Abstract
Background
Effects of silver nanoparticles (AgNP) on the intestinal virome/phage community are mostly unknown. The working hypothesis of this study was that the exposure of pharmaceutical/nanomedicine and other consumer-use material containing silver ions and nanoparticles to the gastrointestinal tract may result in disturbance of the beneficial gut viruses/phages.
Methods
This study assesses the impact of AgNP on the survival of individual bacteriophages using classical virology cultivation and electron microscopic techniques. Moreover, how the ingested AgNP may affect the intestinal virus/phages was investigated by conducting whole-genome sequencing (WGS).
Results
The viral cultivation methods showed minimal effect on selected viruses during short-term exposure (24 h) to 10 nm AgNP. However, long-term exposure (7 days) resulted in significant reduction in the viral/phage population. Data obtained from WGS were filtered and compared with a nonredundant viral database composed of the complete viral genomes from NCBI using KRAKEN (confidence scoring threshold of 0.5). To compare the relative differential changes, the sequence counts in each treatment group were normalized to account for differences in DNA sequencing library sizes. Bioinformatics techniques were developed to visualize the virome comparative changes in a phylogenic tree graph. The computed data revealed that AgNP had an impact on several intestinal bacteriophages that prey on bacterial genus Enterobacteria, Yersinia and Staphylococcus as host species. Moreover, there was an independent effect of nanoparticles and released ions.
Conclusion
Overall, this study reveals that the small-size AgNP could lead to perturbations of the gut microbial ecosystem, leading to the inactivation of resident phages that play an important role in influencing gastrointestinal health.
Keywords: bacteriophages, intestine, microbiome, nanoparticle, virome, WGS, intestinal content, silver nanoparticles
Introduction
The recent explosion of pharmaceutical and consumer products that incorporate nanoparticles with antimicrobial properties, such as the widely used silver nanoparticles (AgNP), has ignited a safety concern to the microbial component of the human body.1–6 The intestinal microbiome contains bacteria, archaea, fungi, viruses, phages and other eukaryotic organisms. Most of the earlier studies were focused on the safety assessment of AgNP to the commensal bacteria residing in the gut.2,5,7 There are several studies that demonstrate the antiviral effects of AgNP on various pathogenic species that belong to RNA virus (HIV-I and hepatitis C virus), DNA virus (monkeypox virus), ssRNA virus (respiratory synticial virus) or dsDNA virus (Herpes Simplex virus and Influenza virus).8–10 However, the effect of AgNP exposure on bacteriophages, which are the dominant members of the viral component of the body associated and environmental virome, is not assessed in detail.
Recent viral metagenomic studies indicated that bacteriophages are probably the most abundant and genetically diverse biological entities in nature and have been consistently found in every environment where their bacterial hosts are present.11,12 It is being predicted that there are about 1031 phage particles on earth, exceeding the total number of bacteria by an order of magnitude1 indicating that phages play important biological roles.13,14 It is also worth to note that bacteriophages have been used as alternatives to antibiotics in the form of bacteriophage therapy to control bacterial pathogens.15 The rise in antimicrobial resistance has particularly revived interest in bacteriophage research for controlling infection and food contamination.16,17
Similar to other animals, the human body thrives in commensal association with diverse bacterial, fungal and viral species, which altogether influence host physiology and development of diseases.18 To this effect, there have been concerted efforts to understand the microbial ecology of the human gastrointestinal tract. The accumulated evidence on the bacterial component of the gut biome indicates a commensal association that confers substantial benefits to the host, including development of robust immune system, protection from pathogens and digestion of food that otherwise is not digested by the host enzymes. However, information on viruses as component of the commensal gut microbial community and the interaction therein is just emerging.19 The available evidence suggest that bacteriophages are not only the most abundant members of the human gut virome but also are stable residents in the gut in a way similar to that of the symbiotic gut bacterial population and likely involve in modulating host physiology and immunity.19 Bacteriophages are also known to horizontally transfer genetic material from one bacterium to the other in the transduction process. So, it is not entirely surprising that enteric phages play a role in driving the population and composition of the gut microbiota.20
In addition to bacteriophages, the beneficial role of enteric eukaryotic viruses in promoting healthy gut structure and immune development has recently been reviewed.21 Taken together, these reports indicate that bacteriophages and some eukaryotic viruses, considered as resident gut viruses collectively termed virome/virobiota, are likely to complement the commensal microbiome in modulating gut health and disease. According to Nanotechnology Consumer Products Inventory, the number of consumer products claimed to have nanomaterials increased from 54 to more than 1,800 from 2005 to 2014. Overall, silver accounts for 24% of the nanomaterials.22 The increased use of AgNP and many other engineered nanoparticles in pharmaceutical and consumer products may ultimately result in increased human exposure particularly for the gastrointestinal system and homeostasis of the resident bacteria and viruses that are essential to host health. Given the fact that commensal gut microbes are crucial for the normal functioning of the host, there is an urgent need to assess and understand the effects of widespread application of metallic nanoparticles, such as AgNP, gold nanoparticles and copper nanoparticles that are used in preparation and packaging of human food, animal feed, supplements and several pharmaceutical products and medical devices. Furthermore, AgNP can reduce the mucin secretion by intestinal epithelial cells–a niche where several commensal bacteria and bacteriophages thrive.23 In the first phase of this study, bacteriophages MS2, phiX174, PP7 and the respective bacterial host strains Escherichia coli C3000, E. coli K12 and Pseudomonas aeruginosa were tested to address the antiviral/antiphagic properties of AgNP. Next, as a proof of concept, fecal samples were incubated with AgNP (test agent, representing nanoparticles) or silver acetate (AgOAC, positive control, representing silver ions) to determine its effect on the global community of bacteriophages by whole-genome sequencing (WGS) of viral genomic material. Novel bioinformatics approach was used to visualize the virome comparative changes in a phylogenic tree graph to further aid in interpretation of results.
