Abstract
The mitochondrial lipid cardiolipin (CL) contributes to the spatial protein organization and morphological character of the inner mitochondrial membrane. Monolysocardiolipin (MLCL), an intermediate species in the CL remodeling pathway, is enriched in the multisystem disease Barth syndrome. Despite the medical relevance of MLCL, a detailed molecular description that elucidates the structural and dynamic differences between CL and MLCL has not been conducted. To this end, we performed comparative atomistic molecular dynamics studies on bilayers consisting of pure CL or MLCL to elucidate similarities and differences in their molecular and bulk bilayer properties. We describe differential headgroup dynamics and hydrogen bonding patterns between the CL variants and show an increased cohesiveness of MLCL’s solvent interfacial region, which may have implications for protein interactions. Finally, using the coarse-grained Martini model, we show that substitution of MLCL for CL in bilayers mimicking mitochondrial composition induces drastic differences in bilayer mechanical properties and curvature-dependent partitioning behavior. Together, the results of this work reveal differences between CL and MLCL at the molecular and mesoscopic levels that may underpin the pathomechanisms of defects in cardiolipin remodeling.
Introduction
Cardiolipin (CL) is an unusual phospholipid with a headgroup consisting of two phosphatidic acid moieties bridged by a central glycerol and four acyl chain tails. In eukaryotes, CL localizes predominantly to the inner mitochondrial membrane (IMM) to a concentration of up to 20 mol% (1). CL plays a role in many organellar functions and has been shown to bind to a variety of mitochondrial proteins (2). Two important roles of CL involve maintaining the highly curved morphology of mitochondrial cristae (3, 4) and aiding the assembly of respiratory supercomplexes (5).
Under certain physiological conditions, CL can assume a negative spontaneous curvature that results from an inverted conical geometry because of a small headgroup size relative to the large volume occupancy of its four acyl chains. In prokaryotic cell membranes, this leads to enrichment of CL at the highly curved polar regions and fission sites of rod-like bacteria (6, 7, 8). The ionization state of the two phosphate moieties of CL has been a debated topic. Recent studies suggest that CL is doubly deprotonated at physiological pH and carries a charge of −2 (9, 10, 11, 12), though other literature indicates circumstances in which CL can carry a −1 charge (13, 14, 15, 16). The tuning of CL’s charge state will affect the headgroup charge repulsion and modulate molecular geometry. Indeed, the phase behavior and intrinsic curvature of CL depend on both pH and ionic concentration (17, 18, 19, 20, 21, 22, 23).
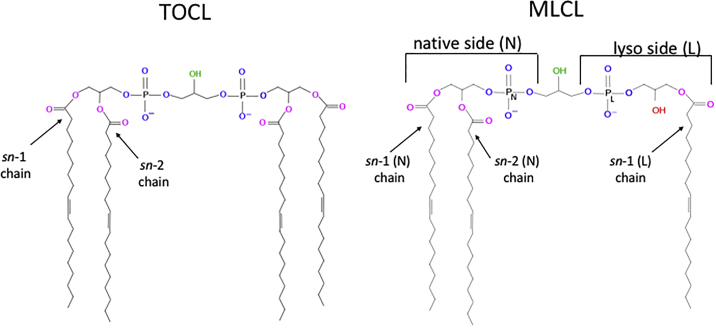
After the de novo biosynthesis of nascent CL within the IMM, the lipid undergoes a remodeling process to yield the mature CL fatty acid composition, consisting of four unsaturated hydrocarbon chains (24). CL is first deacylated by a CL-specific phospholipase (25, 26) that cleaves an acyl chain from a headgroup glycerol, rendering the triacyl species monolysocardiolipin (MLCL) with a hydroxyl group at the site of cleavage (Fig. 1). Multiple enzymes have been identified that catalyze the reacylation of MLCL. Most MLCL remodeling is driven by tafazzin, an MLCL transacylase encoded by the nuclear X-linked TAZ gene. TAZ deficiency causes accumulation of MLCL and abnormal fatty acid profiles of tetraacyl CL, which results in aberrant IMM morphology and disruption of the assembly and spatial organization of IMM protein complexes (3, 4). In humans, mutations in the TAZ gene lead to Barth syndrome, a multisystem disorder that clinically presents with cardiomyopathy, skeletal muscle weakness, growth retardation, and possibly death in young males (27).
Figure 1.
Structures of TOCL and MLCL. Hydrogen bond acceptors are colored (pink: ester oxygens, blue: phosphate oxygens, green: central hydroxyl, red: lyso hydroxyl of MLCL). MLCL is partitioned into the native (N) side with full acyl chain complement, and the lyso side (L) at which the sn-2 acyl chain is lysed. To see this figure in color, go online.
CL-containing bilayers have been well-characterized through a variety of biophysical studies, yielding information about ionization states, polymorphic phase behaviors, and lipid-protein interactions. In contrast, there is a relative dearth of experimental studies on the structural and dynamic properties of MLCL despite its clinical relevance. It has been shown that MLCL can modulate the activity of the apoptotic factor tBid (28) in a different manner than CL, and that MLCL-containing bilayers have a stronger preference for lamellar phases than those containing CL (29). Computational studies have also focused on the more prominent CL species. A number of computational molecular modeling and molecular dynamics (MD) studies have explored the structural and dynamic properties of tetraacyl CLs (30, 31, 32, 33, 34, 35), whereas to our knowledge only a single MD study has interrogated the physical characteristics of MLCL using a coarse-grained (CG) model to investigate the polymorphic phase behavior in homogeneous lipid systems (36).
The molecular scale differences between tetraacyl and triacyl CL species may impact bulk bilayer properties, which may give rise to different morphological characteristics of bilayers with differing CL-MLCL ratios. However, the molecular scale properties are yet to be thoroughly investigated. To study the local effects arising from molecular differences between CL and MLCL, we performed atomistic MD simulations on bilayers composed purely of tetraoleoylcardiolipin (TOCL) or trioleoylmonolysocardiolipin (hereby used synonymously with MLCL). We describe the headgroup orientation and acyl chain ordering resulting from tail number asymmetry. The orientation and hydrogen (H)-bonding partners of the lyso hydroxyl group are quantified. We characterize differences in headgroup cohesion between the species and suggest a mechanism that could explain the effects of MLCL on protein-lipid interactions. To study larger-scale bilayer mechanical properties, we performed CG-MD on heterogeneous bilayers mimicking the composition of the IMM, implementing a previously described buckling protocol (35) to examine trends in critical buckling pressure and curvature-based lipid segregation.
Methods
All-atom simulations
Atomistic MD simulations were performed using the GROMACS 5 package (37) and the CHARMM36 force field (38). Initially, a solvated bilayer consisting of 50 TOCL (−2e charge) molecules per leaflet was generated using the CHARMM-GUI input generator (39). The bilayer was solvated with 8482 water molecules, and 200 sodium ions were added to neutralize the total system charge. The TOCL system was used as the basis to generate the initial MLCL system. This was accomplished by replacing one sn-2 chain in each TOCL molecule by a hydroxyl group, as depicted in Fig. 1. The topology file was modified to accommodate the chain deletion, and the parameters chosen for the new hydroxyl group matched that of the central glycerol hydroxyl. A GROMACS-compatible topology file (.itp) for MLCL is included in the Supporting Materials and Methods.
Energy minimization and equilibration steps were performed according to the CHARMM-GUI guidelines, with steepest-descent minimization for 5000 steps followed by canonical ensemble equilibration for 50 ps with a timestep of 1 fs, followed by isothermal-isobaric ensemble equilibration with position restraints on the headgroup phosphorus atoms for 325 ps at a timestep of 2 fs, with semiisotropic pressure coupling accomplished using the Berendsen barostat. Both TOCL and MLCL bilayers were equilibrated using the same protocol.
