Abstract
In Arabidopsis thaliana, HEAT SHOCK TRANSCRIPTION FACTORA1b (HSFA1b) controls resistance to environmental stress and is a determinant of reproductive fitness by influencing seed yield. To understand how HSFA1b achieves this, we surveyed its genome-wide targets (ChIP-seq) and its impact on the transcriptome (RNA-seq) under non-stress (NS), heat stress (HS) in the wild type, and in HSFA1b-overexpressing plants under NS. A total of 952 differentially expressed HSFA1b-targeted genes were identified, of which at least 85 are development associated and were bound predominantly under NS. A further 1780 genes were differentially expressed but not bound by HSFA1b, of which 281 were classified as having development-associated functions. These genes are indirectly regulated through a hierarchical network of 27 transcription factors (TFs). Furthermore, we identified 480 natural antisense non-coding RNA (cisNAT) genes bound by HSFA1b, defining a further mode of indirect regulation. Finally, HSFA1b-targeted genomic features not only harboured heat shock elements, but also MADS box, LEAFY, and G-Box promoter motifs. This revealed that HSFA1b is one of eight TFs that target a common group of stress defence and developmental genes. We propose that HSFA1b transduces environmental cues to many stress tolerance and developmental genes to allow plants to adjust their growth and development continually in a varying environment.
Keywords: ChIP-seq, development, heat shock factors, long non-coding natural antisense RNAs, RNA-seq, stress, transcription factors
Plants must co-ordinate gene expression to balance growth and stress defence. Arabidopsis HSFA1b influences this by reconfiguring an extensive transcription regulatory network at the onset of heat stress.
Introduction
The heat shock response (HSR) is a highly conserved cellular process, which is regulated at the transcriptional level by the heat shock transcription factors (HSFs; Ohama et al., 2016). In their active state, HSFs form homo- and/or heterotrimers, accumulate in the nucleus, and activate transcription of stress-associated genes by binding to heat shock cis-elements (HSEs). HSEs are three inverted pentameric DNA repeats of 5'-nGAAn-3' on the promoters of stress genes (Scharf et al., 2012; Jaeger et al., 2014; Zheng et al., 2016; Jacob et al., 2017). Importantly, the functions of HSFs go beyond the HSR to regulating responses to diverse abiotic and biotic stresses and the modulation of cell growth and development (Evans et al., 2007; Akerfelt et al., 2010; Hashimoto-Torii et al., 2014; Jacob et al., 2017).
Plant HSFs differ from those in other eukaryotes in three ways. First, plant HSF families are larger (Baniwal et al., 2004; Guo et al., 2016). For instance, there are 21 HSFs in Arabidopsis thaliana (Arabidopsis) compared with only single HSFs in both yeast and fruitfly and four in mammals (Nover et al., 2001; Akerfelt et al., 2010; Scharf et al., 2012; Guo et al., 2016). Secondly, plant HSFs are divided into three structurally distinct classes: A, B, and C. Classes A and C function as transcription activators, whereas members of class B HSFs are transcriptional repressors (Scharf et al., 2012; Guo et al., 2016; Jacob et al., 2017). Thirdly, in addition to post-translation regulation of HSFs, some plant HSF genes, such as HSFA2, A3, B2a, and B2b, are regulated transcriptionally in a stress-dependent manner by clade A1 HSF genes as well as other transcription factors (TFs; Schramm et al., 2008; Scharf et al., 2012; Bechtold et al., 2013; Guo et al., 2016; Jacob et al., 2017). This has led to diversification of tissue and functional specificities (Liu et al., 2011; Guan et al., 2013; Hwang et al., 2014; Perez-Salamo et al., 2014; Jacob et al., 2017). In Arabidopsis, the clade A1 HSF family, which is expressed in all tissues (Miller and Mittler, 2006, Swindell et al., 2007; Bechtold et al., 2013), consists of four genetically redundant members, HSFA1a, HSFA1b, HSFA1d, and HSFA1e. Only a quadruple null mutant hsfA1a/hsfA1b/hsfA1d/hsfA1e (hereafter called qKO) is unable to initiate HSR (Liu et al., 2011; Yoshida et al., 2011). Furthermore, qKO plants show diminished development and growth manifested throughout all growth stages. This is not evident in genotypes with fewer combinations of clade A1 HSF null mutants (Busch et al., 2005; Liu et al., 2011). The qKO mutant is defective in the development of the seed, consequently retarding germination, seedling establishment, and also growth through all vegetative stages. However, qKO mutant plants do complete their life cycle under non-stress (NS) conditions (Liu et al., 2011).
Overexpression of HSFA1b in Arabidopsis and oilseed rape (Brassica napus) not only promotes enhanced resistance to abiotic and biotic stress but also affects development by causing a redistribution of biomass in favour of reproductive structures at the expense of vegetative shoot growth, resulting in increased seed yield (Prändl et al., 1998; Bechtold et al., 2013; Jung et al., 2013). In contrast to the qKO mutant, there is no effect of HSFA1b overexpression on any aspect of seed or seedling viability (Bechtold et al., 2013).
The identity of development-associated genes that are subject to regulation by clade A1 HSFs needs to be determined, as well as the full extent of those genes involved in defence against stress. Therefore, we set out to identify HSFA1b-regulated development-associated genes under NS and heat stress (HS) conditions and to determine how these genes may be regulated in comparison with stress-associated genes. By applying HS for a short period of 30 min, when HSFA1b is active (Busch et al., 2005; Li et al., 2010; Liu et al., 2011), we anticipated that we would detect not only the early events in the induction of stress-defensive genes but also changes in expression of genes implicated in growth and development. This was done by surveying genome-wide binding of HSFA1b to its target genes and combining this with transcriptomics using both wild-type (WT) plants under NS and HS conditions, and those overexpressing HSFA1b under NS conditions.
Materials and methods
Arabidopsis genotypes and transgenes
The 35S:HSFA1b plants (35S:HSFA1b-RFP-B/Col-0), the hsfA1a/hsfA1b (Ws-0) mutant, and the qKO mutant (Col-0/Ws-0) have been described previously (Busch et al., 2005; Liu et al., 2011; Bechtold et al., 2013). To construct the HSFA1bPRO:HSFA1b-eYFP (enhanced yellow fluorescent protein) gene fusion (hereafter called NP:HSFA1b), HSFA1b was PCR-amplified, using Phusion DNA polymerase (Thermo-Fisher, Paisley, UK), from Arabidopsis Col-0 genomic DNA, generating an amplicon containing its promoter and full genomic sequence minus the stop codon using primers 5'-CACCTCGAATAATTGTCAAGCTCAC-3' and 5'-TTTCCTCTGTGCTTCTGAG-3'. The amplicon was inserted into the pENTR plasmid using the D-TOPO cloning kit (Thermo-Fisher). The Gateway LR reaction (Thermo-Fisher) was used to create the HSFA1b-eYFP fusion in the binary Ti vector pGWB40 (Nakagawa et al., 2007) creating pGWB40-HSFA1bPRO:HSFA1b-eYFP, which was transferred to Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis Col-0 (Bechtold et al., 2013). Transformants were selected on Murashige and Skoog (MS) medium containing 25 µg ml–1 hygromycin B and 50 µg ml–1 kanamycin. To select expressing lines, immunoblotting (Prändl et al., 1998) was carried out using anti-green fluorescent protein (GFP) antibody (ab209; Abcam, Cambridge, UK).
Growth phenotypes and HS conditions
All plants were grown in an 8 h day:16 h night under controlled environment conditions (Bechtold et al., 2013). Five-week-old plants were subjected to HS at 37 °C (from 23 °C) for 30 min at 86% relative humidity to maintain the vapour pressure deficit at 1 kPa and therefore avoid a coincident dehydration stress (Fryer et al., 2003; Galvez-Valdivieso et al., 2009).
Rosette expansion of soil-grown 2-week-old seedlings over 11 d was carried out to monitor the effect of HS (2 h at 37 °C) or overexpression of HSFA1b under non-stress conditions. Daily measurements of rosette area were made using a chlorophyll fluorescence imager (Fluorimager, Technologica Ltd, Colchester, UK) as described by Baker (2008).
ChIP-PCR, ChIP-seq, and data analysis
Fully expanded leaf samples from 5-week-old NS and HS NP:HSFA1b and Col-0 plants were used for ChIP experiments. A detailed step-wise protocol can be found as a Supplementary Methodology at JXB online. PCR analysis to detect HSFB2a in ChIP DNA samples was carried out as previously described (Bechtold et al., 2013). Library preparation and massively parallel DNA sequencing of ChIP DNA samples (ChIP-seq) were carried out at The Genome Analysis Centre (now the Earlham Institute; http://www.earlham.ac.uk/) using Illumina TruSeq ChIP Library Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were sequenced on an Illumina HiSeq2000 platform using 100 bp paired ends and a sequencing depth of ≥10 million reads per library. The data from two independent plants in one HS experiment are combined.
