Abstract
Mutations in SNX14 cause the autosomal recessive cerebellar ataxia 20 (SCAR20). Mutations generally result in loss of protein although several coding region deletions have also been reported. Patient-derived fibroblasts show disrupted autophagy, but the precise function of SNX14 is unknown. The yeast homolog, Mdm1, functions in endoplasmic reticulum (ER)-lysosome/vacuole inter-organelle tethering, but functional conservation in mammals is still required. Here, we show that loss of SNX14 alters but does not block autophagic flux. In addition, we find that SNX14 is an ER-associated protein that functions in neutral lipid homeostasis and inter-organelle crosstalk. SNX14 requires its N-terminal transmembrane helices for ER localization, while the Phox homology (PX) domain is dispensable for subcellular localization. Both SNX14-mutant fibroblasts and SNX14KO HEK293 cells accumulate aberrant cytoplasmic vacuoles, suggesting defects in endolysosomal homeostasis. However, ER-late endosome/lysosome contact sites are maintained in SNX14KO cells, indicating that it is not a prerequisite for ER-endolysosomal tethering. Further investigation of SNX14- deficiency indicates general defects in neutral lipid metabolism. SNX14KO cells display distinct perinuclear accumulation of filipin in LAMP1-positive lysosomal structures indicating cholesterol accumulation. Consistent with this, SNX14KO cells display a slight but detectable decrease in cholesterol ester levels, which is exacerbated with U18666A. Finally, SNX14 associates with ER-derived lipid droplets (LD) following oleate treatment, indicating a role in ER-LD crosstalk. We therefore identify an important role for SNX14 in neutral lipid homeostasis between the ER, lysosomes and LDs that may provide an early intervention target to alleviate the clinical symptoms of SCAR20.
Introduction
Spinocerebellar Ataxia, Autosomal Recessive 20 (SCAR20; OMIM 616354) is a distinct cerebellar ataxia that is caused by mutations in the gene encoding Sorting Nexin 14 (SNX14) (1,2). To date, at least 45 individuals from 24 families have been reported, with 18 different point mutations or deletions, with most resulting in either truncation or complete loss of the SNX14 protein (1–6) (Fig. 1A). SCAR20 is characterized by a progressive early onset cerebellar hypotrophy leading to ataxia with severe intellectual disability, with many patients exhibiting relative macrocephaly, progressively coarsening facial features and absent speech. Additional features such as epilepsy, deafness and skeletal abnormalities have also been reported in some individuals (1,2,7). A recessive splice donor mutation has been reported in the Hungarian Vizsla dog breed, which also presented with a progressive cerebellar ataxia, which was associated with primary Purkinje neuronal degeneration and loss (8).
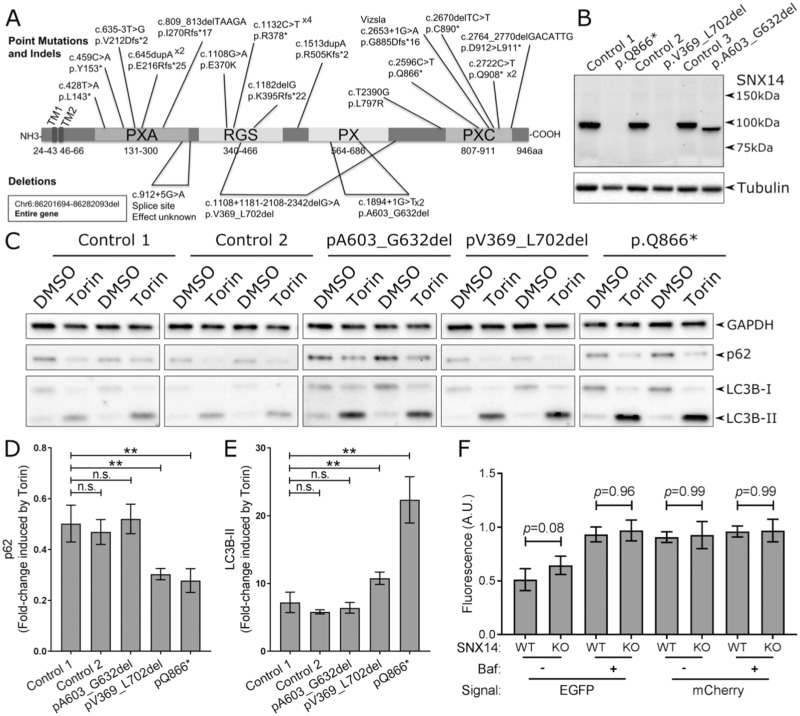
Figure 1.
SNX14 mutations enhance response to autophagy induction. (A) Mutations in the SNX14 gene reported to cause SCAR20. (B) SNX14 protein expression in control and SCAR20 patient dermal fibroblasts. Mutations in the SNX14 gene result in either no detectable SNX14 protein (c2596C > T; p.Gln866* and c.1108 + 1181_2108–2342del; p.Val369_Leu702del) or a truncated SNX14ΔPX protein (c1894 + 1G > T; p.A603_G632del). (C) Control and SCAR20 patient-derived fibroblasts were cultured for 6 h with or without autophagy inducing conditions (250 nm Torin1 or 0.1% DMSO) which affect levels of p62 and LC3B-II. (D) Fold-change of p62 expression in response to Torin1 treatment. (E) Fold-change of LC3B-II expression in response to Torin1 treatment. (D, E) n = 6, error bars = SEM, **P ≤ 0.01, ns P ≥ 0.05, Student’s t-test. (F) Quantification of EGFP and mCherry fluorescence intensity in SNX14WT and SNX14KO HEK293 cells cultured under autophagy inducing conditions with or without inhibition of lysosome-autophagosome fusion with bafilomycin A (50 nm, 6 h) treatment. N = 4–7 (Cells), error bars = STD, **P ≤ 0.01, ANOVA.
SNX14 belongs to the Sorting Nexin SNX-RGS family of Phox homology (PX)-domain containing proteins consisting of SNX13, SNX14, SNX19 and SNX25 (9,10). In addition to the PX domain, SNX13, SNX14 and SNX25 also have a regulator of G-protein signalling (RGS) domain. SNX-RGS members also contain two hydrophobic N-terminal predicted transmembrane helices, along with ‘PXA’ and ‘PXC/C-Nexin-C’ domains that are proximally located towards the N- and C-terminals, respectively (11). SNX proteins generally localize to endosomal membranes via the PX domain, which binds to phosphoinositides (PtdIns), which are maintained on the endosome surface (9,10). Interestingly, one SNX14 splice donor mutation identified in two unrelated families (1,6) results in exon skipping that removes 30 amino acids of the PX domain, suggesting it is essential for SNX14 function. However, recent studies reveal that the SNX14 PX domain manifests weak or negligible affinity for PtdIns (2,11,12), creating uncertainty as to the precise role of this domain.
Observations of SCAR20 patient-derived fibroblasts in vitro and to a lesser degree in skin biopsies revealed an excessive accumulation of cytoplasmic vacuoles containing electron dense material (1). A later study showed accumulation of vacuoles in neural precursor cells generated from SNX14 patient induced pluripotent stem cells (2). In the latter study, the authors reported an engorgement of lysosomes and showed an elevation of LC3B-II (indicative of autophagosome membranes) during autophagy induction. Other PX domain proteins such as SNX18 and HS1BP3 have been reported to positively and negatively regulate autophagy, respectively (13,14). In a siRNA screen targeting PX domain proteins in GFP-LC3 expressing HEK293 cells in nutrient depleted medium, SNX14 was also identified. However, they focused on SNX18 and HS1BP3 since the impact of SNX14 knockdown could not be validated in a secondary screen (13,14). Over-expression of SNX14 has been shown to positively regulate the degradation of 5-HT6R (Serotonin/5-Hydroxytriptamine subtype 6 receptor) (12). This appears to be mediated by autophagy as inhibition with chloroquine blocked this effect. Collectively, these results implicate a possible role for SNX14 in autophagy.
The yeast SNX14 homolog Mdm1 was recently found to have an unexpected role in inter-organelle crosstalk (15). Mdm1 functions as a molecular tether, physically bridging two organelles: the endoplasmic reticulum (ER) and yeast vacuole, the latter being equivalent to mammalian lysosomes/late endosomes (LE) (15). Indeed, the importance of ER-endolysosomal communication has become increasingly well established for its essential role in normal metabolism and disease (16). More recently Mdm1 was found to function in lipid metabolism at ER-vacuole/lysosome junctions, where it associates with lipid droplets (LDs) that cluster at the NVJ prior to their autophagy-mediated degradation in the vacuole/lysosome (16). Furthermore, yeast ER-vacuole/lysosome contact sites (also denoted as nuclear ER-vacuole junctions, NVJs) serve as sites for a specialized form of autophagy known as piecemeal autophagy of the nucleus (PMN) (17). Although Mdm1 was not required for functional PMN (15), its presence at ER-lysosome junctions implied a potential role for SNX14 in autophagy, and a potential conserved function for SNX14 in inter-organelle communication.
