Abstract
CRISPR adaptive immune systems protect bacteria from infections by deploying CRISPR RNA (crRNA)-guided enzymes to recognize and cut foreign nucleic acids. Type VI-A CRISPR-Cas systems include the Cas13a enzyme, an RNA-activated ribonuclease (RNase) capable of crRNA processing and single-stranded RNA degradation upon target transcript binding. Here we present the 2.0 Å resolution crystal structure of a crRNA-bound L. bacterium Cas13a (LbaCas13a), representing a recently discovered Cas13a enzyme subtype. This structure and accompanying biochemical experiments define for the first time the Cas13a catalytic residues that are directly responsible for crRNA maturation. In addition, the orientation of the foreign-derived target RNA-specifying sequence in the protein interior explains the conformational gating of Cas13a nuclease activation. These results describe how Cas13a enzymes generate functional crRNAs and how catalytic activity is blocked prior to target RNA recognition, with implications for both bacterial immunity and diagnostic applications.
INTRODUCTION
A repertoire of defensive mechanisms evolved to protect bacteria against phage and other mobile genetic elements 1. Unique among these mechanisms are CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR-associated) adaptive immune systems, which use RNA-guided nucleases to direct cleavage of invading DNA or RNA 2,3. The programmability of DNA-targeting CRISPR-Cas enzymes has driven a revolution in technologies for gene editing, transcriptional control, and genomic imaging 4–6. Similarly, RNA-targeting CRISPR-Cas systems offer the potential for RNA-programmed RNA detection and development as scaffolds for designer RNA binding proteins 7–14.
Type VI-A RNA-targeting CRISPR-Cas systems are defined by the single effector protein Cas13a (formerly C2c2), which forms a target surveillance complex with a CRISPR RNA (crRNA). The crRNA, encoded by the genomic CRISPR locus, contains a defined 5′ hairpin structure preceding a 3′ foreign-derived variable region (or 3′ spacer) 8,15. In Type VI-A CRISPR-Cas systems, Cas13a catalyzes maturation of the transcribed pre-crRNA using a metal ion-independent active site that has not been well defined 10,16. Once assembled with its mature crRNA, Cas13a is competent for foreign single-stranded RNA (ssRNA) target surveillance. Recognition of a spacer-complementary ssRNA transcript (ssRNA-activator) activates the HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domains of the enzyme to create a composite general RNase active site 8,16,17, distinct from that responsible for crRNA maturation 10. Across the Cas13a family, this composite HEPN active-site is functionally diverse, in terms of both nucleotide cleavage preference and turnover efficiency 14. Two distinct subfamilies, named for their cleavage preference (adenosine (A)-cleaving or uridine (U)-cleaving), exist within the Cas13a protein family and can be harnessed in parallel for RNA detection 14.
The molecular cues of Cas13a HEPN nuclease activation are unknown, limiting the development of Cas13a-based tools. Recent structural studies on the apo and binary complex of Leptotrichia shahii Cas13a established the HEPN domains form a surface exposed composite active site16. While an attractive possibility, multiplexed RNA detection from the two Cas13a subfamilies is limited by an incomplete mechanistic understanding of the different crRNA binding specificities and catalytic efficiencies, especially in light of substantial sequence divergence within the family14.
To determine the structural basis for the divergent activities of the A-cleaving Cas13a subfamily, we solved a 2.0 Å resolution crystal structure of Lachnospiraceae bacterium Cas13a (LbaCas13a). This structure and accompanying biochemical experiments revealed the active-site for pre-crRNA processing in the A-cleaving Cas13a subfamily. The structure also showed that LbaCas13a shields the spacer region of the crRNA from target hybridization, providing clues to the conformational rearrangement required to activate the HEPN nuclease for general RNase activity. Finally, structural comparisons to the previously modeled U-cleaving subfamily member, LshCas13a16, rationalize how these enzyme subfamilies recognize unique sets of crRNAs that enable orthogonal RNA detection in complex mixtures. Together, these findings provide new insights into the mechanisms controlling crRNA generation by, and target RNA activation of, Cas13a.
