Abstract
Genome editing using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) predominantly induces non-homologous end joining (NHEJ), which generates random insertions or deletions, whereas homology-directed repair (HDR), which generates precise recombination products, is useful for wider applications. However, the factors that determine the ratio of HDR to NHEJ products after CRISPR/Cas9 editing remain unclear, and methods by which the proportion of HDR products can be increased have not yet been fully established. We systematically analyzed the HDR and NHEJ products after genome editing using various modified guide RNAs (gRNAs) and Cas9 variants with an enhanced conformational checkpoint to improve the fidelity at endogenous gene loci in HEK293T cells and HeLa cells. We found that these modified gRNAs and Cas9 variants were able to enhance HDR in both single-nucleotide substitutions and a multi-kb DNA fragment insertion. Our results suggest that the original CRISPR/Cas9 system from the bacterial immune system is not necessarily the best option for the induction of HDR in genome editing and indicate that the modulation of the kinetics of conformational checkpoints of Cas9 can optimize the HDR/NHEJ ratio.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) has revolutionized our ability to edit the genome in basically all species (1–3). The CRISPR/Cas9 system, which consists of a nuclease (Cas9) and two short single-strand RNAs (crRNA and tracrRNA), was originally a bacterial adaptive immune system. These two RNAs can be fused for genome editing as single-guide RNA (sgRNA) (For simplicity, we use the term ‘guide RNA (gRNA)’ to mean sgRNA in this manuscript) (2). Cas9 and a gRNA form a ribonucleoprotein complex and bind to genomic DNA. The Cas9–gRNA complex scans the genome to identify a protospacer adjacent motif (PAM) and then a genomic DNA sequence adjacent to PAM that matches the gRNA sequence to cleave it (4). This scanning process depends on three-dimensional gRNA-dependent and gRNA-independent interactions of the Cas9–gRNA complex to DNA. The gRNA-dependent interaction is derived from the base-paring between a gRNA and genomic DNA. In contrast, the gRNA-independent interactions take place between genomic DNA and the amino acid residues of Cas9, including the PAM recognition (5). Based on these findings, two groups have independently developed SpCas9 variants in which this gRNA-independent DNA interaction is altered (eSpCas9(1.1) and SpCas9-HF1) and shown that these variants have fewer off-target effects than wild-type (WT) Cas9 (6,7).
An alternative approach to improve the specificity of Cas9 is to modify gRNAs by removing 5′ nucleotides from the standard 20 nt gRNAs (especially, 17 nt gRNAs have been proposed to be the best as tru-gRNAs) or adding extra 5′ guanines to compromise the optimum interaction between the target DNA and the Cas9–gRNA complex (8,9). Recently, a conformational change from an inactive state to an active state of the HNH nuclease domain of Cas9 was shown to be a checkpoint of mismatches between target DNA and a gRNA, and these Cas9 variants and the modified gRNAs improved the fidelity of Cas9 by making the threshold for this checkpoint higher than that of WT Cas9 (10,11). It has also been shown that the REC3 domain of Cas9 recognizes the mismatches between the target DNA and a gRNA to control the conformational change of the HNH domain, and mutagenesis in the REC3 domain led to the creation of yet another Cas9 variant with higher fidelity, HypaCas9 (10). Thus, the conformational checkpoint of Cas9 for proof-reading is based on the interactions between the REC3 domain and the gRNA/DNA heteroduplex that allosterically determine whether the HNH nuclease domain transitions from the inactive state to the active state. Therefore, different thresholds for the conformational checkpoint can affect the overall kinetics of how Cas9 cleaves and dissociates from DNA.
When a conformational change in the HNH domain occurs by matched base-pairing between a gRNA and target DNA, Cas9 generates a double-strand break (DSB) at the target site to activate two endogenous DNA repair pathways in the cell: homology-directed repair (HDR) and non-homologous end joining (NHEJ). HDR is based on DNA recombination between the genomic DNA and homologous template DNA, which facilitates precise genome editing when the exogenous donor DNA is provided to the cells. In contrast, NHEJ is a template-free, error-prone repair mechanism in which the broken ends of DNA are joined together, often with random insertions or deletions (indels) (12). Thus, NHEJ is often used to disrupt genes by creating frameshift mutations.
In general, CRISPR/Cas9 is very efficient at inducing NHEJ and has already been established as an excellent tool for knocking-out genes in various species (13). Several studies are currently investigating the clinical applications of exploiting the high NHEJ-induction activity of CRISPR/Cas9—such as the disruption of CCR5 and PD-1 for HIV and cancer therapy, respectively (14–17). When donor DNA is provided, both HDR and NHEJ can be activated concurrently, even within a single cell. In general, however, NHEJ is predominant.
When precise genome editing based on HDR is needed, the random indels produced by NHEJ represent harmful byproducts (18). The inefficiency of CRISPR/Cas9 in inducing HDR without NHEJ is currently limiting its potential for even wider applications, such as gene correction therapy by HDR. Significant efforts have been made to enhance HDR and suppress NHEJ. The synchronization of the cell cycle and the timed delivery of the CRISPR/Cas9 system as a ribonucleoprotein complex (rather than a plasmid) enhances HDR while minimizing NHEJ (19). The ligase 4 inhibitor SCR7 can suppress NHEJ (20,21), and the inhibition of 53BP1 is effective for enhancing HDR (22). Asymmetric oligonucleotide donor DNA can also enhance HDR (23). Base editors that exchange nucleobases without creating DSBs are available for certain single-nucleotide substitutions (24,25). However, the factors that determine the balance of HDR and NHEJ induced by CRISPR/Cas9 remain unclear, hampering efforts to induce HDR more frequently.
To analyze the balance of HDR and NHEJ, we must detect and quantify these activities. However, no assays that can efficiently accomplish this task have yet been developed. We recently developed a droplet digital polymerase chain reaction (ddPCR)-based method that can be used to efficiently quantify genome editing outcomes at endogenous gene loci (18,26). Our method allows for the systematic evaluation of the HDR and NHEJ activities of various genome editing conditions to clarify what determines the balance of HDR and NHEJ. Therefore, we tested various modified gRNAs and Cas9 variants with modified interactions with DNA, including eSpCas9(1.1), SpCas9-HF1, and HypaCas9 to address the impact of the altered conformational checkpoint on the balance between HDR and NHEJ. Our findings provide new insight into how the balance of CRISPR/Cas9-induced HDR and NHEJ is determined.
MATERIALS AND METHODS
Statistical analyses
The transfection experiments in HEK293T cells and HeLa cells were performed in triplicate (three biological replicates). A two-tailed Student's t-test was performed to analyze the differences between WT Cas9 and the Cas9 variants, and the standard gRNA and the gRNAs with different-length. No samples were excluded.
