Abstract
Promoter escape involves breaking of the favourable contacts between RNA polymerase (RNAP) and the promoter to allow transition to an elongation complex. The sequence of DNA template that is transcribed during promoter escape (ITS; Initially Transcribed Sequence) can affect promoter escape by mechanisms that are not yet fully understood. We employed a highly parallel strategy utilizing Next Generation Sequencing (NGS) to collect data on escape properties of thousands of ITS variants. We show that ITS controls promoter escape through a combination of position-dependent effects (most prominently, sequence-directed RNAP pausing), and position-independent effects derived from sequence encoded physical properties of the template (for example, RNA/DNA duplex stability). ITS often functions as an independent unit affecting escape in the same manner regardless of the promoter from which transcription initiates. However, in some cases, a strong dependence of ITS effects on promoter context was observed suggesting that promoters may have ‘allosteric’ abilities to modulate ITS effects. Large effects of ITS on promoter output and the observed interplay between promoter sequence and ITS effects suggests that the definition of bacterial promoter should include ITS sequence.
INTRODUCTION
Bacterial RNA polymerase (RNAP) initiates transcription by binding to the promoter DNA which is followed by melting of DNA duplex in the vicinity of transcription start site to form initiation-competent open complex. Synthesis of an RNA product of 9–15 nt length leads to breaking of the RNAP–promoter contacts established in the open complex (promoter escape) and formation of a stable elongation complex capable of processive transcription of long stretches of template DNA (1–3). All of the above steps of transcription initiation could in principle be limiting to the overall output of the gene and could also be a target of the regulatory signals (4). Discovery of core promoter elements (-10 and -35 elements) that allow recognition of promoter DNA by RNAP provided initial insight into the essential role of DNA template sequence in directing transcription initiation. Subsequently, several additional sequence conserved promoter elements (for example, the UP element (5) or discriminator (6,7)) were discovered, firmly establishing DNA template sequence control of open complex formation. Indeed, specific contacts of RNAP with promoter elements are sufficient for the formation of the open complex in most cases in the absence of any additional accessory factors.
In contrast to well-established crucial role of DNA template sequence in directing RNAP to a functional open complex, such a role in promoter escape is less clear. Promoter escape takes place while RNAP transcribes ∼20 bp of template sequence (Initially Transcribed Sequence, ITS) that immediately follows the transcription start site. Abortive initiation (8,9) is an important aspect of promoter escape whereby RNAP undergoes repeated cycles of synthesis and release of short transcripts (typically 2–15 nt long) before a productive escape and formation of stable elongation complex occurs. A relatively simple correlation between the strength of promoter-RNAP contacts and the kinetics of promoter escape was observed (10–12). Stronger promoter contacts lead to slower escape (11) and higher abortive yield (12) consistent with the idea that the energetic cost of breaking favorable RNAP–promoter contacts established during formation of the open complex is one important determinant of promoter escape. During initial transcription RNAP remains bound to the promoter while the active site of the enzyme translocates resulting in enlargement of the transcription bubble and scrunching of single-stranded DNA strands of the bubble as the template DNA is pulled into the enzyme (13,14). These observations lead to a model in which the scrunching provides a mechanism for accumulating energy that serves to offset the stability of the open complex at the escape point (13–17). However, experiments aimed at testing the scrunching as a major force driving force of escape did not support such role for scrunching (11,18).
The ability of ITS to affect promoter escape is well documented (10,12,19–22) and intriguingly, the ability of ITS to influence functional properties of the transcription elongation complex far downstream of the promoter was observed as well (23). The dependence of escape on ITS is difficult to decipher since no obvious sequence motifs within ITS were identified although a correlation between purine content and productive yield was observed (10). It was first noted over thirty years ago by Kammerer et al. (22) that changing ITS could affect in vivo promoter strength >10-fold. Extensive studies of the effects of ITS on the relative amounts of abortive and productive transcript yields (10,12,19–21) showed that both parameters could be greatly affected by ITS. No simple correlation between escape properties of a given ITS and substrate binding affinities for the positions within the ITS was found (10) confirming that some intrinsic DNA signals embedded in ITS largely determine the ratio of abortive to productive products and define escape kinetics. In an effort to relate ITS to the escape kinetics, a three-pathway kinetics model of transcription initiation was proposed (24). While the model had some success in partially reproducing behaviors of some promoters, the insights to the mechanism of escape were limited and its generality is unknown. The work by Skancke et al. (25) on ITS variants of T5 N25 promoter demonstrated a strong reverse correlation between escape efficiency and RNA sequence encoded bias for the pretranslocated state of RNAP enzyme (26,27). In this state the active site is still occupied by the base at the 3′-end of the RNA and thus unable to bind incoming NTP for the next base addition. The mechanistic explanation for this observation was that ITS that bias RNAP to the pretranslocated state would increase the probability of backtracking and the release of abortive product reducing the escape efficiency (25). While this work was limited to a single promoter and a relative small number of ITS variants, it is probably the most clear to date example of a correlation between ITS encoded physical property and promoter escape.
ITS could also modulate promoter escape by affecting partitioning of the open complex between productive and unproductive pathways for the escape. The branched mechanism of initiation involving formation of inactive RNAP–promoter complexes that were unable to escape was observed for several promoters (15,28–32). Recent single molecule studies allowed direct observation of long-lived paused or paused-backtracked RNAP complexes further arguing for the existence (at least in some promoters) of the branched pathway of promoter escape (33,34).
There are many ways by which the sequence of ITS could possibly affect promoter escape. RNA/DNA heteroduplex stability and DNA/DNA duplex stability during initial transcription are determined by ITS. Both of these parameters contribute to overall stability of RNAP–promoter complex during initial transcription and thus could affect escape efficiency. DNA scrunching energetics that was proposed to play important role in escape could be also DNA template sequence dependent. There could be also sequence specific interactions of RNAP with downstream duplex, RNA/DNA heteroduplex, single stranded elements of the transcription bubble or with RNA exiting the enzyme. The multiplicity of these potential contributions to the dependence of escape on ITS makes the task of understanding the mechanism of this phenomenon daunting. We hypothesized that solving this puzzle would be facilitated by collecting the data on escape properties of a very large number of ITS variants. Contributions of specific effects to the escape could be then potentially extracted from the data by averaging out (facilitated by large data sets) the confounding competing contributions. Towards this goal, we employed a highly parallel strategy utilizing Next Generation Sequencing (NGS) as a biochemical quantitative readout (35,36) to obtain escape kinetics data on a large set of ITS variants in a context of four promoters. Analysis of these data confirmed that escape kinetics could be profoundly affected by ITS. ITS often can function as an autonomous entity exerting its effect on escape in a promoter-context independent manner. However, strong promoter specific effects were also observed. Overall, the data are consistent with a picture where ITS affects promoter escape through a combination of position dependent effects (most prominently, template directed pausing signals) and position independent effects derived from sequence encoded physical properties of the template.
MATERIALS AND METHODS
Materials
ATP, UTP, GTP and CTP (NTP’s), and heparin were purchased from Sigma (St. Louis, MO, USA). Cy3 NHS ester was from GE Healthcare (Piscataway, NJ, USA). All synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). Escherichia coli core RNAP with a His-tag on the C-terminus of the β' subunit was expressed in BL21(DE3) cells using the polycistronic expression vector (pVS10; a gift from Dr Irina Artsimovitch, The Ohio State University, Columbus, OH) and purified as described in (37). σ70 was expressed and purified as previously described (38). Purified GreB protein was a gift from Dr. Irina Artsimovitch (The Ohio State University, Columbus, OH, USA).
DNA template constructs
DNA duplexes were prepared by one of the following three approaches: (a) PCR amplification of the synthetic oligonucleotide corresponding to full-length nontemplate strand of the desired duplex. This approach was used to prepare the constructs containing randomized segments of ITS. (b) PCR amplification of products of ligation reaction of synthetic oligonucleotides corresponding to promoter (–75 to –1) and transcribed regions (+1 to +60) of the construct. This approach was used to prepare template libraries where different promoters were attached to 96 ITS variants and the libraries of all single base mutants of selected ITS. (c) Extension by PCR of partial duplexes obtained by hybridizing appropriate synthetic oligonucleotides containing complementary overlapping sequences at their 3′ ends (as described in (36)). This approach was used to prepare the constructs labelled with Cy3 at –4 position. Experimental protocols for DNA construct preparation and sequences of all synthetic oligonucleotides used (Supplementary Table S1) are provided in Supplementary Information.
NGS-based analysis of promoter escape
Experimental design for these experiments is illustrated by Supplementary Figure S1 (Supplemental Information). Typically, DNA template (120 nM) and RNAP holoenzyme (200 nM) were mixed in 100 μl transcription buffer (20 mM Tris, 100 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA, 0.1 mM DTT and 5% glycerol) and incubated for 10 min in room temperature to allow formation of the open complex. Transcription was initiated by addition of NTPs (100 μM) and heparin (0.2 mg/ml). Reactions were stopped after 10 s and 10 min with 25 mM EDTA. RNA products were purified using Zymo-Spin RNA Clean & Concentrator-5 (Zymo Research). The 5′-triphosphate of the transcripts was converted to 5′-monophosphate to allow subsequent RNA adaptor ligation. RNA 5′-triphosphate conversion was carried out with RNA-5′-Polyphosphatase (Epicentre) according to manufacturer instructions. Processed RNA was purified using Zymo-Spin RNA Clean & Concentrator-5 and was ligated to RNA adaptor (O247, Table S1e, Supplementary Information) in a 40 μl reaction mixture containing 250 nM adapter and T4 RNA ligase (10 units) for 30 min at 37°C according to manufacturer protocol. Reaction mix was purified on Zymo-Spin RNA Clean & Concentrator-5. Purified ligated RNA products were reverse transcribed with O248 primer (Table S1A, Supplementary Information) in 20 μl reaction using AccuScript High Fidelity Reverse Transcriptase (Stratagene, La Jolla, CA, USA). A 2 μl sample of reverse transcription reaction was used as a template in 20 μl PCR amplification (20–25 cycles) using O249 and O250 primers (Table S1E, Supplementary Information). These primers added sequencing barcodes and the ends compatible with Ion Torrent sequencing. Barcoded PCR products were purified on Wizard SV Gel and PCR Cleanup Kit (Promega, Madison, WI, USA) and their concentrations were determined using Qubit ds DNA BR Assay fluorescence assay kit (Invitrogen, Carlsbad, CA, USA). Barcoded libraries were mixed (to allow multiplexed sequencing of many samples on a single sequencing chip) in equimolar amounts and the mix was purified on Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA). Libraries were sequenced by St. Louis University Department of Biochemistry and Molecular Biology Genomics core facility on Ion Torrent Proton™ (Life Technologies, Carlsbad, CA, USA).
