Abstract
Clostridium difficile, a major human enteropathogen, must cope with foreign DNA invaders and multiple stress factors inside the host. We have recently provided an experimental evidence of defensive function of the C. difficile CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) system important for its survival within phage-rich gut communities. Here, we describe the identification of type I toxin–antitoxin (TA) systems with the first functional antisense RNAs in this pathogen. Through the analysis of deep-sequencing data, we demonstrate the general co-localization with CRISPR arrays for the majority of sequenced C. difficile strains. We provide a detailed characterization of the overlapping convergent transcripts for three selected TA pairs. The toxic nature of small membrane proteins is demonstrated by the growth arrest induced by their overexpression. The co-expression of antisense RNA acting as an antitoxin prevented this growth defect. Co-regulation of CRISPR-Cas and type I TA genes by the general stress response Sigma B and biofilm-related factors further suggests a possible link between these systems with a role in recurrent C. difficile infections. Our results provide the first description of genomic links between CRISPR and type I TA systems within defense islands in line with recently emerged concept of functional coupling of immunity and cell dormancy systems in prokaryotes.
INTRODUCTION
All living organisms need to survive in changing environments by adapting their physiology. Horizontal gene transfer contributes to the acquisition of new adaptive traits important for survival. However, these foreign DNA elements can be deleterious and even lead to cell death in the case of phage infection. The constant need to maintain the balance between DNA uptake and defense processes would drive the genome evolution. To cope with the presence of invaders, prokaryotes have developed efficient defense systems including recently discovered CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems (1).
The CRISPR-Cas systems are found in about half of sequenced bacterial genomes and in almost all archaeal genomes (2). These prokaryotic adaptive immunity systems provide defense against foreign nucleic acids (3,4). CRISPR loci are arranged in arrays of almost identical direct repeats of ∼30 bp separated by similarly sized variable sequences called spacers. Some spacers match viral or plasmid DNA and have been acquired during prior encounters with mobile genetic elements in ‘adaptation’ step (5). CRISPR arrays are transcribed as single RNA transcripts (pre-crRNA) that are processed to generate small CRISPR RNAs (crRNAs). In complex with Cas proteins, these crRNAs interfere with bacteriophage infection and plasmid transfer by recognizing foreign nucleic acids. This complementary base-pairing recognition leads to the destruction of targeted nucleic acids during an ‘interference’ process, thus protecting cells from the invasion by foreign genetic elements. The Cas proteins are involved in all stages of CRISPR-Cas activity. Universal Cas1 and Cas2 components are required for adaptation, while Cas6 proteins are necessary for crRNA processing. Either a single Cas protein or a multisubunit Cas proteins complex together with the mature crRNA achieve the interference step (1). During bacterial infection, vegetative cells survive in phage-rich gut communities and the bacteria could control the genetic exchanges favoured within this environment by relying on efficient anti-invader defense systems including CRISPR-Cas (6–8).
The human pathogen Clostridium difficile is an anaerobic spore-forming bacterium constituting the major cause of antibiotherapy-associated nosocomial diarrhoea in adults (9). This enteropathogen can lead to a variety of pathologies ranging from diarrhoea to pseudomembranous colitis, a potentially lethal disease. Transmission of C. difficile is mediated by contamination of the gut by spores. Anti-microbial therapy disturbs the colonic microflora allowing colonization of the intestinal tract by C. difficile from pre-existing or acquired spores (10,11). After spore germination and multiplication of vegetative cells, the pathogen produces either one or both of the two toxins (TcdA and TcdB) that are the major virulence factors. These two large toxins induce alterations in the actin cytoskeleton of intestinal epithelial cells (12,13). Yet, many aspects of C. difficile pathogenicity and its regulation still remain poorly understood. Regulatory RNAs may contribute to several steps during infection. It is increasingly recognized that bacterial non-coding RNAs (ncRNAs) play a critical role in adaptive responses and in various metabolic, physiological and pathogenic processes (14). By combining in silico analysis, RNAseq and genome-wide promoter mapping, we have recently identified more than 200 ncRNAs in C. difficile (15). This includes riboswitches, trans-acting riboregulators and cis-acting antisense RNAs, but also CRISPR RNAs that are among the most abundant RNAs revealed by our deep sequencing analysis.
We have recently provided experimental evidence for the function of CRISPR-Cas system in C. difficile (16). The C. difficile CRISPR-Cas system is characterized by the presence of an unusually large set of CRISPR arrays (an average of 8.5 per genome), the presence of two sets of cas genes conserved in almost all sequenced C. difficile strains and the prophage location of several CRISPR arrays (16–18). Both complete and partial C. difficile cas gene operons belong to the less characterized I-B subtype. Phage genome sequencing and CRISPR spacer homology analysis revealed a correlation with host range of several newly sequenced C. difficile phages (16,18). We demonstrated the role of cas genes in an heterologous host, Escherichia coli, and the defensive function of the C. difficile CRISPR-Cas system in an active interference process by analysis of plasmid conjugation efficiency in C. difficile (16). CRISPR arrays location within prophages in C. difficile is rather unique. High transmissibility of CRISPR systems and their association with plasmids, megaplasmids and in some cases prophages have been suggested (19). However, why CRISPR arrays are located within phages and plasmids remains unknown. One hypothesis would be the stabilization of loci against loss and competition with other invaders (19).
We report here that most of the CRISPR arrays are co-localised with toxin–antitoxin (TA) systems in the C. difficile genome. TA modules encode two-component systems consisting of a stable ‘toxin’ and an unstable ‘antitoxin’ (20). The overexpression of toxin either kills cells or confers growth stasis. TA systems have been initially discovered on plasmids where they confer stability of maintenance through post-segregation killing (21). Plasmid loss results in a rapid decrease in levels of the unstable antitoxin, which allows the stable toxin to kill the plasmid-free cells. TA systems have also been found on bacterial and archaeal chromosomes, sometimes in great numbers but their function remains largely unclear. Among suggested functions are prophage maintenance, chromosomal region stabilization, prevention of phage infection, stress response and persister formation (20,22–27).
TA systems are classified into six types depending on the nature and action of the antitoxin that can be either a protein or a small antisense RNA (20). In type I systems, the antitoxin is a small antisense RNA that forms RNA duplex with the toxin-encoding mRNA (28,29). Most studies are devoted to type II TA systems, in which the protein antitoxin sequestering the toxin is more easily defined than the RNA antitoxin of type I TA (30). Numerous identified TA modules, generally of type II, are part of the mobilome including phages, plasmids, transposons and integrative and conjugative elements that can be shared by distant bacteria thus contributing to bacterial evolution (21,31). RNA antitoxins belong to the largest and most extensively studied set of sRNA regulators that act by modulating the translation and/or stability of their mRNA targets. Most of type I toxins are small hydrophobic proteins of <60 amino acids containing a potential transmembrane domain and charged amino acids at the C-terminus (32). In many cases, they seem to act like phage holins by inducing pores into cell membranes and thus impairing adenosine triphosphate synthesis (29). Replication, transcription and translation are consequently inhibited, which leads to cell death. However, alternative mechanisms of action have been also suggested for both membrane-associated toxins like BsrG and cytoplasmic toxins for example RalR with nuclease activity (33–35).
In this work, we describe the first identification of type I TA systems in C. difficile and highlight their association with CRISPR-Cas defense system. Through the analysis of deep-sequencing data for the strain 630Δerm, six potential TA loci were identified in the close proximity of transcribed CRISPR arrays. Three of these TA loci have been selected and the structure of their overlapping transcripts confirmed by Northern blot, 5′/3′RACE and reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR) analyses. The small proteins of unknown function in the proximity of CRISPR arrays have all the characteristic sequence features of type I toxins. We provide experimental evidence for the membrane localization and the toxic nature of these small proteins. Inducible toxin overexpression led to growth arrest of C. difficile, which was abolished by the co-expression of the RNA antitoxin in cis and in trans. Using half-life measurements to assess the toxin mRNA and the RNA antitoxin stability, we show that the CRISPR-associated TA loci encode a rather stable toxin and unstable antitoxin RNA. By RNA band shift analysis we show an efficient duplex formation between TA transcripts in vitro. Finally, our results suggest that both the CRISPR-Cas genes and the type I TA modules could be co-regulated by stress- and biofilm-related factors, supporting a possible link between these two systems. The genomic analysis of more than 2,500 C. difficile strains revealed the general co-localisation of CRISPR arrays with potential TA modules, some of them located within the prophage regions, expanding our conclusions to the majority of the sequenced C. difficile strains.
MATERIALS AND METHODS
Plasmid and bacterial strain construction and growth conditions
C. difficile and E. coli strains and plasmids used in this study are presented in Supplementary Table S1. C. difficile strains were grown anaerobically (5% H2, 5% CO2 and 90% N2) in TY (36) or Brain Heart Infusion (BHI, Difco) media in an anaerobic chamber (Jacomex). When necessary, cefoxitin (Cfx; 25 μg/ml) and thiamphenicol (Tm; 15 μg/ml) were added to C. difficile cultures. E. coli strains were grown in LB broth (37), and when needed, ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml) was added to the culture medium. The non-antibiotic analog anhydrotetracycline (ATc) was used for induction of the Ptet promoter of pRPF185 vector derivatives in C. difficile (38). Strains carrying pRPF185 derivatives were generally grown in TY medium in the presence of 250 ng/ml ATc and 7.5 μg/ml Tm for 7.5 h. Growth curves were obtained using a GloMax plate reader (Promega).
All routine plasmid constructions were carried out using standard procedures (39). All primers used in this study are listed in Supplementary Table S2. For inducible expression of C. difficile genes, we used the pDIA6103 derivative of pRPF185 vector expression system lacking a gusA gene (15,38). The CD2517.1 gene (−89 to +178 relative to the translational start site), the CD2907.1 gene (−84 to +223 relative to the translational start site), CD2517.1-RCd8 TA region with RCd8 promoter (−306 to +504 relative to the translational start site of CD2517.1) and CD2907.1-RCd9 TA region with RCd9 promoter (−294 to +456 relative to the translational start site of CD2907.1) were amplified by PCR and cloned into StuI and BamHI sites of pDIA6103 vector under the control of the ATc-inducible Ptet promoter giving pDIA6319, pDIA6195, pDIA6202 and pDIA6196, respectively.
The knockdown antisense system on pRPF185 vector derivative was used to deplete the C. difficile 630Δerm strain for the specific ribonucleases RNase III, RNase J and RNAse Y. The rncS gene fragment comprising part of 5′ untranslated region (UTR) and the beginning of the rncS coding part (−39 to +188 relative to the translational start site) was amplified by PCR on C. difficile 630Δerm strain genomic DNA and cloned into StuI and BamHI sites of pRPF185 vector in antisense orientation under the control of the ATc-inducible Ptet promoter giving pDIA6126. Similar strategy was used to construct plasmids pDIA5975 and pDIA5977 for inducible expression of antisense RNA for RNase J and RNase Y genes (+7 to +217 and +55 to +210 relative to the transcriptional start site (TSS) identified by deep sequencing, respectively).
For subcellular localization of toxins we used reverse PCR approach to construct CD2517.1-HA and CD2907.1-HA-expressing plasmids on the basis of corresponding pDIA6103-derivatives with primers designed to introduce the HA-tag sequence at the C-terminal part of coding toxin regions, directly upstream the stop codon (Supplementary Table S2). To compare the action of short and long forms of antitoxins on cognate and non-cognate toxins when co-expressed either in cis or in trans (from a site distant from the vector MCS), we used reverse PCR approach and Gibson assembly to construct different plasmids on the basis of the corresponding pDIA6103-derivatives (pT) (Supplementary Tables S1, S2 and Supplementary methods). DNA sequencing was performed to verify plasmid constructs using pRPF185-specific primers IMV507 and IMV508. The resulting derivative pRPF185 plasmids were transformed into the E. coli HB101 (RP4) and subsequently mated with C. difficile 630Δerm (40) (Supplementary Table S1). C. difficile transconjugants were selected by sub-culturing on BHI agar containing Tm (15 μg/ml) and Cfx (25 μg/ml).