Unrestrained MD simulations were run for 500 ns for both bilayers using a timestep of 2 fs. Temperature was maintained at 303 K using a Nose-Hoover thermostat (40, 41) with a coupling time constant of 1 ps. A semiisotropic pressure coupling scheme was implemented using the Parrinello-Rahman barostat (42) with a coupling constant of 5 ps. Bilayer compressibility was set to 4.5 × 10−5 bar−1, and the reference pressures both in the bilayer plane (X-Y) and normal (Z) to the bilayer were set to 1 bar to generate a zero-surface-tension ensemble. Van der Waals interactions were cut off at 1.2 nm, with the interactions modified using the force-switch method between 1.0 and 1.2 nm. Long-range electrostatics were calculated using the particle mesh Ewald (PME) method, with a real-space cutoff of 1.2 nm. The first 200 ns of unrestrained MD was considered further equilibration, leaving 300 ns for production MD to be analyzed. Bilayers were visually inspected using VMD 1.9 (43). Analyses of simulations were performed using a combination of GROMACS tools and in-house MATLAB (The MathWorks, Natick, MA) scripts. Acyl chain order parameters were calculated with the following equation:
| (1) |
in which θ for a given acyl carbon is defined as the angle between the bilayer normal and the vector originating at the acyl carbon and terminating at the acyl hydrogen.
Interdigitation of monolayer acyl chains was calculated by integrating the dimensionless overlap parameter ρov (44),
| (2) |
and
| (3) |
where ρt and ρb are the acyl densities along the z coordinate of the top and bottom leaflets respectively. Integrating the overlap parameter (Eq. 3) over the box z dimension (Lz) yields an interdigitation length, λov, which characterizes the extent of interleaflet interdigitation.
Lateral pressure profiles (LPPs) were calculated using GROMACS-LS (45), a modified version of GROMACS 4.5.5. The calculation of the LPPs are based on 100 ns of simulation data, with a save rate for position and velocities every 5 ps. Although the simulations were run using PME, GROMACS-LS does not permit PME and electrostatics must be calculated with a straight cutoff. The recommended cutoff for GROMACS-LS is 2.2 nm (45), but we found that a cutoff of 3.2 nm was necessary to observe convergence. Further discussion of GROMACS-LS parameter choices and convergence is presented in the Supporting Materials and Methods and Figs. S1–S3.
The output parameters of GROMACS-LS are the components of the stress tensor, σ, as a function of the z dimension (bilayer normal). The lateral and normal components of the pressure profile are and , respectively, and the LPP is given by the difference in these components, . The first moment of π(z) links the LPP to the bending modulus (kc) and spontaneous curvature (c0):
| (4) |
where the bilayer is centered at z = 0.
All reported error estimates are standard errors, calculated using block averaging. For each observable, appropriate block lengths were determined by scanning a range of block sizes and observing the effect of block size on estimated error.
Bilayer defect analysis
The hydrophobic core accessibility was analyzed in the atomistic simulations by a method similar to Vamparys et al. (46). The simulation box was divided into square grids with a grid spacing of 0.5 Å. The occupancy of each grid voxel was determined by first projecting the volume occupancy of the bilayer atoms, calculated with the volmap tool in VMD, onto the grid. Each X,Y grid position was then scanned from the top down until an occupied voxel was reached. The X,Y grid position was then assigned as either headgroup (if the first occupied grid encountered was a headgroup atom) or tail (if an acyl carbon). The headgroups included linking glycerols and all carbons bonded to oxygen, including the ester carbon that begins the acyl chains (see Fig. 1). This allowed us to construct a two-dimensional (2D) map indicating if the first grid box contacted would be occupied by the headgroup or tail. From this we could determine the fraction of the bilayer surface allowing access to acyl chains. To then discriminate small gaps in the headgroup coverage from larger “defects,” each grid point assigned as tail on the 2D surface was treated as the centerpoint of a spherical probe with a specific radius. If the grid point and all neighboring grid points within this radius were classified as tails, those grid points were then assigned as defects. In this way, only hydrophobic patches with sizes and shapes that could accommodate the spherical probe were classified as true defects.
CG simulations
CG simulations were conducted in GROMACS 5 using the Martini force field (47). A heterogeneous bilayer consisting of palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoylphosphatidylethanolamine (POPE), and TOCL was generated using the CHARMM-GUI Martini bilayer generator (48). To generate a model for MLCL, a single sn-2 connected chain was removed from the TOCL model. The glycerol bead type that was connected to the deleted tail was changed from Na to P1, matching the form that represents other lysolipids in the Martini lipid repository. The Martini representation of the TOCL headgroup consists of a central glycerol bead connected on both sides by a phosphate bead followed by two glycerol beads, each with one acyl chain attached (the “sn-2” equivalent chain attached to the glycerol bead proximal to the phosphate, and the “sn-1” equivalent chain attached to the terminal glycerol bead). To generate MLCL, the sn-2 chain on one side was removed and the attached glycerol bead type converted to a P1 polarity level, which matches other lysolipids found in the Martini lipid repository.
The original Martini parametrization of CL (taken from (30)) contains several harmonic angle potentials at the headgroup level, with three-atom angle combinations of phosphate-central glycerol-phosphate (PGP, 105°), central glycerol-phosphate-proximal glycerol (GPG, 110°), and phosphate-proximal glycerol-distal glycerol (PGG, 120°). We assumed the only angle potential that could be affected by removal of an acyl chain to be the PGG angle. Therefore, we ran equilibrium simulations of our CG systems both with and without the PGG potential and assessed the effect of the angle potential on interphosphate distances and headgroup tilt. We found no significant differences arising from removal of the potential (Fig. S4) and thus decided to keep the PGG potential for our CG model of MLCL. A GROMACS-compatible topology file (.itp) describing the Martini MLCL model is included in Supporting Materials and Methods.
CG simulations were run with a timestep of 30 fs. Lennard-Jones interactions were shifted to 0 at a cutoff of 1.1 nm using the potential-shift-Verlet implementation. Electrostatics were shifted to 0 at the same cutoff using the reaction field method. Temperature coupling was accomplished using the v-rescale thermostat, with a reference temperature of 303 K and a time constant of 1 ps. The Parinello-Rahman barostat was used to maintain constant pressure with a time constant of 12 ps. Reported simulations times are multiplied by 4 as per Martini convention (47).
CG buckling protocol
To examine mechanical stability and curvature-based partitioning of MLCL-containing CG bilayers, we applied a previously described methodology (35). Briefly, a bilayer consisting of 40% POPC, 40% POPE, and 20% MLCL is generated with dimensions of 30 nm × 10 nm in the XY (bilayer) plane. The specific lipid concentrations were chosen to allow for direct comparison to our previous study involving TOCL. Systems were fully solvated (>15,000 water beads in every simulation) and 324 sodium ions were added to the TOCL and MLCL-containing systems to neutralize the total charge. Lateral pressure in the long dimension is incrementally increased by 1 bar every 9 μs while the short lateral dimension is kept constant and the normal direction pressure maintained at 1 bar. At a certain applied lateral pressure, the bilayer adopts a highly curved buckled state to relieve the strain from the applied pressure. This 1-bar increment is then simulated again at 0.1-bar increments for 36 μs each to allow finer-tuned inspection of the buckling process. The extent of bilayer buckling was quantified by calculating the lateral compressional strain:
| (5) |
in which L is the length of the simulation box in the dimension of applied pressure and Lo is the original box length at 1 bar. Snapshots from the buckling process are then extracted and simulated in the canonical ensemble, allowing the box size to be fixed and control the curvature. The system is simulated at fixed curvatures for 9 μs. To determine the extent of curvature-based partitioning, the concentration of lipid constituents within each monolayer are then calculated as a function of the local curvature environment. The calculation of curvature is described in our previous work (35). Briefly, the headgroup coordinates of each monolayer are separately fit to a cubic smoothing spline function, and the curvature (C) at any point along the curve direction (y) is given by
| (6) |
where f′ and f″ are the first and second derivative of the monolayer height function with respect to the Y-dimension, respectively; C has units of inverse distance.