Quality control of raw fastq files was carried out using in-house programs (available upon request). Using GSNAP (Wu and Nacu, 2010), ChIP-seq reads were mapped to the Arabidopsis genome (TAIR10) allowing one mismatch and output files in SAM format. Removal of unmapped reads and duplicates followed by conversion of SAM format to the binary form (BAM), sorting and indexing of BAM files was done using SAMtools (Li et al., 2009). Library normalization followed by conversion to signal tracks in BedGraph format was performed using BEDtools (Quinlan and Hall, 2010). Normalized BedGraph files were visualized using the Integrated Genome Browser (IGB; Nicol et al., 2009). Peaks were called using MACS2 (Zhang et al., 2008) with the options –g 1.2e8 –f BAMPE –q 0.05 –B –trackline –FE 2. Peaks within pericentromeric regions and broad peaks on gene bodies of highly transcribed genes were considered as false positives regardless of their q and FE values (Nix et al., 2008; Chen et al., 2012). K-means clustering of ChIP-seq signals on the regions occupied by HSFA1b and generation of density heat maps were carried out using seqMINER (Ye et al., 2011). Annotation of the closest genomic features to the regions bound by HSFA1b was carried out with ChIPpeakAnno (Zhu et al., 2010) using the batch annotation function and a dedicated function for peaks on bidirectional promoters. Overlap between annotated target genomic features was determined and Venn diagrams were generated using Jvenn (Bardou et al., 2014). Gene Ontology (GO) analysis on the target genomic features was carried out using the Singular Enrichment Analysis (SEA) tool in the AgriGO database (Du et al., 2010). Sequences of the regions bound by HSFA1b were used for de novo motif discovery using MEME (Bailey et al., 2009) with a cut-off P<0.0001 and using default options.
The published ChIP-seq data analysed in this study were downloaded from the NCBI Gene Expression Omnibus (GEO) database and subjected to the same analysis using MACS2 and criteria as described above. The GEO accession codes for each TF are as follows: HBI1, GSE53078; LFY, GSE24568; PIF5/PIF4, GSE68193; PRR5, GSE36361; PRR7, GSE49282; and SEP3, GSE46987.
RNA-seq data analysis
RNA was extracted from NS WT, HS WT, and NS 35S:HSFA1b plants (three biological replicates of each) as described previously (Bechtold et al., 2013) and analysed using massively parallel sequencing (RNA-seq) as follows: synthesis of cDNA, library preparation, and sequencing were carried out at Earlham Institute on the Illumina HiSeq2000 platform using 50 bp paired-end sequencing with minimum read depth ≥25 million reads per library.
Quality control of raw fastq files was carried out as for the ChIP-seq data with the following modifications: filtered RNA-seq reads were mapped against the Arabidopsis transcriptome using GSNAP with the known splices option for RNA-seq (five mismatches allowed). Transcript assembly and differential expression analysis were carried out using Cufflinks and Cuffdiff (Trapnell et al. 2012) followed by the geometric library normalization method with threshold q≤0.05.
Assembly of a hierarchical TF network was done by downloading all connections for selected TFs from the Cistrome Atlas (O’Malley et al., 2016) or BZIP28 ChIP-seq data (Zhang et al., 2017) and assembling all pairwise interactions in Excel manually in the form Gene A (PD) Gene B, using an exemplar template available from a previously published analysis (Bechtold et al., 2016). The resulting file containing the connections was uploaded into Cytoscape 3.3.1 (Shannon et al., 2003; www.cytoscape.org) and visualized initially using default settings.
Identification of lincRNAs and cisNAT RNAs
The sequenced reads were aligned using TopHat 2 (Kim et al., 2013) on the TAIR10 DNA sequence. The GFFProf script included with RNAprof (Tran et al., 2016) was used to predict all new transcriptional units compared with Araport11 gene annotations (Cheng et al., 2017). Only the transcriptional units >200 nucleotides were kept. The coding potential was estimated using existing annotation (repTAS; Liu et al., 2012) and CANTATAdb (Szcześniak et al., 2016) and, if absent, it was predicted using COME (Hu et al., 2017). Using R (http://www.R-project.org/), all annotated target genomic features of HSFA1b were intersected with transcribed genomic features to generate normalized FPKM (fragments per kilobase of transcript per million mapped reads) values. FPKM values=0 in all conditions were discarded. The differential expression of HSFA1b target genes was determined based on q-value and fold change. Genes were designated as up-regulated and down-regulated based on expression values of HSFA1b target genes in the WT under HS and 35S:HSFA1b under NS relative to NS WT.
qRT-PCR
The method and primers for APX2, MBF1c, HSFA2, and HSFB2b have been described previously (Bechtold et al., 2013). All other primers used in this study are given in Supplementary Table S2.
Accession number
The ChIP-seq and RNA-seq data have been deposited in the NCBI GEO database under code GSE85655.
Results
HSFA1b preferentially binds to downstream and intragenic regions of its target genes under NS conditions
Genome-wide HSFA1b target genes were identified by ChIP-seq from NP:HSFA1b plants expressing a C-terminal fusion of HSFA1b to eYFP under the control of its native promoter (see the Materials and methods). C-terminal fusions to clade A1 HSFs, including HSFA1b, do not affect their function (Prändl et al., 1998; Liu et al., 2008; Jung et al., 2013; Bechtold et al.,2013). The transgenic line chosen (NP:HSFA1b_6) had the least immunodetectable protein of the seven lines surveyed (Supplementary Fig. S1A) and was validated as suitable for ChIP-seq by carrying out ChIP-PCR to show binding of HSFA1b–eYFP to the promoter of HSFB2b (Supplementary Fig. S1B, C; see the Materials and methods). ChIP-seq was performed on 5-week-old plants grown under NS conditions and subjected to HS treatment (37 °C for 30 min). The control for these experiments was Col-0 (WT) plants treated in the same way. HSFA1b, along with HSFA1a, regulates the initial phase (<1 h) of the HSR; thereafter, stress-inducible HSF genes take over (Busch et al., 2005; Li et al., 2010; Liu et al., 2011). This was confirmed by a ≥30 min delay of HS-inducible gene expression in hsfA1a/hsfA1b compared with WT plants (Supplementary Fig. S2).
The minimum exposure to 37 °C that affected growth was 2 h, at which a slight but significant (P≤0.05; Student’s t-test) reduction in the rate of rosette expansion was measured 4 d and 5 d post-stress in Col-0 (Supplementary Fig. S3).
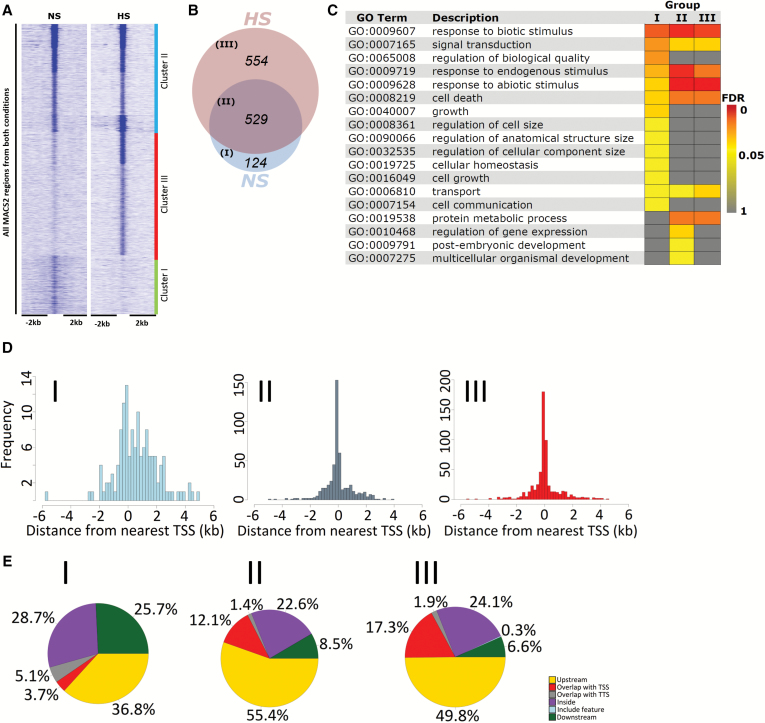
Peak calling from ChIP-seq data (see the Materials and methods) identified 709 and 1083 HSFA1b-binding sites [q≤0.05; fold enrichment (FE) ≥2] under NS and HS, respectively (Supplementary Data S1), comprising 1207 HSFA1b target genes. K-means clustering of the binding regions identified three groups (Fig. 1A; Supplementary Data S1): unique to NS (Group I), common to NS and HS (Group II), and unique to HS (Group III; Fig. 1B). Examples of HSFA1b binding near genomic features in Groups I–III are shown in Fig. 2A–C. The target genes in each group (Supplementary Data S1) were intersected with loci mapped to genome-wide DNase I-hypersensitive sites in NS and HS seedlings (Supplementary Fig. S4; Sullivan et al., 2014). Open chromatin in NS conditions showed enrichment for Group I genes (P=4.31E-08) but less so for Group II and III genes (P=0.03 for both comparisons), while the opposite was observed for HS seedlings (Group I; P=0.91; Supplementary Fig. S4).