Light microscopy
For light microscopy, bacterial cells were observed at 100× magnification on an Axioskop Zeiss Light Microscope. Cell length was estimated for more than 100 cells for each strain using ImageJ software (41).
RNA extraction, quantitative real-time PCR, northern blot and 5′/3′RACE
Total RNA was isolated from C. difficile strains grown 7.5 h in TY medium containing 7.5 μg/ml of Tm and 250 ng/ml of ATc as previously described (42). For biofilm samples C. difficile 630Δerm strain was grown for 72 h in TY medium using continuous-flow microfermentor culture system (43). The 24-h planktonic culture in TY medium was used for comparative analysis. The cDNA synthesis by reverse transcription and qRT-PCR analysis were performed as previously described (44). In each sample, the relative expression for a gene was calculated relatively to the 16S rRNA gene or dnaF gene (CD1305) encoding DNA polymerase III or ccpA gene encoding catabolite control protein. The relative change in gene expression was recorded as the ratio of normalized target concentrations (ΔΔCt) (45). Northern blot analysis and 5′/3′RACE experiments were performed as previously described (15).
RNA band-shift assay and in vitro processing by RNase III
Templates for the synthesis of RNA probes were obtained by PCR amplification using the Term and T7 oligonucleotides (Supplementary Table S2). RNAs were synthesized by T7 RNA polymerase with [α-32P] UTP as a tracer and were then gel purified. RNA concentrations were monitored by counting out the radioactivity and the RNA samples were stored until use (46). This gives a 298-nt long CD2907.1 and a 132-nt long RCd9 transcripts with three additional G at the 5′ extremity. Just before use, RCd9 RNA was 5′-radiolabeled and incubated with increasing concentrations of CD2907.1 mRNA under two different conditions referred as N (Native) and F (Full RNA duplex) conditions, respectively. Radiolabeled RCd9 was incubated either alone or with the unlabeled CD2907.1 RNA to allow them to anneal in Tris-Mg-acetate-Na-acetate (TMN) buffer for 5 min at 37°C (20 mM Tris acetate, pH 7.5, 10 mM magnesium acetate, 100 mM sodium acetate). Alternatively, after denaturation at 90°C for 2 min, labeled RCd9 RNAs in 1×TE were incubated 30 min at 37°C with the unlabeled CD2907.1 to allow them to anneal. The complexes were immediately loaded on native polyacrylamide gels to control for hybridization efficiency (47) or submitted to in vitro processing by RNase III of E. coli. RNase III digestion of free or complexed RCd9 was performed at 37°C in TMN buffer containing 1 μg tRNA from 1 min to 15 min with 0.05 units of RNase III (Epicentre). After precipitation, addition of loading buffer and heat denaturation, samples were analyzed on 8% polyacrylamide-Urea gels.
Subcellular localization of HA-tagged toxins by cell fractionation and western blotting
The C. difficile cultures were inoculated from overnight grown cells in 10 ml of TY medium at OD 600 nm of 0.05, allowed to grow for 3 h before addition of 250 ng/ml ATc and incubation for 90 min followed by centrifugation and protein extraction. Cell lysis, fractionation and protein analysis were performed as previously described (48). Coomassie staining was performed for loading and fractionation control. Western blotting was performed as previously described (49) with anti-HA antibodies.
Measurement of RNA decay by rifampicin assay
For determination of toxin and antitoxin RNA half-lives the C. difficile strains were grown in TY medium supplemented with 250 ng/ml ATc and 7.5 μg/ml Tm for 7.5 h at 37°C. Samples were taken at different times after addition of 200 μg/ml rifampicin (0, 2, 5, 10, 20, 40, 60 and 120 min) and subjected to RNA preparation and northern blotting.
In silico screening for potential new TA genes and CRISPR arrays co-localization
The raw sequencing read data of 2,584 C. difficile strains were downloaded for this genomic analysis (16,50). For each strain, we realized an assembly with Spades (51) and an automatic annotation using PROKKA (52). Then we selected small proteins from 40 to 60 amino acids in length, adjacent to CRISPR arrays and performed an orthology analysis using proteinortho5 (53). Multiple alignment was done using ClustalW (54).
RESULTS
Identification of toxin–antitoxin system candidates in C. difficile genome
We have revisited our previously reported deep sequencing data (15) and observed an unusual transcriptional unit organization in the close proximity of CRISPR loci in the genome of C. difficile strain 630Δerm (Supplementary Figure S1). The presence of several overlapping transcripts was detected by comparison of Tobacco Acid Pyrophosphatase treated (TAP+) and non-treated (TAP−) samples for TSS mapping. This analysis combined with the RNA-seq data for whole transcript coverage revealed that the majority of CRISPR RNA loci are associated with potential antisense RNAs of genes encoding small proteins of unknown function (Figure 1). A more detailed analysis of the nature of the overlapping convergent transcripts allowed us to identify candidates for six type I TA systems that co-localized with CRISPR 3/4, CRISPR 6, CRISPR 7, CRISPR 11, CRISPR 12 and CRISPR 16/15 arrays in C. difficile (Figure 1 and Supplementary Table S3) (32). An additional pair of antisense RNA and small protein gene near CRISPR 9 array had divergent sequence without common type I TA features. Supplementary Table S3 summarizes the data on the candidate antitoxin RNAs including the 5′-end determination of the transcripts by global TSS mapping and the position of the 3′-ends deduced from RNA-seq data.
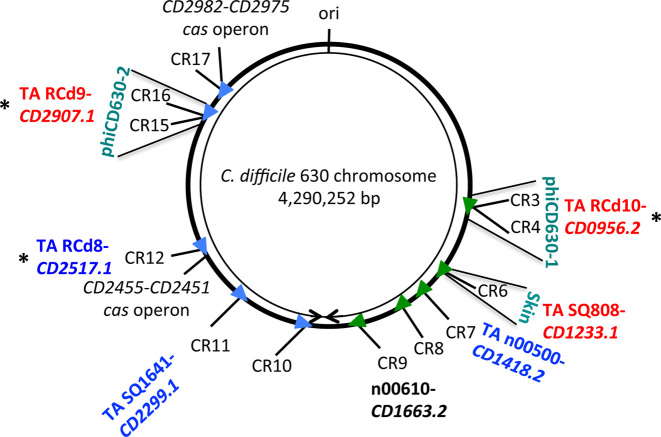
Figure 1.
Genomic map of potential type I TA loci in association with CRISPR arrays in Clostridium difficile strain 630. Schematic view of the genomic location of expressed CRISPR arrays in strain 630. CRISPR arrays are numbered according to CRISPRdb database (2). Arrowheads indicate the array position and the transcriptional orientation. The location of the associated TA modules, the cas operons, the prophage regions and the replication origin (ori) are indicated. The right and left replichores are shown by arrows. The n00610 antisense RNA overlaps the CD1663.2 gene, which encodes a small protein with a divergent sequence associated with CRISPR 9 array. The CRISPR-associated TA modules within prophage regions are RCd9-CD2907.1, RCd10-CD0956.2 and SQ808-CD1233.1. ‘*’ indicates the three TA modules that were selected for detailed analysis.
Interestingly, three potential TA modules together with associated CRISPR arrays are located within prophage regions (Figure 1). The pathogenicity-island location of type I TA modules has been reported in Staphylococcus aureus (55,56). The CRISPR 3/4 array, the associated potential toxin gene CD0956.2 and the antitoxin gene are located within the phiCD630-1 prophage region while the identical CRISPR 16/15 array and the potential TA module containing CD2907.1 toxin and antitoxin genes are located within the phiCD630-2 prophage region (Figure 1). Similar to the txpA/RatA type I TA module in Bacillus subtilis (57), the CD1233.1/SQ808 pair is located within the skin element of C. difficile strain 630 (58), yet there is no sequence homology between the two loci. This CD1233.1/SQ808 pair is located near the CRISPR 6 array in the C. difficile skin element.
We have chosen three representative type I TA modules for further detailed analysis. The RCd8-CD2517.1 module is located near the CRISPR 12 array, which is associated with a partial cas operon. The RCd9-CD2907.1 and RCd10-CD0956.2 modules are located near the CRISPR 16/15 and CRISPR 3/4 arrays. They lie respectively within the phiCD630-1 and phiCD630-2 prophage regions, which have identical sequences and are thus indistinguishable from each other through gene expression analysis. These two highly similar prophages phiCD630-1 (1088001-1143874) and phiCD630-2 (3377033-3434358) of 55.9 and 57.3 kb in length, respectively, are located in an inverted orientation on different replichores of the C. difficile chromosome. The regions encoding TA modules and CRISPR arrays are identical. We assigned the names RCd8 (previously named SQ1781), RCd9 (previously named CD630_n01000) and RCd10 (previously named CD630_n00370) to the putative antitoxin RNAs (Figure 1). We have previously demonstrated the active expression of the CRISPR arrays associated with the chosen modules and their functionality for interference with plasmid DNA (16).
We first mapped by 5′/3′RACE analysis the transcriptional start and termination sites of the genes corresponding to the potential toxin and the antitoxin RNAs for selected loci. Figure 2 shows the chromosomal organization of these genes and the position of 5′ and 3′ ends of overlapping transcripts identified by 5′/3′RACE (Table 1, Supplementary Table S3 and Figure S2). The alignment of the TA genomic regions revealed the presence of conserved sequences upstream of the TSS for both the putative toxin and antitoxin genes and allowed the identification of consensus elements for Sigma A-dependent promoters upstream of their TSS (Supplementary Table S3, indicated in blue and red in Supplementary Figure S2). Moreover, the consensus sequence promoters recognized by the alternative Sigma factor of the general stress response, Sigma B, could be identified upstream of the TSS of both the potential antitoxin and toxin genes (indicated in green in Supplementary Figure S2).
Figure 2.
Schematic representation of potential type I TA loci in C. difficile chromosome: (A) CRISPR 12 region, (B) CRISPR 16/15 (CRISPR 3/4) regions. The horizontal arrows below the double-stranded sequences show the toxin, antitoxin and CRISPR transcripts, the direction of transcription is indicated by arrowheads. The transcriptional start sites for sense and antisense transcripts identified by 5′/3′RACE and TSS mapping are indicated by vertical arrows with their genomic location. Line thickness corresponds to the proportion of observed extremities. The genomic location of 5′- and 3′-ends of the transcripts are indicated above the sequence. The inverted repeats at the position of transcriptional terminators are indicated by black arrows. The positions of Sigma A-dependent promoter −10 and −35 elements of antitoxin (AT) and toxin (T) are shown in boxes. The positions of ribosome binding site, translation initiation codon and stop codon of toxin (T) mRNA are underlined. The positions of Sigma B-dependent promoter elements are shown in boxes for both TA genes.