Plasmids and oligonucleotides
The plasmids for expression of the Cas9 variants that we created are available via Addgene (Addgene #108292–108302). We used pX459 V2.0 (Addgene #62988) for the transient coexpression of WT SpCas9 and the puromycin-resistant gene. The Cas9 sequence in pX459 V2.0 was replaced by that of eSpCas9(1.1) (Addgene #71814) for the expression of eSpCas9(1.1). The N497A, R661A, Q695A, and Q926A mutations and the N692A, M694A, Q695A, and H698A mutations were introduced into pX459 V2.0 for SpCas9-HF1 and HypaCas9, respectively. The Y450A and R661A mutations in SpCas9-HF1 were introduced and reverted, respectively, to generate the SpCas9-HF1 variants. The K848A, K810A, and K855A mutations were introduced into WT Cas9 and eSpCas9(1.1) to generate the eSpCas9(1.1) variants. The oligonucleotide donors were all 60 nt; point mutations were present in the middle of the oligonucleotide donors, as described previously (18). Sense and anti-sense oligonucleotides were annealed by a thermal cycler and purified by ethanol precipitation, so they were all double-stranded unless otherwise noted. The sequences of the oligonucleotide donors and gRNAs that were used in this study are summarized in Supplementary Tables S1 and S2. All of the oligonucleotide donors and oligonucleotides for gRNA cloning, which were purified by standard desalting, were purchased from FASMAC, Japan. The knock-in experiment was done with pX330 (for WT Cas9) (Addgene #42230), eSpCas9(1.1), pX330 with the N497A, R661A, Q695A, and Q926A mutations (for SpCas9-HF1), and N692A, M694A, Q695A and H698A mutations (for HypaCas9). AAV-CAGGS-EGFP (Addgene #22212) as a targeting vector and AAVS1 gRNA-T2 were used for targeting (3,27). For the genotyping PCR in the AAVS1 targeting experiment, we used the AAV 5′ J F, Puro J R and AAV WT R primers from Mandegar et al. (28) and designed the AAV WT F2 primer (Supplementary Table S3).
HEK293T cell and HeLa cell culture, transfection and selection of transfected cells
HEK293T cells and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) with high glucose, sodium pyruvate, and L-glutamine (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). For transfection, 3 × 104 cells and 2 × 104 cells were plated into each well of a 96-well plate for HEK293T cells and HeLa cells, respectively. One day later, the cells were transfected with 90 ng of a plasmid for nucleases and 10 ng of oligonucleotide donor DNA using 0.3 μl of Lipofectamine 2000 (Thermo Fisher Scientific) per well for HEK293T cells and 0.3 μl of Lipofectamine 3000 and 0.2 μl of P3000 reagent (Thermo Fisher Scientific) for HeLa cells per well, according to the manufacturer's instructions. The media was exchanged for the same media supplemented with 5 μg/ml and 0.5 μg/ml of puromycin for HEK293T cells and HeLa cells, respectively, 1 day after transfection. We found that these conditions were optimum for eliminating cells that were not transfected while keeping transfected cells alive (Supplementary Figure S1). Genomic DNA was extracted from the cells as described previously (18) 2 and 4 days after the addition of puromycin for HEK293T cells and HeLa cells, respectively. Genomic DNA was resuspended in 30 μl of water per well.
AAVS1 targeting of by reverse transfection of HEK293T cells
Lipofectamine 2000 complex for reverse transfection was prepared in a 1.5-ml tube using 100 ng of pX330 or a pX330-based plasmid for a Cas9 variant, 100 ng of AAV-CAGGS-EGFP, and 0.6 μl of Lipofectamine 2000 in 20 μl of OPTI-MEM (Thermo Fisher Scientific) according to the manufacturer's instructions. HEK293T cells were detached from a plastic dish and resuspended in DMEM with high glucose, sodium pyruvate, and l-glutamine supplemented with 10% fetal bovine serum at 3 × 105 cells/ml. We transferred 200 μl of the cell suspension (6 × 104 cells) into the 1.5 ml tube and mix the cells with the complex. The mixture was incubated at room temperature for 30 min. We plated 110 μl of the mixture (3 × 104 cells) in a well of a 96-well plate to monitor the NHEJ activity 3 days after transfection. The rest of the cells were resuspended in 10 ml of the media and plated in an entire 96-well plate (300 cells/well). The media was changed to the same media supplemented with 0.5 μg/ml puromycin, which was exchanged once a week. Pictures of the plates were taken, the EGFP and Hoechst 33342 fluorescence was observed by using BZ-X700 (Keyence, Japan), and the genomic DNA was extracted from the surviving cells 19 days after transfection. PrimeSTAR Max DNA Polymerase (Takara Bio, Japan) and Phusion High-Fidelity DNA Polymerase (New England Bio-Labs, USA) were used for genotyping PCR. The sequences of the primers and the detailed protocol for PCR are summarized in Supplementary Table S3. The frequencies at which HEK293T cells acquired puromycin-resistance were calculated from the number of wells with the resistant cells as follows:
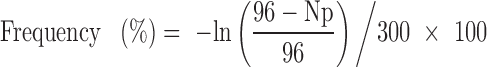 |
where Np = the number of wells with yellow media color due to the presence of surviving cells. This is based on the fact that the number of puromycin-resistant cells per well follows Poisson distribution, and the same strategy was described before (29).
ddPCR assay to detect HDR and NHEJ
Most of the ddPCR assays for RBM20, GRN, and ATP7B were described and validated in our previous publication (18). The protocols for the ddPCR assays were the same as previously described. For some gRNAs, we designed new hydrolysis probes to detect NHEJ at the cut sites (Supplementary Figure S2). The ddPCR assay for AAVS1 to measure NHEJ was carried out using HindIII to digest the genomic DNA and the same thermal cycles as for the RBM20 assays. The full information on all of the probes and primers used in the ddPCRs is shown in Supplementary Tables S4 and S5. Typical 2D ddPCR plots are shown in Supplementary Figures S3 and S6B.