Results and Discussion
Headgroup geometry
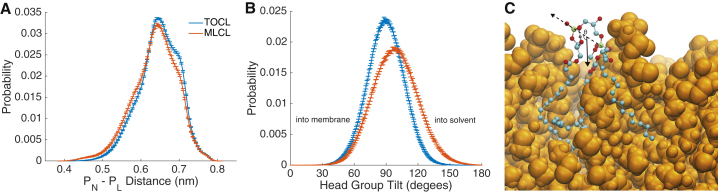
We begin with an investigation of the structural and dynamic molecular properties of MLCL in comparison to CL in homogeneous bilayers. The interphosphate distance has been used previously as a measure of the compactness of the CL central glycerol and has been used to parametrize the Martini CG CL model (30, 36). Fig. 2 A shows the interphosphorus atom distance distribution. The profiles are very similar, with MLCL displaying a slight shift toward shorter interphosphate distances.
Figure 2.
Geometry and orientation of headgroups. (A) The intramolecular distance between phosphorus atoms is given. (B) Headgroup tilt measured as the angle between the bilayer normal and the vector pointing from the PN phosphorus to the PL phosphorus is given. The legend in (A) also applies to (B), and error bars represent block-averaging-based standard errors. An example of an obtusely tilted (107.2°) headgroup is shown in (C). The dotted line is the PN-PL vector and the solid line is the downward Z-vector. All hydrogens are removed except for the lyso hydroxyl hydrogen, which is depicted in gray. The coloring of other atoms is as follows: phosphorous = lime, carbon = cyan, oxygen = red. The orange spheres represent other lipids in the bilayer. To see this figure in color, go online.
To determine if the acyl chain asymmetry of MLCL causes changes to the headgroup orientation, we calculated the angle between the phosphorous-phosphorous vector (P) and the bilayer normal, pointing into the membrane (Fig. 2 B). We defined the directionality of the P vector as initiating from the PN phosphorus (native side) and terminating at the PL phosphorus (lyso side); thus, an angle less than 90° indicates the lyso side tilts down into the membrane, and conversely, an obtuse angle indicates the lyso side tilts out of the membrane toward the solvent. The tilt angle definition and an example of a lipid with an obtuse tilt angle is presented in Fig. 2 C. As TOCL lacks a lyso side, the “lyso” phosphorous was assigned to be on the side from which an acyl chain was cleaved when converting to MLCL. As expected, the TOCL angle distribution was found to be symmetric around 90° (mean = 90.1° ± 0.1°, |skew| = 0.01). In contrast, the MLCL distribution was shifted toward obtuse angles (mean = 98.1° ± 0.2°, |skew| = 0.12), indicating the lyso side phosphate orients away from the hydrophobic core of the bilayer.
Lyso OH characterization
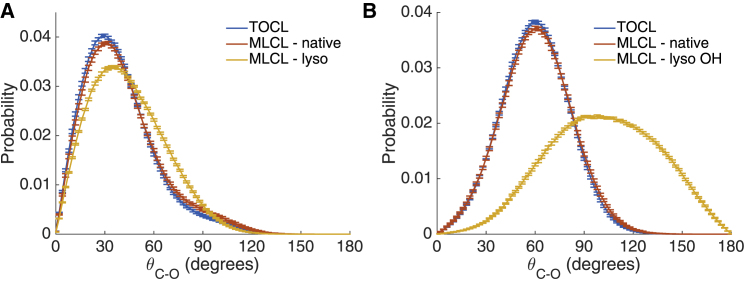
We next examined the behavior of the lyso hydroxyl group of MLCL and its interactions with proximal molecular components. Replacing a fatty-acid group with a hydroxyl group has several potential implications. First, the small hydroxyl group of MLCL has less steric bulk than the acyl chain of TOCL, which may allow for greater local conformational flexibility. Second, the polar hydroxyl group may tend to orient itself more toward the polar headgroup region rather than the hydrophobic core. Third, the addition of an additional hydrogen bond donor has potential implications for altering the intermolecular H-bonding network, which may result in changes to the structure and dynamics of the lipid, as the only H-bond donor in TOCL is the central glycerol hydroxyl. To determine the orientational preference of the hydroxyl group, we calculated the angle between the glycerol C-O bond vector (pointing from C to O) and the inward-pointing bilayer normal for both the sn-1 and sn-2 glycerol carbons on the lyso and native sides of TOCL and MLCL. A typical TOCL sn-1 C-O bond points sharply into the bilayer at an average angle of around 30° (Fig. 3 A), whereas a typical TOCL sn-2 C-O bond extends into the bilayer at an average angle of 60° (Fig. 3 B). The native side of MLCL displays a nearly identical distribution to that of TOCL for both sn-1 and sn-2 bonds. On the lyso side, the sn-1 angle is slightly shifted to higher angles, pointing less directly down into the membrane. As expected, the lyso hydroxyl (Fig. 3 B, yellow line) does not maintain a similar angle distribution to its counterpart ester bond on the native side. Instead, the distribution is very broad, reflecting an increase in rotational freedom of the significantly smaller hydroxyl group, and is centered around 100°, pointing toward the water-headgroup interface of the associated monolayer.
Figure 3.
Orientation of glycerol sn-1 and sn-2 C-O bond vectors. The angles between glycerol C-O bond vectors and bilayer normal for sn-1 chains (A) and for sn-2 chains and MLCL lyso OH (B) are shown. Error bars represent block-averaging-based standard errors. To see this figure in color, go online.
We determined the hydrogen bonding partners of the lyso hydroxyl group using the gmx hbond program. The results are displayed in Table 1 and are separated into self (intramolecular) and nonself (intermolecular) interactions. 25 ± 4.1% of the lyso hydroxyl groups formed intramolecular hydrogen bonds, primarily with the oxygens of the adjacent phosphate group, with smaller contributions from the linking ester groups. 28.1 ± 3.6% of lyso hydroxyl groups were found to create intermolecular hydrogen bonds with other MLCL molecules. A majority (17.5 ± 3.6%) of these H-bonds were formed with phosphate oxygens. The asymmetry between bonding to nonself PO4N and PO4L (7.1 ± 1.7% to 10.4 ± 3.2%) may be caused by headgroup tilting (Fig. 2 B), which positions the P04L groups closer to the water interface. Hydrogen bonds to water compose 31.4 ± 0.2% of lyso hydroxyl H-bonds. In sum, MLCL lyso OH groups participated (as donors) in intra- and intermolecular H-bonds 84.5 ± 5.5% of the time.
Table 1.
Hydrogen Bond Partners of Lyso OH
| H-Bond Acceptor | Self | Nonself | Total |
|---|---|---|---|
| lyso OH | N/A | 3.3 ± 0.2% | 3.3 ± 0.2% |
| Central OH | 0.9 ± 0.2% | 0.4 ± 0.2% | 1.3 ± 0.3% |
| PO4N | 0.9 ± 0.9% | 7.1 ± 1.7% | 8.0 ± 1.9% |
| PO4L | 20 ± 4% | 10.4 ± 3.2% | 30.4 ± 5.1% |
| ester O | 3.2 ± 0.1% | 6.9 ± 0.2% | 10.1 ± 0.2% |
| Water | N/A | 31.4 ± 0.2% | 31.4 ± 0.2% |
| Total | 25.0 ± 4.1% | 59.5 ± 3.6% | 84.5 ± 5.5% |
SEs were estimated using block averaging.
