Abstract
Translation-dependent mRNA quality control systems protect the protein homeostasis of eukaryotic cells by eliminating aberrant transcripts and stimulating the decay of their protein products. Although these systems are intensively studied in animals, little is known about the translation-dependent quality control systems in plants. Here, we characterize the mechanism of nonstop decay (NSD) system in Nicotiana benthamiana model plant. We show that plant NSD efficiently degrades nonstop mRNAs, which can be generated by premature polyadenylation, and stop codon-less transcripts, which are produced by endonucleolytic cleavage. We demonstrate that in plants, like in animals, Pelota, Hbs1 and SKI2 proteins are required for NSD, supporting that NSD is an ancient and conserved eukaryotic quality control system. Relevantly, we found that NSD and RNA silencing systems cooperate in plants. Plant silencing predominantly represses target mRNAs through endonucleolytic cleavage in the coding region. Here we show that NSD is required for the elimination of 5′ cleavage product of mi- or siRNA-guided silencing complex when the cleavage occurs in the coding region. We also show that NSD and nonsense-mediated decay (NMD) quality control systems operate independently in plants.
INTRODUCTION
In eukaryotic cells, various quality control systems function to identify and degrade aberrant mRNAs thereby preventing the accumulation of aberrant proteins (1). Normal eukaryotic mRNAs are single-stranded monocistronic transcripts, which have a cap at the 5′ end, a polyA tail at the 3′ end, and contain characteristic 5′ and 3′ untranslated regions (UTR). Quality control systems recognize an mRNA as an aberrant transcript if any of these features is lacking or is very unusual.
Eukaryotic mRNAs form stable ring-like structures as the eIF4G component of the 5′ cap binding complex binds to the polyA Binding Protein (PABP). This structure stabilizes the mRNA and stimulates its translation. Degradation of mRNAs is tightly regulated (1,2). The rate-limiting step of normal mRNA decay is deadenylation, which finally leads to the dissociation of PABP. Subsequently, 3′-5′ exonuclease complexes can degrade the transcript and/or the 5′ cap is removed, and then the decapped mRNA is decayed by the XRN 5′-3′ cytoplasmic exonuclease (3,4). RNA quality control systems quickly degrade the identified aberrant transcript by circumventing the rate-limiting deadenylation step of normal mRNA decay.
At least three quality control mechanisms, the nonsense-mediated decay (NMD), the nonstop decay (NSD) and the no-go decay (NGD) systems monitor the quality of mRNAs during translation (5,6). NMD identifies premature termination codon containing mRNAs, NSD recognizes mRNAs lacking an in-frame stop codon, while NGD targets mRNAs, which contain elongation inhibiting features such as secondary structures or modified nucleotides. These translation coupled quality control systems have to (i) degrade the faulty transcripts to prevent further production of the aberrant proteins, (ii) rescue and recycle the ribosomes from the transcripts and (iii) eliminate the aberrant protein products (6). Translation coupled quality control systems accelerate the degradation of aberrant mRNAs by stimulating deadenylation-independent decapping, triggering endonucleolytic cleavage or by accelerating deadenylation. Ribosome dissociation and recycling is coupled to normal or quality control specific translation termination (see below), whereas the proteins synthesized from aberrant transcripts might be eliminated by the Ribosome-associated Quality Control system (referred to as RaQC instead of the frequently used RQC abbreviation because the latter one is also used for plant RNA quality control system) (7). Relevantly, in addition to eliminating faulty transcripts, these quality control mechanisms are also involved in the regulation of several wild-type mRNAs. Although these systems have been intensively studied in yeast and animals, plant NMD is poorly understood and neither NSD nor NGD was studied in plants.
NSD and NGD are closely related quality control systems. The Pelota [in mammals, and Dom34 (Duplication Of Multilocus region) in yeast] and the Hbs1 (Hsp70 subfamily B Suppressor) proteins play important role in both systems (8). NSD targets two types of stop-codon free mRNAs, nonstop mRNAs, which contain polyA sequences at the 3′ end, and stop codon-less mRNAs, which lack the polyA tail (9). Nonstop mRNAs are generated by premature polyadenylation, while an endonucleolytic cleavage in the coding region leads to the production of stop codon-less mRNAs. NSD identifies stop-codon free mRNAs at the termination step of translation (8,10). When a translating ribosome reaches a stop codon of a normal mRNA, the eRF1-eRF3–GTP termination complex binds to the A-site. GTP-hydrolysis by eRF3 leads to dissociation of eRF3 and conformation changes of eRF1. Upon accommodation of eRF1, the ATPase ABCE1 (ATP-Binding Cassette Sub-Family E Member 1) is recruited. Finally, the synthesized peptide is released and the ribosome is dissociated and recycled (7,11). By contrast, when a ribosome translates a nonstop or a stop codon-less mRNA, it can reach the 3′ end of the transcript. The A-site will be empty, thus the canonical termination complex fails to bind it. According to the current model of animal NSD, the Pelota–Hbs1–GTP (NSD) complex is recruited to the empty A-site (6,12). The Hbs1 resembles to eRF3, while the Pelota is similar to eRF1 but lacks the sequences required for stop codon identification and peptide release. Thus, the NSD complex can bind the A-site codon-independently and terminate translation without peptide release. Hbs1-mediated GTP hydrolysis leads to dissociation of Hbs1, conformation changes of Pelota and finally to the recruitment of ABCE1. The ABCE1 with the Pelota triggers unusual termination. The subunits are dissociated, the mRNA is dropped but the peptidyl-tRNA at the P-site remains associated with the 60S subunit. The NSD target transcript could be quickly degraded by the SKI-exosome 3′-5′ cytoplasmic exonuclease complex or after decapping by the XRN1 exonuclease (13,14). The exosome complex consists of ten proteins, while the associated SKI complex is comprised by the SKI2, SKI3 and SKI8 proteins. The non-canonical translation termination generated peptidyl-tRNA-60S complex is identified by the RaQC system, which leads to ubiquitination and degradation of the faulty protein (7).
In yeast, the SKI7 protein, which is part of the yeast SKI complex and resembles to eRF3, also plays a role in the degradation of nonstop transcripts (15,16). It might bind to the empty A-site and recruits the SKI-exosome complex to degrade the transcripts. Recently, it was shown that in certain fungi alternative splicing products of Hbs1 encode SKI7 protein (17). In vertebrates, the HBS1 end SKI7 encoding mRNAs are also generated by alternative splicing from a single transcript. However, vertebrate SKI7 is not involved in NSD, it plays only role in the general mRNA decay (18).
NGD degrades transcripts when the elongation is inhibited (19). Stalling of the elongating ribosome triggers the endonucleolytic cleavage of the mRNA by an unknown nuclease, close to the stalled ribosome. The 3′ cleavage product is degraded by the XRN 5′-3′ exonuclease. Relevantly, the 5′ cleavage product is a stop codon-less transcript, thus the translating ribosome can run to the 3′ end of it, and then the translation is terminated by the NSD machinery (6). As polyA sequences are translated very inefficiently, translation of the polyA tail of a nonstop transcript frequently leads to ribosome stalling. This might trigger NGD-type endonucleolytic cleavage of the mRNA, and then the 5′ cleavage product is eliminated by the NSD machinery (20).
Other RNA quality control mechanisms, such as the plant RNA silencing amplification pathway that targets non-capped and non-polyadenylated aberrant mRNAs (21,22), might operate independently from translation. RNA silencing is triggered by double-stranded RNA (dsRNA) and causes degradation (or translational repression) of the RNAs homologous to the trigger. dsRNAs are processed by a member of the DICER enzyme family (in plants, DICER-LIKE proteins, DCLs) into 21–24 nt small RNA (sRNA) (23,24). In plants, microRNAs (miRNA) and small interfering RNAs (siRNA) are the most important sRNAs. In Arabidopsis thaliana, miRNAs are cleaved by DCL1 from hairpin-structured single-stranded transcripts, while siRNAs are produced by DCL2, DCL3 and DCL4 from long dsRNAs (25). sRNAs are loaded onto one of the AGO proteins, the key components of the RNA-Induced Silencing Complex (RISC) (26). In plants, RISC exerts its negative regulation on target mRNAs mainly by cleavage and at a lesser extent by translation inhibition (27,28). The cleavage occurs mostly within the coding region. RISC-cleavage generated products are 5′ non-polyadenylated and 3′ uncapped RNA fragments. In plants, these cleavage products can be degraded by exonucleolytic pathways or become substrates to RNA-dependent RNA polymerase (RDR)-mediated silencing amplification (29–31). In Arabidopsis, the 5′ (and much less efficiently the 3′) cleavage fragments of RISC can be converted into dsRNA by RDR6 (32,33), and then secondary siRNAs are generated from these dsRNAs by DCL2 and DCL4 (34,35). Finally, the secondary siRNAs guide RISC complexes to target transcripts, thereby amplifying the silencing response. This RDR-mediated silencing amplification plays a critical role in antiviral responses. Relevantly, the cleavage products of miRNA targets are efficiently degraded by non-silencing pathways. This prevents the uncontrolled amplification of silencing (32,36). It was shown that RDR6 can also generate secondary siRNAs from endogenous miRNA targets (called rqc- or ct-siRNAs) if both the XRN4-mediated 5′-3′ and the SKI-exosome 3′-5′ cytoplasmic exonuclease pathways are inactivated, and that these rqcRNAs cause dramatic phenotypes (36–38). Thus the efficient decay of silencing cleavage products by the exonucleolytic pathways is essential to protect the cell from the accumulation of detrimental rqcRNAs (32,36,37,39). In Arabidopsis, the 3′ cleavage fragments of RISC are eliminated by XRN4 (31,40,41), while the 5′ cleavage fragments are overaccumulated in xrn4 mutants and in plants, in which the SKI-exosome main 3′-5′ exonuclease complex is inactivated (31,37,41). Moreover, recent results have shown that 5′ cleavage fragments are also overaccumulated if RICE, a RISC complex associated specific 3′-5′ exonuclease is depleted (42). It was proposed that HESO1-mediated uridylation of the 5′ cleavage fragments facilitate both XRN4 and RICE pathways in Arabidopsis (41). Thus in Arabidopsis, both 5′-3′ and 3′-5′ exonucleases are important for the degradation of 5′ cleavage fragments and for the prevention of unregulated silencing amplification (43). It is not known why RDR6 generates secondary sRNAs from 5′ cleavage products of viral siRNAs but not from 5′ cleavage fragments of endogenous miRNAs.
