Abstract
Aim:
Variants of TIM-3/HAVCR2 3′UTR miRNA binding sites are significantly associated with cancer; however, roles in post-transcriptional regulation have not been elucidated.
Methods:
The regulatory and coding region single nucleotide polymorphisms (SNPs) of TIM-3/HAVCR2 were identified using an online database. Single nucleotide polymorphism Function Prediction was used to predict potential functional relevance of miRNA binding sites.
Results:
The analysis indicated rs9313439, rs4704846, rs3087616 and rs1036199 affect possible miRNA binding sites in TIM-3/HAVCR2 3′UTR. We used additional data on genotypes and limited minor allele frequency >5% in the HapMap populations. Only rs3087616 and rs4704846 were significantly associated with TIM-3/HAVCR2.
Conclusion:
Both rs3087616 and rs4704846 could be putative variants mediating post-transcriptional regulation of the TIM-3/HAVCR2. Deeper understanding of how 3′UTR variants influence the activity by TIM-3/HAVCR2 for therapy against cancer.
Keywords: : genetic, miRNA, polymorphism, TIM-3/HAVCR2, variant
Lay abstract
TIM-3/HAVCR2 is involved in the pathogenesis of malignant tumors and progression of various types of cancer. Deeper understanding of how 3′UTR variants influence activity by TIM-3/HAVCR2 is useful for research into cancer therapy. In this study, single nucleotide polymorphism Function Prediction was used to predict potential functional relevance of miRNA binding sites. We found rs3087616 and rs4704846 were of interest. Our findings indicate that the 3′UTR variants regulate the activity of TIM-3/HAVCR2 and could aid in targeting the TIM-3/HAVCR2 pathway for cancer treatment.
Cancer is a common, potentially fatal disease resulting from complex interactions between environmental and genetic factors [1]. Studies are increasingly focused on the role of gene polymorphisms in the etiology of cancers. Growing evidence indicates that single nucleotide polymorphisms (SNPs) play an important role in carcinogenesis; gene polymorphisms have been implicated in response to methotrexate toxicity in high-grade osteosarcoma [2,3]. Cancer immunotherapy has produced impressive clinical results in recent years. The TIM-3 mediates immune tolerance in tumor immunity. Thus, targeting TIM-3 has emerged as a promising approach for enhancement of current immunotherapy.
The TIM-3/HAVCR2 gene is located in the chromosomal region 5q33.3 in humans [4]. Despite a large amount of experimental data showing an immunosuppressive function of TIM-3 in vivo, the exact mechanisms of this action are not well understood. The genetic variations of TIM-3/HAVCR2 may contribute to the development of cancers [5,6]. Overexpression of TIM-3/HAVCR2 is associated with a poor prognosis in squamous cell carcinoma, and in colorectal, gastric and breast cancers [7–10]. Additionally, TIM-3/HAVCR2 is intimately involved in the pathogenesis of malignant tumors and progression of various types of cancer.
It is well known that variants of miRNA binding sites can alter gene function. miRNAs can regulate the activity of TIM-3/HAVCR2, and dysregulation of miRNA is implicated in neoplasms. Fooladinezhad and colleagues report that TIM-3 is a specific surface marker for leukemic stem cells and is highly expressed on leukemic stem cells compared with normal bone marrow cells and hematopoietic stem cells. Studies indicate that miRNAs can affect the progression of acute myeloma leukemia by targeting the expression of different genes, such as that of TIM-3. Bioinformatics assessments indicate that miR-330-5p may robustly inhibit the expression of TIM-3, and miR-330-5p has been shown to have a strong inhibitory effect on the expression of TIM-3 in an acute myeloma leukemia cell line [11]. In this study, we performed a bioinformatics analysis, using the HapMap database, to examine whether the TIM-3/HAVCR2 3′UTR variants are associated with tumorigenesis and tumor progression.
Materials & methods
The SNPs of TIM-3/HAVCR2 were identified, in the regulatory and coding regions, using an online database [12]. The bioinformatics tool SNP Function Prediction (FuncPred; [13]) was used to predict the potential functional relevance of the miRNA binding sites. Additionally, we limited our analysis to SNPs with minor allele frequency (MAF) >5% in the HapMap populations, ‘Utah residents with Northern and western European ancestry (CEU)’ and calculated pairwise linkage disequilibrium (LD) values of all the SNPs in the same gene. Then, we selected SNPs that were not in LD (r2 <0.8) and constructed the LD maps of TIM-3/HAVCR2 SNPs using a web service [13].
