Abstract
Genomic association studies of common or rare protein-coding variation have established robust statistical approaches to account for multiple testing. Here, we present a comparable framework to evaluate rare and de novo noncoding single nucleotide variants, insertion/deletions, and all classes of structural variation from whole-genome sequencing (WGS). Integrating genomic annotations at the level of nucleotides, genes, and regulatory regions, we define 51,801 annotation categories. Analyses of 519 autism spectrum disorder families did not identify association with any categories after correction for 4,123 effective tests. Without appropriate correction, biologically plausible associations are observed in both cases and controls. Despite excluding previously identified gene-disrupting mutations, coding regions still exhibited the strongest associations. Thus, in autism the contribution of de novo noncoding variation is probably modest compared to de novo coding variants. Robust results from future WGS studies will require large cohorts and comprehensive analytical strategies that consider the substantial multiple testing burden.
Keywords: autism spectrum disorder, noncoding, missense, loss-of-function, whole-genome sequencing, de novo variation, structural variation, deletion, duplication, inversion, translocation, genetic risk, constraint, conservation, chromatin state, regulatory elements, enhancers, gene set enrichment indel, insertion, deletion, CWAS, category wide association study
The rapid progression of genomics technologies, coupled with expanding cohort sizes, have led to significant progress in characterizing the genetic architecture of complex disorders1–6. To date, studies have mainly focused on genotyping array technologies to survey common variants and large rare copy number variations (CNVs), as well as whole-exome sequencing (WES) to scan rare protein coding variants. Common variant genome-wide association studies (GWAS) have been particularly successful in adult-onset disorders and most loci discovered are in the noncoding genome7. In early-onset disorders with reduced fecundity, including autism spectrum disorder (ASD)8, discovery has been largely driven by the identification of extremely rare, gene-disrupting, de novo mutations that exert considerable risk4,5,9.
Whole genome sequencing (WGS) offers the opportunity to assay the contribution of rare variation in the noncoding genome, a potentially large and hitherto unexplored class of variation. Since noncoding variants mediate the specificity of gene expression at particular developmental stages, tissues, and cell types, identifying such variants could provide important insights into the biology underlying complex disorders10–12. However, interpreting WGS in the noncoding genome presents considerable challenges13. We do not have reliable estimates of the number of loci that could mediate risk, the extent of such risk, nor the genomic characteristics of such loci – keys to predicting the success of such an endeavor. Moreover, we lack a noncoding equivalent to the triplet code in protein coding regions14, which has been critical for predicting of which coding nucleotides will alter gene function when mutated. Any serious exploration of rare variants in the noncoding genome must acknowledge this uncertainty and account for the inevitable multiple comparisons that result, because failure to do so virtually assures the detection of false-positive associations and erroneous biological conclusions. Therefore, WGS association studies will require the same unbiased approaches and statistical rigor that have been applied to linkage, GWAS, or WES-based gene discovery.
Here, we present such an analytical framework and apply it to a family-based cohort. These analyses focus on ASD families with both affected and unaffected cases due to the well-documented contribution of de novo mutations and the existing genomic data that allow us to target families without known genetic risk factors4. Specifically, we examine 519 ASD cases, their unaffected sibling controls, and both parents (2,076 individuals, Supplementary Table 1) from the Simons Simplex Collection (SSC)15. De novo mutations were annotated at the level of nucleotides, genes (Fig. 1), and regulatory regions to define 51,801 annotation categories (Fig. 2). In a category-wide association study (CWAS), no annotation category achieved statistical significance; furthermore, many biologically plausible noncoding categories were enriched in controls at equivalent levels of significance as those enriched in cases (Fig. 3 and 4). We did not observe evidence of a noncoding category comparable to de novo loss-of-function coding mutations in terms of both effect size and frequency (Fig. 5). We have made this analytical framework publically available, along with the necessary annotation data.
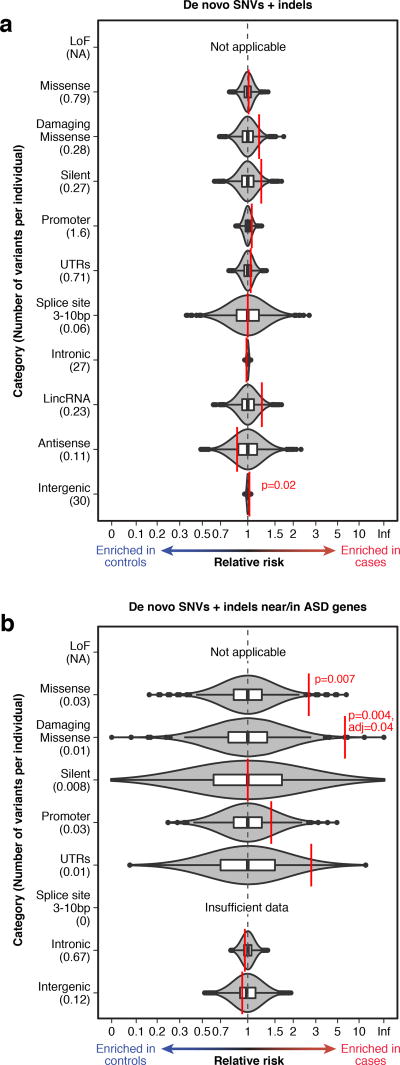
Figure 1. Burden analyses for gene-defined annotation categories.
a) The observed relative risk of de novo mutations in cases vs. controls is shown by the red line against grey violin plots representing the kernel density estimation of relative risk from 10,000 label-swapping permutations of case-control status for 11 gene-defined annotation categories. Box plots further illustrate the relative risk from permutations, including the median (center line), first and third quartiles (box), 1.5x interquartile range or the most extreme value (whiskers), and permuted relative risk observations beyond 1.5x interquartile range (outlier points). P-values from a case-control label-swapping permutation analysis and Bonferroni-corrected p-values (10 tests) ≤0.05 are shown. Loss-of-function variants were not analyzed as cases with such mutations were excluded from the cohort. b) The analysis in ‘a’ is repeated considering only de novo mutations in or near 179 ASD genes. Permutation p-values are Bonferroni-corrected for 7 tests. Considering SNVs and indels separately does not alter these findings (Supplementary Fig. 5).
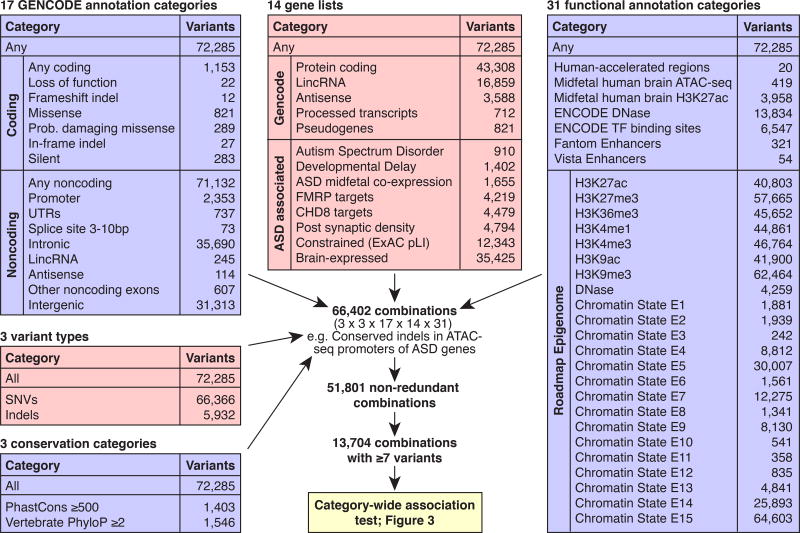
Figure 2. Defining annotation categories.
Five groups of annotations were defined: 1) Conservation across species; 2) Variant type; 3) GENCODE gene definitions; 4) Gene lists; and 5) Functional annotations. Picking one annotation from each group resulted in 66,402 possible combinations of which 51,801 were non-redundant (Supplementary Table 7). The 13,704 annotations categories that included at least seven observed mutations were considered in the category-wide association test.
