Abstract
Background
BRCA1 and BRCA2 (BRCA1/2)-deficient tumors display impaired homologous recombination repair (HRR) and enhanced sensitivity to DNA damaging agents or to poly(ADP-ribose) polymerase (PARP) inhibitors (PARPi). Their efficacy in germline BRCA1/2 (gBRCA1/2)-mutated metastatic breast cancers has been recently confirmed in clinical trials. Numerous mechanisms of PARPi resistance have been described, whose clinical relevance in gBRCA-mutated breast cancer is unknown. This highlights the need to identify functional biomarkers to better predict PARPi sensitivity.
Patients and methods
We investigated the in vivo mechanisms of PARPi resistance in gBRCA1 patient-derived tumor xenografts (PDXs) exhibiting differential response to PARPi. Analysis included exome sequencing and immunostaining of DNA damage response proteins to functionally evaluate HRR. Findings were validated in a retrospective sample set from gBRCA1/2-cancer patients treated with PARPi.
Results
RAD51 nuclear foci, a surrogate marker of HRR functionality, were the only common feature in PDX and patient samples with primary or acquired PARPi resistance. Consistently, low RAD51 was associated with objective response to PARPi. Evaluation of the RAD51 biomarker in untreated tumors was feasible due to endogenous DNA damage. In PARPi-resistant gBRCA1 PDXs, genetic analysis found no in-frame secondary mutations, but BRCA1 hypomorphic proteins in 60% of the models, TP53BP1-loss in 20% and RAD51-amplification in one sample, none mutually exclusive. Conversely, one of three PARPi-resistant gBRCA2 tumors displayed BRCA2 restoration by exome sequencing. In PDXs, PARPi resistance could be reverted upon combination of a PARPi with an ataxia-telangiectasia mutated (ATM) inhibitor.
Conclusion
Detection of RAD51 foci in gBRCA tumors correlates with PARPi resistance regardless of the underlying mechanism restoring HRR function. This is a promising biomarker to be used in the clinic to better select patients for PARPi therapy. Our study also supports the clinical development of PARPi combinations such as those with ATM inhibitors.
Keywords: germline BRCA, PARP inhibitor resistance, homologous recombination, RAD51, TP53BP1, ATM
Key Message
Tumors from germline BRCA1/2-mutation carriers may restore homologous recombination repair (HRR) and acquire PARPi resistance through diverse mechanisms. The direct detection of RAD51 nuclear foci in tumor samples, as a functional biomarker of HRR, may help to refine the selection of patients eligible for PARPi-monotherapy, and to propose new combination therapies for PARPi-resistant patients.
Introduction
BRCA1 and BRCA2 encode essential proteins for DNA homologous recombination repair (HRR) [1]. Loss of function of either gene impairs this high-fidelity DNA repair pathway and results in genetic instability and an increased risk of breast or ovarian cancer in germline BRCA1/2 (gBRCA) mutation carriers [2, 3]. Defective HRR increases sensitivity of gBRCA-mutated tumors to DNA damaging agents including anthracyclines, platinum salts, or to novel agents that block parallel DNA repair pathways, including poly(ADP-ribose) polymerase inhibitors (PARPi) [4–6]. PARP inhibition blocks the repair of DNA single-strand breaks and results in stalling of replication fork progression by trapping PARP on the DNA break [7]. Both contribute to the accumulation of DNA double-strand breaks (DSBs) that HRR-deficient cells cannot repair efficiently.
PARPi are well-tolerated agents and elicit anticancer efficacy in metastatic gBRCA tumors. Their use has been approved for advanced ovarian cancer [olaparib (Lynparza®), rucaparib (Rubraca®) and niraparib (Zejula®)] and for gBRCA breast cancer (BC) [8–10]. Final results from other phase III clinical trials are awaited, both in the early and advanced BC setting (NCT01905592, NCT01945775, NCT02032823).