Results
Selected variants of the TIM-3/HAVCR2 3′UTR & putative miRNA binding sites
In total, 43 SNPs were identified in the TIM-3/HAVCR2 3′UTR; only rs3087616 and rs4704846 have a known MAF value >0.05. According to the bioinformatics analysis, these two SNPs were predicted to affect the activity of the miRNA binding site, as shown in Table 1. The predicted functions of the TIM-3/HAVCR2 3′UTR SNPs, in different ethnic groups are shown in Table 2. The comparisons of LD in common SNPs, among the different populations are listed in Table 3. The most extensively studied SNPs affect the putative binding sites of miRNAs [13].
Table 1. . Selected single nucleotide polymorphisms of TIM-3/HAVCR2 3′UTR and putative miRNA binding sites.
| Name | Alleles | MAF | Putative miRNA binding sites |
|---|---|---|---|
| rs3087616 | C/T | 0.1116 | hsa-miR-1250, hsa-miR-1827, hsa-miR-196a, hsa-miR-196b, hsa-miR-34a, hsa-miR-34c-5p, hsa-miR-449a, hsa-miR-449b and hsa-miR-582-5p |
| rs4704846 | A/G | 0.1581 | hsa-miR-18b, hsa-miR-379, hsa-miR-554, hsa-miR-631, hsa-let-7a, hsa-let-7e, hsa-let-7f, hsa-let-7g and hsa-let-7i |
3′UTR: 3′ Untranslated region; MAF: Minor allele frequency; NA: Not available.
Table 2. . Single nucleotide polymorphisms function prediction results of TIM-3/HAVCR2 3′UTR in different races.
| Name | Alleles | African | Asian | European | African–American | Hispanics |
|---|---|---|---|---|---|---|
| rs3087616 | C/T | 0.822 | 0.991 | 0.804 | 0.833 | NA |
| rs4704846 | A/G | 0.685 | 0.989 | 0.816 | NA | NA |
3′UTR: 3′ Untranslated region; NA: Not available.
Table 3. . Comparison of linkage disequilibrium of common single nucleotide polymorphisms among the populations.
| Name | Alleles | CEU | CHB | CHD | GIH | LWK | MEX | YRI |
|---|---|---|---|---|---|---|---|---|
| rs3087616 | C/T | 0.844 | 0.988 | 0.994 | 0.938 | 0.817 | 0.92 | 0.836 |
| rs4704846 | A/G | 0.816 | 0.989 | NA | NA | NA | NA | 0.704 |
NA: Not available.
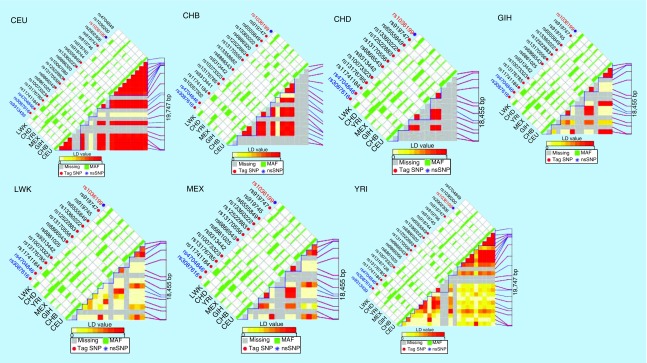
Calculating the LD of all SNPs in the TIM-3/HAVCR2 gene
The bioinformatics tool FuncPred [14] was used to identify the potential functional relevance of the SNPs. To select the SNPs that were not in LD (r2 <0.8), we calculated the pairwise LD values of all SNPs in the same gene and plotted the LD maps of these TIM-3/HAVCR2 SNPs using FuncPred. The pairwise r2 correlations between the two relevant SNPs were represented by each square number. The color of each SNP spot reflects its D′ value; when the D′ value decreases, the color changes from red to white. The haplotype blocks were estimated using the FuncPred software. The MAF of all the above alleles was greater than 0.05. In our study, the polymorphisms, rs3087616 and rs4704846 were the predicted tag SNPs, as indicated by the LD plot of the TIM-3/HAVCR2 region generated using FuncPred. Each square number represents the pairwise r2 correlations between the two relevant SNPs. The color of each SNP spot reflects its D′ value, which changes from red to white as the D′ value decreases. An SNP is shown in Figure 1.