Table 1. The antisense RNA and associated toxin mRNA extremity identification by 5′/3′RACE.
| Name | Description | 5′-end RACE position | 5′-end TSS mapping position | Strand | 3′-end RACE position | Size, nt |
|---|---|---|---|---|---|---|
| RCd10 (CD630_n00370) | Antitoxin of TA associated with CRISPR 3/4 | 1124339 | 1124339 | − | 1124206, 1124058, 1124041 | 134, 283 |
| CD0956.2 | Toxin of TA associated with CRISPR 3/4 | 1124042 | 1124042 | + | 1124339 | 298 |
| RCd9 (CD630_n01000) | Antitoxin of TA associated with CRISPR 16/15 | 3398302 | 3398302 | + | 3398435, 3398583, 3398600 | 134, 283 |
| CD2907.1 | Toxin of TA associated with CRISPR 16/15 | 3398599 | 3398599 | − | 3398302 | 298 |
| RCd8 (SQ1781) | Antitoxin of TA associated with CRISPR 12 | 2907896, 2907991, 2908066 | 2908006, 2908013, 2908124 | + | 2908246 | 130, 255, 350 |
| CD2517.1 | Toxin of TA associated with CRISPR 12 | 2908421 | 2908421 | − | 2908116 | 306 |
The positions of 5′-start and 3′-end of these RNAs were identified by 5′/3′RACE analysis and compared with 5′-end identified by 5′-end RNA-seq analysis (TSS mapping).
We then confirmed the detection of the candidate RNA antitoxin transcripts by northern blot under several growth conditions (Figure 3). Transcript length deduced from TSS mapping, RNA-seq and 5′/3′RACE analysis agreed generally well with the size of RNAs detected by northern blotting. The RCd8 RNA is transcribed in antisense direction to the CD2517.1 gene and overlaps with the CRISPR 12 array (Figure 2A). An abundant transcript of about 140 nt was stably detected by northern blotting under all tested conditions (exponential growth phase, late exponential phase, onset of stationary phase or starvation) (Figure 3). The presence of additional less abundant longer transcripts of about 300 and 400 nt was consistent with 5′/3′RACE results (Table 1) and suggests that they could result from a detected alternative TSS (Figure 2A). The major 140-nt transcript starts with a transcriptional +1 associated with a conserved promoter sequence and stops at the rho-independent terminator (Figure 2 and Supplementary Figure S2).
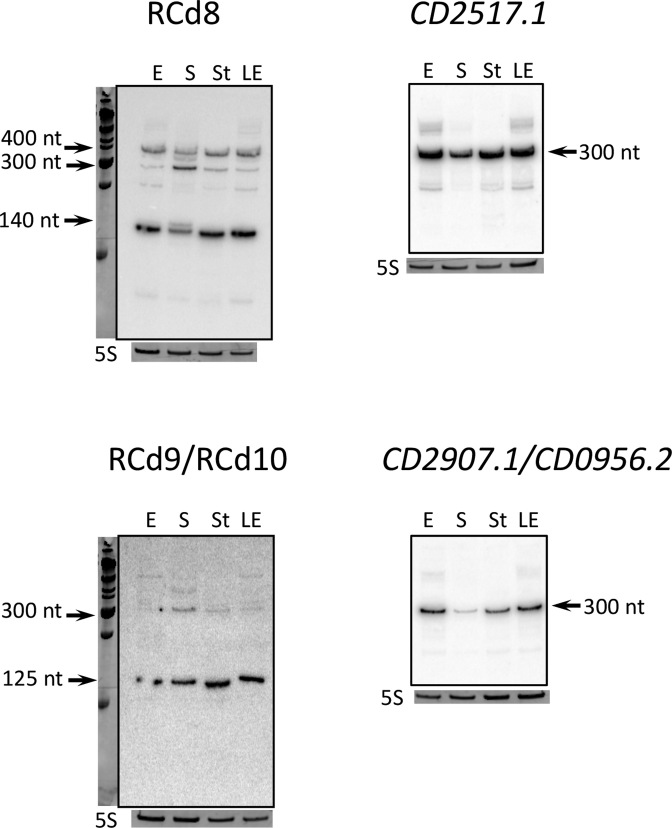
Figure 3.
Detection of antisense RNA and toxin mRNA by northern blot. RNA samples were extracted from 630Δerm strain grown at exponential phase (E, 4 h of growth), late exponential phase (LE, 6 h of growth), entry to stationary phase (S, 10 h of growth) or under nutrient starvation conditions (St). 5S RNA at the bottom serves as loading control. The arrows show the detected transcripts with their size estimated by comparison with RNA molecular weight standards. As indicated at the top, the blots were hybridized either with antitoxin- or toxin-specific probe. The same 5S control panel is shown when reprobing of the same membrane was performed.
The RCd9 RNA is transcribed in antisense direction to the CD2907.1 gene adjacent to CRISPR 16/15 array (Figure 2B). An identical region within the homologous phiCD630-1 prophage encodes the RCd10 RNA transcribed in antisense orientation to the CD0956.2 gene adjacent to CRISPR 3/4 array. An abundant transcript of about 125 nt was detected with RCd9/RCd10-specific probe under all tested conditions by northern blotting in addition to a hardly detectable longer transcript of about 300 nt in accordance with the 134 and 283-nt transcript sizes deduced from the 5′/3′RACE data (Figure 3, Table 1 and Supplementary Table S3). The relative abundance of these transcripts is consistent with the 5′/3′RACE data where most of the 3′ends mapped to the rho-independent terminator position for a short transcript (Figure 2).
To monitor the toxin mRNA expression, we rehybridized the northern blots with the probes matching to CD2517.1 and CD2907.1. In accordance with the 5′/3′RACE data, an abundant transcript of about 300 nt was detected under all tested conditions (Figure 3). A decreased intensity of the toxin mRNA signal was observed for the onset of stationary phase. It is worth noting that a slight decrease in the signal of the antitoxin corresponding to short transcripts was also detected under these conditions with the concomitant appearance of longer transcripts (Figure 3).
From these data we infer that several RNA transcripts for RCd8 of 140, 255 and 350-nt long are detected while the CD2517.1 mRNA is 306-nt long. The RCd8 and CD2517.1 RNAs overlap by 131 nt. The RCd9/RCd10 RNA transcripts are 134 and 283-nt long and overlap the 298-nt long CD2907.1/CD0956.2 mRNAs by 283 or 134 nt for long and short transcripts, respectively. Secondary structure prediction by Mfold revealed that potential antitoxin RCd8 and RCd9/RCd10 RNAs are highly structured (data not shown). Corresponding overlapping mRNA are also highly structured and contain double-stranded secondary structure regions sequestering their ribosome binding sites that could be important for the regulatory process as observed for B. subtilis type I toxin mRNAs (data not shown) (59–61). Moreover, the presence of 5′ UTRs of 88 and 83 nt in length was observed for CD2517.1 and CD2907.1/CD0956.2 mRNAs, respectively (Supplementary Figure S2). The comparison between the secondary structure predictions for the TA pair transcripts suggests potential loop–loop interactions between stem–loop structures that could initiate the RNA duplex formation (data not shown) (59).
We have chosen representative abundant transcripts from TA pair RCd9/CD2907.1 for the analysis of duplex formation between toxin and antitoxin RNA by gel retardation assays. We investigated the interaction between RCd9 and CD2907.1 to determine whether they form a kissing complex as in the case of the antisense RNA, CopA and its target CopT (62). Figure 4A shows that under native conditions RCd9 harbors two conformations both proficient to duplex formation upon incubation with equimolar amount of CD2907.1 transcript suggesting that the primary binding intermediate could involve a loop–loop interaction.
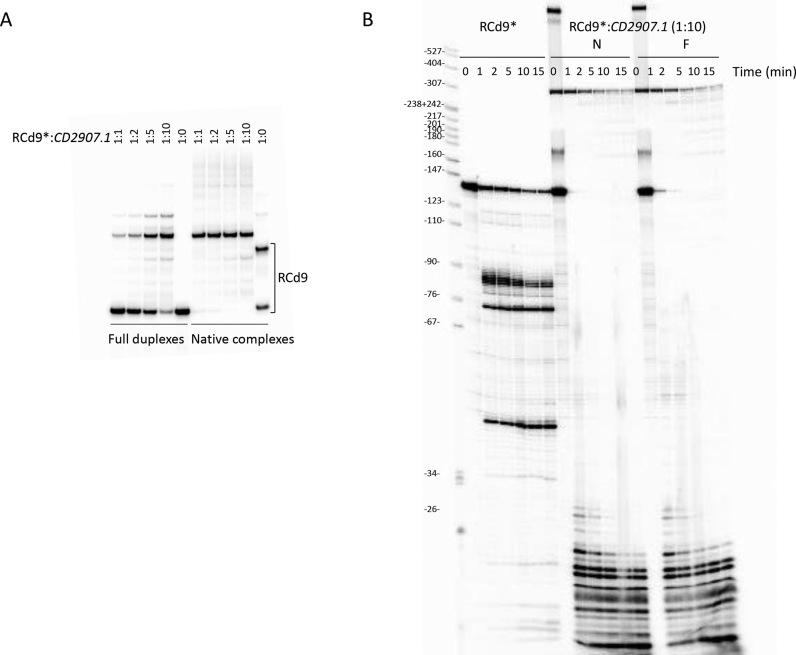
Figure 4.
Analysis of TA RNA duplex formation by RNA band-shift assay and in vitro processing by RNase III. (A) 5′-radiolabeled RCd9 RNA was incubated with increasing concentrations of CD2907.1 under two different conditions referred as N (Native) and F (Full RNA duplex) conditions, respectively. Native RCd9:CD2907.1 complexes were formed at 37°C for 5 min in TMN buffer, and full duplexes were obtained after a denaturation-annealing treatment in TE Buffer (2 min 90°C, 30 min 37°C). The complexes were immediately loaded on native polyacrylamide gels to control for hybridization efficiency or submitted to in vitro processing by RNase III (B). RNase III digestion of free or complexed RCd9 was performed at 37°C in TMN buffer containing 1 μg tRNA from 1 min to 15 min with 0.05 units of RNase III per sample. Samples were analyzed on 8% polyacrylamide-Urea gels.
We then investigated RNase III-dependent cleavages of RCd9 alone and in interaction with CD2907.1 under conditions of native complex formation or after extensive denaturation to define the extent of duplex formation (Figure 4B). RNase III cleaves RCd9 at 3 sites located about 82, 72 and 42 nt from the 5′ extremity. In contrast, RNase III very efficiently and rapidly degrades the duplexes formed under native conditions or after extensive denaturation. This indicates that RCd9 is fully hybridized to the CD2907.1 toxin mRNA and that the kissing intermediate formed under native conditions of hybridization could be converted into a full duplex leading to the RNase III cleavage of the entire duplex.
Functionality of toxin–antitoxin systems in C. difficile
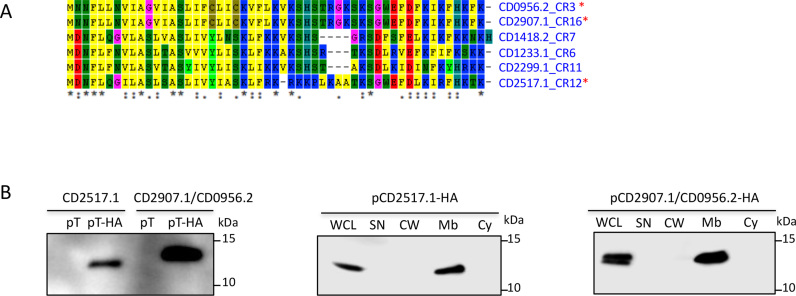
Type I toxins are generally small hydrophobic proteins of <60 amino acids containing a potential transmembrane domain and charged amino acids at the C-terminus (32). The alignment of proteins from the potential TA modules encoded in the proximity of CRISPR arrays revealed that these small proteins have all characteristic features of type I toxins. Indeed, as shown in Figure 5A, the potential toxic proteins are from 50 to 53 amino acids in length, carry a conserved hydrophobic region at their N-terminal part and a lysine-rich, positively charged region at their C-terminal part in agreement with the hydrophobicity profile predictions by Kyte and Doolittle algorithm (data not shown). Transmembrane domain location in N-terminal moiety was predicted by TMHMM program (data not shown). To experimentally identify the expression and localization of these small proteins in C. difficile we constructed plasmids expressing under inducible Ptet promoter either CD2517.1 or CD2907.1/CD0956.2 fused with a HA tag at the C-terminus (Supplementary Table S1). By western blotting with anti-HA antibodies, no signal was detected for whole cell extracts from control strains expressing untagged proteins while a specific signal was detected for strains expressing HA-tagged proteins (Figure 5B). To precise the subcellular localization of these proteins we then performed cell fractionation and examined supernatant, cell wall, membrane and cytosolic fractions by western blotting. As shown in Figure 5B, HA-tagged CD2517.1 and CD2907.1 (CD0956.2) were only detected in the membrane fractions of C. difficile cell extracts suggesting the association of these small proteins with the cell membrane in C. difficile.