Primary resistance to PARPi in a subset of gBRCA patients limits the potential of gBRCA status as the only biomarker of response to that of an enrichment strategy [11]. In addition, acquired resistance in monotherapy responders is a challenge. Previous studies using in vitro models, transgenic mice and human tumor samples have delineated two types of resistance mechanisms to PARPi in gBRCA cells: (i) independent of HRR (cellular extrusion of the PARPi, PARP1 loss, FANCD2 overexpression, SLFN11 inactivation or CHD4 loss) and (ii) dependent on HRR recovery, either by BRCA-independent mechanisms (loss of 53BP1, REV7/MAD2L2, PAXIP1/PTIP, Artemis) or by BRCA-dependent mechanisms [12–22]. The latter include secondary BRCA1/2 mutations that restore the reading frame and the expression of partially functional hypomorphic BRCA1 proteins (BRCA1-11q alternative splice isoform, the RING-less BRCA1 generated by downstream translation initiation, or HSP90-mediated stabilization of BRCA1 C-terminal mutants). Most work in gBRCA clinical samples has focused on ovarian cancer and has established that HRR recovery through secondary BRCA1/2 mutations may act as a resistance mechanism to platinum salts and PARPi. Conversely, little is known about in vivo PARPi resistance mechanisms in gBRCA BC [22].
Patterns of DNA aberrations in the tumors (genomic scars) resulting from HRR deficiency may aid in distinguishing HRR-deficient from HRR-proficient tumors [23–25]. However, genomic scars in gBRCA tumors may persist after restoration of HRR function [26]. In order to improve patient selection for PARPi monotherapy among gBRCA mutation carriers, especially in the metastatic setting, there is a clear need for a functional biomarker of HRR status to be used in the clinic. Previous work by others showed that induction of nuclear foci of the HRR protein RAD51 after neoadjuvant chemotherapy is a measure of HRR functionality in BC biopsies and predicts treatment response [27]. Here, we sought to investigate RAD51 foci as an indicator of functional HRR and its correlation with PARPi resistance in the gBRCA setting. We further explored potential treatment strategies for PARPi-resistant BC.
Methods
Study design
A collection of patient-derived tumor xenograft (PDX) models was generated by implanting tumor samples from patients with a germline BRCA1/2 mutation and breast or ovarian cancer. Their sensitivity to PARPi was evaluated, and the functionality of the HRR pathway was analyzed and compared between the PARPi-sensitive versus the PARPi-resistant PDX samples to find a functional test correlating with response. An exploratory analysis in a set of 20 tumor samples including patients treated with PARPi at our institution was employed to confirm the findings and the potential clinical interest of the functional test. A new therapeutic PARPi combination was tested in vivo in PARPi-resistant PDX models.
See further methods in supplementary material, available at Annals of Oncology online.
Results
gBRCA PDX panel
Fresh tumor samples prospectively collected for implantation into nude mice yielded a total of 12 PDX models (11 gBRCA1 and 1 gBRCA2) (supplementary Table S1, available at Annals of Oncology online). Five models were derived from patients with metastatic disease who had been treated with PARPi, three of which prior to olaparib treatment and two at progression after a sustained partial response (PR) (supplementary Table S1, available at Annals of Oncology online). Persistence of the gBRCA mutations was confirmed in all models but PDX274, and they were associated with loss of heterozygosity, i.e. loss of the wild type allele (supplementary Figure S1, available at Annals of Oncology online).
Olaparib treatment in the gBRCA PDX collection distinguished a subset of PARPi-resistant tumors (Figure 1A) [assessed by modified Response Evaluation In Solid Tumors (mRECIST), see supplementary methods, available at Annals of Oncology online]. Treatment with olaparib exhibited antitumor activity in three gBRCA models: two complete responses (CR) and one stable disease (SD). The remaining nine PDX models were olaparib-resistant (PD). An additional PDX model with acquired resistance (PDX230OR) was generated from its PARPi-sensitive counterpart (PDX230) after >100 days exposure to olaparib (supplementary Figure S2, available at Annals of Oncology online), totaling 13 gBRCA1/2 models. Among the four PDX derived from gBRCA primary tumors, 50% showed CR. The sensitivity to PARPi-treatment in the PDXs from metastatic patients previously treated with olaparib mirrored the patients’ clinical response to olaparib (supplementary Table S1 and Figure S3, available at Annals of Oncology online).
