SUMMARY
DNA damage repair (DDR) pathways modulate cancer risk, progression, and therapeutic response. We systematically analyzed somatic alterations to provide a comprehensive view of DDR deficiency across 33 cancer types. Mutations with accompanying loss of heterozygosity were observed in over 1/3 of DDR genes, including TP53 and BRCA1/2. Other prevalent alterations included epigenetic silencing of the direct repair genes EXO5, MGMT, and ALKBH3 in ~20% of samples. Homologous recombination deficiency (HRD) was present at varying frequency in many cancer types, most notably ovarian cancer. However, in contrast to ovarian cancer, HRD was associated with worse outcomes in several other cancers. Protein structure-based analyses allowed us to predict functional consequences of rare, recurrent DDR mutations. A new machine-learning-based classifier developed from gene expression data allowed us to identify alterations that phenocopy deleterious TP53 mutations. These frequent DDR gene alterations in many human cancers have functional consequences that may determine cancer progression and guide therapy.
In Brief

Knijnenburg et al. present The Cancer Genome Atlas (TCGA) Pan-Cancer analysis of DNA damage repair (DDR) deficiency in cancer. They use integrative genomic and molecular analyses to identify frequent DDR alterations across 33 cancer types, correlate gene- and pathway-level alterations with genome-wide measures of genome instability and impaired function, and demonstrate the prognostic utility of DDR deficiency scores.
INTRODUCTION
DNA damage repair (DDR) genes play key roles in maintaining human genomic stability. Loss of DDR function, conversely, is an important determinant of cancer risk, progression, and therapeutic response (Jeggo et al., 2016). DDR genes can be grouped into functional pathways defined by genetic, biochemical, and mechanistic criteria. Proteins in the same pathway often work in concert to repair specific types of DNA damage (Friedberg et al., 2004). Base excision repair (BER), nucleotide excision repair (NER), and the direct damage reversal/repair (DR) pathways repair DNA base damage, while mismatch repair (MMR) corrects base mispairs and small loops often found in repetitive sequence DNA. Homology-dependent recombination (HR), non-homologous end joining (NHEJ), the Fanconi anemia (FA) pathway, and translesion DNA synthesis (TLS) act alone or together to repair DNA strand breaks and complex events like interstrand crosslinks (Friedberg et al., 2004; Kass et al., 2016). All of the major DDR pathways, with the exception of the FA pathway, have been identified in virtually all organisms. This reflects the universal need to counter the chemical instability of DNA and repair additional damage (Aravind et al., 1999; Eisen and Hanawalt, 1999; Friedberg et al., 2004).
The consequences of DDR deficiency are becoming better understood through analyses of DDR gene alterations in cancer (Alexandrov et al., 2013; Forbes et al., 2017; Garraway and Lander, 2013; Martincorena and Campbell, 2015). For example, frequent TP53 somatic mutations in many cancer types can disrupt the DNA damage response, apoptosis, or senescence pathways active in many early-stage cancers (Bartkova et al., 2005; Fischer, 2017; Gorgoulis et al., 2005; Pfister and Prives, 2017). DDR deficiency may also lead to specific mutational “signatures,” e.g., the short tandem repeat instability linked to the inactivation or silencing of DNA MMR in colorectal, ovarian, or endometrial cancer (Alexandrov et al., 2013; Helleday et al., 2014; Kass et al., 2016)
The therapeutic implications of altered DDR function are becoming better known. Many anti-cancer agents act by generating DNA damage that, if unrepaired, may lead to cell death or senescence. DNA interstrand crosslinks (ICLs) and double strand breaks may be particularly difficult to repair, requiring coordination of the NER, BER, FA, and HR pathways (Duxin and Walter, 2015; Michl et al., 2016; Pearl et al., 2015). The loss of one or more DDR pathway, once recognized, can also be therapeutically targeted through synthetic lethality (Brown et al., 2017; Kaelin, 2005; Srivas et al., 2016). Examples include loss of expression of the DR pathway protein O6-methylguanine-DNA methyltransferase (MGMT), which sensitized cancer cells to alkylating chemotherapy agents (Soll et al., 2017; Weller et al., 2015); and BRCA mutant, HR-deficient breast and ovarian cancers, which are sensitive to inhibition of PARP1, a central protein in the BER pathway (Bryant et al., 2005; Farmer et al., 2005; Lord and Ashworth, 2017). Epigenetic silencing may phenocopy these DNA events.
The Cancer Genome Atlas (TCGA) DNA Damage Repair Analysis Working Group (DDR-AWG) used newly standardized Pan-Cancer Atlas (PanCanAtlas) data to systematically analyze potential causes of loss of DDR function and the resulting consequences across 33 different human cancer types. The loss of specific DDR pathways in cancer, in contrast to other cellular “hallmarks” of cancer (Hanahan and Weinberg, 2011), often generates stable—and thus more readily interpretable—”footprints” in cancer genomes, detected as an increased mutation burden, altered mutational signatures, or copy-number alterations including loss of heterozygosity (LOH). We provide below the most comprehensive analysis to date of DDR pathway gene alterations and their consequences in human cancer. Our results provide a useful resource to guide both mechanistic and therapeutic analyses of the role of DDR in cancer.
RESULTS
We performed analyses with a curated list of 276 genes encompassing all major DNA repair pathways: 208 genes were annotated to one or more specific DDR pathway, with an additional 68 genes annotated to key DDR-related pathways such as nucleotide pool maintenance (e.g., RRM1/2, the regulatory and catalytic subunits of ribonucleotide reductase); critical DNA damage response kinases (e.g., ATM, ATR, CHEK1/2, and WEE1); and genes recurrently mutated in cancer that modulate DDR (e.g., TP53, IDH1, and PTEN). We defined a “core DDR” gene set of 71 DNA repair pathway-specific and 9 DNA damage response genes that were used to facilitate parsimonious pathway representations and analyses (Table S1; Figures S1A and S1B).
Prevalent DDR Alterations across Cancer Types
We first determined the prevalence of DDR alterations across PanCanAtlas cancer types by integrating data on somatic truncating and missense mutations (Figures S1C and S1D), deep copy-number deletions defined by GISTIC (Mermel et al., 2011), and epigenetic silencing events. Binary calls for each event class for the 276 DDR genes across 9,125 PanCanAtlas samples are available through Data and Software Availability. The frequency of these somatic DDR gene alterations is shown in Figures 1A–1C (core DDR genes) and Figures S1E–S1G (all 276 DDR genes). For a complete list of TCGA cancer type abbreviations, please see https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations. Cancer types with a higher global mutation burden (e.g., in UCEC [uterine corpus endometrial carcinoma], COAD [colon adenocarcinoma], and READ [rectum adenocarcinoma] cancers) also had a higher mutation frequency in DDR pathways. Similarly, cancer types with a large number of somatic copy-number alterations (SCNAs; e.g., OV [ovarian serous cystadenocarcinoma], SARC [sarcoma], ESCA [esophageal carcinoma], and STAD [stomach adenocarcinoma]) also had a larger number of SCNAs in DDR pathways.
Figure 1. Cancer Types Display Variable DNA Damage Repair Gene Somatic Alterations.
(A) DDR gene alterations are frequent and non-uniformly distributed by type and frequency across cancer types. Clustered heatmap indicates the percentage (%) of samples in a cancer type (rows, with cancer types listed right, number of samples between parentheses) altered for at least one core gene in a given DDR pathway (columns, with core gene numbers indicated in parentheses for each pathway, bottom). Color intensity indicates the percentage altered, with the percentage given as a number in each cell. RGB color indicates mutations (red), deep deletions (blue), or epigenetic silencing through methylation (green). Gray scale indicates equal contribution from all three alteration types. A “u” symbol in cells indicates a statistically significant enrichment (FDR [false discovery rate] < 10%, difference in alteration percentages > 2%) in alterations. A “co” or “me” symbol in cells indicates a statistically significant (FDR < 10%) co-occurrence or mutual exclusivity of samples altered by mutation, deep deletion, or silencing. Only “co” relations were observed. The two rightmost columns, Mut.load and SCNA load, indicate average mutation frequency (non-silent mutations/Mb) and copy-number burden (number of copy-number segments) by cancer type.
(B) Mutations and deep deletions contribute disproportionately to alter HR genes across nearly all TCGA cancer types. Color and color intensity provide a visual summary of the relative contribution of alteration types to HR pathway variation. The vertical position of each cancer type symbol indicates the percentage altered samples. M+D, mutation and deletion; D+S, deletion and silencing.
(C) Multiple genes contribute to enrichment of DDR pathway alterations. Heatmap depicts for each core DDR pathway (columns) statistically enriched alteration frequencies for genes with >2% alterations. Color intensity indicates percentage altered, with the percentage given in each cell. Specific cancer examples representing gene and pathway associations are listed under each column.
(D) The top 50 most frequently mutated genes among 276 DDR genes. Genes are listed in order of frequency of non-synonymous mutations (y axis left, blue rectangles), together with the fraction of concurrent mutations and LOH events (y axis right, red bars). See also Figure S1 and S2.
Pathway enrichment analysis based on the core DDR gene list revealed several DDR pathways that were statistically enriched for alterations within a specific cancer type, e.g., HR pathway alterations in OV and BRCA (breast invasive carcinoma) cancers (Figure 1A). Nearly three-quarters (20/28, 71%) of associations among DDR pathways and cancer types were also observed using our more inclusive DDR gene set (Figure S1E). Some DDR genes were affected predominantly by one type of alteration, e.g., ALKBH3 and MGMT by epigenetic silencing. Other genes were altered in two or more ways, e.g., mutations in TP53, PTEN, PER1, and BRCA1/2 were frequently accompanied by LOH (Figure 1D). Gene-level mutation frequencies varied widely by cancer type and subtype. For example, mutations in TP53, PTEN, ERCC5, and IDH1 were highly enriched, while other genes such as SOX4, SLX1A, and GTF2H2 were much less frequently mutated (Figure S1J).
We next investigated the association of DDR gene alterations with overall mutation burden. It is already well established that cancers with somatic POLE and POLD1 mutations in their exonuclease domains or with microsatellite instability (MSI) exhibit a substantially higher mutation burden (Barbari and Shcherbakova, 2017; Ionov et al., 1993; Shinbrot et al., 2014; Thibodeau et al., 1993). Here, we observed only two DDR genes (TP53 and POLE) that demonstrated a substantially different association between alterations and overall mutation burden when compared against a background of all other non-DDR genes (Figure S1K).