Figure 5.
Potential type I toxin proteins alignment and analysis. (A) Proteins alignment using ClustalW. ‘*’ on the right indicates toxins from three TA modules selected for detailed analysis. ‘*’ at the bottom indicates conserved residues. (B) Western-blot detection and localization of HA-tagged small proteins in the membrane fraction of C. difficile cell extracts. WCL: whole cell lysate; SN: supernatant; CW: cell wall; Mb: membrane; Cy: cytosolic fraction. Immunoblotting with anti-HA antibodies detected a major polypeptide of ∼10 kDa in whole cell lysates of the strain carrying Ptet-T(CD2517.1 or CD2907.1/CD0956.2)-HA (pT-HA) construct grown in the presence of the 250 ng/ml ATc inducer but not in extracts of strains expressing non-tagged toxins (pT) (left panel). The culture of strains carrying Ptet-T-HA plasmids induced with 250 ng/ml ATc was fractionated into cell wall (CW), membrane (Mb) and cytosolic (Cy) compartments and immunoblotted with anti-HA antibodies (middle and right panels). Proteins were separated on 12% Bis-Tris polyacrylamide gels in MES buffer.
To show the toxic nature of these small proteins, we analyzed the effect of their overexpression on the growth of C. difficile cells in liquid and solid media. HA-tagged proteins CD2517.1 and CD2907.1/CD0956.2 conserved their toxic activity on cell growth when overexpressed from plasmids used for determination of their subcellular localization by western blotting (Supplementary Figure S3 and Figure 5B). This result suggests that despite the presence of HA-tag these small proteins remain active for cell growth inhibition.
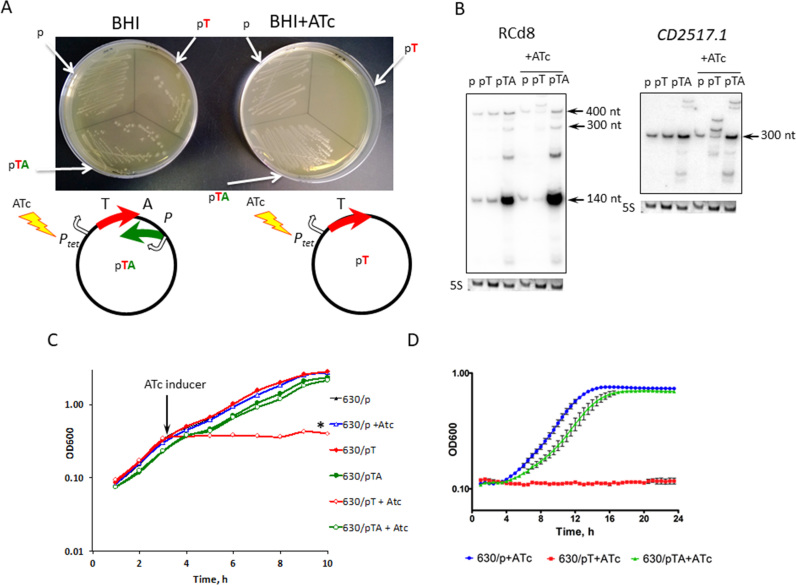
We then generated plasmids allowing either inducible overexpression of an untagged version of one of the small, potentially toxic proteins or simultaneous expression of both the potential toxin and the antisense RNA for the TA modules near the CRISPR 12 and CRISPR 16/15 (CRISPR 3/4) arrays. For this purpose, we cloned either the small protein-coding region with its ribosome-binding site (RBS) (CD2517.1 or CD2907.1/CD0956.2) under the control of the inducible Ptet promoter (pT) or the entire potential TA module (pTA). pTA constructs allow both the inducible overexpression of the putative toxin under the control of the Ptet promoter and the expression of the antisense RNA from its own strong promoter (Figure 6A). C. difficile strain 630Δerm carrying an empty vector (p) was used as a control. No growth difference was observed for any of the three strains on BHI plates in the absence of AnhydroTetracycline (ATc) inducer for both potential TA modules (Figures 6A and 7A). By contrast, a dramatic growth defect was observed on BHI plates in the presence of ATc inducer for the strain overexpressing the genes CD2517.1 or CD2907.1/CD0956.2 (Figures 6A and 7A). Co-expression of these potential toxins with the associated RNA antitoxins led to the full or partial reversion of the growth defect for both TA modules (Figures 6A and 7A). Consistently, northern blotting with RCd8 and RCd9/RCd10-specific probes using RNA extracted from the strain 630Δerm carrying pT or pTA confirmed the important overexpression of the antitoxin RNA from the pTA constructs as compared to the level of expression from their chromosomal location in control strain carrying an empty vector (p) (Figures 6B and 7B).
Figure 6.
Effect of inducible toxin and TA overexpression for CD2517.1-RCd8 TA module near CRISPR 12 on growth in solid (A) and liquid medium (B–D). (A) Growth phenotype of C. difficile strains CDIP369 (630/p), CDIP357 (630/pT) and CDIP332 (630/pTA) on BHI agar plates supplemented with Tm alone (on the left) or with the addition of 500 ng/ml of ATc inducer (on the right) after 24 h of incubation at 37°C. Schematic representations of pT and pTA constructs are shown. The 630 strain carrying an empty vector (p) is used as a control. (B) Detection of RCd8 and CD2517.1 transcripts. For northern blot analysis, RNA samples were extracted from 630/p control strain, from 630/pT strain overexpressing the CD2517.1 toxin and from 630/pTA strain overexpressing the entire TA module grown at late exponential growth phase in the presence of 250 ng/ml ATc (+ATc) or the absence of inducer. As indicated at the top, the blots were hybridized either with antitoxin- or toxin-specific probe. The same 5S control panel is shown when reprobing of the same membrane was performed. (C) Growth of 630/p strain (triangles), 630/pT strain (diamond) and 630/pTA strain (circle) in TY medium at 37°C in the presence (open symbols) or absence (closed symbols) of 250 ng/ml ATc. The time point of ATc addition is indicated by an arrow. (D) Growth curves for 630/p strain, 630/pT strain and 630/pTA strain in TY medium at 37°C in the presence of 250 ng/ml ATc using a GloMax plate reader (Promega). The mean values and standard deviations are shown for three independent experiments.
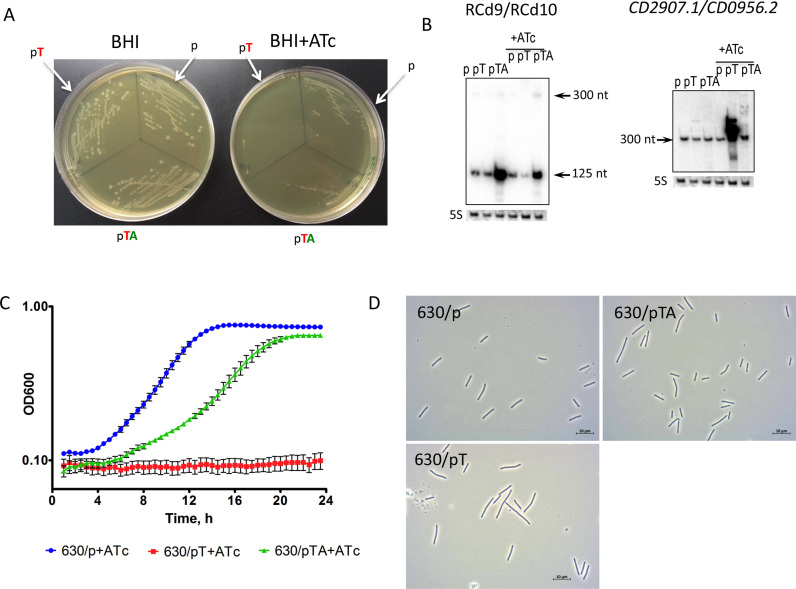
Figure 7.
Effect of inducible toxin and TA overexpression for CD2907.1-RCd9/CD0956.2-RCd10 TA module near CRISPR 16/15 (CRISPR 3/4) on growth in solid (A) and liquid (B–D) medium. (A) Growth of C. difficile strains CDIP369 (630/p), CDIP317 (630/pT) and CDIP319 (630/pTA) on BHI agar plates supplemented with Tm alone (on the left) or with the addition of 500 ng/ml of ATc inducer (on the right) after 24 h of incubation at 37°C. Under inducing conditions 630/pT strain overexpresses CD2907.1 toxin and 630/pTA strain overexpresses entire TA module. The 630 strain carrying an empty vector (p) is used as a control. (B) Detection of RCd9/RCd10 and CD2907.1/CD0956.2 transcripts. For northern blot analysis, RNA samples were extracted from 630/p control strain, 630/pT strain overexpressing CD2907.1 and 630/pTA strain overexpressing the entire TA module grown at late exponential growth phase in the presence of 250 ng/ml ATc (+ATc) or the absence of inducer. As indicated at the top, the blots were hybridized either with antitoxin- or toxin-specific probe. The same 5S control panel is shown when reprobing of the same membrane was performed. (C) Growth of 630/p strain, 630/pT strain and 630/pTA strain in TY medium at 37°C in the presence of 250 ng/ml ATc using a GloMax plate reader (Promega). The mean values and standard deviations are shown for three independent experiments. (D) Selected images from light microscopy observation of 630/p, 630/pT and 630/pTA strains grown in TY medium at 37°C after 1 h of 250 ng/ml ATc addition.
The overexpression of toxins from selected TA modules (Figures 6B and 7B) also induced rapid growth arrest in liquid culture. As shown in Figure 6C for CD2517.1-RCd8 TA module, the addition of ATc inducer after 3h of exponential growth led to rapid growth arrest for the strain carrying the pT plasmid but allowed near normal growth of the C. difficile 630Δerm strain carrying pTA. Similar deleterious growth effects were observed for the strain carrying the pT plasmid when strains pre-grown overnight in the absence of inducer and then diluted in an ATc-containing medium were allowed to grow for 24 h in an automatic plate reader (Figure 6D). For the CD2907.1-RCd9/CD0956.2-RCd10 TA module, we observed only a partial reversion of the growth defect in liquid culture associated with the toxin gene expression when both toxin and antitoxin were co-expressed on pTA plasmid (Figure 7C). This partial restoration of growth could be due to an unbalance in the relative level of toxin and antitoxin expression. Interestingly, northern blotting revealed a reverse correlation between the relative toxin and antitoxin transcript abundance under inducing conditions. Toxin overexpression after ATc induction led to more than 2-fold decrease in the amount of antitoxin transcript expressed from chromosomal location compared to the strain 630Δerm containing the vector alone (lanes ‘pT’ versus ‘p’ in Figures 6B and 7B).