Figure 1. . A linkage disequilibrium plot of the TIM-3/HAVCR2 region generated using the single nucleotide polymorphism Function Prediction software (FuncPred).
Each square number represents the pairwise r2 correlations between the two relevant SNPs. The color of each SNP spot reflects its D′ value, which changes from red to white as the D′ value decreases.
MAF: Minor allele frequency; SNP: Single nucleotide polymorphism.
Discussion
Numerous studies, such as multiple tissue gene expression, animal models and clinical trials suggest that TIM-3/HAVCR2 plays an important role in carcinogenesis. Various groups have analyzed the link between TIM-3/HAVCR2 polymorphisms and the risk of cancer [7–10]. An increased level of TIM-3 is correlated with poor survival in patients with cancer. The soluble form of TIM-3 has been shown to reduce the antigen-specific T-cell response and downregulate the antitumor activity in vivo [15]. Another study revealed that blockade of TIM-3 can reverse the impaired phenotype of natural killer cells in patients with metastatic melanoma [16], highlighting the potential of TIM-3-targeted therapy. Nevertheless, despite all the promising data shown in preclinical models, the role of TIM-3 has not been evaluated in clinical trials, possibly because of insufficient evidence on the role of TIM-3 in clinical patients with cancer.
Many studies confirm that SNPs, in cancer-related genes, can contribute to individual susceptibility to cancer by affecting gene expression and function [17,18]. In recent study, the results have confirmed the bioinformatics prediction of suppressive effect of miR-125a-3p on TIM-3 with miRWalk and TargetScan softwares [19]. Our study supports the notion that mutations in miRNA binding regions contribute to altered gene function. Although our findings indicate that rs9313439, rs4704846, rs3087616 and rs1036199, in the TIM-3/HAVCR2 3′UTR, affect the potential miRNA binding sites, we found that rs3087616 and rs4704846 were significantly associated with TIM-3/HAVCR2 in the lymphoblastoid cell lines. The polymorphisms rs3087616 and rs4704846 were the predicted tag SNPs in our study, suggesting that these variants may contribute to the TIM-3/HAVCR2 post-transcriptional regulation. Our results indicate a mechanism, underlying the roles of rs3087616 and rs4704846, in the regulation of TIM-3/HAVCR2 expression, and provide an explanation for the susceptibility to tumor formation associated with these SNPs. It is possible that the genetic variants in the TIM-3/HAVCR2 3′UTR modulate its expression, and the variants affecting the TIM-3/HAVCR2 miRNA binding sites are associated with carcinogenesis.
Conclusion & future perspective
Our results suggest that the SNPs, rs3087616 and rs4704846 of TIM-3/HAVCR2, involved in the pathways of immunoblastic escape, can be used as predictive factors for the clinical outcome in anticancer therapy. Our study contributes to the understanding of how the miRNA variants of the TIM-3/HAVCR23′UTR may regulate the expression of mRNA. Translation of pharmacogenetic predictors into clinical practice may lead to an improved planning and outcome of cancer treatment. In addition, further functional analysis is necessary to validate the promoter CpG islands and SNPs in the 3′UTR to allow for investigation of TIM-3/HAVCR2 gene regulation as a potential therapy for cancer.
TIM-3/HAVCR2 variants influence post-transcriptional regulation. miRNA-mediated regulation is important in cancer-associated gene expression, particularly with respect to the development of prognostic and diagnostic markers of malignancy. The 3′UTR variants could regulate the activity of TIM-3/HAVCR2 and may aid in targeting the TIM-3/HAVCR2 pathway for cancer treatment. However, the mechanism underlying the association of TIM-3/HAVCR2 transcriptional activity with the variants in the 3′UTR requires validation by functional analysis.
Executive summary.
The single nucleotide polymorphisms (SNPs) of TIM-3/HAVCR2 were identified by using an online database.
The bioinformatics tool SNP Function Prediction was used to predict the potential functional relevance of the miRNA binding sites.
Selected variants of the TIM-3/HAVCR2 3′UTR and putative miRNA binding sites.
Calculated the linkage disequilibrium of all SNPs in the TIM-3/HAVCR2 gene.
Our study contributes to the understanding of how the miRNA variants of the TIM-3/HAVCR2 3′UTR may regulate the expression of mRNA.
