Abstract
Single-molecule force spectroscopy makes it possible to measure the mechanical strength of single noncovalent receptor-ligand-type bonds. A major challenge in this technique is to ensure that measurements reflect bonds between single biomolecules because the molecules cannot be directly observed. This perspective evaluates different methodologies for identifying and reducing the contribution of multiple molecule interactions to single-molecule measurements to help the reader design experiments or assess publications in the single-molecule force spectroscopy field. We apply our analysis to the large body of literature that purports to measure the strength of single bonds between biotin and streptavidin as a demonstration that measurements are only reproducible when the most reliable methods for ensuring single molecules are used.
Introduction
Single-molecule force spectroscopy (SMFS) has provided the means to examine force-dependent properties of biomolecular complexes. Applications include but are not limited to the determination of the strength of receptor-ligand bonds (1, 2, 3), viscoelastic properties of molecules such as protein unfolding or polymer stretching (4, 5), and the characterization of force generation by molecular motors (6, 7). A major challenge for the reliability of these measurements is that the molecules being tested cannot be seen directly, so it is not directly evident if the strength of a single molecule or multiple molecules is being measured. Early in the development of SMFS, some investigators used a method we refer to as “low-adhesion probability,” in which most of the tests report no adhesive force, and therefore, no molecules being tested, so statistical theory argues that the remaining measurements should reflect mostly single molecules (1, 8). Others tried to extract parameters describing single molecules from data on many molecules. It rapidly became clear that the low-adhesion probability method was valid, whereas extraction from multiple molecules was not (9). In the decades since then, however, many additional methods have been proposed to determine single-molecule characteristics in SMFS. This review analyzes the reliability of these methods when they are used to test the strength of single receptor-ligand bonds. This should help researchers both plan their own SMFS experiments and critically evaluate publications.
Use of SMFS to measure bond strength
SMFS is usually performed with the atomic force microscope (AFM), optical trap (OT), magnetic tweezer (MT), or biomembrane force probe (BMFP). The first three of these instruments have been well reviewed (10, 11), and a description of the BMFP can be found in Gourier et al (12). Although these instruments have many differences in form and operation, the procedures for gathering single-bond measurements have common principles.
In general, a probe is coated with a biomolecule or receptor, and a separate surface is coated with a complimentary biomolecule or ligand. Ideally, the biomolecules will be strongly attached to the probe or surface through a known anchor point. Physical adsorption and covalent binding through ubiquitous functional groups like free amines provide high mechanical strength but only provide a known anchor point if the structure of interest is part of a much larger molecule or complex. Anchoring via noncovalent binding to a specific epitope provides a known anchor point, but the anchor may break under the forces needed to test the biomolecule of interest. Both strength and a known anchor point are provided by genetic fusion of the protein of interest with anchors such as cysteine, which covalently binds maleimide (13), or proteins such as SNAP (14) or Halo (15) that enzymatically attach themselves to a target. Regardless of the method of attachment, nonspecific interactions must be blocked (16), and negative controls should be performed to demonstrate that interactions are specific to the immobilized biomolecules.
In each force cycle, the probe is pressed to the surface to allow bonds to form and then retracted until bond rupture (Fig. 1). In dynamic force spectroscopy (DFS), the measured force increases at a loading rate that depends on the retraction velocity and the elastic properties of the probe and molecules. In constant force spectroscopy (CFS), a constant position or a force-feedback loop is used to maintain a constant force. The AFM, OT, and BMFP traditionally use DFS, in which the probe position is controlled while the force on the probe is measured, although a closed feedback loop can be used to control force. In contrast, the MT uses a magnetic field to apply a known force to magnetic beads (probes) that easily supports CFS and can support DFS if the magnetic field can be dynamically changed.
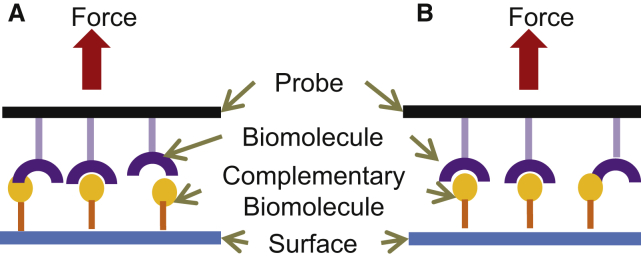
Figure 1.
(A) A conceptual illustration showing how single (A) or multiple (B) bonds might form when a probe touches a surface. To see this figure in color, go online.
Because single-molecule behaviors are intrinsically stochastic, hundreds or even thousands of force cycles must be analyzed at each condition of force or loading rate to characterize the molecular response to force. Biophysical models are then fit to these large data sets to extract a concise description of the properties of the single biomolecules.