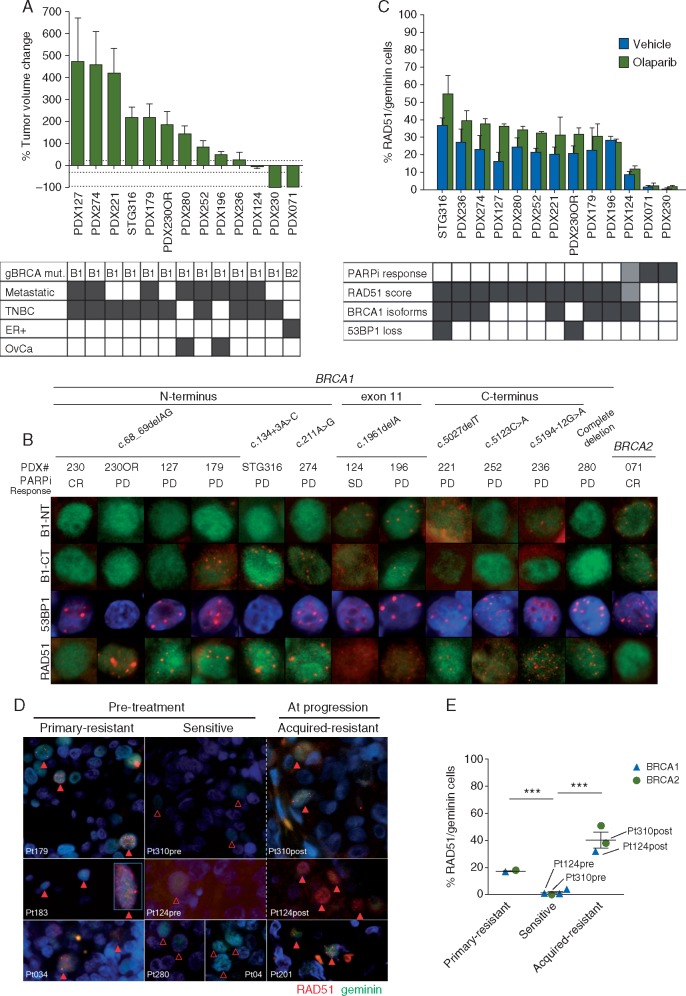
Figure 1.
Homologous recombination repair markers and PARPi response. (A) Antitumor activity of olaparib in gBRCA patient-derived tumor xenografts (PDXs). Best response to olaparib is plotted as the percentage of tumor volume change after at least 21 days of treatment. +20%, –30% and –95% are marked by dashed lines to indicate the range of CR (complete response), PR (partial response), SD (stable disease) and PD (progressive disease). Mut, mutation; B1, mutation in BRCA1; B2, mutation in BRCA2; Metastatic, PDX derived from a metastatic lesion (otherwise, derived from a primary tumor); TNBC, triple negative BC; ER+, estrogen receptor positive BC; OvCa, ovarian cancer. (B) Immunofluorescence staining of BRCA1, 53BP1 and RAD51 across the PARPi-sensitive and PARPi-resistant gBRCA PDX models. Detection of BRCA1 [with an antibody towards the N-terminus of BRCA1 (B1-NT) or C-terminus (B1-CT)], 53BP1 and RAD51 nuclear foci in olaparib-treated PDX models. CR, PD and SD are indicated. For BRCA1, the location of the mutation within the gene is indicated. DAPI staining is shown in blue. Green nuclei indicate geminin-positive cells (S/G2 phase of the cell cycle). (C) RAD51 nuclear foci formation discriminates PARPi-resistant tumors. Quantification of geminin positive cells with RAD51 nuclear foci detected by immunofluorescence in FFPE samples from tumors treated with vehicle (black bars) or olaparib (green bars). The graph displays mean ± SEM from three independent tumors. The association with PARPi-response is shown in the supplementary Table S1, available at Annals of Oncology online. Dark grey, CR; light gray, SD; white, PD. For RAD51, dark gray means high RAD51 score; light gray, intermediate RAD51; white, low RAD51. Expression of hypomorphic BRCA1 isoforms and loss of 53BP1 is depicted in dark gray. (D) RAD51 in patients’ tumors is associated with PARPi clinical response. IF of RAD51 and geminin in the pretreatment setting using a pretreatment tumor sample (or the most recent metastatic sample). Samples from three PARPi-resistant patients (Pt179, skin metastasis of TNBC; Pt183, dermal lymphatic carcinomatosis of ovarian cancer; Pt034, lymph node metastasis of ER+ BC) and four PARPi-sensitive patients (Pt310pre, liver metastasis of ER+ BC; Pt124pre, primary TNBC; Pt280, peritoneal implant of ovarian cancer; Pt04, lymph node metastasis TNBC) are shown. For acquired resistance, samples obtained from three patients at PARPi progression (Pt310post, liver metastasis; Pt124post, skin metastasis; Pt201, skin metastasis of ER+ BC) are shown. Empty arrowheads show geminin-positive cells devoid of RAD51 nuclear foci. Solid arrowheads indicate RAD51/geminin-positive cells. DAPI staining is shown in blue. (E) Quantification of RAD51/geminin-positive cells from tumor samples shown in panel D. Pt183 was not scored as the tumor did not contain 100 geminin-positive cells. Unpaired t test: *P < 0.05; **P < 0.01.
