Abstract
Early embryogenesis relies on the translational regulation of maternally stored mRNAs. In sea urchin, fertilization triggers a dramatic rise in translation activity, necessary for the onset of cell division. Here, the full spectrum of the mRNAs translated upon fertilization was investigated by polysome profiling and sequencing. The translatome of the early sea urchin embryo gave a complete picture of the polysomal recruitment dynamics following fertilization. Our results indicate that only a subset of maternal mRNAs were selectively recruited onto polysomes, with over-represented functional categories in the translated set. The increase in translation upon fertilization depends on the formation of translation initiation complexes following mTOR pathway activation. Surprisingly, mTOR pathway inhibition differentially affected polysomal recruitment of the newly translated mRNAs, which thus appeared either mTOR-dependent or mTOR-independent. Therefore, our data argue for an alternative to the classical cap-dependent model of translation in early development. The identification of the mRNAs translated following fertilization helped assign translational activation events to specific mRNAs. This translatome is the first step to a comprehensive analysis of the molecular mechanisms governing translation upon fertilization and the translational regulatory networks that control the egg-to-embryo transition as well as the early steps of embryogenesis.
INTRODUCTION
Protein synthesis is a fundamental process required for cell proliferation, cell differentiation, and cell response to stress and environmental cues. The level of a protein within a cell depends on the regulation of messenger RNA (mRNA) abundance via transcriptional regulation, but also on the rate of protein synthesis (on the translational machinery) and of protein degradation. Importantly, translational control is critical for protein production in response to a number of physiological and pathological situations, including development (1). The term ‘translatome’ that emerged in 2011 characterizes the subset of mRNAs present in the cell that are actively translated, i.e. that are associated with polysomes, thereby providing a very accurate picture of the functional protein readout of the genome at a specific time point (2). Translatome analysis is made possible by carrying out polysome profiling coupled with high-throughput sequencing (3–5). This approach leads to (a) the identification of the sets of mRNAs that are translated under specific developmental, physiological or pathological conditions, (b) determination of the processes of selection and recruitment of mRNAs onto polysomes, and (c) elucidation of translational regulatory networks.
Translational control is the main driver of gene expression in early stages of development, more specifically during the egg-to-embryo transition. From fertilization to the onset of zygotic transcription, maternal mRNAs in the egg drive the first mitotic cell cycles, independently of mRNA transcription and ribosome biogenesis (6–8). Sea urchin eggs have reached meiotic maturity, and are blocked in a G1-like stage of cell division. Fertilization triggers an increase in protein synthesis, necessary for the resumption of mitotic cell division. The stimulation of cap-dependent translation depends on the release of the cap-binding protein eIF4E from its translational repressor 4E-BP, and its association with the scaffolding protein eIF4G, which depends on mTOR signaling (9–12). Some studies suggest that activation of the translation machinery upon fertilization triggers the unmasking of stored maternal mRNAs, leading to a global increase in protein synthesis, with few changes in the protein synthesis pattern (6,13,14). Nonetheless, selective translation occurs soon after fertilization (15). Several mRNAs are associated with polysomes after fertilization, encoding proteins necessary for cell cycle progression, such as cyclins and the ribonucleotide reductase small subunit R2 (15–18), but to date no large-scale analysis of the translated mRNAs has been undertaken on the egg-to-embryo transition in sea urchin. In this paper, we analyzed the recruitment of maternal mRNAs onto polysomes upon fertilization, using RNA sequencing and comparing the mRNAs present in the polysome fractions between unfertilized eggs and one-cell post-fertilization embryos in the sea urchin Paracentrotus lividus. The determination of the embryo translatome demonstrated that only a fraction of maternal mRNAs is recruited just after fertilization, with enrichment for several biological functions. Furthermore, we showed that the polysomal recruitment of mRNAs just after fertilization varies upon mTOR signaling inhibition, suggesting selective cap-independent translation after fertilization in sea urchin.
MATERIALS AND METHODS
Handling and treatment of eggs and embryos
Sea urchins (Paracentrotus lividus) were collected in the Bay of Crozon (Brittany, France) and maintained at the CRBM facility (Roscoff Marine Station). Animals were induced to spawn by intracoelomic injection of 0.1 M acetylcholine; gametes were collected in filtered sea water (FSW) and kept as a 5% dilution in FSW. Eggs were dejellied 40 s in FSW supplemented with 0.7 mM citric acid and rinsed in fresh FSW. For mTOR inhibition, PP242 [10 μM] was added to the indicated samples 10 min before fertilization (at egg stage). Eggs were fertilized and embryos were cultured at 16°C under constant stirring. Puromycin [300 μg/ml] was added to the indicated samples 20 min before sample collection (at 40 min after fertilization). In all samples, for polysome preparation, emetine [100 μM] was added 5 min before harvesting to freeze the elongating polysomes on translated mRNAs. Both control and treated embryos were harvested 60 min after fertilization for polysome preparation. In vivo protein synthesis activity, p13suc1-sepharose pull-down of CDK1 protein and western blot analysis are described in Supplementary methods.
Polysome preparation
Equivalent numbers of eggs or embryos were collected and polysome preparation was performed as described in (5). Briefly, cells were resuspended in polysome lysis buffer (10 mM Tris pH 7.4, 250 mM KCl, 10 mM MgCl2, 25 mM EGTA, 0.4% Igepal, 5% sucrose, 1 mM DTT, 10 μg/ml aprotinin, 2 μg/ml leupeptin, 40 U/ml RNasin, 100 μM emetine), lysed by 10 strokes in a Dounce homogenizer, clarified by centrifugation at 16 000 × g for 10 min. Lysates were then centrifuged through a 15–40% sucrose gradient in gradient buffer (10 mM Tris pH 7.4, 250 mM KCl, 10 mM MgCl2, 25 mM EGTA, 1 mM DTT) for 2.5 h at 38 000 rpm in a SW41Ti rotor (Beckman). Puromycin-treated samples were incubated with 0.5 M KCl for 15 min at 4°C then for 15 min at 37°C, before applying them on a sucrose gradient. Gradients were collected with an ISCO gradient fractionator, coupled with an optical density recorder, in 21 equivalent fractions. RNA was extracted from each fraction using one volume of 1:1 phenol (pH 4):chloroform (v/v), and isopropanol precipitated in presence of a glycogen carrier. The RNA pellet was resuspended in RNase-free water. One-tenth of the RNA was used for quality control on an agarose gel. RNA was kept at −80°C until further use.
RNA-Seq libraries and sequencing
RNA-Seq data were generated from three independent polysome profiling experiments, each comprising eight different samples as follows: cytoplasm and polysomal fractions from unfertilized eggs and 1-h (one-cell) post-fertilization embryos, in the absence or presence of puromycin. All eight samples from a biological replicate were taken from eggs or embryos arising from a single female and male pair. The independent experiments correspond to three independent sets of parents. RNA purification and NGS libraries construction are described in Supplementary methods. Description of the 24 libraries and corresponding data generated for this study are provided in Supplementary Table S1.
Translatome analysis
The maternal transcriptome was generated from the cytoplasmic RNAs corresponding to unfertilized and fertilized eggs (three independent samples for each condition) on a de novo assembly pipeline (Supplementary methods). The de novo transcriptome was annotated using the Strongylocentrotus purpuratus gene database Echinobase (genome version 3.1) (http://echinobase.org; (19,20)). The transcriptome was filtered, selecting transcripts with the best BLASTn hit values <10−5 and a transcript count (FPKM) greater than 5, leading to a final set of 14 002 transcripts.
The translation status of each transcript was established using the following analysis. Normalization and differential analysis were carried out using the generalized linear model framework according to the edgeR model and R package (21–23), using the raw read counts from the 24 libraries (Supplementary Table S1) to generate three log2-fold change values to evaluate the translation status in unfertilized eggs (UnF_vs_UnFpuro), in fertilized eggs (F_vs_Fpuro) and the recruitment into or exit from polysomal fractions induced by fertilization (F_vs_UnF). For each gene g, we assumed that the observed count 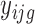 from polysomal fraction of female i in group j follows a negative binomial distribution with a mean parameter
from polysomal fraction of female i in group j follows a negative binomial distribution with a mean parameter 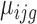 . This mean parameter depends on the sequencing depth for sample corresponding to female i in condition j
. This mean parameter depends on the sequencing depth for sample corresponding to female i in condition j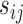 , on the number of pooled polysome gradients
, on the number of pooled polysome gradients 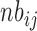 , on the relative abundance of enriched polysomal mRNA g
, on the relative abundance of enriched polysomal mRNA g 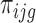 and on the observed counts in the associated cytoplasmic mRNA
and on the observed counts in the associated cytoplasmic mRNA 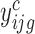 normalized by the corresponding sequencing depth
normalized by the corresponding sequencing depth 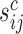 and the number of gradients
and the number of gradients 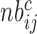 .
.
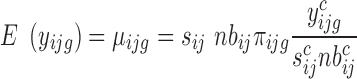 |
The model is as follows:
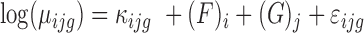 |
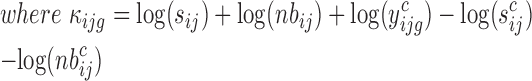 |
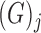 is the main effect of group j (j = UnF, UnFpuro, F, Fpuro),
is the main effect of group j (j = UnF, UnFpuro, F, Fpuro), 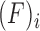 is the main effect of female i (i = 1,2,3) and
is the main effect of female i (i = 1,2,3) and 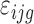 is a random error term that is assumed to be independent between observations.
is a random error term that is assumed to be independent between observations.