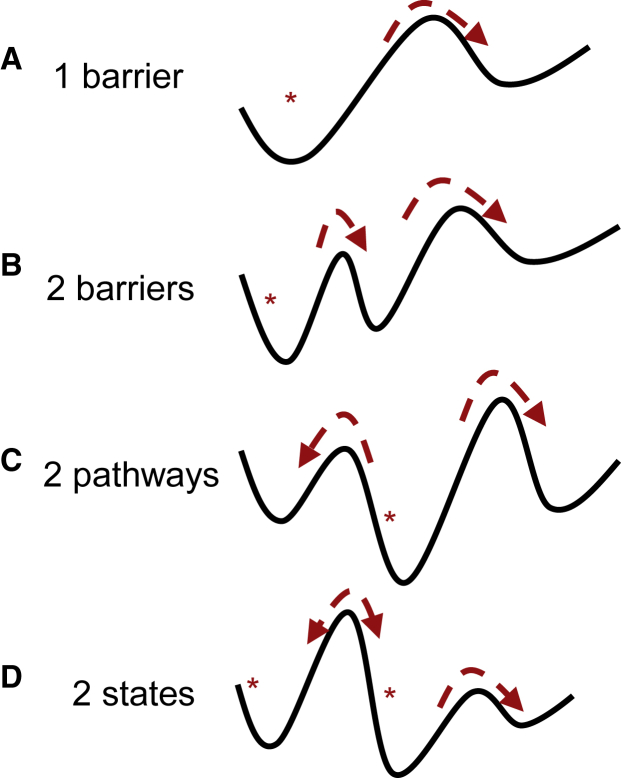
The simplest “one barrier” model for bond rupture assumes that the bond must overcome a single energy barrier to unbind (Fig. 2 A). The rate of transition over the energy barrier is exponentially enhanced by force, and methods to determine the parameters of the energy barrier from DFS or CFS data are well described elsewhere (8, 17, 18). However, it has become clear that molecular bonds often have more complex energy landscapes. Even molecules such as biotin-streptavidin that are not regulated by force in vivo may have a complex response to force (1) that can be explained by more than one energy barrier on the unbinding pathway (1, 19) (Fig. 2 B), whereas other molecules form catch bonds that are activated by force and can have two unbinding pathways (20, 21) (Fig. 2 C), and proteins may exhibit two slowly exchanging bound states because of allostery (22, 23) (Fig. 2 D). Indeed, even a single pathway may be distorted by force in a way that provides complex mechanical responses (24). Similar complexities may also occur for protein unfolding (25, 26). Mathematical models for these more complex energy landscapes include differential equations with terms representing force-dependent transitions for each energy barrier (27, 28, 29). Models for complex landscapes often predict multiple force peaks in DFS or multiple lifetimes in CFS (27, 28, 29). This complexity in the data can complicate efforts to ensure that measurements reflect single bonds. Indeed, we argue that if a method is developed to reduce the effects of multiple bonds, assuming a simple energy landscape such as that of Fig. 2 A, it cannot be assumed to be valid for a new biomolecule with an unknown energy landscape.
Figure 2.
Energy landscapes for bond rupture, or other force-regulated molecular transitions. In (A)–(D), the lines indicate the energy landscapes, with the length of the bond indicated by the horizontal direction and the energy indicated by the vertical. The asterisks (∗) indicate the bound state(s) of the bond that are observed before the application of force. The dashed arrows highlight the transitions that the bond can undergo with or without force; transitions with arrows pointing to the right involve elongation and are thus induced by force, whereas transitions toward the left are inhibited by force. To see this figure in color, go online.
Low adhesion probabilities
The most commonly used method to ensure that measurements reflect single bonds is to use conditions in which the fraction of force cycles that result in measurable adhesion, referred to as the adhesion probability, or , is much less than one. If each bond forms independently, then Poisson statistics can be used to estimate , the probability of forming exactly one bond in any given force cycle, i.e., the efficiency, from the measured adhesion probability (30):
| (1) |
The error in these measurements is the probability of forming multiple bonds in a force cycle, :
| (2) |
The accuracy of the measurements is the fraction of measured adhesive events that are actually single bonds, or .
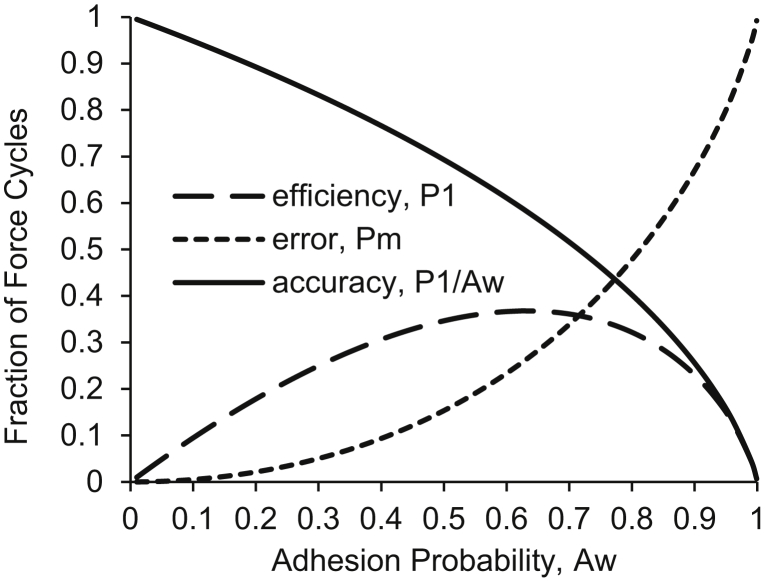
To evaluate or design SMFS experiments, it is useful to consider how the efficiency , the error , and the accuracy vary with the adhesion probability . As illustrated in Fig. 3, the peak efficiency is at an adhesion probability of 63%, where 37% of force cycles result in single bonds, but 26% result in multiple bonds, for an accuracy of only 58%. It may be considered acceptable to reduce the adhesion probability to 35%, where 28% of force cycles form single bonds and only 7% form multiple bonds, for an accuracy of 80%. More desirable would be an adhesion probability of 20% such that single bonds represent 18% of force cycles, and multiple bonds represent 2%, for an accuracy of nearly 90%.
Figure 3.
Prediction using Poisson statistics of the efficiency , error , and accuracy of SMFS experiments as a function of adhesion probability .
The key assumption in Poisson statistics is that bonds form independently, meaning that the formation of one bond does not increase the chance that a second will form. This assumption can fail if low adhesion probabilities are obtained by reducing the site density of multivalent biomolecules. An implicit assumption is that all bonds result in a measurable interaction. This can fail for weak bonds and, in some force loading conditions, for bonds with complex energy landscapes, like catch bonds (31). For this reason, the prevalence of multiple bonds should be estimated by using the force-loading conditions that result in the highest apparent adhesion probability. If these two concerns are addressed, low adhesion probabilities continue to provide a simple and robust technique for SMFS.