BRCA1/2 sequencing, BRCA1 expression and nuclear foci formation in gBRCA PDX samples
No frameshift-correction or genetic reversion of the inherited mutation—the so-called secondary BRCA mutations—occurred in the gBRCA1 PARPi-resistant PDXs. BRCA1 mRNA expression was variable across models and absent in PDX280, a model with a large deletion encompassing the complete BRCA1 gene (supplementary Figure S4, available at Annals of Oncology online). To investigate the potential expression of hypomorphic BRCA1 isoforms and their recruitment to DNA damage sites, we set up immunostaining assays for the DNA damage response (DDR) proteins: BRCA1, RAD51 (as functional HRR marker) and γH2AX (as DNA damage marker), with geminin (as S/G2-cell cycle marker) (supplementary Figure S5, available at Annals of Oncology online).
PDX124 and PDX196 harbor a c.1961delA mutation in BRCA1 exon 11 and express the BRCA1-Δ11q splice isoform (p.Ser264_Gly1366del) [20] which forms nuclear foci detected with both B1-NT and B1-CT antibodies, as expected (Figure 1B and supplementary Figure S6, available at Annals of Oncology online). BRCA1 nuclear foci were also detected in five additional gBRCA PDX models: PDX179, STG316, PDX274, PDX221 and PDX236 (supplementary Table S1, available at Annals of Oncology online and Figure 1B). Western blot confirmed the expression of BRCA1 isoforms at the respective predicted sizes [Δ11q, RING-less, and C-terminal truncated mutant proteins (supplementary Figure S7, available at Annals of Oncology online)]. In summary, hypomorphic BRCA1 isoforms were detected by immunofluorescence (IF) to form nuclear foci in seven PDX models, six with primary or acquired resistance to PARPi and one model showing disease stabilization (PDX124).
Analysis of 53BP1 loss and exome sequencing in gBRCA PDX samples
The assessment of 53BP1 nuclear foci by IF in olaparib-treated PDX samples identified 53BP1 loss in two PARPi-resistant models: PDX230OR and STG316 (Figure 1B). Exome sequencing unveiled somatic mutations in TP53BP1 in both models (supplementary Table S1, available at Annals of Oncology online). The PD model PDX280 harbors a non-previoulsy reported missense mutation in the PARPi resistance gene SLFN11 p.H661D. The SD model PDX124 displays a focal RAD51 amplification and high protein expression (supplementary Figure S8A and B, available at Annals of Oncology online). Mutations in other known PARPi resistance genes (PARP1, REV7/MAD2L2, PAXIP1/PTIP, Artemis, CHD4) were not identified.
Nuclear foci formation of the HRR protein RAD51
The observed recruitment of hypomorphic BRCA1 isoforms to DNA damage sites and/or 53BP1 loss in PARPi-resistant PDXs may help restore their ability to accomplish HRR. As a functional surrogate of HRR, we sought to detect RAD51 nuclear foci in geminin-positive cells and nuclear co-localization with BRCA1. RAD51 nuclear foci were detected in 11 PDXs in olaparib-treated samples, including all models expressing hypomorphic BRCA1 isoforms and/or lacking 53BP1 (Figure 1B). RAD51 foci co-localized with BRCA1 foci in all PDX models expressing hypomorphic BRCA1 isoforms (supplementary Figure S9, available at Annals of Oncology online). The three PARPi-resistant models that lacked hypomorphic BRCA1 isoform expression or 53BP1 loss (PDX127, PDX252 and PDX280) exhibited RAD51 foci suggesting that recovery of HRR occurs via BRCA1-independent mechanisms in these models (Figure 1B). Olaparib-treated samples from PARPi-resistant PDXs showed higher percentage of RAD51-positive cells versus those from PARPi-sensitive models (36 ± 2% in PARPi-resistant versus 5 ± 3% in PARPi-sensitive, P = 0.0017) (Figure 1C). Analysis of γH2AX foci ruled out PARPi pharmacodynamic differences as the reason for this differential response (supplementary Figure S10, available at Annals of Oncology online). The unexpected evidence of endogenous DNA damage and repair markers in untreated samples (supplementary Figure S5 and S10, available at Annals of Oncology online) prompted us to also score RAD51 foci in untreated tumors (Figure 1C). Importantly, untreated samples of PARPi-resistant tumors showed a significantly higher baseline percentage of RAD51-positive cells compared with PARPi-sensitive tumors (24 ± 2% versus 3 ± 2%, P = 0.0025). Thus, HRR functionality was frequent and associated with PARPi resistance in this gBRCA PDX panel.