The scaling factors 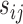 and
and 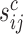 were calculated using the trimmed mean of M-values (TMM) (24). We used the edgeR Bioconductor package with a matrix of offsets
were calculated using the trimmed mean of M-values (TMM) (24). We used the edgeR Bioconductor package with a matrix of offsets 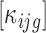 to fit a negative binomial model per gene with a genewise dispersion as calculated in (22,23). The normalization as pointed out in (25) has an important impact on all the downstream analyses. The matrix of offsets was assumed to account for all normalization issues (here sequencing depth, numbers of polysome gradients and cytoplasmic values). We computed GLM likelihood ratio tests for UnF vs UnFpuro differences, F vs Fpuro differences and F vs UnF differences within females, to select for mRNAs translated in unfertilized eggs and in 1-h embryos and for mRNAs for which translation was modified by fertilization. The R code for translatome analysis is available upon request. Raw P-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure (26), which controls the false discovery rate. Genes with an adjusted P-value lower than 0.05 were considered significant. All statistical analyses were performed using R software (27) with Bioconductor (28) packages.
to fit a negative binomial model per gene with a genewise dispersion as calculated in (22,23). The normalization as pointed out in (25) has an important impact on all the downstream analyses. The matrix of offsets was assumed to account for all normalization issues (here sequencing depth, numbers of polysome gradients and cytoplasmic values). We computed GLM likelihood ratio tests for UnF vs UnFpuro differences, F vs Fpuro differences and F vs UnF differences within females, to select for mRNAs translated in unfertilized eggs and in 1-h embryos and for mRNAs for which translation was modified by fertilization. The R code for translatome analysis is available upon request. Raw P-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure (26), which controls the false discovery rate. Genes with an adjusted P-value lower than 0.05 were considered significant. All statistical analyses were performed using R software (27) with Bioconductor (28) packages.
RT-PCR analysis
PCR primer pairs were designed within the ORF of the mRNAs to analyze (Supplementary Table S2). The relative amounts of each mRNA in the polysomal pool (fractions #18–21) and in the cytoplasmic samples from unfertilized eggs and 1-h embryos were determined by quantitative RT-PCR (RT-qPCR), on the same three biological replicates as the ones used for the RNA-Seq analysis, and were run in three experimental replicates. mRNA distribution along the polysome gradient was analyzed by RT-PCR using an equal volume of RNAs from each fraction, as described in (5). Statistical analyses were done using the two-tailed Student's t test. See Supplementary methods for details.
RESULTS
Identification of mRNAs translated upon fertilization
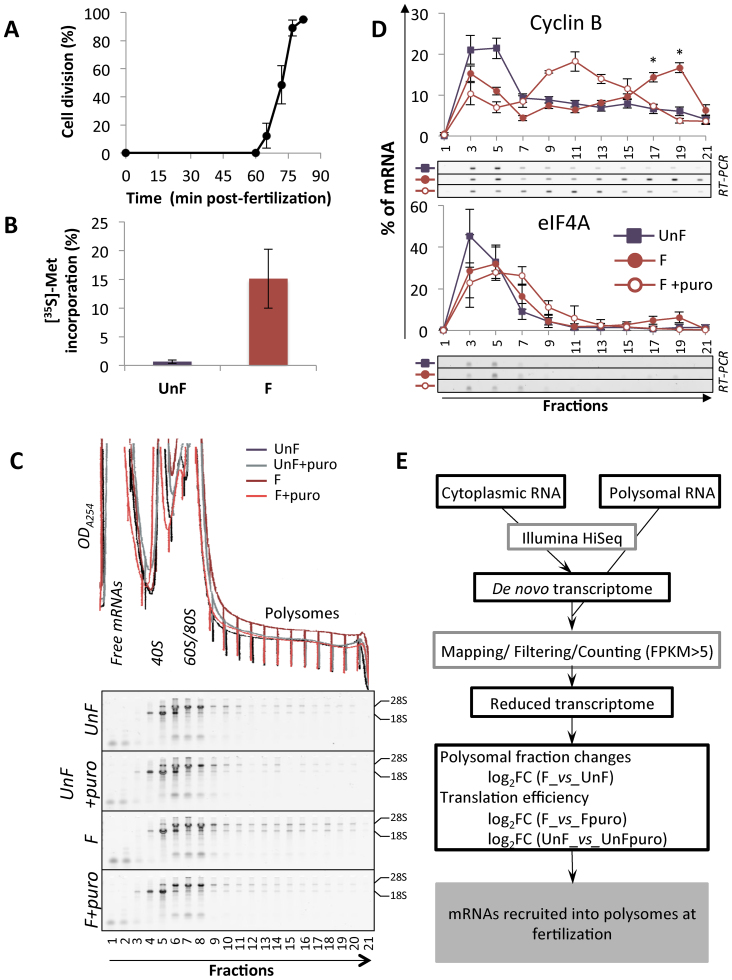
In P. lividus, fertilization induces an increase in protein synthesis, leading to the first embryonic cell division as early as 70 min after fertilization (Figure 1A and B). As noted in different sea urchin species (13,16,29), in P. lividus newly synthesized proteins increased shortly after fertilization, but the SDS-PAGE profile did not vary dramatically across the different time points (Supplementary Figure S1A). To identify the mRNAs that are recruited early on at the egg-to-embryo transition, we performed polysome profiling analysis in unfertilized eggs (UnF) and in 1-h post-fertilization (F) embryos. To do so, we used a polysome gradient, which allows the separation of an mRNA according to its translational status, coupled with RNA-sequencing to identify the translated mRNAs that encode these proteins newly synthesized just after fertilization. As described previously (5), polysome profiles between these two stages showed few differences; nonetheless, ribosomal RNAs were present in larger amounts in the polysomal fraction of fertilized embryos, consistent with the puromycin-sensitive formation of polyribosomes after fertilization (Figure 1C). Furthermore, mRNAs encoding cyclin B were recruited onto active polysomes following fertilization (Figure 1D). In unfertilized eggs, cyclin B mRNAs were associated with the light fractions (#1–7) of the polysome gradient, in agreement with their untranslated status before fertilization. Fertilization induced an association of cyclin B mRNAs with heavy polysome fractions (#18–21). The translation elongation inhibitor puromycin made it possible to distinguish between translated mRNAs and co-migrating mRNPs, because it disrupts only elongating polysomes (30,31) and shifts the translated mRNAs from the heavy fractions to the middle fractions of the gradient (32). Upon puromycin treatment, global protein synthesis was affected (Supplementary Figure S1B), and cyclin B mRNAs were relocated toward the middle of the gradient, demonstrating that this mRNA species was associated with bona fide translating polysomes after fertilization (Figure 1D). Similar behavior was observed for cyclin A and ribonucleotide reductase small subunit R2 mRNAs (Supplementary Figure S1C) (5,15,18). In contrast, eIF4A mRNA distribution in the polysome gradient was unchanged following fertilization (Figure 1D). Based on the polysome distribution in control and puromycin-treated fertilized embryos, we observed that only a fraction of a particular mRNA species was recruited onto translating polysomes after fertilization: 26.8% (SD = 3.3, n = 3), 25.9% (SD = 3.9, n = 3) and 19.1% (SD = 8.5, n = 3) of all cyclin B, cyclin A and R2 mRNAs, respectively, were present on polysomes. We selected the heavy polysome fractions for subsequent RNA sequencing and translatome analysis to identify new maternal mRNAs strongly recruited onto polysomes just after fertilization, with recruitment behavior similar to that of cyclin B, cyclin A and R2 mRNAs.
Figure 1.
(A) Cell division kinetics in Paracentrotus lividus from two independent experiments, error bars represent standard deviation. (B) Protein synthesis activity measured by 15-min pulse-labeling in unfertilized eggs (UnF) and in fertilized embryos (F) performed at 1 h post-fertilization. The results are expressed as the percentage incorporation of [35S]-methionine into protein over total radioactivity taken up by the cells in three independent experiments. Error bars represent standard deviation. (C) Optical density profiles (ODA254) of polysome gradient profiles (top) and corresponding RNA profiles are shown for unfertilized eggs and fertilized embryos treated with puromycin or left untreated. The RNAs from each fraction of the polysome gradient were separated on 2% agarose-TBE gels. The positions of the 18S and 28S ribosomal RNAs are indicated. (D) Distribution on a 15–40% sucrose gradient of mRNAs coding for cyclin B (CycB, positive control) and initiation factor 4A (eIF4A, negative control) before (UnF) and after (F) fertilization. mRNAs were detected by RT-PCR amplification in each fraction (a representative experiment shown). Distribution of the mRNA along the gradient is shown as a percentage of total mRNA, error bars represent SEM on five biological replicates (UnF vs F: * P-value < 0.05). Presence of the mRNA in active polysomes was assessed by treating embryos in vivo with puromycin before polysome gradient fractionation (F+puro in vivo, n = 3). (E) Diagram of the translatome analysis, performed on three independent polysome profiling datasets.
To obtain the full set of mRNAs present in eggs or embryos, cytoplasmic RNAs isolated from unfertilized eggs and 1-h post-fertilization embryos (Supplementary Table S1) were sequenced. Because a reliable reference genome is not currently available for P. lividus, sequences were de novo assembled using the Trinity suite to generate a maternal reference transcriptome. There was a good correlation between the three independent biological replicates (Pearson correlation R2 above 0.80; Supplementary Figure S2). Comparison between unfertilized eggs and 1-h post-fertilization embryos showed no significant differences (Supplementary Figure S3), suggesting that there was no variation in the abundance of the transcripts between these two stages. An estimation of the repertoire of transcripts present in the maternal transcriptome was done in comparison with data available for the American sea urchin Strongylocentrotus purpuratus, for which a genome is available (19,20,33). Annotation against the 23 000 gene predictions of the S. purpuratus genome assigned a best BLASTn hit with an e-value<10−5 for 14 002 transcripts, corresponding to 7685 Trinity unique genes and to 6518 S. purpuratus gene predictions. The maternal transcriptome thus represented roughly 28% of the total gene number, which is within the range of other species (20–45% in mouse, 42% in S. purpuratus, 55% in Drosophila (8,33)).