NGS sequencing data were processed using Galaxy (https://usegalaxy.org) FASTX tools. The reads were filtered to include only the reads for transcripts that initiated at +1 position (to eliminate the possible effects of alternative transcription start sites). Filtered reads were trimmed to the region of interest only and read counts for each unique sequence were calculated using FASTX Collapse command. Galaxy text manipulation tools were then used to format the data into text files listing all unique sequences and their corresponding read counts. R script written by us (available on request) was then used to extract from these raw data files read counts for all relevant sequences of interest. Sequences with read counts <10 were filtered out. Read counts were normalized by dividing them by the sum of all read counts and were used to calculate enrichment for each sequence (the ratio of normalized read counts for each sequence at 10 s and 10 min).
Real-time fluorescence assays for promoter escape kinetics
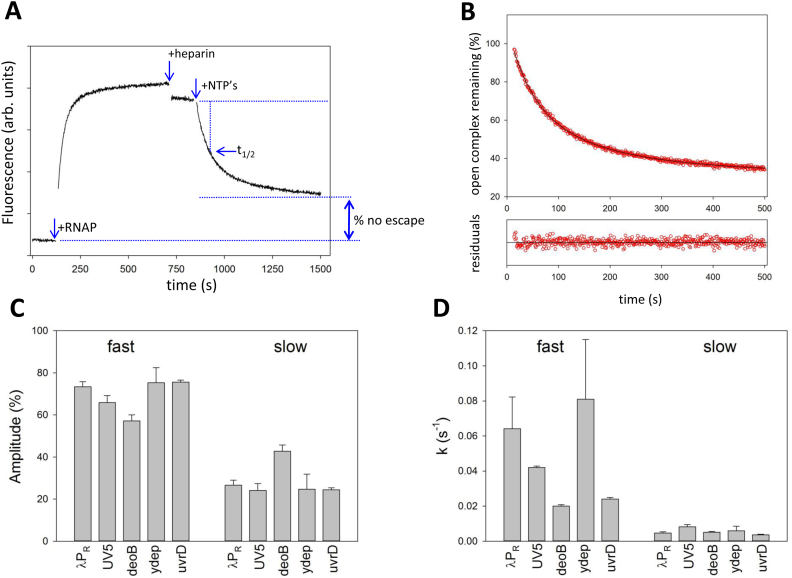
The kinetics of escape for all promoter/ITS constructs was measured in transcription buffer (20 mM Tris 100 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA, 0.1 mM DTT and 5% glycerol) in 200 μl cuvette at 25°C. Emission of Cy3-labelled constructs was recorded at 570 nm (excitation at 540 nm) as a function of time on Aminco-Bowman AB2 spectrofluorometer. Typically, fluorescence of DNA construct (10 nM) was monitored for ∼2 min before RNAP holoenzyme was added to 100 nM. Open complex formation was monitored for ∼10 min after which heparin was added to 0.2 mg/ml. Fluorescence was monitored for ∼2 min after which transcription was initiated by adding the NTPs (100 μM). Fluorescence was monitored until completion of promoter escape reaction (15–30 min). GreB, if present, was at 125 nM. Calculations of t1/2 and % of open complexes that did not escape are explained in Figure 2A. Nonlinear regression fitting of real time fluorescence promoter escape curves was performed using SigmaPlot (Systat Software Inc.).
Figure 2.
(A) Fluorescence assay for real-time monitoring of promoter escape. The data shown are for deoB promoter with deoB ITS. Fluorescence of Cy3 at –4 of the nontemplate strand of the promoter construct was recorded as a function of time. At times indicated by the arrows RNAP, heparin and NTP’s were added. The calculations of t1/2 involved determining the time that was needed for fluorescence signal to reach 50% of the difference between the signal after heparin addition and the signal after completion of escape (obtained from the two-exponential fit illustrated in panel B). Fraction of open complexes that did not escape was calculated as a ratio of the difference between signal at completion of escape and a signal for free DNA to the difference between the signal after heparin addition and the signal for free DNA. (B) Promoter escape follows double exponential kinetics. Portion of the curve from panel A corresponding to promoter escape was fitted to a double exponential kinetic equation. The two rate constants were 0.021 s−1 (54% amplitude) and 0.0056 s−1 (46% amplitude). Residuals plot shows normalized differences between the data and nonlinear fit curve. (C) Amplitudes (averages and standard deviations of minimum 3 repeats) of fast and slow kinetic components for constructs containing deoB promoter and five ITS (indicated by labels in the plot). (D) Rate constants (averages and standard deviations of minimum 3 repeats) of fast and slow kinetic components for constructs containing deoB promoter and five ITS sequences (indicated by labels in the plot).
Calculation of ITS-encoded physical properties
Custom scripts in R (available upon request) were written to calculate DNA/DNA duplex, RNA/DNA duplex, base-stacking and posttranslocated state bias energies for ITS of interest. These scripts allow calculation of total energies for entire sequences as well as for specific segments of the sequences (defined by the 5′ position and the length of the segment). DNA/DNA duplex and RNA/DNA duplex stabilities were calculated using nearest-neighbor model (39) using the parameters from (39) and (40), respectively. Base-stacking energies were calculated using parameters described in (41). Parameters for posttranslocated state bias energy calculations were from (25,26).
RESULTS
Design of highly parallel NGS-based strategy for the analysis of ITS effect on promoter escape kinetics
The main goal of this work was to collect data on escape properties of a large number of ITS variants to enable discovery of relationships between DNA template sequence and promoter escape kinetics that were impossible to detect with limited amount of data previously available. To accomplish this goal, we applied an in vitro NGS-based strategy (Figure S1, Supplementary Information) that allowed parallel studies of escape kinetics of large number of ITS variants (from hundreds to many thousands). This strategy starts with preparing DNA template library where promoter sequence (–75 to –1) is fused with many variants of downstream DNA containing desired ITS variants. Such libraries can be conveniently prepared from synthetic oligonucleotides (as described in Supplemental Information). Promoter escape reaction is then performed on such library and RNA products are collected at different time points of the reaction (Figure S1, Supplementary Information). Promoter escape reactions are performed from preformed open complexes and under single round of transcription condition to eliminate the effect of promoter melting and RNAP rebinding to the promoter. NGS-compatible library prepared from these full-length transcripts is sequenced. Analysis of sequencing data produces read counts for each ITS variant in the library which represent relative amounts of the transcripts. This allows calculating enrichment factor (ratio of relative amounts of transcript for each ITS at 10 s and 10 min of escape reaction) for each ITS. Enrichment factor is a simple parameter that characterizes relative differences in escape kinetics among ITS variants present in the library. Enrichment values for ITS that promote fast escape will be high (>1) because they will be relatively enriched at 10 s compared to 10 min. Conversely, enrichment values for ITS that direct slow escape will be low (<1). We have previously used similar approach to analyse sequence dependence of kinetics of promoter melting (35,36).
ITS can greatly modulate the output of a promoter
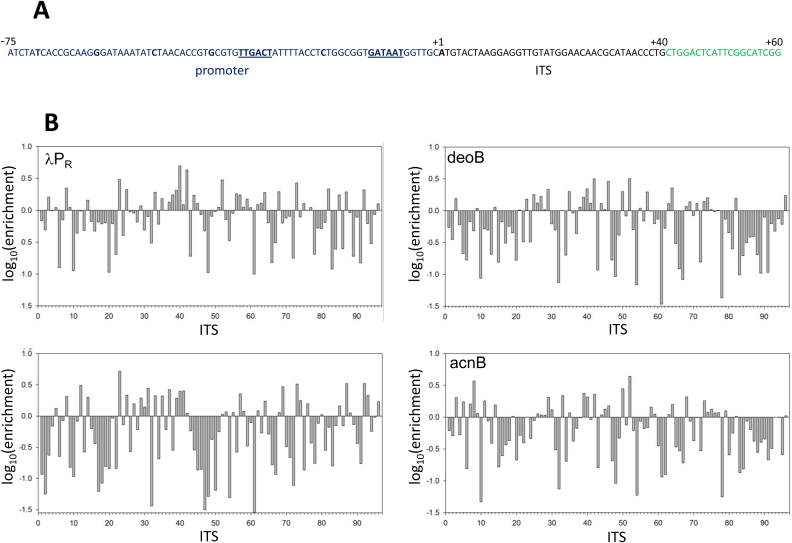
We first investigated to what extent promoter escape kinetics could be modulated by different ITS since a large range of effects of ITS on escape kinetics, if found, would support the potential biological significance of these effects. To answer this question, we examined relative escape kinetics for 42 examples of ITS from E. coli promoters with low (ITS # 7-21, Figure 1B), high (ITS # 22-36&6, Figure 1B) and undetectable (ITS # 37-46&4, Figure 1B) transcript levels in vivo (42). Additional 30 natural E. coli ITS were selected in random and 20 more ITS were generated as random DNA sequence. We expected that this diverse set of ITS variants will allow robust examination of the range of ITS effects on escape. Escape kinetics for all these ITS variants was tested when fused to 4 different promoters to further enhance the diversity of possible ITS effects. The four promoters chosen for the studies included two commonly used model promoters (λPR and UV5), deoB promoter (for which promoter escape was identified by genome wide studies (42) as rate limiting) and acnB promoter (which showed robust transcription in genome wide studies and no signs of slow promoter escape (42)). The ITS of these four promoters were also included in the analysis (ITS # 1–3&5, Figure 1B). A total of 384 promoter-ITS combinations were investigated and results are shown in Figure 1B which shows relative enrichment for each ITS (calculated as a ratio of 10 s reads and 10 min reads, each normalized to the total reads in a given dataset). A wide range of enrichment values was observed (50-fold, 106-fold, 144-fold and 93-fold range between the lowest and the highest value for λPR, deoB, UV5 and acnB, respectively). The data in Figure 1B confirm a strong ability of ITS to modulate escape kinetics and change dramatically the overall output of a promoter.