Footnotes
Authors’ contributions
F Chen designed the study, performed data analysis and interpreted the manuscript. Z Zhang performed the experiments and analyzed the data. F Pu wrote the manuscript. P Xia edited the manuscript for language quality. All the authors contributed to the study, and have read and approved the final manuscript.
Financial & competing interests disclosure
This work is supported by Grants from the Clinical Medical Research Project of Wuhan (number WX17Q38) and the Funded Research Project of Wuhan No.1 Hospital, Wuhan Integrated TCM & Western Medicine Hospital (number 2017Y01). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
- 1.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. 2004;4(11):850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 2.Zeljic K, Supic G, Jovic N, et al. Association of TLR2, TLR3, TLR4 and CD14 genes polymorphism with oral cancer risk and survival. 2014;20(4):416–424. doi: 10.1111/odi.12144. [DOI] [PubMed] [Google Scholar]
- 3.Serra M, Hattinger CM. Polymorphisms of genes related to methotrexate response and toxicity in high-grade osteosarcoma. 2017;13(1):123. doi: 10.1080/17425255.2017.1260300. [DOI] [PubMed] [Google Scholar]
- 4.Du W, Yang M, Turner A, et al. TIM-3 as a target for cancer immunotherapy and mechanisms of action. 2017;18(3) doi: 10.3390/ijms18030645. pii:E645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorman JV, Colgan JD. Regulation of T cell responses by the receptor molecule Tim-3. 2014;59(1–3):56–65. doi: 10.1007/s12026-014-8524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X, Yang J, He Y, Zhang J. Quantitative assessment of TIM-3 polymorphisms and cancer risk in Chinese Han population. 2016;7(24):35768–35775. doi: 10.18632/oncotarget.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Sun J, Gao W, et al. Preoperative Tim-3 expression on peripheral NK cells is correlated with pathologic TNM staging in colorectal cancer. 2017;15(6):3810–3818. doi: 10.3892/mmr.2017.6482. [DOI] [PubMed] [Google Scholar]
- 8.Shen P, Yue R, Tang J, et al. Preferential Tim-3 expression on Treg and CD8(+) T cells, supported by tumor-associated macrophages, is associated with worse prognosis in gastric cancer. 2016;8(8):3419–3428. [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Liu X, Wang X, et al. Polymorphisms in TIM-3 and breast cancer susceptibility in Chinese women: a case–control study. 2016;7(28):43703–43712. doi: 10.18632/oncotarget.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng G, Li M, Wu J, et al. Expression of Tim-3 in gastric cancer tissue and its relationship with prognosis. 2015;8(8):9452–9457. [PMC free article] [PubMed] [Google Scholar]
- 11.Fooladinezhad H, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. Negative regulation of TIM-3 expression in AML cell line (HL-60) using miR-330–5p. 2016;73(3):129–133. doi: 10.1080/09674845.2016.1194564. [DOI] [PubMed] [Google Scholar]
- 12.NCBI dbSNP Short Genetic Variations. www.ncbi.nlm.nih.gov/SNP/
- 13.NIH SNP Function Prediction. http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi
- 14.NIH SNP Function Prediction tool. http://snpinfo.niehs.nih.gov/snpfunc.htm
- 15.Geng H, Zhang GM, Li D, et al. Soluble form of T cell Ig mucin 3 is an inhibitory molecule in T cell-mediated immune response. 2006;176(3):1411–1420. doi: 10.4049/jimmunol.176.3.1411. [DOI] [PubMed] [Google Scholar]
- 16.Zhang G, Liu Z, Duan S, et al. Association of polymorphisms of programmed cell death-1 gene with chronic hepatitis B virus infection. 2010;71(12):1209–1213. doi: 10.1016/j.humimm.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Huang YJ, Niu J, Wei S, et al. A novel functional DEC1 promoter polymorphism −249T>C reduces risk of squamous cell carcinoma of the head and neck. 2010;31(12):2082–2090. doi: 10.1093/carcin/bgq198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank B, Hemminki K, Wirtenberger M, et al. The rare ERBB2 variant Ile654Val is associated with an increased familial breast cancer risk. 2005;26(3):643–647. doi: 10.1093/carcin/bgh342. [DOI] [PubMed] [Google Scholar]
- 19.Emamdoost F, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. The miR-125a-3p inhibits TIM-3 expression in AML cell line HL-60 in vitro . 2017;33(3):342–347. doi: 10.1007/s12288-016-0733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]