To assess the translational changes globally and establish the translatome following fertilization, heavy polysome-associated mRNAs were sequenced from eggs and 1-h post-fertilization embryos. RNAs remaining in the heavy fractions after in vivo puromycin treatment were sequenced in parallel to account for mRNP-associated mRNAs. For the translatome analysis, the reads generated from the 24 samples (cytoplasmic and polysomal samples; see Supplementary Table S1) were mapped independently against the filtered maternal transcriptome comprising 14 002 transcripts. The translatome pipeline is presented in Figure 1E. To quantify each transcript in equivalent numbers of eggs or 1-h-post fertilization embryos, the counts in the polysome samples were corrected by the number of pooled gradients used in each condition. A negative binomial generalized linear model was fitted to quantify each individual transcript in the heavy fractions of the gradient, on three independent biological replicates, taking into account the cytoplasmic abundance of the transcript and the paired nature of the samples (see Methods). Pairwise comparison in unfertilized and fertilized samples allowed the selection of transcripts for which the entry into the heavy polysome fractions was modified after fertilization. Transcripts with significant differences (BH adjusted P-value < 0.05) between unfertilized eggs and fertilized embryos were selected. Data were then filtered for translated mRNAs in both stages to distinguish between bona fide translated mRNAs and co-migrating mRNPs. The translation efficiency for each transcript was assessed by comparing puromycin-treated and untreated samples. Transcripts with a positive fold-change and significant difference between puromycin-treated and untreated polysomes (BH adjusted P-value <0.05) were considered as translated. The polysome data obtained from fertilized samples showed a good correlation between the three independent replicates (R2 above 0.60; Supplementary Figure S2), and no log2FC(F/Fpuro) threshold was set. In contrast, we observed a lower correlation (R2 above 0.40) between the three biological replicates of the unfertilized polysome samples, consistent with the lower translation activity in unfertilized eggs (Figure 1B). Therefore, a threshold for the unfertilized data was set at log2FC(UnF/UnFpuro) > 1, to ensure that only relevant translated transcripts were selected.
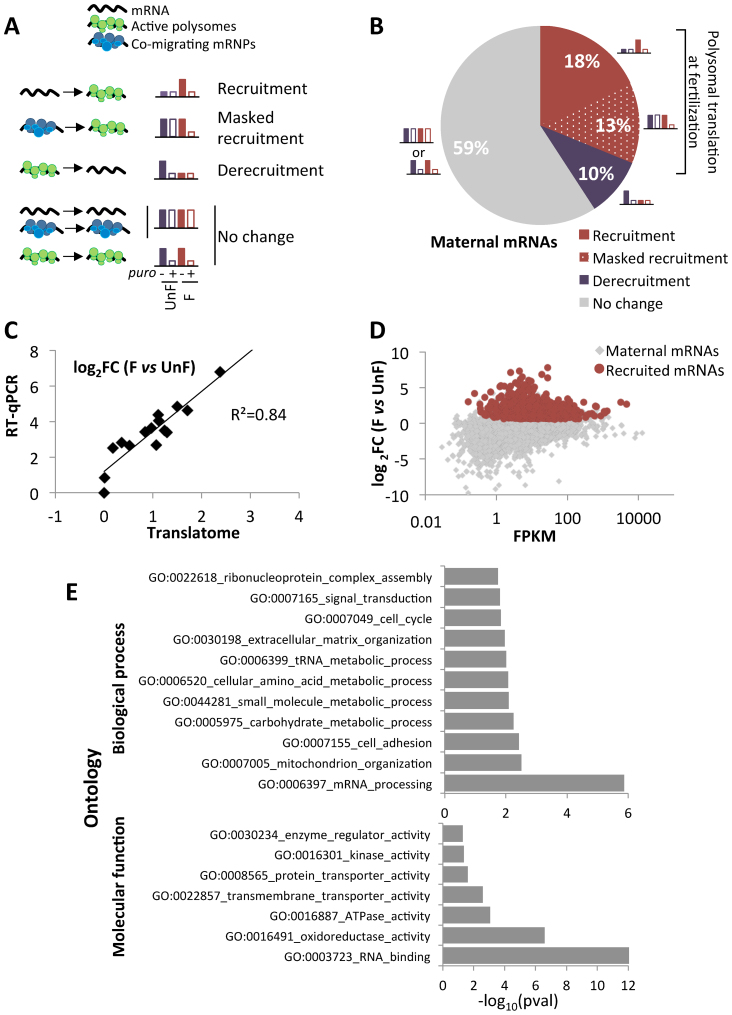
The maternal mRNAs were classified according to their recruitment into the heavy fractions of the gradient and their translation efficiency in unfertilized eggs and fertilized embryos. The different possibilities are summarized Figure 2A; the full overview of the mRNA recruitment into active polysomes is shown Supplementary Figure S4, recruitment and translation efficiency data for each transcript are presented in Supplementary data 2. The majority (59%) of the maternally deposited mRNAs in the egg showed no translational changes following fertilization (Figure 2B, ‘no change’). Among them, 52% of the maternal transcripts (7319/14 002) were declared untranslated in both conditions, whereas 7% (967/14 002) were translated both before and after fertilization, but showed no change in their polysomal distribution. Next, 1351 transcripts (10%) were determined as significantly translated before fertilization and released from polysomes at fertilization (Figure 2B, ‘derecruitment’). Interestingly, some transcripts (1851/14,002; 13%) showed no statistically significant difference between unfertilized and fertilized heavy fractions, but were efficiently translated after fertilization (log2FC(F/Fpuro) > 0, P < 0.05), but not before fertilization (log2FC(UnF/UnFpuro) < 0), suggesting that these transcripts correspond to mRNAs stored in heavy mRNPs or stalled polysomes in unfertilized eggs, and activated for translation after fertilization (Figure 2B, ‘masked recruitment’). These data strongly suggest that puromycin treatments are necessary to fully identify actively translated mRNAs. Finally, 2514 transcripts (18% of the maternal set) were significantly recruited onto active polysomes after fertilization (Figure 2B, ‘recruitment’). The mRNAs encoding cyclin B, cyclin A and ribonucleotide reductase small subunit R2 belonged to this set of 2514 transcripts translated after fertilization. The majority of these mRNAs were translated de novo, however a small proportion (319 transcripts; 2%) was already translated before fertilization, and showed a further polysomal recruitment upon fertilization. Altogether, the mRNAs translationally activated at fertilization corresponded to 31% of the maternal set (recruitment and masked recruitment).
Figure 2.
(A) Schematic diagram of the different possible configurations of maternal mRNA upon fertilization. (B) Pie chart of maternal mRNAs according to their polysomal behavior just after fertilization as determined by the log2FC (F vs UnF), corrected by the puromycin control at both timepoints. (C) Comparison of log2FC values between unfertilized (UnF) and fertilized (F) polysomal mRNA for 15 genes obtained by RNA-Seq analysis (translatome) and by RT-qPCR. The best fitting linear regression is plotted. (D) Log2FC between F and UnF polysomal contents were plotted against the FPKM counts for each mRNA. All maternal mRNAs are shown in light gray, maternal mRNAs recruited upon fertilization are in red. (E) Functional categories and GOterms associated with the 18% recruited mRNAs. The numbers of transcripts obtained in the translated set for each category were compared with expected numbers assuming random representation (binomial test, P-value < 0.05). Bar charts represent the enriched biological processes (top) and molecular functions (bottom) associated with the translated mRNAs.
The polysomal recruitment of a subset of 15 genes (translated and untranslated, high and low abundance) was analyzed by RT-qPCR (see primers in Supplementary Table S2), using polysomal and cytoplasmic mRNAs. Quantification for each mRNA was done relative to a reference mRNA encoding MT1 (metallothionein 1). Polysomal recruitment was then calculated as the ratio of (polysF/cytoF) to (polysUnF/cytoUnF). The fold-change recruitment obtained by RT-qPCR of pooled polysomal vs cytoplasmic RNAs was consistent with the recruitment fold-change found by RNA-Seq analysis (R2 > 0.8; Figure 2C).
Recruitment onto polysomes is not correlated with transcript abundance in the maternal transcriptome
Our translatome analysis showed that only a fraction of the maternal mRNAs enters active polysomes at 1-h post-fertilization, with these transcripts covering a wide range of abundance (Figure 2D). However, fertilization triggering the activation of the translation machinery may lead to translation of all mRNAs proportionally to their abundance in the maternal stock. We then tested whether there was a bias for translation toward abundant mRNAs. Comparing mRNA abundance with their corresponding recruitment index (log2FC(F/UnF)) showed no correlation between these two parameters, and the mean FPKM value of recruited mRNAs was similar to the value obtained for the maternal mRNAs. We further analyzed abundant maternal transcripts with a FPKM>500 from the RNA-Seq data and checked for their translational status. Among the 38 most abundant transcripts, only nine of them were significantly recruited onto polysomes just after fertilization (Supplementary Table S4), including cyclin B, cyclin A and ribonucleotide reductase small subunit R2 mRNAs (15,17,18). These data suggest that the translational status of an mRNA following fertilization is not correlated with its abundance, arguing for a selective translation process upon fertilization.