Figure 1.
(A) Example sequence of a promoter construct used for experiments in panel B. The example shown is for λPR promoter with λPR ITS. Blue font depicts promoter sequence (–75 to –1) and underlined sequence indicates –35 and –10 promoter elements. ITS sequence (+1 to +40) is in black font with +1 position indicated by bold font. Green font sequence corresponds to a common to all constructs target sequence for downstream PCR primer (O105, Table S1A, Supplementary Information). (B) Promoter escape kinetics for 96 ITS variants in the context of λPR, deoB, UV5 and acnB promoters. The numbers on x-axis correspond to following ITS: 1:deoB, 2:UV5, 3:λPR, 4:argF, 5:acnB, 6:cyoA, 7:nfo, 8:rfe, 9:cho, 10:proVp1, 11:proVp2, 12:proVp3, 13:yfiRp5, 14:pstS, 15:spy, 16:ansBp2, 17:parCp4, 18:hipB, 19:yfiEp7, 20:cvrAp6, 21:acsp2, 22:rpoS, 23:serAp1, 24:fliAp1, 25:secG, 26:fnr, 27:ssrAp2, 28:ssrAp, 29:cysKp1, 30:fimA, 31:ptsHp2, 32:serC, 33:purA, 34:slp, 35:frrp3, 36:frrp, 37pheV, 38:yliEp6, 39:yehLp3, 40:yhflp5, 41:yhflp4, 42:ydeP, 43:yneEp7, 44:mtrp2, 45:yiaYp4, 46:yiaYp6, 47:caiF, 48:phoA, 49:cspEp1, 50:smtA, 51:ycdT, 52:ydeEp1, 53:pykAp2, 54:znuC, 55:yeiGp6, 56:yffSp4, 57:yfiRp3, 58:lpoAp2, 59:def, 60:yiaDp3, 61:uvrD, 62:rrsB, 63:lptFp5, 64:yjtDp6, 65:dksAp3, 66:proS, 67:fepA, 68:gltA, 69:ompA, 70:rihB, 71:greA, 72:envR, 73:damp2, 74:gntK, 75:yiaYp4, 76:hypTp4, 77:rnd1, 78:rnd2, 79:rnd3, 80:rnd4, 81:rnd5, 82:rnd6, 83:rnd7, 84:rnd8, 85:rnd9, 86:rnd10, 87:rnd11, 88:rnd12, 89:rnd13, 90:rnd14, 91:rnd15, 92:rnd16, 93:rnd17, 94:rnd18, 95:rnd19, 96:rnd20. Sequences of these ITS are listed in Table S1A, Supplementary Information.
Enrichment parameters properly represent relative differences in escape kinetics despite the underlying kinetic complexity of the process
Enrichment parameter is a simple, convenient but rather crude representation of escape kinetics that informs about overall differences in escape kinetics between ITS variants but neglects the kinetic details behind these differences. To gain insights into the details of escape kinetics and to test if enrichment parameter derived from NGS-based approach is a proper representation of relative differences in escape kinetics, we employed fluorescence assay (11) to follow escape kinetics of selected ITS variants in real-time. We selected five ITS (λPR, UV5, deoB, ydep, uvrD) spanning a range of enrichment values (Figure 1B) for these kinetic studies. Real-time escape assay employs fluorescence probe (Cy3) attached to a nontemplate strand base near transcription start site (11,43). Fluorescence intensity of the probe increases ∼2-fold upon formation of the open complex (Figure 2A) and disappearance of this high fluorescence state upon promoter escape by RNAP can be used to follow the escape kinetics (Figure 2A). Escape kinetics for all five ITS examined required two exponentials to fit experimental data (Figure 2B). The signal upon completion of the escape reaction did not return the level for free DNA (Figure 2A) indicating that some open complexes failed to escape. The fraction of these complexes could be calculated as illustrated in Figure 2A. The faster kinetic component of the escape was dominant as its amplitude was ∼55–80% of the total fluorescence change (Figure 2C). The differences in overall escape kinetics between different ITS were mostly due to changes in the rate constant of the fast kinetic component (Figure 2D). The simplest interpretation of such kinetic behavior is the existence of at least two populations of initially transcribing complexes differing in escape kinetics. Such interpretation is consistent with many reports in literature where heterogeneity in initially transcribing complexes was observed (15,28–30,32–34), including observations of parallel pathways where either RNAP pausing or backtracking leading to slow escape was detected (33,34). To test if backtracking is a factor under our experimental conditions, we measured escape kinetics in the presence of GreB (transcript cleavage factor (44,45)) (Figure S2, Supplementary Information). GreB can rescue backtracked RNAP by catalyzing cleavage of RNA that is misplaced into the secondary channel of the backtracked enzyme rendering the enzyme inactive (44,45). GreB reduced the amplitude of slow kinetic component without changing the slow rate constants and increased the amplitude of fast component increasing its rate constant ∼ 2-fold (Figure S2 A&B, Supplementary Information). These changes produced ∼50% increase in the overall escape rate as measured by reaction half-time (t1/2) (Figure S2C, Supplementary Information). The fact that slow kinetic component was not completely eliminated by GreB (Figure S2A and B, Supplementary Information) may indicate presence of additional mechanisms for delaying escape that do not respond to GreB. Recent report on promoter escape kinetics (46) that used real-time escape assay utilizing fluorescence signal of a probe incorporated into σ subunit also found that generally GreA and GreB increased the rate of escape but on some templates GreB could also inhibit escape. While the observed effects of GreB on escape rates are consistent with the presence of significant amounts backtracking complexes during escape under our experimental conditions, a detailed study of GreA/GreB effects on escape (and template-sequence dependence of these effects) will be needed to fully understand these effects. For example, it was reported that GreB could decrease stability of the open complex (47) which could also affect the rates of promoter escape.
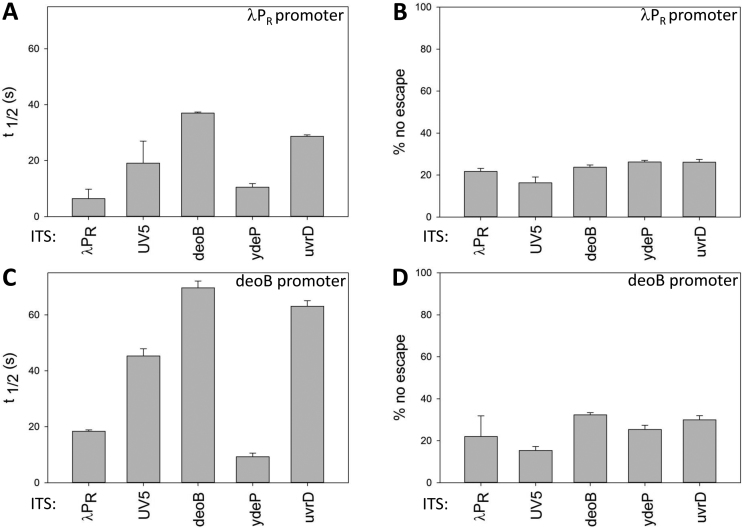
Since escape did not follow a simple kinetics, we chose to use a simple parameter (reaction half-time (t1/2, Figure 2A)) as a convenient way to describe overall escape kinetics with a single parameter (Figure 3A and C). The data in Figure 3A and C illustrate well how the overall escape rate reflects a combination of the effects of promoter sequence and the ITS. Different ITS produced a very similar pattern of t1/2 values when tested in λPR promoter (Figure 3A) or deoB promoter (Figure 3C). However, all t1/2 were two to three times longer in deoB promoter compared to λPR reflecting slower escape kinetics imposed by deoB promoter sequence.
Figure 3.
Effects of five selected ITS on overall promoter escape kinetics (half-time of escape reaction (panels A and C) and the amount of unproductive open complexes (panels B and D) of two promoters: λPR (panels A and B) and deoB (panels C and D).
Real-time escape experiments allowed also the estimation of the fraction of open complexes that failed to escape (Figure 3B and D). Shimamoto proposed the existence of such ‘moribund’ complexes (28–30) which were permanently trapped in abortive initiation cycle and were unable to escape. Recent studies suggested that more than a half of open complexes could be trapped in a state that did not escape (15). If the fraction of these inactive complexes would depend on ITS, this could add an additional level of regulation of escape by ITS, in addition to escape kinetics. The data in Figure 3B and D show that fraction of open complexes that were unable to escape was ∼15–30% and no clear pattern of dependence on ITS could be detected. In the presence of GreB a modest reduction of the fraction of complexes that did not escape could be observed (Figure S2C, Supplementary Information), in agreement with previously proposed role of backtracking in the formation of non-productive initiation complexes (31) and recent kinetic investigation (46). Decrease of open complex stability (47) could also contribute to the observed effect of GreB on fraction of complexes that did not escape.
To test if enrichment parameter derived from NGS-based approach is a proper representation of relative differences in escape kinetics, we compared the values of t1/2 for the five ITS tested using real-time fluorescence escape assay with their corresponding enrichment values from NGS-based experiment. The correlation observed (correlation coefficients = 0.81, Figure S3 A&B, Supplemental Information), supports the notion that despite the underlying kinetic complexity of promoter escape, NGS-derived enrichment parameters provide imperfect but proper representation of relative differences in the overall escape kinetics of ITS variants. This conclusion is also supported by comparing enrichments from NGS-based experiment and t1/2 from kinetic experiment where time-dependent formation of full-length transcript was measured using molecular beacon assay (Figure S3C, Supplemental Information). We noted that the range of enrichment values for the five ITS tested was larger compared to the range of corresponding t1/2 values (Figure S3, Supplemental Information) indicating a nonlinear relationship between these two parameters. Nonlinear relationship between enrichment and t1/2 values that we observed is consistent with complex escape kinetics revealed by our real-time escape kinetics experiments discussed above and with single molecule observations (33,34,48) (Dulin, D.B. et al. bioRxiv 199307, 2017; doi:https://doi.org/10.1101/199307).