Toxins from TA modules in B. subtilis and Enterococcus faecalis have been reported to affect cell envelope biosynthesis, nucleoid condensation, cell division and chromosome segregation (35,63). To assess whether the changes in cell morphology could be induced by toxin overexpression in C. difficile, we analyzed by light microscopy liquid cultures of strain 630Δerm carrying the vector, pT or pTA 1 h after ATc addition. For both TA modules (CD2517.1 and CD2907.1/CD0956.2), the overexpression of the toxins in strain 630/pT led to a significant increase in cell length for about 9 and 5.4% of the cells, respectively. The length of these cells was above the value of 630/p mean length with two standard deviations (10.5 μm) (Figure 7D and Supplementary Figure S4). For control strain 630/p the length of only 1.7% of cells exceeded this value. Co-expression of the entire TA module (pTA) led to a partial reversion of this phenotype to the control culture morphology.
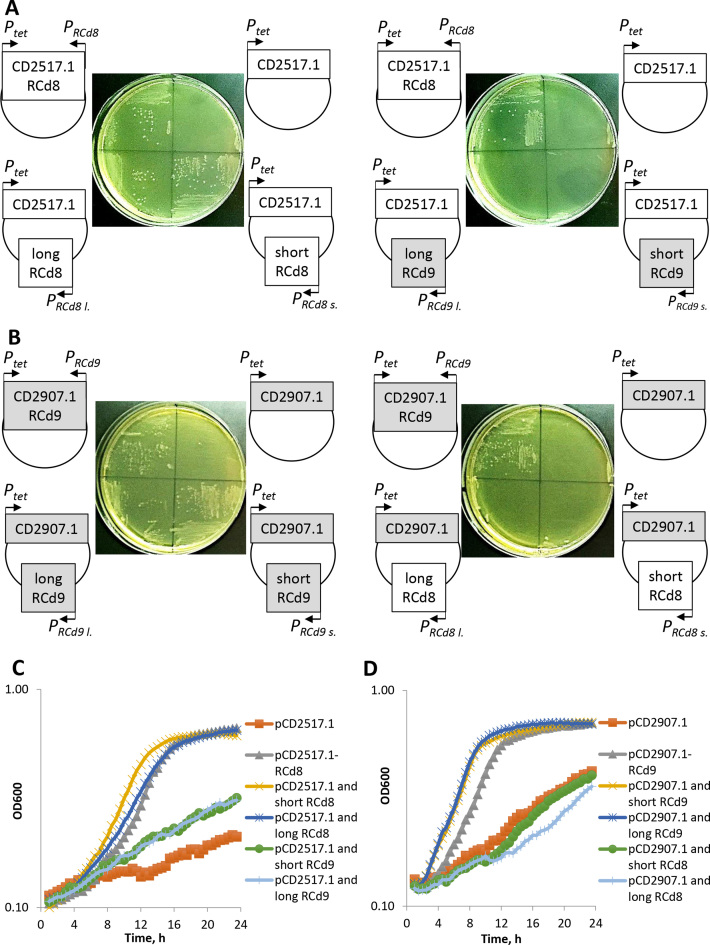
To get further insights into the requirements for repression and the function of abundant short and less abundant full-length antitoxin transcripts, we co-expressed in trans different antitoxin transcripts from their own promoter with the native or HA-tagged toxin proteins under the control of inducible Ptet promoter for both TA modules (Supplementary Table S1). For each combination, toxin and antitoxin were co-expressed from the same plasmid but from distant locations. In the presence of ATc inducer, both short and full-length antitoxins expressed in trans were able to rescue the strains from the toxicity associated with over-expression of the cognate native toxin both on plates and in liquid culture (Figure 8). The reversion of growth defect was similar to that observed with a control strain carrying the plasmid co-expressing the toxin from the inducible Ptet promoter and the cognate antitoxin in cis from the native convergent configuration (Figure 8). Cognate short antitoxin co-expression in trans also led to the reversion of growth defect induced by the overexpression of the HA-tagged toxin (Supplementary Figure S5). These results suggest that the abundant short form of antitoxins is sufficient to repress the associated toxin and that the C-terminal HA-tag does not interfere with antitoxin action. By contrast, a dramatic growth defect was still observed when the non-cognate antitoxins were co-expressed with the toxins (Figure 8). These results demonstrate that RCd8 and RCd9/RCd10 antitoxins act on a highly specific manner to repress their associated toxin not only when they are expressed from the native convergent TA configuration (Figures 6–8) but also when expressed in trans from a distant plasmid location (Figure 8).
Figure 8.
Effect of short and full-length antitoxin co-overexpression with native toxin in trans for CD2517.1-RCd8 and CD2907.1-RCd9/CD0956.2-RCd10 TA modules on growth in solid (A and B) and liquid (C and D) medium. (A) Growth of C. difficile strains CDIP1191 (630/pCD2517.1-RCd8), CDIP357 (630/pCD2517.1), CDIP1161 (630/pCD2517.1 and long RCd8), CDIP1130 (630/pCD2517.1 and short RCd8), CDIP1162 (630/pCD2517.1 and long RCd9) and CDIP1131 (630/pCD2517.1 and short RCd9) on BHI agar plates supplemented with Tm and 2.5 ng/ml of ATc inducer after 24 h of incubation at 37°C. (B) Growth of C. difficile strains CDIP1192 (630/pCD2907.1-RCd9), CDIP317 (630/pCD2907.1), CDIP1164 (630/pCD2907.1 and long RCd9), CDIP1133 (630/pCD2907.1 and short RCd9), CDIP1163 (630/pCD2907.1 and long RCd8) and CDIP1132 (630/pCD2907.1 and short RCd8) on BHI agar plates supplemented with Tm and 3 ng/ml of ATc inducer after 24 h of incubation at 37°C. (C) Growth of the same C. difficile strains as in (A) in TY medium at 37°C in the presence of 2.5 ng/ml ATc using a GloMax plate reader (Promega). The mean values are shown for three independent experiments. (D) Growth of the same C. difficile strains as in (B) in TY medium at 37°C in the presence of 3 ng/ml ATc using a GloMax plate reader (Promega). The mean values are shown for three independent experiments.
Altogether our data demonstrate that functional type I TA modules are present in the proximity of CRISPR arrays in C. difficile.
Stability of the RNAs of the toxin and antitoxin genes
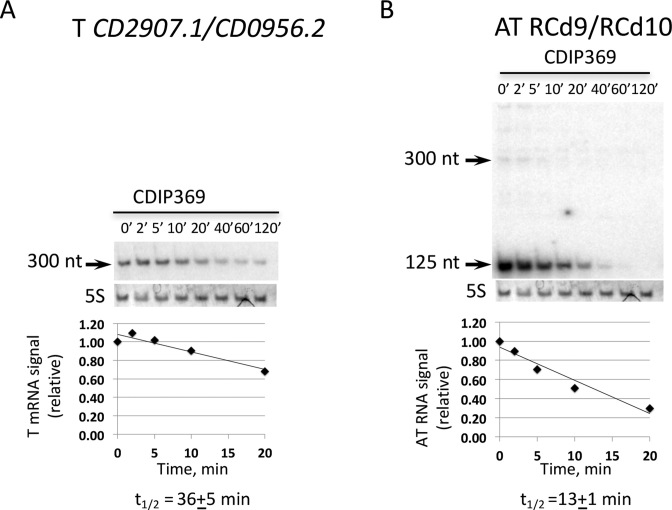
To determine the half-lives of toxin and antitoxin RNAs, C. difficile strains were grown in TY medium until the late-exponential growth phase and rifampicin was added to block transcription. Samples were taken at different time points after rifampicin addition for total RNA preparation and Northern blots were performed using probes targeting the toxin or antitoxin RNAs of one representative TA module. We have used a control strain 630Δerm carrying an empty vector (CDIP369) to allow further comparison with strains depleted for ribonucleases (see below). In this strain, quantification of the northern blots with corresponding probes allowed us to estimate the half-life of the mRNA of the CD2907.1/CD0956.2 toxin gene to about 35 min (Figure 9A). The half-life of the 125-nt transcript for RCd9/RCd10 antitoxin RNA was estimated to be about 13 min (Figure 9B). These results further confirm that the identified type I TA modules produce a rather stable toxin mRNA and a less stable antitoxin RNA.
Figure 9.
Expression and stability of CD2907.1/CD0956.2 toxin (A) and RCd9/RCd10 antitoxin (B) transcripts by northern blot. For determination of half-lives samples were taken at the indicated times after addition of 200 μg/ml rifampicin. RNAs were extracted from strain CDIP369 (630/p). 5S RNA at the bottom of each northern blot autoradiogram serves as loading control. The same 5S control panel is shown when reprobing of the same membrane was performed. The relative intensities of the bands from northern blot analysis via autoradiography were quantified using ImageJ software.
In model Gram-positive bacterium B. subtilis, the double-strand-specific enzyme RNase III plays an essential role in the degradation of toxin mRNA from prophage-encoded type I TA modules (59,64) while the single-strand specific endoribonuclease RNase Y and the 5′-3′ exoribonuclease RNase J1 participate in antitoxin RNA degradation (59). To analyse the possible contribution of these ribonucleases to the degradation of toxin and antitoxin RNAs, we tested the effect of RNase depletion on the stability of the corresponding transcripts. We also evaluated the effect of the depletion for the RNA chaperone protein Hfq. C. difficile 630 encodes CD1289 (rnj), CD1329 (rny) and CD1248 (rncS) proteins homologous to B. subtilis RNase J, RNase Y and RNase III. No transposon insertions were identified in a previously reported TraDIS (transposon-directed insertion site sequencing) experiment for these RNase genes suggesting their crucial role for C. difficile physiology (65). We have previously reported the use of a knock-down strategy based on the expression of an inducible antisense RNA targeting the 5′ part of the coding region of the hfq gene leading to Hfq depletion in C. difficile (49). Here, we used a similar strategy for the construction of C. difficile strains in which we can deplete for RNase J, RNase Y and RNase III using an antisense RNA targeting each of these genes expressed under the control of a Ptet inducible promoter (Supplementary Table S1). The efficient depletion conditions in the presence of ATc were confirmed by western blotting (Supplementary Figure S6).
The half-lives of analyzed toxin and antitoxin RNAs were similar in the wild-type and in the Hfq depleted strain suggesting that neither of the two RNAs is stabilized by Hfq (Supplementary Figure S7). We observed a moderate stabilization of the CD2907.1/CD0956.2 toxin mRNA in the strain depleted for RNase III but also, at a higher level, in the strains depleted for RNase J or RNase Y (Supplementary Figure S7A). These results suggest the possible contribution of the RNase J, RNase Y and in less extent of RNase III to the degradation of toxin mRNA. No major changes in the estimated half-life values were observed for the abundant transcript of antitoxin RNA RCd9/RCd10. However, RNase depletion resulted in observable changes in the degradation pattern of the antitoxin RNA RCd9/RCd10. We observed the appearance of a biphasic degradation curve and the accumulation of longer species especially for RNase Y depleted strain (Supplementary Figure S7B). Generally, the half-lives of longer transcripts increase under RNase depletion conditions. This result suggests that at least RNase Y could in some way contribute to the antitoxin RNA degradation.