Functional enrichment of mRNAs translated upon fertilization
When the translated mRNAs were classified from lowest to highest P-values, cyclin B mRNA ranked first, indicating a strong biological constraint on its polysomal recruitment. Consistent with the fact that the greatest biological change triggered by fertilization is the progression into embryonic cell cycles, many other mRNAs encoding proteins involved in cell cycle regulation were found in this dataset. To estimate the enriched functions in the translatome, GOterms were retrieved from the Trinotate annotation of the maternal transcriptome, and functional classes were compared between the translated mRNA and the maternal mRNA datasets. We used a binomial test to compare the observed number of translated genes with the expected number for each functional class, assuming random representation of the maternal mRNAs in the translated set. Over-represented biological processes in the translatome were ‘mRNA processing’, ‘metabolic processes’, ‘cell cycle’ and ‘signal transduction’; over-represented molecular functions were ‘RNA-binding’, ‘transporter’ and ‘kinase’ activities (binomial test, P-value < 0.05; Figure 2E). We further classified the maternal mRNAs according to their translational status upon fertilization (recruited, translated from mRNP stocks, released from polysomes and unchanged). Half of the maternal mRNAs belonging to the ‘cell cycle’ and the ‘RNA-binding’ functional categories were in the recruited set. A striking feature of the mRNAs from the signaling functional class was their abundance in the translated pools from mRNP stocks (Supplementary Figure S5). Altogether, these data demonstrate that polysomal recruitment at fertilization is selective.
Distribution of identified newly translated mRNAs on polysome gradients
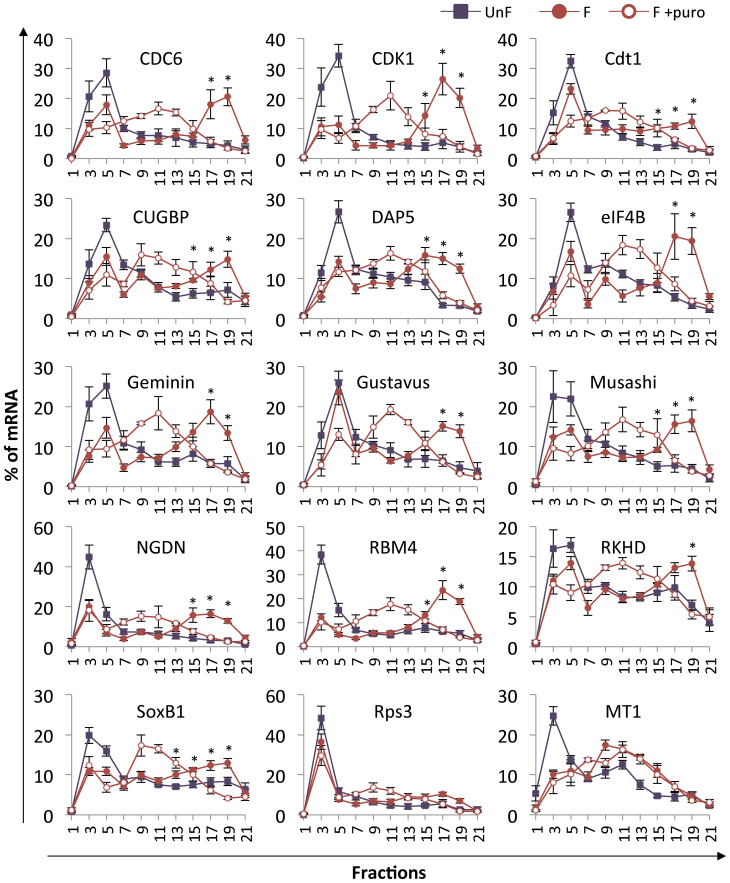
To validate the mRNAs newly identified by the translatome approach as bona fide translated mRNAs after fertilization, we analyzed the distribution of selected mRNAs on polysome gradients prepared from unfertilized eggs and 1-h post-fertilization embryos. We focused on mRNAs belonging to two enriched functional groups, cell cycle and RNA-binding proteins (RNA-BPs), and in two groups involved in regulation of early development (maternal determinants and translation regulation). We chose to test mRNAs spanning a wide spectrum of abundance and fold-change within these groups (Table 1). The polysome gradient distribution was determined for at least five biological replicates; all conditions for each biological replicate were obtained from sibling eggs and embryos of a single female and male pair and were processed in parallel. Our results show that the mRNAs in unfertilized eggs were mainly present in the light fractions of the gradient (#3–7). Fertilization triggered a shift of the mRNAs toward the heavy fractions of the gradient (#18–21). As expected, following puromycin treatment, the heavy polysomes disassembled and relocated to the middle of the gradient, suggesting that mRNAs present in the heavy fractions are actively translated after fertilization. These results demonstrate the recruitment of the tested mRNAs into active polysomes after fertilization (Figure 3). Two mRNAs identified as untranslated by the translatome analysis, encoding respectively MT1 and ribosomal protein rps3 (Supplementary Table S3) were also tested on polysome gradients. The mRNA distribution showed no differences between control and puromycin-treated fertilized samples, indicating that the MT1 and rps3 mRNAs were not translated. Altogether, these data validate the translatome analysis by RNA-Seq described in this study as a means to uncover newly translated mRNAs after fertilization and identify new biological actors strongly regulated at the egg-to-embryo transition.
Table 1. Identification of mRNAs translated after fertilization from RNA-Seq analysis of polysomal mRNAs. Each mRNA, associated with a transcript ID from the de novo assembly of the maternal transcriptome, is identified by its best BLASTn hit against a Strongylocentrotus purpuratus gene model (Echinobase; http://echinobase.org; (19,20)). The mRNA translation index (log2FC(F/Fpuro)) and its polysome fractions after fertilization (log2FC(F/UnF)) are indicated with their respective adjusted P-values (padj). Abundance of the transcript is expressed as fragments per kilobase million (FPKM). The mRNAs further verified in the polysome gradient analysis in Supplementary Figures S1C and S3 are shown in bold. The full translatome dataset is presented in Supplementary data 2.
| Transcript ID | Gene | F_vs_UnF | F_vs_Fpuro | FPKM | SPU best BLAST Hit | ||
|---|---|---|---|---|---|---|---|
| logFC | padj | logFC | padj | ||||
| Cell-cycle related genes | |||||||
| comp77341_c2_seq1 | Cyclin B | 2.964 | 2.14E-19 | 1.631 | 2.33E-07 | 3202.66 | SPU_015285 |
| comp80046_c1_seq2 | Ribonucleotide reductase small subunit R2 | 2.645 | 1.67E-12 | 1.195 | 7.15E-04 | 4679.83 | SPU_024933 |
| comp76547_c0_seq2 | Regulator of chromosome condensation 1 RCC1 | 2.426 | 9.07E-17 | 1.687 | 2.51E-09 | 103.06 | SPU_023992 |
| comp79240_c1_seq1 | Cyclin A | 2.339 | 5.65E-13 | 1.513 | 1.81E-06 | 649.47 | SPU_003528 |
| comp69502_c0_seq1 | 14–3-3 epsilon | 2.223 | 1.96E-10 | 0.864 | 1.17E-02 | 64.30 | SPU_003825 |
| comp79495_c0_seq1 | Cyclin dependent kinase 1 CDK1 | 1.583 | 8.41E-07 | 1.933 | 8.15E-10 | 62.15 | SPU_002210 |
| comp78419_c0_seq1 | Early mitotic inhibitor EMI1 | 1.417 | 2.56E-05 | 0.953 | 4.28E-03 | 14.66 | SPU_008889 |
| comp78987_c0_seq3 | DNA-replication factor cdt1 | 1.393 | 5.09E-07 | 1.192 | 1.30E-05 | 72.55 | SPU_002046 |
| comp77014_c0_seq1 | Polo-like kinase 1 PLK1 | 1.342 | 1.22E-06 | 1.033 | 1.51E-04 | 77.47 | SPU_017949 |
| comp79482_c0_seq1 | Cyclin B3 | 1.071 | 3.16E-04 | 1.400 | 1.25E-06 | 190.16 | SPU_006444 |
| comp79997_c3_seq5 | Geminin | 0.970 | 1.73E-03 | 1.211 | 5.63E-05 | 150.56 | SPU_005762 |
| comp79170_c0_seq2 | Cell division control protein 6 cdc6 | 0.944 | 5.07E-03 | 1.482 | 6.35E-06 | 154.35 | SPU_010595 |
| Maternal determinant of developmental patterning | |||||||
| comp77921_c0_seq1 | Gustavus | 2.582 | 2.03E-13 | 1.533 | 5.46E-06 | 75.98 | SPU_004717 |
| comp79094_c0_seq1 | Alk2 | 2.474 | 1.01E-12 | 1.859 | 2.63E-08 | 17.77 | SPU_016008 |
| comp76027_c0_seq8 | Transforming growth factor beta Univin | 2.311 | 1.16E-11 | 1.799 | 7.47E-08 | 18.28 | SPU_000668 |
| comp77686_c1_seq1 | SoxB1 | 1.713 | 3.74E-06 | 1.700 | 1.87E-06 | 1320.41 | SPU_022820 |
| comp73990_c1_seq1 | Beta-catenin | 1.490 | 1.49E-04 | 1.172 | 2.00E-03 | 194.50 | SPU_009155 |
| comp79158_c0_seq1 | Smad4 | 1.447 | 2.21E-06 | 1.401 | 3.58E-06 | 14.88 | SPU_004287 |
| comp79473_c0_seq3 | Transforming growth factor beta receptor | 1.131 | 1.64E-03 | 1.284 | 3.16E-04 | 6.38 | SPU_027380 |
| RNA-binding proteins (RNA-BPs) | |||||||
| comp70206_c1_seq4 | RBM4 | 3.427 | 3.15E-16 | 1.597 | 3.84E-05 | 100.80 | SPU_022878 |
| comp76265_c0_seq4 | Histone RNA hairpin-binding protein SLBP | 2.121 | 3.72E-08 | 1.246 | 8.67E-04 | 62.95 | SPU_009593 |
| comp68546_c0_seq1 | hnRNP K | 1.849 | 2.21E-09 | 1.666 | 3.54E-08 | 90.53 | SPU_008011 |
| comp66342_c1_seq4 | DAZAP Musashi | 1.757 | 6.76E-08 | 1.294 | 3.29E-05 | 85.10 | SPU_024306 |
| comp79981_c0_seq9 | CUG-BP | 1.401 | 3.26E-07 | 1.189 | 1.24E-05 | 7.74 | SPU_015850 |
| comp62631_c0_seq1 | hnRNP A | 1.191 | 2.06E-04 | 1.825 | 6.83E-09 | 115.70 | SPU_015676 |
| comp78371_c0_seq1 | Nova | 1.103 | 2.74E-04 | 1.592 | 8.14E-08 | 10.04 | SPU_003114 |
| comp76987_c1_seq1 | RKHD/Pem-3/Mex3B homolog | 1.072 | 4.34E-04 | 1.164 | 9.54E-05 | 854.27 | SPU_003290 |
| Translation regulation | |||||||
| comp73250_c0_seq1 | eIF4E binding protein Neuroguidin | 1.862 | 3.19E-04 | 1.366 | 6.29E-03 | 4.79 | SPU_019210 |
| comp78490_c0_seq1 | Termination factor eRF1 | 1.376 | 1.55E-05 | 1.847 | 1.61E-09 | 11.40 | SPU_023948 |
| comp79103_c1_seq1 | DAP5 | 1.339 | 4.26E-04 | 1.398 | 1.68E-04 | 87.12 | SPU_023932 |
| comp64074_c0_seq1 | Initiation factor eIF6 | 1.267 | 2.71E-02 | 1.554 | 4.25E-03 | 7.94 | SPU_012909 |
| comp78411_c2_seq1 | eIF4B | 1.111 | 6.02E-04 | 0.958 | 2.71E-03 | 94.72 | SPU_004840 |
Figure 3.