Henne et al. (2015) demonstrated that overexpression of mdm1 mutants either containing point mutations (analogous to disease causing SNX14 alleles) or completely lacking the PX domain mis-localize throughout the ER network (1,15), and disrupt the contact site between the yeast vacuole/lysosome and the nuclear ER. In addition, over-expression of these loss-of-function mdm1 mutants resulted in intracellular pathology, as manifested by a hyper-sensitivity to the sphingolipid synthesis inhibitor myriocin. Since the ER and/or lysosome are major sites of lipid synthesis and homeostasis for the cell, this led to speculation that SNX14 mutations might interfere with normal ER-endolysosomal inter-organelle communication and normal lipid metabolism. Indeed, this is not unlike the events noted for other neurological diseases such as Niemann–Pick type C (NPC; OMIM 257220), which manifest defects in cholesterol homeostasis between the ER and lysosomes. Like SCAR20, in Niemann Pick Type C there is also Purkinje cell loss (18), implying a common pathological mechanism.`
Further evidence for a potential role for SNX14 homologs in lipid and or cholesterol metabolism comes from Snazarus (Snz), the Drosophila ortholog of Mdm1, which was identified in a fat body enhancer trap screen associated with longevity testing (19). Snz is strongly expressed in the fly fat body, an important organ in fly lipid metabolism. Also, in a study designed to identify regulators of low-density lipoprotein (LDL)-cholesterol transport, SNX14 was identified as a gene linked to cholesterol transport out of the endolysosomal pathway (20). These data provide preliminary evidence that SNX14 may share functionally similar roles with Mdm1 in regulating lipid metabolism and/or transport. This is consistent with the finding accumulation of lipids (observed as electron dense material) in autophagic vacuoles (1) seen in SCAR20 patient-derived fibroblasts and suggests a potential defect in the normal intracellular lipid distribution.
To better understand the role of mammalian SNX14 in cellular physiology and resulting loss of function in patients, we set out to: (a) investigate the implied role of SNX14 in autophagy dysfunction; (b) identify the location of SNX14 in mammalian cells relative to subcellular membrane networks and (c) dissect any potential roles for SNX14 in lipid homeostasis and/or trafficking. Here we show that, similar to yeast Mdm1, SNX14 is associated with the ER. We also find that autophagy remains intact in SNX14-deficient cells. However, SNX14-deficient cells display defects in cholesterol homeostasis, manifested by altered filipin staining in lysosomal compartments, and decreases in cholesterol-ester levels normally associated with ER-derived LDs. Consistent with this, we show that SNX14 sub-cellular distribution is dynamic, with SNX14 accumulating adjacent to LDs following oleate-stimulated LD biogenesis. Our results indicate a specific role of SNX14 in ER-LD crosstalk and sterol metabolism downstream of the endolysosomal pathway, implying an important role for SNX14 in cellular maintenance and function that is disrupted in SCAR20.
Results
Loss of SNX14 enhances the response to autophagy induction. Since previous studies (1,2) indicated that SNX14-deficient cells display defects in autophagy, we focused on characterizing the precise role of SNX14 in autophagic regulation. Autophagy can be induced by Torin1, which can then be monitored by changes in levels of LC3B-II and p62 (Supplementary Material, Fig. S1). We identified optimum conditions for Torin1 induction of human fibroblasts at a concentration of 250 nm for 6 h (Supplementary Material, Fig. S2A), whereby cytosolic LC3B-I was processed into autophagosomal membrane associated LC3B-II and p62 was degraded (Fig. 1C–E; Supplementary Material, Fig. S2B).
SCAR20 patient-derived fibroblasts with no detectable SNX14 expression (p.Q866* and p.V369_L702del) (Fig. 1B) show an increased response to Torin1-mediated autophagy induction (Fig. 1D and E). In contrast, fibroblasts derived from a patient with a PX domain-specific SNX14 truncation (p.A603_G632del) responded to Torin1 treatment at similar levels to control cell lines (Fig. 1D and E). The elevated response of LC3B-II expression in SNX14-deficient cells may indicate either an enhanced up-regulation of autophagosome formation or blockage in the autophagy pathway. However, enhanced degradation of p62 indicates unimpeded formation of autolysosomes (Fig. 1C and D). To monitor autolysosomes in SCAR20 patient fibroblasts we investigated the colocalization of the autophagosome marker Cyto-ID (cationic amphiphilic tracer dye) and the lysosomal marker Lysotracker (acidotropic probe) as previously reported (21). Consistent with the degradation of p62 in response to autophagy induction, we observe a very strong colocalization of autophagosome and lysosome associated markers indicating the efficient formation of autolysosomes in SCAR20 mutant fibroblasts (Supplementary Material, Fig. S3).
Bafilomycin A1 blocks autophagic flux by inhibiting the fusion of lysosomes with autophagosomes (22) (Supplementary Material, Fig. S1). We observed an increase in LC3B-II expression and simultaneous block of p62 degradation in response to Torin-1 treatment with the addition of Bafilomycin A1 but not chloroquine (Supplementary Material, Fig. S4). To directly monitor autophagosome-lysosome fusion we transfected SNX14KO HEK293 cells (Supplementary Material, Fig. S5) with an LC3-mCherry-EGFP expression construct that is incorporated into the autophagosome membrane (23) (Supplementary Material, Fig. S1). Following fusion of lysosomes with the autophagosome, the pH-sensitive EGFP is quenched, leaving the non-sensitive mCherry signal to indicate the formation of a functional autolysosome (Fig. 1F; Supplementary Material, Figs. S1 and S5). As expected, this effect was inhibited by Bafilomycin A1 (Fig. 1F; Supplementary Material, Figs. S1 and S5).
The formation of autolysosomes in SNX14KO cells was not statistically different from SNX14WT cells, indicating that loss of SNX14 did not impact autophagosome-lysosome fusion (Fig. 1F; Supplementary Material, Fig. S5). We note a small trend towards a reduction in the fusion of lysosomes with autophagosomes approaching statistical significance (P = 0.08), but this was not comparable to the effects of Bafilomycin A1. Additionally, SNX14 was rarely identified at puncta positive for lysosome-associated LAMP2 (Supplementary Material, Fig. S6). Therefore, we conclude that SNX14 does not have a direct role in regulating the formation of autolysosomes in the cell lines tested.
SNX14 is located on the endoplasmic reticulum. Since SNX14 appeared dispensable for general lysosome-mediated macroautophagy, we next investigated its precise sub-cellular location. Almost all SNX protein family members are soluble proteins that associate with intracellular membranes via their lipid-binding PX domains to membrane-embedded PtdIns lipids (10). In contrast, SNX14 and its other three RGS-PX homologs all contain predicted N-terminal transmembrane helices in addition to PX domains, implying that their membrane localization may be mediated through both integral membrane anchoring and PX-mediated PtdIns binding. However, recent structural studies on the SNX14 PX domain indicated that key residues necessary for PtdIns binding were altered, with key residues surrounding the PtdIns binding pocket naturally mutated so as to sterically block lipid-binding (11). Mas et al. went on to confirm that the SNX14 PX domain does not display detectable PtdIns-mediated membrane association using two complementary in vitro methods: liposome pelleting assays and nuclear magnetic resonance spectroscopy. In contrast, they found that the closely related SNX19 PX domain, which differs at the proposed binding residues, does bind to PtdIns3P (11). To investigate this further in an in vivo cellular environment, we monitored the sub-cellular localization of human SNX14 and SNX19 PX domains in living yeast. In agreement with Mas et al. (11), we found that the SNX19 PX domain clearly localizes at the PtsIns3P-containing yeast vacuole membrane, whilst the SNX14 PX domain remained in the cytoplasm (Supplementary Material, Fig. S7).
We next sought to characterize SNX14 localization in human cells via immunofluorescence imaging. Although we clearly detect SNX14 by Western blot (Fig. 1B), we were unable to detect endogenous SNX14 with immunocytochemistry using the same antibody. To circumvent this, we mildly overexpressed untagged SNX14, which we could detect with the anti-SNX14 antibody. We observed an unambiguous subcellular pattern reminiscent of the ER (Fig. 2A), which was confirmed by co-localization with the ER marker HSP90B1 (Fig. 2A–C). To define which regions of SNX14 mediate this ER localization, we generated U20S cell lines expressing full length SNX14 as well as N- and C-terminal truncations, all C-terminally FLAG-tagged (Fig. 2D). Following microsomal fractionation, full length SNX14-FLAG and the N-terminal region of SNX14 containing the predicted transmembrane region (but lacking the PX domain onward) was clearly detected in the membrane-enriched fractions (Fig. 2E). In contrast, the C-terminal region of SNX14 (containing the PX domain) was enriched in non-membrane fractions (Fig. 2E). These experiments demonstrated that membrane binding of SNX14 is primarily mediated by sequences N-terminal to the PX domain.