However, the marginal 80–90% accuracy of the method limits interpretation, especially with complex energy landscapes. For example, the biphasic strength of bonds with two states may be mistaken for the biphasic strength of single versus multiple bonds or vice versa. Even if one is willing to sacrifice efficiency, accuracy is limited by nonspecific adhesion, which is not addressed in Fig. 3. That is, when the adhesion probability is reduced to the level of nonspecific adhesion, accuracy decreases again. For these reasons, researchers and readers should ask whether the expected prevalence of multiple bonds might affect the conclusions drawn about single bonds.
Filtering using mechanical fingerprints
It would be clearly desirable to identify and filter out the individual force cycles that result in multiple bonds. Some investigators have filtered out the highest force ruptures up to the number of multiple bonds predicted by Poisson statistics (30). This may be valid for molecules with simple energy landscapes but would be inappropriate for molecules with two states because data reflecting the stronger state would be filtered out. This method should therefore not be used for molecules with unknown properties. A better approach is to filter out force cycles with multiple rupture events (3, 32, 33). However, this assumes that two rupture events will be spatially separated, which often fails when the molecules are short or when adhesion probability is high (34, 35). Nevertheless, this simple filtering method can improve the accuracy of experiments obtained with low adhesion probabilities, especially if the fraction of data filtered out correspond with the statistical predictions.
To provide a more distinct mechanical fingerprint to separate single from multiple bonds, the biomolecule of interest may be coupled to a marker molecule with a known force-extension signature (36, 37, 38, 39). Because the marker molecule extends a relatively long distance, the biomolecule of interest can bind far from the surface, which reduces or allows identification of nonspecific interactions (40, 41, 42). Three different configurations for using mechanical fingerprints are discussed below.
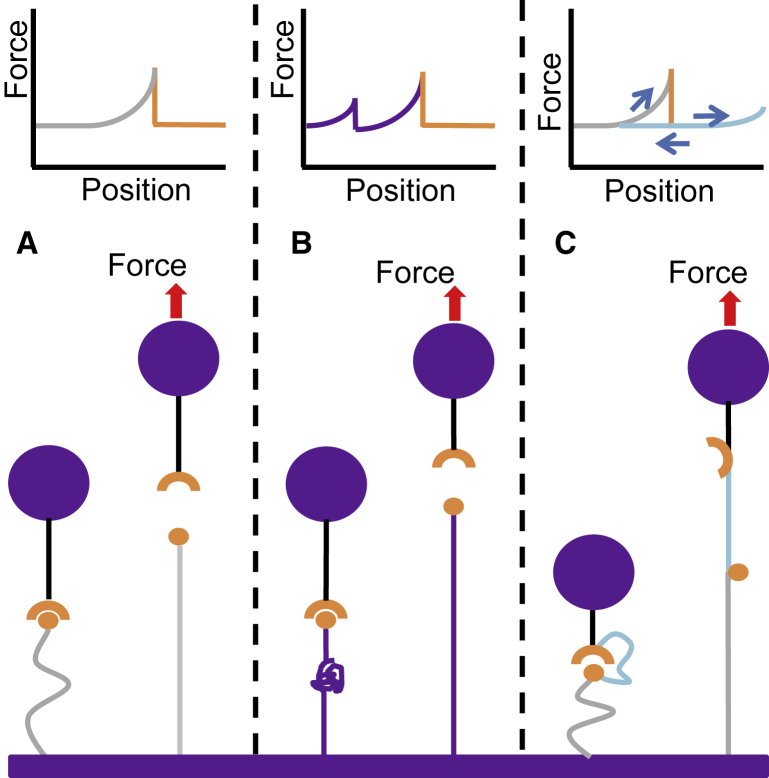
In the simplest configuration (Fig. 4 A), a long, flexible linker is used to covalently bind one molecule of interest to the surface. Polyethylene glycol (43) and DNA (44) handles are commonly used and commercially available with a number of different functional groups at each end. A worm-like chain or freely-jointed chain model can then be fitted to the force-position data to estimate the contour length and the stiffness of the linker (38). Multiple molecules can be distinguished from one molecule by increased stiffness or by spatial separation of multiple rupture events (42, 43).
Figure 4.
Mechanical fingerprint configurations. (A) Using a long, flexible linker such as polyethylene glycol is the simplest configuration. (B) A marker protein that unfolds with a known force signature is linked to a bond-forming protein. (C) A looped linker using a molecular tether allows repeated rupture of the same bond. To see this figure in color, go online.
In a second configuration (Fig. 4 B), genetic engineering is used to express the molecule of interest as a fusion protein with a marker molecule that unfolds under a known force. This configuration is very helpful for measuring protein unfolding (38, 45), but not for measuring unbinding because it would filter out any bonds that rupture before the marker molecule unfolds.
The third configuration is a looped linker (42, 46, 47, 48) in which two biomolecules of interest are attached via a molecular tether (Fig. 4 C), effectively converting a bond rupture event to an elongation event. This provides the unique opportunity of repeatedly testing the same biomolecular pair, which can increase experimental throughput and allow the study of heterogeneity in the bond population (47). The linkers may even contain multiple loops to acquire multiple unbinding measurements from a single force cycle (49, 50).
The use of flexible and looped linkers will increase accuracy significantly by decreasing both nonspecific adhesion and the percent of multiple bonds in the data set. However, some multiple bonds will still rupture at the same length and not be removed, especially if higher adhesion frequencies are used. A major shortcoming is that long, flexible linkers create nonlinear ramps in force that require corrections to obtain accurate parameter estimates (51, 52, 53). These corrections reduce the efficiency of the data analysis, and it remains unclear whether they can be modified to analyze bonds with complex energy landscapes.