Distribution on polysome gradients of selected mRNAs identified in the translatome analysis. mRNAs were detected by RT-PCR amplification in each fraction of the polysome gradient from unfertilized eggs (UnF), 1-h post-fertilization embryos (F) or puromycin-treated embryos (F+puro). Amplicons were run on agarose gels and quantified using ImageJ software. Distribution is shown along the gradient as a percentage of total mRNA. Fraction #1 corresponds to the top of the gradient (free mRNAs) and #21 corresponds to the bottom of the gradient. Values are shown as a mean of at least five independent biological replicates, error bars represent SEM (UnF vs F: *P-value < 0.05)
mRNAs recruited onto polysomes upon fertilization are differentially sensitive to the mTOR pathway
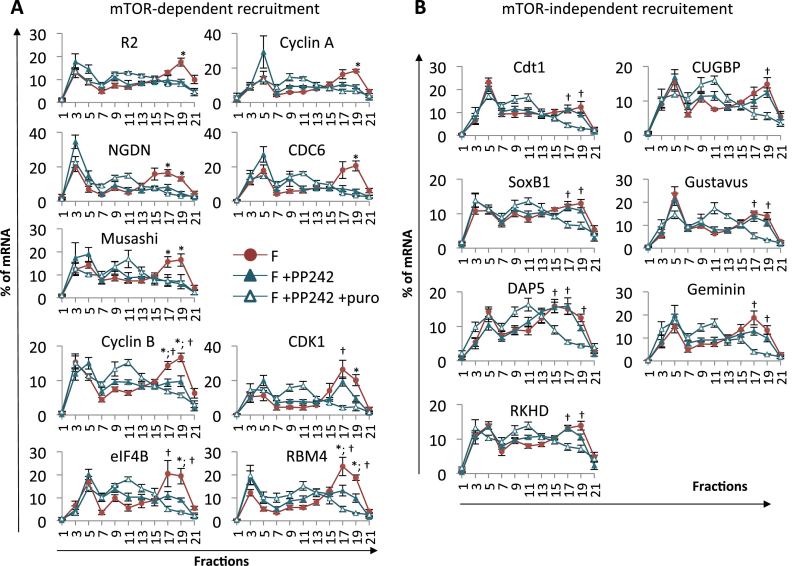
In sea urchin eggs, the translation inhibitor 4E-binding protein (4E-BP) is bound to the cap-binding protein eIF4E, impinging on the formation of an active initiation complex and thus on cap-dependent translation. Fertilization triggers 4E-BP hyperphosphorylation and degradation depending on mTOR signaling pathway activity (10,11). The release of eIF4E from its inhibitor renders it available for the formation of an active initiation complex with eIF4G (12). We have previously demonstrated that the pan-mTOR inhibitor PP242 inhibits the increase in protein synthesis triggered by fertilization, delays progression through the cell cycle and affects the translation of cyclin B, which is partially dependent on mTOR pathway activity (17). Given that we identified mRNAs newly recruited upon fertilization (Figure 3), we investigated whether the mTOR pathway affects the polysomal recruitment of these mRNAs. We first checked that the incubation of sea urchin embryos in the presence of PP242 inhibits 4E-BP degradation as well as the increase in the protein synthesis triggered by fertilization ((11,17) and Supplementary Figure S6). We then compared the mRNA distribution in polysome gradients in control and PP242-treated embryos harvested 1-h post-fertilization, in at least five independent experiments (Figure 4). The tested mRNAs fell into two major classes according to the dependence of their polysomal recruitment on mTOR activity. The proportion of mRNA in polysome fractions between control and PP242-treated embryos was statistically different (F vs F+PP242, P-value < 0.05, Figure 4), indicating that recruitment was affected by mTOR inhibition. Recruitment can be either completely or partially dependent on mTOR activity, depending on the statistical difference in the distribution of mRNAs in polysome gradients in PP242-treated embryos compared with PP242-puromycin-treated embryos (complete dependence: non-significant; partial dependence: F+PP242 vs F+PP242+puro, P-value < 0.05, Figure 4). In contrast, when the proportion of mRNA in polysomal fractions in the presence of PP242 was similar to that displayed in fertilized embryos, recruitment was classified as mTOR-independent. Further comparison of the mRNA distribution along the gradient between unfertilized eggs and PP242-treated fertilized embryos (Supplementary Figure S7) confirms the classification.
Figure 4.
Impact of mTOR inhibition on mRNA polysomal recruitment. PP242 was added 10 min before fertilization and puromycin 40 min after fertilization, in the indicated samples. Control and treated embryos were harvested at 60 min after fertilization for polysome analysis. mRNAs exhibit mTOR-dependent (A) or mTOR-independent (B) polysomal recruitment. Distribution on polysome gradients was monitored as described in Figure 3 from 1-h post-fertilization embryos (F), in the presence of the PP242 inhibitor (F+PP242) or in the presence of PP242 and puromycin (F+PP242+puro). Values are shown as a mean of at least five independent biological replicates, error bars represent SEM (F vs F+PP242: *P-value < 0.05 and F+PP242 vs F+PP242+puro: †P-value < 0.05) The translation of the mRNAs in the corresponding puromycin-treated embryos (F+puro) is shown in Supplementary Figure S8.
The mTOR-dependent class comprises mRNAs for which fertilization-induced polysome recruitment was inhibited by PP242 (F vs F+PP242, P-value < 0.05, Figure 4A). Polysomal recruitment was found to be totally dependent on mTOR for the ribonucleotide reductase small subunit R2, cyclin A, NGDN, CDC6 and Musashi mRNAs: no difference was detected between PP242-treated and PP242-puromycin-treated embryos. Furthermore, the mRNA distribution along the gradient was similar in unfertilized eggs and in PP242-treated fertilized embryos (Supplementary Figure S7). For cyclin B, CDK1, eIF4B and RBM4 mRNAs, the proportion of mRNA in polysome fractions between PP242-treated embryos and PP242-puromycin-treated embryos was also statistically different (F+PP242 vs F+PP242+puro: P-value<0.05, Figure 4A). The cyclin B, CDK1, eIF4B and RBM4 mRNAs showed a significant difference in the polysome fractions between unfertilized eggs and PP242-treated fertilized embryos (P-value < 0.05, Supplementary Figure S7). Some mRNAs showed low residual translation in presence of PP242 (such as cyclin B and eIF4B), whereas others were less affected by mTOR inhibition (CDK1, RBM4). These data suggest that a fraction of these mRNAs was still actively translated despite global protein synthesis inhibition after PP242 treatment, and that their translation is partially dependent on mTOR activity.
Interestingly, we showed the existence of mRNAs for which polysomal recruitment completely escaped the inhibition of cap-dependent translation by PP242-mediated mTOR inhibition (Cdt1, CUGBP, SoxB1, Gustavus, DAP5, Geminin, and RKHD; Figure 4B). In the presence of PP242, the mRNA distribution across the polysome gradient was similar to that displayed in untreated fertilized embryos. Puromycin treatment in the presence of PP242 triggered the disassembly of heavy polysomes and relocation of the mRNAs to the middle of the gradient, suggesting that the mRNAs still present in the heavy fractions of PP242-treated embryos were actively translated (F+PP242 vs F+PP242+puro: P-value<0.05, Figure 4B). In agreement with these data, there was a significant difference in the polysomal fractions between unfertilized eggs and PP242-treated fertilized embryos (P-value < 0.05, Supplementary Figure S7) for these mRNAs. These results strongly suggest the existence of an mTOR-independent translation pathway regulating specific mRNAs in the early sea urchin embryo.
The CDK1 protein is translated de novo after fertilization
As expected, cyclin B mRNA was among the newly translated mRNAs identified in the translatome approach. Surprisingly, the mRNA encoding CDK1, the kinase associated with cyclin B, was also found in the translated set. In the sea urchin S. granularis, CDK1 is an abundant maternal protein and its total level is not modified upon fertilization or by the inhibition of protein synthesis (34). In light of our translatome data, we investigated the translation of the P. lividus CDK1 protein.