Promoter context can modulate ITS effects on escape
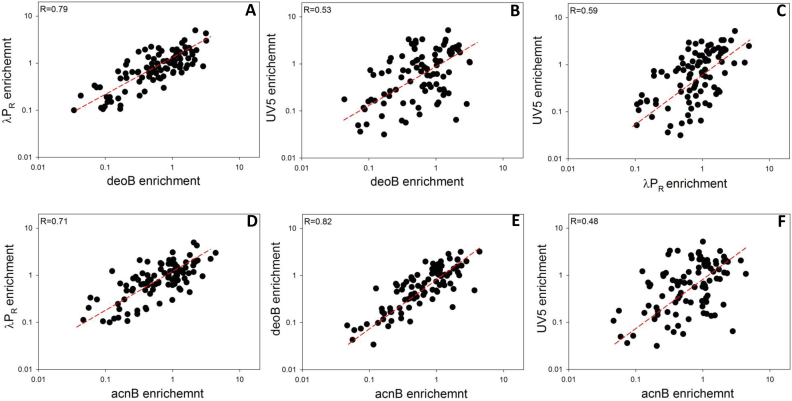
Since interactions of RNAP with core promoter elements involve parts of the enzyme (mostly σ subunit) that are not likely to have direct contact with ITS, we expected that effects of ITS on escape would not depend on promoter context. ITS and promoter in this view would affect escape independently and the overall escape rate would depend on a simple summation of both contributions. Our approach to address this question was to probe correlation between effects of ITS on escape in a context of four promoters used in experiments shown in Figure 1B (Figure 4A–F). Correlation coefficients ranged from 0.82 (deoB versus acnB promoter context; Figure 4E) to 0.48 (UV5 versus acnB promoter context; Figure 4F). The good correlation between most promoter pairs indicated that in general ITS could operate as independent units affecting promoter escape in similar manner regardless of the promoter from which transcription proceeds. Moderate correlations observed for some promoter pairs suggested however that promoter dependence of ITS effects could also occur. Correlation coefficients involving UV5 promoter were the lowest (Figure 4B, C and F) suggesting the strongest promoter-specific effects for this promoter. Correlation coefficient between acnB and deoB promoters was the highest and these two promoters produced similar correlations when paired with the remaining promoters indicating no significant differences between ITS effects in these two promoters. Accordingly, we have chosen to limit further analyses to λPR, deoB and UV5 promoters since the outcomes in acnB promoter seemed redundant with deoB promoter.
Figure 4.
Correlation between the effects of 96 ITS variants on promoter escape in a context of four promoters indicated by axis labels in panels A–F.
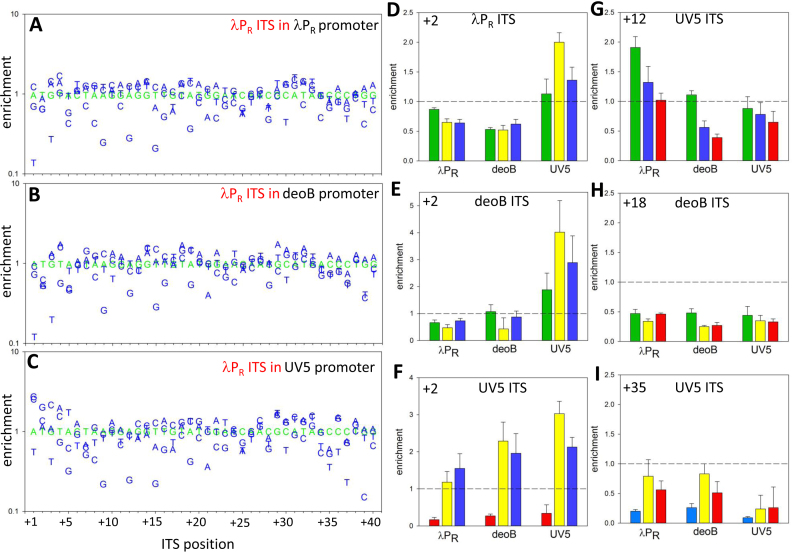
To further probe promoter-context dependence of ITS effects on escape, for three ITS (λPR, deoB and UV5) we prepared DNA template libraries containing all single base substitutions at each position of 40 bp ITS (120 sequence variants for each ITS). We used these templates to determine if the effects of these substitutions on escape depended on promoter context (Figure 5 and Figure S4, Supplementary Information). Many single base substitutions resulted in significant changes in escape kinetics as measured by changes in enrichment values for corresponding ITS (Figure 5 and Figure S4, Supplementary Information) illustrating again the importance of ITS for the overall output of the promoter. Base-substitutions more often were inhibitory (58.5% of all substitutions in nine combinations of promoter-ITS examined). The average (from nine combinations of promoter-ITS) maximum fold change was higher for inhibitory substitutions (7.1-fold versus 2.9-fold for stimulatory substitutions). This could suggest that natural ITS may have evolved to avoid strong inhibitory sequences as previously suggested (10,12,21). In agreement with the data in Figure 4, correlation plots for the effects of single base substitutions on escape in different promoter contexts revealed a range of correlations coefficients (from 0.82 to 0.35; Figure S5, Supplementary Information) indicating both the existence of promoter context dependent and independent effects of base substitutions. While the majority of base substitutions produced similar outcomes regardless of promoter from which transcription originated, clear examples of promoter dependent effects could also be seen. Mutations at transcription start site (+1) had large effect on escape (Figure 5 and Figure S4, Supplementary Information) which was expected due to previously described strong preferences for the initiating NTP. It was not expected though to observe that the same base substitutions at this position in the same ITS, could have completely opposite effects on escape in a context of different promoters (for example, Figure S4B and C, Supplementary Information). Position +2 is another site of clear promoter specific effects of base substitutions. Base substitutions at this position in λPR or deoB ITS had radically different effects in λPR or deoB promoter contexts compared to UV5 promoter context (Figure 5D and E) suggesting that UV5 promoter imposes specific base requirements at +2 position of ITS. Interestingly, base changes at position +2 in UV5 ITS produced similar effects in all three promoter contexts (Figure 5F) suggesting that UV5 promoter specific base preferences at +2 can also depend on sequence context of the ITS. Promoter-specific effects were not limited to ITS positions at or very near transcription start-site. For example, base changes at position +12 of UV5 ITS produced significantly different outcomes in λPR promoter context compared to deoB or UV5 promoter context (Figure 5G). We expected that significant effects of base substitutions in ITS on escape would not extend beyond ∼15 bp of transcribed sequence because the abortive products are rarely longer that ∼15 nt (typically 10 nt or less). Although indeed the first ∼10 bp of ITS exhibited the highest sensitivity to mutations (Figure S6, Supplementary Information), significant effects of mutations were observed all the way up to position +40 (Figure 5 and Figure S4, Supplementary Information). Figure 5H and I show examples of such effects at position +18 and +35, respectively. While it is possible (in fact, likely) that base substitutions at these far away positions influenced the production of the transcript by affecting post-escape events, it is nevertheless revealing to see that a single base substitution in DNA template could have relatively large effect on promoter output. Taken together, the main impact of the data shown in Figures 4 and 5 and in Supplementary Figure S4 (Supplementary Information) is a demonstration of unexpected ‘allosteric’ abilities of a promoter to modulate ITS effects on escape.
Figure 5.
Effects of single base changes in 40 bp λPR ITS on promoter escape in a context of λPR promoter (A), deoB promoter (B) and UV5 promoter (C). Wt sequence is shown in green. Examples of promoter context dependence of base changes at a specific position (+2, D–F; +12, G). Examples of significant effects of base changes on promoter escape at positions far away from +1 (+18, H; +35, I). Bar colours in panels D–I depict identity of the base: green—A; yellow—G; red—T; blue—C.
In-depth analysis of the effect of first 10 bp of ITS on promoter escape
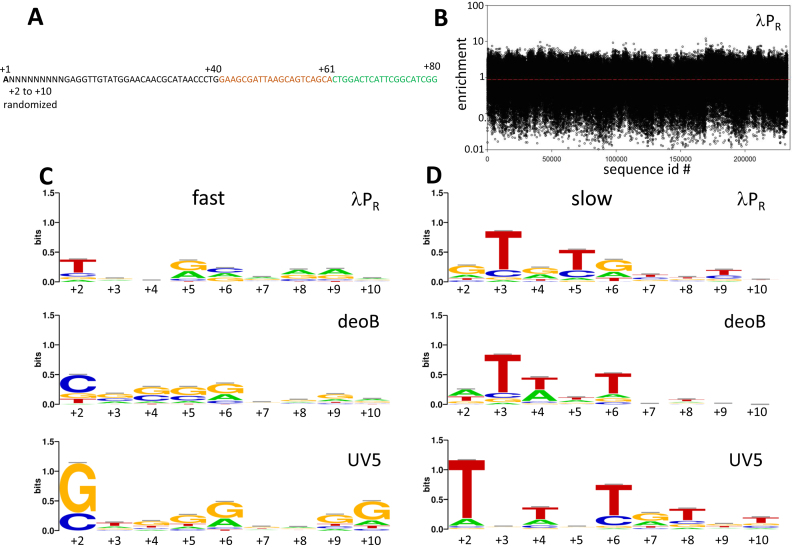
Since the identity of the first ∼10 bp of the transcribed region had the highest impact on escape kinetics (Figure S6, Supplementary Information), in the next series of experiments we targeted this region for detailed analysis. We designed DNA templates coding for libraries of ITS variants containing all possible (262,144) base combinations at each positions of +2 to +10 of ITS (Figure 6A). We prepared three such constructs where λPR, deoB and UV5 ITS containing fully randomized sequence from +2 to +10 were combined with their own promoters (i.e. λPR-λPR(+2 to +10 RND), deoB-deoB(+2 to +10 RND) and UV5-UV5(+2 to +10 RND) constructs). In all these constructs we kept the wt base (A) at +1 position to eliminate strong effects of the identity of initiating nucleotide. We used NGS-based approach to determine ratio of the amounts of each sequence variants at 10 s and 10 min of escape reaction (enrichment, Figure 6B). The range of enrichment values in Figure 6B (also, Figure S8, Supplemental Information) covers ∼3 orders of magnitude, again emphasizing the large potential of ITS for modulating the output of a promoter. Enrichment patterns observed were most similar between λPR and deoB constructs (correlation coefficient = 0.58) and less similar between UV5 and deoB (correlation coefficients = 0.39) or between UV5 and λPR (correlation coefficient = 0.19). This is consistent with UV5 promoter having the strongest promoter specific bias of ITS effects, as already seen in the experiments with 96 ITS variants (Figure 4 and Figure S5, Supplemental Information).