Materials and methods
Bacteriophages and host bacterial stock
Bacteriophage MS2 (ATCC 15597-B1) and its bacterial host E. coli C3000 (ATCC 15597) were procured from American Type Culture Collection (Manassas, VA, USA), and bacteriophages PP7, phiX174 and their respective bacterial hosts E. coli K12 and P. aeruginosa were obtained from the Center for Drug Evaluation and Research (CDER; Silver Spring, MD, USA). The bacterial hosts were cultured overnight in 3% tryptic soy broth (TSB). The following day, fresh TSB was inoculated with the overnight culture (1% v/v) and incubated at 37°C (approximately for 4 h) until the optical density reached 0.5, equivalent to a mid-log-phase. The bacteriophages were propagated and titrated in their host cells following standard double-agar-overlay procedures.24 Briefly, a serially diluted 100 μL phage and 900 μL of the corresponding host bacterial cells grown to log phase were mixed with 4 mL melted soft agar (0.75%). The mix was poured on a tryptic soy agar (1.5%) and incubated overnight at 37°C. The following day, the bacteriophages forming plaques were harvested from the agar plates by washing the surface of the plates with PBS followed by spinning to sediment bacterial cell and agar debris. The resulting suspension was further centrifuged at 4,000 g for 20 min at 4°C and the supernatant was passed through a 0.22 μm filter and stored at −80°C as the phage stock.
Bacteriophage/bacterial titration
The double-agar-overlay plaque assay was used to enumerate the phage titer.24 Briefly, 100 μL of serially diluted phage samples were added to 900 μL of host bacterial cells and mixed with 4 mL soft tryptic soy agar (0.75% TSA). This soft agar was layered on top of a 1.5% TSA plates followed by overnight incubation at 37°C. Phage plaques formed as clear zones on the plates were counted and phage titers were expressed as plaque forming units per mL (PFU/mL). Similarly, bacterial titer was determined by standard streak plate method and colonies were counted and recorded as colony forming units per mL (CFU/mL).
AgNP characterization
The AgNP used in all experiments were procured from NanoComposix (San Diego, CA, USA) as citrate capped form and characterized by the NanoCore Facility at National Center for Toxicological Research (NCTR). Transmission electron microscopy (TEM) and inductively coupled plasma mass spectrometry were used to determine particle size, shape, silver mass and ionic concentrations, while dynamic light scattering was used for size distributions and dispersive index. The AgNP characterization results were similar to the reports provided by the manufacturers.
Effect of AgNP exposure on bacteriophage survival
Effect of the 10 nm size AgNP was assessed on the survival of enteric bacteriophages MS2, PP7 and phiX174 after 24 h of exposure at 25, 50 and 100 μg/mL concentrations. These concentration were previously shown to have dose-depended antiviral effects.10 PFU of bacteriophages were enumerated following the standard double-agar-layer method in the respective bacterial hosts.10,25
Short- and long-term phage survival
Sterile SM buffer was mixed with 10 nm AgNP to a final concentration of 25, 50 and 100 μg/mL. MS2 bacteriophage was seeded into a bottle at an initial titer of 1010 PFU/mL. After a thorough mixing, the suspension was distributed into Eppendorf tubes, sealed and stored at 4°C and 37°C up to 28 days. Three tubes were removed at days 0 (baseline), 1, 2, 7, 14, 21 and 28. The surviving bacteriophages in these samples were enumerated following the double-agar-layer plaque assay (US EPA 2001).
Transmission electron microscopy
In total, 100 μg/mL AgNP of the sizes 10, 75 and 110 nm were mixed with 100 μL MS2 and incubated at 37°C for 2 h. In total, 20 μL of samples of the virus and AgNP mixture were applied to copper grids coated with carbon and glow discharged prior to use. After absorption for 10 min, samples on the copper grids were stained by 1% uranyl acetate and images were recorded under a TEM operated at 200 kV (JEM-2100, Gatan US4000 CCD camera).
Effect of AgNP on the bacteriophage population present in fresh fecal sample from Rhesus monkeys
Pools of naturally excreted fresh fecal material were collected from 18 healthy Rhesus monkeys that were housed in the primate facility of NCTR (Protocol S-0020). Samples were immediately transferred to −80°C freezers and used within 2 days. A 20% fecal suspension was prepared in sterile and deoxygenated Brain–Heart Infusion broth in anaerobic chambers. The suspension was divided into aliquots of 200 mL each (control, 10 nm AgNP treatment and AgOAC treatment) for virome/bacteriophage analysis. AgNP and AgOAC were added to the bottles containing the fecal microbes supplemented with a low-concentration carbohydrate medium, followed by incubation in anaerobic chamber for 7 days.26
Preparation of genomic material for virome/bacteriophage analysis
After the incubation with the AgNP or AgOAC, the primate fecal samples were homogenized in SM phage buffer, centrifuged and the supernatant portion was used for further extraction of genomic material for virome/bacteriophage analysis. After passing the supernatant through sequential 0.45 and 0.22 μm filters, the filtrate was treated with a cocktail of RNase and DNase to exclude free nucleic acids followed by ultracentrifugation to concentrate the virus-like particles in the primate fecal samples.27
From the concentrate, the total viral nucleic acids were extracted and stored at −80°C until processed. The viral DNA was amplified using the GenomiPhi V2 DNA amplification kits, and the quality and quantity were checked by NanoDrop. After quantification of the products, equal concentrations of the amplified DNA from each experimental group were sequenced using next-generation sequencing platforms. Prior to sequencing, the extracted genomic DNA was fragmented with M220 Ultrasonicator. Fragmented DNA was used for preparing uniquely indexed sample library using the TruSeq DNA PCR-Free Sample Preparation kit (Illumina), as recommended by the manufacturer and the final libraries were quantified with Qubit and 2100 Bioanalyzer. The final libraries were size selected with BluePippin for a desired size range of 600–900 bp. The size selected final libraries were quantified with Qubit 2.0 fluorometer and Agilent 2100 Bioanalyzer. The libraries were pooled, and PhiX control was added to 1% of the final library content. In total, 1.4–1.5 pmol of the pooled library was loaded onto the Illumina NextSeq500 to generate 50 M (±20%) 150PE reads for each sample. Sequence reads generated by Illumina machine were demultiplexed and trimmed of adapters on BaseSpace. FASTQ data files were further analyzed as described below.