Filtering with probe-position fluctuations
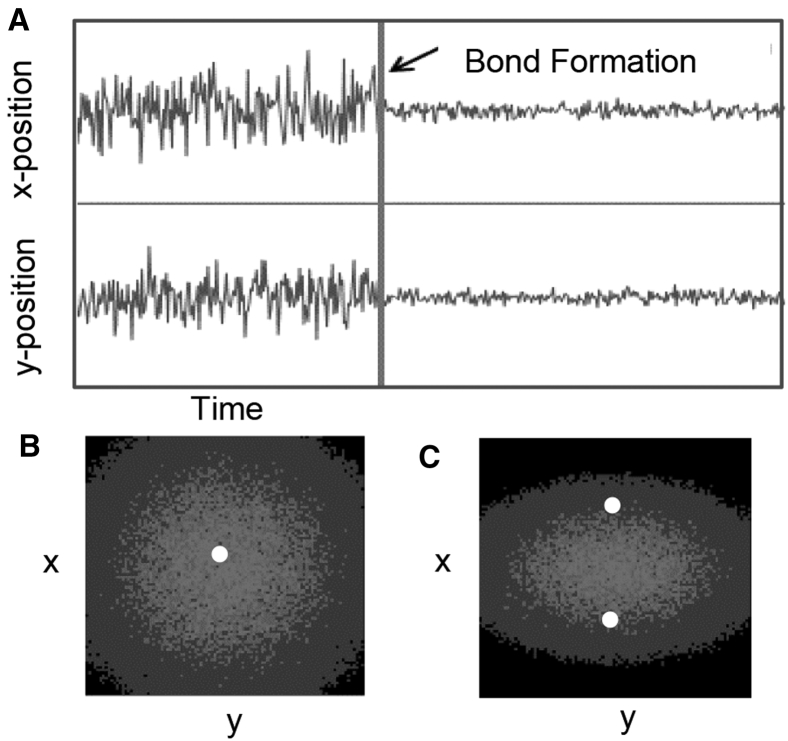
Bartsch et al. (54) showed that the position fluctuation pattern of the probe in an OT can be used to determine if one or more bonds are anchoring the probe to the surface. As long as the probe can be tracked with sufficient spatial sensitivity, this method can also be used with the BMFP and MT (55). In all cases, before the probe attaches to the surface, thermal fluctuations will cause the probe to fluctuate in a random pattern in three dimensions. However, once the probe forms a bond to the surface, the variation in position will be reduced (Fig. 5 A). If the probe is tethered through a single location, then a histogram of the probe position in the x-y plane (where z is normal to the surface) will show a circular pattern around the tether attachment point (Fig. 5 B). However, if two bonds form between the probe and the surface, the movement of the probe along the axis between the two attachment points will be restricted, resulting in a histogram with an elliptical pattern (Fig. 5 C).
Figure 5.
Simulated probe-position data. (A) Probe-position fluctuations due to thermal noise are reduced when the probe is tethered by a bond. (B) After a bond forms, the probe position will fluctuate around the attachment point of the bond (white dot), forming a circular position histogram when viewed from the z-direction. (C) If multiple bonds have formed (white points), the probe will fluctuate less along the axis between the bonds, resulting in an elliptical histogram.
This method assumes nothing about the energy landscapes but does require that the bonds are sufficiently long-lived relative to the data acquisition rate to create accurate histograms of probe position and that multiple bonds are far enough apart relative to the spatial resolution to discern the elliptical shape. These requirements greatly restrict the use of this method but will not affect the validity as long as the requirements are empirically analyzed.
Controlled molecular spacing
Regardless of how the data are filtered, the efficiency remains less than 40% when biomolecules are placed randomly on the surface (Fig. 3). If either the probe or surface could be reliably functionalized with a single molecule in the contact zone, then there would be no mechanism for multiple bonds, ensuring 100% efficiency and accuracy. However, there is currently no method to control probe or surface functionalization at the nanoscale. Microcontact printing is still limited to creating spots that are hundreds of nanometers across. This has been used to increase the multiplexing ability of MT by using arrays of small spots to control microscale bead spacing, but it has not been able to control nanoscale molecular spacing within each spot, so it did not eliminate multiple bonds (56).
Other studies have functionalized probes with low numbers of biomolecules and then counted the number of molecules in the contact zone. For example, AFM cantilever tips were functionalized with just a few biomolecules by functionalizing the probe only in the contact zone through contact-dependent chemistry (57, 58). In one study, the number of fluorescent molecules on the probe was determined by counting photobleaching events (57), but a few biomolecules were still observed in the contact zone.
The reliable use of molecular spacing requires no assumptions about the energy landscapes, so it can be highly robust; thus, these methods warrant continued development. The unsuccessful approaches above have relied on random immobilization of biomolecules within reduced contact zones. An alternative would be the use of any type of nanostructured material that spaces each functional group and therefore biomolecule too far apart for the two to simultaneously bind the probe.
Analyzing high-adhesion probability data
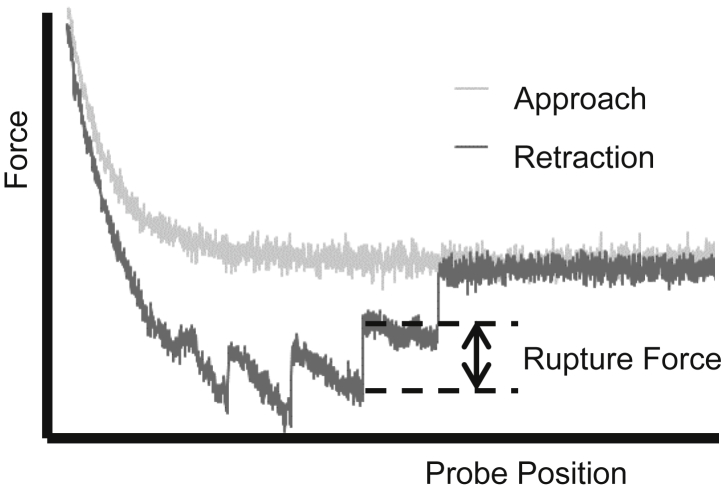
Experiments will be even more efficient if single-molecule characteristics can be extracted from force curves with multiple ruptures. Fig. 6 shows an example of a force cycle with multiple bonds, in which each jump in force represents one or more bond ruptures. Some studies have analyzed this force data by only considering the last rupture event in the force cycle (59, 60), but this again assumes that two rupture events must be spatially separated. This assumption fails especially when there are many rupture events, resulting in hidden multiple interactions (34, 35). Another shortcoming is that these methods assume prior knowledge about the appearance of single molecule data and have not been shown to be reliable for the complex energy landscapes described above. These methods are therefore unreliable for extracting single-molecule measurements from highly multiple bond data as intended.