Figure 6.
(A) Example of the sequence of promoter construct for experiments with randomized sequence from +2 to +10 (only +1 to +80 region is shown). The example shown is for λPR promoter. ITS sequence (+1 to +40) is in black font with +1 position indicated by bold font. Green font sequence corresponds to a common to all constructs target sequence for downstream PCR primer (O105, Table S1A, Supplementary Information). Orange font depicts a sequence complementary to the molecular beacon (O1251, Table S1A, Supplementary Information) used in some experiments to measure full-length transcript. Sequences of all other constructs used are in Supplementary Table S1D (Supplemental Information). (B) Enrichment values for experiment with λPR promoter. Sequence logos for fast (highest enrichment values) 500 sequences (C) and slow (lowest enrichment values) 500 sequences (D) in the context of λPR, deoB or UV5 promoters. Enrichment value ranges for fast 500 ITS were 4.7 to 11.9, 3.4 to 24.0 and 5.7 to 17.4 for λPR, deoB or UV5 promoters, respectively. Enrichments value ranges for slow 500 ITS were 0.008 to 0.031, 0.019 to 0.061 and 0.017 to 0.048 for λPR, deoB or UV5 promoters, respectively.
The data such as those illustrated by Figure 6B provide an opportunity to discover correlations between escape and ITS that previously might not have been obvious due to insufficient volume of data. Many DNA template sequence dependent effects could contribute to escape control by ITS obscuring the clarity of the impact of each of these effects. With large volume of data, the possibility of identifying contributions of individual DNA sequence-dependent contributions should be enhanced. The simplest and the most obvious question that could be asked using the data in Figure 6B is if there are any specific sequence patterns that correlate with fast or slow escape. No such obvious conserved sequence patterns were identified in the past for ITS, but with the large datasets that we obtained in experiments with templates illustrated in Figure 6A, it was worth revisiting this question. Figure 6C and D shows sequence logos for fast escaping ITS (top 500 sequences with highest enrichment) and slow escaping ITS (bottom 500 sequences with lowest enrichment), respectively. For each of the constructs tested, significant preferences for specific bases at various positions within the +2 to +10 region of ITS were observed. These preferences were more pronounced for slow escaping sequences (Figure 6D) and while there was some similarity of these base preferences between different promoters, there were also clear promoter-specific preferences. For example, there was a strong preference for T at +2 and G at +2 for slow and fast escaping ITS variants, respectively, that was unique to UV5 promoter (Figure 6C and D). These strong base preferences at +2 for UV5 promoter are in agreement with the results of single base replacement experiments in UV5 (Figure S4D–F, Supplementary Information). Additional kinetic experiments (Figure S7, Supplementary Information) further illustrate how the identity of the base at +2 in ITS that exhibit fast or slow escape kinetics in the context of λPR promoter can produce an opposite behaviour when placed in the UV5 promoter context.
Slow escaping sequences exhibited a characteristic pattern of 2–3 T’s, most often spaced 2 bp from each other (Figure 6D). These T’s appeared mostly within the first ∼7 bp of the template and in the case of λPR were often followed by a G. While these patterns of T’s were seen in all promoter contexts, the exact positions of these T’s depended on the promoter context. For example, there was a strong preference among slow escaping ITS for a T at +3 in λPR and deoB but not in UV5 (Figure 6D). Also, there was a preference for T at +6 in deoB and UV5 but not in λPR. The existence of both the similarities and promoter context-dependent differences in the data in Figure 6 is consistent with the overall picture where ITS on one hand could affect the escape as independent unit regardless of the promoter from which transcription proceeds but on the other hand, some preferences for specific bases at different positions depending on promoter context are also possible.
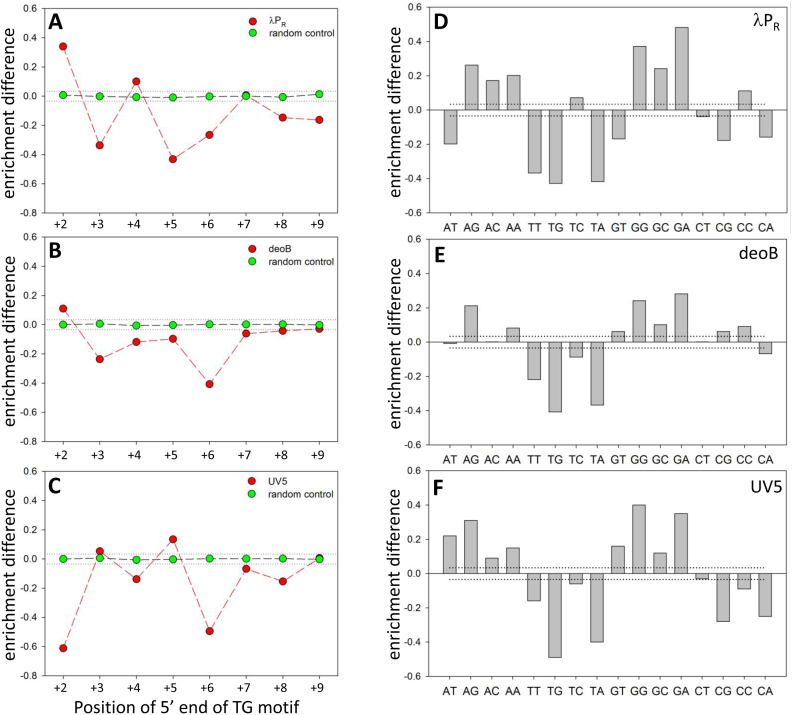
Recent in vivo studies have identified G–10(T/C–1)G+1 (subscripts describe relative positions with –1 corresponding to 3′ end of RNA) motif as a strong elongation pause-inducing signal (49–51). Very similar sequence was identified as elongation pausing signal in in vitro single molecule study (52). The (T/C-1)G+1 part of this motif exhibits some resemblance to the logos for slow escaping sequences (for example, +5/+6 positions in λPR or +6/+7 positions in UV5, Figure 6D). Furthermore, recent single molecule studies have shown that RNAP can pause during transcription initiation (33,34). Taken together, these observations suggest that specific base preferences illustrated by logos for slow escaping ITS (Figure 6D) could reflect the pausing inducing potential of such sequences. Increased probability of pausing would be detected in our experiments as slower escape. To investigate the effect of TG motif (the strongest signal in Figure 6D) on promoter escape in more detail, we analysed the average effect of the TG motif on promoter escape at each position within +2 to +9 region (Figure 7A–C). For this purpose, we calculated the difference between the average of enrichment values for all 16 384 sequences containing TG motif at a given position (i.e. all sequences with a TG at a given position and with all possible sequence variations at the remaining seven positions) and the average enrichment of the entire dataset. This enrichment difference is thus a robust depiction of template position dependence of the overall impact of TG motif on promoter escape. One way to view enrichment differences in Figure 7 is to consider them as reflecting an ability of a given dinucleotide motif at a specific position to move the mean of 16 384 enrichments away from the mean of randomly selected 16 384 sequences. For this to happen, the motif has to have a consistent and significant effects on the enrichment values in a context of many sequences. Compared to random control, TG motif plots exhibit a characteristic pattern for each promoter tested with a maximum negative enrichment difference at +6 in the case of deoB and UV5 (Figure 7B and C) and +5 in case of λPR (Figure 7A). The enrichment differences at +2 position are most likely due to already discussed strong preferences for specific bases at this position and are unrelated to the putative pausing activity of TG motif. Observed enrichment differences were highly statistically significant as illustrated by the dotted lines in Figure 7 that depict the boundary for statistical significance (P-value < 0.0001) of enrichment differences over random control. The effects illustrated in Figure 7A–C were specific to TG motif since much smaller enrichment differences were observed when TC or GT motifs were examined and a characteristic pattern of differences observed for TG motif was not observed (Figure S9, Supplementary Information). Previous studies have concluded that pausing during initiation occurs near position +6 because the steric clash between growing RNA chain and sigma region 3.2 that blocks RNA exit channel begins with 6 nt long RNA product (33). The fact that we observed the maximal effect of TG motif also near position +6 further supports the idea that initiation pausing is the explanation for the position specific base preferences illustrated in Figure 6D. However, our data suggest that the exact position where the maximum pausing could occur may not be limited to +6 and it could be also promoter context dependent (Figures 6 and 7).
Figure 7.
Effect of TG motif at a specific position within +2 to +10 region in a context of λPR promoter (A), deoB promoter (B) and UV5 promoter (C) on escape kinetics averaged over all base combinations at remaining positions (16,384 sequences for each TG position). Enrichment difference (y axis) corresponds to a difference between calculated averaged enrichment with TG at a given position and the average of all enrichment values in the dataset. Random control (green symbols) corresponds to calculations performed on 16,384 randomly selected sequences. Dinucleotide sequence preferences at +5 (λPR promoter; panel D) or +6 (deoB promoter, panel E; UV5 promoter, panel F). Dotted lines depict boundary for statistical significance of enrichment differences over the random control (i.e. enrichment differences between and outside the dotted lines have P-values >0.0001 and <0.0001, respectively).
We further analysed base preferences for the template positions where maximum negative effect on escape of TG motif was observed (+5 in case of λPR and +6 in case of deoB and UV5) by calculating enrichment differences for all 16 dinucleotide combinations (Figure 7D–F). TG motif had the largest negative impact on escape followed by TA. In two promoter contexts (λPR and UV5), CG and CA had also significant negative impact on escape (Figure 7D and F) consistent with consensus pausing sequence for elongating RNAP (49,50). GG and GA had the largest opposite effect (i.e. they favoured escape; Figure 7D–F) suggesting their anti-pausing activity. The preference for GG or GA near +6 could be also seen in the logos for fast escaping sequences (Figure 6C). The repeat of TG motif (TGTG) had a much stronger negative effect on escape (Figure S10, Supplementary Information) compared to a single TG demonstrating the additivity of TG escape inhibitory activity.