One intriguing question is whether DDR genes would be identified as cancer drivers using mutation frequency-based prediction methods. To address this question, we analyzed 276 DDR genes in non-hypermutated cancer samples using five driver prediction tools: 20/20+ (Tokheim et al., 2016), Mut-Sig2CV (Lawrence et al., 2014), OncoDriveFML (Mularoni et al., 2016), MuSiC2 (Dees et al., 2012), and CompositeDriver (https://github.com/khuranalab/CompositeDriver). Our analysis used data and best practices from the PanCanAtlas Drivers/Essentiality Working Group (Bailey et al., 2018) and identified 48 DDR genes as potential drivers by at least one driver identification algorithm. Among these, 8 putative DDR driver genes were unique to a cancer type, and 18 were identified only as part of a PanCanAtlas analysis. The remaining 23 putative drivers were identified in analyses of one or more individual cancer type as well as in PanCanAtlas analyses (Table S2). These identifications are intriguing, though tentative in light of the challenge of identifying drivers against a background of biological heterogeneity. For example, TCEB1, which forms a complex with VHL, was identified as a prominent driver gene in KIRC (kidney renal clear cell carcinoma) (Sato et al., 2013).
In order to assess potential genomic consequences of DDR gene alterations, we performed a mutation signature correlation analysis focusing on loss-of-function alterations in the 48 genes we identified as putative cancer drivers (Table S2). This analysis used 21 mutational signatures derived by the PanCancer Signature group (Covington et al., 2014). Significant associations are displayed in Figure S2A. We confirmed that mutational signature #19 (POLE signature, corresponding to COSMIC [Catalog of Somatic Mutations in Cancer] signature #10) is increased by over 4-fold in POLE-altered cancers (p = 1.0e–6) at the PanCanAtlas level, and in UCEC with greater significance (fold change = 10.1 and p = 4.8e–7, Figure S2B). We also confirmed the association of signature #15 (temozolomide signature, corresponding to COSMIC signature #11) with MGMT alterations (Figure S2C).
Frequent Epigenetic Silencing of DDR Genes and Pathways
Epigenetic silencing was identified as an alternative prominent mechanism leading to recurrent gene deficiency. Stringent calling criteria (detailed in STAR Methods) led to the identification of 12 DDR genes exhibiting strong and consistent methylation-driven transcriptional silencing (Figures 2A and 2B). The most frequently silenced DDR genes were core DR (direct repair) pathway genes MGMT (11% of all the samples) and ALKBH3 (8%), followed by the core MMR genes MLH3 (5%) and MLH1 (4%) (Figure 2C). Epigenetic silencing was less often observed for genes in the HR and FA pathways, e.g., BRCA1, RAD51C, NSMCE3, and FANCF. Methylation silencing was the dominant alteration in MGMT (92.4% of all alterations) and was significantly associated with signature #15 through mutation signature analysis (Figure S2C).
Figure 2. Epigenetic Silencing of DDR Genes and Pathways in Cancer.
(A) Gene/probe pairs showing evidence of silencing. Gene expression for gene/probe pairs (x axis) was Z score-transformed based on probe methylation level then plotted as a mean Z score among samples within a methylated group. Negative false discovery rate (FDR)-corrected log10-transformed p values are plotted on the y axis. Green dashed lines indicate the cutoffs for mean Z scores and FDRs. Genes meeting cutoffs for evidence of silencing have red labels, with specific probes listed in parentheses (see STAR Methods for additional details).
(B) Gene expression and methylation are inversely correlated for silenced genes. Scatterplots show silenced gene/probe pairs for MGMT (two probes), EXO5, RAD51C, MLH1, and FANCF. Gene expression level is plotted on the y axis and methylation on the x axis with red dots representing silenced samples.
(C) Silenced genes are variably distributed across cancer types. Left: oncoprint plot displays the overall frequency of deleterious mutations, deletions, and epigenetic silencing events for each significantly silenced DDR gene (rows, with gene names listed to the right) across 8,739 PanCanAtlas samples. Cancer type is shown in the color key to the right. Frequencies were calculated over the entire cohort, with only altered samples plotted. Right top scale indicates the number of events by molecular type, with the distribution of alterations across cancer types.
(D) Heatmap depicting variable frequency of epigenetic silencing events across 33 cancer types and DDR pathways. Cancer types (rows, shown using the same color code as in C) and 12 significantly silenced DDR genes (columns). Bar plots (right) summarize the frequency of silencing events by pathway: DR (ALKBH3 and MGMT), HR(BRCA1, RAD51C, and NSMCE3), and MMR (MLH1, MLH3, and PMS2). Numbers (x axis) below each bar graph indicate the proportion of samples by cancer type with at least one epigenetically silenced gene annotated to that pathway.
(E) EXO5 silencing shows cancer subtype variation. Scatterplots as in (B) display the same silenced samples, now color-coded according to cancer subtypes as indicated by the dot color code bottom left. Grey dots represent samples that were expressed/not silenced. See also Figure S2 and S3.
The high frequency of EXO5 silencing (94.5% of all EXO5 alterations) was both unexpected and intriguing. Loss of function of this single strand-specific DNA exonuclease sensitizes cells to DNA base adduct and crosslink damage and UV, but not ionizing radiation, and leads to chromosomal instability (Sparks et al., 2012). EXO5 silencing was frequent in GBM (glioblastoma multiforme) (~46%) though not in LGG (brain lower grade glioma) (~4.5%) (Figure 2D). When broken down by cancer subtype, EXO5 silencing was exclusively observed in IDH wild-type GBM and LGG (50% and ~27%, respectively) and in a small fraction of HNSC (head and neck squamous cell carcinoma) human papillomavirus (HPV)-negative cancers (~4%). Other cancer subtypes with a high frequency of EXO5 silencing included STAD (HM-indel, i.e., MSI, ~57% and Epstein–Barr virus [EBV]-positive ~57%), HM-indel, i.e., MSI, COAD (44%), CIN (chromosomal instability) ESCA (~29%), and MSI UCEC (~28%) (Figure 2E). Of note, both EBV-positive (Cancer Genome Atlas Research Network, 2014) and MSI (Cancer Genome Atlas Network, 2012; Kandoth et al., 2013) cancer subtypes have been tightly linked to an extensive CpG island methylator (CIMP) phenotype (Hinoue et al., 2012; Toyota et al., 1999), whereas IDH wild-type, CIN, and HPV-negative cancer subtypes were not CIMP associated. Furthermore, EXO5 deficiency was significantly linked to signature #1 (Figure S2E). The consistent observation of EXO5 silencing in IDH wild-type brain cancers suggests EXO5 may play a role in the pathogenesis of subsets of these cancers.
Frequencies of epigenetic silencing also varied at the pathway level, as defined by silencing of at least one gene in a pathway. Frequent silencing of core DR, HR, and MMR pathway genes was observed in gastrointestinal cancers (ESCA, COAD, STAD, and READ), HNSC, and GBM (Figure 2D). We also observed frequent DR pathway silencing in DLBCL (lymphoid neoplasm diffuse large B cell lymphoma) and in UCEC (with MGMT + ALKBH3 silencing in >60% or >40%, respectively). SKCM (skin cutaneous melanoma), ACC (adrenocortical carcinoma), LGG, and UCEC were characterized by a high frequency of MMR pathway silencing, whereas OV, UCS (uterine carcinosarcoma), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), and TGCT (testicular germ cell cancers) all showed high-frequency DR and HR pathway silencing that was reflected in altered gene expression data (Figures S3A and S3B) and in limited reverse-phase protein array (RPPA) data on 23 DDR genes (Figures S3C–S3E).
Genomic Instability Linked to HR Deficiency and Prognosis
We observed positive correlations among six different SCNA scores that were used to characterize the extent of aneuploidy, LOH, and homologous recombination deficiency in PanCancer Atlas samples (Figure 3A). We also found moderate, statistically significant positive correlations between mutation burden and SCNA scores, contrary to a previously reported inverse correlation between SCNA and mutation frequency in 12 cancer types (Ciriello et al., 2013). A likely explanation for this discrepancy is our use of a much larger and more inclusive set of samples across more cancer types and including all mutation and SCNA data, as opposed to the restricted subset of ~500 selected functional events as in the previous study. SCNA burden (determined from the frequency of SCNA segments) and mutation burden (measured as non-silent mutations per Mb) were positively correlated across all PanCanAtlas samples (ρ = 0.36, p < 1e–16, Figure S4A), while hypermutated samples and hypersegmented samples were largely mutually exclusive.
Figure 3. Somatic Copy-Number Alteration Scores in Relation to Clinical Outcomes and DDR Gene Alterations.
(A) Matrix heatmap of mean Spearman correlation between SCNA scores and mutation load across 33 cancer types.
(B) Forest plot of association between homologous recombination deficiency (HRD) score and progression-free interval (PFI). Results are shown for 28 cancer types with valid outcomes data. Cancer type symbols to the left are followed by sample number (N) included in the model and the number of PFI events. Hazard ratios (HR) and HR 95% confidence intervals are shown to the right. The “P Value” represents the Cox proportional hazards model p value for differences in survival between high versus low HRD score samples. *, a statistically significant association after applying a false discovery correction (threshold 10%).
(C) Representative Kaplan-Meier (KM) survival curves for PFI of four cancer types as a function of high versus low HRD Scoring. Cancer samples in GBM, ESCA, PRAD, and ACC were defined as high HRD scoring if the HRD score was above the median within a cancer type. Log rank test p values are displayed in the top right-hand corner of each plot.
(D) Volcano plot of significance and magnitude of DDR gene ridge regression coefficients. Our ridge regression model fitted the alteration status of 276 DDR genes to HRD score across 8,464 cancer samples. Homologous recombination repair (HR) genes above a significance threshold of FDR < 0.2 are plotted and labeled in red.
(E) HRD scores of two cancer types stratified by BRCA1 and RAD51B alteration status. The two cancer types with the largest number of BRCA1 or RAD51B alterations are plotted to show HRD score distributions as a function of gene alteration status. Mann-Whitney U test p values are displayed above the bracket for each cancer-type-specific comparison. See also Figures S4 and S7.