RESULTS
ddPCR-based assay to quantify the HDR and NHEJ activities in endogenous gene loci
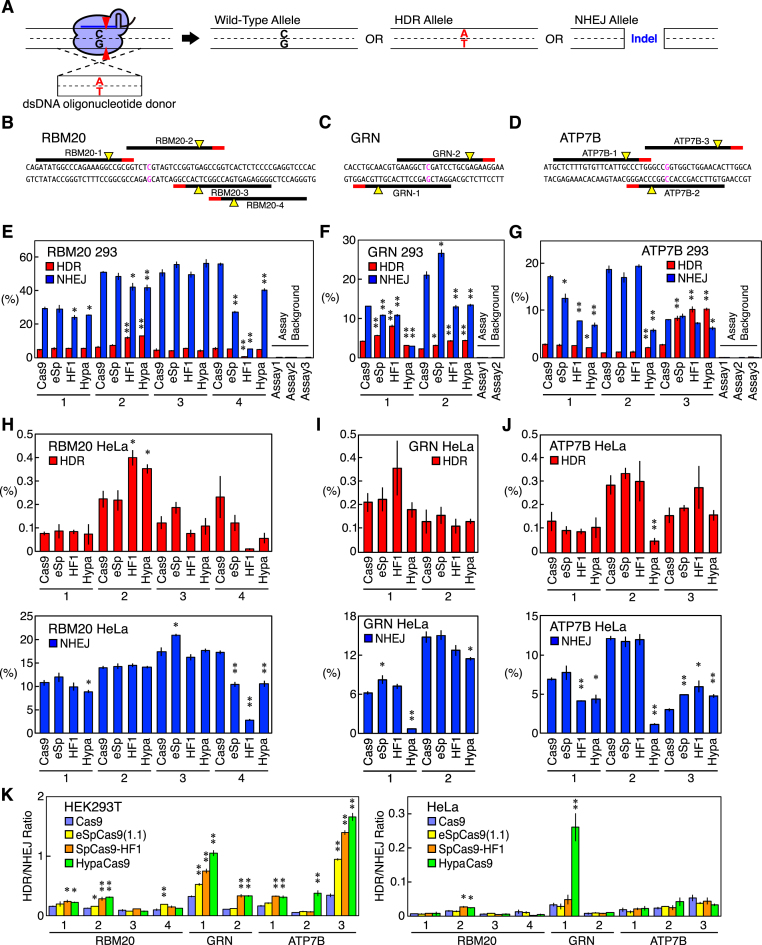
To quantify the HDR and NHEJ activities induced by Cas9 in endogenous gene loci, we introduced three pathogenic point mutations—RBM20 R636S, GRN R493X and ATP7B R778L (Supplementary Table S6)—by HDR using donor oligonucleotide DNA (Supplementary Table S1). The allelic frequencies of the desired point mutation and the indels were monitored as the HDR and NHEJ activities, respectively (Figure 1A). We chose these three targets because we had already established a ddPCR-based system with allele-specific hydrolysis probes to precisely quantify the allelic frequencies in these three genes (Supplementary Figures S2 and S3) (18). As we used the human U6 promoter to drive the gRNA expression, the gRNAs required a guanine at the 5′ end for their optimum expression. We therefore first designed a total of nine perfectly matched gRNAs with an endogenous 5′ guanine (RBM20-1-4, GRN-1 and 2, and ATP7B-1-3) (Figure 1B–D and Supplementary Table S2). We tested 60-nt sense, anti-sense, and annealed double-stranded donor oligonucleotide DNAs with three gRNAs (RBM20-2, GRN-2, and ATP7B-g5, which was a gRNA with an extra 5′ guanine; see below), combined with WT Cas9 and found that the double-stranded donors gave the highest HDR-to-NHEJ ratio for all three gRNAs (Supplementary Figure S4). Therefore, we decided to use the annealed double-stranded donors for subsequent experiments.
Figure 1.
The HDR and NHEJ activities of Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 with perfect-match gRNAs in RBM20, GRN, and ATP7B in HEK293T cells and HeLa cells. (A) The design of the assay to quantify the HDR and NHEJ activities of a Cas9–gRNA complex. A Cas9–gRNA complex was designed to cleave the genome DNA adjacent to a mutation site. Double-stranded oligonucleotide donor DNA was used to induce a point mutation (C→A in this example). The frequencies of the resulting three alleles (wild-type, HDR, and NHEJ) were quantified by ddPCR to measure the HDR and NHEJ activities. (B–D) Perfect-match gRNAs tested in RBM20 (B), GRN (C) and ATP7B (D). The PAMs, cleavage sites, and point mutation sites are represented by red lines, yellow triangles, and magenta characters, respectively. (E–J) The HDR and NHEJ activities in HEK293T cells (E–G) and HeLa cells (H–J) of Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 with perfect-match gRNAs in RBM20 (E and H), GRN (F and I), and ATP7B (G and J). The frequencies of the HDR alleles (red) and NHEJ alleles (blue) among the total alleles ± S.E. are shown (n = 3). The assay backgrounds (which are negligible) of the probes used in this study are also shown on the right in (E–G). Student's t-test was used to evaluate the difference in the HDR and NHEJ activities between WT Cas9 and the three Cas9 variants. *P< 0.05 and **P< 0.01. (K) The HDR/NHEJ ratio induced by Cas9 and its variants in HEK293T cells and HeLa cells. The values ± S.E. calculated from (E–J) are shown (n = 3). Student's t-test was used to evaluate the difference between WT Cas9 and the three Cas9 variants. *P< 0.05 and **P< 0.01.
Genome editing activities of Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9
We hypothesized that the altered conformational checkpoint of the high-fidelity Cas9 variants would affect not only the fidelity of genome editing but also the HDR and NHEJ activities, as it changes the kinetics of the DNA cleavage and potentially the Cas9–DNA interaction after the cleavage, which would influence how the DNA repair machinery and donor DNA interact with the genomic DNA. We therefore introduced the disease-associated point mutations by WT Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 with identical gRNAs in RBM20, GRN and ATP7B, in HEK293T cells and HeLa cells.
To eliminate variations in the transfection efficiency, we puromycin-selected transfected cells based on the coexpression of the puromycin-resistant gene together with Cas9 and a gRNA. We confirmed that the background noise of the assays was negligible (<0.1%) (Figure 1E–G) and found that the three Cas9 variants showed distinctive activities from WT Cas9 (Figure 1E–J). Overall, the three variants increased HDR and/or decreased NHEJ with many gRNAs. Only RBM20-3 in HEK293T cells induced HDR and NHEJ basically at the same level with WT Cas9 and the Cas9 variants. The enhancement in HDR was most evident with ATP7B-3 in HEK293T cells, as all the three Cas9 variants induced more than three times more HDR than WT Cas9. We also observed that the three variants behaved differently in some target sites. For example, each variant showed quite unique HDR and NHEJ activities with RBM20-4, while only HypaCas9 induced more HDR and less NHEJ than WT Cas9 with ATP7B-2. No clear difference in the susceptibility to the Cas9 variants was observed between gRNAs on the sense and anti-sense strands (Figure 1E–J). We also calculated the HDR/NHEJ ratio in HEK293T cells and HeLa cells in Figure 1E–J. Of the nine gRNAs we tested, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 gave HDR/NHEJ ratios higher than those of WT Cas9 with four, six and seven gRNAs, respectively (Figure 1K). The positive effects on HDR of the Cas9 variants were less evident in HeLa cells than in HEK293T cells, but SpCas9-HF1 and HypaCas9 still increased the HDR/NHEJ ratio over that of WT Cas9 with one and two gRNAs out of the nine gRNAs, respectively (Figure 1K). Notably, most of the conditions with the three Cas9 variants did not reduce the HDR/NHEJ ratio to below that of WT Cas9 in either HEK293T or HeLa cells (Figure 1K).
Taken together, these results indicated that the altered conformational checkpoint of Cas9 strongly affects its HDR and NHEJ activities, which can increase the HDR/NHEJ ratio by up to 6.9-fold (with HypaCas9 and ATP7B-2) and 7.7-fold (with HypaCas9 and GRN-1) in HEK293T cells and HeLa cells, respectively (Figure 1K). Therefore, Cas9 variants with perfect-match gRNAs can help enhance HDR while exerting reduced off-target effects. Of particular note, HypaCas9 showed the greatest impact on improving the HDR/NHEJ ratio.