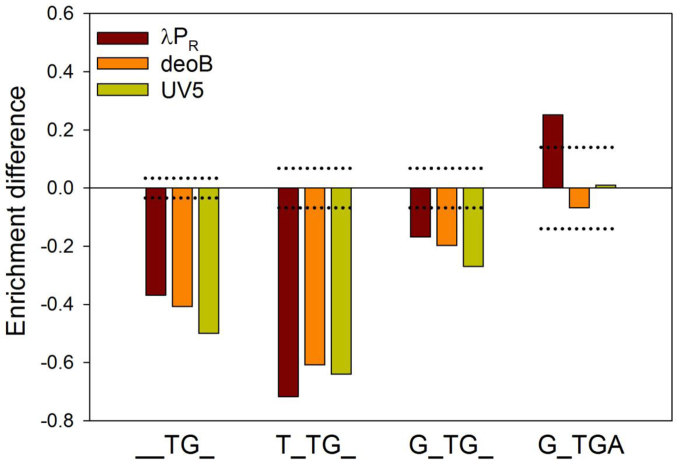
While TG motif at +6 (or +5) on average had a negative impact on escape, its effect was sequence context dependent, i.e. it could be enhanced, decreased or even eliminated depending on specific sequence at remaining positions in +2 to +10 region. This is illustrated by logos for fast and slow escaping sequence containing TG at +5 (λPR) or +6 (deoB or UV5) (Figure S11, Supplementary Information). Sequences encoding faster escape exhibited some preference for A immediately after TG motif and a G 2 bp upstream of TG (Figure S11A, Supplementary Information), whereas sequences encoding slower escape exhibited preference for T 2 bp upstream of TG (Figure S11B, Supplementary Information). The logos in Supplementary Figure S11 (Supplementary Information) showed similarity with logos in Figure 6 suggesting that impact of flanking sequences on TG effects mirrors their overall effects on promoter escape observed with all data. The possibility of strong impact of sequence context on the effect of TG motif is further demonstrated by comparing average enrichment difference (Figure 8) due to the presence of TG and expanded motifs (T_TG, G_TG, and G_TGA) derived from the logos in Supplementary Figure S11 (Supplementary Information). The enhancement of TG effect by a T 2bp upstream and its elimination (or even reversal as in case of λPR) in case of G_TGA motif is apparent.
Figure 8.
Inhibitory effect of TG at +5 (λPR) or +6 (deoB or UV5) can be enhanced or eliminated by its sequence context. Enrichment difference for each promoter for TG motif alone is compared with TNTG, GNTG and GNTGA. Dotted lines depict boundary for statistical significance of enrichment differences over the random control (i.e. enrichment differences between and outside the dotted lines have P-values >0.0001 and <0.0001, respectively).
Sigma-dependent promoter proximal pausing is a well-established property of promoters containing –10 like sequence in the transcribed region near the promoter (53,54). We were thus curious, if a potential inhibitory effect of such sequence on escape could be also detected in our NGS-based analysis. Supplementary Figure S12 (Supplementary Information) shows that indeed the presence of such sequence in +2 to +10 region inhibited escape with the peak of the effect observed when 5′ end of the sequence was at +3 or +4. This provides further support to the idea that template sequence directed pausing of RNAP during escape is an important mechanism by which ITS could affect promoter escape.
Taken together, it appears that template position specific base preferences illustrated by Figures 6–8 (and Figures S9–S12, Supplementary Information) for fast or slow escaping sequences are best explained by sequence-dependent modulation of RNAP pausing during initiation. The exception here are strong base preferences at position +2 that seem to have a different underlying biophysical mechanism.
Correlations between sequence encoded physical properties of DNA template and escape
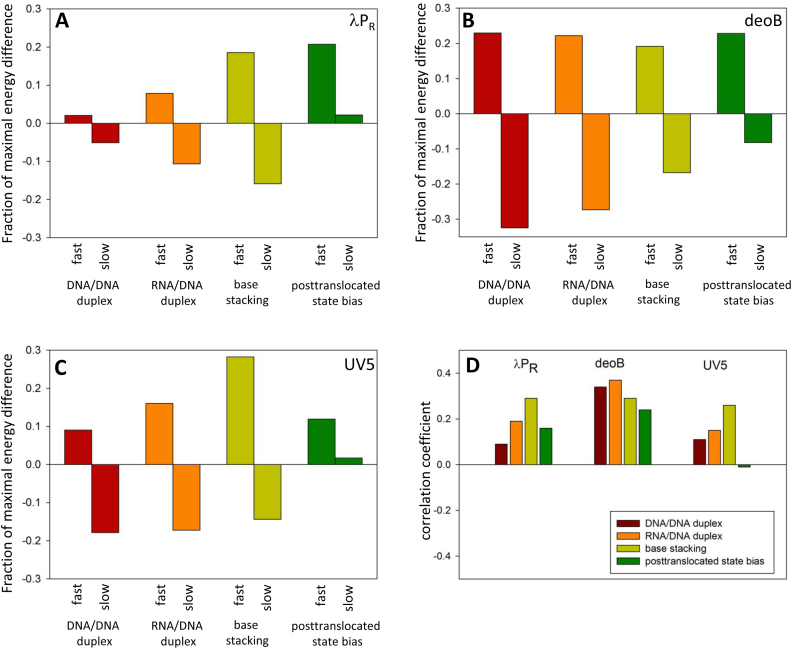
ITS sequence determines various physical characteristics of the DNA or RNA encoded by ITS. Some of these characteristics could affect promoter escape providing another possible mechanism for template sequence dependence of the escape. For example, ITS encodes thermodynamic stability of RNA/DNA heteroduplex which could affect the probability of short transcript dissociation from the template which in turn could be a factor in escape kinetics. Similarly, ITS encodes thermodynamic stability of DNA/DNA duplex that needs to be unwound for transcription through the ITS. The energetic cost of duplex unwinding could also affect the escape kinetics. These template sequence dependent but position independent effects are challenging to identify but with our data describing escape properties of a large number of ITS variants it was worthwhile to probe this issue. Our approach was to calculate some of these sequence-encoded properties for all sequence variants probed experimentally in experiments with templates randomized at +2 to +10 and to examine if any bias in these properties could be detected between fast and slow escaping sequences. The most obvious candidates for a role in promoter escape are the stability of DNA/DNA or RNA/DNA duplex for the reasons described above. Both of these properties exhibited a specific bias (Figure 9A–C) where fast and slow escaping sequences on average had higher and lower than average duplex stability, respectively. Similar trends are also detectable in the entire data set where small (indicating a trend rather than strong relationship) but statistically significant correlation coefficients between the enrichment parameter (i.e. escape kinetics) and duplex stability were observed when all 262 144 sequence variants were examined (Figure 9D). The parameters that are used to calculate DNA/DNA and RNA/DNA duplex stabilities are very similar (39,40). Similar patterns observed (Figure 9) for these two ITS encoded physical properties are thus not surprising. However, these parameters are not identical and we noted that RNA/DNA stability in general produced more pronounced bias for fast and slow escaping sequences (Figure 9A–C) and higher correlation coefficients for entire data sets (Figure 9D) compared to DNA/DNA duplex stability.
Figure 9.
DNA/DNA duplex stability, RNA/DNA duplex stability, base stacking energy and posttranslocated state bias for fast (top 500 enrichment values) and slow (bottom 500 enrichment values) sequences for λPR (A), deoB (B) and UV5 (C). The Y-axis depicts a difference between the average value for fast or slow sequences and the average value for all 262 144 variants of 9 nt sequence. This difference is expressed as a fraction of maximal energy difference from the average within all variants of 9 nt sequence. (D) Correlation between enrichment values and DNA/DNA duplex stability, RNA/DNA duplex stability, base stacking energy and posttranslocated state bias. All energy differences between fast and slow sequences in panels A–C were statistically significant (P-value < 0.0001). All correlation coefficients in panel D (with the exception of posttranslocated state in case of UV5) were statistically significant (P-value < 0.0001).
We also examined base stacking energy as a possible ITS-encoded physical property that could play a role in escape. This analysis was motivated by work from Murakami laboratory (55) that identified a stacking interaction between initiating NTP and –1 base of DNA template strand as an important contributor to efficient initiation. We reasoned that the role of base stacking might not be limited just to the initiating NTP but could also extend to stacking between the 3′ end of the growing RNA chain and incoming NTPs at subsequent positions of the template. Such stacking could both stabilize the short RNA/DNA heteroduplex (decreasing the probability of dissociation of short RNA product) as well as stabilize the initial binding of the NTP. Base stacking between 3′ end of RNA and the incoming NTP could occur very early on in the transcription reaction cycle (before the chemical processes of making the new phosphodiester bond) which could be especially beneficial when RNA product is still very short. Base stacking exhibited a clear bias for higher than average stacking energy associated with fast escaping sequences and lower than average base stacking energy associated with slower escaping sequences (Figure 9 A-C). There were also small but statistically significant correlations between the enrichment and base stacking within entire data sets (Figure 9D). In two out of three promoters, base stacking energy bias for fast and slow escaping sequences and the correlations within entire data sets for base stacking energy were stronger than for DNA/DNA or RNA/DNA duplex stability (Figure 9).
It was previously reported (25) that the ratio of abortive to productive transcripts for variants of N25 ITS correlated well with experimentally determined RNA sequence dependent energy describing the bias between pretranslocated and posttranslocated states of RNAP (26). We thus examined if a correlation between the bias towards posttranslocated state and escape kinetics (enrichment parameter) could be also detected in our datasets containing very large number of ITS variants. We found that differences between posttranslocated state bias for fast and slow escaping sequences (Figure 9A–C) where the least consistent among the physical parameters tested in Figure 9. Analysis of correlation between posttranslocated state bias and escape kinetics for entire datasets showed that posttranslocated state bias had the lowest correlation coefficient in deoB and UV5 among physical properties tested and no statistically significant correlation was found in UV5 (Figure 5D).
DISCUSSION
Molecular mechanisms that define promoter escape are likely complex and involve composite molecular underpinnings. Our data relates sequence of many variants of ITS to the relative kinetics of promoter escape they encode. Such data on its own does not identify exact molecular basis of how a given ITS affects escape. To resolve these questions, more detailed mechanistic studies combining rapid kinetics studies and single-molecule investigations will be needed. Previously unknown relationships between template sequence and escape kinetics identified in our work will provide guidance for selecting ITS for such studies. Nevertheless, several new insights into the mechanism by which ITS could affect promoter escape can be made by analysing our data with the aid of previously described observations.