Genomic scarring with large-scale genome instability has been attributed to homologous recombination deficiency (HRD) (Watkins et al., 2014). We calculated a HRD score, combined from HRD-LOH (Abkevich et al., 2012), LST (large-scale state transitions) (Popova et al., 2012), and NtAI (number of telomeric allelic imbalances) scores (Birkbak et al., 2012), for all PanCanAtlas samples using SCNA calls generated from ABSOLUTE (Figure S4B). This analysis also substantially extends earlier analyses using 15 cancer types (Marquard et al., 2015). HRD scores varied widely across cancer types, with the highest scores in ovarian cancer (OV) and the lowest in KICH (kidney chromophobe), KIRP (kidney renal papillary cell carcinoma), LAML (acute myeloid leukemia), and THCA (thyroid carcinoma). Many cancer types also had small subsets of samples with high HRD scores.
HRD scores were significantly associated with progression-free interval (PFI) in eight cancer types (Figures 3B and 3C). Higher HRD scores were often associated with shorter PFI (Figure 3C). Notable exceptions were higher HRD scores associated with better clinical outcomes in GBM and OV (Figure 3C), and with overall survival in OV (Figures S7B and S7C). This may reflect the use of potent, DNA-damaging platinum-based compounds as standard-of-care therapy for OV (Figure S7) (Mills et al., 2016). The association of a higher HRD score with better outcomes in GBM may be linked to IDH mutations. These have been associated to higher HRD (data not shown; Sulkowski et al., 2017) and better outcomes (Brat et al., 2015) than IDH wild-type GBM cancers. We identified additional contributions of DDR gene alterations to HRD scores by using a Bayesian ridge regression to model HRD scoring as a function of DDR gene alterations while controlling for cancer type as a covariate (Table S3). This analysis, performed in 8,464 samples, excluded potentially confounding POLE mutants and MSI-high cancers (Figures 3B–3D).
HR pathway genes with significant positive HRD associations included BRCA1, BRCA2, RAD51B, and RAD51C. BRCA1 alterations had the largest positive weight for predicting higher HRD scores. Alterations in either BRCA1 or RAD51B increased HRD score in most cancer types including the top two cancer types with the greatest number of alterations in either gene (Figure 3E). TP53 alterations were also associated with higher HRD scores, consistent with previous observations of a higher SCNA burden in TP53-mutated cancers (Ciriello et al., 2013). Other alterations in several MMR and NER genes had significant negative associations with HRD score, e.g., MLH1, TCEB3, and MSH2, suggesting a potential mutually exclusive relationship between HR and MMR or NER deficiencies.
Gene fusions with the potential to disrupt DDR function were identified using ChimerDB 3.0 (Lee et al., 2017) and TCGA Fusion Gene Data Portal (Yoshihara et al., 2015) then manually checked for read alignments to identify 188 high-confidence fusion events that involved 108 DDR genes in 205 cancer samples (Figure S4C). The most frequently affected DDR genes were SMARCA4, PTEN, RAD51B, and SMARCA1 (Figure S4D). A majority of the fusions, including 18 in HR and 5 in MMR genes, had breakpoints in DDR functional domains that were predicted to disrupt function with readily predictable therapeutic consequences, e.g., for the HR gene fusions involving RAD51B and BRCA1 (Figure S4E). Fusions also often had multiple partners, as well as breakpoints: e.g., RAD51B had three recurrent breakpoints with five different fusion partner genes (CEP170, ENOX1, NPC2, ZFYVE26, and PCNX) (Figure S4F).
Protein Structure Modeling of Recurrent DDR Mutations
We further examined potential functional consequences of relatively rare, recurrent mutations in 22 DDR genes by protein structural analyses and modeling. These included MGMT, PARP1, TOP3A, BLM, ERCC2, HFM1, POLE, POLD1, and POLQ, which collectively harbored 1,380 unique rare, recurrent non-synonymous mutations. Many of these mutations were found at domain interfaces or along solvent-accessible surfaces, and a substantial fraction (n = 370 or 26.8%) were predicted to be strongly destabilizing (ΔΔGfold ≥ 3 kBT) (Figures 4A and 4B; Figure S5). An additional small number (n = 26 or 1.9%) were predicted to be strongly stabilizing (ΔΔGfold ≤ −3kBT) and thus might affect function by restricting protein conformational changes (Figure 4C).
Figure 4. Rare, Recurrent DDR Gene Mutations Differentially Alter Protein Structural Stability.
Key DDR genes (listed in C as column labels) were selected for protein modeling based on the frequency of rare and recurrent mutations and experimentally determined structures that covered a majority of amino acid residues.
(A) POLE mutations are clustered in protein functional domains. Spheres represent mutations colored by mutation frequency (boxed key top right) overlaid on a POLE structural model. 3D mutation hotspots are present in both the exonuclease (blue) and polymerase (green) domains.
(B) Most MGMT somatic mutations likely alter protein structure. The location of mutations shown on a structural model as spheres, colored by predicted effect. Mutations with protein folding energy (ΔΔGfold) ≥ 3 kBT are predicted to be strongly destabilizing, whereas those altering stability by less than |ΔΔGfold| < 1 kBT were considered not significant (NS).
(C) Many DDR gene somatic mutations are predicted to destabilize protein structure. Structure-based calculations of the effect of 1,380 mutations on ΔΔGfold are plotted, with the number of unique mutations/proteins given in parentheses below each protein name.
(D) Altered protein stability is associated with greater burden of genomic alterations. Plot uses standardized Z scores (see STAR Methods) across cancer types to compare samples harboring strongly destabilizing versus non-destabilizing mutations. Association strength depended on the DDR gene, e.g., destabilizing mutations in POLB were associated with lower SCNA and higher mutation burdens, while mutations in PARP1 were associated with a higher SCNA and lower mutation burden.
(E) Altered stability in four proteins was associated with a large shift (Z > 0.5) in SCNA burden. Split violin plots show the different distributions of SCNA burden among samples with a destabilizing versus non-destabilizing mutations in each gene.
(F) Altered stability in five proteins was associated with a large shift (Z > 2.0) in mutation burden. See also Figure S5.
POLE exonuclease domain mutations are positively associated with a higher mutation burden (p = 0.02) and negatively associated with SCNA burden (p = 0.006). Structure-based modeling predicted an additional subset of POLE mutations that may destabilize POLE structure and reduce catalytic efficiency. In similar fashion, most MGMT somatic mutations are likely to affect MGMT activity by destabilizing protein structure. Molecular dynamics simulations of a subset of mutations (n = 86) in six proteins (POLE, POLD1, POLQ, ERCC2, HFM1, and BLM) revealed many additional mutations that were not predicted to be destabilizing but did alter protein dynamics (Figure S5). Thus, molecular modeling and protein dynamics in concert may reveal mechanisms by which somatic mutations alter DDR protein function.
In order to link other destabilizing DDR mutations to genomic instability, we compared both mutation and SCNA burden scores for genes with predicted strongly destabilizing versus non-destabilizing mutations (Figure 4D). The majority of predicted destabilizing mutations showed a positive association with either higher mutation and SCNA or mutation burden scores. Many genes displayed an inverse relationship between these two burden scores, with, e.g., destabilizing mutations in POLB associated with higher mutation and lower SCNA burden, whereas destabilizing PARP1 mutations were associated with lower mutation and higher SCNA burden (Figures 4D–4F). Thus, predicted destabilizing mutations may be directly influencing downstream burden scores by altering or abolishing DDR function.
Machine-Learning-Derived Expression Signature Predicts TP53 Inactivation
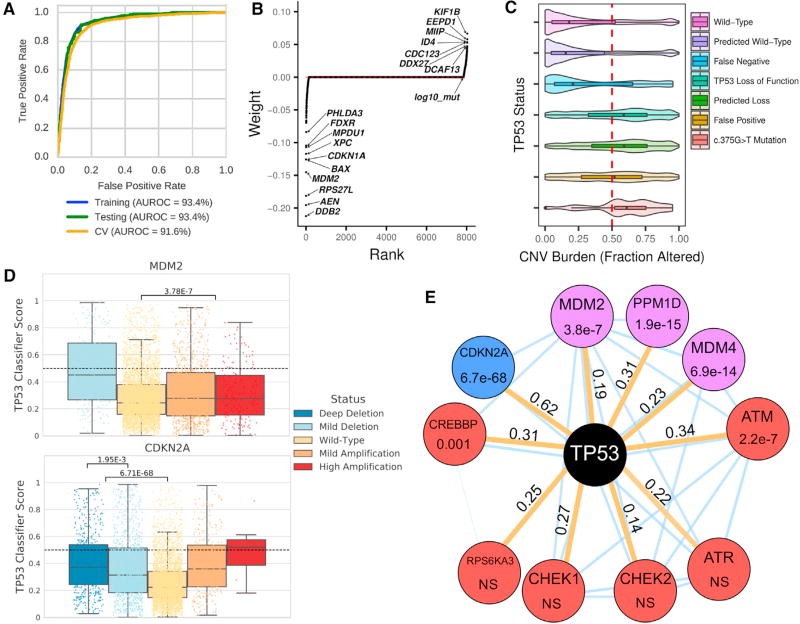
The loss of TP53 function across many cancer types has significant functional consequences as measured by genomic instability in association with a higher SCNA burden and increased HRD scores (Figures 1D and 3D). Cancer-associated TP53 mutations may promote these consequences through simple loss of function, as well as by altering transcription or through dominant-negative, gain-of-function mechanisms (Bouaoun et al., 2016; Kastenhuber and Lowe, 2017; Olivier et al., 2010; Pfister and Prives, 2017; Stracquadanio et al., 2016). A subset of these consequences can also be phenocopied by other genomic alterations. In order to better predict the consequences of TP53 inactivation and identify potential phenocopies of TP53 loss, we constructed a TP53 classifier that predicts inactivation status from RNA sequencing expression data, adjusted for cancer type and mutation burden (details in STAR Methods), then used this to analyze cancer types with comparable numbers of TP53 alterations. The resulting classifier was highly sensitive and specific (Figure 5A), and when trained using PanCanAtlas data, it outperformed individual cancer models in 14 out of 19 cases (Figures S6A and S6B).