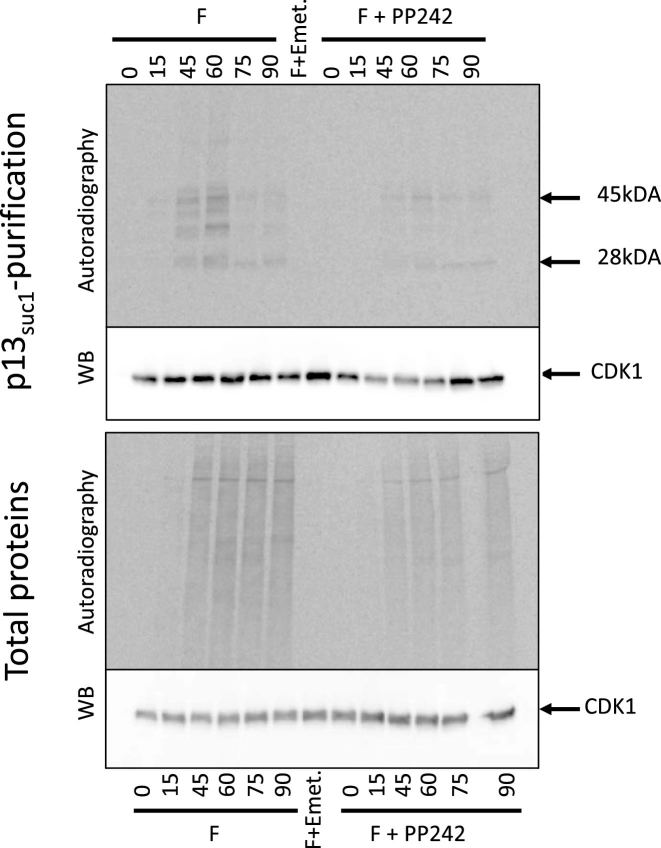
The protein p13suc1 binds CDK1, both as a free protein and as a complex with cyclin B (35). To visualize newly synthesized CDK1, p13suc1-sepharose beads were used to affinity-purify CDK1 from [35S]-methionine labeled embryos; bound proteins were detected by autoradiography followed by western blot using an anti-PSTAIR antibody (directed against a conserved peptide of CDK1, Sigma P7962). As shown in Figure 5 (top panel), a 28 kDa radiolabeled protein migrating at the expected size for CDK1 was pulled down by p13suc1-sepharose beads after fertilization whereas no labeled protein was detected before fertilization nor in the presence of emetine. Furthermore, the amount of the pulled-down, newly synthesized CDK1 was lower in PP242-treated embryos than in control embryos. Western blot analysis of the pulled-down proteins revealed the CDK1 protein in all conditions, consistent with its presence as a maternal protein. Western blotting and autoradiography of crude lysates showed that the level of total P. lividus CDK1 protein did not increase after fertilization, nor was significantly altered by emetine or PP242 (mTOR inhibition) (Figure 5, bottom panel), suggesting that the contribution of the newly synthesized CDK1 is negligible compared with the amount of maternal protein already present in the egg. A radioactive 45 kDa band was also pulled down by the p13suc1-sepharose beads, this protein was not detected before fertilization nor in the presence of emetine. We therefore suspect that this protein was cyclin B because the radiolabeled protein has the correct expected size in sea urchin. However, we have no direct evidence of its identity because no antibody cross-reacting with the P. lividus protein is available for western blotting (as shown in (17)).
Figure 5.
CDK1 protein is newly synthesized after fertilization. [35S]-methionine labeled proteins from embryos collected at the indicated times after fertilization were incubated on p13suc1-sepharose beads; affinity-purified proteins were resolved on SDS-PAGE and transferred to a nitrocellulose membrane. After exposure on a PhosphorImager screen (autoradiography), the membrane was incubated with anti-PSTAIR antibodies directed against the CDK1 protein (WB). Autoradiography and western blot on total proteins are shown in the bottom panel. Fertilized (left) and PP242-treated (right) embryos are shown. As a control, [35S]-methionine labeled embryos were cultured in presence of emetine, which was added 5 min after fertilization to inhibit protein synthesis, and the lysate prepared from embryos harvested at 90 min was used for p13suc1-sepharose bead purification (F+emet). The experiment was performed twice.
At the protein level, the CDK1 protein is translated de novo after fertilization, consistent with the polysome recruitment data.
DISCUSSION
Coupling polysome gradient optimization in sea urchin (5) with high-throughput sequencing, we re-examined the translational changes occurring upon fertilization by carrying out a large-scale translatome analysis in the sea urchin P. lividus. To our knowledge, this is the first time that such a dataset has been produced in a model system in which egg-to-embryo transition occurs independently of meiotic maturation resumption. Here, precise normalization to correct for the technical effects affecting translatome data analysis was included in a generalized linear model framework. This approach to modeling and analyzing the data demonstrated that fertilization triggers the selective translation of maternal mRNAs, and also revealed an alternative model to classical cap-dependent translation in early development.
Translation of a subset of maternal mRNAs upon fertilization
The repertoire of mRNAs present in the egg and the 1-h embryo was sequenced from RNA-Seq libraries constructed after poly(A) selection. Various comparisons of the transcript repertoire showed no variation in the abundance of transcripts. These data suggest that no transcripts are produced nor degraded during this short lapse of time, in accordance with the transcription initiation of early zygotic genes and early degradation of maternal transcripts occurring at ∼4 h post-fertilization (36,37). Therefore, the observed protein synthesis changes triggered by fertilization arise solely from the differential use of maternally deposited mRNAs. Our study further suggests that the recruitment of mRNA onto active polysomes upon fertilization is not correlated with their abundance. For example, the abundant polyadenylated mRNAs encoding the cleavage stage histones detected in the maternal transcriptome are not loaded onto polysomes in our analysis, in agreement with a report showing that histone H3 mRNA moves onto polysomes only after the first cleavage (38).
In this study, we analyzed the set of mRNAs that are recruited 1 h post-fertilization, in the one-cell embryo, before the occurrence of the first cell division. We showed that only a limited subset of the maternally stored mRNAs is efficiently translated at this early timepoint after fertilization. This timing corresponds to the egg-to-embryo transition, which is the first key developmental stage in the change from a differentiated germ cell into a totipotent embryo (8). Not surprisingly, we recaptured mRNAs previously identified as translated upon fertilization, such as cyclin B and the ribonucleotide reductase small subunit (15,17,18), providing a proof-of-concept for our translatome data. We also demonstrated the validity of translatome analysis for the identification of newly translated mRNAs after fertilization. Our study revealed that there is no massive recruitment of mRNAs following fertilization in sea urchin; instead there is strong selection for polysomal recruitment among the stored maternal mRNAs, with the enrichment of certain functional classes in the newly synthesized proteins.
In addition to fertilization, parthenogenetic treatments can also activate protein translation. Treating unfertilized eggs with the calcium ionophore A23187 can activate comparable protein synthesis; similarly, increasing the intracellular pH with ammonia treatment results in the partial activation of protein synthesis. More specifically, cyclin B synthesis is strongly and similarly induced by calcium ionophore and ammonia treatments of unfertilized eggs, suggesting that a common regulatory process is involved in the translation of this gene (16,39). These type of treatments can also be used with translatome analysis to further decipher the regulatory mechanisms involved in mRNA polysome recruitment upon fertilization.
Maternal mRNAs translated upon fertilization are involved in several biological processes
The mRNAs encoding cell cycle regulators were enriched in the translated set of maternal mRNAs. It is well established that the accumulation of some key components of the cell cycle, such as cyclins, are regulated at the translational level in oocytes, eggs and early embryos, as well as in somatic cells (40). Surprisingly, we found mRNAs encoding the CDK1 partner in the translated set of maternal mRNAs. We demonstrated that the CDK1 protein is newly synthesized, further validating our translatome screening at the protein level. The amount of newly synthesized CDK1 is negligible compared with the amount of maternal protein already present in the egg, and the apparent steady state level remains unchanged. This incongruity has been noted for other proteins in Drosophila (41). However, the newly synthesized CDK1 associated with cyclin B may have a role in the auto-amplification loop of the complex (reviewed in (42)). Furthermore, a large number of cell cycle regulators were also translationally activated, giving an additional layer of complexity to the regulation of cell cycle progression.
Determinants for axis specification and endomesoderm formation are encoded by maternal mRNAs (43). A subset of these mRNAs was translated early after fertilization. The establishment of the oral-aboral axis is highly regulated by the spatially localized zygotic expression of Nodal, which depends on maternal factors involved in TGF-β signaling (44,45). Several members of the TGF-β pathway, such as the activin receptor-like kinase receptor ALK2, Vg1/univin and SMAD4, were present among the maternal mRNAs strongly recruited upon fertilization. Interestingly, other mRNAs involved in the TGF-β pathway were predominantly enriched in the fraction of maternal mRNAs that were in mRNPs or stalled polysomes before fertilization (Supplementary Figure S5). Among the maternal determinants of the endomesoderm gene regulatory network (GRN), SoxB1 mRNA was strongly recruited and translated in the one-cell embryo. This early translation is in agreement with data in S. purpuratus, where the SpSoxB1 protein is first detected after the two-cell stage and increases in abundance during the cleavage stages (46). Other maternal mRNAs acting in the GRN (for example Otx or Ets1/2 (43)) are not recruited onto polysomes after fertilization, suggesting that SoxB1 may have an additional role in early cleavage stages. Interestingly, in zebrafish, SoxB1 is one of the most strongly and earliest translated maternal mRNAs, and is involved in maternal mRNA clearance (47). The mRNA clearance occurring at the maternal-to-zygotic transition is essential for proper embryonic development (7). In sea urchin, maternal mRNA clearance involves maternal and zygotic mechanisms (37) in which SoxB1 may be involved. Another example is the E3-ubiquitin ligase specificity receptor Gustavus (GUS) that regulates the accumulation of the Vasa protein in small micromeres. The GUS protein accumulates in the embryo between the egg and the four-cell stage (48), consistent with the strong polysomal recruitment of its mRNA that we detected after fertilization. Translation activation of maternal determinants after fertilization may reveal novel regulatory networks in early development.