RESULTS
Global architecture of LbaCas13a:crRNA complex
The Cas13a enzyme family depends on two distinct nuclease activities for biological function, yet neither nuclease active-site has been well described in terms of mechanism or conformational regulation. To further investigate the mechanisms of pre-crRNA processing and activator driven HEPN nuclease activity, we studied a divergent A-cleaving homolog, LbaCas13a, enabling comparisons to previously investigated U-cleaving subfamily members LshCas13a and Leptotrichia buccalis (Lbu) Cas13a (Fig. 1a). We determined the crystal structure of LbaCas13a (residues 1-1437) in complex with a mature crRNA to 2.0 Å resolution by native single-wavelength anomalous dispersion (SAD) (Fig. 1b–d; Table 1; Supplementary Fig. S1a). To obtain a mature crRNA for binary complex crystallization, LbaCas13a was incubated with a 60-nt pre-crRNA, allowing the enzyme to process the crRNA, before purifying the resulting complex (Supplementary Fig. S1b). The mature crRNA within the structure was resolved as 41 well-ordered nucleotides that includes the 28-nt 5′ direct repeat (henceforth referred to as the 5′ handle) (A(-28)-C(-1)) and 13 nucleotides of the 3′ spacer. Only the first 13 nucleotides (C(1)-G(13)) of the 3′ spacer are well-ordered within the crystal structure with the remaining 11 nucleotides G(14)-C(24) present, but disordered within the structure (Fig. 1b; Supplementary Fig. S1c).
Figure 1. Overall structure of the LbaCas13a:crRNA complex.
a, Phylogenetic tree of all Cas13a family members with colored circles highlighting the A- and U-cleaving subfamilies (adapted from 14). The homolog described in this study, LbaCas13a, is highlighted in yellow. b, Schematic representation of the crRNA repeat scaffold crystallized in this study. Nucleotides at the 5′ and 3′ end that were disordered in the structure are shown in semitransparent text. The ribose and nucleobase of G(14) were disordered. Only the phosphate of G(14) was modeled in the structure and is represented here as a dashed circle. c, Domain organization schematic of LbaCas13a shown with the REC, NUC, NUC1, and NUC2 lobes annotated. The color code used is consistent throughout the manuscript. d, Two views of the LbaCas13a:crRNA complex are shown related by a 180° rotation as cartoon (top) or surface (bottom) representations. The crRNA backbone is shown in orange.
Table 1.
Data collection and refinement statistics
| LbaCas13a:crRNA (24-nt spacer)+ |
LbaCas13a:crRNA (20-nt spacer)+ |
LbaCas13a:pre-crRNA (24-nt spacer)+ |
LbaCas13a:crRNA (24-nt spacer) (SAD)‡ |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | C 1 2 1 | P 1 | C 1 2 1 | C 1 2 1 |
| Cell dimensions | ||||
| a, b, c (Å) | 150.84, 92.21, 115.86 | 88.78, 89.21, 116.31 | 150.84, 92.21, 115.86 | 150.69, 91.45, 115.37 |
| α β γ (°) | 90.00, 92.42, 90.00 | 86.14, 86.97, 66.16 | 90.00, 92.42, 90.00 | 90.00, 92.76, 90.00 |
| Resolution (Å) | 47.12-1.99 (2.02-1.99) * | 48.57-2.20 (2.24-2.20) * | 47.11-1.80 (1.86-1.80) * | 46.94-3.50 (3.83-3.50) * |
| Rmerge | 0.079 (0.841) | 0.110 (1.060) | 0.048 (0.666) | 0.406 (0.534)† |
| I / σI | 15.2 (2.0) | 8.6 (1.3) | 18.1 (2.2) | 36.0 (30.0) |
| CC (1/2) | 0.99 (0.70) | 0.996 (0.466) | 0.99 (0.77) | 0.99 (0.99) |
| Completeness (%) | 98.8 (89.7) | 97.5 (96.6) | 97.3 (95.3) | 100.0 (100.0) |
| Redundancy | 6.9 (6.0) | 3.6 (3.7) | 5.0 (5.2) | 118.6 (120.2) |
| Refinement | ||||
| Resolution (Å) | 47.12-1.99 (2.02-1.99) | 48.57-2.20 (2.24-2.20) | 47.11-1.80 (1.86-1.80) * | |
| No. reflections | 107,256 | 161,267 | 142,815 | |
| Rwork / Rfree | 18.50 / 21.63 | 19.65 / 23.30 | 18.59/ 21.85 | |
| No. atoms | 12,499 | 24, 245 | 12,394 | |
| Protein | 11,646 | 23, 086 | 11,601 | |
| Ligand/ion | 41 | 16 | 26 | |
| Water | 812 | 1,143 | 767 | |
| B-factors | ||||
| Protein | 36.35 | 44.31 | 37.08 | |
| Ligand/ion | 56.91 | 83.97 | 64.51 | |
| Water | 39.68 | 41.30 | 37.95 | |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.003 | 0.002 | 0.006 | |
| Bond angles (°) | 0.482 | 0.415 | 0.830 | |
A single dataset was scaled and merged for each crystal structure.