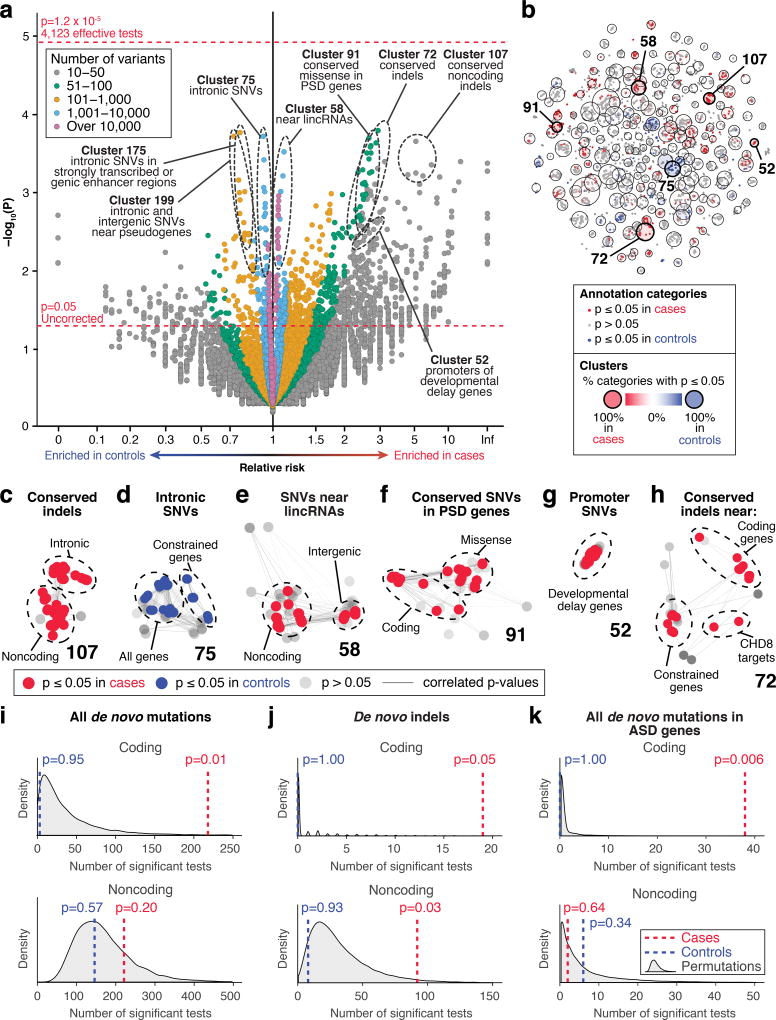
Figure 3. Category-wide association study.
a) The burden of de novo SNVs and indels in n=519 cases vs. n=519 controls for 13,704 annotation categories with ≥7 observed variants are shown as points in the volcano plot (Supplementary Table 7). Permutation p-values were calculated by 10,000 label-swapping permutations of case-control status in each annotation category. No test survives Bonferroni correction for 4,123 effective tests (top horizontal red line). b) Correlations of p-values between annotation categories (small dots) in simulated data are shown by proximity in the first two t-SNE dimensions. The large circles show 200 independent clusters of annotation categories defined by k-means clustering. The circle size represents the degrees of freedom accounted for by the cluster using Eigenvalue decomposition. In total, 4,123 effective tests explain 99% of the variability in p-values (Supplementary Fig. 6). c–h) Six clusters from (b) are shown in greater detail, with cluster number in bold. The edges represent p-value correlation ≥0.4. i–k) The number of nominally significant annotation categories (p≤0.05 from two-sided binomial test) was calculated for cases, controls, and 10,000 permutations to assess whether more annotation categories are enriched for de novo variants in cases than expected in (a). Cases have a greater than expected number of nominally significant categories relating to coding mutations and noncoding indels, but not for all noncoding mutations, nor for noncoding mutations nearest to ASD genes. P-values were calculated as the proportion of permutations in which the same or a greater number of categories had a two-sided binomial test p-value ≤0.05 as in the observed data.
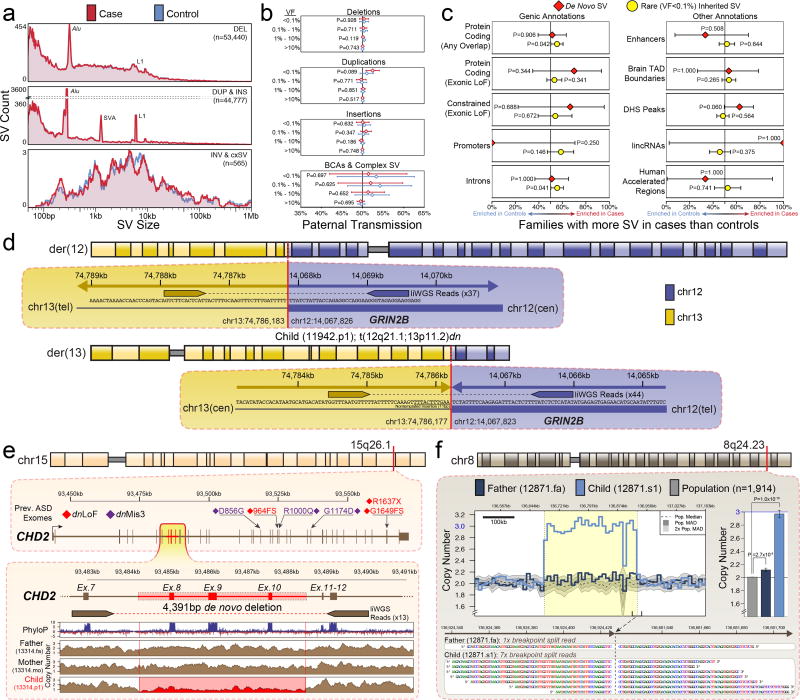
Figure 4. Structural variation in 519 ASD families.
Structural variation (SV) analyses identified an average of 5,863 SVs per genome 171 de novo SVs. a) We observed no difference in distribution of SV sizes between cases (n=519) and sibling controls (n=519) for any class of SV (cxSV = complex SV) at an unadjusted nominal significance threshold (two-tailed Wilcoxon rank-sum test; alpha = 0.05). b) We observed no differences in maternal/paternal transmission rates between cases and sibling controls for any class of SV or any range of variant frequencies (VF) (two-tailed binomial test). Mean paternal transmission rate (dot) and 95% binomial confidence intervals are shown in plot (error bars). c) We observed no significant enrichments for either de novo or rare inherited SV (VF < 0.1%) in genic or noncoding annotations after correcting for multiple comparisons in a two-sided sign test between case and control counts. Error bars represent the 95% confidence intervals. d) Analysis of balanced SV discovered a de novo reciprocal translocation in a case predicted to disrupt GRIN2B (t(12q21.2;13p11.2)), a constrained gene previously implicated in ASD4,21. e) WGS revealed small CNVs undetected by previous analyses, including a 4,391bp de novo deletion of exons 8–10 of CHD2 (GRCh37.63:chr15:g.93484245_93488636del), a gene previously implicated in ASD from de novo coding mutations4. f) Analysis of breakpoint sequences also classified 23 de novo SVs that were predicted to be germline mosaic in the parents, including this 242.8kb paternally transmitted mosaic duplication at 8q24.23 that was previously characterized as de novo in the child (GRCh37.63:chr8:g.136681615_136924426dup). Bar plots represent the means and 95% confidence intervals of estimated copy number in the duplicated locus. All p-values were calculated with a two-tailed t-test of estimated copy numbers in sequential 36.4kb bins.