Fig. 1.
Alteration of HSFA1b binding in response to HS. (A) Heat map with k-means clustering showing the enrichment of ChIP-seq signals from NS and HS samples at a 4 kb window around HSFA1b-bound regions in the genome. (B) Venn diagram showing the number of HSFA1b target genomic features in Groups I–III. (C) GO Slim analysis heat map comparing genomic features of enriched Biological Process terms in Group I–III with Benjamini–Hochberg FDR <0.05. (D) Three histograms showing the frequency of HSFA1b binding relative to the distance from the TSS of target genomic features in Groups I–III. (E) Pie charts showing the distribution of HSFA1b binding on target genomic features in Groups I–III.
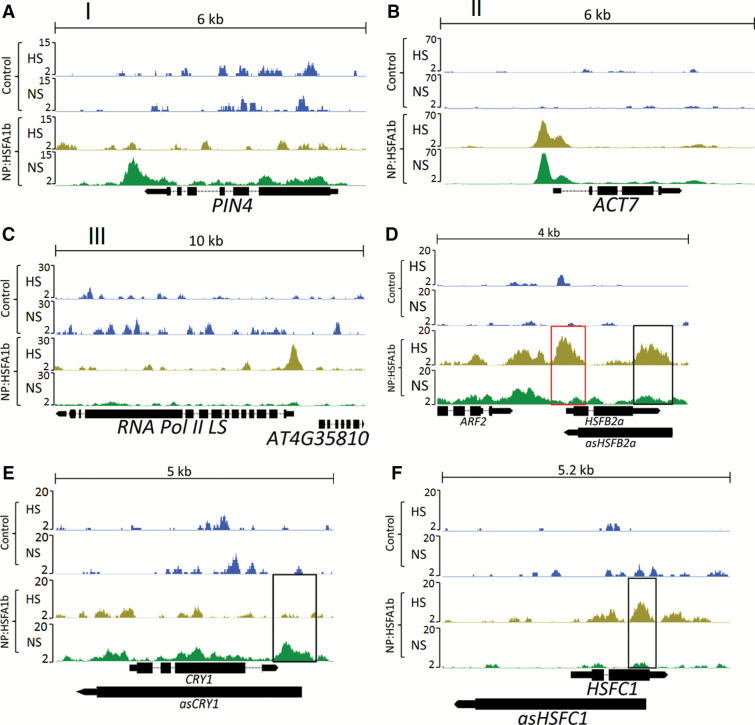
Fig. 2.
Genome browser view of normalized ChIP-seq tags in NP:HSFA1b NS and NP:HSFA1b HS along with controls showing examples of HSFA1b binding to genes in (A) Group I, (B) Group II, and (C) Group III. (D) Examples of HSFA1b targeting the promoter of HSFB2a (red rectangle) as well as a region known to code for an antisense RNA that targets HSFB2a (asHSFB2a; black rectangle). (E and F) HSFA1b targeting a region that codes for asCRY1 and asHSFC1, respectively.
To obtain an overview of the biological functions of proteins encoded by HSFA1b-bound genes in each group, we carried out a GO analysis (see the Materials and methods). In all groups, there was a significant enrichment for Biological Process (BP) GO terms that reflected the role of HSFA1b in plant stress responses (Fig. 1C; Supplementary Data S2). However, there were many enriched BP terms associated with growth and development from Groups I and II but not Group III (Fig. 1C; Supplementary Data S2).
Forty percent of HSFA1b binding events mapped to within ~250 bp of the transcription start site (TSS) of Group II and III genes (Fig. 1D). In contrast, binding of Group I genes by HSFA1b was spread across greater distances (<1% of the binding sites mapped within 250 bp of the TSS; Fig. 1D). A breakdown of binding regions in relation to the main features of genes showed that HSFA1b preferentially targeted inside and downstream of genes in Group I (54%) in contrast to ~30% for Group II and III genes (Figs 1E; 2B, C; Supplementary Data S1).
Detection of long non-coding RNAs
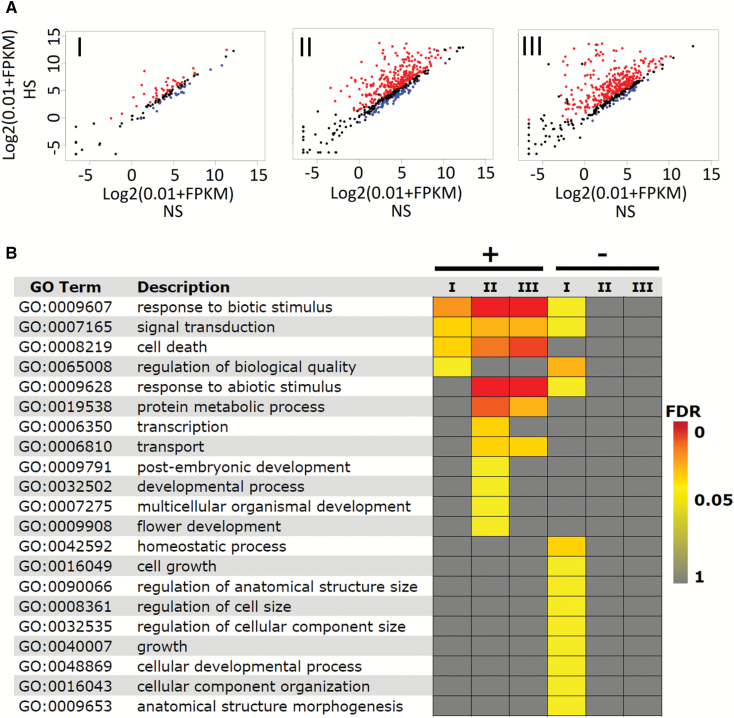
In WT plants under NS and HS, 7137 differentially expressed genes (DEGs; q-value ≤0.05) were identified in response to HS (Supplementary Data S3). Of these, 721 were HSFA1b-bound genes (Fig. 3A; Supplementary Data S4). Enriched GO terms revealed that down-regulated HS-responsive genes were enriched for a number of growth functions (Fig. 3B; Supplementary Data S5).
Fig. 3.
Most HSFA1b target genes are responsive to HS. (A) Scatter plots of transcript abundance of HSFA1b target genes in response to HS. Red and blue dots represent up- and down-regulated genes, respectively (q≤0.05), and black dots show genes with expression q>0.05. The numbers shown in each panel are the number of DEGs in each group. (B) GO Slim analysis heat map comparing the enriched BP terms of up- (+) and down- (–) regulated HS-responsive genes in Groups I–III (Benjamini–Hochberg FDR <0.05).
The prevalence of HSFA1b binding internal to and downstream of protein-coding genes suggested that it binds to genomic regions in or near cis natural antisense long non-coding (cisNAT) RNA genes (Ariel et al., 2015). Using the RNA-seq data on WT (NS and HS) plants, we predicted all transcripts in addition to those present in the Araport11 database (Cheng et al., 2017). The transcripts were then classified as coding transcripts, cisNAT or long intergenic non-coding (linc) RNAs (see the Materials and methods). All binding sites of Groups I–III classified as located internal or distal to the putative sense target gene (cis natural antisense long non-coding; Supplementary Data S1) were intersected with the complete list of NAT gene co-ordinates. Table 1 summarizes this analysis and includes data on the number of NAT genes that also have HSFA1b bound to the corresponding sense gene under NS and/or HS conditions (see also Fig. 2D–F).
Table 1.