To better understand how the silencing generated cleavage fragments are eliminated, we studied the degradation of silencing products in Nicotiana benthamiana model species. As the NSD system eliminates the stop codon-less mRNAs in animals, and the 5′ cleavage fragments of miRISC are stop codon-less mRNAs in plants, we hypothesized that the NSD machinery is involved in the decay of RISC generated 5′ cleavage products in plants. Here we show that plant NSD degrades both nonstop and stop codon-less mRNAs, and that Pelota, HBS1 and SKI2 are required for the elimination of these aberrant transcripts. Then we demonstrate that NSD plays an important role in the elimination of the 5′ cleavage products of miRNA- or siRNA-guided RISC complexes when the cleavage occurs within the coding region. Thus NSD and RNA silencing systems act cooperatively in plants. In contrast, plant NSD and NMD systems operate independently.
MATERIALS AND METHODS
Plasmid constructs
P14, GFP, G-600, U1DN, TRV-P (44,45), TRV-P-XRN4, PPG (40), GFP-171.1, amiR-GFP (46), miR-171c (47), 35S:AtTAS1b and 35S:miR173 (34) clones were previously described. Other constructs and the primers used in this study are described in detail in the Supplementary Data S1 file.
Plant growth conditions
Nicotiana benthamiana plants were grown in the greenhouse. After agroinfiltration or VIGS treatment (see below) the plants were kept in a growth chamber at 23°C under 16 h/8 h light/dark condition. Columbia wild type and pelota1 and hbs1 T-DNA mutants (Pelota1:At4G27650, SAIL_881_B10 and Hbs1: At5G10630, WiscDsLox477-480D24) Arabidopsis plants were grown in a light room, at 16 h/8 h. light/dark condition, at 23°C day and 18°C night temperature.
Agroinfiltration assay
Agroinfiltration is the most efficient transient gene (co-)expression system for plants (48). To co-express different genes, each construct is introduced into an Agrobacterium tumefaciens strain, and then the different Agrobacterium cultures are mixed before infiltration. Agroinfiltration was described previously (49). Wild-type or silenced N. benthamiana leaves were agroinfiltrated with a mixture of bacterium cultures (OD600 of each culture was 0.4, or in the case of P14 0.2). At least two cultures are mixed, one expresses the P14 internal control, whereas the second bacterium expresses the test or the control construct (50). Agroinfiltration induces RNA silencing, thus P14 also served as a silencing suppressor in these assays (the role of P14 is described in detail at Supplementary Data S1). We have previously shown that under these experimental conditions, the P14 does not modify the NMD or the miRNA-based silencing experiments (40). However, as P14 affects the tasiRNA pathway, it was omitted from that assay. Cycloheximide treatment of agroinfiltrated leaves was conducted as described (50).
VIGS-agroinfiltration assay
Virus-induced gene silencing (VIGS) is the most effective transient gene silencing system for plants (51). Combination of VIGS and agroinfiltration (VIGS-agroinfiltration) can be used to identify NSD trans factors. The VIGS-agroinfiltration system is described in detail in the Supplementary Data S1 file and Supplementary Figure S1.
To silence a gene (or a gene family), a segment from the target gene is incorporated into a Tobacco Rattle Virus (TRV) VIGS vector. If a gene family is targeted, a conserved segment is cloned, while specific regions are used for gene-specific inactivation. To silence a gene, N. benthamiana plants were infected with the recombinant TRV VIGS vectors (sequences of the VIGS fragments are shown at Supplementary Data S1). TRV infection induces antiviral RNA silencing response, thus the virus concentration will be very low in the upper leaves. Moreover, the silencing will specifically inactivate the host genes that are homologous to the incorporated sequence. To trigger VIGS, ∼21 days old N. benthamiana plants were co-agroinfiltrated with a mixture of three Agrobacterium cultures. One expressed P14, the second expressed TRV RNA1 and the third expressed TRV RNA2 containing segments from N. benthamiana PDS (phytoene desaturase) gene or from PDS and ∼600 nt long sequence from the coding region of the gene what we want to silence. PDS is used to monitor silencing. When the upper leaves started to bleach (indicating that PDS silencing was efficient and suggesting that the silencing of the gene of interest is also effective), leaves under the bleaching ones were agroinfiltrated (45). Quantitative RT-PCR (qRT-PCR) assays confirmed that the target mRNA levels were significantly reduced (Supplementary Figure S1B). Arabidopsis Pelota1 and Hbs1 genes were used for complementation assays. The domain structures of the predicted proteins and the alignments of their N. benthamiana homologs of Pelota, Hbs1 and SKI2 are shown at Supplementary Figures (Supplementary Figures S2–S5). For complementation assay (Supplementary Figure S6), the silenced leaves were co-infiltrated with three cultures, one expressing P14, the second expressing the NSD reporter and the third one expressing the tested factor.
RNA gel blot, qRT-PCR and oligoT binding assays
RNA gel blot assays were previously described (49). Briefly, total RNA was isolated and separated on denaturing agarose gel. For small RNA gel blots, the RNA samples were separated on UREA-PAGE (47). The gels were blotted and hybridized with radioactively labeled probes.
For qRT-PCR assays, cDNA was synthesized with RevertAid First Strand cDNA Synthesis kit (Thermo Scientific) from DNAse I treated total RNA samples. qRT-PCR assays were carried out with Fast Start Essential DNA Green Master Mix (Roche) in a Light Cycler 96 (Roche) Real-Time PCR machine.
To assess if the NSD reporter transcripts accumulate in deadenylated form, total RNA samples were hybridized with oligo(dT) cellulose (MicroPoly[A]PuristTM Small Scale mRNA Purification Kit, Ambion) according to the manufacturer's protocol. The bound and supernatant fractions were subjected to RNA gel blot assays.
RNA-sequencing and data processing
For Arabidopsis RNA-seq assays, leaves of 30 days plants were used. For RNA-seq1 and RNA-seq2 experiments, PDS-silenced and P-Pelota-silenced N. benthamiana plants were separately generated and were differently selected. For RNA-seq1, P-Pelota-silenced plants were selected, in which the SCL6-IV 5′cleavage fragments overaccumulated moderately (and similarly), while for RNA-seq2, P-Pelota samples were selected, in which the SCL6-IV 5′ fragment overaccumulated dramatically (also see figure legends for Supplementary Figures S8 and S11).
For RNA sequencing, libraries were prepared from total RNA samples with Illumina® Ribo-Zero™ rRNA Removal (Plant Leaf) and TruSeq™ v2 RNASeq Library Preparation Kits. At Seq2 ScriptSeq™ v2 RNASeq Library Preparation Kit was used. Briefly, cDNAs were transcribed with random primers from rRNA-depleted samples. Sequencing was performed as a paired-end at RNA-seq1 and Arabidopsis RNA-seq or as a single-end at RNA-seq2 on Illumina HiSeq 2500 platform, resulting in 30–50 M (for paired-end) and ∼20 M (for single-end) read per sample (library preparation and sequencing were conducted as a service by Genewiz, Inc. and UD-Genomed Medical Genomics Technologies LTD). Read alignment, transcript assembly, MA and PCA Plots and differential gene expression (DGE) were conducted with the ‘new Tuxedo’ protocol using Hisat2 v2.1.0, StringTie 1.3.3b, DeSeq2 v1.18.1 software versions (52,53). For the DGE analysis the draft genome of N. benthamiana was downloaded from the Sol Genomics Network's FTP server (54). For the Arabidopsis DGE analysis TAIR10 genome annotation was downloaded from the Phytozome database (55,56). N. benthamiana and Arabidopsis degradome data were retrieved from NCBI BioProject database (accession numbers: PRJNA288746 and PRJNA407271, respectively). For GO analysis the AgriGO v2 server was used (57). To define miRNA cleavage positions in leaves, published degradome and sRNA libraries were downloaded and analyzed with The UEA sRNA workbench tools (UEA sRNA Workbench v3.2) (58). Our RNA-seq data are available at NSBI GEO database (accession number: GSE99805).
For qRT-PCR validation, we isolated RNAs from newly grown plants or used the same RNA samples that were subjected to RNA-seq. For each miRNA target transcript, two pairs of primers were used, one pair to amplify an upstream and another to amplify a downstream region relative to the predicted cleavage site. (The primers are listed in the Supplementary Data S1). The 5′ and 3′ cleavage product levels were compared.
Sucrose gradient centrifugation
Native extracts were prepared from agroinfiltrated plant material using ribosome buffer supplemented with cycloheximide (100 mM Tris pH 8.5, 50 mM KCl, 5 mM EGTA, 0.5% Nonidet P-40 and 10 μg/ml cycloheximide) or EDTA (100 mM Tris 8.5, 50 mM KCl, 5 mM EGTA, 0.5% Nonidet P-40 and 30 mM EDTA). Extracts were cleared by centrifugation at 4°C 12 000 rpm/10 min twice. After saving 10% input samples, cleared lysates were loaded onto 10–50% sucrose gradient columns. Sucrose gradient separation was done on Sorwall Ultracentrifuge Combi OTD, rotor AH650 at 32 000 rpm for 3 h at 4°C. Fractions of 0.5 ml each were collected and used for RNA gel blot assays.
RESULTS
SKI2 but not XRN4 is required for the elimination of 5′ cleavage fragments of vsiRISC in N. benthamiana
To better understand the degradation of silencing products, we studied the role of XRN4 and SKI2 in eliminating the 5′ cleavage fragments in N. benthamiana plants. To this aim, our previously developed VIGS-agroinfiltration transient assay system was used (45) (for details, see Materials and Methods, Supplementary Data S1 and Supplementary Figure S1). Briefly, a Tobacco Rattle Virus-based VIGS vector was used to silence PDS (as a control), or PDS and XRN4, or PDS and SKI2 genes in N. benthamiana plants (referred to as PDS-, P-XRN4- and P-SKI2-silenced plants, respectively). The leaves of silenced plants were agroinfiltrated with the mixture of two agrobacteria. One expressed a PDS siRNA-sensor construct (40), in which a 100 nucleotides long segment from the PDS links the PHA (phytohemagglutinin) and GFP genes (PPG, for PHA-PDS-GFP fusion construct, Figure 1A). The second agrobacterium expressed the P14 internal control (the role of P14 is described in detail in Materials and Methods and Supplementary Data S1). In all three plants, PDS siRNAs generated from the recombinant TRV viruses guide the RISC to cleave the PPG transcript in the PDS linker region. Three days post-agroinfiltration, RNA samples were collected from the infiltrated patches, and RNA gel blot assays were conducted using P14 and PHA probes. For simplicity, although P14 was co-agroinfiltrated in almost all assays, it will not be mentioned later in the main text. However P14 is shown on the figures.