Influence of guanines at the 5′ end of gRNAs on the outcomes of genome editing
When driving the expression of gRNAs by the human U6 promoter, it is necessary to add an extra 5′ guanine when the gRNA does not have an endogenous one. It was also demonstrated that the addition of extra 5′ guanines affects the interaction of the Cas9–gRNA complex with DNA, which results in better proof-reading, most likely via a higher threshold for the conformational checkpoint of Cas9, as shorter gRNAs (a similar approach) have been shown to modulate the checkpoint (9,30).
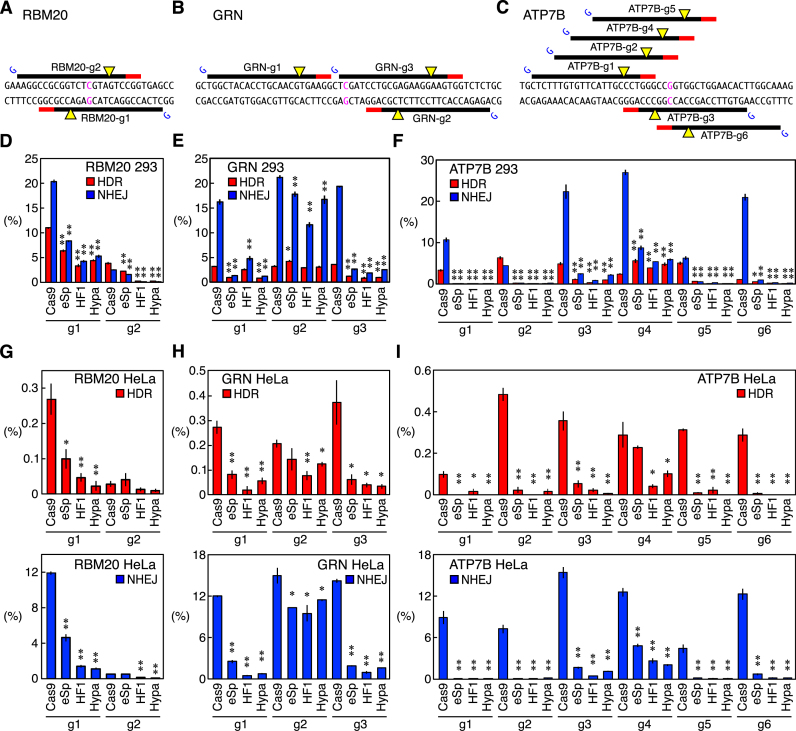
As we noted that changes in the conformational checkpoint of Cas9 strongly influenced the genome editing outcomes (Figure 1), we designed a total of 11 gRNAs with an extra 5′ guanine (RBM20-g1 and RBM20-g2, GRN-g1-3 and ATP7B-g1-6) (Figure 2A–C and Supplementary Table S2) and induced disease-associated mutations using WT Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9. For most of the gRNAs with an extra 5′ guanine that we tested, all of the Cas9 variants showed substantially lower activities for both HDR and NHEJ in HEK293T cells and HeLa cells compared to WT Cas9 (Figure 2D–I). This was probably because the compound effect of the Cas9 variants and the mismatched guanine in gRNAs raised the threshold for the conformational checkpoint too excessively. GRN-g2 and ATP7B-g4 were the only exceptions in HEK293T cells, with which the three Cas9 variants were able to induce HDR at a comparable or higher level while inhibiting NHEJ compared to WT Cas9 (Figure 2E and F). With the other nine gRNAs, the three Cas9 variants induced only one third or less HDR or NHEJ in HEK293T cells or HeLa cells compared to WT Cas9 on average. At present, how GRN-g2 and ATP7B-g4 differ from the other nine gRNAs is unclear, but the same trend was observed in HeLa cells as well (Figure 2H and I). Kleinstiver et al., Kim et al., and Zhang et al. also observed the same suppressive effect on the NHEJ activity of gRNAs with a mismatched 5′ guanine combined with Cas9 variants (6,31,32). These results indicate that the Cas9 variants combined with a gRNA with an extra 5′ guanine tend to have a threshold for the conformational checkpoint that is so stringent that genome editing events can be induced only at a low level.
Figure 2.
The HDR and NHEJ activities of Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 with gRNAs with an extra 5′ guanine in HEK293T cells and HeLa cells. (A–C) gRNAs with an extra 5′ guanine tested in RBM20 (A), GRN (B) and ATP7B (C). PAMs, cleavage sites, and point mutation sites are represented by red lines, yellow triangles and magenta characters, respectively. For each gRNA, an extra guanine was added at the 5′ end to allow the expression by the U6 promoter, which is represented by blue Gs. (D–I) HDR and NHEJ activities of Cas9, eSpCas9(1.1), SpCas9-HF1 and HypaCas9 in HEK293T cells (D–F) and HeLa cells (G–I) with gRNAs with an extra guanine in RBM20 (D and G), GRN (E and H) and ATP7B (F and I). The frequencies of the HDR alleles (red) and NHEJ alleles (blue) among the total alleles ± S.E. are shown (n = 3). Student's t-test was used to evaluate the difference in the HDR and NHEJ activities between WT Cas9 and the three Cas9 variants. *P< 0.05 and **P< 0.01. In general, gRNAs with an extra 5′ guanine combined with the Cas9 variants exhibited very low activities compared to WT Cas9.