Analysis of bacteriophage metagenomics
For the analysis of bacteriophages present in the fecal samples, first the complete viral metagenome was analyzed. To analyze the viral metagenomes, raw sequence paired-end reads from each sample (control, AgNP, AgOAC) were filtered for low-quality signals or ambiguous characters (<30 out of 40 quality). Reads were assigned to their corresponding samples according to their barcode-tagged primer sequences. Primers, linkers and adaptors were trimmed from the sequences and sequences shorter than 50 bp were removed. Removal of sequences of eukaryotic origin was performed following a MegaBLAST search against the human/monkey genome database from NCBI. Low-complexity filters were applied to remove highly repetitive sequences. Next, filtered reads were compared with a nonredundant custom-built viral database composed of the complete viral genomes from NCBI (http://www.ncbi.nlm.nih.gov/genome/viruses/) by employing the methods in the KRAKEN software package.28 This custom database was built to remove low-complexity sequences, which are a primary source of false-positive classification hits.29 KRAKEN achieves high sensitivity and high speed by utilizing exact alignments of k-mers and a novel classification algorithm. A preliminary search was used to retrieve only those viruses that matched KRAKEN’s default criteria in which the taxon is assigned based on the taxon having the highest number of matches for the given k-mer. This produced a large number of taxonomy matches for each sample with reasonably good confidence (~95% according to KRAKEN). To obtain higher confidence in the matches, a second search was conducted by employing the confidence scoring threshold of 0.5 within KRAKEN’s filtering routine, which is reported to improve precision to ~99%.
Results
Effects of AgNP on bacterial hosts and bacteriophages
The 10 nm AgNP at the lowest dosage (25 μg/mL) showed killing of phage hosts (P. aeruginosa, E. coli C3000, E. coli C, E. coli K12 and E. coli B) within 24 h (Figure 1). The bacterial titer reduction ranged from 3 to 8 log10 CFU/mL. The AgNP had various levels of antibacterial activity on different bacterial hosts–had the maximum antibacterial activity on Pseudomonas (PP7) as compared to other bacterial hosts.
Figure 1.
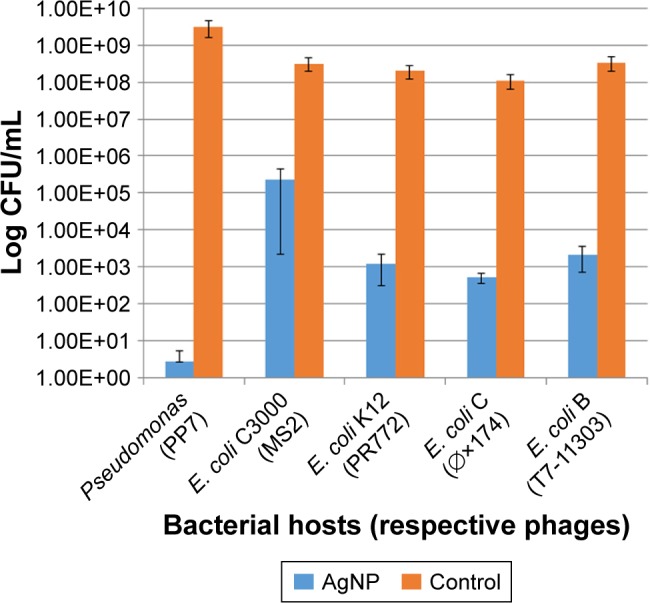
Effect of the 10 nm AgNP (25 μg/mL) on phage hosts (P. aeruginosa, E. coli C3000, E. coli K12, E. coli C, E. coli B) within 24 h. Bar graphs represent mean and SD from three independent experiments.
Abbreviations: AgNP, silver nanoparticles; CFU, colony forming units.
In contrast to the bacterial hosts, the bacteriophages were not affected by exposure to the AgNP in a 24-h exposure time. Only the highest dose (100 μg/mL) of the 10 nm AgNP showed a modest reduction in viral load measured as PFU/mL within 24 h (Figure 2). The reduction in PFU ranged from 1 to 3 log10.
Figure 2.
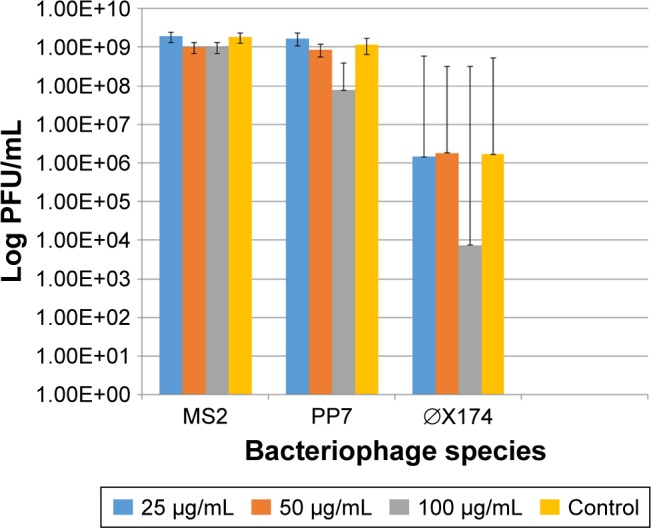
Effect of the 10 nm AgNP on the survival of enteric bacteriophages MS2, PP7 and phiX174 after 24 h of exposure at 25, 50 and 100 μg/mL concentrations. PFU of bacteriophages were determined following the standard DAL method in the respective bacterial hosts. Bar graphs represent mean and SD from three independent experiments.
Abbreviations: AgNP, silver nanoparticles; PFU, plaque forming units; DAL, double agar layer.