Figure 5. Machine Learning to Predict TP53 Inactivating Mutations in Cancer.
(A) Robust classifier performance by receiver operating characteristic (ROC) and area under the ROC curve (AUROC). Training data, cross validation assessment, and held out test set (10%) for 19 cancer types were used.
(B) Model-derived gene weighting. Classifier weights indicate individual gene influence on classification accuracy. Negative weights indicate increased gene expression in TP53 wild-type samples.
(C) SCNA burden is correlated with known/predicted TP53 status. Plots show SCNA/CNV burden as fraction altered for known or predicted TP53 status. The SCNA profile for TP53 mutation c.375G>T in TP53 exon 4 appears similar to other TP53 loss events.
(D) SCNA in TP53-interacting genes MDM2 and CDKN2A phenocopies TP53 loss. Results shown are for PanCanAtlas TP53 wild-type samples.
(E) TP53 network gene alterations phenocopy TP53 deficiency. Mutations were manually curated and selected a priori. All mutation tests including only TP53 wild-type/non-hypermutated cancers are indicated by orange edges. Node color indicates event class (red, mutation; blue, copy-number loss; and purple, copy-number amplification); edge values indicate Cohen’s d effect size. Thin blue edges indicate predicted interactions from the STRING database. NS is “not significant” with p > 0.005. See also Figure S6.
Individual weights of the TP53 classifier identified 10 top negative-weighted genes, of which 9 are confirmed TP53 target genes (Kastenhuber and Lowe, 2017) (Figure 5B). The remaining gene, MPDU1, may have been identified by virtue of being located ~80 kb downstream of TP53 and thus sensitive to TP53 copy loss. Of note, our classifier was able to predict TP53 deficiency independent of cancer type with a high AUROC (area under the receiver operating characteristic curve; 0.94), and in samples initially removed from training. These included cancer types with few TP53 events (THCA and UVM [uveal melanoma] cancers), as well as those dominated by TP53 events (OV and UCS cancers) (Figures S6C–S6F). The classifier was also able to distinguish TP53 mutant from wild-type BRCA and UCEC, with nearly all basal-subtype BRCA cancers predicted to be TP53 deficient (Figures S6G and S6H). An analogous approach has been used to predict RAS pathway activation in PanCanAtlas cancers (Way et al., 2018).
The classifier enabled the identification of phenocopying mutations both in TP53 and in other functionally related genes. Consistent with previous pan-cancer analyses (Zack et al., 2013), we observed that predicted TP53 loss-of-function samples, including cancers with synonymous TP53 c.375G>T mutation, had an increased SCNA burden when compared with wild-type samples (Figure 5C). This synonymous mutation may act by altering a splice donor to produce alternatively spliced transcripts that compromise TP53 function (Collado-Torres et al., 2017). Samples with c.375G>T or c.375G>A mutations were also enriched for a 200 base pair truncation in exon 4 when compared with wild-type TP53 samples (OR [odds ratio] = 61.9, p < 2.2e–16). This mutation/truncation pairing was previously observed in a pancreatic cancer cell line and as a SNP (rs55863639) likely pathogenic for Li-Fraumeni syndrome (Leroy et al., 2014).
Significantly increased classifier scores were also noted for cancers with MDM2 copy-number amplification and CDK2NA copy-number deletion in an analysis including only non-hypermutated cancers without deleterious TP53 mutation (Figure 5D). We had observed a copy-number dosage effect for CDK2NA copy-number deletions, where loss of the CDKN2A-encoded P14ARF protein can phenocopy TP53 alterations (Sherr, 2001). Among eight other tested genes, MDM4 and PPM1D copy-number amplification and ATM and CREBBP gene mutations were associated with increased TP53 classifier scores, while ATR, CHEK1/2, or RP6SKA3 mutations were not (Figure 5E). These results suggest the general utility of this approach, even in circumstances where a diversity of molecular events and potential downstream consequences might occur.
DDR Alterations Are Associated with Clinical Outcomes
We computed multiple DDR footprint scores based on quantitative estimates of DNA damage and investigated their association with clinical outcomes. These aggregated 43 DDR footprint scores such as mutation burden and copy-number burden and extended other published scores, e.g., repair proficiency scoring (RPS) (see Data and Software Availability for per-sample estimates). We tested DDR footprint score associations with overall survival (OS) and progression-free interval (PFI) across 28 cancer types by fitting Cox proportional hazards models, using survival outcome data generated by Liu et al. (2018).
SCNA-based scores were more strongly associated with survival outcomes, compared with mutation- and expression-based DDR footprint scores (Figure S7A). LOH burden (number of genomic segments with LOH), combined HRD score, and HRD component scores (HRD-LOH [Abkevich et al., 2012], LST [Popova et al., 2012], and NtAI [Birkbak et al., 2012]) were significantly associated with OS and PFI for six to ten cancer types (ACC, BRCA, ESCA, GBM, KIRP, MESO [mesothelioma], PRAD [prostate adenocarcinoma], OV, UCEC, and UVM) (Figure S7B). In all but two cancer types (GBM and OV; Figures 3B and 3C), higher HRD or LOH burden scores were associated with poorer prognosis (Figure S7B). These associations remained statistically significant in multivariate Cox proportional hazard models after accounting for known covariates such as patient age, cancer type, and stage (Figure S7C).
Fewer mutation-based associations with clinical outcomes were identified: e.g., a high mutation burden was associated with better prognosis in UCEC and STAD cancers, but worse prognosis in LGG and ACC cancers (Figure S7C). Of note, expression-derived DDR footprint scores (e.g., expression Cumulative Density Function trAnsform of Rank Distribution or eCARD scores [Zimmermann et al., 2016]) derived for a single cancer type (OV) had significant prognostic associations in 7 cancer types for OS and for 6 cancer types for PFI (Figure S7A) that were the opposite of associations observed in OV and STAD (Figure S7B). Low repair proficiency scores were associated with a worse OS as previously reported (Pitroda et al., 2014), though for PFI in only a subset of cancer types (Figure S7B). These associations between DNA damage consequences and survival across 28 human cancer types confirm previously reported survival associations and provide a rationale for extending these analyses to additional cancer types.
DISCUSSION
We used TCGA PanCanAtlas data to systematically analyze the prevalence, nature, and consequences of DDR gene and pathway alterations across 9,125 samples representing 33 different cancer types. DDR gene alterations were ubiquitous: approximately 1/3 of TCGA PanCanAtlas cancer types showed significant enrichment of somatic mutations in DDR genes, often accompanied by SCNA/LOH events. The functional consequences of these alterations could often be readily inferred. For example, the HR pathway was altered in nearly 40% of cancers, e.g., in BRCA1/2-mutated ovarian and triple-negative breast cancers, where HR is a key determinant of platinum chemotherapy as well as PARP inhibitor response.
DNA methylation-dependent epigenetic silencing was also surprisingly frequent—though more variable—than mutation or deletion calls and encompassed one-third of TCGA cancer types. Nearly three-quarters (20 of the 28, or 71%) of statistically significant associations observed using our comprehensive annotation of DDR pathways (Figure S1E) were driven by silencing of genes in the MMR, HR, and DR pathways. Some of the recurrently silenced genes we identified have been previously identified, e.g., MGMT (Esteller et al., 1999), MLH1 (Cancer Genome Atlas Research Network, 2014; Kuismanen et al., 2000; Simpkins et al., 1999), MLH3 (Lhotska et al., 2015), BRCA1 (Esteller et al., 2000), and HLTF (SMARCA3) (Moinova et al., 2002). Epigenetic silencing of DR pathway genes in gastrointestinal, central nervous, and lymphoid cancers were all associated with a high mutation burden.
We also identified new, epigenetically silenced genes including DDB2 and EXO5. DDB2 is a NER pathway gene necessary for UV damage repair, whereas EXO5 (DEM1) is a little-studied single-stranded exonuclease that has been linked to genetic instability and DNA damage hypersensitivity especially to DNA crosslinking agents (Sparks et al., 2012). These findings highlight the role of epigenetics in shaping the DDR deficiency landscape in cancer and may provide useful biomarkers for enhanced response to, e.g., alkylating agent therapy. Three additional significant pathway enrichments were also identified when we excluded epigenetic silencing: the DR pathway in LGG cancers, the NER pathway in BLCA (bladder urothelial carcinoma) cancers, and the BER pathway in LIHC (liver hepatocellular carcinoma) cancers (Data and Software Availability).
Analyses of loss-of-function alterations within DDR pathways identified co-occurring alterations (largely consisting of mutations and epigenetic silencing) in the MMR pathway in UCEC and STAD cancers, though no pathway by cancer-type-specific, mutually exclusive combinations. We found little evidence for somatic mutation co-occurrence between MMR and NER pathways (Figure S1H), or MMR and HR pathways (Figure S1I). MMR and NER can repair DNA base pair mismatches, bulky DNA base damage, and small DNA loops. Thus, the loss of both MMR and NER might markedly sensitize cancer cells to alkylating agent therapy and provide a starting point to identify effective treatment combinations (Srivas et al., 2016). Of note, 1/5 (22%) of DDR genes participate in more than a single DDR pathway (Table S1; Figures S1A and S1B). Thus, alteration of these “pathway-promiscuous” DDR proteins may have a disproportionately large effect on genomic instability.
The detailed understanding of DDR genes and pathways provides immediately plausible mechanisms by which many DDR gene alterations might increase specific mutation types, as well as overall mutational burden. Mutational signature analyses provide a second way to identify potential mutation sources and mechanisms (Alexandrov et al., 2013; Rogozin et al., 2017). For example, we found a previously undefined signature 8 was strongly associated with BRCA deficiency, especially BRCA1 (Figure S2D). EXO5 deficiency, identified here as an often epigenetically silenced DDR gene, was associated with signature 1 across multiple cancer types (Figure S2E) and has been associated with poor clinical outcomes (Gingras et al., 2016; Totoki et al., 2014). Co-occurrence plots of mutations and SCNA/LOH for the top 50 mutated DDR genes in TCGA samples (Figure 1D) and genome-wide DNA damage scores that further encompass LOH and aneuploidy (Figure 3A) suggest the potential for additional complex interactions among DDR gene alterations.