Only 13.4% ± 0.3 of the lyso hydroxyl hydrogen bonds were satisfied by H-bond acceptors at the lyso glycerol level (ester oxygens and other lyso OH groups), whereas the rest of the hydrogen bond acceptors are geometrically above the glycerol. The hydroxyl group must then point out of the membrane to access those hydrogen bond donors, which provides a plausible explanation for both the tilted headgroup (Fig. 2 B) and shifted angle distribution (Fig. 3 B). An example of a tilted headgroup with intramolecular H-bond between the lyso hydroxyl hydrogen and lyso phosphate oxygen is presented in Fig. 2 C.
Acyl chains
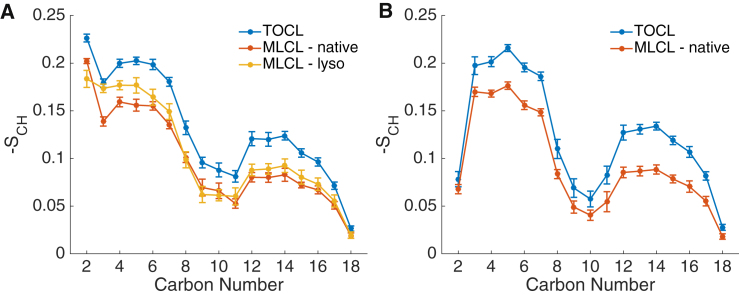
In addition to characterizing the MLCL headgroup structure, we performed analyses of the acyl chains. We focused not only on similarities and differences between TOCL and MLCL, but also on the asymmetry between the native and lyso sides of MLCL. To gain insight into the structure and flexibility of the sn-1 and sn-2 acyl chains, we calculated acyl chain order parameters (SCH, see Eq. 1), which measures the orientational mobility of C-H bonds at different positions along the acyl chain. We found that the SCH values of MLCL chains on the native side of the membrane reveal a depth-dependent pattern similar to those of TOCL, but with an overall decrease in order throughout the chain (Fig. 4).
Figure 4.
Acyl chain order parameters. −SCH order parameters along the sn-1 (A) and sn-2 (B) acyl chains for TOCL and MLCL, calculated using Eq. 1, are given. Error bars represent block-averaging-based standard errors. To see this figure in color, go online.
When comparing the native MLCL sn-1 to TOLC sn-1 (Fig. 4 A) or comparing the sn-2 chains between MLCL and TOCL (Fig. 4 B), SCH follows a similar pattern of decreasing order from the headgroup-proximal end toward the bilayer center. Although the magnitudes of the order parameters are uniformly reduced comparing TOCL to native-side MLCL, the lyso side sn-1 chain of MLCL differs significantly from the other trends. The lyso side sn-1 chain of MLCL displays increased ordering in headgroup-proximal chain positions compared to the native-side chain of MLCL, more closely matching the level of order in TOCL. We attribute this difference to the headgroup tilt shown in Fig. 2 B. As the lyso side of the headgroup tilts out of the headgroup plane, it pulls the acyl chains more into the hydrophilic interfacial region. This may cause the interface-proximal acyl chain components on the lyso side to adopt a more rigid conformation to keep the chain oriented toward the hydrophobic core of the bilayer.
Taken together, the H-bonding, chain order parameters, and MLCL headgroup geometry are likely related. When the lyso OH forms H-bonds between the central glycerol or phosphates, as it does 38.4% of the time, these interactions may cause the lyso side of MLCL to be pulled toward the solvent phase. The enthalpic gain from these additional H-bonds may compensate for increased chain ordering in the lyso side chain compared to the native-side chains in MLCL (Fig. 4 A). These analyses suggest a possible mechanism by which the presence of MLCL could disrupt lipid-lipid and lipid-protein interactions. Namely, the presence of the lyso OH group could disrupt the H-bonding patterns observed between TOCL and PC or PE lipids. Furthermore, the CL phosphates are known to mediate specific ionic contacts with IMM-proteins, which may be diminished/disrupted in the presence of high MLCL concentrations because the phosphate oxygens become partially occupied with H-bonds to the lyso OH groups.
Molecular packing geometry
To understand the molecular shape characteristics of MLCL and TOCL we considered both measurements based upon the bulk membrane properties and molecular scale analyses. We calculated the area per headgroup for both bilayers to determine to what extent this geometric parameter modulated molecular shape (Table 2). The area per lipid of TOCL was found to be 129.0 ± 0.3 Å2, in good agreement with the experimental value of 129.8 Å2 determined by Pan et al. using small-angle x-ray and neutron scattering (49). The area per lipid of MLCL was 109.0 ± 0.2 Å2, a reduction of 15.5%. Although the area per lipid arises from the properties of both the headgroup and tails, we have calculated the average area per acyl chain, which shows MLCL chains occupy a greater area than TOCL (36.3 Å2 ± 0.1 vs. 32.2 Å2 ± 0.1). However, the increase in area/tail in MLCL (13%) is not sufficient to compensate for the reduction in number of tails (25%) for the molecular shape of MLCL to be equivalent to TOCL. The increased area/tail in MLCL is caused by MLCL chains splaying to a greater extent than those of TOCL. The chain splay was analyzed directly by calculating the splay angle, measured between the vector connecting the central glycerol carbon with the terminal acyl chain carbon and the bilayer normal vector. From the splay angles, the splay distance was determined (as the projection of the splay vector onto the X-Y plane, Fig. S5). The average splay distance was increased in MLCL (9.6 ± 0.1 Å) compared to TOCL (9.1 ± 0.1 Å) by 5.5%. Because the tails in MLCL are sampling wider angle distribution, this results in a reduced hydrophobic thickness compared to TOCL (28.3 ± 0.1 to 30.9 ± 0.1 Å), measured as the distance between the average position of the ester carbons at the start of the acyl chains in each leaflet.
Table 2.
Geometric Properties of MLCL and TOCL
| Area per Lipid (Å2) | Area per Acyl Chain (Å2) | Hydrophobic Thickness (Å) | Interdigitation (Å) | c0 (1/Å) | |
|---|---|---|---|---|---|
| TOCL | 129.0 ± 0.3 | 32.2 ± 0.1 | 30.9 ± 0.1 | 5.49 ± 0.03 | −0.0012 ± 0.003 |
| MLCL | 109.0 ± 0.2 | 36.3 ± 0.1 | 28.3 ± 0.1 | 6.54 ± 0.04 | 0.020 ± 0.004 |
Area per lipid is calculated by dividing the average bilayer area by the number of lipids per leaflet. Area per acyl chain is the area per lipid divided by four for TOCL and three for MLCL. Hydrophobic thickness is the average distance between the first carbon in the acyl chains between leaflets. Standard errors were estimated using block averaging.
The increase in area per chain in MLCL is consistent with a decreased density of monolayer chain atoms. This decreased density may generate “hydrophobic gaps” at the interface of the monolayers, allowing for increased interdigitation between the lipid monolayers for MLCL. We calculated the extent of interdigitation for both bilayers (see Methods) and found that the λov increased by 18% from TOCL to MLCL (5.49 ± 0.03 Å and 6.54 ± 0.04 Å, respectively), indicating increased acyl interdigitation in the MLCL bilayer. The increased interdigitation presumably factors into the decreased hydrophobic thickness in MLCL as well.
The large reduction in total tail cross-sectional area may drastically change the shape of MLCL relative to TOCL, if the cross-sectional headgroup area is unchanged. This assumption is supported by the similar interphosphate distances shown in Fig. 2 A; however, the headgroup tilting (Fig. 2 B) would contradict a constant headgroup area assumption. We can estimate the effect of lipid tilt on headgroup area by treating the headgroup as a disk and calculating the projected area of the tilted disk onto a flat plane, where Aproj = Aflat × cos(θ−90°), where θ is the tilt angle. Based upon the probability distributions of θ (Fig. 2 B), TOCL has an Aproj = 0.837 × Aflat and MLCL has an Aproj = 0.733 × Aflat. Under the assumption that TOCL and MLCL have the same Aflat, which is supported by the similar interphosphate distance distributions (Fig. 2 A), the projected area of the MLCL is decreased by 12.5% compared to the projected headgroup area of TOCL. Although the tilting of MLCL does have a substantial effect on reducing the lipid headgroup area, the loss of an acyl chain has a more significant effect on altering the molecular geometry of MLCL (15.5% reduction in tail area vs. 12.5% reduction in headgroup area). Therefore, the reduction in tail area in MLCL is not fully compensated for by headgroup tilting, which we predict will result in a shifting of the MLCL molecular geometry toward a more cylindrical shape than TOCL.