Summary of HSFA1b binding to cisNAT genes and lincRNA genes under NS and HS conditions (q-value <0.05)
| Group I (NS only) | Group II (HS and NS) | Group III (HS only) | |
|---|---|---|---|
| All cisNAT genes bound by HSFA1b (n) | 76 | 364 | 377 |
| Proportion of cisNAT genes whose target sense gene is not bound by HSFA1b under the same conditions | 57% | 58% | 99% |
| All lincRNA genes bound by HSFA1b (n) | 11 | 39 | 29 |
The transcriptome of HSFA1b-overexpressing plants shows an intermediate state between NS and HS wild-type plants
We performed RNA-seq on 35S:HSFA1b plants overexpressing HSFA1b-RFP (see the Materials and methods) under NS conditions. A total of 3306 protein-coding genes showed differential expression in these plants compared with NS WT plants (q≤0.05; Supplementary Data S3), of which 72% were differentially expressed in HS WT plants (Supplementary Data S3). A Pearson correlation test on all the transcriptome data (q≤0.05) showed significant positive correlation between NS 35S:HSFA1b and both NS (r=0.92) and HS WT plants (r=0.88). Moreover, the expression levels of heat shock protein (HSP) genes in 35S:HSFA1b NS plants was intermediate to WT NS and HS plants (Fig. 4A). This suggested that the 35S:HSFA1b plants under NS conditions were poised in a state between growth and stress defence. Consistent with these observations, DEGs in 35S:HSFA1b NS plants showed enrichment for both stress-associated and developmental GO terms, paralleling the enriched BP terms in WT HS plants (Figs 3B, 4B; Supplementary Data S5). In keeping with our previous observations (Bechtold et al., 2013), the rate of rosette expansion over 14–25 d post-germination was markedly reduced in 35S:HSFA1b compared with Col-0 (Supplementary Fig. S5A). At 5 weeks old, rosettes were typically visibly smaller (Supplementary Fig. S5B, C) and flowering time was shorter by 1 d (Supplementary Fig. S5D).
Fig. 4.
35S:HSFA1b plants under NS partially mimic HS WT plants. (A) Heat map comparing normalized FPKM values for HSP genes in NS and HS WT and 35S:HSFA1b NS plants. (B) GO Slim analysis comparing the enriched Biological Process terms (Benjamini–Hochberg FDR <0.05) of up- (+) or down (–)-regulated DEGs in HS WT and NS 35S:HSFA1b plants compared with NS WT plants.
HSFA1b extends its influence by exerting indirect control over gene expression
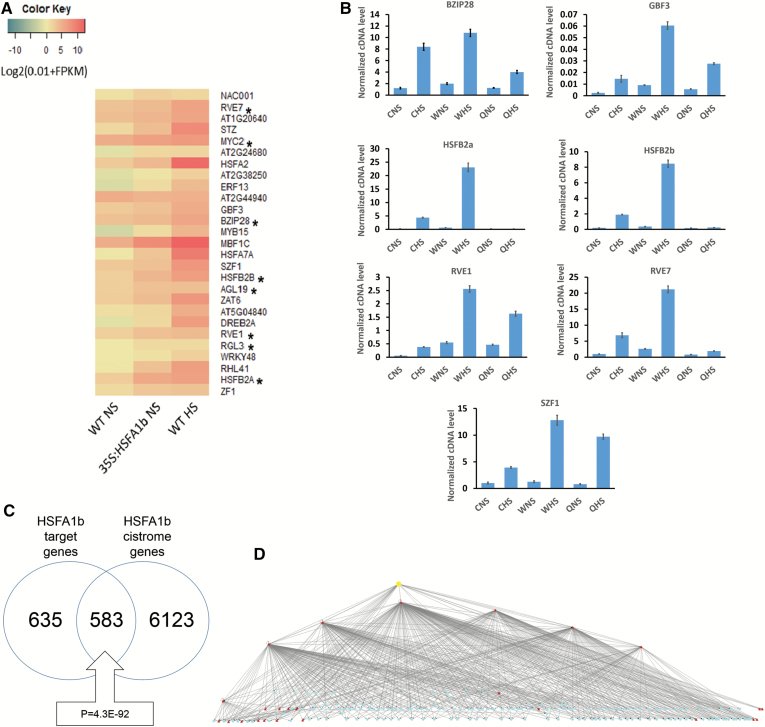
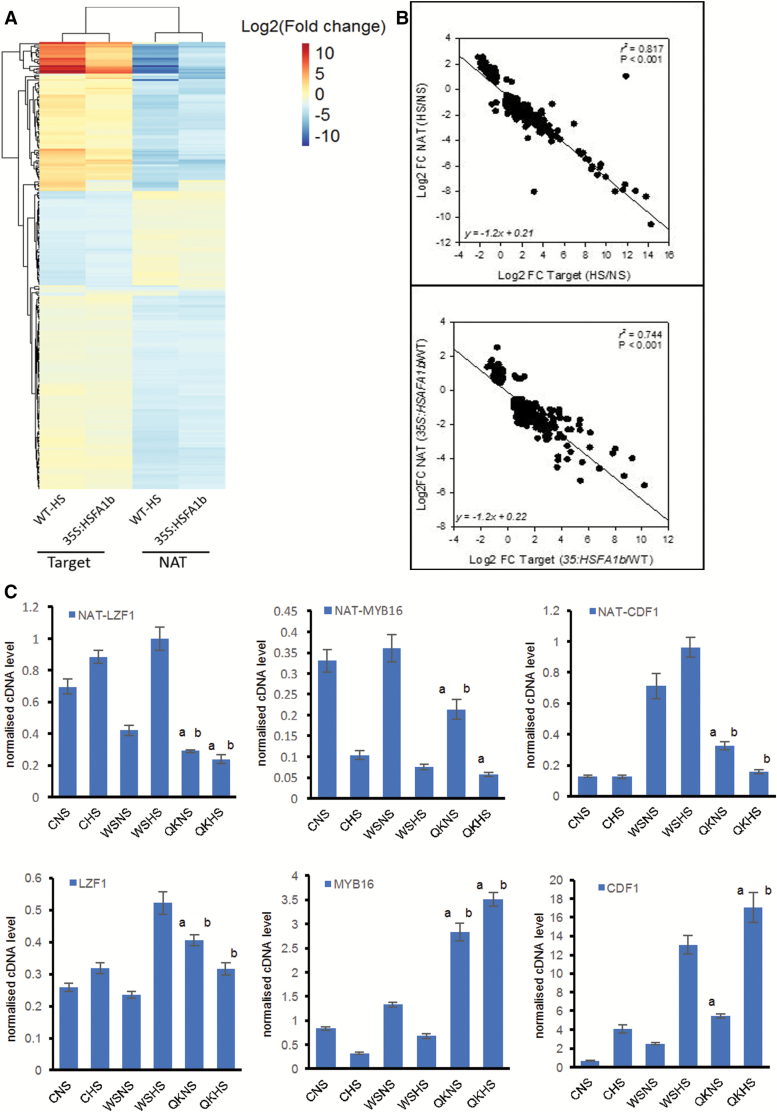
By intersecting the ChIP-seq data from NP:HSFA1b plants under HS and NS conditions (Supplementary Data S1) with the DEGs from NS 35S:HSFA1b plants compared with NS WT plants (Supplementary Data S3), we classified 1821 genes as differentially expressed in WT HS plants and in 35S:HSFA1b NS plants that were not bound by HSFA1b (Supplementary Data S1, S3). These were designated as indirectly regulated by HSFA1b, of which 281 are associated with development (Supplementary Data S7). We reasoned that this indirect regulation is achieved by HSFA1b being able directly to control the expression of other transcription regulatory genes, such as TF genes. We identified 27 TF genes as directly regulated by HSFA1b (Fig. 5A; Supplementary Data S1, S3). Eight of these genes have effects on growth and development (Fig. 5A; Kuno et al., 2003; Achard et al., 2008; Kolmos et al., 2014; Wunderlich et al., 2014; Kang et al., 2015; Xu et al., 2015; Valenzuela et al., 2016). A selection of seven development-associated TF genes directly regulated by HSFA1b were tested for altered expression in NS and HS qKO plants compared with their parental genotypes (Liu et al., 2011). In all examples, the expression of these genes was down-regulated in qKO plants compared with at least one parental genotype under NS and/or HS conditions (Fig. 5B), confirming their regulation by clade A1 HSFs (see also Fig. 6C).
Fig. 5.
HSFA1b can indirectly control expression through its regulation of TF gene expression. (A) Heat map comparing normalized FPKM values for 28 TF genes bound by HSFA1b and differentially expressed in 35S:HSFA1b and WT HS plants. Asterisks (*) indicate development-associated TF genes. (B) Confirmation of the regulation of selected TF genes by clade A1 HSF genes. Quantitative real-time RT–PCR was conducted on RNA from qKO rosettes under NS and HS in comparison with its parental genotypes Col-0 (C) and Ws-0 (W). The suffixes ‘a’ and ‘b’ are where the qKO mutant shows a significant difference (P<0.05) under the same conditions (NS or HS) from Col-0 and Ws-0, respectively. (C) Venn diagram showing the overlap between HSFA1b target genes scored from the ChIP-seq data (Supplementary Data S1) and the target genes bound by HSFA1b (HSF3) from the Arabidopsis Cistrome Atlas (http://neomorph.salk.edu/dev/pages/shhuang/dap_web/pages/index.php). The boxed callout number is the P-value for the significance of the overlap between the two data sets (hypergeometric distribution test). (D) An overview of a Cytoscape-generated HSFA1b hierarchical TF gene network using the data outputs from the Cistrome Atlas with the ChIP-seq and RNA-seq data from this study. The yellow node is HSFA1b, red nodes are TF genes bound by HSFA1b, and blue nodes are differentially expressed TF genes that respond to HS and HSFA1b overexpression, are not bound by HSFA1b, but are scored as binding to the red node TFs. An interactive version of this network is available as an interactive file (Supplementary Cytoscape File S1).