We extended the insights gained from analyzing the type and distribution of DDR gene alterations in cancer by using additional approaches. For example, a combination of protein structural modeling and molecular dynamics simulations were used to predict the functional consequences of rare, recurrent nonsynonymous mutations in 22 DDR genes. We found that POLD1 mutations, despite being less common than the POLE or POLQ mutations that contribute to hereditary colorectal cancer risk (Bellido et al., 2016) and the hypermutated phenotype (Briggs and Tomlinson, 2013; Church et al., 2013), were as strongly associated with genomic instability. Molecular dynamics simulations further identified a subset of DDR gene mutations with the potential to alter protein conformational changes independent of effects on protein stability (Figure S5), raising the provocative question of whether destabilizing mutations alone contribute to genomic instability in cancer.
A second extension of PanCanAtlas genomic data was to use machine learning to predict TP53 inactivation status from gene expression data. This approach identified both TP53 mutant and TP53 mutant phenocopies, as well as potential TP53 tissue-specific roles in, e.g., ESCA and CESC cancers. This approach was developed using TP53 but is general and thus could be applied to other DDR genes. These and additional approaches may have their greatest value in annotating rare mutations where there may be few or no clinical or experimental data from which to predict mutation functional consequences.
In light of the central role played by DDR pathways in ensuring cell survival after DNA damage, we reasoned that DDR deficiency scores might be broadly predictive of both therapeutic response and clinical outcomes. When tested against PanCanAtlas survival data encompassing 28 cancer types (Liu et al., 2018), we identified associations among most DDR footprint scores and clinical outcomes after controlling for covariates such as age, cancer grade, and stage (Figure S7). These associations were consistent in linking a high mutation burden or genomic instability with worse clinical outcomes across almost all cancer types. We also identified HRD-high cancers including subsets of ovarian, uterine, lung squamous, esophageal, sarcoma, bladder, lung adenocarcinoma, head and neck, and gastric carcinomas. Virtually all of these subsets of cancers may have enhanced responsiveness to platinum-based compounds that are given as standard-of-care therapies. This extends the recognition of a bimodal distribution of HRD scores in breast and ovarian cancers and the enrichment of BRCA1/2 mutant or methylated cancers among HRD-high cancers that are more likely to respond to platinum-containing therapy (Telli et al., 2016). These results indicate the potential of HRD scoring to predict both platinum response and PARP inhibitor sensitivity.
The high frequency of DDR gene and pathway alterations in TCGA PanCanAtlas cancer samples identifies opportunities to improve cancer therapy. For example, HR defects are common in many cancer types and may compromise successful DNA replication and genome stability (Macheret and Halazonetis, 2015; Zeman and Cimprich, 2014). Thus, combination therapies that induce or potentiate replication stress or impair replication fork protection may be particularly effective in killing HR-deficient cancers (Ray Chaudhuri et al., 2016; Rondinelli et al., 2017; Taglialatela et al., 2017).
The many different cancer types that show epigenetic silencing of DR pathway genes such as MGMT and ALKBH3 may be especially vulnerable to types of DNA damage normally repaired by these proteins (Soll et al., 2017; Wang et al., 2015). DDR pathway deficiencies are also mechanistically linked to mutation burden and mutational diversity. Thus, DDR pathway deficiencies in cancer may potentiate immune-based therapies by driving neoantigen production to enhance immune recognition and targeting (Balachandran et al., 2017; Germano et al., 2017; Le et al., 2017) and thus may identify subsets of patients with a higher likelihood of responding to immune-based therapies.
Our analysis of DDR pathways across the 9,125 cancers of 33 types included in PanCanAtlas data identified associations among genomic data types; confirmed and extended several previously reported findings; and provided insight into the mechanistic origins and consequences of DDR deficiency in cancer. These results collectively reinforce the importance of DDR gene function in shaping cancer risk, progression, and therapeutic response.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
-
●
KEY RESOURCES TABLE
-
●
CONTACT FOR REAGENT AND RESOURCE SHARING
-
●
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
●
METHOD DETAILS
-
○
DNA Damage Repair Pathway Curation
-
○
Alteration summary for DDR pathways across cancer types
-
○
Filtering and functional annotation of somatic mutations in DDR genes
-
○
DDR gene epigenetic silencing events
-
○
Determination of deep deletions of DDR genes and SCNA-based DDR scores
-
○
Computation of homologous recombination deficiency (HRD) scores
-
○
Ridge Regression Analysis
-
○
Survival Analysis
-
○
Curation and Examination of DDR Fusion Events
-
○
Generating Protein Structural Models at Atomic Resolution
-
○
Molecular Dynamics Simulations
-
○
In-silico expression-based predictor of TP53 inactivation
-
○
-
●
DATA AND SOFTWARE AVAILABILITY
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for additional analysis details should be directed to and will be fulfilled by the Lead Contact: Chen Wang (wang.chen@mayo.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We used TCGA PanCanAtlas cancer samples defined by the whitelist commonly agreed upon by TCGA Analysis Working Groups (AWGs) (Data and Software Availability) for all analyses. These 9,125 samples encompassed 33 different histopathologic cancer types representing most major classes of human adult cancer. Some analyses were performed for 32 cancer types, with a 33rd type, AML (LAML) excluded in selected analyses as indicated. While a complete analysis of this cohort’s overall demographics are beyond the scope of this paper, relevant demographic factors such as biologic gender, patient age and disease characteristics were accounted for and are noted when reporting the results of specific analyses.
METHOD DETAILS
DNA Damage Repair Pathway Curation
DNA damage repair (DDR) gene list including 276 genes was assembled from relevant gene lists including MSigDB v5.0 (see Key Resources Table) an online catalog of DDR genes from recently published resources (Pearl et al., 2015; Key Resources Table), and knowledge-based curation of information on specific DNA repair pathways or subpathways (see, e.g., (Bell and Kowalczykowski, 2016; Kowalczykowski, 2015) on HR and HR-associated subpathways). Three-quarters (n = 211, 76%) of these 276 genes encompassed nine major DDR pathways: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), the Fanconi anemia (FA) pathway, homology-dependent recombination (HR), non-homologous DNA end joining (NHEJ), direct damage reversal/ repair (DR), translesion DNA synthesis (TLS), and nucleotide pool maintenance (NP)(Brown et al., 2017; Friedberg et al., 2004; Jeggo et al., 2016; Pearl et al., 2015; Tubbs and Nussenzweig, 2017). The remaining 65 genes have been linked to more than one DDR pathway, or coordinate cellular and molecular responses to DNA damage, and thus may represent an important focus for DDR pathway-associated therapeutic development (Brown et al., 2017; Pearl et al., 2015). The complete gene list is contained in Table S1.
Alteration summary for DDR pathways across cancer types
In order to provide a comprehensive overview of alterations in DDR genes, we combined three data sources of binary alteration calls in terms of loss-of-function events: 1). Deleterious mutations from exome sequencing; 2). Deep deletions from GISTIC calls; and 3). Epigenetic silencing through methylation versus expression analysis. Aggregation of the three data types and integration with whitelisted samples resulted in binary calls across 9,125 samples (see Data and Software Availability).
Overall alteration scores were computed by merging three binary calls, i.e., a sample was called altered for a gene if it was mutated, deleted and/or epigenetically-silenced. A sample was called altered for a DDR pathway if at least one gene in the pathway was altered. A permutation test was performed to analyze whether the alteration percentages in the DDR pathways were enriched or depleted (higher or lower than expected) using a null model, for which the genes were randomly assigned to each of the pathways. A gene-pathway graph is used to denote gene to pathway membership relationship. A total of 1,000,000 permuted gene-to-pathway graphs were generated using Birewire (Gobbi et al., 2014) Permuted alteration percentages were then compared to the actual percentages using the standard permutation test (Knijnenburg et al., 2009, 2011). Additionally, we analyzed whether alterations of different types tended to be mutually exclusive or co-occurring. For each combination of a cancer type and DDR pathway, we have three binary vectors that indicate for the samples of that cancer type whether at least one gene in a DDR pathway was mutated, deleted or silenced. These vectors were randomly permuted 100,000 times, after which the original alteration percentage is compared with the permuted percentages.
Filtering and functional annotation of somatic mutations in DDR genes
A total of 313,497 somatic mutations in 276 curated DDR genes were selected from the MC3 MAF (v0.2.8) and underwent the following stepwise filtering process (Figure S1C). In brief, “PASS” filter mutations were selected and supplemented manually with mutations called in whole-genome amplified (wga) samples and by gap-filler assays that were not marked with the “PASS” filter tag. Mutations with low mutant allelic fractions and mutations had a low variant coverage were then removed. Thereafter, common variants reported in ExAc, the Phase-3 1000 Genomes Project or in the ESP6500 databases were removed. The intronic mutations, the mutations in 3′ or 5′ UTR regions or UTR flanking regions, silent mutations, and small, in-frame insertions and deletions were also removed. Mutations were removed if samples were duplicate-sequenced, marked as PanCanAtlas “Do not use” samples, or not included in the MC3 exome bam free best pairing samples. Approximately 89% of raw mutation calls were filtered out by this process, leaving a final list of 279,002 mutations that were used for further analysis.
To estimate the probability of missense mutations being damaging, we further annotated these missense mutations using six commonly used functional prediction algorithms (Figure S1D): PolyPhen-2 (Adzhubei et al., 2013), SIFT (Kumar et al., 2009), Mutation Taster (Schwarz et al., 2014), Mutation Assessor (Reva et al., 2011), LR and LRT (Chun and Fay, 2009). The missense mutations that were called as “deleterious” or “damaging” by four or more algorithms were defined as “deleterious” mutations. TP53 hotspot mutation substitutions such as R175H and R273H that didn’t meet this threshold were manually rescued.