LPPs
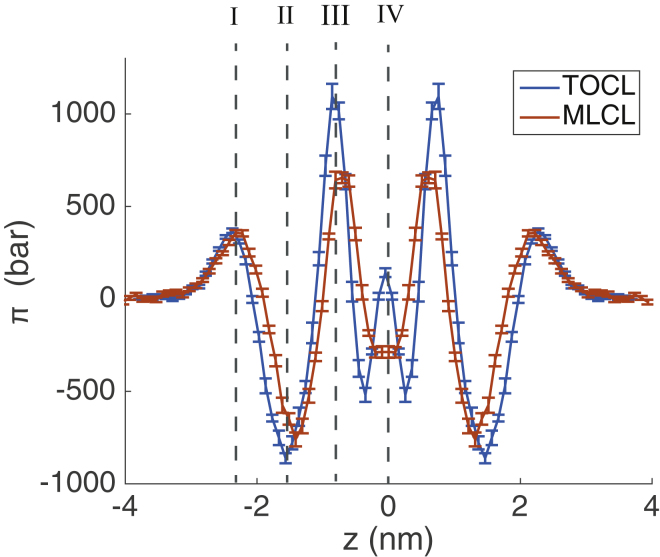
To assess the stress within the bilayers and estimate the sign of the spontaneous curvature, we calculated the LPPs for the MLCL and TOCL bilayers using GROMACS-LS (45). Fig. 5 presents the LPPs of the TOCL and MLCL bilayers, symmetrized about the bilayer center (z = 0). Moving from the bulk solvent toward the bilayer center, both LPPs display a moderate positive pressure peak (I) attributed to the repulsion in the headgroups, with the MLCL peak occurring at slightly lower z-position (closer to bilayer center) because of the difference in bilayer thickness. A large negative peak (II) is seen in both bilayers, indicative of the lipid-water interfacial tension, followed by another positive peak (III) closer to the center of the bilayer, which is typical of unsaturated lipids and has been demonstrated to align with the location of the acyl chain double bound (45). The negative interfacial tension peak (II) and positive double-bond peak (III) of MLCL are both smaller in magnitude than that of TOCL, indicating that the monolayers experience less lateral stress in those regions. In addition, the TOCL LPP has a positive peak at the bilayer center (IV) that MLCL lacks. This difference in lateral pressure can be rationalized in terms of the respective lipid geometries: the inverted conical shape of TOCL would lead to larger stresses at the tip of its acyl chains, whereas the cylindrical MLCL lacks such strain. The increased interdigitation in MLCL is consistent with the slightly negative pressure at the bilayer center as it promotes association of the two monolayer acyl chains.
Figure 5.
Lateral pressure profiles for TOCL and MLCL bilayers. The LPPs are calculated with GROMACS-LS, and the bilayer center is at z = 0. To see this figure in color, go online.
The first moment of the LPP is equal to the product of the monolayer bending modulus (kc) and spontaneous curvature (c0) according to Eq. 4. From our LPPs we calculated kcc0 to be −1.2 × 10−21 ± 3 × 10−21 J/nm for TOCL and 20.4 × 10−21 ± 4 ×10−21 J/nm for MLCL. The bending modulus will always be positive and therefore the sign of co is negative for TOCL and positive for MLCL. co cannot be determined without a confident estimate of the kc, and we caution that the obtained values of the first moment of the LPPs were highly dependent on the electrostatic cutoff used for analysis (see Fig. S3). However, the bending modulus of TOCL has been measured experimentally (49) to be ∼10−19 J, and if we assume this value of kc for both TOCL and MLCL, we can estimate the value c0. With this assumption of kc, we obtain c0s of −1.20 × 10−3 Å−1 and 2.0 × 10−2 Å−1 for TOCL and MLCL, respectively. Our observed c0 of TOCL is consistent within error with experimental measurements of TOCL in the absence of divalent cations (50). It is clear that removal of an acyl chain shifts the spontaneous curvature from a moderate negative value to a highly positive value. We note that the transition from small negative intrinsic curvature to high positive intrinsic curvature has also been observed when comparing spontaneous curvatures of phosphatidic acid and lysophosphatidic acid (51).
Lipid diffusion
We next investigated the spatial dynamics of MLCL to determine if the reduction in acyl chain numbers of MLCL increases lateral mobility. To this end, we calculated the 2D mean-squared displacement of both species at a range of lag times (Fig. S6) and extracted the diffusion coefficients from the slope of the curve using the equation , where d is the dimensionality (two in our case), D is the diffusion coefficient, and τ is the time lag. The lateral diffusion coefficient of TOCL was found to be 4.0 ± 0.01 × 10−8 cm2/s, and the diffusion coefficient of MLCL was 3.8 ± 0.1 × 10−8 cm2/s.
The assumption that lateral diffusion would increase with fewer acyl chains appears to be incorrect. The incorrectness of this assumption may be due to several factors. First, the increased hydrogen bonding capacity of the MLCL headgroup may bridge adjacent lipids, creating temporary networks with decreased motions. Second, we observe tighter packing of MLCL headgroups seen through reduced area per headgroup and greater interdigitation (Table 2). The increased interdigitation and tighter headgroup packing may lead to more drag on the lipids. It should be noted that artifacts due to periodic box size can impact observed diffusion rates, an effect that requires very large system sizes to fully dissipate (52, 53). However, this effect likely impacts observed diffusion rates to the same extent for both systems, and therefore the observed decreased mobility of MLCL should be valid for larger system sizes as well.
Acyl chain accessibility
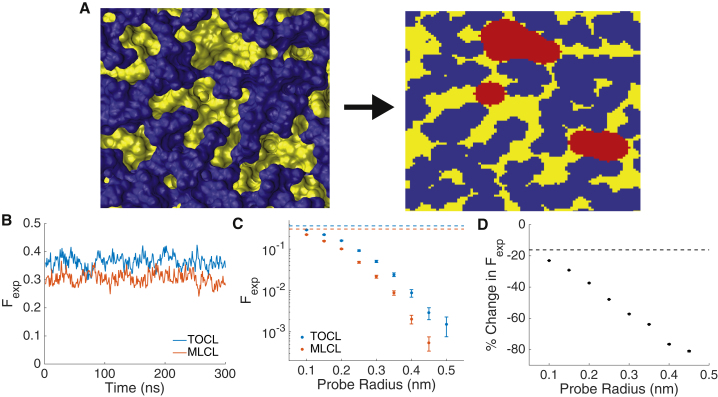
Area per lipid calculations indicate that the headgroups of MLCL pack tighter than those of TOCL. We reasoned that this tighter packing may lead to increased cohesion of the headgroup region and decreased accessibility of solvent to acyl chains. Such an effect may have an important effect on membrane-protein interactions. For example, Vanni et al. (54) showed through a combination of experimental techniques and MD simulations that the binding of the amphipathic packing lipid sensor (APLS) protein is dependent on the formation of “defects” in the headgroups of bilayers that expose the hydrophobic interior. To examine this possibility, we constructed 2D surface maps of our bilayers and assigned the surface as either headgroup or acyl chain, based upon the first atom that was encountered as a probe descended from above the bilayer (Fig. 6 A). A simple measurement of headgroup integrity is the fraction of the surface map that shows exposed acyl chain, Fexp (Fig. 6 B). We find that TOCL consistently displayed a larger Fexp (0.365 ± 0.003) than MLCL (0.306 ± 0.003).