Expression analysis of TA and CRISPR-Cas systems
We wondered whether the chromosomal co-localization of CRISPR arrays and TA modules would imply the possible connection between these systems. As mentioned above, the alignment of CRISPR-associated TA module sequences strongly suggested the presence of both Sigma-A-dependent and Sigma-B-dependent promoters upstream of the TSS of the toxin and antitoxin genes for the six TA modules (Supplementary Figure S2). We have recently demonstrated the crucial role of the alternative Sigma B factor in the adaptive strategies of C. difficile inside the host (66). We then re-examined the transcriptome data for the sigB mutant as compared to the parental strain and observed up to 5-fold decrease in the expression of the entire gene sets for both the partial and complete cas operons (CD2455 and CD2982) of type I-B C. difficile CRISPR-Cas system (Table 2). qRT-PCR analysis validated these transcriptome data (Table 2). In accordance, the search for Sigma-B-dependent promoter sequences revealed the presence of consensus elements GTTTTTA-N12-GGGATTT and TTATAA-N12-GGGTTAA upstream of TSS for cas gene operons CD2455 and CD2982, respectively. These promoter sequences are characterized by the presence of a conserved −10 promoter element associated and a less conserved −35 promoter element. Such a promoter structure suggests the possible implication of other regulatory components controlling these operons together with the Sigma B factor. The high sequence conservation among direct repeats within multiple CRISPR arrays suggests that the same set of Cas proteins processes all expressed pre-crRNA in C. difficile strains (16). Thus, the induction of cas genes under stress conditions would allow the overall activation of CRISPR-Cas defense mechanisms. Transcriptome analysis of the sigB mutant also revealed differential expression of several newly identified TA genes and associated CRISPR arrays (Table 2). To confirm these data, we performed qRT-PCR analysis for selected TA gene pairs and CRISPR arrays (Table 2). In accordance with transcriptome data, we confirmed by qRT-PCR the downregulation of several CRISPR-associated TA genes in the sigB mutant strain as compared to the parental strain even without stress exposure (Table 2).
Table 2. Differential expression of TA and CRISPR-Cas systems revealed by transcriptome and/or qRT-PCR analysis.
| Gene ID | Function | Ratio sigB/630Δerm Microarraya | Ratio sigB/630Δerm qRT-PCR | Ratio biofilm/plankton qRT-PCR |
|---|---|---|---|---|
| CD2982 b | CRISPR-associated Cas6 family protein | 0.19 | 0.22 | 14.7 |
| CD2981 | CRISPR-associated protein, CXXC-CXXC | 0.21 | ||
| CD2980 | CRISPR-associated autoregulator DevR family protein | 0.26 | ||
| CD2979 | CRISPR-associated Cas5 family protein | 0.25 | ||
| CD2978 | CRISPR-associated Cas3 family helicase | 0.30 | ||
| CD2977 | CRISPR-associated Cas4 family protein | 0.33 | ||
| CD2976 | CRISPR-associated Cas1 family protein | 0.37 | ||
| CD2975 | CRISPR-associated Cas2 family protein | 0.37 | ||
| CD2455 c | CRISPR-associated protein | 0.55 | 0.61 | 9.4 |
| CD2454 | Conserved hypothetical protein | 0.55 | ||
| CD2453 | CRISPR-associated negative autoregulator | 0.47 | ||
| CD2452 | CRISPR-associated protein | 0.53 | ||
| CD1233.1 | Toxin of TA associated with CRISPR 6 | 0.51 | ||
| CD2517.1 | Toxin of TA associated with CRISPR 12 | 0.25 | 0.42 | 26.0 |
| RCd8 | Antitoxin of TA associated with CRISPR 12 | 0.67 | 7.3 | |
| CD630_n00860 | CRISPR 12 | 7.7 | ||
| CD2907.1 | Toxin of TA associated with CRISPR 16/15 | 1.8 | ||
| RCd9/RCd10 | Antitoxin of TA associated with CRISPR 16/15 /CRISPR 3/4 | 0.5 | 0.42 | 2.3 |
| CD630_n00990 | CRISPR 16/15 | 0.54 | 9.8 |
Gene names and functions correspond to those indicated in the MaGe database Clostriscope (https://www.genoscope.cns.fr).
aA gene was considered as differentially expressed between the strain 630Δerm and the sigB mutant when the P-value is < 0.05.
bFirst gene of the complete cas operon CD2982-CD2975.
cFirst gene of the partial cas operon CD2455-CD2452. SQ1781 corresponds to RCd8, CD630_n01000 to RCd9 and CD630_n00370 to RCd10.
The induction of CRISPR-Cas-mediated defense capacities within biofilm community or more generally within the gut microbiota, which includes phages, could be important for bacterial survival under conditions promoting gene transfer. In E. coli, type I toxin ralR gene expression is induced during growth in biofilms (67). We thus compared the expression of selected CRISPR-associated TA modules and CRISPR-Cas systems within biofilm and planktonic cultures and observed a strong, up to 20-fold, induction of expression of selected genes (Table 2). Overall these results suggest that the cas operons and the CRISPR arrays could be co-regulated with associated type I TA systems by stress- and biofilm-related factors.
Genomic analysis of TA and CRISPR arrays co-localization
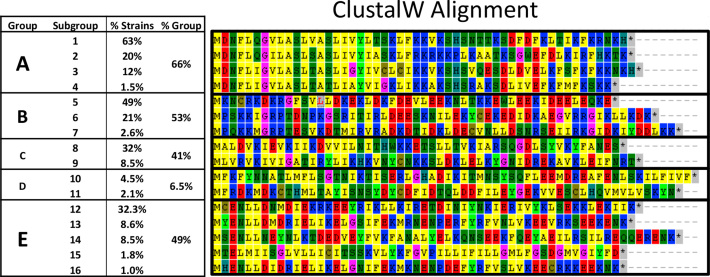
We analyzed the extent of co-localization of potential type I TA with CRISPR arrays in available C. difficile sequences. From more than 2,500 C. difficile genome sequences assembled and automatically annotated, we first found that 98% contain CRISPR arrays (from 1 to 30). In these CRISPR-containing strains, we then searched for the presence, immediately adjacent to CRISPR loci, of open reading frames from 40 to 60 amino acids, as one of the characteristic features of type I toxins is their small size. This search resulted in about 7000 hits. The CRISPR-associated small proteins were only absent in 67 genomes of which 58 lacked cas gene homologs. Then, an orthology analysis identified 16 proteins present each in more than 25 strains (Supplementary Table S4). Figure 10 shows an alignment of these 16 representative small proteins adjacent to CRISPR arrays combined in five major groups (A–E) according to their homology.
Figure 10.
Alignment of small proteins at the near proximity of CRISPR arrays in C. difficile strains. The representative proteins of five major groups are shown and their occurrence within analysed C. difficile strains is indicated. The multiple alignment was done using ClustalW.
The three small proteins characterized in this study (CD2907.1, CD0956.2 and CD2517.1) belong to group A. This group is largely distributed in C. difficile as it is present in two-third of the analyzed strains (Figure 10). CRISPR 16/15 and CRISPR 3/4-associated toxins belong to the most represented subgroup, A1, found in 63% of strains, CRISPR 12 associated toxin belongs to subgroup A2, that is present in 20% of the analyzed strains. Other CRISPR-associated toxins of strain 630 are represented in less extent within the same group. Finally, as two of the characterized toxins are located within prophage regions in strain 630, we wondered whether prophage localisation could be a common feature of CRISPR-associated small proteins. In 13 from 22 known C. difficile phages, we found potential toxins all belonging to the group A that could be part of TA modules. However, the co-localization with CRISPR arrays is detected only in the phi027 prophage of the R20291 strain.
To provide an experimental confirmation of potential TA and CRISPR arrays co-expression in another C. difficile strain, we have looked at the RNA-seq data of the epidemic strain R20291 (68). In this strain, we detected three co-localized CRISPR and TA pairs (Supplementary Figure S8A). One pair was intact (TA and CRISPR), while the two others have mutation in the toxin genes and only antitoxin was detected (A and CRISPR). We confirmed their co-expression in the published R20291 RNA-seq data using the COV2HTML software for visualization (69) (Supplementary Figure S8B).
In summary, we found that (i) CRISPR-associated small proteins are present in the vast majority of C. difficile strains and (ii) their primary orthology group is homologous to newly identified type I TA toxins.
DISCUSSION
Here, we report the first identification of functional type I TA modules in C. difficile 630 chromosome. Deep-sequencing, northern blotting and 5′/3′RACE revealed the presence of overlapping transcripts for type I toxin gene and associated RNA antitoxin in several chromosomal loci. Comparison of the newly identified type I TA systems in C. difficile with previously studied TA systems in other bacteria revealed no sequence homology for small toxin proteins. However, we observed a conservation of their membrane association and the presence of charged amino acids in the C-terminal part (32,35,70). The inducible overexpression of toxin genes strongly impaired the C. difficile growth while co-expression of associated antitoxin RNA in cis or in trans prevented this growth defect. The major short antitoxin transcripts were fully active to rescue the strains from the toxicity associated with their cognate toxins. Interestingly, despite extensive homology between studied TA regions, only cognate antitoxins were able to rescue the strains from the toxicity of corresponding toxins, no growth was observed when non-cognate antitoxins were co-expressed with toxins. Similarly, no cross-interaction between non-cognate TA pairs was found for the Ibs-Sib and Zor-Orz type I TA modules in E. coli (71–74) as well as for 17 type II TA modules in Vibrio cholerae chromosome superintegron (75). Our results provide important data on the specificity of antitoxin action that could explain why multiple TA pairs are maintained in the genome of C. difficile. Through the half-live measurements we demonstrated that these TA modules encode a rather stable toxin mRNA and an unstable antitoxin RNA. In addition, by gel retardation experiments we showed an efficient duplex formation between antitoxin and toxin RNA in vitro.
Mechanisms involved in the regulation of toxin expression and RNA decay within type I TA systems differ between bacteria (29,60,61,64,76–78). Two major modes of antisense RNA antitoxin action on toxin expression have been identified for type I TA. Antitoxin can either inhibit toxin mRNA translation or stimulate mRNA degradation (61). Dual-acting SR4 antitoxin in B. subtilis controls both toxin mRNA decay and translation (60). In addition, the importance of mRNA folding in controlling toxin expression was recently highlighted for TisB and ZorO in E. coli (74,78–80) and for AapA1 in Helicobacter pylori (76). Secondary structure prediction suggests that the RBS of toxin mRNAs within type I TA modules in C. difficile is sequestered within stable secondary structure as usually observed in other type I TA systems. In these cases processing events or other elements are required to initiate translation. The presence of a long 5′ UTR observed in this study that can serve as a target of antitoxin action is also a common feature for many type I toxin mRNAs (61,78). Even in the case of antitoxin RNA action through translational inhibition of toxin mRNA, the RNA duplex could be subjected to degradation by RNase III. This duplex-specific endoribonuclease often cleaves double-stranded RNA regions formed through base-pairing interactions between antitoxin RNA and complementary toxin mRNA as shown for txpA/RatA system within B. subtilis skin element (57,64). Interestingly, the essential role of RNase III in B. subtilis was demonstrated in protecting it from the expression of toxin genes borne by two prophages, skin and SPbeta, through antitoxin RNA (64). Other ribonucleases could be also involved in toxin and antitoxin RNA decay including RNase E in E. coli (81) and RNase Y and RNase J in B. subtilis (29,59,82).