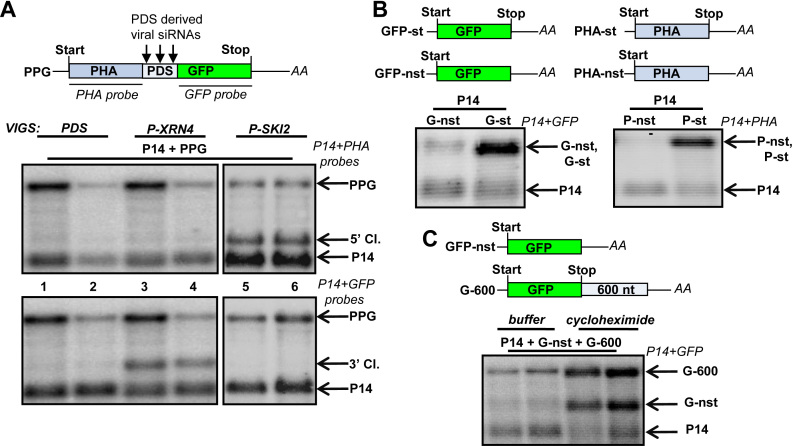
Figure 1.
Nonstop decay operates in plants. (A) In N. benthamiana, the 5′ and the 3′ cleavage fragments of PDS viral siRNA-programmed RISC are eliminated by SKI2- and XRN4-dependent pathways, respectively. VIGS-agroinfiltration assays were conducted to study the degradation of vsiRISC cleavage products. Leaves of PDS-silenced (PDS) negative control, and PDS + XRN4 or PDS + SKI2 silenced test plants (P-XRN4 and P-SKI2, respectively) were agroinfiltrated with P14 internal control and PPG sensor construct (P14 + PPG). Schematic, non-proportional representation of the PPG vsiRISC sensor transcript. Two samples isolated from different plants were run in parallel to illustrate experimental variations. RNA gel blots were hybridized with P14 and PHA (upper panel) or P14 and GFP (bottom panel) probes (probe names are in italics). The stabilized 5′ and 3′ cleavage products of PPG (5′ and 3′ Cl.) are indicated. (B) NSD reporter mRNAs accumulate to low levels in plants. The GFP-nst and PHA-nst NSD reporter mRNAs (G-nst and P-nst) and the corresponding stop codon containing control mRNAs (G-st and P-st) were agroinfiltrated with P14 internal control into the leaves of wild-type N. benthamiana plants. (C) NSD is a translation-dependent system in plants. G-nst NSD reporter was co-expressed with P14 internal control and a cycloheximide sensitive positive control (G-600) in N. benthamiana leaves. The samples were treated with cycloheximide translation inhibitor or with buffer. G-600 mRNA is targeted by NMD, another translation-dependent quality control system therefore sensitive to cycloheximide (also see below at Supplementary Figure S14). Note that both the G-nst and the G-600 mRNAs are overexpressed in the cycloheximide treated samples.
PPG 5′cleavage fragments could not be detected in the PDS-silenced control plants confirming that the 5′ cleavage products are degraded quickly (Figure 1A, upper panel, lanes 1 and 2). Relevantly, the 5′ cleavage fragments of PPG reporter mRNAs were abundant in P-SKI2-silenced plants, while these cleavage products were undetectable in the P-XRN4-silenced lines (Figure 1A, upper panel, compare lanes 5 and 6 to 3 and 4). The RNA samples were also hybridized with GFP probe that detects both the full-length mRNA and the 3′ cleavage product. The 3′cleavage fragment accumulated to high levels in the P-XRN4-silenced but not in the PDS- or P-SKI2-silenced lines (Figure 1A, bottom panel, compare lanes 3 and 4 with 1 and 2 or 5 and 6, respectively).
These results indicate that in N. benthamiana, SKI2 is required for the decay of the 5′ but not the 3′cleavage fragments of viral siRNA-programmed RISC (referred to as vsiRISC). Moreover, these data suggest that in N. benthamiana, XRN4 is not involved in the decay of 5′ cleavage fragments and confirm our previous result that XRN4 is essential for the degradation of 3′ cleavage products of vsiRISC (40).
Nonstop decay operates efficiently in plants
We hypothesized that plant NSD plays a role in the elimination of the silencing generated 5′ cleavage products. However, NSD has not been studied in plants yet. Hence, we first tested if NSD is functional in plants. N. benthamiana leaves were agroinfiltrated with nonstop NSD reporter (GFP-nst and PHA-nst, for GFP-nonstop and PHA-nonstop) constructs, in which the stop codon of the ORF and the in-frame stop codons from the 3′UTR were eliminated. In the control constructs (GFP-st and PHA-st) the stop of the ORFs was retained (Figure 1B). Both NSD reporter mRNAs accumulated to very low levels compared to the stop codon containing controls (Figure 1B). These data indicate that NSD degrades nonstop (polyadenylated but stop codon free) transcripts sequence independently in plants. Alternatively, transcription of nonstop mRNAs is repressed. To distinguish between these two possibilities, the effect of cycloheximide translation inhibitor on NSD reporter expression was studied. NSD is a translation-dependent decay system (10). If the low nonstop reporter transcripts level is due to NSD-mediated accelerated degradation, translation inhibition should lead to enhanced accumulation. Indeed, cycloheximide treatment led to dramatically enhanced expression of the NSD reporter transcripts (Figure 1C). These data show that NSD operates efficiently in plants.
The key NSD factors are conserved in eukaryotes
Pelota and Hbs1 proteins are the central NSD factors in fungi as well as in animals (8,59). Although it was proposed that in Arabidopsis, the putative Pelota and Hbs1 (At4G27650, At5G10630, respectively) homologs are single copy genes (60), our bioinformatical analysis suggests that at least two putative Pelota copies (At4G27650, At3G58390) are present in the genome (Supplementary Data S1, Supplementary Figures S3 and S4). Therefore, silencing assays were used to study the role of Pelota and Hbs1 homologues in plant NSD. As N. benthamiana genome may also encode more than one homolog of Pelota (Supplementary Data S1, Supplementary Figures S2 and S3), the silencing constructs were designed to target the most conserved regions.
To test if the function of Pelota and Hbs1 is conserved in plants, we assessed the expression of the nonstop reporter transcripts in PDS-silenced control plants, and in plants, in which the PDS and the putative Pelota or Hbs1 genes were silenced (P-Pelota- and P-HBS1-silenced plants). qRT-PCR assays confirmed that the Pelota and the Hbs1 mRNA levels were severely reduced in the silenced plants (Supplementary Figure S1B).
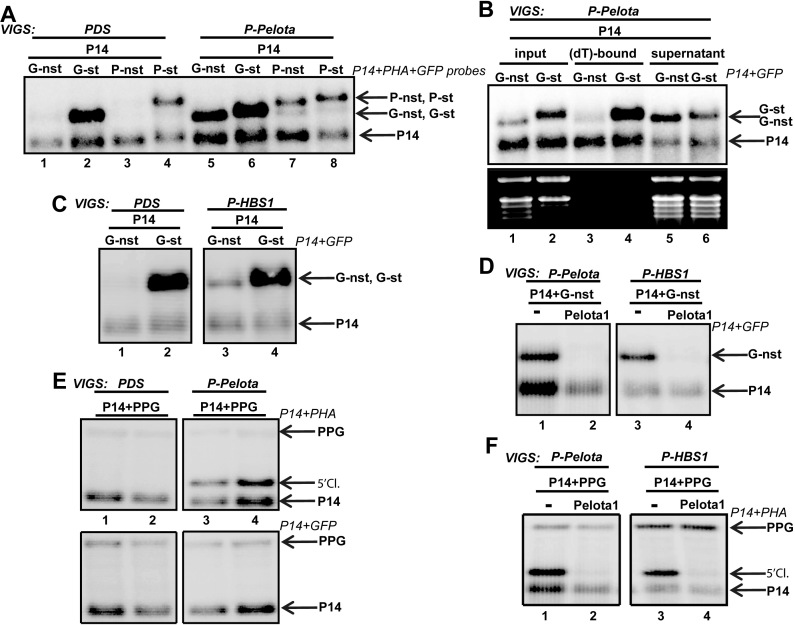
The nonstop reporter mRNAs accumulated to high levels in both P-Pelota- and P-HBS1-silenced plants relative to the PDS control line (Figure 2A, compare lanes 1 to 5, 3 to 7 and Figure 2C, lane 1 to 3), indicating that both proteins are required for NSD in N. benthamiana. Moreover, the nonstop reporter mRNAs migrated slightly faster during electrophoresis than the stop-codon containing controls in the P-Pelota- and P-HBS1-silenced plants (Figure 2A–C, Supplementary Figures S1C and S1D). We assumed that the nonstop transcripts accumulate in deadenylated form in the NSD-deficient plants. To test this, total RNA was isolated from P-Pelota-silenced leaves, in which P14 and G-nst were co-expressed. The RNAs were incubated with oligo(dT) cellulose, and then the bound and unbound fractions were subjected to RNA gel blot assay. Relevantly, the G-nst nonstop reporter mRNA barely bound to the oligo(dT) cellulose, while the P14 transcripts were retained effectively (Figure 2B, compare lane 3 to 5). Thus we concluded that the nonstop mRNAs are mainly present in deadenylated form in NSD-deficient leaves.
Figure 2.
Pelota and Hbs1 are required for plant NSD. (A andB) NSD reporter mRNAs accumulate to high levels in deadenylated form in Pelota-silenced plants. (A) GFP-nst and PHA-nst NSD reporter (G-nst and P-nst) and the corresponding control (P-st and G-st) constructs were agroinfiltrated with the P14 internal control into the leaves of PDS-silenced control (PDS) or PDS+Pelota silenced (P-Pelota) test plants. Note that in the P-Pelota plants, the NSD reporter mRNAs migrate faster than the stop-codon containing control transcripts. (B) NSD reporter mRNA is present in deadenylated form in P-Pelota-silenced leaves. P14 was co-expressed with G-st control or with G-nst NSD reporter mRNAs in P-Pelota-silenced leaves. RNAs were isolated and incubated with oligo(dT) cellulose, and then the (dT)-bound and the unbound (supernatant) fractions were subjected to RNA gel blot assay. Note that the G-nst NSD reporter transcript accumulates mainly in the supernatant, while the P14 and the G-st control mRNAs predominantly accumulate in the (dT)-bound fraction. (C) NSD reporter mRNAs accumulate to high levels in the P-HBS1-silenced plants. G-nst reporter and the G-st control constructs were agroinfiltrated with P14 into PDS– and PDS + HBS1 silenced (P-HBS1) plants. Note that the NSD reporter mRNAs migrate faster relative to the controls in P-HBS1 plants. (D) Pelota overexpression complements both the Pelota- and the Hbs1-deficient lines. G-nst reporter and P14 control constructs were expressed in the absence (–) or presence of the Arabidopsis AtPelota1 (Pelota1) in the leaves of P-Pelota- and P-HBS1-silenced plants. Reduced G-nst level indicates that Pelota1 complemented the NSD deficiency of the silenced plants. (E andF) Pelota and Hbs1 are required for the elimination of the 5′ but not the 3′ cleavage products of vsiRISC. PPG vsiRNA sensor and P14 (-) or PPG, P14 and Pelota1 were co-agroinfiltrated into PDS- and P-Pelota-silenced (E) and into P-HBS1-silenced (F) plants. Note that the 5′ cleavage fragment of vsiRISC accumulates to high levels in both P-Pelota and P-HBS1 plants and that Pelota overexpression complements the absence of endogenous Pelota and Hbs1 in the silenced plants.