Effect of AgNP concentrations, incubation temperature and time duration on the survival of phages
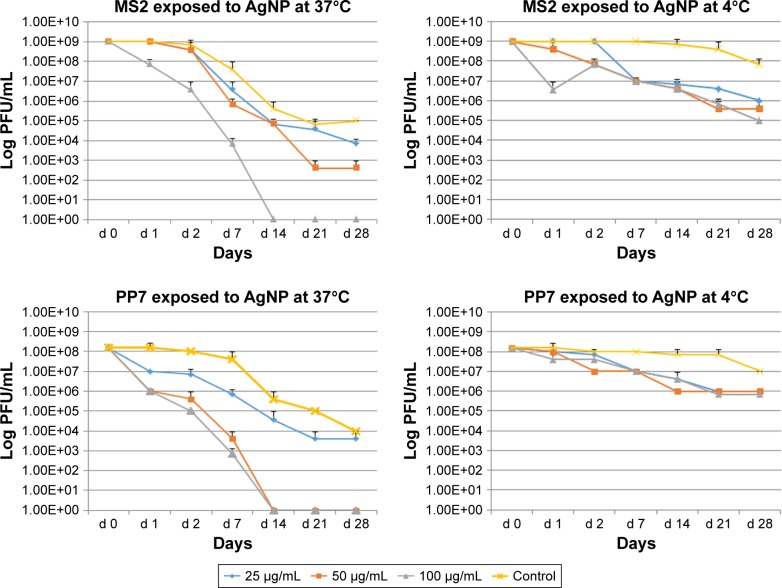
Additional experiments were conducted to assess the long-term (till day 28) effects of AgNP exposure on bacteriophage survival. Viral suspensions of MS2 and PP7 phages were exposed to the three doses (100, 50 and 25 μg/mL) of the 10 nm AgNP and incubated at 4°C and 37°C.
Both phages were more susceptible for the AgNP-mediated killing at 37°C as compared to 4°C (Figure 3). Exposure of MS2 phages to high dosage (100 μg/mL) of AgNP at 37°C resulted in the absence of PFU by day 14, whereas exposure to the medium (50 μg/mL) dose caused a continuous reduction in the phage titer till day 21 (Figure 3, upper panel). The low-dose AgNP exposure (25 μg/mL) followed a similar pattern as the medium dose till day 14. At 4°C, there was no difference in the PFU formation in MS2 during the treatment of any dose of AgNP; however, there was a clear difference between the control and AgNP-treated MS2 phages after day 2.
Figure 3.
Effect of temperature, dose and time of interaction with AgNP on the survival of enteric bacteriophages MS2 and PP7. The values indicate mean and SD from three independent experiments.
Abbreviations: AgNP, silver nanoparticles; PFU, plaque forming units.
The PP7 phages appear to be more susceptible to AgNP at high and medium doses–these phages were completely inactivated at 37°C on day 14 (Figure 3, lower panel). The low dose caused minimal decrease in phage titer (almost 1 log10 virus PFU/mL) reduction as compared to control at any time (except day 28). At 4°C, only 1–2 log10 decrease of the PP7 PFU/mL was observed.
Effects of short-term (24 h) and long-term (7 days) exposure to AgNP (10 nm) on bacteriophages as visualized by TEM
The TEM images of control phages showed a very clear envelope surface of the each viral particle (Figure 4A). By contrast, the TEM images of short-term (24 h) AgNP-treated phages showed a minimal effect on the structural integrity of outer envelope of viral particle (Figure 4B). The structural integrity of surface envelope of the viral particle disintegrated during long-term exposure to AgNP (Figure 4C).
Figure 4.
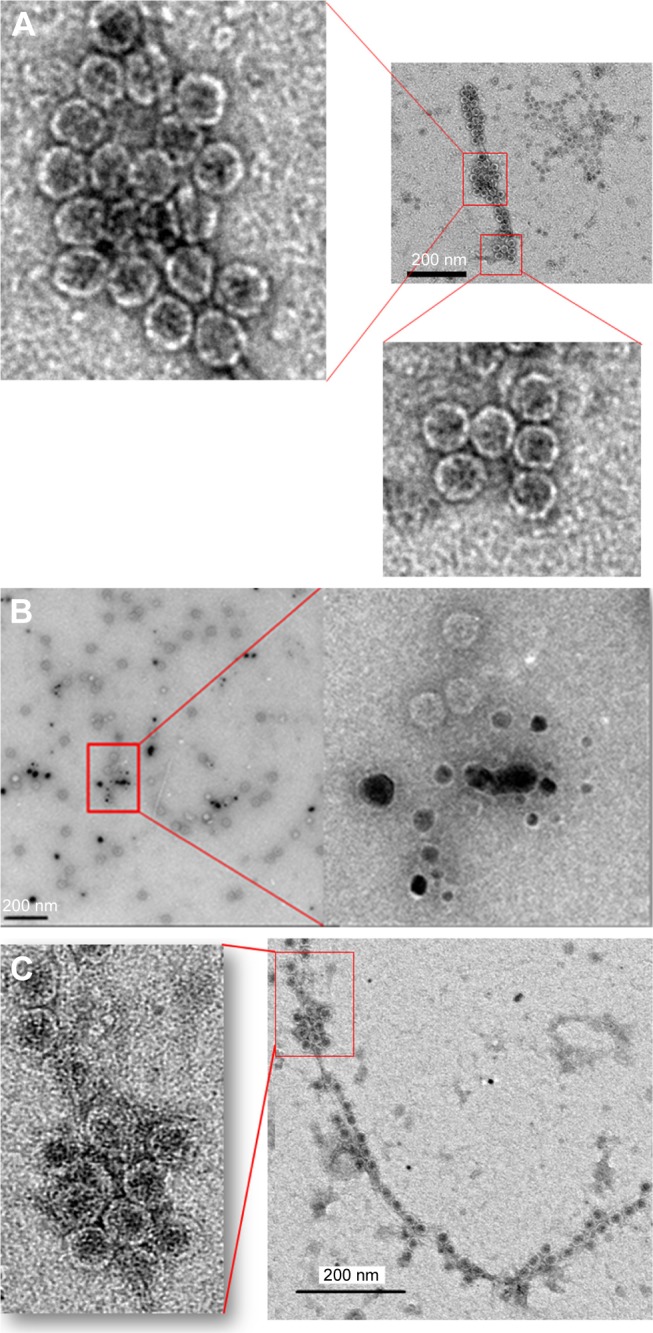
Transmission electron microscopy images of MS2 phage. (A) MS2 control and (B) MS2 treated with 10 nm silver nanoparticles at 100 μg/mL concentration for 24 h. (C) MS2 treated with 10 nm silver nanoparticles at 100 μg/mL concentration for 7 days.