Figure 6.
A typical force-displacement curve for an AFM. Each instantaneous change in force represents one or more bond ruptures.
An alternative approach is to assume that the histograms of the data set reflect mixtures of single and multiple bonds and to extract single-bond properties from multiple bond data. The method of autocorrelation of force histograms (59, 61) has been found to be inaccurate because the peaks in the data are as likely to reflect noise as different numbers of bonds, except when adhesion probabilities are low (9). This approach would fail for two-state bonds even with low-probability data because it assumes that all single bonds rupture at similar forces. Fitting the data to models while assuming mixtures of single and multiple bonds (62, 63, 64) also fails at high adhesion probabilities, likely also because of overfitting noisy data (9). Other methods have not been empirically tested in comparison with single-molecule data (65, 66), so they should not be considered reliable at this time.
Summary of methods to ensure single bonds
Here, we summarize what we learned about ensuring that SMFS really measures single bonds:
-
1)
Poisson statistics remains a critical technique for measuring single bonds. All publications should report adhesion probabilities and justify the use of Poisson statistics by clarifying how the experimental conditions support the independence assumption, and by using Poisson statistics to predict the expected prevalence of multiple bonds.
-
2)
When relying purely on low adhesion probabilities, the adhesion probability should be below 20–35% (for 90–80% accuracy by Poisson statistics). Because 10–20% error does not meet the generally expected 95% level of certainty, the discussion should address whether the expected prevalence of multiple bonds might affect the conclusions drawn about single bonds.
-
3)
Accuracy can be greatly improved by filtering out most multiple bonds. The filtering may be assessed in comparison with the number of multiple bonds predicted by Poisson statistics. However, filtering does not solve the fundamental problem that efficiency increases only incrementally above an adhesion probability of 35% (Fig. 3) and that many multiple bonds will escape filtering, so filtering should still be used with low adhesion probabilities.
-
4)
Extendable molecular markers have both advantages and disadvantages. Extendable molecular markers increase filtering accuracy and reduce nonspecific adhesion but also create nonlinear force ramps that require corrections that may not be accurate for complex energy landscapes. For these reasons, it should not be considered necessary to use marker molecules.
-
5)
Probe fluctuations are often not an option for measuring bond strength. This method requires long-lived bonds and high-frequency data acquisition.
-
6)
Controlled molecular spacing is promising. However, this approach requires more development to achieve single-molecule contact zones.
-
7)
Extracting single-bond properties from high-adhesion probability data has never been demonstrated to be reliable. We propose that any method using high-adhesion probabilities (including through the use of controlled molecular spacing) should not be considered reliable unless it can be validated by comparison of high versus low adhesion probability data on the same biomolecules.
The biotin-streptavidin bond
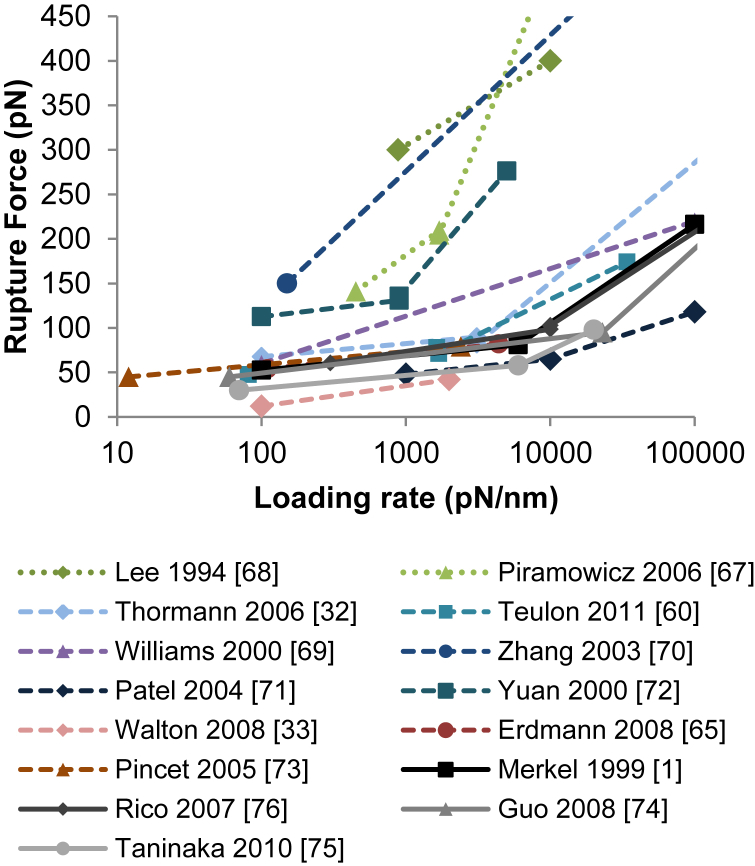
Based on our analysis above, we analyzed the literature on the strength of the biotin-streptavidin bond to determine whether the inconsistencies reported for this bond are greatly reduced when we only consider studies that met our criteria for SMFS. Some argue convincingly that biotin-streptavidin bonds strengthen over time by entering a lower energy state with a large energy barrier (67). Because this provides a valid reason for discrepancies in measurements, we only considered experiments in which newly formed bonds were tested. We then asked whether they used conditions that support independence of bond formation and resulted in adhesion probabilities below 35%. We required this whether or not probe-position fluctuations or mechanical fingerprints were used to eliminate multiple bond data. We also require the description of negative controls because these are standard requirements for any molecular experiments. We found 15 publications discussing the strength of newly formed streptavidin bonds using DFS and a loading rate near 1000 pN/s. The mean rupture force at 1000 pN/s within these publications ranges from 40 to 320 pN. The results from these publications are shown in Fig. 7.