Transient complementation assays (45) were conducted to further prove that the strong expression of NSD reporter transcripts in P-Pelota- and P-HBS1-silenced plants is due to the reduced NSD activity (for details of complementation assay, see Supplementary Data S1 and S6a). NSD reporter constructs were co-agroinfiltrated with a construct that expressed the A. thaliana At4G27650 encoded Pelota (Pelota1) into the leaves of P-Pelota- and P-HBS1-silenced plants. Pelota1 co-infiltration led to dramatically reduced nonstop reporter mRNA levels in both lines (Figure 2D), indicating that overexpressed Pelota1 can complement the absence of Pelota as well as Hbs1. By contrast, co-expression of At5G10630 encoded Hbs1 complemented the P-HBS1 but not the P-Pelota plants (Supplementary Figure S6b). We postulate that in plants, like in yeasts and mammals, Pelota is the key component of the NSD machinery and Hbs1 plays a stimulatory effect on Pelota.
Taken together, our data show that NSD is active in higher plants and that under physiological conditions activities of both Pelota and Hbs1 proteins are required for the efficient elimination of the nonstop transcripts.
NSD eliminates the 5′ cleavage fragments of vsiRISC
The 5′ cleavage products of RNA silencing are stop codon-less truncated RNAs. To test our hypothesis that plant NSD plays a role in the elimination of the silencing generated 5′ cleavage products, the PPG siRNA-sensor (Figure 1A) was agroinfiltrated into PDS-silenced control and P-Pelota- or P-HBS1-silenced test plants, and then the accumulation of the cleavage products was studied. Relevantly, the 5′ cleavage fragment of PPG overaccumulated in both P-Pelota- and P-HBS1-silenced plants (Figure 2E, lanes 3 and 4, Figure 2F, lanes 1 and 3), whereas the 3′ fragments remained at the background level (Figure 2E, bottom panel). Complementation assays confirmed that accumulation of 5′ fragment could be attributed to reduced NSD activity in the P-Pelota- and P-HBS1-silenced lines. Ectopic Pelota1 co-expression led to dramatically reduced 5′ cleavage fragment level in both P-Pelota- and P-HBS1-silenced lines (Figure 2F, lanes 2 and 4). Thus, our results suggest that in plants NSD is required for the degradation of the vsiRISC generated 5′ but not the 3′ cleavage fragments.
NSD eliminates the 5′ cleavage fragments of miRISC when cleavage occurs in the coding region
To verify whether NSD components are also involved in the degradation of cleavage products of miRISC, leaves of P-Pelota-, P-HBS1-, P-XRN4- and PDS-silenced plants were agroinfiltrated with GFP-171.1 miRNA-sensor construct (Figure 3A) alone, or with the GFP-171.1 sensor and either amiR-GFP or pri-miR171c artificial miRNA. These miRNAs target the sensor transcript in different regions. The amiR-GFP and the pri-miR171c cleave the GFP-171.1 sensor mRNA in the coding region and the 3′UTR regions, respectively (46,47).
Figure 3.
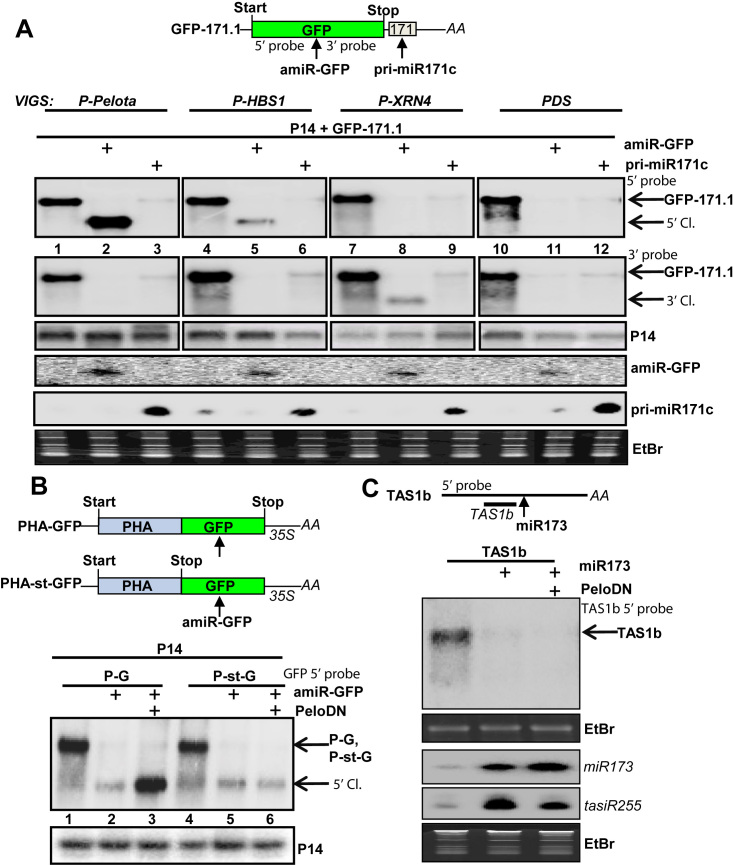
NSD eliminates the 5′ cleavage fragments of miRISC when the cleavage occurs in the coding region. (A) The role of NSD components and XRN4 in the degradation of miRISC-generated cleavage fragments. PDS-, P-XRN4-, P-Pelota- and P-HBS1-silenced plants were co-agroinfiltrated with P14 internal control and GFP-171.1 miRNA sensor or P14 and GFP-171.1 constructs were co-expressed with either amiR-GFP or pri-miR171c constructs. amiR-GFP targets the sensor mRNA within the coding region, while pri-miR171c targets the sensor within the 3′UTR. Note that the weak miR171 signals in the samples without the agroinfiltrated pri-miR171c originates from the endogenous locus. (B) The effect of PeloDN on the accumulation of 5′ cleavage fragments of amiR-GFP-programmed RISC. PHA-GFP fusion (P-G) construct and PHA-st-GFP (P-st-G), a similar sensor construct in which a stop codon is present between the PHA and GFP reporter genes, were co-agroinfiltrated with amiR-GFP, or with amir-GFP and a dominant-negative version of Arabidopsis Pelota1 (PeloDN) (p14 was used as internal control). 5′ cleavage product accumulates only when PeloDN is present and the amir-GFP–directed cleveage occurs within the coding region. (C) NSD does not affect the tasiRNA silencing pathway. N. benthamiana leaves were co-agroinfiltrated with TAS1b mRNA and miR173 expressing constructs, or with TAS1b, miR173 and PeloDN expressing constructs. RNA gel blot assays were conducted to monitor the abundance of miR173, TAS1b and the tasiR255 RNAs. TAS1b is cleaved by miR173-programmed RISC, and then tasiR255 is generated by RDR6 and DCL4 from the 3′ cleavage product. The presence of PeloDN does not stabilize the miR173-generated TAS1b cleavage product. As P14 modifies the tasiRNA pathway, P14 was not co-expressed.
Co-expression of either amiR-GFP or pri-miR171c resulted in strongly reduced GFP-171.1 mRNA level in all plants (Figure 3A, upper panel), confirming that both miRNAs targeted the sensor transcript efficiently. In the PDS-silenced control, neither the 5′ nor the 3′ cleavage fragment could be detected (Figure 3A, lanes 11 and 12) indicating that the cleavage products are quickly degraded. In the P-XRN4-silenced samples, co-expression of amiR-GFP (Figure 3A, lane 8) led to the overaccumulation of 3′ but not the 5′ cleavage fragments (the 3′ cleavage fragment might also be overaccumulated in the pri-miR171c co-expressed sample but this product cannot be detected by our probes). These data confirm our previous result that in N. benthamiana, XRN4 plays a role in the decay of the 3′ cleavage fragments of miRISC (40) and suggest that, it is not required for the elimination of 5′ cleavage products.
In P-Pelota- and P-HBS1-silenced plants, we failed to detect the 3′ cleavage fragments of the miRNA-sensor in either amiR-GFP or pri-miR171c co-expressed sample (Figure 3A, lanes 1–6). Relevantly, the 5′ cleavage fragments accumulated to high levels in P-Pelota- and P-HBS1-silenced plants when amiR-GFP was co-expressed but not when pri-miR171c was co-expressed (Figure 3A, upper panel compare lanes 2 to 3 and 5 to 6). These unexpected results indicate that plant NSD degrades the 5′ cleavage fragments of miRISC when the cleavage occurs in the coding region but it is not required for the elimination of 5′ cleavage products when the miRISC cleaves in the 3′UTR. Alternatively, the type of miRNA defines if the NSD is required for the degradation of 5′ fragment.
To distinguish between these two possibilities, the amiR-GFP miRNA was co-expressed in NSD-deficient cells with either PHA-GFP or PHA-st-GFP sensors. PHA-GFP is a fusion construct, while a stop codon separates the GFP from the PHA in the PHA-st-GFP sensor. Thus, the amiR-GFP targets the PHA-GFP transcript in the coding and the PHA-stop-GFP mRNA in the 3′UTR region (Figure 3B). To inactivate NSD, a dominant-negative mutant (PeloDN) of Arabidopsis Pelota1 was co-expressed. Control experiments suggested that PeloDN, in which the highly conserved K45V46 amino acids were changed to A45A46 residues, acts as a dominant-negative mutant in N. benthamiana leaves (Supplementary Figure S7).
In the presence of PeloDN, the 5′cleavage fragment accumulated when the amiR-GFP cleaved the target mRNA in the coding region but not when it cut the target transcript in the 3′UTR (Figure 3B). Similarly, amiR-GFP 5′ cleavage products accumulated from PHA-GFP but not from PHA-stop-GFP mRNAs in P-Pelota silenced leaves (see below at Figure 5B, lanes 2 and 4).