Fig. 6.
HSFA1b regulates expression of cisNAT genes and their target sense genes. (A) Heat map showing differentially expressed cisNAT genes and their putative sense targets in WT plants in HS compared with NS and in 35S:HSFA1b NS plants compared with WT NS plants. (B) Linear correlation plots showing the relationship between cisNAT and sense target transcript abundance for WT plants subjected to HS compared with NS (top panel) and for 35S:HSFA1b NS plants compared with WT NS plants (bottom panel). (C) Transcript levels determined by qPCR of selected cisNAT genes and their TF gene sense target in HS and NS qKO plants compared with their parental genotypes as in the legend of Fig. 5B. The suffixes ‘a’ and ‘b’ are where the qKO mutant shows a significant difference (P<0.05) under the same conditions (NS or HS) from Col-0 (C) and Ws-0 (W) respectively.
HSFA1b and eight of its direct TF gene targets, BASIC LEUCINE ZIPPER28 (BZIP28), REVIELLE7 (RVE7), SALT INDUCED ZINC FINGER1 (SZF1), HSFB2b, G-BOX BINDING FACTOR3 (GBF3), HSFB2a, RVE1, and ETHYLENE RESPONSE FACTOR13 (ERF13; Fig. 5A) are represented in the Arabidopsis Cistrome Atlas (O’Malley et al., 2016). The Cistrome Atlas is a database of genome-wide TF-binding sites experimentally determined by DNA affinity purification sequencing (DAP-seq) (O’Malley et al., 2016). Cistrome-generated binding data for HSFA1b showed a highly significant overlap, capturing 48% of the sites determined by ChIP-seq (Fig. 5C). To determine if the eight directly HSFA1b-regulated TFs connect to further TF genes, all differentially expressed TF gene targets of these eight TFs were collected. These were classified into those connecting to at least one of the eight directly HSFA1b-regulated TF genes, those also targeted directly by HSFA1b, and, finally, those also connecting to other indirectly HSFA1b-regulated TF genes in the data set. The resulting hierarchical network of direct interactions between TF genes consists of 183 nodes and 546 edges, the top node being HSFA1b because it has no inward connections (Fig. 5D). The network model visually demonstrates that HSFA1b is capable of indirectly regulating gene expression by acting upon the expression of other TF genes at least one or two steps removed from a direct interaction with it. While this paper was being written, a limited ChIP-seq data set became available for BZIP28 (Zhang et al., 2017). Of the 133 BZIP28 target genes in seedlings subject to tunicamycin treatment (see the Discussion), 29 were differentially expressed in 35S:HSFA1b NS leaves, resulting in a simple illustrative network of indirect regulation of the transcript levels of these genes by HSFA1b via BZIP28 (Supplementary Fig. S6 at JXB online.
Indirect regulation of gene expression by HSFA1b could also occur via its interactions with 817 cisNAT genes under NS and/or HS conditions (Table 1; Fig. 2D–F; Supplementary Data S1). RNA-seq data revealed that 413 of these cisNAT genes were differentially expressed in HS WT plants and/or 35S:HSFA1b NS plants (Fig. 6A). These NAT transcripts correspond to 357 putative sense target loci (Supplementary Data S8). GO classification of the cisNAT target genes revealed enrichment only for stress-associated functions (Supplementary Data S8). However, the only significantly enriched GO Molecular Function class was for 39 gene targets coding for transcription regulators [false discovery rate (FDR) ≤0.1; Supplementary Data S8]. Of these, eight [LIGHT REGULATED ZINC FINGER PROTEIN1 (LZF1), RELATED TO AP2.7 (RAP2.7), ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN56 (ANAC056), ANAC078, ANAC083, HOMEOBOX PROTEIN2 (HB2), MYB DOMAIN CONTAINING PROTEIN16 (MYB16),and CYCLING DOF FACTOR1 (CDF1)] have development-associated functions (Supplementary Table S1). There was a high negative correlation between the differential levels of HS- and 35S HSFA1b-responsive NAT RNAs and the transcript levels of their overlapping sense targets (Fig. 6A, B). Reciprocal differential levels between at least one WT genotype and qKO of selected NAT antisense and their sense cognate transcripts under NS and/or HS were also observed (Fig. 6C).
HSFA1b is one of eight TFs that regulate a common set of stress and developmental genes
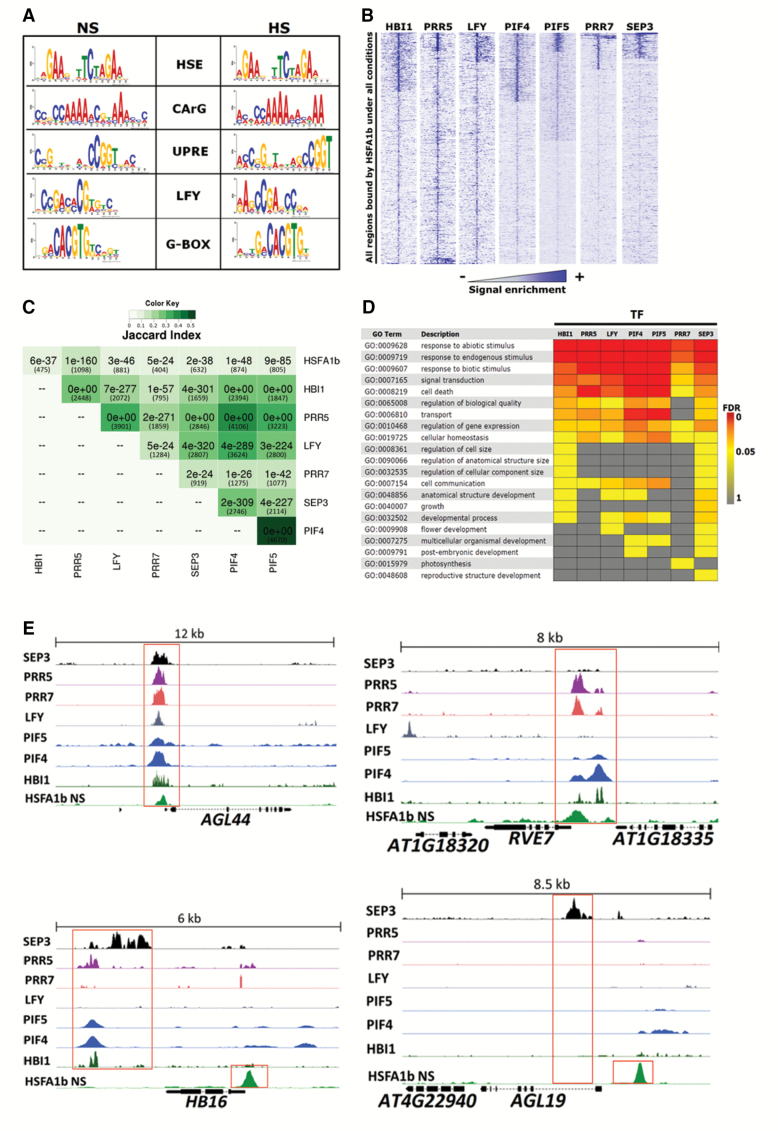
The TF networks depicted (Fig. 5D; Supplementary Fig. S6) do not consider any convergence of signalling from other networks. To determine how HSFA1b could co-operate with other TFs, we searched for enriched TF-binding motifs present within the HSFA1b ChIP-seq peaks (see the Materials and methods). As expected, a consensus heat shock cis-element (HSE) was identified from these enriched peak sequences (Fig. 7A). Reported variant HSEs, such as gapped HSEs (Guo et al., 2008) and HSE1b motifs (Bechtold et al., 2013, were not detected. Likewise, TC-rich elements and STRE motifs bound by HSFA1a in vitro (Guo et al., 2008) were absent from the data set. In addition, four significantly over-represented conserved known motifs were identified in both the NS and HS data sets (Fig. 7A). These were the G-box motif, recognized by various TFs, including BZIP, BASIC HELIX LOOP HELIX (BHLH), and PSEUDO RESPONSE REGULATOR (PRR) TFs (Chawade et al., 2007; Nakamichi et al., 2012), the CArG element bound by MADS-box TFs (Moyroud et al., 2011), the recognition motif of the LEAFY (LFY) TF (Pajoro et al., 2014), and the Unfolded Protein Responsive Element (UPRE) (Martínez and Crispeels, 2003).