Utility of gRNAs of different lengths for identifying the optimum threshold for the conformational checkpoint of Cas9 for inducing HDR and suppressing NHEJ
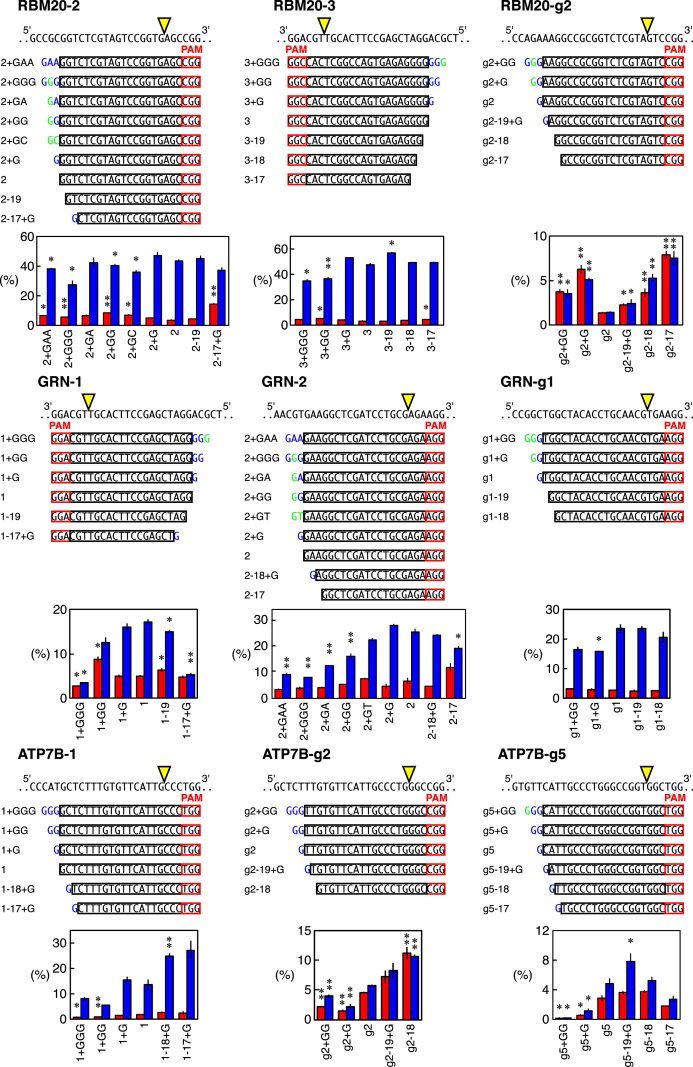
Given our finding that the threshold for the conformational checkpoint of Cas9 strongly affects the genome editing outcomes, we further addressed the relationship between the conformational checkpoint and the HDR- and NHEJ-inducing activities by testing multiple gRNAs of various lengths combined with WT Cas9 in HEK293T cells. As described above, an extra 5′ guanine is expected to increase the threshold for the conformational checkpoint of Cas9 (Figure 2). We therefore designed gRNAs with one, two and three extra 5′ nucleotides (mainly guanines) or extra matched endogenous nucleotides for RBM20-2, RBM20-3, RBM20-g2, GRN-1, GRN-2, GRN-g1, ATP7B-1, ATP7B-g2 and ATP7B-g5 to incrementally increase the threshold for the conformational checkpoint of Cas9. The removal of a few 5′ nucleotides from the standard 20 nt gRNAs is known to be effective for reducing off-target damage in the genome via improved conformational checkpoint (8,11). We therefore also designed several shorter gRNAs to generate a series of gRNAs (Figure 3).
Figure 3.
The HDR and NHEJ activities of gRNAs of different lengths combined with Cas9 in HEK293T cells. The design of the gRNAs with different lengths for RBM20-2, RBM20-3, RBM20-g2, GRN-1, GRN-2, GRN-g1, ATP7B-1, ATP7B-g2 and ATP7B-g5. The genomic sequences are shown above the gRNA series. The gRNA series of RBM20-2 and GRN-2 included ones with extra nucleotides of the endogenous genomic sequence or non-guanine extensions. Extra nucleotides that match and do not match the endogenous sequences are highlighted in green and blue, respectively. The frequencies of the HDR alleles (red) and the NHEJ alleles (blue) among the total alleles ± S.E. induced by WT Cas9 and these gRNA series are shown (n = 3). Student's t-test was used to evaluate the difference in the HDR and NHEJ activities between the original gRNA and the modified gRNAs. *P< 0.05 and **P< 0.01.
As expected, the gRNAs of different lengths exhibited different HDR and NHEJ events. We were able to identify at least one gRNA length that induced more HDR and/or less NHEJ than the original gRNA length with RBM20-2 (2+GAA, 2+GGG, 2+GG, 2+GC, 2-17+G), RBM20-3 (3+GGG, 3+GG, 3-17), GRN-1 (1+GG, 1-19, 1-17+G), GRN-2 (2+GAA, 2+GGG, 2+GA, 2+GG and 2-17), and GRN-g1 (g1+G) (Figure 3). These results showed that the thresholds for the conformational checkpoints to induce the maximum HDR and NHEJ for some gRNAs are different. However, the other four gRNAs, RBM20-g2, ATP7B-1, ATP7B-g2 and ATP7B-g5, exhibited similar trends in both the HDR and NHEJ activities at different lengths (Figure 3).
We also addressed whether or not certain combinations of a Cas9 variant and gRNA of different lengths exerted compound effects on genome editing, as combining these two approaches was expected to further increase the threshold for the conformational checkpoint. To this end, we introduced point mutations in HEK293T cells with gRNA series of RBM20-2 and GRN-2 combined with eSpCas9(1.1). As expected, we found that eSpCas9(1.1) was more sensitive to differences in the gRNA length than WT Cas9, so eSpCas9(1.1) lost most of its activity with several gRNAs (Supplementary Figure S5). However, four gRNAs (RBM20-2+G, RBM20-2-19, GRN-2+G and GRN-2-17) enhanced HDR and/or inhibited NHEJ (Supplementary Figure S5). Therefore, combinations of a Cas9 variant and a gRNA of different lengths may be useful for adjusting conformational checkpoints to achieve the best HDR/NHEJ ratio.
Utility of Cas9 variants with altered interaction with DNA to identify the optimum conformational checkpoint of Cas9 for the induction of HDR
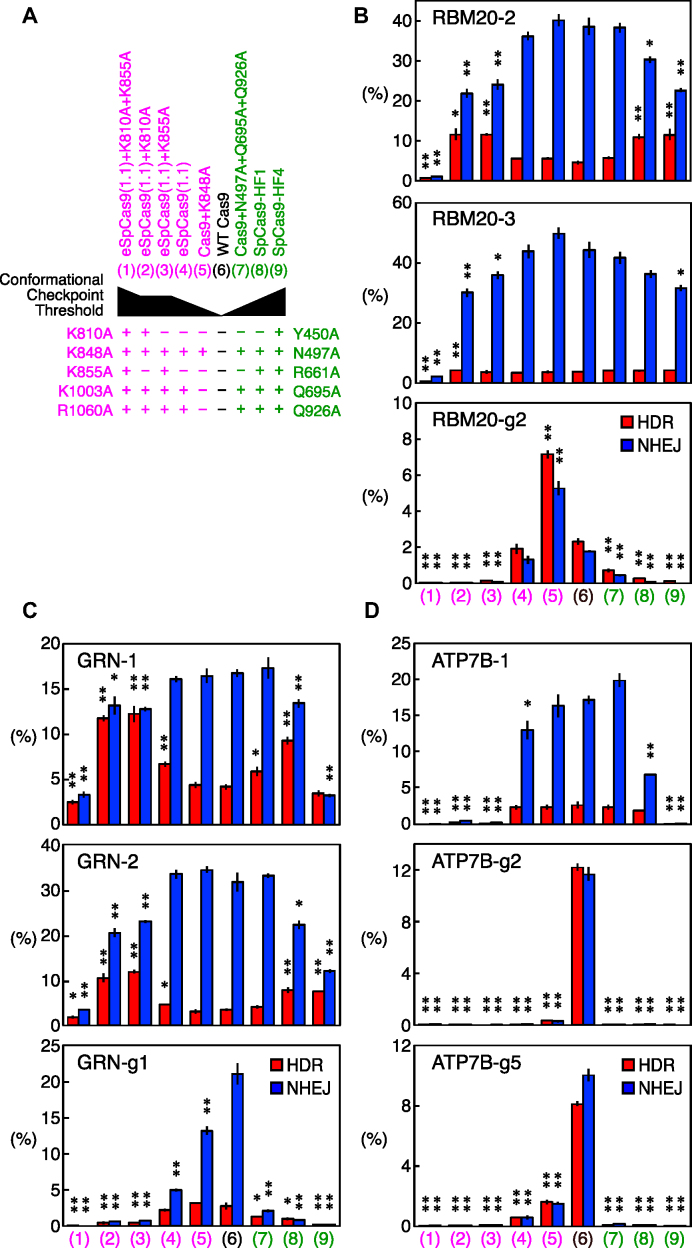
To further confirm the existence of optimum thresholds for the conformational checkpoint of Cas9 for inducing HDR and NHEJ, we next tested a series of Cas9 variants with altered interactions with DNA in addition to eSpCas9(1.1) and SpCas9-HF1 in HEK293T cells. eSpCas9(1.1) and SpCas9-HF1 have three (K848A, K1003A, and R1060A) and four (N497A, R661A, Q695A and Q926A) point mutations, respectively, for modifying the gRNA-independent interaction of Cas9 to DNA, which also results in an increased conformation threshold for the activation of the HNH domain (Figure 4A) (6,7,10,11).