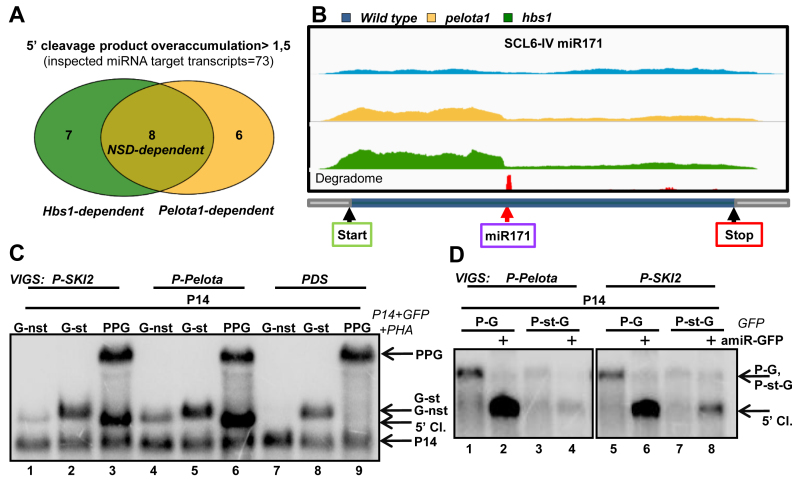
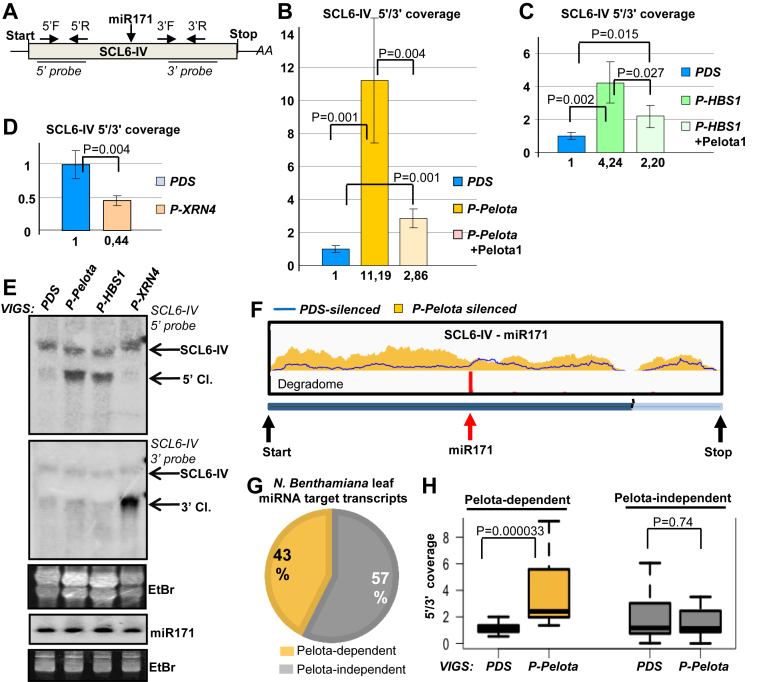
Figure 5.
SKI2 is required for the elimination of NSD target mRNAs. (A andB) NSD plays a role in the decay of 5′ cleavage fragments of miRISC in Arabidopsis. Comparative RNA-seq experiment was conducted from Pelota1 and Hbs1 T-DNA mutants (pelota1, hbs1) and from wild-type Arabidopsis plants. (A) Venn diagram shows the Pelota-, Hbs1- and the NSD-dependent Arabidopsis transcripts. The 5′/3′ coverage ratio of 73 miRNA target transcripts was analyzed. Pelota- or HBS1-dependent mRNAs are defined when the 5′ fragment overaccumulate >1.5-fold in the mutant relative to the wild type. If the 5′ fragment overaccumulate in both mutants, the mRNA was categorized as NSD-dependent. (B) The coverage of the SCL6-IV transcript. Yellow and green columns show the number of reads at a given position from pelota1 and hbs1 Arabidopsis mutants, while blue columns indicate the number of reads from wild type control. The degradome peak (red column) and the miRNA cleavage site (red arrow) are marked. (C) NSD reporter mRNAs accumulate to high levels in SKI2-deficient leaves. GFP-nst (G-nst) nonstop reporter construct was co-agroinfiltrated with P14 into the leaves of PDS-silenced (negative control), P-Pelota-silenced (positive control) and P-SKI2-silenced test plants. As controls, the same leaves were also co-agroinfiltrated with P14+G-st control and with P14+PPG vsiRISC sensor constructs. (D) SKI2 is involved in the decay of 5′ cleavage products of miRISC. Leaves of P-Pelota- and P-SKI2-silenced plants were agroinfiltrated in the presence or in the absence of amir-GFP with P14+PHA-GFP (P-G) fusion constructs, or with P14+P-st-G construct, in which a stop codon separates the PHA and the GFP reporter genes. The blot was hybridized with GFP 5′ fragment probe, stripped, and then reprobed for P14.
Taken together, we concluded that plant NSD is a translation-dependent mechanism, which degrades RISC generated 5′ products when the cleavage occurs within the coding region. In this case, NSD should not be involved in the degradation of 5′ cleavage fragments of the non-translated silencing target transcripts such as TAS ncRNAs. To test this assumption, miR173 was co-expressed with the ath-miR173-targeted AtTAS1b ncRNA in the presence or absence of PeloDN. Previously, it was demonstrated that 5′ cleavage product of TAS1b is quickly degraded, while the 3′ cleavage fragment is substrate for RDR6-dependent tasiRNA production (34). As Figure 3C shows, co-expression of PeloDN did not lead to the overaccumulation of the 5′ cleavage product of TAS1b. We also failed to detect the 5′cleavage fragment of TAS1b in P-Pel (and P-SKI2) silenced plants (Supplementary Figure S7B). These results support our assumption that NSD does not play a role in the decay of non-translated 5′ cleavage products. However, we cannot formally exclude the possibility that the remaining NSD activity is enough to eliminate the 5′ cleavage TAS1b fragment.
NSD is involved in the elimination of the 5′ cleavage products of endogenous miRNA silencing pathways
Transient assays show that NSD plays a critical role in the elimination of vsiRISC and miRISC generated 5′ cleavage fragments of reporter genes (Figures 2 and 3). To analyze if NSD is also involved in the degradation of 5′ cleavage products of endogenous miRNA targets, we studied in details the accumulation of the 5′ product of the SCL6-IV miRNA171 target mRNA (Figure 4A–F). SCL6-IV mRNA was selected because previous studies indicated that miRNA171 efficiently cleaves this transcript in N. benthamiana mesophyll cells (61). We conducted qRT-PCR assays to compare the accumulation of 5′ and 3′ cleavage products of SCL6-IV in PDS-silenced control and P-Pelota-, P-HBS1- and P-XRN4-silenced test plants. Two pairs of primers were designed, one pair detected the 5′ cleavage product and the full-length mRNA, while the other pair detected the 3′ cleavage product and the full-length mRNA. In line with the results of transient assays we found that the 5′ fragment was overaccumulated in both P-Pelota and P-HBS1-silenced lines relative to the PDS-silenced control (Figure 4B and C), while the 3′ fragment of the SCL6-IV endogenous miRNA target mRNA was overaccumulated in the P-XRN4-silenced plants (Figure 4D). Moreover, the 5′ fragment level was massively reduced when Pelota1 was expressed in the P-Pelota- or P-HBS1-silenced leaves, showing that NSD was restored (although not completely) (Figure 4B and C).
Figure 4.
NSD is involved in the degradation of 5′ cleavage fragments of several endogenous miRNA targets. (A) The schematic, non-proportional representation of the N. benthamiana SCL6-IV miRNA171 target transcript. The primer pairs that were used to measure the accumulation of the 5′ (5′F and 5′R) and 3′ (3′F and 3′R) cleavage fragments and the probes that were used for the northern assay are shown. (B and C) The 5′ cleavage fragments of SCL6-IV miR171 target mRNAs are overaccumulated in NSD deficient leaves. qRT-PCR assays were carried out from PDS-, P-Pelota- and P-HBS1-silenced leaves, and from Pelota1-complemented P-Pelota- and P-HBS1-silenced leaves. (D) qRT-PCR experiments show that the 3′ cleavage fragments of SCL6-IV mRNAs are overaccumulated in P-XRN4-silenced leaves. (E) RNA gel blot assay was carried out to study the accumulation of the 5′ and the 3′ cleavage products (5′ Cl. and 3′ Cl.) of SCL6-IV mRNA in PDS-, P-XRN4-, P-Pelota- and P-HBS1-silenced leaves. Note that VIGS did not alter the expression of the miRNA171. (F–H) Comparative RNA-seq assay conducted from PDS- and P-Pelota-silenced plants show that Pelota plays a role in the degradation of a subset of endogenous miRNA target transcripts. (F) The coverage of the SCL6-IV transcript. Yellow columns show the number of reads at a given position from P-Pelota, while blue line indicates number of reads from PDS-silenced control. The degradome peak (red column) and the miRNA cleavage site (red arrow) are marked. (G andH) The 5′/3′coverage ratio (see main text) of miRISC targets from PDS- and P-Pelota silenced plants were compared. Transcript whose 5′/3′ coverage ratio was at least 1.5-fold higher in the P-Pelota–silenced plants were defined as Pelota-dependent.
To further support these results, northern blot assays were conducted to analyze the accumulation of the cleavage product of SCL6-IV in P-Pelota-, P-HBS1- and P-XRN4-silenced N. benthamiana plants. The 3′ cleavage fragments of SCL6-IV transcripts accumulated to very high levels in P-XRN4-silenced plants, while the 5′ cleavage products were overaccumulated in both P-Pelota- and P-HBS1-silenced plants (Figure 4E). The level of miR171 was not affected by the silencing of NSD components or XRN4 (Figure 4E). These results confirm that NSD is involved in the elimination of the 5′ cleavage products of endogenous SCL6-IV transcripts.
Next we conducted an RNA-seq experiment to clarify if NSD is involved in the decay of the 5′ cleavage products of all endogenous miRNA targets or only a subset of miRNA target transcripts is degraded by NSD. To address this question, RNA-seq libraries (in triplicates) were prepared with random primers from ribo-depleted RNA samples, which were isolated from P-Pelota- and PDS-silenced N. benthamiana leaves (called RNA-seq1 experiment). Pelota inactivation had significant effect on gene expression, 10.24% (6035 out of 58 863) of annotated genomic loci showed significantly altered expression. 4.74% of annotated transcripts were up- and 5.5% down-regulated in the P-Pelota silenced plants relative to the PDS-silenced control (Supplementary File1, Supplementary Figures S8 and S9). GO analysis of the up-regulated genes revealed (Supplementary Figure S9B and C) that regulation of transcription (and the closely related categories) was the only significantly enriched cellular process. Relevantly, miRNAs play an important role in the regulation of transcription factors in plants. None of the GO categories was enriched within the down-regulated genes.