Figure 6.
Bilayer defect analysis. (A) A top-down surface representation of a frame in the TOCL bilayer simulation is shown. Blue corresponds to headgroups and yellow to acyl chains. The right panel shows the 2D surface grid using the same color scheme. Red areas indicate defects found using a probe with radius 2.5 Å. (B) A time course of the total fraction of the 2D grid points assigned as acyl chain accessible is given. (C) The fraction of bilayer calculated to be a “defect” as a function of defect probe radius for TOCL and MLCL, is shown. The dotted reference lines are the averages of (B), and error bars represent block-averaging-based standard errors. (D) The percent change in fraction of membrane defined as a defect, from TOCL to MLCL, as a function of probe radius is given. The dotted line is the % change in total acyl accessibility from (B). To see this figure in color, go online.
Although a useful first indicator, total exposure of chains does not take into account the geometry of gaps in the headgroup. A gap may only be relevant for protein binding if it is large enough for the protein to sense. Indeed, Vanni and colleagues show that binding of APLS is dependent upon the insertion of aromatic side chains, and that a gap of sufficient size must be present for insertion (they estimate a defect area of at least 0.2 nm2 for the insertion of phenylalanine). To examine our surface maps for the presence of biologically relevant (larger) defects, at each acyl-exposed grid point, we examined the surrounding grid points in a given probe radius. If all surrounding grid points also had acyl chain exposure, the entire probed region was assigned as a defect (Fig. 6 A, red). The corresponding membrane fractions are presented in Fig. 6 C for a range of probe radii. As with total acyl exposure, the MLCL bilayer surface exposed fewer defects than the TOCL bilayer for all measured probe radii. For larger probe radii, the difference between the two bilayers grows larger. Fig. 6 D presents the percent decrease in defect exposure from the TOCL to MLCL bilayers. As the probe radius increases, the difference between them grows. We estimate that a relevant probe size for protein side-chain insertion is a radius of 2.5 Å (area of 0.196 nm2), at which the MLCL bilayer displays 47.9% fewer defects than TOCL. Thus, moderate differences in headgroup coverage can result in drastic differences in the presentation of defects on scales relevant to protein binding.
CL binds several peripheral proteins in addition to over 60 integral proteins (2). MLCL clearly provides a higher barrier than TOCL for such proteins attempting to interact with the hydrophobic interior of the bilayer. However, not all peripheral proteins interact with membranes via defect sensing. To our knowledge, the only comparative study of protein binding examining both CL and MLCL was with the apoptotic factor tBid, in which the affinity of the protein was greater for MLCL than for CL (28). This suggests that tBid-bilayer interactions may be headgroup-dependent rather than hydrophobic, in which case the modified headgroup dynamics or orientation may play a role in differential CL binding.
Mechanical properties
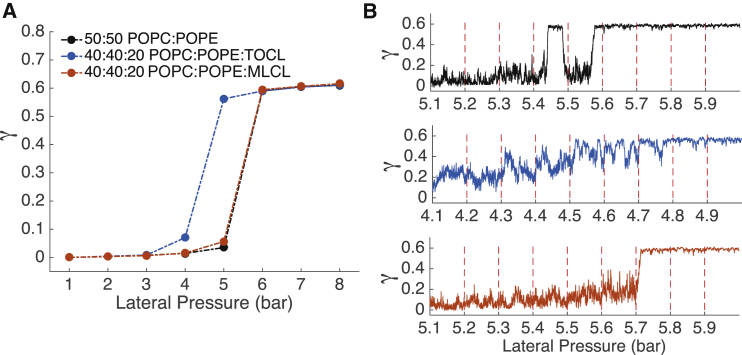
The membrane properties studied to this point have been molecular in scale, but the molecular differences between TOCL and MLCL may also manifest at larger length scales. In a recent study (35), we subjected bilayers containing CL to applied external pressures to induce buckling in a CG (Martini (47)) model. The buckling procedure provided insight into the mechanical properties of these bilayers and provided a platform for studying curvature-based demixing tendencies of the component lipids. To assess the effect of MLCL on these properties, we repeated our previous procedure, replacing TOCL with MLCL in a bilayer containing 40% POPC, 40% POPE, and 20% CL, which serves as a simplified model for the IMM. For reference, we also present data from a bilayer lacking CL, consisting of 50% POPC and 50% POPE. Fig. 7 A shows the effect of applied pressure on box compression, quantified by the compressional strain γ, described in Eq. 5. The changes in γ in the 4–6 bar range indicate buckling events, in which buckled bilayers have γ > 0.5. The TOCL-containing bilayer buckled at a lower pressure than the MLCL bilayer, indicating it is more susceptible to shape deformations from applied pressures.
Figure 7.
Buckling of CG bilayers. (A) The response of bilayers to induced lateral pressure is shown. γ = 0 corresponds to a flat bilayer and γ ∼ 0.6 to a maximally compressed bilayer. (B–D) Time courses of smaller incremental (0.1-bar) pressure increases are presented. Each pressure increment was run for 36 μs. The colors of the trends in (B–D) are consistent with the colors in (A) and therefore indicate the bilayer composition. Data for POPC:POPE and POPC:POPE:TOCL were previously reported in (35) and are reproduced here for comparison to MLCL-containing bilayers. To see this figure in color, go online.
The 1-bar pressure increments in Fig. 7 A led to complete buckling events; none of the simulations persist in an intermediately buckled state. To determine if stable intermediates are present along the buckling pathway, we performed simulations in 0.1-bar pressure increments along the previously determined buckling pathway (Fig. 7 B). We previously observed that TOCL-containing bilayers sampled intermediately buckled states, whereas we now find the MLCL-containing bilayer behaves in a two-state fashion, only sampling the completely flat and completely buckled states. For both buckling procedures, the MLCL-bilayer closely mimicked the properties of the POPC/POPE bilayer.
Bilayer deformability is crucial to maintaining the highly curved cristae membrane. A more rigid bilayer accrues a larger energetic penalty when forced into curved morphologies. The more brittle nature of the MLCL-containing bilayer may partially explain the abnormal morphologies commonly seen in tafazzin-deficient mitochondria. Further, the mitochondrion regulates its morphology and can rapidly effect changes in cristae structure to modulate metabolic rates (55, 56). Our data indicate that MLCL-containing bilayers are insensitive to small applied pressure changes. Although lateral pressure is used here as a computational tool and pressure changes of this magnitude are unlikely to be experienced in vivo, these observations may reveal important differences between MLCL and TOCL in modulating bilayer curvature in response to cellular stimuli. Supporting this idea, a 31P NMR study on CL phase behavior showed that although tetraacyl CL undergoes a lamellar (Lα) to inverted hexagonal phase (HII) transition under high-salt conditions, MLCL is restricted to the lamellar phase under similar conditions (29). The effect of monovalent salt likely causes a reduction in the headgroup area through direct binding and electrostatic screening. The shape of MLCL should be more cylindrical than TOCL, and therefore a greater reduction in the headgroup area would be required to induce an Lα to HII conversion. Furthermore, the orientation of the MLCL headgroup, with the phosphates adopting positions out of the bilayer-water interface plane, may result in less sensitivity to ionic screening.