Effect of AgNP on the bacteriophage population present in the intestine
Pooled fecal samples from 18 Rhesus monkeys were treated with 10 nm AgNP or AgOAC or kept as untreated control. Though the specific extraction process was used to highly enrich the virome genetic material, however, the sequencing output data showed a contamination of host sequence genetic materials. Sequences of eukaryotic origin were filtered following a MegaBLAST search against the human/monkey genome database from NCBI. Low-complexity filters were applied to remove highly repetitive sequences. Total numbers of reads were highest in control followed by AgNP- and AgOAC-treated samples, respectively (Table 1). Data were further filtered so that the virus sequence counts in the control condition had to be greater than 10 for it to be included in the differential fold analysis. This removed the singletons and provided a better confidence on the data set (Table S1). Applying this filter reduced the assigned viral read matches to ~2,278,000 in each group. This clearly indicated that the control group had about 80% sequences that were less than 10.
Table 1.
Richness of total viral reads in untreated control and experimental groups
| Output | Untreated controls | AgNP treated | AgOAC treated |
|---|---|---|---|
| Total no of reads (sequences) | 52,135,927 | 49,619,587 | 44,035,973 |
| Read length (single) | 50–150 bp | 50–150 bp | 50–150 bp |
| Viral read matches Assigned (KRAKEN default threshold) | 15,643,484 | 2,794,759 | 2,478,357 |
Abbreviations: AgNP, silver nanoparticles; AgOAC, silver acetate.
Next, we employed another scoring method offered by KRAKEN application to filter the classified k-mer matches to have higher coverage. This scoring method is based on the description in the KRAKEN online manual (https://ccb.jhu.edu/software/kraken/MANUAL.html). Basically, it computes a ratio of how many k-mers match for a given sequence to the number not matched. This score is included in Table S2 (in the column next to the sample counts called K-mer Score). To generate this table, we chose to filter the sequence counts to have a 0.5 score for the sequence to be counted in to the viral species. This score can be relaxed for filtering to a lower acceptance. The data generated by this method provide a very high confidence of correct classifications to a given virus genome (99%+).28 However, this filtering method reduced the number of viruses in the population as well as reduced the number having high fold change between samples. So, the fold changes are fairly accurate as well, which provides high confidence and may help overcome the challenge of having single sample from the pooled virome. To compare the relative differential changes between samples for each taxon, the sample counts were normalized to account for differences in DNA sequencing library sizes. A simple global target mean (GM) derived from the average of all three samples’ total matched counts [TC(i)] was computed and a normalization factor [NF(i) = GM/TC(i)] was found for each sample. The normalized taxon counts for each sample [CNtaxon(y)(i)] was then found by multiplying each taxon count by Ctaxon(y), by NF(i). Using these normalized values, the log2 fold changes between treated and untreated samples were computed for each taxon based on the filtered results (Table S3). P-values for the fold differences were determined by modeling the fold data as a log2 normalized distribution function. There were 52 significantly altered species (P-value ≤ 0.05), as shown in Table S3. The top 5 significantly altered viral species for the two treatment conditions are shown in Table 2.
Table 2.
Top 5 significantly altered viral species by treatment type
| Taxon ID | Name | Controls counts | AgNP treatment counts | Log2 fold (AgNP/control) | P-value (AgNP/control) |
|---|---|---|---|---|---|
| 10760 | Enterobacteria phage T7 | 855 | 39,281 | 5.52 | 4.21E−12 |
| 110456 | T7likevirus | 58 | 841 | 3.85 | 4.77E−07 |
| 10855 | Spiroplasma phage 4 | 12 | 1 | −3.84 | 2.26E−04 |
| 1357713 | Bacillus phage phiCM3 | 12 | 1 | −3.84 | 2.26E−04 |
| 1340830 | Mycobacterium phage KayaCho | 12 | 1 | −3.84 | 2.26E−04 |
|
| |||||
| Controls counts | AgOAC Treatment counts | Log2 fold (AgOAC/control) | P-value (AgOAC/control) | ||
|
| |||||
| 12417 | Lactobacillus phage phi adh | 29 | 192 | 2.72 | 4.32E−04 |
| 1340830 | Mycobacterium phage KayaCho | 12 | 1 | −3.66 | 0.0025 |
| 10679 | Enterobacteria phage P2 | 12 | 1 | −3.66 | 0.0025 |
| 578234 | Lactobacillus phage Lv-1 | 71 | 338 | 2.24 | 0.0025 |
| 10271 | Rabbit fibroma virus | 47 | 216 | 2.21 | 0.0027 |
Abbreviations: AgNP, silver nanoparticles; AgOAC, silver acetate.
Most prominent finding from this analysis is that there is a distinct effect of AgNP verses silver ions. During AgNP treatment, there is an increase in the abundance of phages that have the lysogenic property and harbor on the pathogenic bacteria (Enterobacteria phage T7 and T7likevirus), whereas there was a decrease in the abundance of phages that harbor on the commonly known pathogenic bacteria (Bacillus or mycobacteria). By contrast, during AgOAC treatment, there was a higher abundance of lysogenic phages (Lactobacillus phage phi adh and Lactobacillus phage Lv-1) that harbor on the commensal bacteria Lactobacilli–the predominant microbial flora in gut. Phages that harbor on the pathogenic bacteria were reduced during AgOAC treatment also.