Next we studied if the 5′ cleavage fragments of miRNA targets are overexpressed in P-Pelota silenced leaves. First, we identified transcripts that are efficiently cleaved by a conserved miRNA in the N. benthamiana leaves. Using previously published miRNA and degradome data (mRNA 3′ cleavage product library, also called PARE) (61), we could select 17 conserved miRNAs which are abundant in N. benthamiana leaves, and 57 of their potential target transcripts, which have a clear degradome peak at the putative miRNA cleavage site (Figure 4 and Supplementary Figure S9c). All these cleavage sites are present in the coding region. We assumed that these target mRNAs are efficiently cleaved by the given conserved miRNA in N. benthamiana leaves. These selected miRNA-targets were further analyzed.
If Pelota plays a role in the decay of the 5′ cleavage product, the 5′ fragments of miRISC cleaved mRNAs will be overaccumulated in the Pelota-silenced plants (Figure 4F and Supplementary Figure S10). Thus, the ratio of total read numbers upstream and downstream from the miRNA cleavage site (referred to as 5′/3′ coverage ratio) will be higher in the P-Pelota silenced samples relative to the PDS-silenced controls. Indeed, we found that in ∼40,4% of the miRISC targets (at 23 out of 57), the 5′/3′ coverage ratio was at least 1.5-fold higher in the P-Pelota samples (defined as Pelota-dependent miRNA targets, while the rest will be defined as Pelota-independent targets) (Figure 4G and Supplementary Figure S9C). Relevantly, only 8.8% (5 transcripts out of 57) showed 1.5-fold increased 3′/5′ coverage ratio in the P-Pelota relative to the PDS-silenced samples. These data strongly suggest that Pelota plays a role in the elimination of the 5′ cleavage products of a significant subset of endogenous miRNA targets.
The results of the high-throughput experiment was confirmed by (i) repeating the RNA-seq assay (RNA-seq2, Supplementary Figure S11) and (ii) by conducting qRT-PCR assays. Different set of P-Pelota- and PDS-silenced plants were used for the RNA-seq1, RNA-seq2 and the qRT-PCR assays. Although the plant samples for the two RNA-seq assays were differently selected (see Materials and Methods, Supplementary File1, and Supplementary Figures S8A and S11A), highly overlapping subsets of endogeneous miRNA targets were identified as Pelota-dependent in both experiments. In RNA-seq1 23, in RNA-seq2 27 (out of 57) Pelota-dependent mRNAs were identified, and 19 were defined as Pelota-dependent in both experiments (Supplementary Figure S12).
To further support these results, seven mRNAs, which were defined as Pelota-dependent in both RNA-seq assays and two mRNAs, which were identified as Pelota-independent in both RNA-seq were further analyzed by qRT-PCR. The results of the qRT-PCR were consistent in 8 out of 9 samples with the RNA-seq data (Supplementary Figure S13). Thus, RNA-seq and qRT-PCR assays strongly suggest that NSD plays an important role in the decay of 5′ cleavage fragments of a significant subset (40–50%) of endogenous miRNA targets in N. benthamiana leaves.
To confirm that the NSD system is also involved in the degradation of 5′ miRNA cleavage fragments in other plants, RNA-seq assays were conducted from leaf samples of wild-type and pelota1 and hbs1 T-DNA mutant Arabidopsis plants (Supplementary File1, Supplementary Figure S14). Although Pelota is present in two copies in Arabidopsis, and we do not know whether these lines are null or hypomorph mutants, we assumed that the NSD activity is impaired in both mutants. If it is true and NSD is also involved in the degradation of 5′ cleavage fragments in Arabidopsis, the 5′ cleavage products of endogenous miRNA targets should be overaccumulated in these mutants. Indeed, we found that 5′ cleavage fragments of 14 transcripts (out of 73) overaccumulated in pelota1, 15 in hbs1 and 8 in both mutants (Figure 5A and B, Supplementary Figures S15 and S16). Thus NSD is also involved in the decay of 5′ cleavage fragments of a subset of endogenous Arabidopsis miRNA targets. To further confirm this conclusion, qRT-PCR assays were carried out (Supplementary Figure S17). We selected five putative NSD-dependent mRNAs (mRNAs whose 5′ cleavage fragments were overaccumulated >1.5-fold in both mutants) for qRT-PCR studies. qRT-PCR showed that the 5′ cleavage fragments of 4 (out of 5) mRNAs were significantly overaccumulated in both pelota1 and hbs1 mutants relative to the control. Thus, these are genuine NSD-dependent transcript. qRT-PCR results also suggest that NSD might also play a minor role in the elimination of 5′ cleavage fragments of mRNAs, which were categorized (based on RNA-seq experiments) as NSD-independent (for instance Supplementary Figure S17, ARF16). Taken together, RNA-seq and qRT-PCR assays confirm that NSD is involved in the elimination of 5′cleavage fragments of certain endogenous miRISC targets in Arabidopsis as well as in N. benthamiana.
However, while in N. benthamiana 40–50% of the leaf miRNA target mRNAs are Pelota-dependent, in Arabidopsis only 10–20%. Further studies are needed to unravel why NSD is important for the decay of only certain miRISC targets and to clarify why NSD plays a more important role in the decay of 5′ miRISC cleavage products in N. benthamiana. It is possible that 5′ cleavage degradation is more redundant in Arabidopsis (for instance, no obvious orthologs of the RICE 3′-5′ exonucleases can be identified in N. benthamiana) or it is the consequence of different experimental approaches. Nevertheless, our data clearly show that NSD is involved in the decay of miRISC 5′ cleavage fragments in both plants.
SKI2 is required for NSD
SKI2 is required for the elimination of the vsiRISC generated 5′ cleavage fragments (Figure 1A). To analyze if SKI2 also plays a role in the degradation of other NSD targets, the accumulation of the nonstop reporter transcripts and the 5′ cleavage products of miRISC were studied in P-SKI2-silenced test and in P-Pelota-silenced positive and PDS-silenced negative control plants.
Relevantly, the NSD reporter transcripts expressed to easily detectable levels in both P-SKI2-silenced test and P-Pelota-silenced positive control plants but not in the PDS-silenced negative control (Figure 5C, compare lanes 1 and 4 to 7), indicating that both SKI2 and Pelota are involved in the degradation of nonstop mRNAs. Thus, we propose that the nonstop mRNAs are mainly degraded by the SKI-exosome complex in plants. Indeed, the NSD reporter transcripts were barely detectable in P-XRN4-silenced lines (Supplementary Figure S18).
To study the role of SKI2 in the elimination of 5′ cleavage products of miRISC, the PHA-GFP and PHA-stop-GFP sensor constructs were co-expressed with amiR-GFP in the leaves of P-SKI2- and P-Pelota-silenced plants. When the cleavage occurred in the coding region (PHA-GFP mRNA), the 5′ cleavage products accumulated to very high levels in both plants (Figure 5D, lanes 2 and 6). By contrast, when the RISC cleaved in the 3′UTR (PHA-stop-GFP mRNA), the 5′ cleavage fragments were slightly overaccumulated only in the P-SKI2-silenced lines (Figure 5D, compare lane 4 to 8). Thus, SKI2 may contribute to the degradation of the RISC generated 5′ cleavage fragment even when the mRNA is targeted in the 3′UTR region. These data are consistent with the results of a recent report that analyzed the role of SKI2 in suppression of RNA silencing (32).
We also analyzed the accumulation of the 5′ cleavage fragments of SCL6-IV endogenous miRNA target. We found that in P-SKI2-silenced plants, like in P-Pelota-silenced lines, the 5′ cleavage fragments of SCL6-IV were overaccumulated (Supplementary Figure S19).
Taken together, SKI2 is required for the elimination of all NSD targeted transcripts including the nonstop mRNAs and the 5′ fragments generated by vsi- or miRISC cleavage in the coding region. However, SKI2 may also be involved in the decay of silencing generated stop codon-less transcripts if the cleavage occurs in the 3′UTR.
In NSD-deficient leaves, the 5′ cleavage products of miRISC accumulate in the polysomal fractions
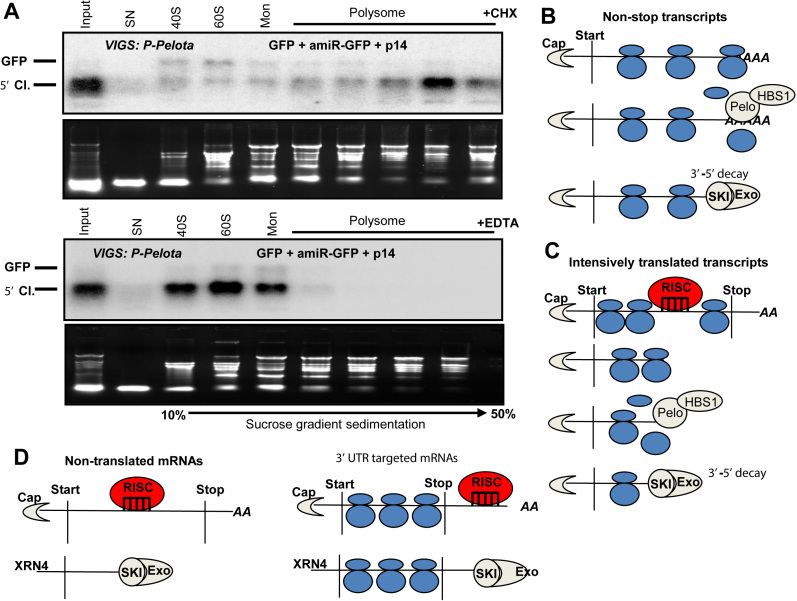
In yeast and animals, the NSD factors and ABCE1 play a critical role in the ribosome dissociation from the 3′end of the stop codon-less transcripts (5,7). We hypothesized that plant NSD is also involved in the dissociation of ribosomes from the RISC generated 5′ cleavage fragments. If this assumption is correct, in NSD-deficient cells the RISC generated 5′ cleavage products should be associated with ribosomes. Indeed, when GFP was co-expressed with amiR-GFP (that cleaves GFP in the coding region) in P-Pelota-silenced leaves, the 5′ cleavage fragments accumulated predominantly in the polysomal sucrose gradient fractions (Figure 6A, upper panel). Similarly, when GFP was co-expressed with amiR-GFP and PeloDN in wild-type N. benthamiana leaves, the 5′ cleavage products were also associated with ribosomes (Supplementary Figure S20A). Moreover, in the P-Pelota silenced leaves the 5′ cleavage fragment of SCL6-IV endogenous miRISC target transcript was also associated with ribosomes (Supplementary Figure S20C). We noted that the 5′ cleavage fragments accumulated in approximately half of the experiments in the heavy polysomal and in other half of the experiments in the light polysomal fractions (compare Supplementary Figure S20A and B). Further studies are required to clarify what determines whether the 5′ cleavage fragments accumulate in the light or the heavy polysomal fractions.
Figure 6.