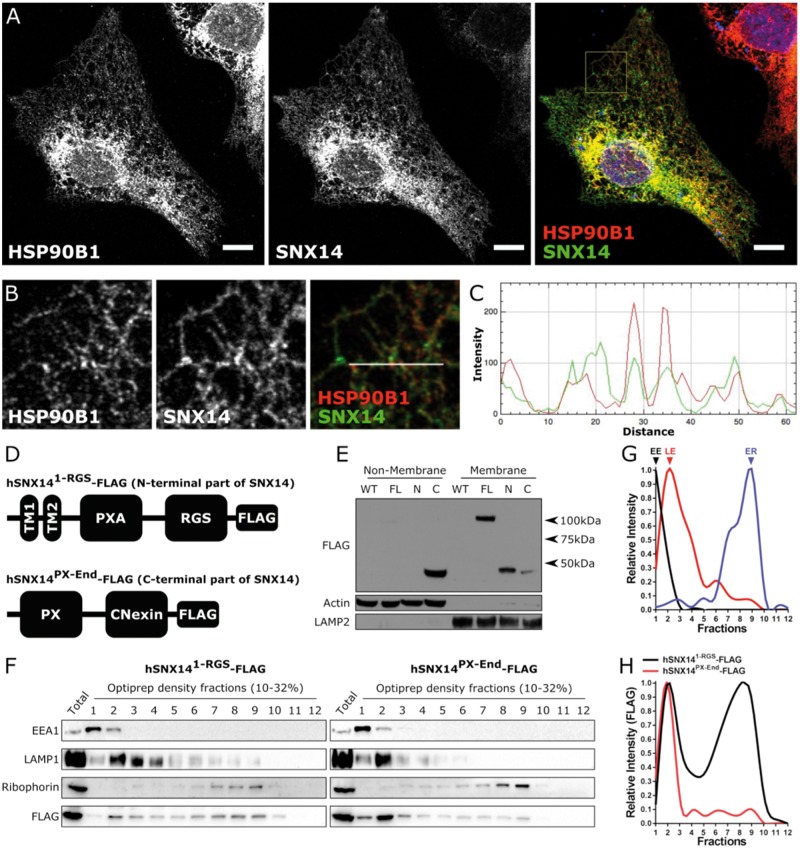
Figure 2.
SNX14 is localized to the endoplasmic reticulum membrane. (A, B) Overexpressed SNX14 in U2OS cells display a similar subcellular distribution to ER associated HSP90B1 (Scale bar = 10 µm). (C) Line plot of the region designated in (B) showing the relationship between SNX14 (Green) and HSP90B1 (Red) intensity. (D) U2OS cell lines express either N- or C-terminal segments of SNX14-FLAG. (E) The N-terminal segment of SNX14-FLAG is predominantly responsible for membrane association. (F) Organelle fractionation reveals different distributions between N- and C-terminal SNX14 segments. (G) Relative intensity of early endosome (EE) associated EEA1, late endosome (LE) associated LAMP1 and endoplasmic reticulum (ER) associated Ribophorin of hSNX141-RGS-FLAG expressing U2OS cells. (H) Relative intensity profile of N- and C-terminal segments of SNX14 show different distribution profiles.
To test our prediction that the N-terminal region of SNX14 is required for ER localization, we conducted organelle fractionation using density centrifugation. Endosomal/lysosomal organelles were enriched in lower density fractions compared to the ER, which was enriched in higher density fractions (Fig. 2F and G). The N-terminal region of SNX14 displayed enrichment in both lysosomal and ER membrane-containing fractions (Fig. 2H, black). In contrast, the C-terminal part of SNX14 was not enriched in ER fractions (Fig. 2H, red), providing further evidence that sequences in the N-terminal portion are important for its membrane/ER association.
To further dissect the sub-cellular localization of native full length SNX14, and the contribution of the PX domain, we fractionated a fibroblast cell line from one parent of a patient with SCAR20, that was heterozygous for the truncating allele SNX14ΔPX (c1894 + 1G > T; p.A603-G632del). This mutation results in an expressed but truncated protein that lacks part of the PX domain. Western blotting readily detects both full length and truncated SNX14 variants (Supplementary Material, Fig. S8A). In both parental cell lines, full length SNX14WT was expressed at a higher level than the truncated SNX14ΔPX protein (Supplementary Material, Fig. S8A). Nevertheless, both forms of SNX14 displayed a similar fractionation profile of two peaks, indicating enrichment in lower and higher density fractions (Supplementary Material, Fig. S6B). This SNX14 fractionation profile was most similar to ER-associated Calnexin, which also displays two distinct peaks in the same fractions (Supplementary Material, Fig. S8B and C). Interestingly, the lower density peak for SNX14 was not shared with the peak enrichment for any of the LE/lysosomal associated markers tested (Supplementary Material, Fig. S8B and C). Altogether, these data indicate that native SNX14 localizes primarily to the ER network and a functional PX domain is not required for this association.
SNX14 mutations disrupt intra-cellular cholesterol homeostasis. The observation that SNX14 associates with the ER membrane led us to consider what consequences might occur from its loss-of-function. The ER is the primary lipid biosynthesis organelle in eukaryotes, and the site of synthesis and homeostasis for cholesterol (24). As previously described, SCAR20 patients share some clinical similarity to patients with NPC, where cholesterol transport from the autolysosome to the plasma membrane and ER network is reported to be disrupted (25). We therefore examined the subcellular distribution of cholesterol in SCAR20 patient fibroblasts.
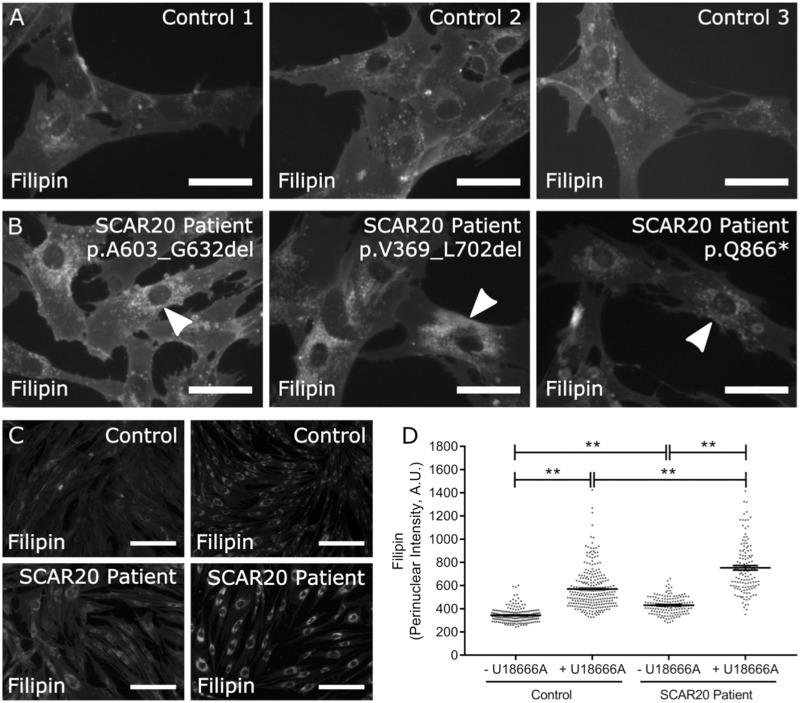
Filipin forms a complex with unesterified cholesterol allowing observation with ultraviolet excitation. In addition to labelling the cell membrane, we observed that filipin was most intense as a punctate pattern around the perinuclear region (Fig. 3A). Perinuclear filipin intensity appeared greater in SCAR20 patient fibroblasts compared to controls (Fig. 3B, arrow heads). As a positive control, we examined filipin distribution in cells treated with U18666A, which mimics the loss of NPC1 protein by inhibiting transport of cholesterol from LE and lysosomes (26,27). Again, perinuclear filipin intensity was more pronounced in SCAR20 patient cells compared to controls (Fig. 3C and D). Intriguingly, the addition of U18666A appeared to enhance the difference between patient and control cells suggesting combined loss of both NPC1 and SNX14 function was additive (Fig. 3C and D).
Figure 3.