RNAP pausing during initiation is one of the primary means by which promoter escape kinetics is modulated by ITS
Analysis of escape kinetics of a large number of ITS variants confirmed that the outcome of transcription initiation for a promoter could be profoundly affected by the sequence of ITS. Clear position-specific base preferences that we observed (Figure 6) were similar to previously described elongation pausing signals (49–52). Recent single molecule studies demonstrated formation of long-lived pauses during initiation (33,34,48) (Dulin, D.B. et al. bioRxiv 199307, 2017; doi:https://doi.org/10.1101/199307) and implicated YG sequence motif in directing the formation of such long pauses (48). Taken together, these observations demonstrate that RNAP pausing during initiation is one of the primary means by which promoter escape kinetics is modulated by ITS. (T/C)G step appears to be particularly difficult for RNAP to transcribe through and in the presence of additional stress, this can lead to pausing. In the case of elongation pausing, the additional stress is due to RNAP interaction with G–C pair at the upstream end of RNA-DNA hybrid (or due to unwinding of a stable G–C base pair positioned 10 bp upstream of (T/C)G motif in concert with transcription through (T/C)G) (49–52). In the case of initiation, the additional stress likely results from the need to displace the polypeptide (sigma region 3.2) blocking the RNA channel when transcription reaches ∼ +6 position (18,31,55–57). Single molecule studies on initiation pausing support this view (33,34,48) (Dulin, D.B. et al. bioRxiv 199307, 2017; doi:https://doi.org/10.1101/199307) and suggest that the paused state at +6 could function as a checkpoint directing RNAP either to productive or non-productive pathways of initiation (Dulin, D.B. et al. bioRxiv 199307, 2017; doi:https://doi.org/10.1101/199307). The sequence near +6 position will thus have a strong effect on escape as illustrated by Figure 7D–F. However, while we observe the maximum effect of (T/C)G motif at or around +6, we can also see its effect at other positions (for example, +3 in case of λPR (Figure 7A). Furthermore, the enlarged motifs consisting of repeats of (T/C)G motif (for example, TGTG or TGCG, Figure S9, Supplementary Information) inhibit escape more effectively and at essentially any position of ITS. These observations suggest that general relative instability(stress) of initial transcription complexes resulting from DNA scrunching or oversized transcription bubble could also be sufficient to induce some pausing on (T/C)G motif resulting in significant effects on escape kinetics. The fact that the position of the maximum effect of (T/C)G motif can be different depending on promoter context (Figure 7A–C) is in agreement with other observed promoter-context effects (e.g. at +2 (Figure 5)). This further reinforces the surprising conclusion that promoter sequence can ‘allosterically’ affect escape events that are controlled by ITS. Previously, a simple correlation between the strength of promoter contacts and promoter escape has been described (11). However, the communication between promoter sequence and ITS effects described here is qualitatively different phenomenon, indicating that different promoter sequences could induce different conformations of open complexes where the perturbations in RNAP and/or DNA produced by RNAP–promoter contacts are transmitted to RNAP active site or its vicinity.
How the (T/C)G motif facilitates RNAP pausing is an interesting mechanistic question. Imashimizu et al. (51) proposed a detailed kinetic and structural model for elongation pausing in vivo on G–10C–1G+1 motif. These authors proposed that the increased flexibility of sugar-phosphate backbone in DNA or RNA encoded by (T/C)G motif could interfere with proper positioning of template base for incoming NTP or a misalignment of 3′ end RNA resulting in pausing. The model predicts possibility of pausing both in pre- and posttranslocated states as well as the formation of backtracked pauses. Possible role of interaction between RNAP and the ‘core recognition element’ in elongation pausing was proposed (50). The same mechanisms could apply to pausing during initiation except that the G–10 at the upstream end of RNA-DNA hybrid that is a part of elongation pause signal is not present in case of initiation. As discussed above, in initiation, the stress due to steric clash of growing RNA with RNAP polypeptides around +6 or a more general stress of the initially transcribed complex resulting from DNA scrunching or an oversized transcription bubble is likely the functional equivalent of G–10. It provides an additional barrier to translocation that facilitates pausing at (T/C)G motif. More studies will be needed to fully understand the mechanisms by which (T/C)G motif induces pausing during initiation.
Previous studies reached conflicting conclusions regarding the involvement of backtracking in elongation pausing on G–10(T/C–1)G+1 motif (49,51). Our analysis of escape kinetics using real-time fluorescence assay was consistent with the presence of backtracked intermediates during escape (Figure S2, Supplementary Information). Similarly, backtracked paused initiation intermediates were observed in single molecule experiments (34). Transcript cleavage factors GreA/B will be normally present in vivo reducing the impact of such backtracked complexes on promoter escape. In preliminary experiments, we tested the effect of GreB on the enrichment pattern observed for the library of 96 ITS variants in a context of λPR promoter (Heyduk, E. and Heyduk, T., unpublished) and found no significant change in the pattern compared to no GreB experiment. This suggests that backtracked intermediates, while they were present under our experimental conditions and could affect the kinetics of escape, they were not a major factor affecting the correlations between ITS sequence and escape kinetics. Nevertheless, a detailed analysis of ITS dependence of the effects of GreA/B on escape is in order (and currently under way in our laboratory using NGS-based approach) in light of observed striking template sequence dependent differences between effects of GreA and GreB on escape (46).
Promoter context dependent effects at position +2
In addition to (T/C)G motif, other position-specific effects of ITS on escape were observed. The strongest effects were observed at position +2 (Figure 6). UV5 promoter at position +2 exhibited strong preference for G and T in fast and slow escaping sequences, respectively (Figure 6). The G at position +2 of nontemplate strand is a preferred base of core recognition element (CRE) that is recognized through unstacking and insertion into a pocket formed by residues of β subunit of RNAP (58). This interaction contributes to the thermodynamic and kinetic stability of the ‘open’ complex (58,59). A simple mechanism whereby the identity of a base at position +2 affects escape kinetics by modulating open complex stability is inconsistent with our data. For example, G at +2 is strongly preferred in UV5 promoter context for fast escape. This is opposite to what one would expect for a base that makes open complex more stable since reverse correlation between open complex stability and escape kinetics would be expected and was experimentally observed (11). Also, the preference for G at +2 for fast escaping sequences was not observed for the two other promoters tested (Figure 6C). If in all promoters that we tested, base preferences at position +2 would be derived from their effect on open complex stability, this would mean that the relative strength of CRE interaction with different bases at +2 would depend on promoter context. There is no experimental data yet to suggest that. While it has been suggested that CRE interaction with +2 counteracts elongation pausing (50), the ability to both stimulate and counteract pausing was also observed (59). This variability of the effects of +2 interactions on elongation pausing or promoter escape is very intriguing and requires further investigation.
More generally, it would be possible that many effects of ITS on escape could be due to ITS dependence of open complex stability. Our experimental conditions included heparin competitor in the reaction mixture which could further exacerbate open complex stability effects. However, no significant effects of ITS on open complex stability (with the exception of +2 position discussed above) have been reported. Also, we did not observe significant dependence of open complex stability on ITS in our preliminary investigation using NGS-based approach on λPR promoter containing randomized +2 to +10 (Heyduk, E. & Heyduk, T., unpublished). It is thus unlikely that modulation of open complex stability by ITS is a major means by which ITS modulate escape kinetics.
ITS-encoded physical properties of DNA template provide position-independent ‘force’ that contributes to promoter escape kinetics
The bias of RNA/DNA (or DNA/DNA) duplex stability energy that we observe for fast and slow escaping ITS (Figure 9) suggests that in addition to position specific effects discussed above, there is also a more general position-independent ITS dependent ‘force’ that contributes to the control of escape by ITS. Higher duplex stability energy within the first ∼10 bp of ITS promotes faster escape. This is a significant but not a dominating factor since the observed correlation coefficients, while highly statistically significant, were low. While both RNA/DNA and DNA/DNA duplex energy show correlation with escape or bias for fast and slow escaping sequences, it is likely that only one of these factors is truly relevant for escape. There are only relatively small differences between RNA/DNA and DNA/DNA parameters for near-neighbor models of duplex stability (39,40). The correlation coefficient between duplex stability energy for all sequence variants of 9 bp RNA and DNA duplexes is 0.88 (Table S2, Supplementary Information) suggesting that functional correlation with one duplex energy (either RNA/DNA or DNA/DNA) with escape would produce also a correlation with escape for the other that might not be functionally relevant. RNA duplex stability is a more likely candidate for functionally relevant correlation. More stable short RNA/DNA hybrids at the beginning of transcription could enhance escape by reducing probability of transcript dissociation or misalignment of 3′ end of the transcript with the active site. More stable DNA/DNA duplex would present a higher barrier for transcription bubble enlargement at the beginning of transcription (when RNAP is still bound to the promoter) and would thus be expected to inhibit escape (contrary to what we observe). Furthermore, we have previously demonstrated that reducing the energetic gain of transcription bubble collapse during escape or reducing the energetic cost of melting DNA duplex in front of the growing bubble did not affect the escape kinetics (11). We interpret the correlations and energy bias illustrated in Figure 9 as result of a functional correlation between escape kinetics and RNA/DNA heteroduplex stability. The observed correlation between DNA/DNA duplex stability and escape is likely a secondary effect that reflects high correlation between sequence dependence of RNA/DNA and DNA/DNA duplex stability. This is not the only example of the effect of sequence encoded duplex energy on the events during transcription initiation. We have previously shown a strong correlation between duplex stability energy of –10 promoter element and the rate of promoter melting (35,36).
We also observed significant bias for base stacking energy between incoming NTP and the 3′ end of the RNA product for slow and fast escaping sequences (Figure 9). In fact, this was overall a stronger correlation than between RNA/DNA duplex stability and escape. It is unlikely that this correlation is a secondary effect of a strong correlation for RNA/DNA duplex energy since correlation between duplex energies and base stacking energies is only moderate (correlation coefficients of 0.37 and 0.06 for RNA/DNA and DNA/DNA duplex energy, respectively; Table S2, Supplementary Information). We propose that favorable stacking energy between incoming NTP and the 3′ end of the RNA could stabilize short RNA bound to a template which in turn could favor escape the same way as favorable RNA/DNA duplex stability could. The stabilization would occur through bridging interactions of NTP where NTP base interacts with RNA end through base stacking and with RNAP through interactions with its binding pocket. The added benefit of stabilization through base-stacking is that it could happen early on during reaction cycle, before the new phosphodiester is formed.