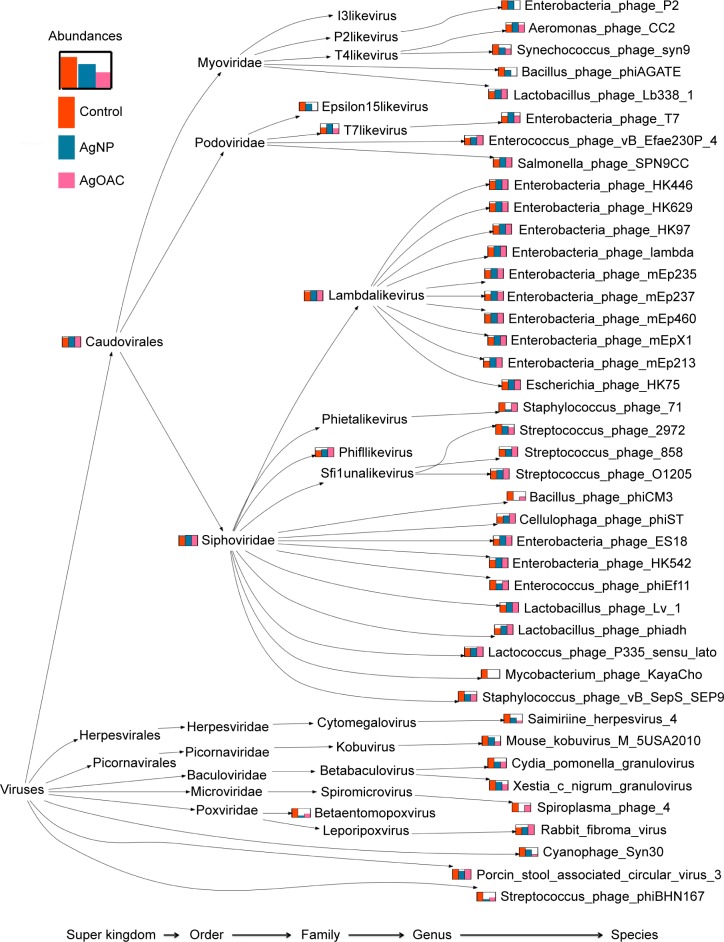
To better visualize the data obtained in Table S3, a program was written to create a phylogenic tree, as shown in Figure 5, which shows the species comparison abundances for the significantly altered species by treatment type. Furthermore, this phylogenetic tree shows the taxonomy profile comparison between the three sample types.
Figure 5.
Phylogenetic tree showing differences in the composition of viral/phage species in control (red), AgNP (blue) and AgOAC (pink) fecal samples after 7 days of treatment.
Abbreviations: AgNP, silver nanoparticles; AgOAC, silver acetate.
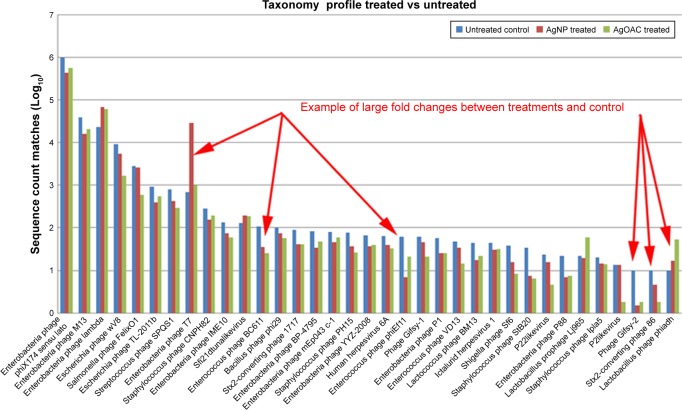
Employing the filtered data counts, the top 35 (by total sequence counts) viral species were compared. Figure 6 represents the relative abundance differences between the three samples (control, AgNP and AgOAC). Several examples of large fold changes are illustrated in this figure.
Figure 6.
Abundance of viral species in control, AgNP and AgOAC fecal samples after 7 days of treatment.
Abbreviations: AgNP, silver nanoparticles; AgOAC, silver acetate.
In general, AgNP had an impact on several intestinal bacteriophages that prey on bacterial genus Enterobacteria, Yersinia and Staphylococcus as host species. Interestingly, all analysis results consistently revealed that Enterobacteria phage T7 has high abundance in the AgNP treatment group and was found to be significantly altered (P-value = 0.0004). Moreover, this effect appears to be nanoparticle specific, because the silver ion did not show similar effect.