Curvature-based partitioning
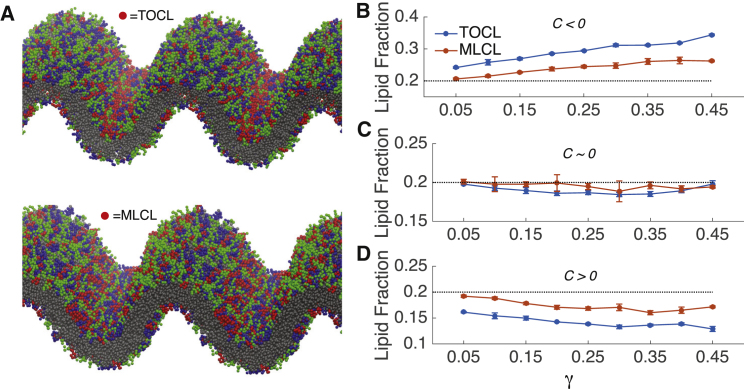
Our analysis of bilayer buckling is a valuable tool to study curvature-dependent lipid sorting, as buckled membranes contain regions of both positive and negative curvature. Previously we have shown that TOCL partitions strongly to regions of negative curvature (35). We extracted frames from the buckling trajectories of the systems containing 40% POPC, 40% POPE, and 20% TOCL/MLCL and simulated them without pressure coupling, fixing the box size and therefore the extent of curvature. The simulations were run for 9 μs, and the concentrations of lipids at positive and negative curvature (C) were determined. Fig. 8 displays a visual representation of the TOCL/MLCL partitioning in a buckled bilayer, as well as quantification of the concentrations of TOCL and MLCL in different curvature regimes. The bilayer snapshots in Fig. 8 A, illustrate the strong partitioning of TOCL to negative curvature regions, whereas MLCL does not exhibit the same degree of partitioning. Curvature-based lipid partitioning is determined in negative-curvature regions (C < −0.05 nm−1, Fig. 8 B), in flat regions (0.05 nm−1 > C > −0.05 nm−1, Fig. 8 C), and positive-curvature regions (C > 0.05 nm−1, Fig. 8 D). MLCL partitioning to regions of negative curvature is greatly reduced at all buckling magnitudes (Fig. 8 B), and depletion of MLCL is reduced at regions of positive curvature (Fig. 8 D) when compared to TOCL. We observe that the fraction of TOCL or MLCL in the flat regions (Fig. 8 C) match the bulk concentration of 20% for both systems.
Figure 8.
Curvature-driven segregation of TOCL and MLCL. (A) Snapshots from the final frame of the fixed-box γ = 0.30 simulation illustrate partitioning propensities. TOCL and MLCL headgroups are colored red, POPC headgroups are colored green, POPE headgroups are colored blue, and all tail beads are colored gray. The compositions of the bilayers contain 40% POPC, 40% POPE, and 20% TOCL (top) or 20% MLCL (bottom). (B–D) Concentrations of TOCL and MLCL in bilayers fixed at compression levels ranging from γ = 0.05 to γ = 0.45 are given. (B) Concentrations in regions of high negative curvature (C < −0.05 nm−1), (C) concentrations in regions with low curvature (C > −0.05 nm−1, C < 0.05 nm−1), and (D) concentrations in regions of high positive curvature (C > 0.05 nm−1) are shown on the bottom. The dotted line is a reference to the bulk concentration (20%) of TOCL/MLCL in these bilayers. Error bars represent block-averaging-based standard errors. To see this figure in color, go online.
Molecular geometry is the main determinant of lipid spontaneous curvature; therefore, reduction of curvature-based partitioning in our CG system suggests that MLCL is more cylindrical than TOCL, in agreement with calculations from the atomistic simulations. Furthermore, we do not believe these are model-dependent observations, as the molecular geometry is well-preserved in the CG model. The lipid-headgroup-tilt distributions are highly similar between the atomistic and CG models for both TOCL and MLCL (Fig. S4). Membrane bending energetics depend on both the pliability (bending modulus) and spontaneous curvature of the lipid bilayer constituents, so both aspects examined in these CG simulations may play a role in differential mitochondrial morphology.
The effects of MLCL on cristae curvature may also go beyond simple bending energetics. A variety of proteins shape the IMM, including ATP synthase, which self-organizes into rows of dimers along the cristae tips where regions of high curvature are apparent (57). This organization is disrupted in tafazzin-deficient mitochondria (4). CL may be essential in maintaining these dimer rows, in which case the impact of MLCL extends to disrupting curvature-generating proteins. Alternatively, the organization of ATP synthase may depend on preexisting curved morphologies which are maintained by CL.
Conclusions
In summary, we performed MD simulations on multiple scales to elucidate molecular- and bulk-scale similarities and differences between TOCL and MLCL. Through atomistic simulations, we characterized differences in headgroup tilt, acyl chain dynamics, and solvent accessibility to the hydrophobic bilayer core, and determined the orientation and H-bonding partners of the lyso hydroxyl group of MLCL. The potential implications of these molecular-scale effects may include disruption of specific lipid-lipid patterning and direct lipid-protein interactions. Molecular geometry analysis was performed on the atomistic simulations, which suggested that the molecular shape of MLCL is more cylindrical than the inverted-conical shape of TOCL. However, the change in the molecular geometry of MLCL may be slighter than what would be predicted based solely on the loss of an acyl chain. The headgroup tilting of MLCL results in a smaller headgroup area and partially compensates for the tail-area reduction from the acyl chain reduction. Nonetheless, the effect of replacing TOCL with MLCL is profound, as we observed in CG simulations. The CG simulations showed that MLCL-containing bilayers are more resistant to shape changes upon applied lateral pressure, and that in buckled bilayers MLCL showed reduced partitioning to regions of negative curvature. The differences in the molecular and bilayer-scale properties of MLCL compared to TOCL may be important factors in presentation of aberrant mitochondrial morphologies associated with disease states such as Barth syndrome.
Author Contributions
K.J.B., N.N.A., and E.R.M., designed the research. K.J.B. performed the research. K.J.B. and E.R.M. analyzed the data. K.J.B., N.N.A., and E.R.M. wrote the manuscript.
Acknowledgments
This research has been supported by the National Institutes of Health through grants R35-GM1197623 (E.R.M.) and 1R01-GM113092 (N.N.A.) and the National Science Foundation through a graduate research fellowship, grant #1247393 (K.J.B.). Computational resources for this project were provided by the University of Connecticut University Information Technology Services through access to the Hornet High Performance Computing cluster.
Editor: Emad Tajkhorshid.
Footnotes
Supporting Materials and Methods, six figures, and two data files are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30436-3.