Interestingly, we observed enrichment in mRNAs encoding RNA-BPs in the translated set of maternal mRNAs. The RNA-BP RBM4 was found in the top five mRNAs recruited after fertilization (log2FC > 3; P-value < 10−5; Table 1). RBM4 is involved in specific translation in hypoxia (49), and in internal ribosome entry site (IRES)-dependent translation under stress conditions in mammalian cells (50). The Drosophila RBM4 homolog LARK is required for development (51). These data suggest that RBM4 may play a role in specific translation in early sea urchin development. Several of the other translated mRNAs encode RNA-BPs that are involved in the translation repression of specific mRNAs. For example, CUGBP is an RRM-domain containing RNA-BP first identified in Xenopus embryos for its ability to bind specifically to a GU-rich element (embryonic deadenylation element EDEN) located in the 3′-UTRs of some mRNAs that are rapidly deadenylated and translationally repressed after fertilization in early development (52). Another example is RKHD/MEX3B, a KH- and ring-domain containing RNA-BP, involved as a translation repressor in embryonic cell fate (reviewed in (53)). In sea urchin, RKHD has been identified as an ubiquitous maternal and localized zygotic mRNA (54), but its function has not yet been investigated. In our study, CUGBP and RKHD were strongly recruited onto polysomes after fertilization. No data is available on the maternal presence of the proteins encoded by these mRNAs, but the finding that they are newly synthesized after fertilization suggests that control of mRNA fate (stability, localization, translation, etc.) may be set up or modified at the egg-to-embryo transition, and gives insights into possible mechanistic regulations. Interestingly, these two mRNAs were translated even in PP242-treated embryos, and also before fertilization in some individuals (data not shown). In both conditions, the translation inhibitor protein 4E-BP was present and inhibited cap-dependent translation, suggesting an alternative translation initiation pathway for CUGBP and RKHD mRNAs.
Identification of mRNAs showing mTOR-independent polysome recruitment in sea urchin
In mammalian cells, the mTOR pathway preferentially regulates the translation of mRNAs containing a 5′ terminal oligopyrimidine (TOP) tract, such as the ones coding for ribosomal proteins and some translation factors (55–57). Strikingly, in our study, despite the activation of the mTOR pathway upon fertilization, the transcripts coding the ribosomal proteins were significantly under-represented in the translated set of mRNAs, and were shown to be mainly untranslated (Supplementary Figure S5). Before fertilization, the egg already contains an excess of ribosomes, that will be recruited onto functional polysomes as development proceeds (6), suggesting that the translation of new ribosomal proteins may not be necessary at this early stage, and that activation of the mTOR pathway upon fertilization may impact the translated set of mRNAs differentially in a developmental context. A translatome analysis of PP242-treated embryos will help apprehend the full spectrum of residual translation when cap-dependent translation is impaired.
In this study, we showed that impairing the mTOR pathway differentially impacts the polysomal recruitment of mRNAs upon fertilization. When embryos are treated with PP242 or rapamycin, overall protein synthesis is inhibited, but some proteins are still newly synthesized ((17) and Figure 5). For the first time in sea urchin, we identified mRNAs that are recruited onto polysomes in PP242-treated embryos, thus independently of mTOR activity. We can rule out that these mRNAs are present in a stalled polysome conformation, because they were sensitive to puromycin treatment and therefore present in actively translating polysomes in the presence of PP242. The mechanisms behind their specific translation regarding the mTOR pathway, and more precisely cap-independent or IRES mechanisms, were not addressed in this study. However, these mRNAs can now be used to uncover cis- and trans- regulatory mechanisms in further experiments. Noteworthily, DAP5, an eIF4G homolog unable to bind eIF4E, was still translated in PP242-treated embryos. DAP5 mRNA is known to be translated in an IRES-dependent manner and encodes a known IRES-transacting factor (58,59). DAP5 knockout mice die at an early stage of development (60), and recently DAP5 has been shown to be involved in the control of embryonic cell differentiation (61). Therefore, cap-independent translation and IRES elements may be involved upon fertilization. We cannot rule out that other mechanisms are involved in mTOR-independent translation (62,63). For example, an alternative non-canonical cap-dependent translation machinery is mediated by the eIF4E homolog eIF4E2, activated in hypoxic cells, when eIF4E is inhibited by 4E-BP (49). Epitranscriptomic marks such as mRNA methylation in 5′ UTRs have recently been shown to allow translation of some mRNAs in a eIF4F- or cap-independent manner (64–66). Our results suggest evolutionarily conserved use of an alternative translation initiation in the context of early development. Whether the above molecular mechanisms are involved in these regulations in the sea urchin embryo remains to be seen.
CONCLUSION
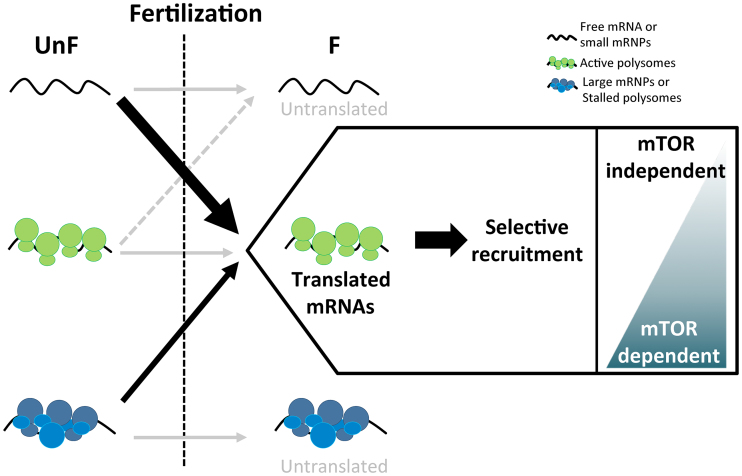
The polysomal recruitment dynamics and the mRNAs identified in this study made it possible to propose a model for selective translation events following fertilization in the sea urchin egg-to-embryo transition (Figure 6). The activation of the translational machinery upon fertilization (9) leads to the polysomal recruitment of a specific subset of maternal mRNAs, independently of their abundance. Remarkably, these mRNAs predominantly belong to certain functional categories, coding for proteins that may be involved in cell cycle transitions or in the later events of development. Further work is needed to decipher their involvement after fertilization. Interestingly, the impairment of the mTOR pathway physiologically activated at fertilization did not affect the newly translated mRNAs equally, suggesting that an alternative initiation model may be involved upon fertilization. Finally, this dataset gives cues for future work on the molecular mechanisms involved in the regulation of translation triggered by fertilization, by linking events of translation activation to specific mRNAs and deciphering the translation regulation network (TRN) required for the fine-tuned control of the egg-to-embryo transition.
Figure 6.
Model of polysomal recruitment dynamics upon fertilization in sea urchin. Before fertilization, translation activity is generally repressed. Fertilization triggers the selective recruitment of a new subset of mRNAs onto polysomes, with over-represented functional categories such as cell cycle, RNA-binding and signaling (recruitment from free mRNAs or small mRNPs before fertilization, large black arrow). The recruitment of some mRNAs depends on the mTOR pathway, either completely or partially, whereas polysomal recruitment of other mRNAs are independent of mTOR (box). Some mRNAs present in stalled polysomes or stored in large mRNPs before fertilization are activated for translation after fertilization (thin black arrow). Fertilization also triggers the selective repression of a small subset of mRNAs translated in the egg before fertilization (dashed gray arrow), but most maternal mRNAs do not change their polysomal behavior (gray arrow).