Fig. 7.
Seven TFs co-ordinate with HSFA1b. (A) All significant motifs within HSFA1b peaks discovered by MEME (P<0.0001) in the NS and HS data sets. (B) Density heat maps showing enrichment of ChIP-seq signals of the seven TFs on 10 kb around the regions bound by HSFA1b. (C) Combined Jaccard index and pHYPER correlation matrix showing the significance of overlaps between the target genes of the seven TFs and HSFA1b under NS conditions only. Numbers of genes in each overlap are in parentheses. (D) GO Slim analysis comparing the enriched Biological Proess terms of the common targets between HSFA1b and the other seven TFs. (E) Genome browser view of normalized ChIP-seq tags from the ChIP-seq data of the eight TFs showing examples of target genes bound by HSFA1b and up to seven other TFs.
Published ChIP-seq data from seven TFs known to bind to the co-occurring motifs significantly overlapped with HSFA1b target regions under NS and HS (Fig. 7B, Supplementary Data S9; see the Materials and methods). The TFs were HOMOLOG OF BEE2 INTERACTING WITH IBH1 (HBI1; a BHLH TF; Fan et al., 2014), LFY (Pajoro et al., 2014), SEPALLATA3 (SEP3; a MADS-box TF; Pajoro et al., 2014), PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 (BHLH TFs; Pedmale et al., 2016), PRR5, and PRR7. Furthermore, there was considerable overlap in target genes not only between HSFA1b and the seven TFs but also between the seven TFs (Fig. 7C; Supplementary Data S9), examples of which are shown in Fig. 7E. Since the published ChIP-seq data sets were from NS plants, we scored for co-occurrence of the seven TF motifs only in Groups I and II (i.e. under NS conditions; Supplementary Data S9). In Group I genomic features, shared sites were between 3% and 6.4% and in Group II between 15.5% and 27.8%.
The target genes shared between HSFA1b and each of the seven TFs (Supplementary Data S9) showed significant enrichment of GO terms for response to stress and endogenous stimuli (Fig. 7D; Supplementary Data S10). Also, apart from the PRR TFs, there was significant enrichment of GO terms for developmental processes (Fig. 7D; Supplementary Data S10). The expression of PRR5, HBI1, PRR7, and PIF5 was responsive to HS (q=0.0003) but not to HSFA1b overexpression (Supplementary Data S3), while PIF4, LFY, and SEP3 were responsive to neither HS nor overexpression of HSFA1b. There are 76 targets for all eight TFs, of which 15 are associated with developmental processes (Table 2). Thirty-nine of these (including 10 developmental genes) responded to HS, and 18 (including two developmental genes) showed significant changes in transcript abundance in response to HSFA1b overexpression (Table 2; Supplementary Data S3).
Table 2.
Target genes common to HSFA1b (NS and HS), PRR5, PRR7, HBI1, LFY, SEP3, PIF4, and PIF5
| Locus | Gene symbol | Locus | Gene symbol | Locus | Gene symbol |
|---|---|---|---|---|---|
| AT1G05370 | AT1G05370 | AT2G45820 | AT2G45820 | AT4G28230 | AT4G28230 |
| AT1G07580 | AT1G07580 | AT2G45960 | PIP1B | AT4G37260 | MYB73 |
| AT1G13245 | RTFL17 | AT3G10985 | SAG20 | AT4G39838 | AT4G39838 |
| AT1G13250 | GATL3 | AT3G11415 | AT3G11415 | AT5G01600 | FER1 |
| AT1G14920 | GAI | AT3G11700 | FLA18 | AT5G08139 | AT5G08139 |
| AT1G17990 | AT1G17990 | AT3G14440 | NCED3 | AT5G11740 | AGP15 |
| AT1G18740 | AT1G18740 | AT3G15200 | AT3G15200 | AT5G13100 | AT5G13100 |
| AT1G21380 | AT1G21380 | AT3G15210 | ERF4 | AT5G21940 | AT5G21940 |
| AT1G29640 | AT1G29640 | AT3G15770 | AT3G15770 | AT5G22940 | F8H |
| AT1G32640 | MYC2 | AT3G15790 | MBD11 | AT5G24530 | DMR6 |
| AT1G32920 | AT1G32920 | AT3G16240 | DELTA-TIP | AT5G25220 | KNAT3 |
| AT1G72450 | JAZ6 | AT3G22380 | TIC | AT5G47220 | ERF2 |
| AT1G77280 | AT1G77280 | AT3G24518 | AT3G24518 | AT5G47225 | AT5G47225 |
| AT1G78070 | AT1G78070 | AT3G24520 | HSFC1 | AT5G48250 | BBX8 |
| AT1G80440 | AT1G80440 | AT3G49790 | AT3G49790 | AT5G49520 | WRKY48 |
| AT2G14210 | AGL44 | AT3G50750 | BEH1 | AT5G53400 | BOB1 |
| AT2G22426 | AT2G22426 | AT3G59060 | PIL6 | AT5G57660 | COL5 |
| AT2G23290 | MYB70 | AT3G59940 | AT3G59940 | AT5G58070 | TIL |
| AT2G23430 | ICK1 | AT4G00360 | CYP86A2 | AT5G60680 | AT5G60680 |
| AT2G28550 | RAP2.7 | AT4G01250 | WRKY22 | AT5G61970 | AT5G61970 |
| AT2G29660 | AT2G29660 | AT4G01720 | WRKY47 | AT5G62000 | ARF2 |
| AT2G41890 | AT2G41890 | AT4G23630 | BTI1 | AT5G62430 | CDF1 |
| AT2G41900 | OXS2 | AT4G26700 | FIM1 | AT5G65305 | AT5G65305 |
| AT2G41940 | ZFP8 | AT4G27260 | WES1 | AT5G67300 | MYBR1 |
| AT2G44810 | DAD1 | AT4G27510 | AT4G27510 | AT5G67420 | LBD37 |
| AT2G45660 | AGL20 |
Summary of genes responsive to HSFA1b overexpression under NS and HS (bold) and HS in the WT (italics) are from Supplementary Data S3 (q≤0.05). Underlined loci are developmental genes.
Discussion
HSFA1b regulates growth- and development-associated genes as well as stress resistance genes
Genome-wide binding and transcript profiling has shown how HSFA1b directly and indirectly regulates the expression of genes coding for resistance to abiotic and biotic stress (Figs 1C, 3C, 4C; Supplementary Data S2, S5, S6). This is consistent with previous studies and confirms that both WT and HSFA1b-overexpressing plants in this study responded typically to this moderate HS treatment (Prändl et al., 1998; Guo et al., 2008; Chauhan et al., 2011; Nishizawa-Yokoi et al., 2011; Bechtold et al., 2013; Jung et al., 2013; Guo et al., 2016; Jacob et al., 2017).
HSFA1b also targets and influences the expression of up to 354 genes involved in plant growth and development (Figs 1C, 3C, 4C; Supplementary Data S2, S5, S6; Supplementary Table S1). The molecular functions of these genes are very diverse (Figs 1C, 3C, 4C, 5A; Table 2; Supplementary Table S1; Supplementary Data S2, S5–S7; S10). These range from cell integrity-associated chaperones engaged in chloroplast development, hormone metabolism (auxins and brassinosteroids), photoreceptors, components of photomorphogenesis signalling, cell wall synthesis enzymes, development-associated TFs, and to the defective alleles of genes associated with growth and development phenotypes (Supplementary Table S1). Therefore, we could not identify a discrete group of genes that would explain the phenotypic effects of HSFA1b overexpression or the effect of a moderate HS on growth of WT plants. Instead, our data show that many genes of diverse function are affected. We suggest that the wide-ranging but subtle effects that moderate HS and HSFA1b overexpression has on plant growth (Supplementary Figs S3, S5A–D; Bechtold et al., 2013) is the net consequence of this effect upon widely diverse cellular functions.
Reconfiguration of HSFA1b genome-wide binding going from NS to HS
There was a clear difference in HSFA1b binding profiles to genomic regions under HS and NS conditions such that three groups (I–III) could be distinguished (Figs 1A, B, D, E; Supplementary Data S1). Therefore, within the first 30 min of HS, HSFA1b ceases to engage with 124 of these genes (Group I) that are predominantly associated with growth and development functions and targets 553 protein-coding genes (Group III) of which a substantial number are associated with defence against environmental stress (Supplementary Data S2). Group II HSFA1b target genes, bound under both conditions, have enrichment for GO terms in both stress responses and growth and development (Supplementary Data S2). It has been previously shown that there is a substantial genome-wide shift in the distribution of open chromatin in transitioning from NS to HS, which changes accessibility of HSEs (Sullivan et al., 2014). This suggests that this re-configuration of HSFA1b binding targets is associated with a distinct distribution of genome-wide DNase I-hypersensitive sites (Supplementary Fig. S4).