Figure 7.
Published studies examining the strength of the biotin-streptavidin bond. The models used to fit the data are plotted instead of the raw data to provide a more readable plot. Publications that did not use reliable methodologies to ensure single bonds are shown as dashed lines. Publications that used reliable methodologies to ensure single bonds are shown as solid lines. To see this figure in color, go online.
Two of these publications reported high-adhesion probabilities (>50%) (67, 68), and the results from these are shown as dotted lines, as they are expected to be unreliable. The results are shown as dashed lines when the reliability of the study cannot be determined from the publication. Six publications provided too little information to assess adhesion probabilities (32, 60, 69, 70, 71, 72). Three did not demonstrate the specificity of their measured adhesions with a negative control (33, 65, 73). In the four publications that described low-adhesion probabilities (<35%) and negative controls, the mean rupture force at 1000 pN/s ranged from 45 to 70 pN according to both raw data and models (Fig. 7) (1, 74, 75). This small discrepancy might be explained by differences in instrument calibration or temperature (33, 76). Similar forces were reported in the publications that used low-adhesion probabilities but did not describe results of negative controls. In contrast, the four highest forces were reported in the two studies with high-adhesion probabilities and two of the six studies that did not report adhesion probabilities (Fig. 7). Therefore, the body of work on biotin-streptavidin bond measurements is reproducible when low adhesion probabilities (<35%) are reported, but not when they are not. This demonstrates that SMFS studies should always include in the methods section a description of adhesion probabilities.
Future directions
Our analysis demonstrates that it is still necessary to use low-adhesion probabilities to obtain reliable data on single bonds with SMFS. Therefore, SMFS has remained remarkably low throughput in an era of high-throughput experimentation. Instead of developing methods to get reliable measurements from increased adhesion probabilities, a better approach is to obtain data more efficiently by using methods such as MT (74) or centrifugal force microscopy (77) that can perform SMFS on many molecules simultaneously. An even better approach may be to develop methods to avoid SMFS altogether. Although most single-molecule methods are used to identify population variations in behavior, SMFS has been necessary simply to apply the same conditions to all the molecules being tested. It would be an enormous stride forward to have a method for quickly obtaining average position data on many bonds or molecules subjected to the same force or average force data on many molecules stretched to the same length without the extra effort needed to determine population variation.
Author Contributions
K.C.J. and W.E.T. each contributed to the literature research and perspectives and wrote the manuscript.
Acknowledgments
Support for this work comes from NIH grants 1R01 HL117639, 1R01 Al119675, and 1R01 Al106987.
Editor: Brian Salzberg.
References
- 1.Merkel R., Nassoy P., Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 2.Yago T., Lou J., Zhu C. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J. Clin. Invest. 2008;118:3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yakovenko O., Sharma S., Thomas W.E. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J. Biol. Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels B.R., Masi B.C., Wirtz D. Probing single-cell micromechanics in vivo: the microrheology of C. elegans developing embryos. Biophys. J. 2006;90:4712–4719. doi: 10.1529/biophysj.105.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 6.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 7.Block S.M., Goldstein L.S., Schnapp B.J. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 8.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 9.Williams P.M. Analytical descriptions of dynamic force spectroscopy: behaviour of multiple connections. Anal. Chim. Acta. 2003;479:107–115. [Google Scholar]
- 10.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seol Y., Neuman K.C. SnapShot: force spectroscopy and single-molecule manipulation. Cell. 2013;153:1168–1168.e1. doi: 10.1016/j.cell.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 12.Gourier C., Jegou A., Pincet F. A nanospring named erythrocyte. The biomembrane force probe. Cell. Mol. Bioeng. 2008;1:263–275. [Google Scholar]
- 13.Francis M.B., Carrico I.S. New frontiers in protein bioconjugation. Curr. Opin. Chem. Biol. 2010;14:771–773. doi: 10.1016/j.cbpa.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Fichtner D., Lorenz B., Franz C.M. Covalent and density-controlled surface immobilization of E-cadherin for adhesion force spectroscopy. PLoS One. 2014;9:e93123. doi: 10.1371/journal.pone.0093123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popa I., Kosuri P., Fernandez J.M. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat. Protoc. 2013;8:1261–1276. doi: 10.1038/nprot.2013.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanoss C.J. Hydrophobicity of biosurfaces - origin, quantitative-determination and interaction energies. Colloid Surface B. 1995;5:91–110. [Google Scholar]
- 17.Dudko O.K., Mathé J., Hummer G. Extracting kinetics from single-molecule force spectroscopy: nanopore unzipping of DNA hairpins. Biophys. J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudko O.K., Hummer G., Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best R.B., Paci E., Dudko O.K. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J. Phys. Chem. B. 2008;112:5968–5976. doi: 10.1021/jp075955j. [DOI] [PubMed] [Google Scholar]