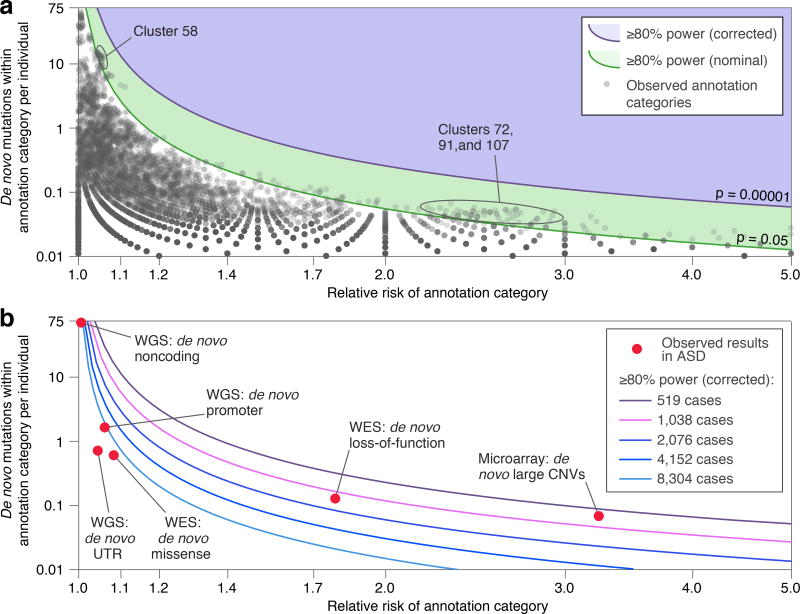
Figure 5. Effective number of tests in CWAS and power calculation.
a) The green line shows the threshold to achieve 80% power at nominal significance across the range of relative risks of a category (log10 scaled x-axis) and number of de novo mutations per individual within the category (log10 scaled y-axis). The purple line shows the 80% power corrected for 4,123 effective tests. The grey dots represent the observed results for de novo mutation burden in 519 families for the 13,704 annotation categories with ≥7 mutations. b) The lines show the threshold of 80% power across the range of relative risks and category sizes as sample size increases (correcting for correspondingly more effective tests, see Supplementary Information). For reference, the relative location for six classes of variation are shown.
Results
Cohort selection and characteristics
All 519 ASD cases were selected from the SSC based on the absence of de novo loss-of-function mutations or large de novo CNVs in prior WES and microarray data, with the objective to enrich this sample for undiscovered de novo variation. The majority of cases (92%, N=479/519) were selected randomly after this exclusion, while the remaining 8% were selected for a pilot study,16 and were enriched for factors associated with increased de novo burden: older fathers, female cases, and cases with nonverbal IQ ≤70 (Supplementary Table 1).
Identification of single nucleotide variants and insertion-deletions
Single nucleotide variants (SNVs) and small insertion-deletions <50 bp (indels)17 were discovered using the Genome Analysis ToolKit (GATK)18; family structure was leveraged to define high quality calls (Supplementary Fig. 1, 2). Overall, we identified 3.7 million autosomal variants per individual, including 3.4 million SNVs and 0.3 million indels. Six algorithms were employed to detect de novo SNVs and indels outside of low complexity regions and 1,638 previously validated de novo mutations (1,477 SNVs; 161 indels) were used to distinguish high confidence mutations (Supplementary Figs. 1, 2; Supplementary Table 2). Using independent experimental validation, confirmation rates compared favorably with published literature for both SNVs (96.8%, 212/219) and indels (82.4%, 145/176) (Online Methods and Supplementary Table 3)16. Both WGS and WES data were available for 990 children. In GENCODE-defined, autosomal coding regions, 1,116 de novo variants (1,075 SNVs, 41 indels) were detected by WGS compared to 896 de novo variants (869 SNVs, 27 indels) by WES (Supplementary Table 4). Of the 896 de novo WES variants, 870 were detected in the WGS data (97%; 849 SNVs, 21 indels) and 768 of these met our quality criteria (88% of 870; 754 SNVs, 14 indels). WGS identified an additional 348 de novo mutations (321 SNVs, 27 indels), in large part due to limited coverage in the WES data (Supplementary Table 4), including 19 predicted to result in loss-of-function and 58 missense predicted to be probably damaging by PolyPhen2. Considering variants not detected in exome analysis in all 1,038 children, we observed 24 de novo loss-of-function mutations, including three case mutations in genes predicted to be loss-of-function intolerant (CLCN3, FNBP4, PHIP), and 259 missense mutations, including seven probably damaging case mutations in genes predicted to be loss-of-function intolerant (Supplementary Table 4).
In WGS data, we observed a median of 64 de novo SNVs and 5 de novo indels per child across autosomes, for a total of 72,298 variants (66,366 SNVs; 5,932 indels) (Supplementary Table 5). We saw a slight excess of mutations in cases compared to their sibling controls that remained after applying linear regression to adjust for quality metrics influencing de novo mutation detection (RR = 1.024, p = 0.0006 for all mutations; RR = 1.024, p = 0.001 for noncoding mutations alone; one-sided binomial test; Supplementary Fig. 3). However, when we corrected for the effect of paternal age, which is known to affect mutation rates19,20, no significant difference in de novo burden remained (RR = 1.006, p = 0.2 for all variants; RR = 1.005, p = 0.24 for noncoding mutations alone; Supplementary Fig. 4). Correction for all covariates, including paternal age and sequencing quality, was applied to all subsequent tests of de novo mutation burden.
Case-control association tests of SNVs and indels
The sheer diversity and complexity of noncoding functional annotations necessitates a strategy to interpret the multiple parallel hypotheses they evoke. For gene-based analyses, we used GENCODE gene definitions and surveyed four coding categories (e.g. missense) and seven noncoding categories (e.g. UTRs) (Fig. 1a). In all analyses, we compared the number of de novo mutations that map to these regions in cases compared to sibling controls, and then assessed the significance of these comparisons using 10,000 within-sibship case/control label-swapping permutations. P-values were calculated as the proportion of permutations with RR as or more extreme than in the observed data, taking into account the direction of the observed RR (case burden, RR>1; control burden RR<1). For categories with empirical p < 0.01, we ran another 90,000 permutations for accuracy. This analytical approach is used throughout the manuscript, unless otherwise noted. After correcting for multiple comparisons, no significant excess of de novo variants in any gene-defined category was observed (Fig. 1a). Repeating the analysis considering SNVs and indels separately and considering only variants within or near to one of 179 genes associated with ASD (Supplementary Fig. 5) at a liberally defined false discovery rate (FDR < 0.3; Supplementary Table 6)4, only an excess of predicted damaging de novo missense mutations is apparent, though both promoter regions and UTRs showed a trend towards enrichment in cases (Fig. 1b). Neither constrained genes21 nor mRNA targets of Fragile X Mental Retardation Protein (FMRP)22 yielded nominally significant categories. Similar results were obtained when considering only variants at nucleotides conserved across species.
We next extended our analyses to include noncoding variation and designed a category-wide association study (CWAS) to assess multiple hypotheses. We integrated five approaches to annotation: 1) Gene sets implicated in ASD biology (e.g., targets of FMRP); 2) functional annotation (e.g., chromatin state); 3) conservation across species; 4) type of variant (e.g., SNVs, indel); and 5) GENCODE gene definitions. In total, we surveyed 51,801 distinct combinations of annotation categories (Fig. 2, Supplementary Table 7), comparing the burden of de novo mutations in cases vs. controls for each category (Fig. 3a). Eschewing a priori hypotheses, we treated all tests equally. Illustrating the risks in testing a limited set of investigator-selected annotation categories without appropriate correction, we observed equivalent p-values in the top categories enriched in either cases or controls, many of which would yield strong biological hypotheses (Table 1; Supplementary Table 7). For example, the top category enriched among cases was from conserved indels near protein coding genes within regions of weak transcription (chromatin state 5)23, while the top category enriched in controls was noncoding SNVs near loss-of-function intolerant genes within genic enhancers (chromatin state 6; Table 1).
Table 1.