The indirect regulation of gene expression by HSFA1b through a network of TF genes
Of the 2121 genes that show differential transcript levels in HS-exposed WT and 35S:HSFA1b plants under NS, 84% were not targets for HSFA1b binding (Supplementary Data S1, S3). Such genes were classified as indirectly regulated by HSFA1b and include 281 development-associated genes (Supplementary Data S6, S7). Therefore, HSFA1b could exert its influence over stress defence and growth-associated processes through an extensive transcriptional regulatory network. We depicted this network as hierarchical because HSFA1b transcript levels do not vary substantially in response to environmental stress (Nishizawa et al., 2006; Swindell et al., 2007; Bechtold et al., 2013; Sullivan et al., 2014). This is in contrast to the transcriptional regulation of all the other TF genes considered here. We identified 27 TF genes that were direct targets of HSFA1b that showed differential expression in response to HS in WT plants, overexpression of HSFA1b, and, for a sample of seven of them, altered expression in the qKO mutant (Fig. 5A, B). The regulation by HSFA1b of the expression of these many TF genes implies considerable complexity even in a network only one step removed from direct regulation and shows how large such networks could be (Fig. 5D; Supplementary Cytoscape File S1). However, the TF binding data from the Cistrome Atlas used to generate the network overestimates the number of binding events that would occur in vivo, which is the case for HSFA1b (Fig. 5C). Furthermore, for some TF families this assay does not work (O’Malley et al., 2016). Nevertheless, despite these limitations, the resulting network clearly depicts the potential for layers of indirect regulation of gene expression by HSFA1b. The recent availability of a ChIP-seq data set for BZIP28 (Fig. 5A; Zhang et al., 2017) from seedlings undergoing a tunicamycin-induced unfolded protein response (UPR) confirmed this notion of indirect regulation by HSFA1b of other TF genes (Supplementary Fig. S6).
A potentially confounding factor in the classification of direct and indirect regulation of gene expression by HSFA1b is the reliance upon 35S:HSFA1b plants under NS conditions. However, the 50-fold HSFA1b overexpression in the 35S:HSFA1b line chosen (Supplementary Data S3; Supplementary Fig. S1; Bechtold et al., 2013) did not alter transcript levels over and above the levels encountered in WT plants subject to HS (Fig. 4A). Nevertheless, 28% of DEGs in the 35S:HSFA1b plants were not responsive to HS (see the Results and Supplementary Data S3). This is to be expected since HSFA1b does control responses to stresses other than HS. These include resistance to infection by Pseudomonas syringae and the oomycete Hyaloperonospora arabidopsidis, oxidative stress, high light stress, and drought stress (Bechtold et al., 2013; Jung et al., 2013). Therefore, not all genes whose expression is altered in 35S:HSFA1b plants would necessarily be expected to respond to HS in WT plants. However, we cannot rule out that some genes are aberrantly expressed because of high HSFA1b overexpression levels, although some surety was also provided by confirming altered responses of selected genes in the qKO mutants (Fig. 5B; see the Results).
HSFA1b control of cisNAT gene expression
Genome-wide binding of HSFA1b showed a preference for binding internal to or downstream of the TSS of protein-coding genomic loci under NS conditions and less so under HS (Fig. 1D, E). This was associated with HSFA1b targeting 817 cisNAT RNA genes and 79 lincRNA genes (Supplementary Data S1), and 51% of these were differentially expressed under HS and/or in 35S:HSFA1b plants (Table 1; Supplementary Data S8). In general, lincRNA levels are differentially regulated in response to abiotic and biotic stress, and cisNAT RNAs form 10–30% of the total non-coding RNA complement (Liu et al., 2012, 2015a, b; Yu et al., 2013; Wang et al., 2014; Ariel et al., 2015; Bouchard et al., 2015; Muthusamy et al., 2015; Shafiq et al., 2016). In contrast, the fact that HSFA1b regulates the expression of many more cisNAT genes than lincRNA genes suggests that it specifically targets them for regulation (Table 1; Supplementary Data S1, S8). This greatly extends observations made on the regulation of HSFB2a and its asHSFB2a cisNAT gene, which show reciprocal transcript levels (Wunderlich et al., 2014) and is termed discordant expression (Wang et al., 2014). From the transcriptomics analysis of the 412 HS- and HSFA1b-regulated cisNATs, >98% of them and their partner sense transcript showed discordant expression (Fig. 6A, B; Supplementary Data S8), contrasting with de-etiolating seedlings where ~55% showed this pattern (Wang et al., 2014). Indirect regulation by HSFA1b of gene expression could occur via its direct regulation of cisNAT RNA levels and the eventual silencing (si)RNAs generated from them. However, in de-etiolating Arabidopsis seedlings, siRNAs play no role in the light regulation of cisNAT–target gene pairs (Wang et al., 2014). Instead, a correlation was noted between cisNAT gene expression and histone H3 acetylation in dark and light conditions. Acetylation of histones is mediated by non-coding RNAs (Groen and Morris, 2013; Wang et al., 2014) and, in animals, is co-ordinated with that of HSFs (Erkina and Erkine, 2006; Petesch and Lis, 2008; Akerfelt et al., 2010; Guertin and Lis, 2010). We speculate that the same could happen in plant cells undergoing a transition from NS to HS and could be how co-ordinated changes in chromatin condensation and HSFA1b binding occur (Supplementary Fig. S4). In addition, cisNATs have been shown to enhance translation of the target mRNA (Jabnoune et al., 2013; Bazin et al., 2017). Therefore, the HSFA1b-mediated control of cisNAT gene expression may lead to altered translation of specific transcripts under HS conditions.
HSFA1b is one route for the transmission of environmental cues to a core set of stress-responsive and development-associated genes
In Arabidopsis, genomic regions are occupied by multiple TFs and enriched for genes involved in development and stimulus responses (Heyndrickx et al., 2014). The notion of co-operation with other TFs is supported by the clear enrichment of co-occurring binding motifs in promoter regions of HSFA1b-bound genes under NS and HS conditions (Fig. 7A). In the ChIP-seq data sets available to us from NS plants, we compared them with our NP:HSFA1b NS data only. This revealed 76 gene targets common to the eight TFs (Fig. 7B, C, E; Supplementary Data S9). These eight TF genes have all been implicated in the control of growth and development and responses to environmental stress. For example, HBI1 is important in poising plants between growth and the level of immunity to pathogens (Fan et al., 2014). PRR5, PRR7, PIF4, PIF5, LFY, and SEP3 regulate genes responsive to cold stress, oxidative stress, light quality, and photoperiod, as well as playing developmental roles (Moyroud et al., 2011; Nakamichi et al., 2012; Pajoro et al., 2014; Pedmale et al., 2016). We suggest that HSFA1b is one of at least eight and probably many more TFs that transduce a variety of endogenous and environmental signals controlling different combinations of genes drawn from a core group of development- and stress-associated genes that control the plant’s multiple physiological responses to the highly variable environment it encounters.
Supplementary data
Supplementary data are available at JXB online.
Methodology. A detailed step-by-step protocol for the preparation of ChIP-seq samples from Arabidopsis leaves.
Fig. S1. Properties of the HSFA1b–eYFP line used for ChIP-seq.
Fig. S2. Time series qRT-PCR results comparing the activation time of four heat-responsive genes.
Fig. S3. The impact of heat stress on growth of Arabidopsis shoots.
Fig. S4. The degree of overlap between Groups I–III HSFA1b target genes and the nearest transcription start site (TSS) loci in genome-mapped DNase I-hypersensitive sites.
Fig. S5. Phenotype of 35S:HSFA1b plants compared with Col-0.
Fig. S6. Hierarchical network showing interaction between HSFA1b and BZIP28.
Table S1. Experimentally confirmed developmental genes bound by HSFA1b
Table S2. Primers used in the qPCR analyses.
Data S1. All genomic sites bound by HSFA1b under NS and HS
Data S2. Significant GO terms of HSFA1b target genes in Groups I, II, and III.
Data S3. All differentially expressed genes in WT HS versus WT NS, 35S:HSFA1b versus WT NS, and 35S:HSFA1b versus WT HS.
Data S4. Expression of all Group I, II, and III genes in WT HS compared with WT NS.
Data S5. All significant GO terms of up-regulated and down-regulated Group I, II, and III genes in response to HS.
Data S6. All significant GO terms of up- and down-regulated Group I, II, and III genes in 35S:HSFA1b NS.
Data S7. Genes whose expression is indirectly regulated by HSFA1b.
Data S8. Target genomic loci and their cisNAT RNAs that show differential expression.