- 20.Marshall B.T., Long M., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 21.Pereverzev Y.V., Prezhdo O.V., Thomas W.E. The two-pathway model for the catch-slip transition in biological adhesion. Biophys. J. 2005;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas W., Forero M., Vogel V. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W., Lou J., Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J. Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki Y., Dudko O.K. Biomolecules under mechanical stress: a simple mechanism of complex behavior. J. Chem. Phys. 2011;134:065102. doi: 10.1063/1.3533366. [DOI] [PubMed] [Google Scholar]
- 25.Zhuravlev P.I., Hinczewski M., Thirumalai D. Force-dependent switch in protein unfolding pathways and transition-state movements. Proc. Natl. Acad. Sci. USA. 2016;113:E715–E724. doi: 10.1073/pnas.1515730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan G., Le S., Chen H. Elasticity of the transition state leading to an unexpected mechanical stabilization of titin immunoglobulin domains. Angew. Chem. Int. Ed. Engl. 2017;56:5490–5493. doi: 10.1002/anie.201700411. [DOI] [PubMed] [Google Scholar]
- 27.Bartolo D., Derényi I., Ajdari A. Dynamic response of adhesion complexes: beyond the single-path picture. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;65:051910. doi: 10.1103/PhysRevE.65.051910. [DOI] [PubMed] [Google Scholar]
- 28.Thomas W.E., Vogel V., Sokurenko E. Biophysics of catch bonds. Annu. Rev. Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 29.Dudko O.K., Filippov A.E., Urbakh M. Beyond the conventional description of dynamic force spectroscopy of adhesion bonds. Proc. Natl. Acad. Sci. USA. 2003;100:11378–11381. doi: 10.1073/pnas.1534554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans E., Kinoshita K., Leung A. Long-lived, high-strength states of ICAM-1 bonds to beta2 integrin, I: lifetimes of bonds to recombinant alphaLbeta2 under force. Biophys. J. 2010;98:1458–1466. doi: 10.1016/j.bpj.2009.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans E., Leung A., Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thormann E., Hansen P.L., Mouritsen O.G. Dynamic force spectroscopy on soft molecular systems: improved analysis of unbinding spectra with varying linker compliance. Colloids Surf. B Biointerfaces. 2006;53:149–156. doi: 10.1016/j.colsurfb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Walton E.B., Lee S., Van Vliet K.J. Extending Bell’s model: how force transducer stiffness alters measured unbinding forces and kinetics of molecular complexes. Biophys. J. 2008;94:2621–2630. doi: 10.1529/biophysj.107.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayyas E., Bernardo M., Hoffmann P.M. Dissociation kinetics of an enzyme-inhibitor system using single-molecule force measurements. Biomacromolecules. 2010;11:3352–3358. doi: 10.1021/bm100844x. [DOI] [PubMed] [Google Scholar]
- 35.Getfert S., Reimann P. Hidden multiple bond effects in dynamic force spectroscopy. Biophys. J. 2012;102:1184–1193. doi: 10.1016/j.bpj.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann T., Dougan L. Single molecule force spectroscopy using polyproteins. Chem. Soc. Rev. 2012;41:4781–4796. doi: 10.1039/c2cs35033e. [DOI] [PubMed] [Google Scholar]
- 37.Ott W., Jobst M.A., Nash M.A. Single-molecule force spectroscopy on polyproteins and receptor-ligand complexes: the current toolbox. J. Struct. Biol. 2017;197:3–12. doi: 10.1016/j.jsb.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Hughes M.L., Dougan L. The physics of pulling polyproteins: a review of single molecule force spectroscopy using the AFM to study protein unfolding. Rep. Prog. Phys. 2016;79:076601. doi: 10.1088/0034-4885/79/7/076601. [DOI] [PubMed] [Google Scholar]
- 39.Scholl Z.N., Li Q., Marszalek P.E. Single molecule mechanical manipulation for studying biological properties of proteins, DNA, and sugars. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014;6:211–229. doi: 10.1002/wnan.1253. [DOI] [PubMed] [Google Scholar]
- 40.Janissen R., Berghuis B.A., Dekker N.H. Invincible DNA tethers: covalent DNA anchoring for enhanced temporal and force stability in magnetic tweezers experiments. Nucleic Acids Res. 2014;42:e137. doi: 10.1093/nar/gku677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riquelme M.V., Zhao H., Agah M. Optimizing blocking of nonspecific bacterial attachment to impedimetric biosensors. Sens. Biosensing Res. 2016;8:47–54. [Google Scholar]
- 42.Kim J., Zhang C.Z., Springer T.A. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466:992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulchek T.A., Friddle R.W., Noy A. Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc. Natl. Acad. Sci. USA. 2005;102:16638–16643. doi: 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cecconi C., Shank E.A., Bustamante C. DNA molecular handles for single-molecule protein-folding studies by optical tweezers. Methods Mol. Biol. 2011;749:255–271. doi: 10.1007/978-1-61779-142-0_18. [DOI] [PubMed] [Google Scholar]
- 45.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 46.Wiita A.P., Ainavarapu S.R., Fernandez J.M. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc. Natl. Acad. Sci. USA. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halvorsen K., Schaak D., Wong W.P. Nanoengineering a single-molecule mechanical switch using DNA self-assembly. Nanotechnology. 2011;22:494005. doi: 10.1088/0957-4484/22/49/494005. [DOI] [PubMed] [Google Scholar]