Figure 4.
The HDR and NHEJ activities of the Cas9 variants with altered thresholds for the conformational checkpoint in HEK293T cells. (A) The Cas9 variants tested in this study. WT Cas9 and eight eSpCas9(1.1) and SpCas9-HF1 variants were tested. The point mutations for those variants and the estimated thresholds for the conformational checkpoint of these variants are shown. (B–D) The HDR and NHEJ activities of the Cas9 variants with the RBM20 (B), GRN (C) and ATP7B (D) gRNAs. The frequencies of the HDR alleles (red) and NHEJ alleles (blue) among the total alleles ± S.E. are shown (n = 3). Student's t-test was used to evaluate the difference in the HDR and NHEJ activities between WT Cas9 and the Cas9 variants. *P< 0.05 and **P< 0.01. With RBM20-g2, ATP7B-g2 and ATP7B-g5, the HDR and NHEJ frequencies were very well correlated, and no variant was able to increase the HDR/NHEJ ratio. With all six of the other gRNAs, at least two variants increased the HDR/NHEJ ratio, suggesting the existence of optimum thresholds for the conformational checkpoint.
Based on the results of previous studies, we added or reverted mutations to incrementally modify the interaction between Cas9 and DNA. For eSpCas9(1.1), we introduced either or both the K810A and K855A mutations (eSpCas9(1.1)+K810A, eSpCas9(1.1)+K855A, and eSpCas9(1.1)+K810A+K855A). The eSpCas9(1.1)+K810A and eSpCas9(1.1)+K855A variants were expected to have a similar interaction profile to DNA, whereas eSpCas9(1.1)+K810A+K855A were expected to have the fewest interactions with DNA. We also reverted the K1003A and R1060A mutations in eSpCas9(1.1) (Cas9+K848A). For SpCas9-HF1, we added the Y450A mutation to remove this residue's interaction with DNA (published as SpCas9-HF4) (6). We also reverted the R661A mutation in SpCas9-HF1 to create a Cas9 variant (Cas9+N497A+Q695A+Q926A) with an intermediate interaction between WT Cas9 and SpCas9-HF1 (Figure 4A). With these modifications, the generated Cas9 series were expected to have different thresholds for the conformational checkpoint (Figure 4A).
We tested these Cas9 series with RBM20-2, RBM20-3, RBM20-g2, GRN-1, GRN-2, GRN-g1, ATP7B-1, ATP7B-g2 and ATP7B-g5. The incremental alteration of Cas9–DNA interaction highlighted the existence of a minimum interaction between Cas9 and DNA required to edit the genome. eSpCas9(1.1)+K810A+K855A induced extremely little HDR and NHEJ with all nine tested gRNAs (Figure 4B–D). We speculate that this was because this variant did not generate sufficient interaction with DNA to induce genome editing. The gRNAs with an extra 5′ guanine generally generated less HDR and NHEJ with the Cas9 variants than the perfect-match gRNAs. However, ATP7B-1 exhibited almost no genome editing activity with eSpCas9(1.1)+K810A, eSpCas9(1.1)+K855A, and SpCas9-HF4, despite it being a perfect-match gRNA, highlighting the gRNA sequence-dependency of the threshold for the minimum interaction between Cas9 and DNA (Figure 4D). With the other four perfect-match gRNAs (RBM20-2, RBM20-3, GRN-1 and GRN-2), eSpCas9(1.1)+K810A, eSpCas9(1.1)+K855A and SpCas9-HF4 were able to enhance HDR and/or inhibit NHEJ, suggesting that these variants can be good options for perfect-match gRNAs (Figure 4B–D).
These results again demonstrate that the threshold level of the conformational checkpoint of Cas9 strongly influences genome editing, which can occur at different extents for HDR and NHEJ. There seems to be an optimum level of conformational checkpoint to achieve the best HDR/NHEJ ratio, which is also dependent on the sequence of gRNAs.
Effects of Cas9 with improved proof-reading on the integration of a larger DNA fragment into the genome via HDR
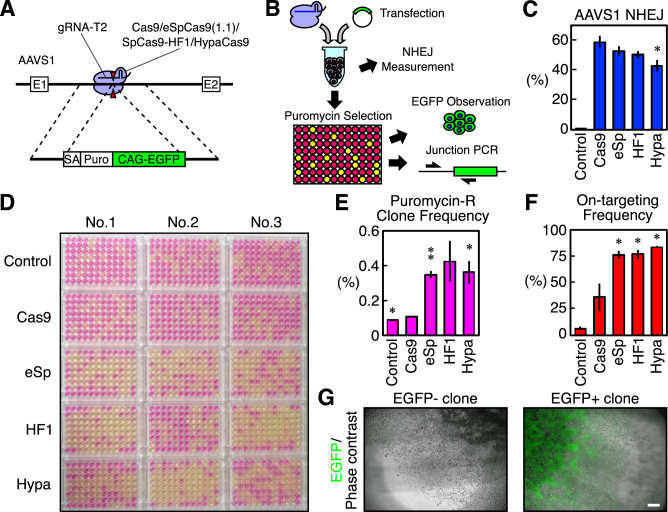
We next addressed whether or not the Cas9 variants with improved proof-reading also have positive effects on knock-in of a larger DNA fragment into the genome via HDR in addition to single-nucleotide substitutions, as we had already clarified in our ddPCR-based assay (Figures 1–4). For this purpose, we knocked an ∼4.2-kb DNA fragment consisting of a splicing acceptor, puromycin-resistant gene, and EGFP gene driven by the CAG promoter into the adeno-associated virus integration site (AAVS)1 (27) using the AAVS1 gRNA-T2 (a perfect-match gRNA, Supplementary Table S2) (3) combined with WT Cas9, eSpCas9(1.1), SpCas9-HF1 and HypaCas9 (Figure 5A).
Figure 5.