Burden results for most significant annotation categories from CWAS.
| Variant type | Most significant categories within level of analysis | Variants per child (adjusted) |
Relative risk |
p-value uncorrected |
Number of comparisons |
p-value corrected |
|---|---|---|---|---|---|---|
| Cases – lowest p-value per cluster in CWAS for top five clusters | ||||||
| De novo indels | Conserved indels near protein coding genes within chromatin state 5 (Weak transcription) regions (Cluster 72) | 0.05 | 2.93 | 0.0002 | 4,123 | 0.66 |
| De novo SNVs | Conserved coding SNVs within post synaptic density genes (Cluster 91) | 0.06 | 2.63 | 0.0002 | 4,123 | 0.82 |
| De novo indels | Conserved intronic indels within chromatin state 15 (Quiescent) regions (Cluster 107) | 0.03 | 5.00 | 0.0002 | 4,123 | 0.91 |
| De novo SNVs | Intergenic SNVs near lincRNAs underlying H3K36me3 (Elongating) peaks (Cluster 58) | 4.69 | 1.11 | 0.0003 | 4,123 | 1.00 |
| De novo SNVs and indels | Conserved coding variants in post synaptic density genes within chromatin state 6 (Genic enhancer) regions (Cluster 23) | 0.01 | Inf | 0.0005 | 4,123 | 1.00 |
| Controls – lowest p-value per cluster in CWAS for top five clusters | ||||||
| De novo SNVs | Noncoding SNVs near constrained genes within chromatin state 6 (Genic enhancer) regions (Cluster 175) | 0.47 | 1.36 | 0.0002 | 4,123 | 0.70 |
| De novo SNVs | Intergenic SNVs near pseudogenes underlying H3K9me3 (Constitutive repression) peaks (Cluster 199) | 0.41 | 1.44 | 0.0002 | 4,123 | 0.78 |
| De novo SNVs | Intronic SNVs in constrained genes within chromatin state 5 (Weak transcription) regions (Cluster 75) | 6.04 | 1.09 | 0.0002 | 4,123 | 0.78 |
| De novo SNVs | Noncoding SNVs near constrained genes within chromatin state 5 (Weak transcription) regions (Cluster 20) | 7.10 | 1.07 | 0.001 | 4,123 | 1.00 |
| De novo SNVs and indels | Intergenic variants near genes co-expressed in midfetal brain within chromatin state 5 (Weak transcription) regions (Cluster 121) | 0.08 | 1.86 | 0.004 | 4,123 | 1.00 |
De novo SNVs and indels from n=519 cases and n=519 controls were compared using a case-control label-swapping permutation analysis as described in Results. P-values were Bonferroni-corrected for 4,123 effective tests.
Similar to the correlation structures seen in other forms of genome-wide analyses, many of these annotation categories encompass overlapping sets of variants and are thus dependent (Fig. 3b), raising the question of what constitutes an appropriate correction for multiple comparisons. To estimate this correction we generated 20,000 simulated datasets of annotated mutations (Supplementary Information) and assessed the correlation of p-values for the 51,801 categories across the simulations. Eigenvalue decomposition estimated 4,123 effective tests (Supplementary Fig. 6) leading to a category-wide significance threshold of 1.2×10−5 (Fig. 3a). No annotation category was within an order of magnitude of this threshold. K-means clustering of the simulated p-values was used to identify the 200 most independent clusters of annotation categories (Fig. 3b–h). Testing the single category within each cluster with the most mutations per individual, a metric independent of cluster enrichment, also failed to identify a category within an order of magnitude of a significance threshold corrected for these 200 tests (2.5×10−4; Supplementary Fig. 6).
We next considered whether there was evidence of a tendency towards enrichment of the 51,801 categories in cases, suggesting an underlying signal. We therefore counted the number of nominally significant categories and compared this to expectation (Fig. 3i–k). In coding regions, we observed more significant tests than expected in cases for SNVs and indels together (p = 0.01), but not in noncoding regions, either overall (p = 0.20) or near ASD genes (p = 0.64). Notably, categories restricted to noncoding de novo indels showed a greater number of nominally significant results than expected (p = 0.03, Fig. 3h).
To explore the concept of an underlying signal further, we developed a polygenic risk score based on de novo variants, akin to similar scores developed previously for common and rare variants24,25. The rate of de novo mutations in cases and controls was weighted based on the category RR and adjusted for p-value correlation structure (Fig. 3b). Cross validation was used to select annotation categories that best predicted case-control status. The resulting model included annotation categories relating to overall de novo burden (e.g. all variants, all intergenic variants) and conservation scores across vertebrate species, but not coding regions or other functional annotations. The derived score accounted for only 0.31% of the variability in case status, which was not significantly different from zero.
Finally, we explored the impact of rare inherited SNVs and indels in the 405 families of European ancestry (Supplementary Table 1)26. Since runs-of-homozygosity (ROH) blocks often contain multiple homozygous variants inherited simultaneously, we counted only one variant per ROH block and excluded variants in ROH blocks that overlapped coding regions. No significant excess of rare homozygous (≤1% allele frequency) or heterozygous (≤0.1% allele frequency) SNVs and indels was observed overall or separately for maternally or paternally inherited variants, and no category reached significance in a CWAS (Supplementary Figs. 7, 8).
Identification of structural variants
We next assessed whether structural variants (SVs), which rearrange large segments of the genome and can yield strong functional consequences, might demonstrate a noncoding signal. While much of the focus in SV detection from WGS has concentrated on CNVs alone, we previously demonstrated the importance of translocations, inversions, and inversion-mediated complex SVs in ASD and congenital anomalies27–29. We thus characterized all classes of SV accessible to short-read WGS. Our SV discovery pipeline integrated eight algorithms to capture changes in read-depth, clusters of reads with abnormal alignments, and mobile element insertions (Online Methods). We then developed an SV filtering pipeline to correct for the limited concordance among individual algorithms and several de novo prediction modules (Supplementary Figs. 9, 10). Statistically significant CNV segments were integrated with predicted balanced SVs using a series of breakpoint linking methods to identify signatures of 20 canonical, balanced, and complex SV classes (Supplementary Table 8)27,30.
These analyses identified 98,785 SV sites and a median of 5,843 SVs per individual (Supplementary Fig. 11). These variants resulted in 101 likely loss-of-function and 30 whole-gene copy gain SVs per person; 4.1% of all SVs altered coding sequence compared to 2.2% of SNVs and indels. We observed >99% sensitivity and a 2.5% FDR for CNVs from WGS compared to 1,087 CNVs previously reported from microarray data in these families (>40 kb; Supplementary Fig. 12)4. We relied on higher resolution long-insert WGS (3.5 kb inserts, 102x median physical coverage) on 456 cases to validate SVs below microarray resolution (10kb – 40kb) and found a 5.2% overall FDR for 986 SVs (Supplementary Fig. 12).
We detected 171 de novo SVs from these 519 families, including 158 germline and 13 predicted somatic mosaic SVs. To facilitate reproducibility between studies, Supplementary Table 9 and Supplementary Data provide localization and visualization of each predicted de novo SV that can be evaluated by independent researchers. Validation assays could be designed for 168 de novo SVs using five complementary approaches, which revealed a 97.0% validation rate (163/168; Supplementary Table 9), including five subjects with sex chromosome aneuploidies (0.7% of cases and 0.2% of controls; Supplementary Fig. 13). We also observed 23 SVs initially predicted to arise de novo that demonstrated evidence of germline mosaicism in a parent (Supplementary Table 10). Collectively, this catalogue of de novo SVs achieved high specificity, almost certainly at the cost of sensitivity for SV detection from short-read WGS, though there are few gold-standard datasets to estimate accurate SV mutation rates at present. Notably, a study from Turner et al. published during revision of this manuscript identified 88 de novo SVs from 476 of these quartets with an estimated 87.5% confirmation rate using microarray validation31.
Association analyses of SV
Given the rarity of de novo SVs, there were limited data to derive insights comparable to de novo SNVs and indels (Fig. 3; Supplementary Fig. 14). There was no significant difference in overall de novo SV burden between cases and controls (RR = 1.14, p = 0.47; Fig. 4a–c). There was a non-significant enrichment in cases for de novo loss-of-function SVs (1.3% in cases, 0.6% in controls; RR = 2.33; p = 0.34), which was more pronounced by removing mosaic SVs (p = 0.07), and included two SVs that disrupted ASD-associated genes: exonic deletion of CHD2 (GRCh37.63:chr15:g.93484245_93488636del) and a balanced translocation of GRIN2B (t(12q21.2;13p11.2); Fig. 4d–e). The GRIN2B translocation emphasizes the importance of surveying all SV classes. Four other cases harbored SVs that disrupted constrained genes (pLI > 0.9)21 that were not associated with ASD (LNPEP, CYFIP1, SAE1, ZNF462), while three occurred in siblings (USP34, NUCKS1, STS). No significant enrichments were detected in any class of noncoding variation or from CWAS-based analyses of SVs after correction for multiple testing (Supplementary Table 11), nor were significant associations detected from analyses of 19,643 rare inherited SVs (MAF <0.1%) and 441 rare homozygous deletions (MAF <1%).