DDR gene epigenetic silencing events
Illumina Infinium DNA methylation bead arrays, including both HumanMethylation27 (HM27) and Human Methylation450 (HM450), were used to assay 9,106 cancer samples (i.e., 8,586 primary solid cancers, 361 metastatic cancers and 159 blood malignancies) across 33 different cancer types, together with 1,066 adjacent normal samples. We excluded 19 normal prostate samples for potential label switching as identified by the Working Group and confirmed by pathology re-review. An external HM450 brain dataset containing 58 sorted neuronal and glial cells (Guintivano et al., 2013) from post-mortem frontal cortex of normal individuals was introduced as a normal control as GBM had only two adjacent normal samples, as were 60 sorted blood samples from 6 healthy individuals examined by HM450 (Reinius et al., 2012)). Data from HM27 and HM450 were then combined and normalized using a probe-by-probe proportional re-scaling method to yield a common set of 22,601 probes with comparative methylation levels. Briefly, we modeled the difference between HM27 and HM450 by two different technical replicates (TCGA-07-0227/TCGA-AV-A03D, measured 44/198 times and 12/169 times on HM27 and HM450, respectively), and applied a proportional rescaling method to remove platform effects.
Probes located within potential promoter regions (upstream and downstream 1500bp flanking regions of Transcription Start Sites (TSSs) of all annotated transcripts by UCSC) of the 276 DDR genes were examined for evidence of epigenetic silencing. We started with such probes that are consistently unmethylated (median beta value < 0.2) in each of the normal tissue types as well as sorted blood cells. Within each cancer type, for each probe/gene pair, the gene expression was first Z-score transformed using the mean and standard deviation calculated with the unmethylated cancer samples (i.e., sample with a beta value of (0, 0.1)). Samples across all cancer types were then pooled together. We chose the probes that exhibited epigenetic silencing using the following criteria: 1)at least 5 samples were observed with a beta value of 0.3 or above (defined as the methylated group); 2) mean Z score of the methylated group was lower than −1.65; 3) FDR-corrected p value according to one-side t test on Z scores was lower than 1e–2 between unmethylated and methylated group; and 4) the maximum beta value of the methylated group was higher than 0.75. Probes meeting these standards were retained to summarize epigenetic silencing events at the gene level. For genes with only one retained probe, a beta value cutoff of 0.3 was applied to call their silencing status, while genes with multiple probes left, the cutoff was relaxed to 0.2 but requiring that greater than half of the probes consistently silenced for that gene.
For DDR gene RAD51C, there were no common probes located in the promoter region. However, probe cg14837411 on HM27 and probe cg27221688 on HM450 were only 100bp apart and both correlated with gene expression. We manually added these back based on a beta value cutoff of 0.2 or above and combining both probes to call RAD51C epigenetic silencing.
Determination of deep deletions of DDR genes and SCNA-based DDR scores
Binary deep deletion calls were made using output from a PanCan GISTIC2.0 run on the samples (Mermel et al., 2011). GISTIC calls of −2 (indicating a loss of more than half of baseline ploidy) were called as deep deletions. Deep deletion calls were transformed into binary matrix format and made available in the (see Data and Software Availability).
The two SCNA burden scores (number of segments and fraction of genome altered) were computed using relative copy number segment data (see Data and Software Availability). For the first score, we counted the number of segments present in the copy number profile for each TCGA sample, and for the second score, we took the percentage of base pairs present in the copy number profile for each sample that belonged to segments with either greater than 0.1 or less than −0.1 log2 fold-change from baseline ploidy. All SCNA burden scores are available in Data and Software Availability.
The aneuploidy score reports the total number of arm-level amplifications and deletions in each cancer and was computed using ABSOLUTE (Carter et al., 2012) and an arm-level clustering algorithm, as described in Taylor et al. (2018). The two LOH scores (total number of segments with LOH events and fraction of genome containing LOH events) were computed directly using output from ABSOLUTE (Carter et al., 2012). All aneuploidy and LOH scores are available online (see Data and Software Availability).
Computation of homologous recombination deficiency (HRD) scores
We calculated HRD scores following previous published 3 components of HRD/genome scarring scores: HRD-LOH (Abkevich et al., 2012), LST (Popova et al., 2012), NtAI (Birkbak et al., 2012) and the implementation of a sum of the three (Marquard et al., 2015). Segment LOH and SCNA were generated by TCGA Network Aneuploidy AWG using ABSOLUTE (Carter et al., 2012)
Ridge Regression Analysis
Bayesian ridge regression was performed on 276 DDR genes using alteration status and encoding tumor type as 33 additional binary variables. HRD scores were modeled for 8464 tumor samples (MSI-high and POLE mutant samples were excluded from this analysis). Maximally uninformative gamma-distributed priors with shape and rate parameters equal to 0.1 were used for the precision of coefficients weighting and regularization factor. Coefficient significances were computed by first dividing the coefficient values by the square roots of the diagonal elements of the variance-covariance matrix of the weights to obtain coefficient t-statistics, and then by performing two-tailed t tests with 8326 degrees of freedom on these values. The regression was performed in Python-3.6.3 using the linear_model module in scikit-learn-0.19.1.
Survival Analysis
We evaluated clinical outcomes associations using newly developed, well-validated clinical data for each cancer type by the PanCanAtlas Survival Working Group’s paper and following their recommendations (Liu et al., 2018). Their analysis was based on clinical outcomes data from primary sources that were analyzed with extensive quality control and validation.
We considered two primary clinical outcomes: overall survival (OS), defined as “the period from date of diagnosis until death from any cause”; and progression free interval (PFI), defined as “the period from date of diagnosis until the occurrence of an event in which the patient with or without the tumor does not get worse.” Within each cancer type, we dichotomized the cohort according to the median score of each DNA damage footprint score to create two groups (high versus low) for survival comparisons. For a given cancer type, we fit univariate and multivariate Cox proportional hazard models using the DDR footprint score group as a predictor. For multivariate models, we considered age, tumor grade, and tumor American Joint Committee on Cancer pathology stage as covariates. We divided cancer grade into two categories of “low-grade” for grades 1–2, and “high-grade” for 3–4. Cancer stages were grouped as “early” for stages I and II, and “late” for stages III and IV. Additional covariates such as subtype, gender and race were considered but not used because of sparsity or convincing evidence for inclusion. For each set of DDR footprint Cox models, we applied the Benjamini-Hochberg correction to control the false discovery rate for DNA damage footprint scores.
Curation and Examination of DDR Fusion Events
We downloaded 30,001 fusion gene (FG) transcript candidates from the ChimerSeq module of ChimerDB 3.0 (December 2016) (Lee et al., 2017). By overlapping these putative fusion genes with 276 DDR genes, we identified 889 fusions involving 224 DDR genes. Among these, 464 FGs including 173 DDR genes were derived from TCGA cancer samples. To reduce false positives, we used read count information together with FusionScan, TopHat-Fusion, and PRADA. Following their filtering criteria, we used fusion transcripts having ≥ 2 seed/junction reads for FusionScan and PRADA analyses, and those with ≥ 100 spanning pairs for TopHat-Fusion analyses that identified 289 FGs. We were able to augment these with 343 additional FGs involving 141 DDR genes by downloading putative FGs from TCGA Fusion Data Portal using our 276 DDR genes as a query (September 2016) (Yoshihara et al., 2015). Combining these two FG datasets we identified 488 FGs involving 174 DDR genes in 477 cancer samples.
We next checked the read alignments for each DDR fusion gene. RNA-seq data of 477 samples were downloaded using gdc-client. BAM files were processed to obtain unmapped reads only using Bowtie2. We created fusion transcript composed of 100 bp sequences before and after break points using nibFrag, one of the BLAT Suite of programs. After creating a 200 bp length index of each fusion transcript, we aligned the unmapped reads to this fusion sequence index, then manually checked the read alignments. If a FG had at least one 20nt-20nt seed spanning read at the break point with a stair-shaped mapping of all reads we considered this a high likelihood gene fusion. This analysis identified 192 high likelihood FGs involving 108 DDR genes in 209 cancer samples.
Generating Protein Structural Models at Atomic Resolution
We began our structure-based analysis from UniProt canonical isoform sequences, and searched the PDB (Berman et al., 2000) for existing experimental structures. Experimental structures exist for human POLQ domains including the DEAD-box, Helicase, Sec63, and polymerase. The first three domains were solved in a single crystal structure, 5A9J (Newman et al., 2015). The polymerase domain is available in 4X0P (Zahn et al., 2015). We will abbreviate these POLQ structures as DHS and Pol, respectively, that together encompass 65% of the POLQ protein. No experimental structure exists for full-length human POLE. Thus we utilized the high-resolution experimental structure from yeast (4M8O (Hogg et al., 2014); 57% sequence identity) to generate our initial model (Roy et al., 2010) of the first 1155 amino acids of POLE that encompasses the polymerase and exonuclease domains.
Experimental structures for full-length human POLD1 and HMF1 are also lacking. Thus we utilized homology modeling using as templates 3IAY (Swan et al., 2009) (52% identical), and 4KIT (Mozaffari-Jovin et al., 2013) (31% identical), respectively. The structure of human BLM was taken from 4CGZ (Newman et al., 2015) and TOP3A from 4CGY (Bocquet et al., 2014). ERCC2 has been solved (5IVW [He et al., 2016]), but at 94% sequence identity. Thus homology modeling was used to update the experimental structure to the target wild-type sequence.
The remaining proteins for which we applied molecular modeling (Figure 4)are available in the PDB at > 95% sequence identity. We used homology modeling to revert protein to wild-type sequences and to fill in missing loops. FoldX (Schymkowitz et al., 2005; Van Durme et al., 2011) was used to perform in silico mutagenesis and side chain rotamer optimization, and to calculate ΔΔGfold for each variant. Results are summarized and visualized in Figure 4 and Figure S5.
Molecular Dynamics Simulations
In order to provide a detailed assessment for the most recurrent variants in selected DDR proteins (n = 86; observed in at least 2 samples for POLQ or 3 samples for others), we utilized Generalized Born implicit solvent molecular dynamics (MD) simulations, which were carried out using NAMD (Phillips et al., 2005) and the CHARMM27 with CMAP (Mackerell et al., 2004) force field. We utilized an interaction cutoff of 12Å with strength tapering (switching) beginning at 10Å, a simulation time step of 1fs, and conformations recorded every 2ps. Each initial conformation was used to generate 2 replicates that were each independently energy minimized for 10,000 steps, followed by heating to 300K over 0.5ns via a Langevin thermostat and a further 6ns of simulation generated at 2fs/step with the final 5ns analyzed. Implicit solvation accelerates system kinetics due to lack of explicit motion by the solvent required for solute motion. For POLE we studied in greater depth the recurrent V411A/D/I/L/M and P286R substitutions. For each, 30ns of simulation trajectory was generated after minimization and heating, for each variant in triplicate, and the final 20ns analyzed. Across the 6 variants plus the WT, 640ns of MD trajectory was generated. All proteins were modeled without substrate (as apo structures). In total, 1118ns of MD trajectory was generated and used to better understand the differences in effect of different protein variants on dynamics.