Accumulation of cholesterol in SCAR20 patients. (A, B) Filipin staining of dermal fibroblasts derived from (A) control and (B) SCAR20 patients show increased perinuclear intensity is indicated by arrowheads (Scale bar = 50 µm). (C) Filipin stained dermal fibroblasts derived from a control and SCAR20 patient (p.Q866*) show increased perinuclear distributions of filipin intensity following 24 h of U18666A treatment (Scale bar = 200 µm). (D) Quantification of filipin intensity in the perinuclear region of each cell. Perinuclear accumulation of cholesterol in SNX14 mutant fibroblasts is exacerbated by U18666A treatment. Each dot represents a single cell, n ≥ 143, bars = mean, error bars = SEM, **P ≤ 0.01, one-way ANOVA.
Investigations on patient-derived fibroblasts were variable and limited by the small number of cell lines available. Filipin labelling has traditionally been used to diagnose patients with NPC disease, despite much heterogeneity in measurements both within and between cell lines (28,29). Therefore, we decided to generate SNX14 knockout cells using CRISPR-Cas9 on the immortalized, morphologically homogenous cell line HEK293. Clonal cell lines were selected following Western blotting to identify clones with and without SNX14 protein, which were then confirmed by DNA sequencing (Fig. 4A and B).
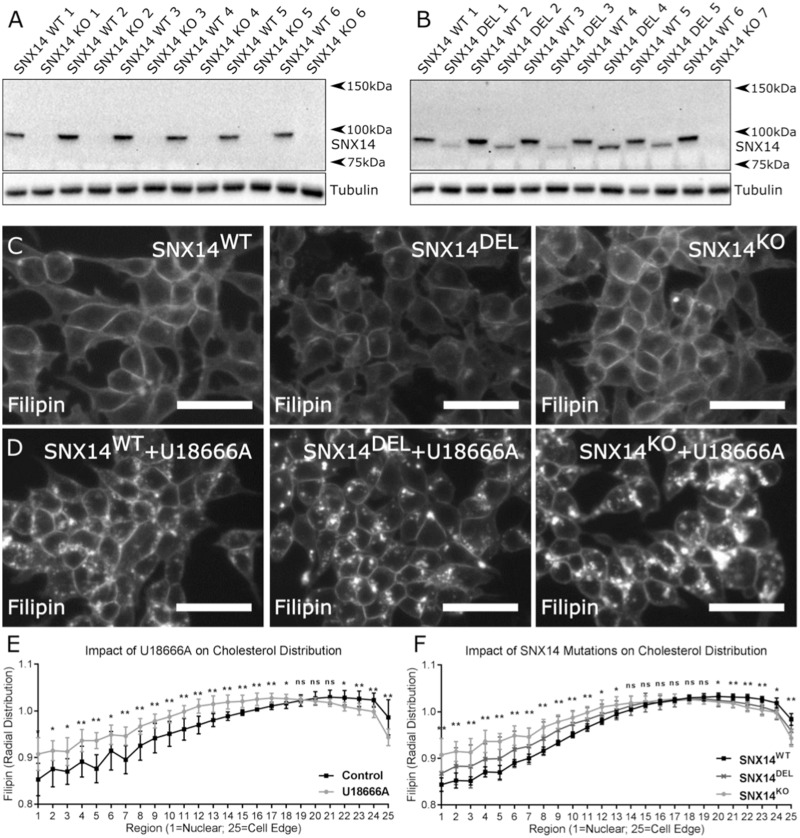
Figure 4.
Perinuclear accumulation of cholesterol in SNX14 mutant cells. SNX14 protein in cell lysates from HEK293 clones generated from single cell sorting following CRISPR-Cas9 mediated targeting of the SNX14 gene. (A, B) Clones display either full length SNX14 protein (SNX WT 1–6), no SNX14 protein (SNX14 KO 1–7) or truncated SNX14 protein (SNX14 DEL 1–5). Cells were cultured (C) with or (D) without 23.5 µm U18666A for 24 h. SNX14 mutant cells showed greater accumulation of cholesterol with U18666A treatment. (E) U18666A increased perinuclear distribution of cholesterol. (F) SNX14 mutant clones treated with U18666A displayed an increased perinuclear distribution of cholesterol compared to SNX14WT clones. (C, D) Scale bar = 50 µm. (E, F) The mean radial distribution of filipin signal intensity plotted from nuclear to peripheral regions, N = 6 (SNX14WT clones), N = 5 (SNX14DEL clones), N = 7 (SNX14KO clones), dots = mean, error bars = SD, **P ≤ 0.01, *P ≤ 0.05, n.s. P ≥ 0.05, Student’s t-test.
Since we noted that U18666A treatment differentially affected SCAR20-derived fibroblasts, we investigated whether simultaneous blocking of NPC1 and loss of SNX14 function would similarly impact SNX14KO HEK293 cells. Without U18666A, filipin staining revealed a continuous nucleus-to-periphery distribution, with lowest signal in the central/nuclear region (Fig. 4C). In contrast, treatment with U18666A resulted in an accumulation of intense filipin-positive LE/lysosome population (Fig. 4D and E; Supplementary Material, Fig. S9). This intensity was quantified from nuclear-to-peripheral regions, and showed that U18666A treatment increased filipin intensity in perinuclear regions but decreased it in peripheral regions (Fig. 4E). Comparison of untreated SNX14KO and SNX14WT cells showed no difference in their nuclear-peripheral filipin intensity distributions (Supplementary Material, Fig. S9). However, when treated with U18666A, a clear difference in the distribution profile of SNX14KO and SNX14WT cells was detected while SNX14DEL clones displayed an intermediate effect (Fig. 4F). This confirms the combinatorial effect of NPC1 and SNX14 seen in patient cells (Fig. 3).
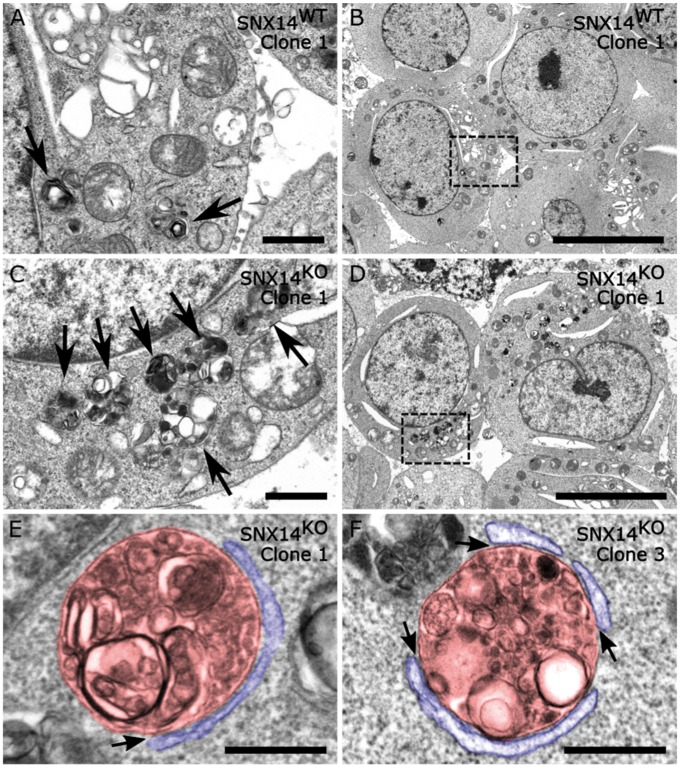
Close examination of filipin puncta in U18666A-treated conditions revealed that they were generally larger and more intense in SNX14DEL and SNX14KO cells (Fig. 4D). We used electron microscopy to further examine these structures in three SNX14WT and SNX14KO clones. Under standard culturing conditions we observed very few distinct LE/lysosomal structures (Supplementary Material, Figs. S10 and S11). In contrast, U18666A treatment resulted in a marked increase of various LE/lysosomal structures containing electron dense material (Supplementary Material, Figs. S12 and S13). This is consistent with the induction of filipin and LAMP1 positive puncta by U18666A treatment (Supplementary Material, Fig. S14). Interestingly, we observed many more clusters of LE/lysosomal structures in U18666A-treated SNX14KO cells compared to U18666A-treated SNX14WT cells (Fig. 5A–D; Supplementary Material, Figs. S12 and S13, arrows). These clusters likely reflect the accumulation of lipids and other cellular debris in response to U18666A and the resulting defects in lysosome homeostasis. Intriguingly, we identified the presence of membrane contact sites between LE/lysosomal compartments and ER membranes in SNX14KO cells, suggesting that SNX14 was not an absolute requirement for this inter-organelle interaction (Fig. 5E and F). No evidence of ER stress in SNX14KO cells was detectable (Supplementary Material, Fig. S14).
Figure 5.