We observed 9 cases (1.7%) and 10 controls (1.9%) with large balanced chromosomal anomalies >3 Mb, as well as 35 CNVs >40 kb not detected by microarray (Supplementary Fig. 12). Consistent with our previous analyses27, rare SVs were more likely to cause genic loss-of-function than common SVs (odds ratio = 1.59; p=1.33×10−30), particularly of constrained genes (odds ratio = 2.28; p = 2.26×10−9). However, there was no significant difference between cases and controls in overall SV size, percent of genome rearranged, or distribution of complex SVs (Fig. 4; Supplementary Fig. 11). We also did not detect any changes in SV burden in proximity to genes, or any signal when surveying up to 1 Mb from the transcription start site of all genes (minimum p = 0.50), constrained genes (minimum p = 0.06), or ASD-associated genes (minimum p = 0.28; Supplementary Fig. 15). Thus, despite dramatically improved access to the SV spectrum from WGS, we found no significant differences in the rate of rare inherited SV, nor did we observe evidence of significant biased transmission from either parent for any annotation category (Supplementary Table 12).
Power calculation
To estimate the required sample sizes, we performed a power calculation across estimates of RR and numbers of mutations per annotation category. Due to the complex correlation structure between categories, we used eigenvector analysis to estimate the effective number of tests conducted. This number increases with sample size, due to increased likelihood of observing sufficient de novo mutations in any given annotation category to achieve significance: the number of effective tests increases from 4,123 at 519 families to ≈7,600 at 4,000 families and approaches an asymptote of ≈10,000 (Fig. 5 and Supplementary Fig. 6). The multiple testing burden produces a threshold for statistical significance on the order of 5×10−6. In this setting, over 8,000 families would be necessary to discover a noncoding element equivalent to missense variation. Further samples would likely be needed to hone in on a specific locus.
Discussion
We present an analytical framework for testing association between cases and controls in WGS data. Unlike the coding regions, we do not have a clear hypothesis of which noncoding regions harbor human disease-causing rare variants, nor do we understand which specific alleles are intolerant to mutation within those regions. To identify robust results, we have thus reasoned that WGS analyses must follow the same principles of other genomic analyses, acknowledging, defining, and correcting for the multiple comparisons that have been conducted, either explicitly or implicitly32–35.
By selecting annotations at the level of nucleotides, genes, and regulatory regions, we define 51,801 annotation categories and develop methods to test their association with ASD risk by de novo mutation burden. Considering the correlation structure of p-values in simulated datasets, we determine the number of effective tests conducted, which increases with sample size but plateaus around 10,000 tests (Supplementary Fig. 6). These simulated data also allow us to define and test categories selected from independent annotation clusters, thereby permitting the use of a multiple testing correction that depends only on the number of clusters and does not change with sample size (Supplementary Fig. 6). This CWAS approach is extensible to WGS association designs using different annotations. Our methods to accomplish this framework can be replicated using code hosted on Amazon Web Services on a publicly available Amazon Machine Image (for the most current AMI ID and SV pipelines see https://github.com/sanderslab/WGS-pipeline and https://github.com/talkowski-lab/SV-Adjudicator, respectively).
Applying this framework to 519 families with a child affected by ASD, we are unable to demonstrate a rare noncoding variant contribution to ASD risk. Specifically, we do not observe association in 10 gene-defined categories (Fig. 1a), 200 independent annotation clusters (Supplementary Fig. 6), or 51,801 annotation categories (Fig. 3a). Furthermore, we do not observe an excess of nominally significant noncoding categories in cases (Fig. 3i) and could not develop an accurate predictor of case status using cross validation. In contrast, the same techniques identified association of missense mutations in ASD genes (Fig. 1b), deleted exons in ASD genes (Fig. 4e), and an excess of nominally significant coding categories (Fig. 3i). Considering these results in the context of a power analysis (Fig. 5) gives important insight into genomic architecture, since it is unlikely that there is a class of noncoding variation equivalent to coding loss-of-function mutations in terms of both mutation frequency and effect size. These analyses also suggest that UTRs and promoter regions are likely to demonstrate association equivalent to, or weaker than, that of missense mutations. Finally, regulatory loci in intergenic and intronic regions, such as enhancers, are likely to be even harder to associate with ASD.
Prior to this analysis, this lack of power was predicted, but not a foregone conclusion due to the lack of equivalent systematic analyses of WGS data, the previous detection of ASD association in 225 families from WES19,34,35, and previously reported nominally significant associations in ASD from WGS cohorts of fewer than 100 families16,36. Our estimates suggest that over 8,000 families would be required to demonstrate signal in a CWAS analysis such as performed here (Fig. 5). Improved characterization of the noncoding functional genome, including RNA-seq37, ChIP-seq38, Hi-C39, and massively parallel reporter assays40, could marginally bring this number down. Moreover, there are numerous WGS initiatives underway that will achieve such sample sizes in the near future13 and necessitate an openly accessible, adaptable, and reproducible analysis framework to compare results across studies.
Our analyses provide some preliminary insights into the most likely noncoding risk factors that will emerge from larger samples. Our gene-defined analysis suggests that UTRs and promoters of ASD genes could be the first categories to demonstrate noncoding risk (Fig. 1b). The CWAS analysis highlights the role of conserved indels, both in the 51,801 categories (Fig. 3a) and the 200 independent clusters (Supplementary Fig. 6). The burden analysis also identifies noncoding indels as a potential contributor (Fig. 3j), while the polygenic risk further implicates conservation across vertebrate species. Of note, by disrupting regulatory elements to a greater degree than SNVs while occurring far more often than SVs, indels could represent a sweet spot of statistical power for interrogating the noncoding genome.
Arguably, an alternative approach to WGS association designs might involve a priori prediction of which regulatory elements of the noncoding genome are important for disease risk, thereby limiting the number of tests evaluated and consequent statistical corrections. In terms of establishing a robust, unbiased framework to interpret disease association, we find this argument wanting. Perhaps the simplest way to understand why is by analogy to candidate gene studies of complex disorders, which have had a miserable record regarding replication41, with a plethora of false positive and a paucity of true positive results42. This history should make us highly skeptical of methods based on investigator-selected a priori hypotheses in the noncoding genome. Continuing the analogy, instead of candidate genes, the field would be substituting “candidate annotations” with all likelihood of poor outcomes, due to myriad combinations of annotations, cell types, brain regions, and developmental stages. Several ASD studies have selected different regions of the noncoding genome on which to focus16,31,36,43,44, and associations from initial small studies have failed to replicate in larger datasets (Supplementary Fig. 15), a trend likely to persist if nominal significance is the threshold chosen for exploring genomes. The excess of missense mutations in postsynaptic density genes seen here is illustrative (Cluster 91, Fig. 3a). We observe over 2.5-fold enrichment of these mutations in cases versus controls; however, exome data of 1,288 independent families45 reveals a much more modest 1.2-fold enrichment (p=0.27). This highlights the “winner’s curse” in which the effect size in the discovery sample is likely to be greatly inflated46, even for true associations.
Refinements in DNA sequencing, computing capability, and statistical analyses now permit simultaneous evaluation of the coding and noncoding genome, and will eventually precipitate a sea change in how we interpret the impact of rare variation on disease risk. Yet, the complexity of the noncoding genome complicates interpretation for both de novo and inherited variation, and there are perils in underestimating its complexity. Large-scale functional assays will continue to provide increasingly refined annotation of the regulatory genome and perhaps eventually a noncoding equivalent to the triplet code will emerge. Until that time, we recommend the GWAS path for WGS studies: rigorous evaluation of multiple hypotheses and appropriate correction for that multiplicity, as we have outlined here. If we hold to these standards, it will require very large sample sizes to make headway, but we predict that the ensuing inferences will be sound and replicable.
ONLINE METHODS
Sample selection
519 quartet families (2,076 samples) with no known de novo rare CNVs, de novo loss-of-function mutations, or inherited rare CNVs at known ASD loci in the proband were selected from the Simons Simplex Collection (SSC; Supplementary Table 1). The first 40 families were additionally selected for high paternal age, low IQ, and female sex while the second 479 were selected at random. All families had pre-existing microarray data4 and WES (472 quartets and 47 proband trios)45. All subjects were consented for participation in genomic studies, data were de-identified by SFARI prior to sharing with researchers, and data access and analyses were approved by the UCSF IRB, Partners Healthcare IRB, and the University of Utah IRB.