Knock-in of a DNA fragment into AAVS1 via HDR induced by Cas9 and the Cas9 variants in HEK293T cells. (A) A schematic representation of the AAVS1 targeted with an approximately 4.2-kb cassette of a puromycin-resistant gene and EGFP driven by the CAG promoter. Cas9 and the Cas9 variants combined with the AAVS1 gRNA-T2 were used. (B) A schematic representation of transfection and an analysis of the cells. HEK293T cells were reverse transfected with a plasmid for the dual expression of Cas9 or a Cas9 variant and a gRNA, and the targeting donor plasmid. Half of the transfected cells were used to monitor NHEJ by ddPCR. The other half of the cells were plated in a 96-well plate and selected for puromycin-resistant cells. The survival of the cells in each well was monitored by the color change induced by the presence of phenol-red in the media. The surviving cells were analyzed by EGFP observation and 5′ junctional PCR for the correct integration of the fragment. (C) The NHEJ activities of Cas9 and the Cas9 variants at AAVS1. The frequencies of the NHEJ alleles among the total alleles±S.E. analyzed by ddPCR are shown (n = 3). Student's t-test was used to evaluate the difference between WT Cas9 and the Cas9 variants or the control. *P< 0.05. (D) The color of the media in the 96-well plates after puromycin selection. (E) The frequencies of HEK293T cells that acquired puromycin-resistance. The values ± S.E. are shown (n = 3). Student's t-test was used to evaluate the difference between WT Cas9 and the Cas9 variants or the control. *P< 0.05 and **P< 0.01. (F) The frequencies at which a well containing puromycin-resistant cells was also positive on 5′ junctional PCR for the correct targeting. The values ± S.E. are shown (n = 3). Student's t-test was used to evaluate the difference between WT Cas9 and the Cas9 variants or the control. *P< 0.05. (G) Examples of puromycin-resistant cells that were negative and positive for EGFP. More than 87% of the wells positive on junctional PCR were also positive for EGFP, and we noted no significant differences in this frequency between WT Cas9 and the Cas9 variants (see also Supplementary Figure S6D). The scale bar is 300 μm. The two images are shown at the same scale.
We reverse-transfected HEK293T cells in a 1.5-ml tube and plated them into 96-well plates (Figure 5B). One well per sample was used to monitor NHEJ events at the AAVS1 by ddPCR (Supplementary Figure S6A and B, and Supplementary Table S4), and we found that only HypaCas9 induced a lower amount of NHEJ than WT Cas9 (Figure 5B and C). We selected HEK293T cells that had acquired puromycin-resistance in 96-well plates and visualized the wells with the surviving cells based on the color of the culture media, which contained phenol-red (Figure 5B and D). We confirmed that the color of the media well reflected the survival of the cells (Supplementary Figure S6C). We observed a striking difference between the WT Cas9 and the Cas9 variants. Based on the number of wells with puromycin-resistant cells, we calculated the frequency at which cells acquired resistance. Some cells acquired puromycin-resistance even after transfected with an unrelated gRNA (control), probably due to random integration of the targeting vector (Figure 5D and E). WT Cas9 did not show a markedly improved frequency of acquisition of puromycin-resistance, although the difference between the control and WT Cas9 was still statistically significant (Figure 5E). To our surprise, eSpCas9(1.1), SpCas9-HF1, and HypaCas9 generated markedly higher frequencies of acquisition of puromycin-resistance than WT Cas9, although the difference between WT Cas9 and SpCas9-HF1 was not statistically significant (P = 0.052 by Student's t-test) due to a relatively large variation in the triplicates of SpCas9-HF1 (Figure 5D and E).
We examined whether or not these puromycin-resistant cells were correctly targeted at the AAVS1 by 5′ junction PCR of the integrated DNA fragment and confirmed that more than 75% of the wells with surviving cells contained correctly targeted cells with all the three Cas9 variants, whereas the frequency was only approximately 36% for WT Cas9 (Figure 5B, F and Supplementary Figure S7).
We also examined the fluorescence of EGFP in the junction PCR-positive wells to discriminate partial integration of the cassette (Figure 5G). We found that more than 87% of the junction PCR-positive wells were also EGFP-positive with all WT Cas9, eSpCas9(1.1), SpCas9-HF1, and HypaCas9, and there was no marked difference in the frequency of partial integration of the cassette containing EGFP (Supplementary Figure S6D). Taken together, these data showed that the threshold for the conformational checkpoint of the Cas9–gRNA complex is a general and fundamental determinant of the HDR- and NHEJ-inducing activities of CRISPR/Cas9, not only for single-nucleotide substitutions but also for knock-in of larger DNA fragments via HDR.
DISCUSSION
Given that the CRISPR/Cas9 system was originally an adaptive immune system in bacteria, it is unsurprising that the authentic CRISPR/Cas9 components are not necessarily ideal for genome editing of mammalian cells. Modifications will be required for certain applications of the CRISPR/Cas9 system, including therapeutic and experimental purposes.
We hope to find ways to enhance HDR and/or inhibit NHEJ, as the authentic CRISPR/Cas9 system predominantly induces NHEJ. The modified conformational checkpoint threshold, which was originally meant to improve the fidelity of CRISPR/Cas9, changes the kinetics of DNA cleavage and potentially the interaction of the Cas9–gRNA complex with the target DNA, which then affects the interaction of the cleaved genomic DNA with the donor DNA and DNA repair mechanisms. Therefore, we systematically addressed the HDR- and NHEJ-inducing activities of various Cas9 variants and modified gRNAs with enhanced conformational checkpoint.
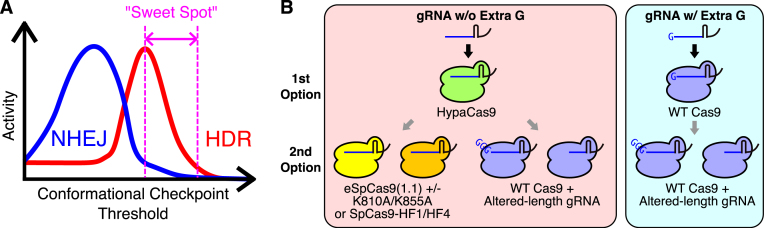
Although the exact mechanism is unclear at present, we found that an altered conformational checkpoint was able to enhance the HDR-inducing activity of Cas9. Interestingly, HDR and NHEJ require different levels of the conformational checkpoint for their maximum induction at many target sites, resulting in no correlation between the HDR- and NHEJ-inducing activities, as we observed in our previous study (18). Therefore, we can shoot for ‘sweet spots’ for the highest HDR/NHEJ ratio by modifying the Cas9 and/or gRNA at these target sites (Figures 3, 4, and 6A). Interestingly, however, there were some targets at which the HDR- and NHEJ-inducing activities of Cas9 were well-correlated, such as RBM20-g2 and ATP7B-g2 (Figures 3 and 4). At these targets, changes in the conformational checkpoint were still effective in enhancing HDR, but NHEJ was also enhanced to a similar extent, keeping the HDR/NHEJ ratio basically constant (Figures 3 and 4). This suggests the existence of two types of target sites: One results in non-correlated HDR and NHEJ, while the other results in well-correlated HDR and NHEJ. Differences in the gRNA sequence or the targeted DNA strand may cause the observed differences between these two types of targets, but further studies will be required to clarify the underlying mechanism.