All MD trajectories were first aligned to the initial wild-type conformation of each protein using Cα atoms. Root Mean Squared Deviation (RMSD) was calculated using Cα atoms. Principal Component (PC) analysis was performed in Cartesian space. Analysis was carried out using custom scripts, leveraging VMD (Humphrey et al., 1996) and the Bio3D R package(Grant et al., 2006). Protein structure visualization was performed in PyMol (The PyMOL Molecular Graphics System. Version 1.5.0.3. Schrödinger, LLC) and VMD.
We used wild-type simulations as a benchmark for determining if variants altered the intrinsic dynamics of each protein (ΔPC). We first identified the region of PC space sampled by the densest 90% of each protein WT simulation (Figure S5). If the median PC coordinate for a variant was outside of this region, we considered the variant to have altered the activation of the corresponding PC motion. Differences in folding energy were classified as moderately (ΔΔGfold ≥ 1 kBT ≈ 0.6 kcal/mol) or strongly (ΔΔGfold ≥ 3 kBT ≈ 1.8 kcal/mol) destabilizing.
In-silico expression-based predictor of TP53 inactivation
We trained a classifier to use RNA-seq expression data to predict TP53 functional status. In brief, we trained a logistic regression classifier with an elastic net penalty using the Scikit-learn implementation of stochastic gradient descent (Pedregosa et al., 2010). The labels (y) for the supervised task included samples with MC3 annotated deleterious TP53 mutations (samples with silent mutations were considered TP53 wild-type) and samples with TP53 deep copy number loss as predicted by the GISTIC2.0 algorithm (Mermel et al., 2011). We included cancer-types in the model that had greater than 15 samples in each class, and between 5% and 95% of samples in both classes. Other samples were removed (see Figure S6A). The features (X) consisted of the 8,000 most variably expressed genes by median absolute deviation (MAD). We dropped expression of TP53 itself from the features to prevent the model from relying on target gene data. MAD genes were z-scored and concatenated with binarized dummy variables for all cancer types and mutation burdens (total log10 mutation count) to adjust for potential confounding factors. To reduce the effect of mutation burden confounding, we also removed outlier samples with the most extreme hypermutation phenotypes (> 5 standard deviations above the mean log10 mutation count). The goal of the classification scheme was to determine the weights (w) that minimize the following objective function:
Where i indexes samples, p indexes genes, λ and l are regularization and elastic net mixing hyperparameters, respectively. We selected optimal hyperparameters by balanced 5-fold cross validation with the goal of inducing a sparse solution. We also used a balanced 10% held out set to test the performance of the classifier on data that was not used for training or hyperparameter optimization. We fit the final model on the remaining 90% of the data and report performance using receiver operating characteristic (ROC) curves and area under the ROC curve (AUROC) metrics.
We manually selected an a priori set of genes known to interact with TP53 for our phenocopying experiment (L.A. Donehower, unpublished data). We tested MDM2, MDM4, and PPM1D amplifications, CDKN2A deletions, and ATM, ATR, CHEK1, CHEK2, CREBBP, and RPS6KA3 mutations. For the copy number tests, we included both deep and shallow alterations in the altered set compared to cancers with wild-type profiles only. We removed tumors with deleterious TP53 mutations or deep copy number loss (n = 4,037). From the remaining 5,629 tumors, we removed 219 hypermutated cancers leaving an analytic set of 5,410 cancer samples. We performed independent t tests and calculated Cohen’s D effect sizes comparing the assigned TP53 classifier scores for wild-type against altered cancers. We considered variants significant if they were less than a Bonferroni adjusted p value (p > 0.005). We visualized the results in a network diagram presented in Figure 5E. The underlying interaction network was downloaded from the STRING database (version 10.5) (Szklarczyk et al., 2017). The thickness of edges in the STRING network display interaction confidence and were generated by experimental data. Note that there are no direct interaction edges between RPS6KA3 and TP53 and PPM1D and TP53.
We provide materials under an open source license to reproduce and expand upon this analysis at https://github.com/greenelab/pancancer. Additional details, including benchmarking analyses, are provided in Way et al. (2018).
Supplementary Material
KEY RESOURCES TABLE
Highlights.
DNA damage repair (DDR) gene alterations are prevalent in many human cancer types
Homology-dependent recombination (HR) and direct repair were most frequently altered
Loss of DDR function is linked to frequency and types of cancer genomic aberrations
Altered HR function can be associated with better or worse outcomes by cancer type
Acknowledgments
This work was supported by the following grants from the NIH: U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, P30 CA016672, U24 CA210949, and U24 CA210950. Investigators were supported in part by P01CA077852 (T.A.K., N.C., I.S., and R.J.M.), U01 CA217883 (T.A.K, N.C., and I.S.), R35GM124952 (Y.S., M.K., and H.C.), GBMF 4552 (C.S.G.), T32 HG000046 (G.P.W.), R01LM012806 (Z.Z., P.K., and P.J.), DOD W81XWH-12-2-0050, HU0001-16-2-0004 (J.L. and H.H.), W81XWH-16-1-0237 (R.A.), U24 CA210990 (C.Y.), CA190635 and P01CA193124 (L.L.), P50 CA136393 (C.W.), and the Mayo Clinic Center for Individualized Medicine (M.T.Z. and C.W.). We are grateful to Dr. Allison Kudla (Institute for Systems Biology) for her contributions in forming the graphical abstract.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.076.
AUTHOR CONTRIBUTIONS
DDR genes and pathways, T.A.K., L.W., L.L., G.B.M., P.W.L., D.A.W., L.S., R.J.M., and Y.X. Mutations, L.W., C.M., E.S., S.Z., and D.A.W. SCNAs, G.F.G., B.F., Y.X., and A.D.C. DNA methylation/gene expression, N.C., H.F., H.S., and P.W.L. RPPA, Y.L., G.B.M., R.A., and J.N.W. DDR footprint scores, T.A.K., L.W., M.T.Z., N.C., G.F.G., A.D.C., G.P.W., C.S.G., Y.L., R.A., B.F., C.Y., and D.W. Protein structure, M.T.Z., Y.S., M.K., and H.C. Fusions, P.K., P.J., and Z.Z. TP53 classifier, G.P.W., C.S.G., and L.A.D. Driver analysis, M.H.B. and L.D. Clinical associations, N.C., J.L., and H.H. Editing team, T.A.K., L.W., M.T.Z., N.C., G.F.G., A.D.C., H.F., R.J.M., Y.X., and C.W. Project leadership, R.J.M., Y.X., and C.W.
DECLARATION OF INTERESTS
Michael Seiler, Peter G. Smith, Ping Zhu, Silvia Buonamici, and Lihua Yu are employees of H3 Biomedicine, Inc. Parts of this work are the subject of a patent application: WO2017040526 titled “Splice variants associated with neomorphic sf3b1 mutants.” Shouyoung Peng, Anant A. Agrawal, James Palacino, and Teng Teng are employees of H3 Biomedicine, Inc. Andrew D. Cherniack, Ashton C. Berger, and Galen F. Gao receive research support from Bayer Pharmaceuticals. Gordon B. Mills serves on the External Scientific Review Board of Astrazeneca. Anil Sood is on the Scientific Advisory Board for Kiyatec and is a shareholder in BioPath. Jonathan S. Serody receives funding from Merck, Inc. Kyle R. Covington is an employee of Castle Biosciences, Inc. Preethi H. Gunaratne is founder, CSO, and shareholder of NextmiRNA Therapeutics. Christina Yau is a part-time employee/consultant at NantOmics. Franz X. Schaub is an employee and shareholder of SEngine Precision Medicine, Inc. Carla Grandori is an employee, founder, and shareholder of SEngine Precision Medicine, Inc. Robert N. Eisenman is a member of the Scientific Advisory Boards and shareholder of Shenogen Pharma and Kronos Bio. Daniel J. Weisenberger is a consultant for Zymo Research Corporation. Joshua M. Stuart is the founder of Five3 Genomics and shareholder of NantOmics. Marc T. Goodman receives research support from Merck, Inc. Andrew J. Gentles is a consultant for Cibermed. Charles M. Perou is an equity stock holder, consultant, and Board of Directors member of BioClassifier and GeneCentric Diagnostics and is also listed as an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays. Matthew Meyerson receives research support from Bayer Pharmaceuticals; is an equity holder in, consultant for, and Scientific Advisory Board chair for OrigiMed; and is an inventor of a patent for EGFR mutation diagnosis in lung cancer, licensed to LabCorp. Eduard Porta-Pardo is an inventor of a patent for domainXplorer. Han Liang is a shareholder and scientific advisor of Precision Scientific and Eagle Nebula. Da Yang is an inventor on a pending patent application describing the use of antisense oligonucleotides against specific lncRNA sequence as diagnostic and therapeutic tools. Yonghong Xiao was an employee and shareholder of TESARO, Inc and is employed by Genospace. Bin Feng is an employee and shareholder of TESARO, Inc. Carter Van Waes received research funding for the study of IAP inhibitor ASTX660 through a Cooperative Agreement between NIDCD, NIH, and Astex Pharmaceuticals. Raunaq Malhotra is an employee and shareholder of Seven Bridges, Inc. Peter W. Laird serves on the Scientific Advisory Board for AnchorDx. Joel Tepper is a consultant at EMD Serono. Kenneth Wang serves on the Advisory Board for Boston Scientific, Microtech, and Olympus. Andrea Califano is a founder, shareholder, and advisory board member of DarwinHealth, Inc. and a shareholder and advisory board member of Tempus, Inc. Toni K. Choueiri serves as needed on advisory boards for Bristol-Myers Squibb, Merck, and Roche. Lawrence Kwong receives research support from Array BioPharma. Sharon E. Plon is a member of the Scientific Advisory Board for Baylor Genetics Laboratory. Beth Y. Karlan serves on the Advisory Board of Invitae.