Model of plant NSD. (A) In NSD-deficient cells, the 5′ cleavage fragments of miRISC are associated with ribosomes. Extracts were isolated with cycloheximide (CHX) or EDTA containing buffer (upper and bottom panel) from P-Pelota-silenced (P-Pelota) leaves, in which GFP, amiR-GFP and P14 were co-expressed. These extracts were subjected to sucrose gradient sedimentation, and then the fractions were analyzed by RNA gel blot assays. The blots were hybridized with GFP 5′ fragment probe. Note that the 5′ cleavage fragment (5′ Cl.) accumulates in the polysomal fractions in the CHX containing samples, while polysomal migration of 5′ fragment is disrupted by the addition of EDTA. Mon indicates monosomal fraction. (B) The role of NSD in the decay of nonstop and (C) RISC cleavage generated stop codon-less transcripts. It is not known whether SKI-exosome mediated decay occurs when the mRNA is still associated with ribosomes or SKI-exosome degrades mRNAs, from which the ribosomes have already removed. (D) NSD is not required for the 5′ fragment decay when si/miRISC cuts outside of the coding region or if the target transcript is not translated. For details, see the main text.
Taken together, association of miRISC 5′cleavage fragments with ribosomes strongly suggests that plant NSD system is required for the efficient release of ribosomes from the miRISC generated 5′ cleavage fragments.
NSD and NMD quality control systems operate independently in plants
NMD is a translation termination coupled, conserved eukaryotic quality control system that eliminates premature termination codon containing aberrant mRNAs (62,63). NMD recognizes a stop codon as a premature one if the translation termination is inefficient because the 3′ UTR is unusually long or the 3′UTR contains an intron more than 50 nucleotide downstream from the stop codon. Slow termination leads to the binding of UPF1 NMD key factor to the terminating ribosome. UPF1 recruits additional factors to form a functional NMD complex, which initiates the rapid degradation of the mRNA.
As NMD inactivation affects amplification of RNA silencing (36) and because NSD and silencing systems are connected (this study), we wanted to examine whether the NSD and NMD translation coupled quality control systems are also connected in plants. NSD and NMD reporter transcripts were expressed in P-Pelota-silenced (NSD-deficient) and P-UPF1-silenced (NMD-deficient) test plants and in PDS-silenced control line. The G-600 NMD reporter mRNA, which has a long NMD-inducing 3′ UTR (45), accumulated to high levels in P-UPF1-silenced leaves but expressed to very low levels in the PDS- and P-Pelota-silenced plants (Supplementary Figure S21). In contrast, the GFP-nst NSD reporter transcripts were barely detectable in the PDS- and P-UPF1-silenced samples, while it was abundant in P-Pelota-silenced leaves. These data suggest that Pelota is not required for NMD and that UPF1 key NMD factor is not required for NSD. In line with this latter finding, the NSD reporter mRNA also accumulated to very low levels (Supplementary Figure S21C) when the NMD was impaired by expressing a dominant-negative mutant of UPF1 (50). Thus we concluded that the NSD and NMD systems function independently in plants. By contrast, a recent study has shown that NSD and NMD systems operate cooperatively in C. elegans (64). This finding is consistent with previous reports that plant NMD degrades target transcript exclusively by exonucleolytic pathways (65), while in animals, NMD machinery also endonucleolytically cleaves target transcripts upstream from the premature termination codon.
DISCUSSION
Here, we have demonstrated that NSD operates in plants and identified the key components of plant NSD. Moreover, we have showed that plant NSD and RNA silencing systems act cooperatively but did not find connection between NSD and NMD.
It is supposed that NSD-mediated mRNA decay is important in eukaryotes, as it rescues the stalled ribosomes and prevents the accumulation of truncated proteins from the target transcripts (6). We speculate that in plants NSD has an additional quality control function: it suppresses silencing amplification initiated from the stop codon-less mRNAs, thereby protecting the regulatory sRNA homeostasis of the cell.
NSD is an ancient and conserved eukaryotic quality control system
We demonstrated that NSD reporter transcripts are eliminated in a translation-dependent manner in plants, and that Pelota, Hbs1 and SKI2 (but not XRN4 or UPF1) proteins are required for the degradation of NSD target mRNAs (Figures 1, 2, 5 and Supplementary Figures S18, S19 and S21).
As homologs of Pelota and Hbs1 can be identified in all eukaryotic lineages, it was proposed that NSD system was already active in the stem eukaryotes (last common ancestors of extant eukaryotes) and that its mechanism is well conserved (60). Plants diverged very early from the other eukaryotic lineages. Thus our findings that NSD functions in plants, and that, the key components of NSD are identical in plants and animals, strongly supports this hypothesis (60).
The NSD and the ribosome-associated quality control (RaQC) systems cooperate in fungi and animals (7). The stop codon-less transcripts are targeted for decay by the NSD machinery, while their protein products are ubiquitinated and degraded by the RaQC system. In yeasts, the nonstop transcripts are decayed very effectively, while in mammals, the nonstop mRNAs accumulate to relatively high levels but their protein products are quickly degraded (10,16,66). The NSD reporter mRNAs were barely detectable in wild-type plants indicating (Figure 1B) that in plants, like in yeast, the NSD machinery degrades the stop codon-less transcripts very efficiently. However, as homologs of the RaQC system can be identified in plants, one can speculate that the protein products of NSD target transcripts are also ubiquitinated and degraded in plants.
Targets of plant NSD might be abundant
We showed that plant NSD efficiently eliminates both nonstop and stop codon-less mRNAs (Figures 1–3). Nonstop transcripts are generated when premature polyadenylation occurs in the coding region (67). In Arabidopsis, 11% of mRNAs are subjected to alternative polyadenylation in the coding region indicating that nonstop aberrant transcripts are frequently generated in plants. NSD-mediated elimination of nonstop mRNAs could be important to ensure protein homeostasis in plants.
The role of NSD in degrading stop codon-less plant mRNAs might be even more important. As plant miRNAs regulates thousands of genes, and miRISC mainly cleaves within the coding region, the 5′ cleavage fragments of miRISC might be the most important sources of stop codon-less NSD target transcripts. Indeed we found that transcription related gene expression is significantly altered in P-Pelota-silenced plants (Supplementary Figure S9) and that NSD is involved in the elimination of 5′ cleavage products of a significant subset of miRISC target transcripts in both Arabidopis and N. benthamiana plants (Figures 4, 5 and Supplementary Figures S9–S17). It is an interesting question why only certain miRISC 5′ cleavage fragments are degraded in an NSD-dependent manner. The miRISC 5′ cleavage products could be decayed by alternative pathways in plants, by the NSD-SKI-exosome pathway (this study and 32), by XRN4-mediated 5′-3′ decay or by RICE–mediated 3′-5′ exonuclease pathways (41,42). Although these pathways could be partially redundant, the different degradation pathways targets miRISC cleavage fragments with certain selectivity. The composition of the miRISC complexes or specific features of the target mRNAs could define which mRNA degradation pathway is recruited to the 5′ cleavage products. We postulate that NSD is especially important for the decay of intensively translated targets. NSD sensitivity of the cleavage fragment could also be affected by the localization of miRISC-target complex.
Model of plant NSD
As the mechanism of NSD appears to be conserved, we propose a model how NSD functions in plants. Nonstop transcripts are barely detectable in wild-type cells but accumulate to high levels in a deadenylated form in Pelota-deficient (and likely in HBS1- and SKI2-deficient) plant cells (Figures 2 and 5). We postulate that the elongating ribosome is stalled when it runs into the polyA tail of a nonstop transcript. In wild-type cells, the Pelota/Hbs1 NSD complex binds to the A-site of the stalled ribosome and stimulates the dissociation of the ribosome and (directly or indirectly) recruits the SKI-exosome complex to degrade the aberrant mRNA (Figure 6B). In the lack of NSD, the polyA tail is degraded but the nonstop transcript is stabilized, as the stalled ribosome protects the mRNA form the 3′-5′ exonucleases (5′ end is protected by the cap).
When RISC cuts a translated mRNA in the coding region, the generated 5′ cleavage fragment is a stop codon-less transcript. Plant NSD also plays a critical role in the degradation of these fragments (Figures 1–4). We propose that the elongating ribosome runs to the 3′ end of the 5′ cleavage fragment in wild-type as well as in NSD-deficient cells. In the absence of NSD, the ribosome might protect the 3′ end transcript. Indeed, in the NSD-deficient cells, the 5′ cleavage products accumulate to high levels and are associated with ribosomes (Figure 6A and Supplementary Figure S20). In wild-type cells, the NSD complex could be recruited to the empty A-site at the 3′ end resulting in ribosome dissociation and SKI-exosome mediated transcript degradation (Figure 6C).
NSD is not required for the decay of 5′ cleavage products, when the cleavage occurs in a non-coding region (3′UTR) or if a non-translated transcript is cleaved (Figure 3). The 3′ ends of these 5′ cleavage products are not protected by a ribosome. We postulate that these unprotected 5′ cleavage products are degraded in an NSD-independent manner (Figure 6D).
Can plant NSD play a role in regulating silencing amplification?
NSD is involved in the elimination of silencing generated 5′ cleavage fragments. We hypothesize that it plays a role in the control of RDR6-mediated silencing amplification.
Silencing amplification should be strictly regulated in plants. It is required for effective antiviral response, while hyperactive amplification might lead to the generation of deleterious rqc-siRNAs from 5′ cleavage fragments of endogenous miRISC (32,36–38). To prevent production of unregulated rqcRNAs, the XRN4 and SKI-exosome complexes should be efficiently recruited to the 5′ cleavage products of miRISC. We speculate that NSD facilitates the efficient recruitment of SKI-exosome complex to the NSD-dependent subset of miRISC 5′ cleavage products, thereby reducing the generation of detrimental rqc-siRNAs (Supplementary Figure S22). Thus NSD protects the sRNA homeostasis in plant cells. We hypothesize that in NSD-deficient cells, the 5′ cleavage fragments of miRISC accumulate and (at least a fraction of it) finally might enter into the RDR6-mediated silencing amplification pathway (Supplementary Figure S22).
NSD-mediated suppression of silencing amplification might be important for normal development. However, silencing amplification is necessary for the efficient antiviral response. NSD also stimulates the degradation of 5′ cleavage products of vsiRISC (Figure 2). Thus our model that NSD suppresses silencing amplification pathway predicts that NSD reduces the efficacy of the RDR6-mediated antiviral responses. Indeed, mutation in the Pelota ortholog of tomato resulted in enhanced resistance to a Begomovirus (68). Very recently, it has been shown that in rice, mutation in one of the Pelota orthologs leads to enhanced salycilic acid levels and consequently to increased resistance against Xanthomanas oryzae bacterial pathogen. However, as RNA silencing is also involved in antibacterial defense, it is possible that more efficient silencing also contributed to the resistance (69).