Whole genome sequencing
Whole blood-derived DNA from all individuals was transferred from the Rutgers University Cell and DNA Repository (RUCDR) to the New York Genome Center (NYGC). Twenty-one families were excluded for low DNA quality and the remaining 519 families were submitted for WGS. Data for the first 40 families were generated by PCR-based library preparation and Illumina Hi-Seq 2000, and PCR-free library preparation and Illumina Hi-Seq X Ten were used for the remaining 479 families. All sequencing used 150 bp paired-end cycles with a median insert of 423 bp. These data had a 99.3% median alignment rate, 0.50 strand balance, 0.11% duplication rate, and 37.8X median coverage per individual.
Data processing
Using the NYGC processing pipeline, FASTQ reads were aligned to the GRCh37.63 reference using BWA-mem v0.7.8-r455. Reads were sorted and duplicates removed with Picard version 1.83. Indel realignment, base quality score recalibration, and variant calling were performed using GATK haplotype caller (GATK v3.1-1-g07a4bf8 for 19 batch 1 families, v3.2-2-gec30ce for 21 batch 1 families, and v3.4-0-g7e26428 for all 479 batch 2 families.
The BAM and gVCF files for all 2,076 samples were transferred to Amazon Web Services (AWS) S3 storage system where they are accessible with approval from the Simons Foundation Autism Research Initiative (SFARI Base, https://sfari.org/resources/sfari-base). For downstream steps on AWS, we deployed CfnCluster on the Lustre cluster system using multiple m4.10xlarge instances. Using the GATK best practices protocol, (https://software.broadinstitute.org/gatk/best-practices/; GATK v3.4-46-gbc02625), we merged individual gVCF files into a combined VCF and ran SNP and indel recalibration. Variant Quality Score Recalibration (VQSR) metrics were created from a training set of validated resources: dbSNP build 138, HapMap 3.3, 1000 Genomes OMNI 2.5, and 1000 Genomes Phase 1. For the following analysis, we excluded variant calls located in low-complexity regions47 or with VQSR tranche 99.9–100%, as these calls have high error rate or unusual characteristics47,48. Indels were realigned using left-normalization, and multiple-allelic variants were split into individual VCF lines using BCFtools49.
Detection of high quality SNVs and indels
As we had no established best practices for filtering rare variants in WGS data, we developed an optimized set of quality metric thresholds to detect rare SNVs and indels. For this, we compared two sets of rare variants with distinct quality metrics: 1) private transmitted variants observed in one family with no frequency information in 1000 Genomes or ExAC (likely true variants), and 2) Mendelian violations in at least one child but also observed in an unrelated individual (likely false positives). We used receiver operating characteristic (ROC) curves to assess the ability of individual quality metrics to distinguish these true and false calls (Supplementary Figs. 1, 2; Supplementary Table 2). The metric and threshold that yielded the maximum increase in specificity and the minimum decrease in sensitivity were selected and applied as a filter to the training set. We repeated this process until we no longer observed improvement in sensitivity and specificity (details in Supplementary Information).
Detection of high quality de novo SNVs and indels
De novo SNVs were detected by four algorithms run on the default settings: TrioDeNovo50, DenovoGear51, PlinkSeq (https://atgu.mgh.harvard.edu/plinkseq/), and DenovoFlow. For de novo indels, DenovoGear was replaced with Scalpel52. DeNovoFlow is a custom script that parses all possible Mendelian violations from each family, given GATK quality metrics. The union of these four algorithms predicted 86,921 Mendelian violation SNVs and 5,726 indels per child.
These numbers are large, suggesting a high false positive rate. To identify high quality de novo variants, we applied the same sequential ROC approach as above with true positive calls defined by PCR Sanger validation of de novo mutations from prior work (1,302 selected SNVs; 95 selected indels), and with all variant- and individual-level quality metrics for the child and both parents (Supplementary Figs. 1, 2). Using 3 additional metrics for SNVs, this analysis predicted 87.3% sensitivity and 98.8% specificity, and using 4 additional metrics for indels, 86.3% sensitivity and 93.0% specificity (Supplementary Table 2).
Validation of high quality de novo SNVs
From the 66,366 high quality de novo SNVs, 250 mutations were selected at random, conditional on available DNA, for validation in the child and both parents using PCR amplification and high-throughput sequencing on an Illumina MiSeq. In all analyses, we sought to both validate the presence of the putative variant and confirm de novo status based on absence of the variant in both parents. By investigation of off-target coverage, we determined that ≥ 50X depth was required for highly accurate genotyping of variants and we used this threshold for all validation experiments. From the initial 250 variants, 13 either failed PCR amplification or MiSeq coverage thresholds in the proband, and an additional 18 variants failed coverage in at least one parent. Among the remaining 219 variants with successful assays, 212 were successfully validated and all validated variants were confirmed to have arisen de novo (212/219; 96.8% confirmation rate; Supplementary Table 3).
Validation of high quality de novo indels
We performed indel validation in two stages. In the initial exploratory analyses, 250 noncoding indels (125 deletions, 125 insertions) were selected at random from 9,961 high quality de novo indel predictions for validation using the same PCR and MiSeq approach. Variants larger than 50bp were excluded from the analysis (16 variants). Among the remaining 234 variants in the exploratory study, 22 variants were filtered due to failed PCR or insufficient MiSeq coverage in the probands (14 variants) or a parent (8 variants) (<50X depth). The remaining 212 putative mutations were examined with VarDict53. Among these, 137 validated in the proband and six were determined to be inherited, for an overall confirmation rate in the exploratory analyses of 61.8% (131/212 variants with sufficient coverage in parents and child; Supplementary Table 3).
Based on these exploratory analyses, de novo indel prediction was refined (Supplementary Information), identifying 5,932 mutations overall, and a second round of validation was performed on 200 randomly selected variants <50bp. From this final validation set, 176 were successfully assayed and achieved adequate coverage in the child and parents; 148/176 variants validated (84.1%), though three were determined to be inherited, yielding a final confirmation rate of 145/176 (82.4%) for de novo indel predictions, a significant improvement over the exploratory analyses (Supplementary Table 3).
Validation of mutations near ASD-associated genes
Four putative mutations near known ASD-associated genes also all validated as de novo: one SNV in the promoter of ADNP (GRCh37.63:chr20:g.49548007A>G), two SNVs near GABRB3 (GRCh37.63:chr15:g.26327365A>G, GRCh37.63:chr15:g.26327513C>T), and one indel in the promoter of NRXN1 (GRCh37.63:chr2:g.51259258delG).
SNV and indel annotation and statistical burden analyses
Variants were annotated to five annotation groups (Table 1) using Annovar54 and Bamotate4:
1) Variant type
Each variant was first classified by type, including SNV, indel (<50 bp), or SV ( ≥50 bp; deletions, duplications, insertions, inversions, and complex events).
2) Gene-defined annotation
Gene definitions from GENCODE (wgEncodeGencodeCompV19)55 were obtained from the UCSC table browser (https://genome.ucsc.edu/) and variants annotated using Bamotate; where multiple annotations were possible, variants were assigned in the following priority: coding, intron, promoter, UTRs, and intergenic. Promoters were defined as 1kb upstream of a transcription start site (TSS). The nearest TSS was identified for intergenic variants.
3) Species conservation
Variants were annotated to two conservation metrics: phastCons 46-way scores, and phyloP scores from a 46-way vertebrate comparison from the UCSC table browser56,57.