RAD51/geminin score and response to PARPi in patients’ samples
We confirmed the feasibility of detecting RAD51 and γH2AX foci in FFPE tumor samples from patients, by firstly staining for RAD51 and geminin in 20 patients’ tumor samples including some matched with the gBRCA PDX models (n = 7) (supplementary Figure S11A–C, available at Annals of Oncology online). These results prompted us to further assess the potential clinical utility of the RAD51/geminin score as a functional biomarker of PARPi treatment in patients treated with various PARPi at our institution (n = 10 tumors), including two paired pre-/post-PARPi samples. This cohort included eight patients with germline mutations in BRCA1/2 (BRCA1, n = 5; BRCA2, n = 3; diagnosis of BC, n = 6; ovarian cancer, n = 2). The samples had been collected prior to (n = 7) or at progression to treatment with a PARPi (n = 3). We stained and scored for RAD51 (Figure 1D and E). Importantly, PARPi-resistant tumor samples showed an inverse relationship between the RAD51 score and clinical efficacy of the PARPi. Exome sequencing identified a BRCA2-secondary mutation in one tumor with acquired resistance and RAD51 foci (supplementary Figure S12, available at Annals of Oncology online).
Platinum salts in olaparib-resistant tumors
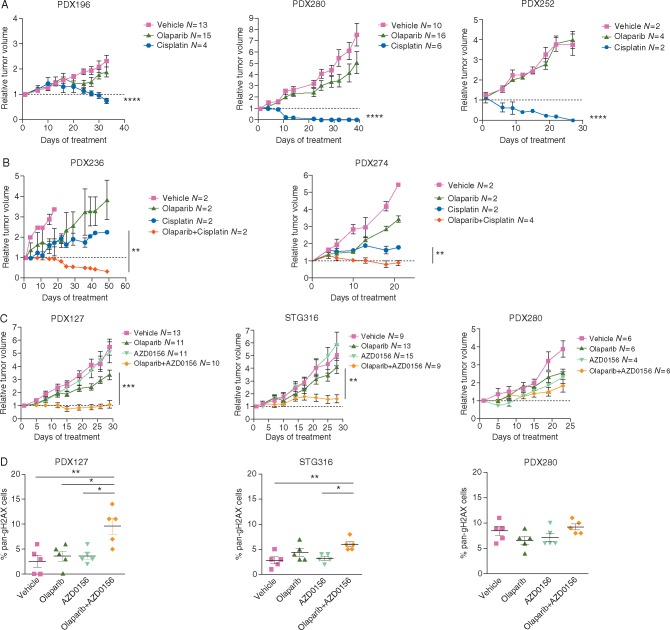
HRR recovery/retention that limits PARPi efficacy may not imply resistance to platinum-based treatments in gBRCA cancers. A previous study in gBRCA ovarian cancer showed a 40% response rate to platinum chemotherapy in the setting of resistance to olaparib [28]. We next assessed the efficacy of cisplatin in the two HRR proficient, RAD51-positive ovarian cancer PDX models (PDX196 and PDX280) (Figure 2A). Response to cisplatin was confirmed in both PDX models and in the clinic for Pt280 (data not available for Pt196 due to carboplatin hypersensitivity). Next, we assessed the activity of platinum-based chemotherapy in advanced/metastatic BC in the context of PARPi resistance. We previously reported that PDX127 showed resistance to PARPi but response to platinum, in agreement with the clinical response of the patient [29]. Similarly, the PARPi-resistant models, RAD51-positive model PDX252 exhibited significant tumor regression when treated with cisplatin (Figure 2A). In the PARPi-resistant PDX236 and PDX274, cisplatin-only slowed tumor growth as compared with vehicle, while its combination with olaparib achieved PR and SD, respectively (Figure 2B). These results highlight that platinum-based therapies can be active in PARPi-resistant metastatic BC and suggests that RAD51 foci formation does not predict resistance to platinums in this setting.