Discussion
Incorporation of silver and AgNP into health supplements, food packages, baby products and several household items has increased tremendously due to its strong antimicrobial property. Furthermore, availability of such products in open market is advertised for their potential to enhance the immunity against microbial attack. The most likely targeted site for these potentially antimicrobial nanomaterials is the gastrointestinal tract following intentional or accidental ingestion. However, the interaction of AgNP with phages and viruses is a fundamentally unexplored area, and it is more true for the effect of such products on the gastrointestinal commensal phages and viruses.30 To address these lacunae, this study investigated the capacity of the 10 nm AgNP to kill pure phages using several different kinds of bacteriophages by incubating them with variable doses (25, 50 and 100 μg/mL). The selection of this size and dose was based on our previous finding where only small size of AgNP (10 nm) was capable of killing phages; on the other hand, the bigger size of AgNP (75 and 100 nm) failed to show antiviral properties.10 Antiphagic properties were evaluated by their capacity to form the bacteriophage PFU ratio. Only 10 nm AgNP were able to inactivate phages. This further confirmed that the antimicrobial property of AgNP is size dependent, with smaller the size–better the antimicrobial effect.7,10 The antiviral effects were also dose dependent; the higher the dose, the more effective the antiviral property is. Further investigation was carried out to determine the survival of bacteriophages when exposed to variable concentrations of 10-nm sized AgNP at body (37°C) and storage temperatures (4°C). The survival of bacteriophages was dependent on the time of interaction, temperature and doses of AgNP. These observations raised a concern regarding the safety of using such products on human health, especially when these products are intentionally used as a pharmaceutical product or as health supplement. Furthermore, there are studies that show migration of silver from food contact material. The resident gut virus and phages play major roles in host health and diseases.31–33 Additionally, the importance of resident gut viruses, particularly phages, has been recently revisited and are now believed to be stable residents of the virobiota known to be involved in deriving the gut bacteria diversity as well as modulating bacterial virulence.19 Pharmaceutical quality control microbiology mainly focuses on the prevention of infection. However, several pharmaceutical products cause dysbiosis, which may be a result of alteration in the population of the gut commensal bacterial and viral/phage population. Moreover, the importance of interactions between host, bacteria and phages has recently attained much attention. Keeping the integrity of the gastrointestinal environment is critical to maintain the benefits of the commensal gut bacteria and viruses, albeit several factors including several pharmaceutical products, xenobiotics, diet and antimicrobials perturb the microbial population and composition of the gastrointestinal tract. It is therefore essential to monitor the potential impacts of AgNP on the resident commensal gut microbes since nanoparticles are used and continued to be applied by various food and medical industries to inhibit the growth of pathogenic microbes. However, there is an absolute ignorance that the pharmaceutical products used as antimicrobial agents may also inhibit the growth and proliferation of the commensal and beneficial microbes. Here in this study, we demonstrated the effects of nanoparticles as well ion-specific silver. This study primarily focused on the analysis of DNA virus/phages because DNA phage families are the dominant group.34–37 Furthermore, majority of the RNA viruses found in human feces are of transient plant viruses (not residents), possibly ingested with food.37,38
To obtain a more visual interaction of AgNP and AgOAC with virobiome, a phylogenic tree graph was generated using the software programs PAUDA and MEGAN.39–41 This taxonomy tree shows viral groups of significant differences between the control and the AgNP- or AgOAC-treated fecal samples. Overall, there was increased abundance of the Enterobacteria phage T7 and T7likevirus phage population that have the lysogenic property and harbor on the gram-negative bacterial population. By contrast, during AgOAC treatment, there was higher abundance of lysogenic phages (Lactobacillus phage phi adh and Lactobacillus phage Lv-1) that harbor on the commensal bacteria Lactobacilli. This clearly indicates that perturbation of AgNP might have affected the commensal microbiome and thus there is a proliferation of bacteria belonging to the Enterobacteriaceae family, a known effect of AgNP that has been earlier published by our group.7 Due to the higher abundance of Enterobacteria; the population Enterobacteria phage T7 and T7likevirus phages increased. Silver ions treatment caused abundance of phages that harbor on the commensal bacteria (Lactobacillus). Recent studies in humans and animals demonstrated a relatively higher concentration of bacteriophages sticking to the gastrointestinal mucus layer than the immediate surrounding environment, which likely gives the phages an easy access to their bacterial hosts. In such a way, the mucus-associated phages reduce the number of bacteria and potentially protect the host’s mucosal surfaces from bacterial infection.42 The nonhuman primate gut microbiota, as used in the present study, may not exactly mimic the human gut microbiota. Nonetheless, the animal model used in the present study provided strong evidence for the phage-bacteria interaction. Our earlier studies showed that a 13-week oral exposure to AgOAC in Sprague-Dawley rats caused gastrointestinal distress in the animals, thus indicating that the lysogenic phages (Lactobacillus phage phi adh and Lactobacillus phage Lv-1) might be killing the commensal protective bacteria Lactobacillus during AgOAC treatment.7 The present study clearly provides clues that by the virtue of their unique nature, lytic bacteriophages infect their specific bacterial hosts and kill the bacteria as the phages replicate. Earlier studies reported that patients suffering from gastrointestinal disorders tend to have a different structure and composition of gastrointestinal bacteriophages than the healthy controls, suggesting the role of bacteriophages in the development of intestinal disease such as inflammatory bowel disease.43,44 Recently, there is an emergence of microbiome modulator products in the nanomedicine industry. These products are designed to modify bacterial populations in the gastrointestinal tract to aid in prevention or treatment of health conditions. Imbalance in the commensal microbiota is a known cause for many chronic diseases such as Clostridium difficile infections, antibiotic-associated diarrhea, diabetes, irritable bowel syndrome, Crohn’s disease and ulcerative colitis.
This study provides insight into the nanoparticle disruption of microbiome and clearly shows the importance of inclusion of bacterial and viral/phage analysis interaction during safety assessment of xenobiotic products that are predicted to have antimicrobial properties.
In summary, this study demonstrates that the AgNP have the capability to inactivate gastrointestinal phages/virus that is concentration, temperature and time dependent. The bioinformatics techniques were utilized to obtain confidence in the data when the sample size was limited and to visualize the virome comparative changes in a phylogenic tree graph to further aid in the interpretation of results.
Acknowledgments
The authors sincerely thank Dr Scott Lute, CDER, US Food and Drug Administration (US-FDA), for providing phages and their respective hosts. We highly appreciate the guidance provided by Dr Angel Paredes from NanoCore facility, NCTR, for help with the electron microscopy. Assistance provided by the primate care facility personnel to collect monkey fecal samples is highly appreciated. Dr Bekele was a participant of US-FDA Commissioner’s Fellowship Program. This study was supported by the NCTR/US-FDA (E07506).
Footnotes
Disclosure
The findings and conclusions presented in this manuscript are those of the authors and do not necessarily represent the views of the US-FDA. The authors report no other conflicts of interest in this work.