4) Gene sets
Gene lists associated with ASD were selected (e.g. post-synaptic density genes). ASD risk genes (FDR<0.3) were obtained from Sanders et al. (2015).4 Genes co-expressed with ASD risk genes were defined as the union of the two co-expression modules identified by Willsey et al. (2013)58 in human 1) midfetal prefrontal and primary motor-somatosensory cortex and 2) infant mediodorsal thalamic nucleus and cerebellar cortex. Genes associated with developmental delay were downloaded from the Development Disorder Genotype-Phenotype Database (https://decipher.sanger.ac.uk/ddd; Sept 2016)5,59. The 2,156 genes were filtered to: 1) confirmed developmental disorder gene, 2) predicted loss-of-function, and 3) including term “Brain” in the organ specificity list. CHD8 target genes were defined as the union of lists from two ChIP-Seq studies60,61, and FMRP target genes were selected from Darnell et al. (2011)22. Human post-synaptic density (PSD) proteins were downloaded from the Genes2Cognition database (http://www.genes2cognition.org/)62. Constrained genes were defined as probability of loss-of-function intolerant (pLI) score ≥ 0.9 in the ExAC database21. Either the transcript in which the variant was located or the nearest TSS (intergenic variants) was cross-referenced to these gene lists. For all gene lists, see Supplementary Table 6.
5) Regulatory regions
BED files were obtained for multiple regulatory regions. Vista enhancers were downloaded from the UCSC genome browser63 (vistaEnhancers) and pre-defined enhancers from the FANTOM 5 server (http://enhancer.binf.ku.dk/presets/)64. ENCODE-defined transcription factor binding and DNase hypersensitive sites were downloaded from UCSC genome browser (wgEncodeRegTfbsClusteredV2 and wgEncodeRegDnaseClusteredV3). Human accelerated regions (HARs) were obtained from Doan et al. 201665.
We also utilized histone marks and chromatin state data from the NIH Roadmap Epigenome Project66. We merged data from brain tissues (E067 Angular Gyrus, E068 Anterior Caudate, E069 Cingulate Gyrus, E070 Germinal Matrix, E071 Hippocampus Middle, E072 Inferior Temporal Lobe, E073 Mid Frontal Lobe, E074 Substantia Nigra, E081 Fetal Brain Male, E082 Fetal Brain Female), neurospheres (E053 neurosphere cultured cells cortex-derived, E054 neurosphere cultured cells ganglionic eminence-derived), ES-derived neuronal cells (E007 H1-derived neuronal progenitor cultured cells, E009 H9-derived neuronal progenitor cultured cells, E010 H9-derived neuron cultured cells), and astrocytes (E125 NH-A Astrocytes).
We also utilized data sets generated at UCSF from mid-fetal human prefrontal cortex tissue (15–22 gestational weeks), including ATAC-seq (open chromatin) and ChIP-seq for H3K27ac (putative enhancers). Peaks were called by MACS (H3K27ac ChIP-seq) and Homer (ATAC-seq). Identified peaks common to two or more individual samples (1≥ bp overlap) were used for annotation.
Specific to our SV analysis we investigated topologically associating domain (TAD) boundaries identified in fetal lung fibroblasts (IMR90) and embryonic stem cells (ESCs)67. The union of TAD boundaries was compiled from IMR90 and ESCs, and overlapping IMR90 and ESC boundaries were collapsed with BEDTools68 and converted to hg19 coordinates by UCSC liftOver69.
Burden testing for de novo SNVs and indels is described in results. All annotation reference files and software for annotation and burden testing of SNVs and indels can be accessed and implemented via a publicly available customized Amazon Machine Image on AWS (see https://github.com/sanderslab/WGS-pipeline for current AMI ID).
Detection of high quality de novo structural variants
In our SV detection pipeline (Supplementary Fig. 9), we initially maximized sensitivity by integrating four paired-end/split-read (PE/SR) algorithms, three read-depth (RD) algorithms, and a mobile element insertion (MEI) detection pipeline to discover candidate SVs. We then adjudicated each predicted variant with a joint analysis of the cohort that included a series of modules for de novo variant filtering and a statistical test for de novo status. Our pipeline incorporated PE and SR calls from Delly v0.7.370, Lumpy v0.2.1371, Manta v.0.29.672, and WHAM-GRAPHENING v1.7.073, each run jointly on all four family members; read-depth calls generated by NYGC from GenomeSTRiP v2.00.169674, CNVnator v0.3.275, and cn.MOPS v1.8.976; and MEI calls from Melt v2.0.577 (additional details in Supplementary Information). To determine the likelihood of a true SV, we developed an iterative random forest based modeling technique by testing for statistically significance differences between samples with and without SV across four classes of orthogonal evidence types: 1) Discordant PE read pairs, 2) clipped SR, 3) RD, 4) b-allele frequency (BAF) (additional details in Supplementary Information). We used a batch-specific framework to jointly adjudicate SVs (pilot n=160 and Phase 1 n=1,916) to correct for demonstrable RD differences between the datasets (PCR+ and PCR-free, respectively), and further split the samples by sex for SV on allosomes. Metrics computed in the Phase 1 PCR-free samples were used whenever available. Across all passing CNVs, we genotyped homozygous deletions, defined as samples with median normalized RD <0.08. We identified five samples with sex chromosome anomalies: three XXY Klinefelter syndrome and two XYY syndrome (Jacob’s syndrome). These variants were initially detected from our initial depth assessment, then replicated by an independent algorithm78.
In addition to polymorphic and de novo CNVs, we assessed balanced and complex SV in the SSC, as we have done previously in this cohort with large SVs27. We applied the algorithm integration pipeline for PE/SR calls described above to find candidate inversion and translocation breakpoints and resolved the variant structure at these loci by matching the ordering of breakpoints to complex SV signatures previously identified by Collins et al.27. We identified 22,840 observations of 258 inversion-associated CNV between 300 bp and 5 kb that were not found with the CNV discovery pipeline, as they lacked canonical PE/SR evidence and were below RD-only algorithm resolution. In total, we identified 53,440 deletions, 20,782 duplications, 23,995 insertions, 197 inversions, 4 reciprocal translocations, 5 sex chromosome aneuploidies, and 367 complex SV across 8 classes (Supplementary Table 8).
Validation of de novo SVs
We performed extensive validation of all putative de novo SV predictions using combinations of microarray, PCR with Sanger sequencing, PCR and long-read MiSeq sequencing, microarray, long-insert whole-genome sequencing (liWGS), and Digital Droplet PCR (ddPCR). Assays were attempted for at least one validation method on all de novo predictions, and a subset of variants were confirmed by multiple methods (see Supplementary Information for complete validation details). Overall, we successfully designed assays for 168/171 de novo SV predictions (two variants were from individuals that lacked sufficient DNA for confirmation and assays could not be designed for another variant due to repetitive sequences at the breakpoint). We observed an overall validation rate of 97.0% (163/168) across all SV classes. All validation assays are provided in Supplementary Information, including the number of assays performed and split reads from confirmation experiments for each variant (Supplementary Table 9). In addition, a series of visualizations were generated for each de novo SV prediction for each variant to enable visual inspection (see Supplementary Data).
Comparison of SVs to microarray and long-insert WGS
We compared the performance of short-insert WGS (siWGS) SV calls to rare CNVs detected from long-insert WGS (liWGS, “jumping”) libraries on 456 of the 519 cases27 and microarray data from all 2,071 samples with SV4. We performed the following filtering to correct for differences in resolution and sample differences across platforms: 1) microarray size threshold >40 kb; liWGS size range 10–40 kb was used; 2) repeat masking: SV comparisons were retained if ≤30% of the variant region overlapped an annotated segmental duplication region, microsatellite, heterochromatin, or one of our defined multi-allelic regions (Supplementary Table 13); 3) all variant frequencies <10%. These filters resulted in 1,399 siWGS CNVs in the array comparison (Supplementary Fig. 12) and 986 variants in the liWGS comparisons (additional details in Supplementary Information). Overall, we observed a 2.5% FDR and 99.6% sensitivity for microarray data and a 5.2% FDR and 91.9% sensitivity for liWGS (Supplementary Fig. 12).