- 48.Krasnoslobodtsev A.V., Zhang Y., Lyubchenko Y.L. A flexible nanoarray approach for the assembly and probing of molecular complexes. Biophys. J. 2015;108:2333–2339. doi: 10.1016/j.bpj.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han X., Qin M., Wang W. A versatile “multiple fishhooks” approach for the study of ligand-receptor interactions using single-molecule atomic force microscopy. Langmuir. 2012;28:10020–10025. doi: 10.1021/la301903z. [DOI] [PubMed] [Google Scholar]
- 50.Valle F., Zuccheri G., Samorì B. A polymeric molecular “handle” for multiple AFM-based single-molecule force measurements. Angew. Chem. Int. Ed. Engl. 2008;47:2431–2434. doi: 10.1002/anie.200704526. [DOI] [PubMed] [Google Scholar]
- 51.Evans E., Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedsam C., Wehle A.K., Gaub H.E. Dynamic single-molecule force spectroscopy: bond rupture analysis with variable spacer length. J. Phys. Condens. Matter. 2003;15:S1709–S1723. [Google Scholar]
- 53.Ray C., Brown J.R., Akhremitchev B.B. Correction of systematic errors in single-molecule force spectroscopy with polymeric tethers by atomic force microscopy. J. Phys. Chem. B. 2007;111:1963–1974. doi: 10.1021/jp065530h. [DOI] [PubMed] [Google Scholar]
- 54.Bartsch T.F., Fisinger S., Florin E.L. Detecting sequential bond formation using three-dimensional thermal fluctuation analysis. ChemPhysChem. 2009;10:1541–1547. doi: 10.1002/cphc.200900211. [DOI] [PubMed] [Google Scholar]
- 55.del Rio A., Perez-Jimenez R., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Vlaminck I., Henighan T., Dekker C. Highly parallel magnetic tweezers by targeted DNA tethering. Nano Lett. 2011;11:5489–5493. doi: 10.1021/nl203299e. [DOI] [PubMed] [Google Scholar]
- 57.Liu J., Butte M.J. Single molecule labeling of an atomic force microscope cantilever tip. Appl. Phys. Lett. 2012;101:163705. doi: 10.1063/1.4760283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Long F., Cao B., Shahbazian-Yassar R. Modification of a single-molecule AFM probe with highly defined surface functionality. Beilstein J. Nanotechnol. 2014;5:2122–2128. doi: 10.3762/bjnano.5.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Florin E.L., Moy V.T., Gaub H.E. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 60.Teulon J.M., Delcuze Y., Pellequer J.L. Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 2011;24:490–502. doi: 10.1002/jmr.1109. [DOI] [PubMed] [Google Scholar]
- 61.Wong J., Chilkoti A., Moy V.T. Direct force measurements of the streptavidin-biotin interaction. Biomol. Eng. 1999;16:45–55. doi: 10.1016/s1050-3862(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 62.Lo Y.S., Huefner N.D., Beebe T.P. Specific interactions between biotin and avidin studied by atomic force microscopy using the Poisson statistical analysis method. Langmuir. 1999;15:1373–1382. [Google Scholar]
- 63.Lo Y.S., Simons J., Beebe T.P. Temperature dependence of the biotin-avidin bond-rupture force studied by atomic force microscopy. J. Phys. Chem. B. 2002;106:9847–9852. [Google Scholar]
- 64.Stevens F., Lo Y.S., Beebe T.P. Computer modeling of atomic force microscopy force measurements: comparisons of Poisson, histogram, and continuum methods. Langmuir. 1999;15:207–213. [Google Scholar]
- 65.Erdmann T., Pierrat S., Schwarz U.S. Dynamic force spectroscopy on multiple bonds: experiments and model. Europhys. Lett. 2008;81:48001. [Google Scholar]
- 66.Sun L., Cheng Q.H., Zhang Y.W. Effect of loading conditions on the dissociation behaviour of catch bond clusters. J. R. Soc. Interface. 2012;9:928–937. doi: 10.1098/rsif.2011.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Odrowaz Piramowicz M., Czuba P., Szymoński M. Dynamic force measurements of avidin-biotin and streptavdin-biotin interactions using AFM. Acta Biochim. Pol. 2006;53:93–100. [PubMed] [Google Scholar]
- 68.Lee G.U., Kidwell D.A., Colton R.J. Sensing discrete streptavidin biotin interactions with atomic-force microscopy. Langmuir. 1994;10:354–357. [Google Scholar]
- 69.Williams P.M., Moore A., Tendler S.J.B. On the dynamic behaviour of the forced dissociation of ligand-receptor pairs. J. Chem. Soc. Perk. T. 2000;2:5–8. [Google Scholar]
- 70.Zhang X., Moy V.T. Cooperative adhesion of ligand-receptor bonds. Biophys. Chem. 2003;104:271–278. doi: 10.1016/s0301-4622(02)00381-2. [DOI] [PubMed] [Google Scholar]
- 71.Patel A.B., Allen S., Williams P.M. Influence of architecture on the kinetic stability of molecular assemblies. J. Am. Chem. Soc. 2004;126:1318–1319. doi: 10.1021/ja0366991. [DOI] [PubMed] [Google Scholar]
- 72.Yuan C., Chen A., Moy V.T. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 2000;39:10219–10223. doi: 10.1021/bi992715o. [DOI] [PubMed] [Google Scholar]
- 73.Pincet F., Husson J. The solution to the streptavidin-biotin paradox: the influence of history on the strength of single molecular bonds. Biophys. J. 2005;89:4374–4381. doi: 10.1529/biophysj.105.067769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo S., Ray C., Akhremitchev B.B. Effects of multiple-bond ruptures on kinetic parameters extracted from force spectroscopy measurements: revisiting biotin-streptavidin interactions. Biophys. J. 2008;95:3964–3976. doi: 10.1529/biophysj.108.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taninaka A., Takeuchi O., Shigekawa H. Reconsideration of dynamic force spectroscopy analysis of streptavidin-biotin interactions. Int. J. Mol. Sci. 2010;11:2134–2151. doi: 10.3390/ijms11052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rico F., Moy V.T. Energy landscape roughness of the streptavidin-biotin interaction. J. Mol. Recognit. 2007;20:495–501. doi: 10.1002/jmr.841. [DOI] [PubMed] [Google Scholar]
- 77.Yang D., Ward A., Wong W.P. Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 2016;7:11026. doi: 10.1038/ncomms11026. [DOI] [PMC free article] [PubMed] [Google Scholar]