DATA AND SOFTWARE AVAILABILITY
The raw data, processed data and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PancanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The PancanAtlas publication page includes a dedicated data resource related to this manuscript. This resource contains alteration calls for the 276 DDR genes as well as the 43 DDR footprint scores across all TCGA samples. The mutation data can be found here https://gdc.cancer.gov/about-data/publications/mc3-2017). TCGA data can also be explored through the Broad Institute Fire-Browse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). Details for software availability are in the Key Resource Table.
References
- Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, Smith-McCune K, Broaddus R, Lu KH, Chen J, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Curr. Protoc. Hum. Genet Chapter 7. John Wiley & Sons; 2013. Predicting functional effect of human missense mutations using PolyPhen-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173 doi: 10.1016/j.cell.2018.02.060. https://doi.org/10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbari SR, Shcherbakova PV. Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy. DNA Repair (Amst.) 2017;56:16–25. doi: 10.1016/j.dnarep.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bell JC, Kowalczykowski SC. Mechanics and single-molecule interrogation of DNA recombination. Annu. Rev. Biochem. 2016;85:193–226. doi: 10.1146/annurev-biochem-060614-034352. [DOI] [PubMed] [Google Scholar]
- Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, Pons T, González S, Iglesias S, Darder E, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 2016;18:325–332. doi: 10.1038/gim.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Wang ZC, Kim J-Y, Eklund AC, Li Q, Tian R, Bowman-Colin C, Li Y, Greene-Colozzi A, Iglehart JD, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID, Thomä NH. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIa and RMI1. Nat. Struct. Mol. Biol. 2014;21:261–268. doi: 10.1038/nsmb.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, Olivier M. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum. Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, et al. Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S, Tomlinson I. Germline and somatic polymerase ε and d mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 2013;230:148–153. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-L, Rappailles A, Duquenne L, Huvet M, Guilbaud G, Farinelli L, Audit B, d’Aubenton-Carafa Y, Arneodo A, Hyrien O, Thermes C. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, et al. NSECG Collaborators. DNA polymerase ε and d exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Torres L, Nellore A, Kammers K, Ellis SE, Taub MA, Hansen KD, Jaffe AE, Langmead B, Leek JT. Reproducible RNA-seq analysis using recount2. Nat. Biotechnol. 2017;35:319–321. doi: 10.1038/nbt.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington K, Shinbrot E, Wheeler DA. Mutation signatures reveal biological processes in human cancer. bioRxiv. 2014 https://doi.org/10.1101/036541.
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, Mooney TB, Callaway MB, Dooling D, Mardis ER, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr. Opin. Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Second. ASM Press; 2004. [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Germano G, Lamba S, Rospo G, Barault L, Magrì A, Maione F, Russo M, Crisafulli G, Bartolini A, Lerda G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116–120. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- Gingras M-C, Covington KR, Chang DK, Donehower LA, Gill AJ, Ittmann MM, Creighton CJ, Johns AL, Shinbrot E, Dewal N, et al. Australian Pancreatic Cancer Genome Initiative. Ampullary cancers harbor ELF3 tumor suppressor gene mutations and exhibit frequent WNT dysregulation. Cell Rep. 2016;14:907–919. doi: 10.1016/j.celrep.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi A, Iorio F, Dawson KJ, Wedge DC, Tamborero D, Alexandrov LB, Lopez-Bigas N, Garnett MJ, Jurman G, Saez-Rodriguez J. Fast randomization of large genomic datasets while preserving alteration counts. Bioinformatics. 2014;30:i617–i623. doi: 10.1093/bioinformatics/btu474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LVF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CPE, Shen H, Byun H-M, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundström E-B, Sauer-Eriksson AE, Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase ε. Nat. Struct. & Mol. Biol. 2014;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- Kaelin WGJ., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Moynahan ME, Jasin M. When genome maintenance goes badly awry. Mol. Cell. 2016;62:777–787. doi: 10.1016/j.molcel.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijnenburg TA, Wessels LFA, Reinders MJT, Shmulevich I. Fewer permutations, more accurate P-values. Bioinformatics. 2009;25:i161–i168. doi: 10.1093/bioinformatics/btp211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijnenburg TA, Lin J, Rovira H, Boyle J, Shmulevich I. EPEPT: a web service for enhanced P-value estimation in permutation tests. BMC Bioinformatics. 2011;12:411. doi: 10.1186/1471-2105-12-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuismanen SA, Holmberg MT, Salovaara R, de la Chapelle A, Peltomäki P. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am. J. Pathol. 2000;156:1773–1779. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Lee K, Yu N, Jang I, Choi I, Kim P, Jang YE, Kim B, Kim S, Lee B, et al. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017;45(D1):D784–D789. doi: 10.1093/nar/gkw1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. Analysis of TP53 mutation status in human cancer cell lines: a reassessment. Hum. Mutat. 2014;35:756–765. doi: 10.1002/humu.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhotska H, Zemanova Z, Cechova H, Ransdorfova S, Lizcova L, Kramar F, Krejcik Z, Svobodova K, Bystricka D, Hrabal P, et al. Genetic and epigenetic characterization of low-grade gliomas reveals frequent methyl-ation of the MLH3 gene. Genes Chromosomomes Cancer. 2015;54:655–667. doi: 10.1002/gcc.22266. [DOI] [PubMed] [Google Scholar]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high quality survival outcome analytics. Cell. 2018;173 doi: 10.1016/j.cell.2018.02.052. https://doi.org/10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC, Richardson AL, Silver DP, Szallasi Z, Birkbak NJ. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016;35:909–923. doi: 10.15252/embj.201693860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Timms KM, Reid JE, Gutin AS, Krivak TC, Hennessy B, Paul J, Brown R, Lanchbury JS, Stronach EA. Homologous recombination deficiency score shows superior association with outcome compared with its individual score components in platinum-treated serous ovarian cancer. Gynecol. Oncol. 2016;141:2–3. [Google Scholar]
- Moinova HR, Chen W-D, Shen L, Smiraglia D, Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R, et al. HLTF gene silencing in human colon cancer. Proc. Natl. Acad. Sci. USA. 2002;99:4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Sci. 2013;341:80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JA, Savitsky P, Allerston CK, Bizard AH, Özer Ö, Sarlós K, Liu Y, Pardon E, Steyaert J, Hickson ID, Gileadi O. Crystal structure of the Bloom’s syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Res. 2015;43:5221–5235. doi: 10.1093/nar/gkv373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations inhuman cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2010;12:2825–2830. [Google Scholar]
- Pfister NT, Prives C. Transcriptional regulation ion by wild-type and cancer-related mutant forms of p53. Cold Spring Harb. Perspect. Med. 2017;7:a026054. doi: 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chi-pot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitroda SP, Pashtan IM, Logan HL, Budke B, Darga TE, Weichselbaum RR, Connell PP. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008291. 229ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, Delattre O, Sigal-Zafrani B, Bollet M, Longy M, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee J-E, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, Söderhäll C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Goncearenco A, De S, Lada AG, Poliakov E, Panchenko AR, Cooper DN. Mutational signatures and mutable motifs in cancer genomes. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx049. https://doi.org/10.1093/bib/bbx049. [DOI] [PMC free article] [PubMed]
- Rondinelli B, Gogola E, Yücel H, Duarte AA, van de Ven M, van der Sluijs R, Konstantinopoulos PA, Jonkers J, Ceccaldi R, Rottenberg S, D’Andrea AD. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Shinbrot E, Henninger EE, Weinhold N, Covington KR, Göksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, Palazzo JP, Fishel R, Goodfellow PJ. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA alkylation damage repair: lessons and therapeutic opportunities. Trends Biochem. Sci. 2017;42:206–218. doi: 10.1016/j.tibs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Kumar R, Singh M, Wold MS, Pandita TK, Burgers PM. Human exonuclease 5 is a novel sliding exonuclease required for genome stability. J. Biol. Chem. 2012;287:42773–42783. doi: 10.1074/jbc.M112.422444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R, Shen JP, Yang CC, Sun SM, Li J, Gross AM, Jensen J, Licon K, Bojorquez-Gomez A, Klepper K, et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J, Zeron-Medina J, Castro-Giner F, Tomlinson IP, Goding CR, et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat. Rev. Cancer. 2016;16:251–265. doi: 10.1038/nrc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, Liu Y, Sundaram RK, Hegan DC, Fons NR, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2463. https://doi.org/10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. & Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang J-W, Madubata C, Anand R, Levy B, Rabadan R, et al. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell. 2017;68:414–430. doi: 10.1016/j.molcel.2017.09.036. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ, The Cancer Genome Atlas Research Network. Cherniack AD, Beroukhim R, Meyerson M. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33 doi: 10.1016/j.ccell.2018.03.007. https://doi.org/10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- Tokheim CJ, Papadopoulos N, Kinzler KW, Vogelstein B, Karchin R. Evaluating the evaluation of cancer driver genes. Proc. Natl. Acad. Sci. USA. 2016;113:14330–14335. doi: 10.1073/pnas.1616440113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. A graphical interface for the FoldX forcefield. Bioinforma. 2011;27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- Wang P, Wu J, Ma S, Zhang L, Yao J, Hoadley KA, Wilkerson MD, Perou CM, Guan K-L, Ye D, Xiong Y. Oncometabolite D-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 2015;13:2353–2361. doi: 10.1016/j.celrep.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JA, Irshad S, Grigoriadis A, Tutt ANJ. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna A, Sander C, Cherniack AD, Mina M, Ciriello G, et al. Pan-cancer Ras pathway activation in The Cancer Genome Atlas. Cell Rep. 2018;23 doi: 10.1016/j.celrep.2018.03.046. https://doi.org/10.1016/j.celrep.2018.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Tabatabai G, Kästner B, Felsberg J, Steinbach JP, Wick A, Schnell O, Hau P, Herrlinger U, Sabel MC, et al. DIRECTOR Study Group. MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the director trial. Clin. Cancer Res. 2015;21:2057–2064. doi: 10.1158/1078-0432.CCR-14-2737. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RG. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. Mouse ENCODE Consortium. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng C-Z, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. & Mol. Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann MT, Jiang G, Wang C. Single-sample expression-based chemo-sensitivity score improves survival associations independently from genomic mutations for ovarian cancer patients. AMIA Jt. Summits Transl. Sci. Proc. 2016:94–100. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.