Loss of SNX14 results in greater accumulation of autophagic organelles. Electron microscopy images from (A, B) SNX14WT and (C–F) SNX14KO HEK293 clones. Cell lines were cultured in with 23.5 µm U18666A for 24 h before processing. Arrows indicate autophagic structures containing undigested or partially digested material. (E, F) Examples of membrane contact sites (arrows) detected between autophagic organelles (pseudo coloured red) and the endoplasmic reticulum (pseudo coloured blue) in the absence of SNX14. (A, C) Scale bar = 1 µm; (B, D) Scale bar = 10 µm (E, F) Scale bar = 500 nm.
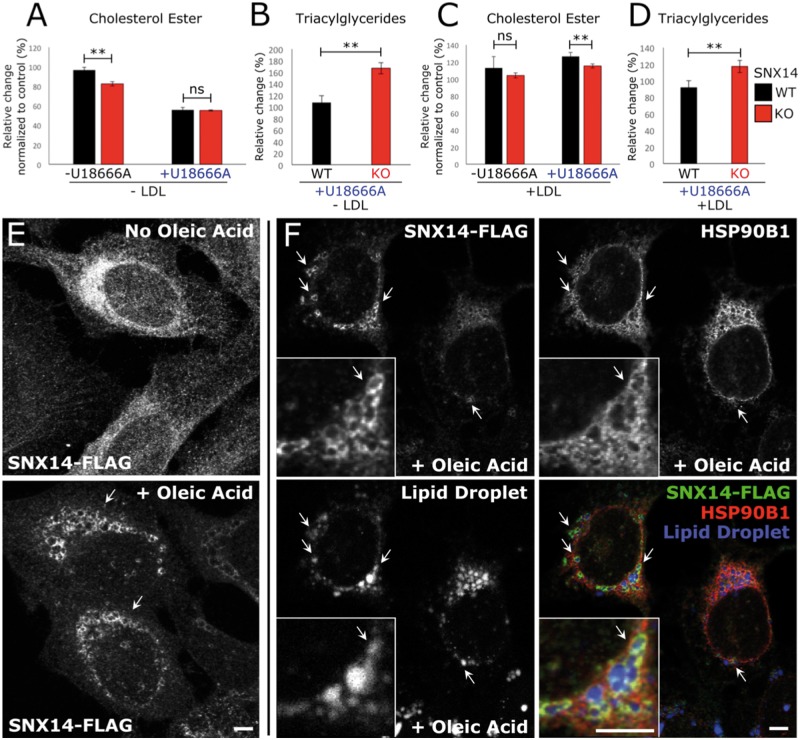
SNX14 associates with oleate-stimulated lipid droplet biogenesis. The increased filipin staining and LE/lysosome morphological defects observed in U18666A-treated SNX14-deficient cells suggested a progressive defect in cholesterol homeostasis. Once at the ER, cholesterol can either be trafficked away to other organelles, or remain and be converted to cholesterol-ester (CE) and packaged with triacylglycerides (TAG) into lipid droplets (LDs) that bud from the ER membrane. Indeed, recent reports indicate that SNX14 homolog Mdm1 functions in LD biogenesis in yeast during nutritional stress, so we next investigated whether SNX14 may function in ER-associated LD homeostasis (16). We found that SNX14 mutant cells displayed slight but significant altered levels of CE (Fig. 6A and C) and TAG (Fig. 6B and D), the two major neutral lipids found within LDs. This was in contrast to phospholipid levels that were not affected (Supplementary Material, Figs. S15 and S16). To determine whether SNX14 played any role in LD homeostasis, we added oleate to SNX14-Flag expressing cells, a condition that stimulates ER-derived LD biogenesis. Immunofluorescent staining revealed a striking redistribution of SNX14 into ring-like structures surrounding the surface of MDH-stained LDs (30) (Fig. 6E and F). Collectively, this suggested that SNX14 may function in ER-LD crosstalk and neutral lipid homeostasis between these two organelles.
Figure 6.
Loss of SNX14 disrupts endoplasmic reticulum-associated neutral lipid metabolism. (A–D) Thin layer chromatography analysis of neutral lipids in SNX14WT and SNX14KO HEK293 cultured without (A, B) or with (C, D) exogenous cholesterol (Low-density lipoprotein; LDL). (A) Cholesterol esters (CE) decreased in SNX14KO cells. (B) Triacylglycerides (TAG) increased in SNX14KO cells on addition of U18666A. (C) CEs decreased when SNX14KO cells were treated with U18666A and LDL. (D) TAG levels increased in SNX14KO cells cultured with U18666A and LDL. N = 2, error bars = SD, **P ≤ 0.01, ns P ≥ 0.05, Student’s t-test. (E) Immunofluorescent images of U2OS cells that were either untreated, or treated with 600 μm oleic acid for 8 h. SNX14-FLAG (anti-FLAG) was observed to accumulate in peri-nuclear ring-like formations following oleic acid treatment (arrows). (F) Co-fluorescent imaging of SNX14-FLAG (Green), ER marker HSP90B1 (Red) and LD stain monodansylpentane (Blue). Scale bar = 5 μm.
Discussion
Loss of SNX14 is associated with progressive cerebellar ataxia, but the cellular and molecular mechanisms underlying this disease have remained elusive (1,12). Akizu et al. (2) described an effect on autophagy induction following SNX14 loss, leading to the idea that SNX14 has a role in autophagosome-lysosome fusion. Our current study confirms that loss of SNX14 does impact autophagy induction, although we found no evidence of a general block in autophagosome-lysosome fusion. Furthermore, autophagy-mediated p62 degradation and autophagosome-lysosome function remain intact in SNX14-deficient cells, indicating that SNX14 is dispensable for general macroautophagy. This contrasts with the effect of other mutations in genes that cause a block in autophagy, which lead to increased levels of both LC3B-II and p62 (14,31). Although we could not detect a direct role for SNX14 in autophagosome-lysosome fusion, SNX14 mutations may impact on the autophagy process in other ways. For example, SNX14 may have a direct role in the formation of autophagosomes as has been reported for PX domain protein HS1BP3 (14). Alternatively, loss of SNX14 may impact autophagy as an adaptive response similar to loss of NPC1, which results in spontaneously increased autophagy (32) as well as disruption to autophagosome fusion (33). One explanation for enhanced autophagic response is that loss of SNX14 primes the ER to promote autophagosome initiation as a stress response. Indeed, we observe SNX14 to be ER-associated, and the ER is a general membrane donor for nascent autophagosomes during early stages of autophagy (34).
Instead, here we propose an alternative function for SNX14: as an ER-associated protein that functions in neutral lipid homeostasis at the ER. This is highlighted by three observations: (1) SNX14 physically localizes to ER membranes, (2) loss of SNX14 effects cholesterol distribution within sub-cellular compartments (as evidenced by filipin accumulation in lysosomes), as well as altered levels of the neutral lipids CE and TAG and (3) SNX14 associates with ER-derived LDs following oleate-stimulated LD biogenesis.
The SNX proteins are typically associated with endosomal membranes via the affinity of the PX domain for PtdIns (9,10). A previous study reported DsRed tagged SNX14 to be localized at lysosomes, and supported this finding with organelle fractionation, as well as showing that SNX14 was partially enriched in the same fractions as LE/lysosome organelles (2). However, recent work revealing the crystal structure of the SNX14 PX domain indicated that it was incapable of PtdIns binding due to the natural R656K substitution within the RYKR consensus sequence, and no appreciable membrane binding could be detected experimentally (11). Consistent with this, we show that the SNX14 PX domain also fails to localize to PtdIns membranes when expressed in yeast, which is in contrast to the SNX19 PX domain that clearly localizes to the PtsIns3P rich vacuole surface. Instead, SNX14 localizes to ER membranes via its predicted N-terminal transmembrane helices. Furthermore, we also show that a SNX14 allele with a loss-of-function mutation in its PX domain has a near identical organelle fractionation profile as wild-type SNX14, indicating that the PX domain is not critical for its sub-cellular distribution. Additionally, we did not detect a significant impact on autophagy induction in skin fibroblasts derived from the patient with the SNX14ΔPX mutation. This suggested that the SNX14 PX domain is dispensable for its role in maintaining normal autophagy and that defective autophagy is unlikely to be the fundamental cause of the clinical symptoms of SCAR20.
We provide strong evidence that, like its yeast homolog Mdm1 (15), SNX14 is an integral membrane protein that associates with ER membranes. This is supported by both immunofluorescence staining of cells, as well as organelle fractionation experiments of endogenous SNX14. Consistent with this, the N-terminal transmembrane domain region, which is necessary for yeast Mdm1 to localize to the ER, is also responsible for SNX14-ER association (15). Also similar to Mdm1, cellular fractionation indicated a partial association of SNX14 with lysosomes as well as the ER. However, we refrain from making solid conclusions on this lysosomal sub-population, as this may represent either a co-purification of ER membranes with the lysosomes (a common occurrence), or a sub-population of SNX14 that is being actively turned over via lysosomal proteases.