SV annotation and statistical burden analyses
Each SV with any predicted overlap with the canonical transcript of 20,156 protein-coding genes (GENCODE v19) was annotated as genic. Deletions were considered loss-of-function (LoF) if they affected any coding sequence, duplications were considered LoF if they affected an exon but did not extend outside the transcript boundary, and inversions were considered LoF if one breakpoint localized to a coding exon or any genic space spanning the coding sequence (but not if the entire coding sequence was inverted). Duplications were considered to be “copy-gain” if they spanned the entirety of a transcript. Intronic variants were required to localize fully to an intron. All variants, including noncoding variants, were additionally annotated with any gene whose UTR or promoter region (<1 kb upstream of TSS) it disrupted. See Supplementary Figure 14 for all SV annotations. Statistical burden testing was also performed using a CWAS design, paralleling the SNV analyses described above. Notably, families were selected after screening for probands harboring large de novo CNVs detected by microarray and de novo coding mutations detected by WES, but families with siblings harboring comparable mutations were not excluded. These analyses can impact estimates of de novo SV association, so we filtered any family where the sibling met similar exclusionary criteria (n=27; Supplementary Table 1). Rare SV analyses were restricted to the 405 families with European ancestry described in the SNV analyses.
Estimation of the number of effective tests in CWAS
We generated 20,000 sets of 72,285 autosomal, simulated variants randomly allocated to cases and controls, with the same proportion of SNVs and indels as in the observed data. These “1x” datasets simulating 519 families were combined at random to yield simulations at 2x, 4x, 8x and 16x (8,304 cases). As with the CWAS analysis we annotated these simulated variants against all 51,801 distinct annotation categories and tested categories for case-control burden using a one-sided binomial test, excluding categories with ≤7 variants in >50% of the simulations. We used Z-scores converted from p-values to estimate correlations between annotation categories. We employed eigen decomposition to estimate the number of effective tests within the genome-wide category space and K-means clustering to identify 200 clusters of correlated annotation categories.
Code availability
Our methods for de novo SNV and indel annotation and statistical analyses in WGS data can be replicated using code hosted on Amazon Web Services on a publicly available Amazon Machine Image (for the most current AMI ID, see https://github.com/sanderslab/WGS-pipeline), and SV analysis pipelines can be found at GitHub (https://github.com/talkowski-lab/SV-Adjudicator).
Data availability
All sequencing and phenotype data are hosted by the Simons Foundation for Autism Research Initiative (SFARI) and are available for approved researchers at SFARIbase (https://base.sfari.org, SFARI_SSC_WGS_1b).
Supplementary Material
Acknowledgments
We are grateful to the families participating in the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC). This work was supported by grants from the Simons Foundation for Autism Research Initiative (SFARI #385110 to N.S., A.J.W., M.W.S., S.J.S.; #385027 to M.E.T., J.D.B., B.D., M.J.D., X.H., and K.M.R.; #388196 to G.B., H.C., A.Q.; and #346042 to M.E.T.), the National Institutes of Health (R37MH057881 and U01MH111658 to B.D. and K.M.R.; HD081256 and GM061354 to M.E.T.; U01MH105575 to M.W.S.; U01MH111662 to M.W.S. and S.J.S. R01MH110928 and U01MH100239-03S1 to M.W.S., S.J.S, and A.J.W.; U01MH111661 to J.D.B.; K99DE026824 to H.B.; U01MH100229 to M.J.D.), Autism Science Foundation to D.M.W., the March of Dimes to M.E.T. Dr. Talkowski was also supported by the Desmond and Ann Heathwood MGH Research Scholars award. We would like to thank the SSC principal investigators (A.L. Beaudet, R. Bernier, J. Constantino, E.H. Cook, Jr, E. Fombonne, D. Geschwind, D.E. Grice, A. Klin, D.H. Ledbetter, C. Lord, C.L. Martin, D.M. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M.W. State, W. Stone, J.S. Sutcliffe, C.A. Walsh, and E. Wijsman) and the coordinators and staff at the SSC clinical sites; the SFARI staff, in particular N. Volfovsky; D. B. Goldstein for contributing to the experimental design; the Rutgers University Cell and DNA repository for accessing biomaterials; the New York Genome Center for generating the WGS data.
Footnotes
Supplementary Information (SI): SI is linked to the online version of the paper at XXXX.
Author Contributions
Experimental design, DMW, HB, JA, MRS, JTG, MJW, XH, NS, BMN, HC, AJW, JDB, MJD, MWS, ARQ, GTM, KR, BD, MET, and SJS; Identified de novo SNVs and indels, DMW, JA, SD, MCG, JDM, LS, AJW, and SJS; Identified structural variants, HB, JA, MRS, JTG, RLC, RML, AF, HZW, XZ, MCG, REH, SK, LS, SAM, ARQ, GTM, and MET; Confirmed de novo variants, DMW, HB, SD, GBS, HZW, BBC, JD, CD, CAE, RY, MFW, and MJW; Annotation of functional regions, DMW, JA, SD, EM, JDM, YL, SP, JLR, NS, MET, and SJS; Generated midfetal H3K27ac and ATAC-Seq data, EM, TJN, ARK, and JLR; Developed genomic prediction score and de novo score, LZ, LK, KR, and BD; Analyzed SNVs and indels (Figs. 1–3), DMW, JA, and SJS; Analyzed SVs (Fig. 4), HB, MRS, JTG, XZ, and MET; Assessment of P-value correlations, effective number of tests, and power analysis (Figs. 3, 5), DMW, JA, LZ, GBS, KR, BD, and SJS; Manuscript preparation, DMW, HB, JA, MRS, LZ, JTG, RLC, SD, BMN, HC, JDB, MJD, MWS, ARQ, GTM, KR, BD, MET, and SJS.
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astle WJ, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429. e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–33. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deciphering Developmental Disorders, S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall CR, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Power RA, et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry. 2013;70:22–30. doi: 10.1001/jamapsychiatry.2013.268. [DOI] [PubMed] [Google Scholar]
- 9.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017 doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata M, Gulden FO, Sestan N. From trans to cis: transcriptional regulatory networks in neocortical development. Trends Genet. 2015;31:77–87. doi: 10.1016/j.tig.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron. 2016;89:248–68. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders SJ, et al. Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci. 2017;20:1661–1668. doi: 10.1038/s41593-017-0017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caskey CT, Tompkins R, Scolnick E, Caryk T, Nirenberg M. Sequential translation of trinucleotide codons for the initiation and termination of protein synthesis. Science. 1968;162:135–8. doi: 10.1126/science.162.3849.135. [DOI] [PubMed] [Google Scholar]
- 15.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Turner TN, et al. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am J Hum Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genovese G, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaste P, et al. A genome-wide association study of autism using the Simons Simplex Collection: Does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry. 2015;77:775–84. doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins RL, et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017;18:36. doi: 10.1186/s13059-017-1158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redin C, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2016 doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand H, et al. Paired-Duplication Signatures Mark Cryptic Inversions and Other Complex Structural Variation. Am J Hum Genet. 2015;97:170–6. doi: 10.1016/j.ajhg.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner TN, et al. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell. 2017;171:710–722. e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 33.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuen RK, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–91. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 37.Cummings BB, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Psych EC, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Berkum NL, et al. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp. 2010 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson EC, et al. No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes. Biol Psychiatry. 2017;82:702–708. doi: 10.1016/j.biopsych.2017.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farrell MS, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20:555–62. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz A, et al. De novo indels within introns contribute to ASD incidence. bioRxiv. 2017 [Google Scholar]
- 44.Brandler WM, et al. Paternally inherited noncoding structural variants contribute to autism. bioRxiv. 2017 [Google Scholar]
- 45.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 47.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–51. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–51. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 49.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Q, et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31:1375–81. doi: 10.1093/bioinformatics/btu839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramu A, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10:985–7. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narzisi G, et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods. 2014;11:1033–6. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright CF, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A. 2014;111:E4468–77. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doan RN, et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell. 2016;167:341–354. e12. doi: 10.1016/j.cell.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 73.Kronenberg ZN, et al. Wham: Identifying Structural Variants of Biological Consequence. PLoS Comput Biol. 2015;11:e1004572. doi: 10.1371/journal.pcbi.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Handsaker RE, et al. Large multiallelic copy number variations in humans. Nat Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klambauer G, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gardner EJ, et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017 doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pedersen BS, Collins RL, Talkowski ME, Quinlan AR. Indexcov: fast coverage quality control for whole-genome sequencing. Gigascience. 2017 doi: 10.1093/gigascience/gix090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing and phenotype data are hosted by the Simons Foundation for Autism Research Initiative (SFARI) and are available for approved researchers at SFARIbase (https://base.sfari.org, SFARI_SSC_WGS_1b).