Figure 2.
Cisplatin or ATM inhibition overcomes PARPi resistance. (A) Relative tumor volume (RTV) of vehicle, cisplatin or olaparib in PDX196, PDX280 and PDX252. Cisplatin was administrated 6 mg/kg weekly unless RTV < 0.5. Olaparib was administrated daily at 50 mg/kg (5 doses/week). Number of tumors per arm is indicated. (B) RTV of vehicle, cisplatin, olaparib or its thereof combination in PDX236 and PDX274. Cisplatin and olaparib were administrated as in panel A. (C) RTV of vehicle, olaparib, AZD0156 or the combination of treatments in PDX127, STG316 and PDX280. Olaparib was administrated daily at 50 mg/kg (5 doses/week) and AZD0156 was administered three times per week at 2 or 2.5 mg/kg. (D) Quantification of pan-nuclear γH2AX-positive cells in PDX127, STG316 and PDX280 treated with vehicle, olaparib, AZD0156 or the combination of drugs at the end point of experiments shown in panel C. All figures show mean and SEM. Statistical P-values are shown when relevant: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-way ANOVA).
Ataxia-telangiectasia mutated blockade plus PARP inhibition in olaparib-resistant tumors
We further explored the potential of DDR inhibitors to enhance PARPi antitumor activity. The ataxia-telangiectasia mutated (ATM) kinase is activated in response to DNA DSBs, signals to cell cycle checkpoints and DNA repair pathways, and is reciprocally synthetic lethal with PARP [30]. As previously suggested, we hypothesized that ATM inhibition is a treatment option for PARPi-resistant BRCA1-deficient tumors that restore HRR through loss of TP53BP1 or REV7/MAD2L2 by enabling ATM-dependent end resection [17, 18]. We tested this hypothesis in three ATM-expressing PDXs (supplementary Figure S13A, available at Annals of Oncology online): STG316, a model that lacks 53BP1, and in PDX127 and PDX280, which are devoid of hypomorphic BRCA1 isoforms, and presumably achieve PARPi resistance by a ‘loss of 53BP1’-like mechanism. In fact, PDX127 harbors a missense mutation in PRCC p.P55T, within the interaction domain with REV7/MAD2L2 (supplementary Figure S13B, available at Annals of Oncology online). The best antitumor activity of the olaparib combination with the ATM inhibitor AZD0156 was achieved in PDX127 (SD) (Figure 2C). We investigated whether ATM inhibition resulted in restoration of HRR deficiency by impairing RAD51 foci formation [17, 18] (supplementary Figure S13C, available at Annals of Oncology online). Unexpectedly, RAD51 foci formation was marginally reduced in combination-treated tumors, arguing that ATM inhibition may exert a broader effect in signaling the olaparib-induced DDR beyond its effects on HRR (supplementary Figure S13C, available at Annals of Oncology online). Quantification of DNA damage by γH2AX staining (pan-nuclear, Figure 2D and foci formation, supplementary Figure S13D, available at Annals of Oncology online) showed a significant increase of pan-γH2AX-positive cells upon ATM plus PARPi as compared with olaparib monotherapy in combination-responders PDX127 and STG316. These results suggest an induction of replication stress in these combination-sensitive models. These results are of interest since an international phase I clinical trial testing the tolerability of olaparib in combination with AZD0156 in solid tumors is currently ongoing (NCT 02588105).
Discussion
There is a need to refine the determinants of PARPi efficacy beyond gBRCA mutations, especially in the metastatic setting. Our analysis of RAD51 foci in a total of 20 BC patient samples, 10 gBRCA1 and 10 gBRCA2, provides new evidence in favor of restoration of HRR functionality as a frequent mechanism of PARPi resistance, and demonstrates the potential of functional biomarkers to discriminate tumors that will fail PARPi monotherapy. The RAD51 foci assay may capture the dynamic changes in DNA repair that occur throughout tumor evolution and may, therefore, more effectively identify the HRR-deficient BRCA1/2-mutated tumors. Unexpectedly, while previous studies reported low levels of baseline DNA damage as a potential limitation to evaluate HRR [27, 31], we were able to detect it and score for RAD51 in untreated samples, which correlated with PARPi response. This highlights that an IF assay for RAD51 staining is feasible in FFPE samples and suggests that testing for RAD51 may be directly transferrable to the clinical setting when a larger study confirms our findings.