Despite similar ER localization topologies, several observations presented here indicate some functional divergence of mammalian SNX14 and yeast Mdm1. Nevertheless, it remains possible that SNX14 retains a weak and potentially transient association with the lysosome surface via interaction with its PX domain, or through protein-protein interactions at the lysosome surface. Indeed, the SCAR20 disease causing SNX14ΔPX mutation (1,6) that includes the first two amino acids (p.R616 and p.Y617) of the RYKR PtdIns binding motif and its differential effect on autophagy suggests that this domain has functional significance other than a generalized loss of function.
Recent reports described engorged endolysosomal structures in SCAR20 cells (1,2). We confirm this in the present study and show that these structures are positive for filipin, indicating the accumulation of unesterified cholesterol. We speculate that SCAR20 may share a similar pathological mechanism to diseases such as Niemann–Pick type C, which result from disruption to ER-endolysosomal inter-organelle communication and normal lipid metabolism. NPC is an autosomal recessive disorder characterized by accumulation of cholesterol in LE/lysosomes, and like SCAR20, affected individuals display progressive cerebellar degeneration (35). Limited post-mortem data from humans and dogs with SCAR20 indicate that Purkinje cells are particularly vulnerable to degeneration (2,8). Such pathology is similar to NPC in which a specific pattern of Purkinje cell death has been carefully characterized in a mouse model of the disease (36). The observation of unambiguous ER-LE contact sites even in SNX14KO cells suggests that SNX14 is not a requirement for tight ER-endolysosomal bridging. The formation of membrane contact sites is mediated by multiple components and this may explain their formation in the absence of SNX14 (37). Nevertheless, this does not preclude the possibility that loss of SNX14 impacts these membrane contact sites at a functional level.
We also provide evidence that SNX14 is involved in ER-localized neutral lipid metabolism. Neutral lipids CE and TAG are made at the ER, but packaged into LDs for long-term storage. In yeast, SNX14 homolog Mdm1 localizes to sites of LD biogenesis on the ER surface adjacent to the yeast vacuole, where these neutral lipids are stored prior to their eventual digestion in the vacuole during long-term starvation (38). In this capacity, Mdm1 functions to promote spatially restricted LD biogenesis at the ER-lysosome interface, and interacts with fatty acyl-CoA synthases to promote this neutral lipid storage in LDs. Similarly, here we find that SNX14 also regulates neutral lipid homeostasis at the ER, and associates with LDs when mammalian cells are treated with oleate to stimulate LD biogenesis. Thus, we propose that SNX14 functions in ER-LD homeostasis similar to its yeast ortholog.
If SNX14 regulates cholesterol and TAG homeostasis at the ER, why do we observe cholesterol accumulation in LAMP1-positive lysosomal compartments? We speculate that this accumulation could be due to either perturbing the efficient flow of lysosome-derived cholesterol from lysosome-to-ER, or potentially by even perturbing the contact sites between the ER and late LEs/lysosomes in mammalian cells. This is evidenced by SNX14KO cells displaying reduced CE and increased TAG, suggesting a decreased delivery of cholesterol to the ER for re-esterification to CE.
In summary, we find that SNX14 is an ER-resident protein that regulates neutral lipid metabolism downstream of endolysosomal lipid trafficking. SNX14 itself associates with LDs during elevated LD biogenesis, and its loss leads to defects in neutral lipid levels, as well as the redistribution of intracellular cholesterol. Consistent with this, SCAR20 causing SNX14 mutations induced the pathological accumulation of cholesterol in late endosomal/lysosomal compartments, which is further perturbed by blocking cholesterol efflux with U18666A. This may particularly impact on Purkinje cells since their size, high metabolic requirements and complex dendritic architecture make them uniquely vulnerable (39). Similar pathologies are observed in other diseases caused by defects in lysosomal cholesterol efflux (18). In contrast with previous literature, we find no evidence that SNX14 has a direct role in autophagosome-lysosome fusion. Taken together, this implies a conserved role for SNX14 in ER-localized lipid metabolism and homeostasis. Resolution of the cellular pathology in patients with SCAR20 disease provides a more rational target for the development of therapies to arrest or alleviate the clinical symptoms, along with a clearer understanding of common mechanisms underlying Purkinje cell loss and associated cerebellar degeneration.
Materials and Methods
Cell culture
Human derived dermal fibroblasts, HEK293 and U2OS cells were cultured in 10% FBS (Thermo Fisher, 10500064) prepared in DMEM (Thermo Fisher, 41966052). Cell lines were passaged with 0.25% Trypsin-EDTA (Thermo Fisher, 25200056) when they approached 80–90% confluency.
Autophagy and autophagosome-lysosome fusion assay
To induce autophagy cells were cultured for 6 h with 250 nm Torin (Merck, 475991). Autophagy was examined by quantification of LC3B-II and p62 levels in response to Torin1 treatment. To monitor autophagosome-lysosome fusion, HEK293 cells were transfected 24 h earlier with pBABE-puro mCherry-EGFP-LC3B (Addgene plasmid #22418). Addition of 50 nm Bafilomycin A1 (Sigma, B1793) was used to block autophagosome-lysosome fusion. To monitor autolysosome formation in human fibroblasts, cells were treated with 1: 500 Cyto-ID (Enzo Life Sciences, ENZ-51031) and 1: 10 000 LysoTracker (ThermoFisher Scientific, L7528) for 30 min, then washed with 10% FBS in PBS prior to imaging. Fluorescent images were taken while the cells were alive. mCherry positive puncta were analysed using CellProfiler (40) with the “Measure Object Intensity” analysis module.
Immunocytochemistry
Cells were washed with PBS before 20 min of fixation with 4% formaldehyde solution (Sigma, F1635). After six washes with PBS, the cells were incubated at 4°C, overnight with primary antibodies prepared in 10% FBS, 0.1% Triton™ X-100 (Sigma, T8787) in PBS at the following concentrations: Mouse anti-CHOP (ThermoScientific, MA1–250) 1: 100; Mouse anti-LAMP1 (Santa Cruz, SC-20011) 1: 100; Rabbit anti-SNX14 (HPA017639) 1: 150; Mouse anti-HSP90B1 (R&D) 1: 300. The primary antibodies were washed away with six washes of PBS before the cells were incubated in the dark for 1 h at room temperature in 10% FBS, 0.1% Triton™ X-100 in PBS with the appropriate secondary antibodies: Donkey anti-mouse AF488 (Thermo Fisher, A21202) 1: 500; Donkey anti-rabbit AF594 (Thermo Fisher, A21207) 1: 500. Following six washes with PBS, the cells were counterstained with Hoechst 33342 (Thermo Fisher, H3570). Nuclear CHOP expression was analysed using CellProfiler (40) with the “Measure Object Intensity” analysis module.
Stably expressing SNX14-FLAG cell lines
The human SNX14 full length and truncated cDNAs (Full length SNX14-FLAG; N-terminal SNX14-FLAG; C-terminal SNX14-FLAG) were generated following PCR amplification (primers available on request) and cloning into the pMXs-3-FLAG vector. U2OS cells were transformed with retroviral vectors expressing Puromycin-expressing plasmids. Stable cell lines were selected via Puromycin-resistance, and stored at −80°C prior to their use in experiments.
Generation of SNX14KO HEK293 cell clones with CRISPR-Cas9
The SNX14 gene was targeted in HEK293 cells using two guide RNAs: 5′-GGAAGATGATTCTCCAGTGG-3′ (Guide A) and 5′-CTATAGACAGACCAATGTTC-3′, cloned into px330 vector (Addgene plasmid #78621). HEK293 cells were transfected with either guide A or guide B RNA, Cas9 and GFP expression vector. GFP+ cells were selected using fluorescence automated cell sorting and seeded as a single cell in one well of a 96 well plate. Colonies were expanded and validated for disruption to the SNX14 protein expression using Western blotting.
Isolation of membranes by microsomal fractionation
Microsomal fractions were isolated using a modified protocol described previously (41). Briefly, cells were grown to 70–80% confluency then dishes were washed once and scraped into 1× PBS, pooled and harvested at 1000g for 5 min at 4°C. The pellet was washed twice with 1×PBS and centrifuged again at 1000g for 5 min at 4°C. The pellet was resuspended in Buffer A (10 mm HEPES-KOH pH 7.2, 250 mm sorbitol, 10 mm KOAc, 1.5 mm MgOAc and protease inhibitor mixture), passed through a 27×1/2-gauge needle 20 times, and centrifuged at 1000g for 5 min at 4°C to pellet cell debris. The post-nuclear supernatant fraction was transferred into a clean tube, and microsomes were sedimented at 6000g for 10 min at 4°C. Harvested microsomes were washed twice with Buffer B (20 mm HEPES-KOH pH 7.2, 250 mm sorbitol, 150 mm KOAc, 0.5 mm MgOAc and protease inhibitor mixture), and the pellet was resuspended in Buffer B.