References
- 1.Williams K, Valencia L, Gokulan K, Trbojevich R, Khare S. Assessment of antimicrobial effects of food contact materials containing silver on growth of Salmonella Typhimurium. Food Chem Toxicol. 2017;100:197–206. doi: 10.1016/j.fct.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Fröhlich EE, Fröhlich E. Cytotoxicity of nanoparticles contained in food on intestinal cells and the gut microbiota. Int J Mol Sci. 2016;17(4):509. doi: 10.3390/ijms17040509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadrup N, Lam HR. Oral toxicity of silver ions, silver nanoparticles and colloidal silver: a review. Regul Toxicol Pharmacol. 2014;68(1):1–7. doi: 10.1016/j.yrtph.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson OD, Klochkov SG, Novikova OV, et al. Toxicity of nanosilver in intragastric studies: biodistribution and metabolic effects. Toxicol Lett. 2016;241:184–192. doi: 10.1016/j.toxlet.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 5.van den Brule S, Ambroise J, Lecloux H, et al. Dietary silver nanoparticles can disturb the gut microbiota in mice. Part Fibre Toxicol. 2016;13(1):38. doi: 10.1186/s12989-016-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Zande M, Vandebriel RJ, Van Doren E, et al. Distribution, elimination, and toxicity of silver nanoparticles and silver ions in rats after 28-day oral exposure. ACS Nano. 2012;6(8):7427–7442. doi: 10.1021/nn302649p. [DOI] [PubMed] [Google Scholar]
- 7.Williams K, Milner J, Boudreau MD, Gokulan K, Cerniglia CE, Khare S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology. 2015;9(3):279–289. doi: 10.3109/17435390.2014.921346. [DOI] [PubMed] [Google Scholar]
- 8.Xiang D, Zheng Y, Duan W, et al. Inhibition of A/Human/Hubei/3/2005 (H3N2) influenza virus infection by silver nanoparticles in vitro and in vivo. Int J Nanomedicine. 2013;8:4103–4113. doi: 10.2147/IJN.S53622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaikwad S, Ingle A, Gade A, et al. Antiviral activity of mycosynthesized silver nanoparticles against herpes simplex virus and human parainfluenza virus type 3. Int J Nanomedicine. 2013;8:4303–4314. doi: 10.2147/IJN.S50070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekele AZ, Gokulan K, Williams KM, Khare S. Dose and size-dependent antiviral effects of silver nanoparticles on feline calicivirus, a human norovirus surrogate. Foodborne Pathog Dis. 2016;13(5):239–244. doi: 10.1089/fpd.2015.2054. [DOI] [PubMed] [Google Scholar]
- 11.Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13(6):278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2(1):63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64(1):69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wommack KE, Nasko DJ, Chopyk J, Sakowski EG. Counts and sequences, observations that continue to change our understanding of viruses in nature. J Microbiol. 2015;53(3):181–192. doi: 10.1007/s12275-015-5068-6. [DOI] [PubMed] [Google Scholar]
- 15.Wittebole X, De Roock S, Opal SM. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence. 2014;5(1):226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11(1):237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endersen L, O’Mahony J, Hill C, Ross RP, McAuliffe O, Coffey A. Phage therapy in the food industry. Annu Rev Food Sci Technol. 2014;5:327–349. doi: 10.1146/annurev-food-030713-092415. [DOI] [PubMed] [Google Scholar]
- 18.Looft T, Allen HK, Cantarel BL, et al. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014;8(8):1566–1576. doi: 10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14(7):654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4(1):4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadwell K. Expanding the role of the virome: commensalism in the gut. J Virol. 2015;89(4):1951–1953. doi: 10.1128/JVI.02966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vance ME, Kuiken T, Vejerano EP, et al. Nanotechnology in the real world: redeveloping the nanomaterial consumer products inventory. Beilstein J Nanotechnol. 2015;6:1769–1780. doi: 10.3762/bjnano.6.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams KM, Gokulan K, Cerniglia CE, Khare S. Size and dose dependent effects of silver nanoparticle exposure on intestinal permeability in an in vitro model of the human gut epithelium. J Nanobiotechnology. 2016;14(1):62. doi: 10.1186/s12951-016-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershey AD, Bronfenbrenner J. Stepwise liberation of poorly sorbed bacteriophages. J Bacteriol. 1943;45(3):211–218. doi: 10.1128/jb.45.3.211-218.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang DX, Chen Q, Pang L, Zheng CL. Inhibitory effects of silver nanoparticles on H1N1 influenza A virus in vitro. J Virol Methods. 2011;178(1–2):137–142. doi: 10.1016/j.jviromet.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Kim BS, Kim JN, Cerniglia CE. In vitro culture conditions for maintaining a complex population of human gastrointestinal tract microbiota. J Biomed Biotechnol. 2011;2011:838040. doi: 10.1155/2011/838040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10(9):607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13(5):1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 30.Rai M, Deshmukh SD, Ingle AP, Gupta IR, Galdiero M, Galdiero S. Metal nanoparticles: the protective nanoshield against virus infection. Crit Rev Microbiol. 2016;42(1):46–56. doi: 10.3109/1040841X.2013.879849. [DOI] [PubMed] [Google Scholar]
- 31.Columpsi P, Sacchi P, Zuccaro V, et al. Beyond the gut bacterial microbiota: the gut virome. J Med Virol. 2016;88(9):1467–1472. doi: 10.1002/jmv.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalmasso M, Hill C, Ross RP. Exploiting gut bacteriophages for human health. Trends Microbiol. 2014;22(7):399–405. doi: 10.1016/j.tim.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci U S A. 2016;113(37):10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackermann HW. Phage classification and characterization. Methods Mol Biol. 2009;501:127–140. doi: 10.1007/978-1-60327-164-6_13. [DOI] [PubMed] [Google Scholar]
- 35.Foca A, Liberto MC, Quirino A, Marascio N, Zicca E, Pavia G. Gut inflammation and immunity: what is the role of the human gut virome? Mediators Inflamm. 2015;2015:326032. doi: 10.1155/2015/326032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A. 2013;110(30):12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4(1):e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC. Methods for comparative metagenomics. BMC Bioinformatics. 2009;(10 Suppl 1):S12. doi: 10.1186/1471-2105-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huson DH, Xie C. A poor man’s BLASTX: high-throughput meta-genomic protein database search using PAUDA. Bioinformatics. 2014;30(1):38–39. doi: 10.1093/bioinformatics/btt254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013;110(26):10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner J, Maksimovic J, Farries G, et al. Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm Bowel Dis. 2013;19(8):1598–1608. doi: 10.1097/MIB.0b013e318292477c. [DOI] [PubMed] [Google Scholar]