A total of 20 datasets were scaled and merged.
Values in parentheses are for highest-resolution shell.
The high Rmerge for the SAD data is due to non-isomorphism between the 20 merged crystals.
LbaCas13a adopts a predominantly α-helical fold comprising six discrete domains that encase the crRNA to form an active surveillance complex (Fig. 1c, d). Consistent with other single-protein CRISPR effectors, its overall architecture is bi-lobed; the N-terminal domain (NTD) and the Helical-1 domain form the crRNA recognition (REC) lobe, and the HEPN1, Helical-2, Helical-3, and HEPN2 domains form the nuclease (NUC) lobe. The crRNA 5′ handle is bound by the REC lobe within a positively charged channel formed between the NTD and Helical-1 domains, while the spacer is channeled into a cavity formed within the NUC lobe (Fig. 1d; Supplementary Fig. 1c). The NUC lobe in turn encompasses two distinct structural domains, NUC1 and NUC2, that “sandwich” the targeting region of the crRNA into a planar orientation (Fig. 1d; Supplementary Fig. 1c). The NUC1 lobe contains the N-terminal half of HEPN1 (HEPN1-I), connected by a large α-helical domain, Helical-2. The NUC2 lobe comprises the C-terminal half of HEPN1 (HEPN1-II), Helical-3 and HEPN2 domains. The overall architecture of this A-cleaving Cas13a is related to the U-cleaving Leptotrichia shahii Cas13a (LshCas13a) 16, with notable differences in the NTD and Helical-3 domain disposition (Supplementary Fig. S2).
Recognition of a compact and divergent crRNA handle
A hallmark of CRISPR interference complexes is highly specific and tight binding between effector proteins and their cognate crRNA 18–22. Multiple sequence alignment of the crRNA repeat across both Cas13a subfamilies suggests that the 5′ handle of Type VI-A CRISPR-Cas systems forms a conserved hairpin structure comprising a 5-nt base-paired stem, an 8–9 nt loop, and a 2-nt bulge within the stem (Fig. 2a) 14. Previous work established that LbaCas13a can process and utilize crRNA handles from all A-cleaving subfamily members but cannot utilize crRNA handles from the related U-cleaving subfamily despite the similarity of their predicted structures (Fig. 2a) 14. We wondered if our structure could inform us on the molecular basis for the restrictions on Cas13a subfamily crRNA exchangeability.
Figure 2. Structure and recognition of the crRNA 5′ handle.
a, Multiple sequence alignment of select type VI-A Cas13a crRNA 5′ handles representing both subfamilies adapted from 14. Nucleotides conserved across all Cas13a homologs, amongst just the U-cleaving, or A-cleaving subfamily are shown in grey, teal or purple respectively. The sites of pre-crRNA processing for each Cas13a homolog are noted by red arrows. LbaCas13a, is highlighted in yellow with the repeat nucleotides numbered negatively from the start of the spacer. b, Side by side comparison of the crRNA crystallized in this study (LbaCas13a, left) and previously 16 (LshCas13a, right). c–f, Highlights of specific interactions between the crRNA 5′ handle and LbaCas13a. Purine stacks within the loop region of the 5′ handle interacting with Helical-1 and the NTD (c–d). Residues involved in conserved contacts between LbaCa13a and the crRNA loop are shown as sticks, with hydrogen-bonding interactions denoted by dashed lines. Binding of the 5′ flank (A(-28)-U(-24)) of LbaCas13a within the shallow Helical-1/HEPN2 groove via F422 and F1300 stacking with G(-26) and A(-25) respectively (e), and sequence-specific recognition of the dinucleotide 3′ AA-bulge (f).