In the case of the newly identified TA pairs in C. difficile, the existence of a long complementary region for TA convergent transcripts could suggest an RNA degradation mechanism for antitoxin action (29). However, our results of transcript half-life measurements show only moderate changes in antitoxin RNA RCd9/RCd10 degradation when RNase III, RNase J or RNase Y were depleted. A slowing down of the degradation process after prolonged rifampicin treatment and an accumulation of longer RNA species were observed in the strains depleted for these ribonucleases. For the rather stable toxin CD2907.1/CD0956.2 mRNA, we observed a further stabilization in the strain depleted for RNase III and an even stronger effect of RNase J and RNase Y depletion on toxin mRNA half-life. We show that TA duplexes could serve as a substrate for efficient degradation by E. coli RNase III in vitro. Hfq depletion does not affect the TA RNA stability with only slight decrease in RCd9/RCd10 antitoxin half-life. Currently, only one type I TA ralR/RalA system in E. coli requires Hfq for antitoxin function (29,34). No need for Hfq for antitoxin control mechanism was reported in B. subtilis (59); however, TA interaction regions were associated with Hfq in co-immunoprecipitation experiments in this bacterium (83).
The present study demonstrates the unique co-localization of the type I TA modules with CRISPR arrays in the bacterial chromosome. Our large genome analysis revealed that this physical genomic link between TA pairs and CRISPR arrays can be extended to the majority of sequenced C. difficile strains. Initially, TA systems were shown to be important for maintenance of plasmids through a post-segregation killing mechanism (20,21,84). The role of numerous chromosomal TA systems remains largely enigmatic, even though their possible implication in stabilization of chromosomal regions has been emphasized. For example, a TA module has been shown to promote the maintenance of an integrative conjugative element STX in V. cholerae (85).
The co-localization of functional type I TA systems with CRISPR arrays that we observed on C. difficile chromosome has never been reported for any other bacterial genome. Nevertheless, several type I TA systems are located within prophage or prophage-like regions both in C. difficile and B. subtilis (29,57,59,64,82,86), even though B. subtilis genome lacks CRISPR arrays (2). The type I TA modules are present within the skin element, which is excised from the chromosome during sporulation, in B. subtilis and C. difficile. As for B. subtilis systems, a role in stabilisation of these chromosomal regions can be hypothesized for TA systems in C. difficile, which carries a high proportion of stable mobile genetic elements in its genome (87).
Based on the observations that prophage-located CRISPR arrays are often associated with type I TA modules in C. difficile, an interesting evolutionary aspect of the C. difficile CRISPR-Cas system can be underlined. Indeed, the TA systems could contribute to the stabilization of the chromosomal regions carrying CRISPR-Cas systems after acquisition of large defense capacities associated with CRISPR arrays. We can hypothesize that TA modules are implicated in maintaining of CRISPR regions, but also in stress response, prophage stability, sporulation control, biofilm formation and other community-associated processes important for this pathogen.
Possible connections between CRISPR and TA systems were highlighted by several recent studies focusing on type II TA (88). Bioinformatics search identified the so-called ‘defense islands’ in bacteria associating immunity and cell death or dormancy functions including CRISPR and type II TA systems (89,90). The original features of C. difficile are that type I toxins were not found in ‘defense islands’. The role of this functional coupling might be the induction of dormancy state in infected or stressed cells to allow the activation of adaptive immunity or specific stress responses. Dormancy was suggested to be a strategy of the last resort when the defense strategies fail face of invaders. Thus, our findings are in line with recently emerged concept on a functional coupling between distinct defense strategies provided by immunity and cell dormancy systems in prokaryotes (88).
The co-regulation of CRISPR-Cas and newly described type I TA systems by the stress-specific factor, Sigma B and the biofilm-related stimuli further suggests the possible connections between these systems in C. difficile. Our findings emphasize additional original features of the recently characterized C. difficile CRISPR-Cas system including the link with community-behavior control, stress response and type I TA systems. Such control of CRISPR-Cas expression in response to stress-related factors could be relevant for the C. difficile infection cycle.
Together with alternative roles of CRISPR-Cas in the control of bacterial physiology and pathogenesis beyond the role in defense against foreign invaders (91,92), stimuli and mechanisms controlling CRISPR-Cas system expression just start to be uncovered. However, multiple connections between TA systems in bacteria and stress response have been reported (22,25). We provide here new data on the co-regulation of type I TA and CRISPR-Cas systems by the general stress response Sigma B factor in C. difficile. Sigma B likely plays a crucial role in the responses to stresses encountered by this pathogen inside the host. Interestingly, the MazEF type II TA module is encoded within the sigB operon in S. aureus with possible regulatory connections (93). Various environmental stimuli including metabolic and genotoxic stresses induce TA gene expression of type I TA systems in B. subtilis, E. coli and S. aureus (29,70,86,94–96). In a multi-stress responsive type I TA system bsrE/SR5 from B. subtilis, the control of antitoxin RNA SR5 by iron limitation stress has been reported to be dependent on the alternative Sigma B factor (82).
Key roles of both type II and type I TA systems have been suggested in bacterial pathogens where they can contribute to virulence, fitness inside the host, persistence, intracellular lifestyle, stress response and biofilm formation (26,27,97,98). More generally, biofilm formation process has been associated in previous studies with bacterial TA systems (26). Recent data suggest that the TxpA type I toxin from the skin element acts to eliminate defective cells and preserve symmetry in B. subtilis biofilms (99). We show here that both the expression of the CRISPR-Cas and the associated TA systems are induced in biofilm conditions in C. difficile. In general, TA systems including well-documented type II TA exist in surprisingly high numbers in all prokaryotes but clostridial TA modules have been only poorly characterized so far. Before this study, no data were available on TA modules in C. difficile with the exception of the recently identified MazEF, a type II TA system member (100). Possible implications of type II TA modules in recurrent C. difficile infection, sporulation and biofilm formation were recently discussed (101). Among the most challenging aspects of C. difficile-associated disease remain the high incidence of recurrent infections and the ability of transition from inert colonization to active infection (102,103). A comparative genomic study showed that the genomes of most dangerous epidemic bacteria are characterized by the accumulation of TA modules (97). Promising perspectives for the applications of TA and CRISPR as a basis for the development of new anti-bacterial strategies could be examined in the future (27,104).
In conclusion, this study provides the first characterization of type I TA modules in the emergent enteropathogen C. difficile. Intriguingly, these chromosomal TA pairs are co-localized with CRISPR array components of bacterial adaptive immunity defense system CRISPR-Cas in the majority of sequenced C. difficile strains. Further investigations will help to precise the biological functions of these widespread chromosomal TA loci for C. difficile physiology and its successful development inside the host, to uncover the molecular mechanisms involved in their regulation and the possible crosstalk between homologous systems, as well as to evaluate their potential for future therapeutic and biotechnological applications in pathogenic bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to J.-M. Ghigo for microfermentor biofilm experiment and L.-C. Fortier for helpful discussions. We thank H. Putzer and C. Condon for anti-RNase J, anti-RNase Y and anti-RNase III antibodies. A.M. research was performed in fulfilment of educational requirements of a dual degree Skoltech-University Paris-Diderot Ph.D. program supported by the Vernadski Fellowship from the French Embassy in Russia and is also partly supported by Skoltech Biomedical Initiative Program grant (SBI RF-0000000136) to Konstantin Severinov.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut Pasteur; University Paris Diderot; Agence Nationale de la Recherche [‘CloSTARn’, ANR-13-JSV3-0005-01 to O.S.]; Institut Universitaire de France (IUF to O.S.); Pasteur-Weizmann Council; University Paris-Sud; Institute for Integrative Biology of the Cell; Vernadski Fellowship (to A.M.); Skoltech Biomedical Initiative [SBI RF-0000000136]; Centre National de la Recherche Scientifique [UMR8261]; ‘Initiative d’Excellence’ Program from the French State [Grant ‘DYNAMO’, ANR-11-LABX-0011 to E.H.]. Funding for open access charge: IUF, Skoltech.
Conflict of interest statement. None declared.
REFERENCES
- 1. Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 2. Grissa I., Vergnaud G., Pourcel C.. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007; 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 4. Bhaya D., Davison M., Barrangou R.. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011; 45:273–297. [DOI] [PubMed] [Google Scholar]
- 5. Amitai G., Sorek R.. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 6. Lopez-Sanchez M.J., Sauvage E., Da Cunha V., Clermont D., Ratsima Hariniaina E., Gonzalez-Zorn B., Poyart C., Rosinski-Chupin I., Glaser P.. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012; 85:1057–1071. [DOI] [PubMed] [Google Scholar]
- 7. Mick E., Stern A., Sorek R.. Holding a grudge: persisting anti-phage CRISPR immunity in multiple human gut microbiomes. RNA Biol. 2013; 10:900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stern A., Mick E., Tirosh I., Sagy O., Sorek R.. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 2012; 22:1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carroll K.C., Bartlett J.G.. Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu. Rev. Microbiol. 2011; 65:501–521. [DOI] [PubMed] [Google Scholar]
- 10. Rupnik M., Wilcox M.H., Gerding D.N.. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009; 7:526–536. [DOI] [PubMed] [Google Scholar]
- 11. Seekatz A.M., Young V.B.. Clostridium difficile and the microbiota. J. Clin. Investig. 2014; 124:4182–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K.. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995; 375:500–503. [DOI] [PubMed] [Google Scholar]
- 13. Vedantam G., Clark A., Chu M., McQuade R., Mallozzi M., Viswanathan V.. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012; 3:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner E.G.H., Romby P.. Chapter three-small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 15. Soutourina O.A., Monot M., Boudry P., Saujet L., Pichon C., Sismeiro O., Semenova E., Severinov K., Le Bouguenec C., Coppée J.-Y.. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013; 9:e1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boudry P., Semenova E., Monot M., Datsenko K.A., Lopatina A., Sekulovic O., Ospina-Bedoya M., Fortier L.C., Severinov K., Dupuy B. et al. Function of the CRISPR-Cas system of the human pathogen Clostridium difficile. Mbio. 2015; 6:e01112–e01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andersen J.M., Shoup M., Robinson C., Britton R., Olsen K.E., Barrangou R.. CRISPR diversity and microevolution in Clostridium difficile. Genome Biol. Evol. 2016; 8:2841–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hargreaves K.R., Flores C.O., Lawley T.D., Clokie M.R.. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. Mbio. 2014; 5:e01045-01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sorek R., Kunin V., Hugenholtz P.. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008; 6:181–186. [DOI] [PubMed] [Google Scholar]
- 20. Page R., Peti W.. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016; 12:208–214. [DOI] [PubMed] [Google Scholar]
- 21. Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003; 301:1496–1499. [DOI] [PubMed] [Google Scholar]
- 22. Gerdes K., Christensen S.K., Lobner-Olesen A.. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005; 3:371–382. [DOI] [PubMed] [Google Scholar]
- 23. Gerdes K., Maisonneuve E.. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012; 66:103–123. [DOI] [PubMed] [Google Scholar]
- 24. Maisonneuve E., Castro-Camargo M., Gerdes K.. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013; 154:1140–1150. [DOI] [PubMed] [Google Scholar]
- 25. Wang X., Wood T.K.. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011; 77:5577–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen Y., Behiels E., Devreese B.. Toxin-antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014; 70:240–249. [DOI] [PubMed] [Google Scholar]
- 27. Yamaguchi Y., Inouye M.. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011; 9:779–790. [DOI] [PubMed] [Google Scholar]
- 28. Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012; 9:1488–1490. [DOI] [PubMed] [Google Scholar]