Figure 6.
A practical guideline for improving the HDR/NHEJ ratio using the CRISPR/Cas9 system. (A) A schematic representation of the relationship between the HDR and NHEJ activities and the threshold for the conformational checkpoint of Cas9. Changing the threshold for the conformational checkpoint often influences the HDR- and NHEJ-inducing activities of Cas9 to different extents. Therefore, there are ‘sweet spots’ where HDR and NHEJ can be upregulated and/or downregulated, respectively. (B) Alterations to improve the HDR/NHEJ ratio from the standard Cas9–gRNA complex. When a target genomic DNA sequence and a gRNA matches the target sequence, HypaCas9 should be used instead of WT Cas9 as the first option. If further improvements are required, eSpCas9(1.1) with and without the K810A or K855A mutation along with SpCas9-HF1 or SpCas9-HF4 can be useful in some cases as a second option. Alternatively, WT Cas9 combined with gRNAs of different lengths can also be tested. When a target sequence requires an extra 5′ guanine for a gRNA, WT Cas9 should be used as the first option, as the other Cas9 variants tend to have much lower activities in such situations. Some gRNAs with altered lengths combined with WT Cas9 can be used as a second option, but this can result in the simultaneous enhancement of NHEJ in many cases.
Generally, when the modification was too excessive, the Cas9–gRNA complex lost its activity for both HDR and NHEJ. This happened with many combinations of a Cas9 variant and a gRNA with an extra 5′ guanine (Figure 2), eSpCas9(1.1) combined with gRNAs with different lengths (Supplementary Figure S5), and eSpCas9(1.1)+K810A+K855A (Figure 4). Under such conditions, the Cas9–gRNA complex may have a threshold for the conformational checkpoint that is too high to transition to the active state, or it cannot stably interact with DNA to allow genome editing to happen. Therefore, the extent of alteration of the conformational checkpoint should be carefully adjusted to enhance HDR (Figure 6A).
Based on our findings, we propose a practical strategy for identifying the best genome editing conditions to induce HDR with less NHEJ using the CRISPR/Cas9 system. When using perfect-match gRNAs, HypaCas9 should be tested first. HypaCas9 increased the HDR/NHEJ ratio compared to WT Cas9 with the most perfect-match gRNAs in HEK293T cells and HeLa cells (Figure 1) and had lower NHEJ activity than WT Cas9 while exhibiting higher HDR activity to knock-in an approximately 4.2-kb DNA fragment at the AAVS1 (Figure 5). eSpCas9(1.1) with and without the K810A and K855A mutations and SpCas9-HF1/HF4 are additional options for further improving the HDR/NHEJ ratio (Figure 4). Alternatively, WT Cas9 can be combined with altered-length gRNAs (Figure 3). Given that these Cas9 variants and gRNAs with altered lengths give improved proof-reading over WT Cas9 and the standard gRNAs as they were originally developed (6–10), there seems to be no down side to applying these options to induce HDR (Figure 6B). However, when an extra 5′ guanine is necessary for a gRNA to be expressed via the U6 promoter, these Cas9 variants tend to increase the threshold for the conformational checkpoint too much to induce genome editing (Figure 2). Therefore, WT Cas9 should be combined with gRNAs with an extra 5′ guanine first. Unfortunately, there are few options for this type of gRNA. The length of gRNAs can be modified to further enhance HDR, but it might increase NHEJ as well (Figure 6B).
CRISPR/Cas9 has been and will be actively applied to basic life science, industrial, and therapeutic purposes. However, if we can induce HDR more efficiently, CRISPR/Cas9 will be able to be utilized in even wider applications. We still do not know exactly how the creation of a DSB by Cas9 leads to HDR or NHEJ, or the factors that determine the balance of the two repair pathways. In this report, we revealed that the threshold for the conformational checkpoint of Cas9 is a key determinant of the balance between HDR and NHEJ. We hope that our findings will lead to further studies to elucidate the mechanisms underlying genome editing by Cas9 and the development of better systems to induce HDR without NHEJ, which will facilitate a wide range of applications, including therapies.
DATA AVAILABILITY
The raw ddPCR data is available at https://figshare.com/authors/Yuichiro_Miyaoka/4958476.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs H. Nishimasu (The University of Tokyo), B.P. Kleinstiver (Massachusetts General Hospital), J.S. Chen and J.A. Doudna (University of California, Berkeley) for the helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Takeda Science Foundation Medical Research Grant (to T.K.); JSPS Grant-in-Aid for Young Scientists (A) [17H04993]; NOVARTIS Research Grant; Mochida Memorial Foundation Research Grant; Uehara Memorial Foundation Research Grant; SENSHIN Medical Research Foundation Grant; Naito Foundation Research Grant (to Y.M.). Funding for open access charge: Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 10. Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A.. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017; 550:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dagdas Y.S., Chen J.S., Sternberg S.H., Doudna J.A., Yildiz A.. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017; 3:eaao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sander J.D., Joung J.K.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014; 32:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doudna J.A., Charpentier E.. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 14. Mandal P.K., Ferreira L.M., Collins R., Meissner T.B., Boutwell C.L., Friesen M., Vrbanac V., Garrison B.S., Stortchevoi A., Bryder D. et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014; 15:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A.. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017; 7:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F. et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016; 6:20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miyaoka Y., Berman J.R., Cooper S.B., Mayerl S.J., Chan A.H., Zhang B., Karlin-Neumann G.A., Conklin B.R.. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 2016; 6:23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin S., Staahl B.T., Alla R.K., Doudna J.A.. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014; 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 21. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Canny M.D., Moatti N., Wan L.C.K., Fradet-Turcotte A., Krasner D., Mateos-Gomez P.A., Zimmermann M., Orthwein A., Juang Y.C., Zhang W. et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018; 36:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 24. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyaoka Y., Chan A.H., Judge L.M., Yoo J., Huang M., Nguyen T.D., Lizarraga P.P., So P.L., Conklin B.R.. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014; 11:291–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009; 27:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandegar M.A., Huebsch N., Frolov E.B., Shin E., Truong A., Olvera M.P., Chan A.H., Miyaoka Y., Holmes K., Spencer C.I. et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016; 18:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander D.C. A rapid method for determining clone frequency in complex populations using PCR and the Poisson distribution. Nucleic Acids Res. 1990; 18:7453–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S., Bae T., Hwang J., Kim J.S.. Rescue of high-specificity Cas9 variants using sgRNAs with matched 5′ nucleotides. Genome Biol. 2017; 18:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang D., Zhang H., Li T., Chen K., Qiu J.L., Gao C.. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017; 18:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw ddPCR data is available at https://figshare.com/authors/Yuichiro_Miyaoka/4958476.