Restoration of HRR can be achieved by secondary BRCA mutations and may be captured by sequencing techniques [32, 33]. We identified a BRCA2 secondary mutation in one BC patient with acquired resistance to olaparib, whereas we did not detect in-frame secondary mutations in any gBRCA1 PDX model. Our data suggest that hypomorphic BRCA1 isoforms contribute to HRR restoration in gBRCA1 BC [20, 21]. Importantly, high RAD51 score predicted poor response to PARPi monotherapy independently of the underlying mechanism of HRR restoration. Further research is needed to establish the RAD51 score cut-off that differentiates responders from nonresponders to PARPi monotherapy and to evaluate the potential impact of RAD51-independent resistance mechanisms that involve replication fork stabilization [14, 16, 34, 35]. A high RAD51 foci score may encourage the use of combination therapies with PARPi, such as those that inhibit HRR [29, 36], or that enhance DNA damage [9, 37, 38]. Here, we propose that a subset of PARPi-resistant gBRCA tumors benefit from combined PARP plus ATM blockade [17, 30, 39].
Our study unveils coexistence of various mechanisms of PARPi resistance in each individual tumor, such as hypomorphic BRCA1 isoforms together with RAD51 amplification or 53BP1 [40] loss. In conclusion, this emphasizes the need of comprehensive functional tests for measuring HRR activity such as the RAD51 assay to better select patients who will benefit most from PARPi monotherapy and those who may benefit from a combination therapy.
Supplementary Material
Acknowledgements
The authors thank Dr Geoffrey Shapiro, Dr Peter Bouwman, Dr Neil Johnson, the Molecular Pathology Group at VHIO and the Breast Cancer and Melanoma Group at VHIO for helpful discussions. The Breast Surgical Unit from Vall d’Hebron Hospital, as well as Cristina Bernadó, Pilar Antón, Maite Calvo, Patricia Cozar and Ambar Ahmed have provided technical support. The authors acknowledge the Cellex Foundation for providing research facilities and equipment.
Funding
Spanish Instituto de Salud Carlos III (ISCIII) funding, an initiative of the Spanish Ministry of Economy and Innovation partially supported by European Regional Development FEDER Funds (FIS PI17/01080 to VS, PI12-02606 to JBal, PI15-00355 to OD, PI13/01711 to SG-E); European Research Area-NET, Transcan-2 (AC15/00063), Asociación Española contra el Cáncer (AECC) (LABAE16020PORTT) and the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) (2014 SGR 1331 and 2017 SGR 540) to VS; Conquer Cancer Foundation of ASCO Young Investigator Award to CC; NIH Grant P30CA008748, the Breast Cancer Research Foundation and the Geoffrey Beene Cancer Research Center to JB. Any opinions, findings and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology or the Conquer Cancer Foundation. We acknowledge ‘Tumor Biomarkers Collaboration’ supported by the Banco Bilbao Vizcaya Argentaria (BBVA) Foundation, GHD, the FERO Foundation and the Orozco Family for supporting this study (to VS and JB). CC is recipient of a Sociedad Española de Oncología Médica (SEOM)—BUCKLER0’0 grant and an AECC grant (AIOC15152806CRUZ). MC-B (LABAE16020PORTT) and SB are recipient of AECC grants. AL-G is recipient of a grant from the Generalitat de Catalunya, Health Department (PERIS, SLT002/16/00477). VS and SGE are supported by the Miguel Servet Program (ISCIII, CP14/00228 and CP10/00617, respectively). The xenograft program in the Caldas laboratory is supported by Cancer Research UK, and also received funding from an EU H2020 Network of Excellence (EuroCAN).
Disclosure
VS declares a noncommercial research agreement with AstraZeneca. JBal is an advisory board member for Clovis, Tesaro and Medivation, and has received speaker bureau honoraria by AstraZeneca. BD, ZL, UP, EC, GJ, WH, CB and MJO are employees of AstraZeneca. All remaining authors have declared no conflicts of interest.