Organelle fractionation
Increasing density fractions (10–32%) were generated by diluting 60% OptiPrep™ Density Gradient Medium (Sigma, D1556) (Supplementary Material, Table S1). Fractions were prepared the day before and stored over night at 4°C before laying the gradient. Density gradients were prepared in 38.5 ml Thinwall, Ultra-Clear™ tubes (Beckman, 344058) by carefully laying solutions of decreasing density. Cell pellets were resuspended in extraction buffer (0.25 M sucrose, 10 mm triethanolamine, 10 mm acetic acid, pH 7.8) and lysed with a Dounce tissue grinder set (Sigma, D9063) until approximately 90% of the cells were lysed (∼100 strokes with the small clearance pestle). The cell lysate was layered on top of the OptiPrep™ Density Gradient (Supplementary Material, Table S1) and was separated using an ultracentrifuge (Beckman), SW32Ti Rotor (Beckman) at 150 000g for 4 h at 4°C. Fractions were extracted using a syringe to pierce the tube at the appropriate locations starting from the top.
Western blotting
Cell pellets were lysed in ice-cold NP-40 buffer (150 mm NaCl, 50 mm pH 8 Tris–HCl, 1% NP-40) containing 1× complete protease inhibitor cocktail (Roche, 11836153001) and 100 µm phenylmethanesulfonyl fluoride (Sigma, 93482). Insoluble contents were pelleted at 13 000g for 30 min at 4°C and the supernatant was prepared in Laemmli sample buffer (Bio-Rad, 161-0747) containing 50 mm dithiothreitol. Samples were heated to 85°C for 15 min before they were run on a polyacrylamide gel with a marker (Bio-Rad, 1610376) for reference. Protein was transferred onto a membrane using Trans-Blot® Turbo™ mini PVDF transfer packs (Bio-Rad, 1704156) with the Trans-Blot® Turbo™ transfer system.
Membranes where blocked with 5% milk, prepared in PBS/0.1%Tween 20® (Sigma, P7949) (PBST) for 1 h at room temperature. They were then incubated on a roller for 15 h at 4°C in PBST with antibodies prepared as follows: Mouse anti-GAPDH (Merck, MAB374) 1:4000; Rabbit anti-α/β-Tubulin (Cell Signaling Technology, 2148) 1: 2000; Rabbit anti-SNX14 (HPA017639) 1: 500; Rabbit anti-LC3B (Abcam, AB48394); Mouse anti-p62 (610833) 1: 500; Mouse anti-EEA1 (BD Biosciences, 610456) 1: 1000; Mouse anti-Complex V alpha-subunit (Invitrogen, 439800) 1:1000; Goat anti-Ribophorin (Santa Cruz, SC-12164) 1:200; Mouse anti-Rab7 (Abcam, AB50533) 1:2000; Rabbit anti-Calnexin (Abcam, AB22595) 1:1000; Rabbit anti-gm130 (Abcam, AB52649) 1:1000; Mouse anti-LAMP1 (Santa Cruz, SC-20011) 1:100; Mouse anti-FLAG (Sigma, F3165) 1:10000.
Blots were washed six times with PBST and incubated in PBST for 2 h at room temperature with the appropriate species of secondary antibody prepared as follows: Goat anti-mouse HRP (DAKO, P0447) 1: 4000; Goat anti-rabbit HRP (DAKO, P0448) 1: 4000; Donkey anti-goat HRP (Santa Cruz, SC-2020) 1: 5000. They were then washed six times with PBST before developing with Clarity™ Western ECL blotting substrate (Bio-Rad, 1705060) and imaging with the ChemiDoc imaging system (Bio-Rad). Relative protein expression was analysed using the “Analyse > Gels” option in Fiji.
Yeast live cell fluorescence microscopy
Living yeast were cultured in SC-dextrose media and imaged using an EVOS FL epi-fluorescence microscopy (Invitrogen). SNX14 and SNX19 Phox homology (PX) domains were cloned into the pBP73A (URA3) yeast expression plasmid via a BamHI/XhoI insertion site, making an N-terminal GFP-fusion construct expressed on the ADH promoter.
Filipin cholesterol distribution, lipid droplet and ER stress assay
HEK293 cells were cultured on mouse Laminin (Invitrogen, 23017-015) coated plastic to prevent them from washing off during in later steps. Autolysosomal cholesterol accumulation was induced with 10 µg/ml (23.5 µm) U18666A (Sigma, U3633) for 24 h. For oleic acid treatment, U2OS cells were treated for 8 h with complete media containing 600 µm oleic acid. ER stress was induced with 1.5 µm Tunicamycin (Sigma, T7765) for 24 h.
Cells were washed three times with PBS before a 20 min fixation with 4% PFA at room temperature. After three washes with PBS, the PFA was quenched with 1.5 mg/ml of glycine (in PBS) for 10 min at room temperature. The cells were then incubated with 50 μg/ml filipin (Sigma, F4767) (in PBS made up with 10% FBS) for 2 h at room temperature in the dark. When a nuclear counter stain was used for quantitation of nuclear-peripheral distribution, 5 μm Syto® 85 orange fluorescent nucleic acid stain (Molecular Probes, S-11366) was included in the filipin staining solution. After 2 h, the cells were washed three times with PBS and stored in the dark at 4°C before imaging using an Olympus IX71 microscope. Nuclear-peripheral filipin distribution was analysed using CellProfiler (40) with the “Measure Object Intensity Distribution” analysis module. Lipid droplets were labelled with monodansylpentane (Abgent, SM1000a) as previously described (30).
Electron microscopy
Cultured fibroblast and HEK293 cells were processed for ultrastructural examination. Samples were fixed in 2.5% glutaraldehyde buffered by 0.1 M sodium cacodylate (pH 7.2), postfixed in 1% osmium tetroxide, dehydrated in ascending grades of alcohol, processed through propylene oxide and embedded in Epon resin. Ultra-thin sections were cut with a diamond knife on a Leica Ultracut UCT Ultramicrotome, placed on copper grids and stained with uranyl acetate and lead citrate. Examination was in a JEOL 1400 transmission electron microscope.
Thin layer chromatography
Lipids were extracted from whole cells using the chloroform/methanol method modified from Bligh and Dyer (1959) (42). All steps were performed in the cold room. Cell pellets were lysed by vortexing in the presence of chloroform. Methanol was added and the suspension was vortexed vigorously. Then, 500 mm NaCl prepared in 0.5% acetic acid was added to get the final concentration of chloroform: methanol: water to 2:2:1.8. Samples were spun at 4000 rpm for 15 min in the cold room and the bottom chloroform layer was recovered. The volume was recorded. Dried lipid pellets were resuspended in chloroform to a final concentration normalized to the initial cell pellet weight. One-dimensional thin layer chromatography was used to separate the extracted lipids using hexane: diethyl ether: acetic acid (80:20:1) as a solvent to separate neutral lipids, and chloroform: acetone: methanol: acetic acid: water (50:20:10:15:5) to separate phospholipids. TLC plates were spray-stained with 3% copper acetate prepared in 8% phosphoric acid. Stained plates were incubated in the oven at 135°C for 30 min to develop the bands (or overnight). Plates were resprayed and reheated as needed to visualize lipids. Stained TLC plates were scanned and then processed for quantification using Fiji (ImageJ). On each plate we ran a serial dilution of standard (neutral or phospho-) lipid mixture of known concentrations to create a standard curve.
Statistics and figure graphics
Data were analysed using either a Student’s t-test or ANOVA as described in the legend of each figure. Figures were generated using Inkscape (https://inkscape.org).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We would like to thank the families with SCAR20 who provided biological samples and clinical information.
Conflict of Interest statement. None declared.
Funding
This work was funded by a grant from Great Ormond Street Hospital Children’s Charity (V4215 to P.S.) and supported by the National Institute for Health Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation and University College London. This work was also sponsored by grants to W.M.H. from the Welch Foundation (I-1873), the National Institute of General Medical Sciences (R35/MIRA R350821601), the Searle Scholars Foundation (SSP-2016-1482) and the University of Texas South Western Endowed Scholars Program.