Biological significance of ITS effects on promoter escape
One possible biological role of a specific ITS of a given promoter could be to regulate promoter output directly through ITS encoded effect on promoter escape kinetics. With such role, one would expect to observe a correlation between transcriptional activity in vivo and promoter escape kinetics measured in vitro. We examined such correlation for ITS from Figure 2 with high or low expression levels in vivo (Figure S12, Supplementary Information). While no strong correlation was observed (it might be unreasonable to expect strong correlation since many other factors could potentially affect transcript levels in vivo), the observed trend with high in vivo transcript showing some preference for faster escape kinetics (Figure S12, Supplementary Information) is intriguing and suggests that such biological role of ITS could be true at least for some ITS. Alternatively, ITS could work indirectly by enabling or inhibiting action of regulatory proteins by modulating time that RNAP spends on escape (i.e. defining time window during which regulation of RNAP in the process of escape could occur). ITS-encoded propensity for pausing during initiation could be especially enticing target for such regulatory interactions.
Profound effects of ITS on-promoter escape that translate into differences in overall promoter output, the functional communication between promoter interactions and ITS effects, and ITS control of RNAP pausing during initiation, they all argue for revising the definition of bacterial promoter to include ITS as a promoter element of similar importance as the standard promoter elements.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Eric Galburt (Washington University) for critical reading and constructive comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R21 AI112919]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Borukhov S., Nudler E.. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008; 16:126–134. [DOI] [PubMed] [Google Scholar]
- 2. Busby S., Kolb A., Buc H.. Where It All Begins: An Overview of Promoter Recognition and Open Complex Formation. 2009; Royal Society of Chemstry [Google Scholar]
- 3. Saecker R.M., Record M.T. Jr, Dehaseth P.L.. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011; 412:754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 5. Ross W., Gosink K.K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R.L.. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993; 262:1407–1413. [DOI] [PubMed] [Google Scholar]
- 6. Haugen S.P., Berkmen M.B., Ross W., Gaal T., Ward C., Gourse R.L.. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006; 125:1069–1082. [DOI] [PubMed] [Google Scholar]
- 7. Feklistov A., Barinova N., Sevostyanova A., Heyduk E., Bass I., Vvedenskaya I., Kuznedelov K., Merkiene E., Stavrovskaya E., Klimasauskas S. et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell. 2006; 23:97–107. [DOI] [PubMed] [Google Scholar]
- 8. Gralla J.D., Carpousis A.J., Stefano J.E.. Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry. 1980; 19:5864–5869. [DOI] [PubMed] [Google Scholar]
- 9. Carpousis A.J., Gralla J.D.. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980; 19:3245–3253. [DOI] [PubMed] [Google Scholar]
- 10. Hsu L.M., Cobb I.M., Ozmore J.R., Khoo M., Nahm G., Xia L., Bao Y., Ahn C.. Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry. 2006; 45:8841–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ko J., Heyduk T.. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem. J. 2014; 463:135–144. [DOI] [PubMed] [Google Scholar]
- 12. Vo N.V., Hsu L.M., Kane C.M., Chamberlin M.J.. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003; 42:3798–3811. [DOI] [PubMed] [Google Scholar]
- 13. Revyakin A., Liu C., Ebright R.H., Strick T.R.. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006; 314:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapanidis A.N., Margeat E., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H.. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006; 314:1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henderson K.L., Felth L.C., Molzahn C.M., Shkel I., Wang S., Chhabra M., Ruff E.F., Bieter L., Kraft J.E., Record M.T. Jr. Mechanism of transcription initiation and promoter escape by E. coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3032–E3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zuo Y., Steitz T.A.. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell. 2015; 58:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winkelman J.T., Winkelman B.T., Boyce J., Maloney M.F., Chen A.Y., Ross W., Gourse R.L.. Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol. Cell. 2015; 59:768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samanta S., Martin C.T.. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J. Biol. Chem. 2013; 288:31993–32003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu L.M. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta. 2002; 1577:191–207. [DOI] [PubMed] [Google Scholar]
- 20. Hsu L.M. Monitoring abortive initiation. Methods. 2009; 47:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu L.M., Vo N.V., Kane C.M., Chamberlin M.J.. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry. 2003; 42:3777–3786. [DOI] [PubMed] [Google Scholar]
- 22. Kammerer W., Deuschle U., Gentz R., Bujard H.. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986; 5:2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deighan P., Pukhrambam C., Nickels B.E., Hochschild A.. Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev. 2011; 25:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue X.C., Liu F., Ou-Yang Z.C.. A kinetic model of transcription initiation by RNA polymerase. J. Mol. Biol. 2008; 378:520–529. [DOI] [PubMed] [Google Scholar]
- 25. Skancke J., Bar N., Kuiper M., Hsu L.M.. Sequence-dependent promoter escape efficiency is strongly influenced by bias for the pretranslocated state during initial transcription. Biochemistry. 2015; 54:4267–4275. [DOI] [PubMed] [Google Scholar]
- 26. Hein P.P., Palangat M., Landick R.. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011; 50:7002–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malinen A.M., Turtola M., Parthiban M., Vainonen L., Johnson M.S., Belogurov G.A.. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012; 40:7442–7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kubori T., Shimamoto N.. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 1996; 256:449–457. [DOI] [PubMed] [Google Scholar]
- 29. Sen R., Nagai H., Shimamoto N.. Polymerase arrest at the lambdaP(R) promoter during transcription initiation. J. Biol. Chem. 2000; 275:10899–10904. [DOI] [PubMed] [Google Scholar]
- 30. Susa M., Sen R., Shimamoto N.. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 2002; 277:15407–15412. [DOI] [PubMed] [Google Scholar]
- 31. Susa M., Kubori T., Shimamoto N.. A pathway branching in transcription initiation in Escherichia coli. Mol. Microbiol. 2006; 59:1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vo N.V., Hsu L.M., Kane C.M., Chamberlin M.J.. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 2. Formation and characterization of two distinct classes of initial transcribing complexes. Biochemistry. 2003; 42:3787–3797. [DOI] [PubMed] [Google Scholar]
- 33. Duchi D., Bauer D.L., Fernandez L., Evans G., Robb N., Hwang L.C., Gryte K., Tomescu A., Zawadzki P., Morichaud Z. et al. RNA polymerase pausing during Initial transcription. Mol. Cell. 2016; 63:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerner E., Chung S., Allen B.L., Wang S., Lee J., Lu S.W., Grimaud L.W., Ingargiola A., Michalet X., Alhadid Y. et al. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6562–E6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heyduk T., Heyduk E.. Next generation sequencing-based analysis of RNA polymerase functions. Methods. 2015; 86:37–44. [DOI] [PubMed] [Google Scholar]
- 36. Heyduk E., Heyduk T.. Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry. 2014; 53:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Artsimovitch I., Svetlov V., Murakami K.S., Landick R.. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 2003; 278:12344–12355. [DOI] [PubMed] [Google Scholar]
- 38. Callaci S., Heyduk E., Heyduk T.. Conformational changes of Escherichia coli RNA polymerase sigma70 factor induced by binding to the core enzyme. J. Biol. Chem. 1998; 273:32995–33001. [DOI] [PubMed] [Google Scholar]
- 39. SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu P., Nakano S., Sugimoto N.. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. 2002; 269:2821–2830. [DOI] [PubMed] [Google Scholar]
- 41. Friedman R.A., Honig B.. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys. J. 1995; 69:1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reppas N.B., Wade J.T., Church G.M., Struhl K.. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006; 24:747–757. [DOI] [PubMed] [Google Scholar]
- 43. Lass-Napiorkowska A., Heyduk T.. Real-time observation of backtracking by bacterial RNA polymerase. Biochemistry. 2016; 55:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borukhov S., Polyakov A., Nikiforov V., Goldfarb A.. GreA protein: a transcription elongation factor from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:8899–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Borukhov S., Sagitov V., Goldfarb A.. Transcript cleavage factors from E. coli. Cell. 1993; 72:459–466. [DOI] [PubMed] [Google Scholar]
- 46. Petushkov I., Esyunina D., Mekler V., Severinov K., Pupov D., Kulbachinskiy A.. Interplay between sigma region 3.2 and secondary channel factors during promoter escape by bacterial RNA polymerase. Biochem. J. 2017; 474:4053–4064. [DOI] [PubMed] [Google Scholar]
- 47. Rutherford S.T., Lemke J.J., Vrentas C.E., Gaal T., Ross W., Gourse R.L.. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 2007; 366:1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bauer D.L.V., Duchi D., Kapanidis A.N.. E. coli RNA polymerase pauses during initial transcription. Biophys. J. 2016; 110:21a [Google Scholar]
- 49. Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S.. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014; 344:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vvedenskaya I.O., Vahedian-Movahed H., Bird J.G., Knoblauch J.G., Goldman S.R., Zhang Y., Ebright R.H., Nickels B.E.. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014; 344:1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Imashimizu M., Takahashi H., Oshima T., McIntosh C., Bubunenko M., Court D.L., Kashlev M.. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015; 16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herbert K.M., La Porta A., Wong B.J., Mooney R.A., Neuman K.C., Landick R., Block S.M.. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006; 125:1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perdue S.A., Roberts J.W.. Sigma(70)-dependent transcription pausing in Escherichia coli. J. Mol. Biol. 2011; 412:782–792. [DOI] [PubMed] [Google Scholar]
- 54. Ring B.Z., Yarnell W.S., Roberts J.W.. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996; 86:485–493. [DOI] [PubMed] [Google Scholar]
- 55. Basu R.S., Warner B.A., Molodtsov V., Pupov D., Esyunina D., Fernandez-Tornero C., Kulbachinskiy A., Murakami K.S.. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 2014; 289:24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kulbachinskiy A., Mustaev A.. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006; 281:18273–18276. [DOI] [PubMed] [Google Scholar]
- 57. Pupov D., Kuzin I., Bass I., Kulbachinskiy A.. Distinct functions of the RNA polymerase sigma subunit region 3.2 in RNA priming and promoter escape. Nucleic Acids Res. 2014; 42:4494–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Petushkov I., Pupov D., Bass I., Kulbachinskiy A.. Mutations in the CRE pocket of bacterial RNA polymerase affect multiple steps of transcription. Nucleic Acids Res. 2015; 43:5798–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.