References
- 1. Roy R, Chun J, Powell SN.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011; 12(1): 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mavaddat N, Peock S, Frost D. et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 2013; 105(11): 812–822. [DOI] [PubMed] [Google Scholar]
- 3. Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 2014; 343(6178): 1470–1475. [DOI] [PubMed] [Google Scholar]
- 4. Bryant HE, Schultz N, Thomas HD. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434(7035): 913–917. [DOI] [PubMed] [Google Scholar]
- 5. Farmer H, McCabe N, Lord CJ. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434(7035): 917–921. [DOI] [PubMed] [Google Scholar]
- 6. Bhattacharyya A, Ear US, Koller BH. et al. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 2000; 275(31): 23899–23903. [DOI] [PubMed] [Google Scholar]
- 7. O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015; 60: 547–560. [DOI] [PubMed] [Google Scholar]
- 8. Robson M, Im SA, Senkus E. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017; 377(6): 523–533. [DOI] [PubMed] [Google Scholar]
- 9. Ledermann J, Harter P, Gourley C. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014; 15(8): 852–861. [DOI] [PubMed] [Google Scholar]
- 10. Kaufman B, Shapira-Frommer R, Schmutzler RK. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015; 33(3): 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tutt A, Robson M, Garber JE. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376(9737): 235–244. [DOI] [PubMed] [Google Scholar]
- 12. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD.. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015; 5(11): 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lord CJ, Ashworth A.. PARP inhibitors: synthetic lethality in the clinic. Science 2017; 355(6330): 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kais Z, Rondinelli B, Holmes A. et al. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep 2016; 15(11): 2488–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murai J, Feng Y, Yu GK. et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016; 7(47): 76534–76550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillemette S, Serra RW, Peng M. et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev 2015; 29(5): 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bunting SF, Callen E, Wong N. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141(2): 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu G, Chapman JR, Brandsma I. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015; 521(7553): 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J, Aroumougame A, Lobrich M. et al. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev 2014; 28(24): 2693–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Bernhardy AJ, Cruz C. et al. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res 2016; 76(9): 2778–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drost R, Dhillon KK, van der Gulden H. et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest 2016; 126(8): 2903–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barber LJ, Sandhu S, Chen L. et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol 2013; 229(3): 422–429. [DOI] [PubMed] [Google Scholar]
- 23. Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Signatures of mutational processes in human cancer. Nature 2013; 500(7463): 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abkevich V, Timms KM, Hennessy BT. et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012; 107(10): 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davies H, Glodzik D, Morganella S. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017; 23(4): 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watkins JA, Irshad S, Grigoriadis A, Tutt AN.. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 2014; 16(3): 211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graeser M, McCarthy A, Lord CJ. et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 2010; 16(24): 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ang JE, Gourley C, Powell CB. et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clin Cancer Res 2013; 19(19): 5485–5493. [DOI] [PubMed] [Google Scholar]
- 29. Johnson SF, Cruz C, Greifenberg AK. et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep 2016; 17(9): 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bryant HE, Helleday T.. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res 2006; 34(6): 1685–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naipal KA, Verkaik NS, Ameziane N. et al. Functional ex vivo assay to select homologous recombination-deficient breast tumors for PARP inhibitor treatment. Clin Cancer Res 2014; 20(18): 4816–4826. [DOI] [PubMed] [Google Scholar]
- 32. Patch AM, Christie EL, Etemadmoghadam D. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015; 521(7553): 489–494. [DOI] [PubMed] [Google Scholar]
- 33. Lheureux S, Bruce JP, Burnier JV. et al. Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly(ADP-ribose) polymerase inhibition. J Clin Oncol 2017; 35(11): 1240–1249. [DOI] [PubMed] [Google Scholar]
- 34. Chaudhuri RA, Callen E, Ding X. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016; 535(7612): 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yazinski SA, Comaills V, Buisson R. et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017; 31(3): 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juvekar A, Burga LN, Hu H. et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov 2012; 2(11): 1048–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee JM, Hays JL, Annunziata CM. et al. Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst 2014; 106(6): dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Balmana J, Tung NM, Isakoff SJ. et al. Phase I trial of olaparib in combination with cisplatin for the treatment of patients with advanced breast, ovarian and other solid tumors. Ann Oncol 2014; 25(8): 1656–1663. [DOI] [PubMed] [Google Scholar]
- 39. Weston VJ, Oldreive CE, Skowronska A. et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood 2010; 116(22): 4578–4587. [DOI] [PubMed] [Google Scholar]
- 40. Bruna A, Rueda OM, Greenwood W. et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016; 167: 260–274. e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.