As both the NSD and silencing systems are ancient, it is tempting to speculate that these systems also act cooperatively in other eukaryotes. Indeed, it was shown that in Drosophila, NSD plays a role in the elimination of 5′ cleavage fragments of a reporter transcript, which was cleaved by transfected dsRNA derived siRNA (70).
DATA AVAILABILITY
The data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE99805).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Z. Kerenyi for creating the PHA-st and PHA-nst clones and for P. Gyula for his useful comments on bioinformatical analyzes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
Hungarian Scientific Research Fund [OTKA K112737 to B.J., K115934 to Cs.T., K116963 and K109835 to S.D., K121287 and K124661 to A.S.]. I. Szadeczky-Kardoss and A. Auber are PhD students at the Georgikon ‘Crop Production and Horticultural Research’ and at the ELTE ‘Classical and Molecular Genetics’ programs, respectively; Janos Bolyai Research Scholarship of the Hungarian Academy of Science (to T.C., T.I.O.). Funding for open access charge: Hungarian Scientific Research Fund [K116963, K109835].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fasken M.B., Corbett A.H.. Process or perish: quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 2005; 12:482–488. [DOI] [PubMed] [Google Scholar]
- 2. Mcmanus J., Cheng Z., Vogel C.. Next-generation analysis of gene expression regulation- comparing the role of synthesis and degradation. Mol. Biosyst. 2015; 11:2680–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parker R., Song H.. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004; 11:121–127. [DOI] [PubMed] [Google Scholar]
- 4. Siwaszek A., Ukleja M., Dziembowski A.. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014; 11:1122–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lykke-Andersen J., Bennett E.J.. Protecting the proteome: eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014; 204:467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shoemaker C.J., Green R.. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandman O., Hegde R.S.. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016; 23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T.. Dom34: Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell. 2012; 46:518–529. [DOI] [PubMed] [Google Scholar]
- 9. Chiabudini M., Tais A., Zhang Y., Hayashi S., Wölfle T., Fitzke E., Rospert S., Wolfle T., Fitzke E., Rospert S.. Release factor eRF3 mediates premature translation termination on polylysine-stalled ribosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2014; 34:4062–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frischmeyer P.A., Van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C.. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002; 295:2258–2261. [DOI] [PubMed] [Google Scholar]
- 11. Jackson R.J., Hellen C.U.T., Pestova T. V.. Termination and Post-termination Events in Eukaryotic Translation. Adv Protein Chem Struct Biol. 2012; 86:45–93. [DOI] [PubMed] [Google Scholar]
- 12. Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim. Biophys. Acta - Gene Regul. Mech. 2013; 1829:634–642. [DOI] [PubMed] [Google Scholar]
- 13. Halbach F., Reichelt P., Rode M., Conti E.. The yeast ski complex: Crystal structure and rna channeling to the exosome complex. Cell. 2013; 154:814–826. [DOI] [PubMed] [Google Scholar]
- 14. Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J.. XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta - Gene Regul. Mech. 2013; 1829:590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kowalinski E., Schuller A., Green R., Conti E.. Saccharomyces cerevisiae Ski7 Is a GTP-Binding protein adopting the characteristic conformation of active translational GTPases. Structure. 2015; 23:1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R.. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002; 295:2262–2264. [DOI] [PubMed] [Google Scholar]
- 17. Marshall A.N., Montealegre M.C., Jiménez-López C., Lorenz M.C., van Hoof A.. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet. 2013; 9:e1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalisiak K., Kuliński T.M., Tomecki R., Cysewski D., Pietras Z., Chlebowski A., Kowalska K., Dziembowski A.. A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic Acids Res. 2016; 45:2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doma M.K., Parker R.. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006; 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guydosh N.R., Green R.. Translation of poly(A) tails leads to precise mRNA cleavage. RNA. 2017; 23:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu A., Saudemont B., Bouteiller N., Elvira-Matelot E., Lepère G., Parent J.-S., Morel J.-B., Cao J., Elmayan T., Vaucheret H.. Second-Site mutagenesis of a hypomorphic argonaute1 Allele Identifies SUPERKILLER3 as an endogenous suppressor of transgene posttranscriptional gene silencing. Plant Physiol. 2015; 169:1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parent J.S., Jauvion V., Bouché N., Béclin C., Hachet M., Zytnicki M., Vaucheret H.. Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 2015; 43:8464–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H.. Partially redundant functions of arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-Acting siRNAs. Curr. Biol. 2005; 15:1494–1500. [DOI] [PubMed] [Google Scholar]
- 24. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 25. Bologna N.G., Voinnet O.. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014; 65:473–503. [DOI] [PubMed] [Google Scholar]
- 26. Fang X., Qi Y.. RNAi in plants: an argonaute-centered view. Plant Cell. 2016; 28:272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O.. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008; 320:1185–1190. [DOI] [PubMed] [Google Scholar]
- 28. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 29. Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R.. A link between rnRNA turnover and RNA interference in Arabidopsis. Science. 2004; 306:1046–1048. [DOI] [PubMed] [Google Scholar]
- 30. Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C.. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell Online. 2007; 19:3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Souret F.F., Kastenmayer J.P., Green P.J.. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 2004; 15:173–183. [DOI] [PubMed] [Google Scholar]
- 32. Branscheid A., Marchais A., Schott G., Lange H., Gagliardi D., Andersen S.U., Voinnet O., Brodersen P.. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 2015; 43:10975–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R., Watanabe Y.. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009; 583:1261–1266. [DOI] [PubMed] [Google Scholar]
- 34. Allen E., Xie Z., Gustafson A.M., Carrington J.C.. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121:207–221. [DOI] [PubMed] [Google Scholar]
- 35. Parent J.-S., Bouteiller N., Elmayan T., Vaucheret H.. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015; 81:223–232. [DOI] [PubMed] [Google Scholar]
- 36. De Alba A.E.M., Moreno A.B., Gabriel M., Mallory A.C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M.D. et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015; 43:2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H., Xia R., Meyers B.C., Walbot V.. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 2015; 27:84–90. [DOI] [PubMed] [Google Scholar]
- 38. Zhang X., Zhu Y., Liu X., Hong X., Xu Y., Zhu P., Shen Y., Wu H., Ji Y., Wen X. et al. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science (80-.). 2015; 348:120–123. [DOI] [PubMed] [Google Scholar]
- 39. Zhang X., Guo H.. mRNA decay in plants: both quantity and quality matter. Curr. Opin. Plant Biol. 2017; 35:138–144. [DOI] [PubMed] [Google Scholar]
- 40. Nyikó T., Sonkoly B., Mérai Z., Benkovics A.H., Silhavy D.. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 2009; 71:367–378. [DOI] [PubMed] [Google Scholar]
- 41. Ren G., Xie M., Zhang S., Vinovskis C., Chen X., Yu B.. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Z., Hu F., Sung M.W., Shu C., Castillo-González C., Koiwa H., Tang G., Dickman M., Li P., Zhang X.. RISC-interacting clearing 3′- 5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC inArabidopsis thaliana. Elife. 2017; 6:e24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chantarachot T., Bailey-Serres J.. Polysomes, stress granules, and processing Bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 2018; 176:254–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Merai Z., Kerenyi Z., Molnar A., Barta E., Valoczi A., Bisztray G., Havelda Z., Burgyan J., Silhavy D., Mérai Z. et al. Aureusvirus P14 is an efficient RNA silencing suppressor that binds Double-Stranded RNAs without size specificity. J. Virol. 2005; 79:7217–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kerényi Z., Mérai Z., Hiripi L., Benkovics A., Gyula P., Lacomme C., Barta E., Nagy F., Silhavy D.. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008; 27:1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O.. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004; 18:2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Csorba T., Bovi A., Dalmay T., Burgyán J.. The p122 subunit of Tobacco Mosaic Virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J. Virol. 2007; 81:11768–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krenek P., Samajova O., Luptovciak I., Doskocilova A., Komis G., Samaj J.. Transient plant transformation mediated by Agrobacterium tumefaciens: principles, methods and applications. Biotechnol. Adv. 2015; 33:1024–1042. [DOI] [PubMed] [Google Scholar]
- 49. Silhavy D., Molnár A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyán J.. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002; 21:3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kertész S., Kerényi Z., Mérai Z., Bartos I., Pálfy T., Barta E., Silhavy D.. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006; 34:6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burch-Smith T.M., Anderson J.C., Martin G.B., Dinesh-Kumar S.P.. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004; 39:734–746. [DOI] [PubMed] [Google Scholar]
- 52. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H. et al. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 2015; 43:D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E.. The arabidopsis information resource: making and mining the ‘gold standard’ annotated reference plant genome. Genesis. 2015; 53:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z.. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017; 45:W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohorianu I., Stocks M.B., Applegate C.S., Folkes L., Moulton V.. The UEA Small RNA Workbench: A Suite of Computational Tools for Small RNA Analysis. Methods in Molecular Biology (Clifton, N.J.). 2017; 1580:193–224. [DOI] [PubMed] [Google Scholar]
- 59. Saito S., Hosoda N., Hoshino S.I.. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013; 288:17832–17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Atkinson G.C., Baldauf S.L., Hauryliuk V.. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 2008; 8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baksa I., Nagy T., Barta E., Havelda Z., Várallyay É., Silhavy D., Burgyán J., Szittya G.. Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genomics. 2015; 16:1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shaul O. Unique Aspects of Plant Nonsense-Mediated mRNA Decay. Trends Plant Sci. 2015; 20:767–779. [DOI] [PubMed] [Google Scholar]
- 63. Fatscher T., Boehm V., Gehring N.H.. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015; 72:4523–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Arribere J.A., Fire A.Z.. Nonsense mRNA suppression via nonstop decay. Elife. 2018; 7:e33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kerényi F., Wawer I., Sikorski P.J., Kufel J., Silhavy D.. Phosphorylation of the N- and C-terminal UPF1 domains plays a critical role in plant nonsense-mediated mRNA decay. Plant J. 2013; 76:836–848. [DOI] [PubMed] [Google Scholar]
- 66. Ito-Harashima S., Kuroha K., Tatematsu T., Inada T.. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007; 21:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., Hunt A.G.. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lapidot M., Karniel U., Gelbart D., Fogel D., Evenor D., Kutsher Y., Makhbash Z., Nahon S., Shlomo H., Chen L. et al. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 2015; 11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang X.-B., Feng B.-H., Wang H.-M., Xu X., Shi Y.-F., He Y., Chen Z., Sathe A.P., Shi L., Wu J.-L.. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018; 60:160–172. [DOI] [PubMed] [Google Scholar]
- 70. Hashimoto Y., Takahashi M., Sakota E., Nakamura Y.. Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem. Biophys. Res. Commun. 2017; 484:1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE99805).