Supporting Material
References
- 1.Horvath S.E., Daum G. Lipids of mitochondria. Prog. Lipid Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Planas-Iglesias J., Dwarakanath H., Klein-Seetharaman J. Cardiolipin interactions with proteins. Biophys. J. 2015;109:1282–1294. doi: 10.1016/j.bpj.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acehan D., Khuchua Z., Schlame M. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9:86–95. doi: 10.1016/j.mito.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acehan D., Malhotra A., Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 2011;100:2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeiffer K., Gohil V., Schägger H. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 6.Kawai F., Shoda M., Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J. Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mileykovskaya E., Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romantsov T., Helbig S., Wood J.M. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 9.Kooijman E.E., Swim L.A., Kagan V.E. Magic angle spinning 31P NMR spectroscopy reveals two essentially identical ionization states for the cardiolipin phosphates in phospholipid liposomes. Biochim. Biophys. Acta. 2017;1859:61–68. doi: 10.1016/j.bbamem.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malyshka D., Pandiscia L.A., Schweitzer-Stenner R. Cardiolipin containing liposomes are fully ionized at physiological pH. An FT-IR study of phosphate group ionization. Vib. Spectrosc. 2014;75:86–92. [Google Scholar]
- 11.Olofsson G., Sparr E. Ionization constants pKa of cardiolipin. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathappa M., Alder N.N. The ionization properties of cardiolipin and its variants in model bilayers. Biochim. Biophys. Acta. 2016;1858:1362–1372. doi: 10.1016/j.bbamem.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hielscher R., Wenz T., Hellwig P. Monitoring the redox and protonation dependent contributions of cardiolipin in electrochemically induced FTIR difference spectra of the cytochrome bc(1) complex from yeast. Biochim Biophys Acta. 2009;1787:617–625. doi: 10.1016/j.bbabio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Kates M., Syz J.Y., Haines T.H. pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 15.Nichols-Smith S., Kuhl T. Electrostatic interactions between model mitochondrial membranes. Colloids Surf. B Biointerfaces. 2005;41:121–127. doi: 10.1016/j.colsurfb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Sidiq S., Verma I., Pal S.K. pH-driven ordering transitions in liquid crystal induced by conformational changes of cardiolipin. Langmuir. 2015;31:4741–4751. doi: 10.1021/acs.langmuir.5b00798. [DOI] [PubMed] [Google Scholar]
- 17.Arslan P., Beltrame M., Muscatello U. Ultrastructural characterization of cardiolipin liquid-crystalline structures in the absence and the presence of divalent cations. Micron. 1980;11:115–125. [Google Scholar]
- 18.Khalifat N., Puff N., Angelova M.I. Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys. J. 2008;95:4924–4933. doi: 10.1529/biophysj.108.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalifat N., Fournier J.B., Puff N. Lipid packing variations induced by pH in cardiolipin-containing bilayers: the driving force for the cristae-like shape instability. Biochim. Biophys. Acta. 2011;1808:2724–2733. doi: 10.1016/j.bbamem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Killian J.A., Koorengevel M.C., de Kruijff B. Effect of divalent cations on lipid organization of cardiolipin isolated from Escherichia coli strain AH930. Biochim. Biophys. Acta. 1994;1189:225–232. doi: 10.1016/0005-2736(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 21.de Kruijff B., Verkleij A.J., Rijnbout H. Further aspects of the Ca2+-dependent polymorphism of bovine heart cardiolipin. Biochim. Biophys. Acta. 1982;693:1–12. doi: 10.1016/0005-2736(82)90464-3. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz A., Killian J.A., Wilschut J. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys. J. 1999;77:2003–2014. doi: 10.1016/S0006-3495(99)77041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seddon J.M., Kaye R.D., Marsh D. Induction of the lamellar-inverted hexagonal phase transition in cardiolipin by protons and monovalent cations. Biochim. Biophys. Acta. 1983;734:347–352. [Google Scholar]
- 24.Lu Y.W., Claypool S.M. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front. Genet. 2015;6:3. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beranek A., Rechberger G., Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J. Biol. Chem. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu Y.H., Dumlao D.S., Dennis E.A. Assessing phospholipase A2 activity toward cardiolipin by mass spectrometry. PLoS One. 2013;8:e59267. doi: 10.1371/journal.pone.0059267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlame M., Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 28.Esposti M.D., Cristea I.M., Dive C. Proapoptotic Bid binds to monolysocardiolipin, a new molecular connection between mitochondrial membranes and cell death. Cell Death Differ. 2003;10:1300–1309. doi: 10.1038/sj.cdd.4401306. [DOI] [PubMed] [Google Scholar]
- 29.Powell G.L., Marsh D. Polymorphic phase behavior of cardiolipin derivatives studied by 31P NMR and X-ray diffraction. Biochemistry. 1985;24:2902–2908. doi: 10.1021/bi00333a013. [DOI] [PubMed] [Google Scholar]
- 30.Dahlberg M., Maliniak A. Mechanical properties of coarse-grained bilayers formed by cardiolipin and zwitterionic lipids. J. Chem. Theory Comput. 2010;6:1638–1649. doi: 10.1021/ct900654e. [DOI] [PubMed] [Google Scholar]
- 31.Dahlberg M., Maliniak A. Molecular dynamics simulations of cardiolipin bilayers. J. Phys. Chem. B. 2008;112:11655–11663. doi: 10.1021/jp803414g. [DOI] [PubMed] [Google Scholar]
- 32.Aguayo D., González-Nilo F.D., Chipot C. Insight into the properties of cardiolipin containing bilayers from molecular dynamics simulations, using a hybrid all-atom/united-atom force field. J. Chem. Theory Comput. 2012;8:1765–1773. doi: 10.1021/ct200849k. [DOI] [PubMed] [Google Scholar]
- 33.Pöyry S., Róg T., Vattulainen I. Mitochondrial membranes with mono- and divalent salt: changes induced by salt ions on structure and dynamics. J. Phys. Chem. B. 2009;113:15513–15521. doi: 10.1021/jp905915m. [DOI] [PubMed] [Google Scholar]
- 34.Róg T., Martinez-Seara H., Vattulainen I. Role of cardiolipins in the inner mitochondrial membrane: insight gained through atom-scale simulations. J. Phys. Chem. B. 2009;113:3413–3422. doi: 10.1021/jp8077369. [DOI] [PubMed] [Google Scholar]
- 35.Boyd K.J., Alder N.N., May E.R. Buckling under pressure: curvature-based lipid segregation and stability modulation in cardiolipin-containing bilayers. Langmuir. 2017;33:6937–6946. doi: 10.1021/acs.langmuir.7b01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahlberg M. Polymorphic phase behavior of cardiolipin derivatives studied by coarse-grained molecular dynamics. J. Phys. Chem. B. 2007;111:7194–7200. doi: 10.1021/jp071954f. [DOI] [PubMed] [Google Scholar]
- 37.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 38.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 41.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 42.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Das C., Noro M.G., Olmsted P.D. Simulation studies of stratum corneum lipid mixtures. Biophys. J. 2009;97:1941–1951. doi: 10.1016/j.bpj.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanegas J.M., Torres-Sánchez A., Arroyo M. Importance of force decomposition for local stress calculations in biomembrane molecular simulations. J. Chem. Theory Comput. 2014;10:691–702. doi: 10.1021/ct4008926. [DOI] [PubMed] [Google Scholar]
- 46.Vamparys L., Gautier R., Fuchs P.F. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 2013;104:585–593. doi: 10.1016/j.bpj.2012.11.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 48.Qi Y., Ingólfsson H.I., Im W. CHARMM-GUI Martini Maker for coarse-grained simulations with the Martini force field. J. Chem. Theory Comput. 2015;11:4486–4494. doi: 10.1021/acs.jctc.5b00513. [DOI] [PubMed] [Google Scholar]
- 49.Pan J., Cheng X., Katsaras J. Structural and mechanical properties of cardiolipin lipid bilayers determined using neutron spin echo, small angle neutron and X-ray scattering, and molecular dynamics simulations. Soft Matter. 2015;11:130–138. doi: 10.1039/c4sm02227k. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y.F., Tsang K.Y., Fan Z.A. Differential dependencies on [Ca2+] and temperature of the monolayer spontaneous curvatures of DOPE, DOPA and cardiolipin: effects of modulating the strength of the inter-headgroup repulsion. Soft Matter. 2015;11:4041–4053. doi: 10.1039/c5sm00577a. [DOI] [PubMed] [Google Scholar]
- 51.Kooijman E.E., Chupin V., Rand P.R. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- 52.Camley B.A., Lerner M.G., Brown F.L. Strong influence of periodic boundary conditions on lateral diffusion in lipid bilayer membranes. J. Chem. Phys. 2015;143:243113. doi: 10.1063/1.4932980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venable R.M., Ingólfsson H.I., Pastor R.W. Lipid and peptide diffusion in bilayers: the Saffman-Delbrück model and periodic boundary conditions. J. Phys. Chem. B. 2017;121:3443–3457. doi: 10.1021/acs.jpcb.6b09111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanni S., Vamparys L., Antonny B. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys. J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cogliati S., Frezza C., Scorrano L. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lizana L., Bauer B., Orwar O. Controlling the rates of biochemical reactions and signaling networks by shape and volume changes. Proc. Natl. Acad. Sci. USA. 2008;105:4099–4104. doi: 10.1073/pnas.0709932105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies K.M., Strauss M., Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.