References
- 1. Thomas A.C., Williams H., Seto-Salvia N., Bacchelli C., Jenkins D., O'Sullivan M., Mengrelis K., Ishida M., Ocaka L., Chanudet E.. et al. (2014) Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am. J. Hum. Genet., 95, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akizu N., Cantagrel V., Zaki M.S., Al-Gazali L., Wang X., Rosti R.O., Dikoglu E., Gelot A.B., Rosti B., Vaux K.K.. et al. (2015) Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat. Genet., 47, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jazayeri R., Hu H., Fattahi Z., Musante L., Abedini S.S., Hosseini M., Wienker T.F., Ropers H.H., Najmabadi H., Kahrizi K. (2015) Exome sequencing and linkage analysis identified novel candidate genes in recessive intellectual disability associated with ataxia. Arch. Iranian Med., 18, 670–682. [PubMed] [Google Scholar]
- 4. Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M.. et al. (2015) Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron, 88, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shukla A., Upadhyai P., Shah J., Neethukrishna K., Bielas S., Girisha K.M. (2017) Autosomal recessive spinocerebellar ataxia 20: report of a new patient and review of literature. Eur. J. Med. Genet., 60, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trujillano D., Bertoli-Avella A.M., Kumar Kandaswamy K., Weiss M.E., Koster J., Marais A., Paknia O., Schroder R., Garcia-Aznar J.M., Werber M.. et al. (2017) Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet., 25, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sousa S.B., Ramos F., Garcia P., Pais R.P., Paiva C., Beales P.L., Moore G.E., Saraiva J.M., Hennekam R.C. (2014) Intellectual disability, coarse face, relative macrocephaly, and cerebellar hypotrophy in two sisters. Am. J. Med. Genet. A, 164, 10–14. [DOI] [PubMed] [Google Scholar]
- 8. Fenn J., Boursnell M., Hitti R.J., Jenkins C.A., Terry R.L., Priestnall S.L., Kenny P.J., Mellersh C.S., Forman O.P. (2016) Genome sequencing reveals a splice donor site mutation in the SNX14 gene associated with a novel cerebellar cortical degeneration in the Hungarian Vizsla dog breed. BMC Genet., 17, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teasdale R.D., Loci D., Houghton F., Karlsson L., Gleeson P.A. (2001) A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J., 358, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Teasdale R.D., Collins B.M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J., 441, 39–59. [DOI] [PubMed] [Google Scholar]
- 11. Mas C., Norwood S.J., Bugarcic A., Kinna G., Leneva N., Kovtun O., Ghai R., Ona Yanez L.E., Davis J.L., Teasdale R.D.. et al. (2014) Structural basis for different phosphoinositide specificities of the PX domains of sorting nexins regulating G-protein signaling. J. Biol. Chem., 289, 28554–28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ha C.M., Park D., Kim Y., Na M., Panda S., Won S., Kim H., Ryu H., Park Z.Y., Rasenick M.M.. et al. (2015) SNX14 is a bifunctional negative regulator for neuronal 5-HT6 receptor signaling. J. Cell. Sci., 128, 1848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knaevelsrud H., Soreng K., Raiborg C., Haberg K., Rasmuson F., Brech A., Liestol K., Rusten T.E., Stenmark H., Neufeld T.P.. et al. (2013) Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J. Cell Biol., 202, 331–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holland P., Knaevelsrud H., Soreng K., Mathai B.J., Lystad A.H., Pankiv S., Bjorndal G.T., Schultz S.W., Lobert V.H., Chan R.B.. et al. (2016) HS1BP3 negatively regulates autophagy by modulation of phosphatidic acid levels. Nat. Commun., 7, 13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henne W.M., Zhu L., Balogi Z., Stefan C., Pleiss J.A., Emr S.D. (2015) Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol., 210, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hariri H., Ugrankar R., Liu Y., Henne W.M. (2016) Inter-organelle ER-endolysosomal contact sites in metabolism and disease across evolution. Commun. Integr. Biol., 9, e1156278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kvam E., Goldfarb D.S. (2007) Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy, 3, 85–92. [DOI] [PubMed] [Google Scholar]
- 18. Lloyd-Evans E., Morgan A.J., He X., Smith D.A., Elliot-Smith E., Sillence D.J., Churchill G.C., Schuchman E.H., Galione A., Platt F.M. (2008) Niemann–Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med., 14, 1247–1255. [DOI] [PubMed] [Google Scholar]
- 19. Suh J.M., Stenesen D., Peters J.M., Inoue A., Cade A., Graff J.M. (2008) An RGS-containing sorting nexin controls Drosophila lifespan. PLoS One, 3, e2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu B.B., Liao Y.C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.H., Li B.L., Song B.L. (2015) Cholesterol transport through lysosome-peroxisome membrane contacts. Cell, 161, 291–306. [DOI] [PubMed] [Google Scholar]
- 21. Oeste C.L., Seco E., Patton W.F., Boya P., Perez-Sala D. (2013) Interactions between autophagic and endo-lysosomal markers in endothelial cells. Histochem. Cell Biol., 139, 659–670. [DOI] [PubMed] [Google Scholar]
- 22. Mauvezin C., Nagy P., Juhasz G., Neufeld T.P. (2015) Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun., 6, 7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy, 3, 452–460. [DOI] [PubMed] [Google Scholar]
- 24. Chauhan N., Farine L., Pandey K., Menon A.K., Butikofer P. (2016) Lipid topogenesis: 35years on. Biochim. Biophys. Acta, 1861, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wojtanik K.M., Liscum L. (2003) The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J. Biol. Chem., 278, 14850–14856. [DOI] [PubMed] [Google Scholar]
- 26. Liscum L., Faust J.R. (1989) The intracellular transport of low density lipoprotein-derived cholesterol is inhibited in Chinese hamster ovary cells cultured with 3-beta-[2-(diethylamino)ethoxy]androst-5-en-17-one. J. Biol. Chem., 264, 11796–11806. [PubMed] [Google Scholar]
- 27. Cenedella R.J. (2009) Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids, 44, 477–487. [DOI] [PubMed] [Google Scholar]
- 28. Vanier M.T., Rodriguez-Lafrasse C., Rousson R., Gazzah N., Juge M.C., Pentchev P.G., Revol A., Louisot P. (1991) Type C Niemann–Pick disease: spectrum of phenotypic variation in disruption of intracellular LDL-derived cholesterol processing. Biochim. Biophys. Acta, 1096, 328–337. [DOI] [PubMed] [Google Scholar]
- 29. Tangemo C., Weber D., Theiss S., Mengel E., Runz H. (2011) Niemann–Pick Type C disease: characterizing lipid levels in patients with variant lysosomal cholesterol storage. J. Lipid Res., 52, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H.J., Hsu C.L., Yang J.Y., Yang W.Y. (2012) Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One, 7, e32693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Renna M., Schaffner C., Winslow A.R., Menzies F.M., Peden A.A., Floto R.A., Rubinsztein D.C. (2011) Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J. Cell Sci., 124, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ordonez M.P., Roberts E.A., Kidwell C.U., Yuan S.H., Plaisted W.C., Goldstein L.S. (2012) Disruption and therapeutic rescue of autophagy in a human neuronal model of Niemann Pick type C1. Hum. Mol. Genet., 21, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A.H., Cassady J.P., Cohen M.A., Chakraborty S., Wang H., Spooner E.. et al. (2013) Impaired autophagy in the lipid-storage disorder Niemann–Pick type C1 disease. Cell Rep., 5, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell. Biol., 11, 1433–1437. [DOI] [PubMed] [Google Scholar]
- 35. Lloyd-Evans E., Platt F.M. (2010) Lipids on trial: the search for the offending metabolite in Niemann–Pick type C disease. Traffic, 11, 419–428. [DOI] [PubMed] [Google Scholar]
- 36. Sarna J.R., Larouche M., Marzban H., Sillitoe R.V., Rancourt D.E., Hawkes R. (2003) Patterned Purkinje cell degeneration in mouse models of Niemann–Pick type C disease. J. Comp. Neurol., 456, 279–291. [DOI] [PubMed] [Google Scholar]
- 37. Eden E.R. (2016) The formation and function of ER-endosome membrane contact sites. Biochim. Biophys. Acta, 1861, 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hariri H., Rogers S., Ugrankar R., Liu Y.L., Feathers J.R., Henne W.M. (2018) Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep., 19, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hekman K.E., Gomez C.M. (2015) The autosomal dominant spinocerebellar ataxias: emerging mechanistic themes suggest pervasive Purkinje cell vulnerability. J. Neurol. Neurosurg. Psychiatry, 86, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J.. et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol., 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim J., Hamamoto S., Ravazzola M., Orci L., Schekman R. (2005) Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J. Biol. Chem., 280, 7758–7768. [DOI] [PubMed] [Google Scholar]
- 42. Bligh E.G., Dyer W.J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol., 37, 911–917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.