Data S9. Shared binding sites between HSFA1b and HBI-1, PRR5, LFY, SEP3, PIF4, PIF5, and PRR7.
Data S10. All significantly enriched GO terms of the genes targeted by HSFA1b and HBI-1, LFY, PRR5, PRR7, SEP3, PIF4, and PIF5.
Cytoscape File S1. A Cytoscape file which allows an interactive view of Fig. 5D but requires first that the open source program is downloaded from http://www.cytoscape.org/.
Acknowledgements
We are grateful to Professor F. Schöffl for the gift of the hsfA1a/hsfA1b mutant and Drs Y.-Y Charng and H.-C. Liu for the qKO mutant. The authors acknowledge the support of a Kingdom of Saudi Arabia Studentship and the Biotechnology and Biological Sciences Research (grant BB/F005822/). This work was supported by the ‘Laboratoire d’Excellence (LABEX)’ Saclay Plant Sciences (SPS; ANR-10-LABX-40) and the ANR grant SPLISIL, France.
References
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. 2008. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Current Biology 18, 656–660. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nature Reviews. Molecular Cell Biology 11, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M. 2015. Battles and hijacks: noncoding transcription in plants. Trends in Plant Science 20, 362–371. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NR. 2008. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annual Review of Plant Biology 59, 89–113. [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, et al. 2004. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences 29, 471–487. [DOI] [PubMed] [Google Scholar]
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J. 2017. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proceedings of the National Academy of Sciences, USA 114, E10018–E10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Albihlal WS, Lawson T, et al. 2013. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. Journal of Experimental Botany 64, 3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Penfold CA, Jenkins DJ, et al. 2016. Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. The Plant Cell 28, 345–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard J, Oliver C, Harrison PM. 2015. The distribution and evolution of Arabidopsis thaliana cis natural antisense transcripts. BMC Genomics 16, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. 2005. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal 41, 1–14. [DOI] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Agarwal P, Khurana P. 2011. Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Molecular Genetics and Genomics 286, 171–187. [DOI] [PubMed] [Google Scholar]
- Chawade A, Bräutigam M, Lindlöf A, Olsson O, Olsson B. 2007. Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics 8, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, et al. 2012. Systematic evaluation of factors influencing ChIP-seq fidelity. Nature Methods 9, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. The Plant Journal 89, 789–804. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina TY, Erkine AM. 2006. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Molecular and Cellular Biology 26, 7587–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TG, Belak Z, Ovsenek N, Krone PH. 2007. Heat shock factor 1 is required for constitutive Hsp70 expression and normal lens development in embryonic zebrafish. Comparative Biochemistry and Physiology. Part A, Molecular & Integrative Physiology 146, 131–140. [DOI] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, et al. 2014. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. The Plant Cell 26, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR. 2003. Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. The Plant Journal 33, 691–705. [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Fryer MJ, Lawson T, et al. 2009. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. The Plant Cell 21, 2143–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen JN, Morris KV. 2013. Chromatin, non-coding RNAs, and the expression of HIV. Viruses 5, 1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. 2013. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. The Plant Journal 74, 840–851. [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. 2010. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genetics 6, e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Chen S, Liu K, Liu Y, Ni L, Zhang K, Zhang L. 2008. Isolation of heat shock factor HsfA1a-binding sites in vivo revealed variations of heat shock elements in Arabidopsis thaliana. Plant and Cell Physiology 49, 1306–1315. [DOI] [PubMed] [Google Scholar]
- Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH. 2016. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Fujimoto M, et al. 2014. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron 82, 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx KS, Van de Velde J, Wang C, Weigel D, Vandepoele K. 2014. A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. The Plant Cell 26, 3894–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Xu Z, Hu B, Lu ZJ. 2017. COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Research 45, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SM, Kim DW, Woo MS, Jeong HS, Son YS, Akhter S, Choi GJ, Bahk JD. 2014. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant, Cell and Environment 37, 1202–1222. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. 2013. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. The Plant Cell 25, 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob P, Hirt H, Bendahmane A. 2017. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnology Journal 15, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger AM, Makley LN, Gestwicki JE, Thiele DJ. 2014. Genomic heat shock element sequences drive cooperative human heat shock factor 1 DNA binding and selectivity. Journal of Biological Chemistry 289, 30459–30469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger AM, Pemble CW 4th, Sistonen L, Thiele DJ. 2016. Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nature Structural and Molecular Biology 23, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H-S, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TD, Pogson BJ, Chory J. 2013. Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proceedings of the National Academy of Sciences, USA 110, 14474–14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, Noh B. 2015. Repression of flowering under a noninductive photoperiod by the HDA9–AGL19–FT module in Arabidopsis. New Phytologist 206, 281–294. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E, Chow BY, Pruneda-Paz JL, Kay SA. 2014. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proceedings of the National Academy of Sciences, USA 111, 16172–16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N, Møller SG, Shinomura T, Xu X, Chua NH, Furuya M. 2003. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. The Plant Cell 15, 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Berendzen KW, Schöffl F. 2010. Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Molecular Biology 73, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Charng YY. 2013. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiology 163, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG. 2008. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. The Plant Journal 55, 760–773. [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. 2011. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant, Cell and Environment 34, 738–751. [DOI] [PubMed] [Google Scholar]
- Liu J, Feng L, Li J, He Z. 2015a Genetic and epigenetic control of plant heat responses. Frontiers in Plant Science 6, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. 2012. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. The Plant Cell 24, 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua NH. 2015b Long noncoding RNA transcriptome of plants. Plant Biotechnology Journal 13, 319–328. [DOI] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. The Plant Cell 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. 2006. Could heat shock transcription factors function as hydrogen peroxide sensors in plants?Annals of Botany 98, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyroud E, Minguet EG, Ott F, et al. 2011. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. The Plant Cell 23, 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy M, Uma S, Backiyarani S, Saraswathi MS. 2015. Genome-wide screening for novel, drought stress-responsive long non-coding RNAs in drought-stressed leaf transcriptome of drought-tolerant and -susceptible banana (Musa spp) cultivars using Illumina high-throughput sequencing. Plant Biotechnology Reports 9, 279–286. [Google Scholar]
- Nakagawa T, Kurose T, Hino T, et al. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. 2012. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proceedings of the National Academy of Sciences, USA 109, 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. 2006. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. The Plant Journal 48, 535–547. [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nosaka R, Hayashi H, et al. 2011. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant and Cell Physiology 52, 933–945. [DOI] [PubMed] [Google Scholar]
- Nix DA, Courdy SJ, Boucher KM. 2008. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics 9, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. 2001. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?Cell Stress and Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Kusakabe K, Mizoi J, et al. 2016. The transcriptional cascade in the heat stress response of arabidopsis is strictly regulated at the level of transcription factor expression. The Plant Cell 28, 181–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, et al. 2014. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biology 15, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, et al. 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Salamó I, Papdi C, Rigó G, et al. 2014. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiology 165, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F. 1998. HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Molecular and General Genetics 258, 269–278. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. 2012. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica et Biophysica Acta 1819, 104–119. [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. 2008. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. The Plant Journal 53, 264–274. [DOI] [PubMed] [Google Scholar]
- Shafiq S, Li J, Sun Q. 2016. Functions of plants long non-coding RNAs. Biochimica et Biophysica Acta 1859, 155–162. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, et al. 2014. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8, 2015–2030. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner MW, Andreas AP. 2007. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szcześniak MW, Rosikiewicz W, Makałowska I. 2016. CANTATAdb: a collection of plant long non-coding RNAs. Plant and Cell Physiology 57, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Vdu T, Souiai O, Romero-Barrios N, Crespi M, Gautheret D. 2016. Detection of generic differential RNA processing events from RNA-seq data. RNA Biology 13, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela CE, Acevedo-Acevedo O, Miranda GS, Vergara-Barros P, Holuigue L, Figueroa CR, Figueroa PM. 2016. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. Journal of Experimental Botany 67, 4209–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH. 2014. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Research 24, 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Gross-Hardt R, Schöffl F. 2014. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Molecular Biology 85, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Guo H, Zhang D, Chen D, Jiang Z, Lin R. 2015. REVEILLE1 promotes NADPH:protochlorophyllide oxidoreductase A expression and seedling greening in Arabidopsis. Photosynthesis Research 126, 331–340. [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. 2011. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Research 39, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, et al. 2011. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Molecular Genetics and Genomics 286, 321–332. [DOI] [PubMed] [Google Scholar]
- Yu X, Yang J, Li X, Liu X, Sun C, Wu F, He Y. 2013. Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biology 13, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-S, Yang H, Ding L, Song Z-T, Ma H, Chang F, Liu J-X. 2017. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. The Plant Cell 29, 1007–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS, Pincus D. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.