 52: 2667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu X, Lin D, Yan G. et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 2010; 54: 4643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lebreton F, Depardieu F, Bourdon N. et al. d-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother 2011; 55: 4606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang S, Sievert DM, Hageman JC. et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 2003; 348: 1342–7. [DOI] [PubMed] [Google Scholar]
- 17. Boyd DA, Mulvey MR.. The VanE operon in Enterococcus faecalis N00-410 is found on a putative integrative and conjugative element, Tn6202. J Antimicrob Chemother 2013; 68: 294–9. [DOI] [PubMed] [Google Scholar]
- 18. Boyd DA, Lalancette C, Lévesque S, Golding GR.. Characterization of a genomic island harbouring a new vanD allele from Enterococcus faecium N15-508 isolated in Canada. J Antimicrob Chemother 2016; 71: 2052–4. [DOI] [PubMed] [Google Scholar]
- 19. Boyd DA, Lévesque S, Picard AC. et al. Vancomycin-resistant Enterococcus faecium harbouring vanN in Canada: a case and complete sequence of pEfm12493 harbouring the vanN operon. J Antimicrob Chemother 2015; 70: 2163–5. [DOI] [PubMed] [Google Scholar]
- 20. Nomura T, Tanimoto K, Shibayama K. et al. Identification of VanN-type vancomycin resistance in an Enterococcus faecium isolate from chicken meat in Japan. Antimicrob Agents Chemother 2012; 56: 6389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S, Lethiec F, Duroux P. et al. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 2005; 33: W557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brockman W, Alvarez P, Young S. et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res 2008; 18: 763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darling AE, Mau B, Perna NT.. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010; 5: e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guy L, Kultima JR, Andersson SG.. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 2010; 26: 2334–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam MM, Seemann T, Bulach DM. et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 2012; 194: 2334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palmer KL, Gilmore MS.. Multidrug-resistant enterococci lack CRISPR-cas. MBio 2010; 4: e00227-10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clewell DB, Weaver KE, Dunny GM. et al. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology. In: Gilmore MS, Clewell DB, Ike Y et al., eds. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014. http://www.ncbi.nlm.nih.gov/books/NBK190430/. [PubMed]
- 28. Shockman GD, Daneo-Moore L, Kariyama R. et al. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microb Drug Resist 1996; 2: 95–8. [DOI] [PubMed] [Google Scholar]
- 29. Sauvage E, Kerff F, Terrak M. et al. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 2008; 32: 234–58. [DOI] [PubMed] [Google Scholar]
- 30. Dahl KH, Mater DD, Flores MJ. et al. Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptide selection. J Antimicrob Chemother 2007; 59: 478–86. [DOI] [PubMed] [Google Scholar]
- 31. Gilmore MS, Ferretti JJ.. The thin line between gut commensal and pathogen. Science 2003; 299: 1999–2002. [DOI] [PubMed] [Google Scholar]
- 32. Ubeda C, Taur Y, Jenq RR. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120: 4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ubeda C, Bucci V, Caballero S. et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81: 965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kinnebrew MA, Ubeda C, Zenewicz LA. et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 2010; 201: 534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brandl K, Plitas G, Mihu CN. et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008; 455: 804–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X, Top J, de Been M. et al. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 2013; 207: 1780–6. [DOI] [PubMed] [Google Scholar]
- 37. Foucault ML, Depardieu F, Courvalin P. et al. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci U S A 2010; 107: 16964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Depardieu F, Foucault ML, Bell J. et al. New combinations of mutations in VanD-type vancomycin-resistant Enterococcus faecium, Enterococcus faecalis, and Enterococcus avium strains. Antimicrob Agents Chemother 2009; 53: 1952–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Casadewall B, Courvalin P.. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol 1999; 181: 3644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Schaik W, Top J, Riley DR. et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 2010; 11: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. García-Solache M, Rice LB.. Draft genome sequence of vancomycin-susceptible, ampicillin-intermediate Enterococcus faecium strain D344RRF. Genome Announc 2016; 4: e01720-15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. García-Solache M, Rice LB.. Genome sequence of the multiantibiotic-resistant Enterococcus faecium strain C68 and insights on the pLRM23 colonization plasmid. Genome Announc 2016; 4: e01719-15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.