- 29. Brantl S., Jahn N.. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015; 39:413–427. [DOI] [PubMed] [Google Scholar]
- 30. Coray D.S., Wheeler N.E., Heinemann J.A., Gardner P.P.. Why so narrow: distribution of anti-sense regulated, type I toxin-antitoxin systems compared with type II and type III systems. RNA Biol. 2017; 14:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nogueira T., Rankin D.J., Touchon M., Taddei F., Brown S.P., Rocha E.P.. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr. Biol. 2009; 19:1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fozo E.M., Makarova K.S., Shabalina S.A., Yutin N., Koonin E.V., Storz G.. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010; 38:3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brielle R., Pinel-Marie M.L., Felden B.. Linking bacterial type I toxins with their actions. Curr. Opin. Microbiol. 2016; 30:114–121. [DOI] [PubMed] [Google Scholar]
- 34. Guo Y., Quiroga C., Chen Q., McAnulty M.J., Benedik M.J., Wood T.K., Wang X.. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014; 42:6448–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jahn N., Brantl S., Strahl H.. Against the mainstream: the membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015; 98:651–666. [DOI] [PubMed] [Google Scholar]
- 36. Dupuy B., Sonenshein A.L.. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 1998; 27:107–120. [DOI] [PubMed] [Google Scholar]
- 37. Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951; 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fagan R.P., Fairweather N.F.. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 2011; 286:27483–27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J., Fritsch E.F., Maniatis T.. Molecular Cloning: a Laboratory Manual, Second Edition. 1989; NY: Cold Spring Harbor Laboratory [Google Scholar]
- 40. O’Connor J.R., Lyras D., Farrow K.A., Adams V., Powell D.R., Hinds J., Cheung J.K., Rood J.I.. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 2006; 61:1335–1351. [DOI] [PubMed] [Google Scholar]
- 41. Collins T.J. ImageJ for microscopy. Biotechniques. 2007; 43:25–30. [DOI] [PubMed] [Google Scholar]
- 42. Andre G., Even S., Putzer H., Burguiere P., Croux C., Danchin A., Martin-Verstraete I., Soutourina O.. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008; 36:5955–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghigo J.M. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001; 412:442–445. [DOI] [PubMed] [Google Scholar]
- 44. Saujet L., Monot M., Dupuy B., Soutourina O., Martin-Verstraete I.. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 2011; 193:3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 46. Folichon M., Allemand F., Regnier P., Hajnsdorf E.. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005; 272:454–463. [DOI] [PubMed] [Google Scholar]
- 47. Hajnsdorf E., Regnier P.. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peltier J., Shaw H.A., Couchman E.C., Dawson L.F., Yu L., Choudhary J.S., Kaever V., Wren B.W., Fairweather N.F.. Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease-mediated cleavage. J. Biol. Chem. 2015; 290:24453–24469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boudry P., Gracia C., Monot M., Caillet J., Saujet L., Hajnsdorf E., Dupuy B., Martin-Verstraete I., Soutourina O.. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J. Bacteriol. 2014; 196:3234–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cairns M.D., Preston M.D., Hall C.L., Gerding D.N., Hawkey P.M., Kato H., Kim H., Kuijper E.J., Lawley T.D., Pituch H. et al. Comparative genome analysis and global phylogeny of the toxin variant Clostridium difficile PCR ribotype 017 reveals the evolution of two independent sublineages. J. Clin. Microbiol. 2017; 55:865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 53. Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J.. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011; 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 55. Pinel-Marie M.L., Brielle R., Felden B.. Dual toxic-peptide-coding Staphylococcus aureus RNA under antisense regulation targets host cells and bacterial rivals unequally. Cell Rep. 2014; 7:424–435. [DOI] [PubMed] [Google Scholar]
- 56. Sayed N., Jousselin A., Felden B.. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 2011; 19:105–112. [DOI] [PubMed] [Google Scholar]
- 57. Silvaggi J.M., Perkins J.B., Losick R.. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005; 187:6641–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Serrano M., Kint N., Pereira F.C., Saujet L., Boudry P., Dupuy B., Henriques A.O., Martin-Verstraete I.. A recombination directionality factor controls the cell type-specific activation of sigmaK and the fidelity of spore development in Clostridium difficile. PLoS Genet. 2016; 12:e1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durand S., Jahn N., Condon C., Brantl S.. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012; 9:1491–1497. [DOI] [PubMed] [Google Scholar]
- 60. Jahn N., Brantl S.. One antitoxin–two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013; 41:9870–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wen J., Fozo E.M.. sRNA antitoxins: more than one way to repress a toxin. Toxins. 2014; 6:2310–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wagner E.G., Blomberg P., Nordstrom K.. Replication control in plasmid R1: duplex formation between the antisense RNA, CopA, and its target, CopT, is not required for inhibition of RepA synthesis. EMBO J. 1992; 11:1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Patel S., Weaver K.E.. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J. Bacteriol. 2006; 188:5374–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Durand S., Gilet L., Condon C.. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012; 8:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dembek M., Barquist L., Boinett C.J., Cain A.K., Mayho M., Lawley T.D., Fairweather N.F., Fagan R.P.. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. Mbio. 2015; 6:e02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kint N., Janoir C., Monot M., Hoys S., Soutourina O., Dupuy B., Martin-Verstraete I.. The alternative sigma factor sigmaB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ. Microbiol. 2017; 19:1933–1958. [DOI] [PubMed] [Google Scholar]
- 67. Domka J., Lee J., Bansal T., Wood T.K.. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 2007; 9:332–346. [DOI] [PubMed] [Google Scholar]
- 68. Maldarelli G.A., Piepenbrink K.H., Scott A.J., Freiberg J.A., Song Y., Achermann Y., Ernst R.K., Shirtliff M.E., Sundberg E.J., Donnenberg M.S. et al. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog. Dis. 2016; 74:doi:10.1093/femspd/ftw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Monot M., Orgeur M., Camiade E., Brehier C., Dupuy B.. COV2HTML: a visualization and analysis tool of bacterial next generation sequencing (NGS) data for postgenomics life scientists. OMICS. 2014; 18:184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sayed N., Nonin-Lecomte S., Rety S., Felden B.. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J. Biol. Chem. 2012; 287:43454–43463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fozo E.M. New type I toxin-antitoxin families from ‘wild’ and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012; 9:1504–1512. [DOI] [PubMed] [Google Scholar]
- 72. Fozo E.M., Kawano M., Fontaine F., Kaya Y., Mendieta K.S., Jones K.L., Ocampo A., Rudd K.E., Storz G.. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008; 70:1076–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Han K., Kim K.S., Bak G., Park H., Lee Y.. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 2010; 38:5851–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wen J., Won D., Fozo E.M.. The ZorO-OrzO type I toxin-antitoxin locus: repression by the OrzO antitoxin. Nucleic Acids Res. 2014; 42:1930–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iqbal N., Guerout A.M., Krin E., Le Roux F., Mazel D.. Comprehensive functional analysis of the 18 Vibrio cholerae N16961 toxin-antitoxin systems substantiates their role in stabilizing the superintegron. J. Bacteriol. 2015; 197:2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arnion H., Korkut D.N., Masachis Gelo S., Chabas S., Reignier J., Iost I., Darfeuille F.. Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: the importance of mRNA folding in controlling toxin expression. Nucleic Acids Res. 2017; 45:4782–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meissner C., Jahn N., Brantl S.. In vitro characterization of the type I toxin-antitoxin system bsrE/SR5 from Bacillus subtilis. J. Biol. Chem. 2016; 291:560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wen J., Harp J.R., Fozo E.M.. The 5′ UTR of the type I toxin ZorO can both inhibit and enhance translation. Nucleic Acids Res. 2017; 45:4006–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berghoff B.A., Hoekzema M., Aulbach L., Wagner E.G.. Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol. Microbiol. 2017; 103:1020–1033. [DOI] [PubMed] [Google Scholar]
- 80. Darfeuille F., Unoson C., Vogel J., Wagner E.G.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 81. Gerdes K., Gultyaev A.P., Franch T., Pedersen K., Mikkelsen N.D.. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997; 31:1–31. [DOI] [PubMed] [Google Scholar]
- 82. Muller P., Jahn N., Ring C., Maiwald C., Neubert R., Meissner C., Brantl S.. A multistress responsive type I toxin-antitoxin system: bsrE/SR5 from the B. subtilis chromosome. RNA Biol. 2016; 13:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dambach M., Irnov I., Winkler W.C.. Association of RNAs with Bacillus subtilis Hfq. PLoS One. 2013; 8:e55156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shen Z., Patil R.D., Sahin O., Wu Z., Pu X.Y., Dai L., Plummer P.J., Yaeger M.J., Zhang Q.. Identification and functional analysis of two toxin-antitoxin systems in Campylobacter jejuni. Mol. Microbiol. 2016; 101:909–923. [DOI] [PubMed] [Google Scholar]
- 85. Wozniak R.A., Fouts D.E., Spagnoletti M., Colombo M.M., Ceccarelli D., Garriss G., Dery C., Burrus V., Waldor M.K.. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009; 5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jahn N., Preis H., Wiedemann C., Brantl S.. BsrG/SR4 from Bacillus subtilis–the first temperature-dependent type I toxin-antitoxin system. Mol. Microbiol. 2012; 83:579–598. [DOI] [PubMed] [Google Scholar]
- 87. Sebaihia M., Wren B.W., Mullany P., Fairweather N.F., Minton N., Stabler R., Thomson N.R., Roberts A.P., Cerdeno-Tarraga A.M., Wang H. et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006; 38:779–786. [DOI] [PubMed] [Google Scholar]
- 88. Koonin E.V., Zhang F.. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. Bioessays. 2017; 39:1–9. [DOI] [PubMed] [Google Scholar]
- 89. Makarova K.S., Wolf Y.I., Koonin E.V.. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013; 41:4360–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Makarova K.S., Wolf Y.I., Snir S., Koonin E.V.. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 2011; 193:6039–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Richter C., Chang J.T., Fineran P.C.. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems. Viruses. 2012; 4:2291–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sampson T.R., Weiss D.S.. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog. 2013; 9:e1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Donegan N.P., Cheung A.L.. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009; 191:2795–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jahn N., Brantl S.. Heat-shock-induced refolding entails rapid degradation of bsrG toxin mRNA by RNases Y and J1. Microbiology. 2016; 162:590–599. [DOI] [PubMed] [Google Scholar]
- 95. Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012; 9:1520–1527. [DOI] [PubMed] [Google Scholar]
- 96. Kawano M., Aravind L., Storz G.. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007; 64:738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Georgiades K., Raoult D.. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS One. 2011; 6:e17962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lobato-Marquez D., Moreno-Cordoba I., Figueroa V., Diaz-Orejas R., Garcia-del Portillo F.. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 2015; 5:9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bloom-Ackermann Z., Steinberg N., Rosenberg G., Oppenheimer-Shaanan Y., Pollack D., Ely S., Storzi N., Levy A., Kolodkin-Gal I.. Toxin-antitoxin systems eliminate defective cells and preserve symmetry in Bacillus subtilis biofilms. Environ. Microbiol. 2016; 18:5032–5047. [DOI] [PubMed] [Google Scholar]
- 100. Rothenbacher F.P., Suzuki M., Hurley J.M., Montville T.J., Kirn T.J., Ouyang M., Woychik N.A.. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012; 194:3464–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gil F., Pizarro-Guajardo M., Alvarez R., Garavaglia M., Paredes-Sabja D.. Clostridium difficile recurrent infection: possible implication of TA systems. Future Microbiol. 2015; 10:1649–1657. [DOI] [PubMed] [Google Scholar]
- 102. Shields K., Araujo-Castillo R.V., Theethira T.G., Alonso C.D., Kelly C.P.. Recurrent Clostridium difficile infection: From colonization to cure. Anaerobe. 2015; 34:59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Smits W.K., Lyras D., Lacy D.B., Wilcox M.H., Kuijper E.J.. Clostridium difficile infection. Nat. Rev. Dis. Primers. 2016; 2:16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lee K.Y., Lee B.J.. Structure, biology, and therapeutic application of toxin-antitoxin systems in pathogenic bacteria. Toxins. 2016; 8:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.