DATA AVAILABILITY
RNA-Seq raw data have been deposited in NCBI BioProject database under the accession number PRJNA288758. The R code for the translatome analysis is available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Service Mer et Observation (Marine Operations department) and the Roscoff Aquarium Service at the Roscoff Marine Station for collecting and maintaining sea urchins. We thank the members of the TCCD research team for their helpful comments and valuable discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Cancer League (La Ligue contre le Cancer, comités Finistère, Côtes d’Armor, Morbihan, Deux-Sèvres et Charente); Brittany Regional Council (Région Bretagne); Finistère Departmental Council [CG29]. H. Chassé was a Ph.D. fellow supported by the Brittany Regional Council (Région Bretagne). Funding for open access charges: CNRS, Sorbonne Université [SU-16-R-EMR-24].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hershey J.W.B., Sonenberg N., Mathews M.B.. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012; 4:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. King H.A., Gerber A.P.. Translatome profiling: methods for genome-scale analysis of mRNA translation. Brief. Funct. Genomics. 2016; 15:22–31. [DOI] [PubMed] [Google Scholar]
- 3. Kuersten S., Radek A., Vogel C., Penalva L.O.F.. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip. Rev. RNA. 2013; 4:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Larsson O., Tian B., Sonenberg N.. Toward a genome-wide landscape of translational control. Cold Spring Harb. Perspect. Biol. 2012; 5:209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chassé H., Boulben S., Costache V., Cormier P., Morales J.. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017; 45:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davidson E.H. The nature and function of maternal transcripts. Gene Activity in Early Development. 1986; NY: Academic Press; 46–125. [Google Scholar]
- 7. Tadros W., Lipshitz H.D.. The maternal-to-zygotic transition: a play in two acts. Development. 2009; 136:3033–3042. [DOI] [PubMed] [Google Scholar]
- 8. Horner V.L., Wolfner M.F.. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev. Dyn. 2008; 237:527–544. [DOI] [PubMed] [Google Scholar]
- 9. Cormier P., Chassé H., Cosson B., Mulner-Lorillon O., Morales J.. Hernández G, Jagus R. Translational control in echinoderms: the calm before the storm. Evolution of the Protein Synthesis Machinery and its Regulation. 2016; Switzerland: Springer International Publishing; 413–432. [Google Scholar]
- 10. Cormier P., Pyronnet S., Morales J., Mulner-Lorillon O., Sonenberg N., Belle R.. eIF4E association with 4E-BP decreases rapidly following fertilization in sea urchin. Dev. Biol. 2001; 232:275–283. [DOI] [PubMed] [Google Scholar]
- 11. Salaun P., Pyronnet S., Morales J., Mulner-Lorillon O., Bellé R., Sonenberg N., Cormier P.. eIF4E/4E-BP dissociation and 4E-BP degradation in the first mitotic division of the sea urchin embryo. Dev. Biol. 2003; 255:428–439. [DOI] [PubMed] [Google Scholar]
- 12. Oulhen N., Salaun P., Cosson B., Cormier P., Morales J.. After fertilization of sea urchin eggs, eIF4G is post-translationally modified and associated with the cap-binding protein eIF4E. J. Cell Sci. 2007; 120:425–434. [DOI] [PubMed] [Google Scholar]
- 13. Brandhorst B.P. Two-dimensional gel patterns of protein synthesis before and after fertilization of sea urchin eggs. Dev. Biol. 1976; 52:310–317. [DOI] [PubMed] [Google Scholar]
- 14. Roux M.M., Radeke M.J., Goel M., Mushegian A., Foltz K.R.. 2DE identification of proteins exhibiting turnover and phosphorylation dynamics during sea urchin egg activation. Dev. Biol. 2008; 313:630–647. [DOI] [PubMed] [Google Scholar]
- 15. Kelso-Winemiller L., Yoon J., Peeler M.T., Winkler M.M.. Sea urchin maternal mRNA classes with distinct development regulation. Dev. Genet. 1993; 14:397–406. [DOI] [PubMed] [Google Scholar]
- 16. Evans T., Rosenthal E.T., Youngblom J., Distel D., Hunt T.. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983; 33:389–396. [DOI] [PubMed] [Google Scholar]
- 17. Chassé H., Mulner-Lorillon O., Boulben S., Glippa V., Morales J., Cormier P.. Cyclin B translation depends on mTOR activity after fertilization in sea urchin embryos. PLoS One. 2016; 11:e0150318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Standart N.M., Bray S.J., George E.L., Hunt T., Ruderman J.V.. The small subunit of ribonucleotide reductase is encoded by one of the most abundant translationally regulated maternal RNAs in clam and sea urchin eggs. J. Cell. Biol. 1985; 100:1968–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cameron R.A., Samanta M., Yuan A., He D., Davidson E.. SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 2009; 37:D750–D754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sodergren E., Weinstock G.M., Davidson E.H., Cameron R.A., Gibbs R.A., Angerer R.C., Angerer L.M., Arnone M.I., Burgess D.R., Burke R.D. et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006; 314:941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Lund A.H., Smyth G.K.. Datta S., Nettleton D.. Differential expression analysis of complex RNA-Seq experiments using edgeR. Statistical Analysis of Next Generation Sequence Data. 2014; NY: Springer [Google Scholar]
- 23. McCarthy D.J., Chen Y., Smyth G.K.. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012; 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson M.D., Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dillies M.-A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., Keime C., Marot G., Castel D., Estelle J. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013; 14:671–683. [DOI] [PubMed] [Google Scholar]
- 26. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995; 57:289–300. [Google Scholar]
- 27. R Development Core Team A language and environment for statistical computing. R Foundation for Statistical Computing. 2017; Vienna [Google Scholar]
- 28. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winkler M.M., Nelson E.M., Lashbrook C., Hershey J.W.. Multiple levels of regulation of protein synthesis at fertilization in sea urchin eggs. Dev. Biol. 1985; 107:290–300. [DOI] [PubMed] [Google Scholar]
- 30. Alexandraki D., Ruderman J.V.. Expression of α- and β-tubulin genes during development of sea urchin embryos. Dev. Biol. 1985; 109:436–451. [DOI] [PubMed] [Google Scholar]
- 31. Blobel G., Sabatini D.. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kang Q., Pomerening J.R.. Punctuated cyclin synthesis drives early embryonic cell cycle oscillations. Mol. Biol. Cell. 2012; 23:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tu Q., Cameron R.A., Davidson E.H.. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2014; 385:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meijer L., Azzi L., Wang J.Y.. Cyclin B targets p34cdc2 for tyrosine phosphorylation. EMBO J. 1991; 10:1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunphy W.G., Brizuela L., Beach D., Newport J.. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988; 54:423–431. [DOI] [PubMed] [Google Scholar]
- 36. Gildor T., Ben-Tabou de-Leon S.. Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLOS Genet. 2015; 11:e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gildor T., Malik A., Sher N., Ben-Tabou de-Leon S.. Mature maternal mRNAs are longer than zygotic ones and have complex degradation kinetics in sea urchin. Dev. Biol. 2016; 414:121–131. [DOI] [PubMed] [Google Scholar]
- 38. Wells D.E., Showman R.M., Klein W.H., Raff R.A.. Delayed recruitment of maternal histone H3 mRNA in sea urchin embryos. Nature. 1981; 292:477–478. [DOI] [PubMed] [Google Scholar]
- 39. Oulhen N., Mulner-Lorillon O., Cormier P.. eIF4E-binding proteins are differentially modified after ammonia versus intracellular calcium activation of sea urchin unfertilized eggs. Mol. Reprod. Dev. 2010; 77:83–91. [DOI] [PubMed] [Google Scholar]
- 40. Tarn W.-Y., Lai M.-C.. Translational control of cyclins. Cell Div. 2011; 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kronja I., Yuan B., Eichhorn S.W., Dzeyk K., Krijgsveld J., Bartel D.P., Orr-Weaver T.L.. Widespread changes in the posttranscriptional landscape at the drosophila oocyte-to-embryo transition. Cell Rep. 2014; 7:1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nigg E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001; 2:21–32. [DOI] [PubMed] [Google Scholar]
- 43. Oliveri P., Davidson E.H.. Gene regulatory network controlling embryonic specification in the sea urchin. Curr. Opin. Genet. Dev. 2004; 14:351–360. [DOI] [PubMed] [Google Scholar]
- 44. Haillot E., Molina M.D., Lapraz F., Lepage T.. The maternal Maverick / GDF15-like TGF- β ligand Panda directs dorsal-ventral axis formation by restricting nodal expression in the sea urchin embryo. PLoS Biol. 2015; 13:e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Range R., Lepage T.. Maternal Oct1/2 is required for Nodal and Vg1/Univin expression during dorsal-ventral axis specification in the sea urchin embryo. Dev. Biol. 2011; 357:440–449. [DOI] [PubMed] [Google Scholar]
- 46. Kenny A.P., Kozlowski D., Oleksyn D.W., Angerer L.M., Angerer R.C.. SpSoxB1, a maternally encoded transcription factor asymmetrically distributed among early sea urchin blastomeres. Development. 1999; 126:5473–5483. [DOI] [PubMed] [Google Scholar]
- 47. Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., Divito K.R., Fleming E.S., Giraldez A.J.. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013; 503:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gustafson E.A., Yajima M., Juliano C.E., Wessel G.M.. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev. Biol. 2011; 349:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uniacke J., Holterman C.E., Lachance G., Franovic A., Jacob M.D., Fabian M.R., Payette J., Holcik M., Pause A., Lee S.. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012; 486:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin J.-C., Tarn W.-Y.. RNA-binding motif protein 4 translocates to cytoplasmic granules and suppresses translation via argonaute2 during muscle cell differentiation. J. Biol. Chem. 2009; 284:34658–34665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McNeil G.P., Zhang X., Roberts M., Jackson F.R.. Maternal function of a retroviral-type zinc-finger protein is essential for Drosophila development. Dev. Genet. 1999; 25:387–396. [DOI] [PubMed] [Google Scholar]
- 52. Paillard L., Omilli F., Legagneux V., Bassez T., Maniey D., Osborne H.B.. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998; 17:278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pereira B., Le Borgne M., Chartier N.T., Billaud M., Almeida R.. MEX-3 proteins: recent insights on novel post-transcriptional regulators. Trends Biochem. Sci. 2013; 38:477–479. [DOI] [PubMed] [Google Scholar]
- 54. Röttinger E., Besnardeau L., Lepage T.. Expression pattern of three putative RNA-binding proteins during early development of the sea urchin Paracentrotus lividus. Gene Expr. Patterns. 2006; 6:864–872. [DOI] [PubMed] [Google Scholar]
- 55. Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012; 485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meyuhas O., Kahan T.. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta. 2014; 1849:801–811. [DOI] [PubMed] [Google Scholar]
- 58. Henis-Korenblit S., Strumpf N.L., Goldstaub D., Kimchi A.. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 2000; 20:496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marash L., Liberman N., Henis-Korenblit S., Sivan G., Reem E., Elroy-Stein O., Kimchi A.. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell. 2008; 30:447–459. [DOI] [PubMed] [Google Scholar]
- 60. Yamanaka S., Zhang X.Y., Maeda M., Miura K., Wang S., Farese R.V. Jr., Iwao H., Innerarity T.L.. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000; 19:5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoffe Y., David M., Kalaora R., Povodovski L., Friedlander G., Feldmesser E., Ainbinder E., Saada A., Bialik S., Kimchi A.. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 2016; 30:1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Truitt M.L., Ruggero D.. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016; 16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shatsky I.N., Dmitriev S.E., Andreev D.E., Terenin I.M.. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit. Rev. Biochem. Mol. Biol. 2014; 9238:1–14. [DOI] [PubMed] [Google Scholar]
- 64. Coots R.A., Liu X.M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.B.. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell. 2017; 68:504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5′ UTR m6A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq raw data have been deposited in NCBI BioProject database under the accession number PRJNA288758. The R code for the translatome analysis is available upon request.