This structure of LbaCas13a revealed that the crRNA preference is determined by a combination of loop conformation and a divergent set of contacts between the 5′ handle and Cas13a (Fig. 2b–f; Supplementary Fig. S3a). LbaCas13a binds to its cognate crRNA with high affinity (Supplementary Fig. S4a) through an extensive network of conserved sugar-phosphate and nucleobase interactions within a cavity formed by the NTD and Helical-1 domains (Fig. 1d; Fig. 2b; Supplementary Fig. S3a). The 5′ handle of LbaCas13a’s cognate crRNA is highly compact, as the loop (A(-18)-A(-11)) is condensed into a helical stack (Fig. 2b–d). Only A(-14) is extruded from the cavity and disordered in the solvent (Fig. 2b; Supplementary Fig. S1c). This conformation is in stark contrast to the structure of U-cleaving LshCas13a’s crRNA, in which A(-13), A(-15), U(-17) and A(-18) are flipped out of the loop (Fig. 2b).
The 5′ handle makes both structure- and sequence-specific contacts with LbaCas13a (Fig. 2c, d, f; Supplementary Fig. S3a). In LbaCas13a, the loop is stabilized by the arrangement of the purines into a quadruple and triple base stack on either side of the single extruded nucleotide, A(-14). The quadruple π-stack (A(-11)-A(-15)) is extended to include Y376 of Helical-1 and is stabilized by base-specific interactions with R369, E410, and E406 (Fig. 2c). Notably, the nucleobases involved in these interactions, G(-12) and A(-15), are strictly conserved within the Type VI-A family (Fig. 2a). The equivalent nucleotides in the Lsh-crRNA (G(-15) and A(-18)) make no base-specific contacts with LshCas13a, consistent with our previous observation that LbaCas13a and LshCas13a cannot functionally exchange crRNA 14. Parallel to the quadruple base stack, nucleotides G(-16)-A(-18) stack between the NTD and Helical-1 domain, forming a triple base stack that is stabilized by R270 hydrogen bonding to backbone phosphates and G(-16) (Fig. 2d). The close proximity of the two sets of stacking nucleotides is further stabilized by a hydrogen bond between the ribose 2′-hydroxyl of G(-12) and nucleobase N3 of A(-17), another contact unique to the compact loop of LbaCas13a.
Beyond stabilizing the crRNA loop, Helical-1 also positions the 5′ flank of the stem-loop in a shallow, surface-exposed groove at the interface with HEPN2 (Fig. 1d; Fig. 2e). crRNAs from the A-cleaving subfamily maintain a 5-nt 5′ flank, which wraps around the exterior of LbaCa13a, with A(-25) and G(-26) stacking with F1300 and F422, respectively (Fig. 2e). Upstream of the 3′ flank, Lba crRNA contains a two-nucleotide bulge, neighboring an A-U base pair that is invariant across the direct repeats of the Type VI-A family. In contrast, the identity of the bulged bases varies as a function of subfamily (Fig. 2a), suggesting that this portion of the sequence may be a crucial feature for crRNA recognition among the two subfamilies. Consistent with this hypothesis, the AA 3′-bulge observed in LbaCas13a (A(-6)-A(-5)) is read sequence-specifically (Fig. 2f; Supplementary Fig. S3a). A(-6) engages in a conserved pocket formed by HEPN1-II and Helical-2, where it stacks with Y928 ordering the nucleobase N7 and amine to hydrogen bond with E921 and Q927, respectively. A(-5) is buried in a pocket formed at the interface of the NTD, Helical-1, and HEPN1-I domains, where it interacts with the backbone carbonyl and side-chain hydroxyl of T21 (Fig. 2f). The set of contacts that recognize A(-6) in LbaCas13a closely resemble those reading out the equivalent A(-8) in LshCas13a 16. In contrast, the second nucleotide of the AC-bulge in LshCas13a (C(-7)) engages in a nucleobase-specific pocket, suggesting the A-cleaving subfamily has diverged to allow for adenosine at this position.
While the REC lobe confers crRNA 5′ handle specificity in both U- and A-cleaving Cas13a subfamilies, the landscape of protein-RNA interactions is markedly different. Our structure has revealed differences in crRNA conformation that could not be predicted through simple RNA-folding algorithms. The structure- and sequence-specific contacts observed in LbaCas13a serve to rationalize the previous finding that LbaCas13a cannot effectively utilize non-cognate crRNA from the U-cleaving subfamily 14.
Spacer recognition and activation of LbaCas13a
Cas13a-crRNA complexes show no general RNase activity in the absence of a complementary ssRNA-activator, suggesting that the enzyme is able to achieve complete conformational inhibition until the activator is bound. The binary LbaCas13-crRNA structure represents the target search complex, allowing us to understand the structural constraints on both target identification and HEPN nuclease activation.
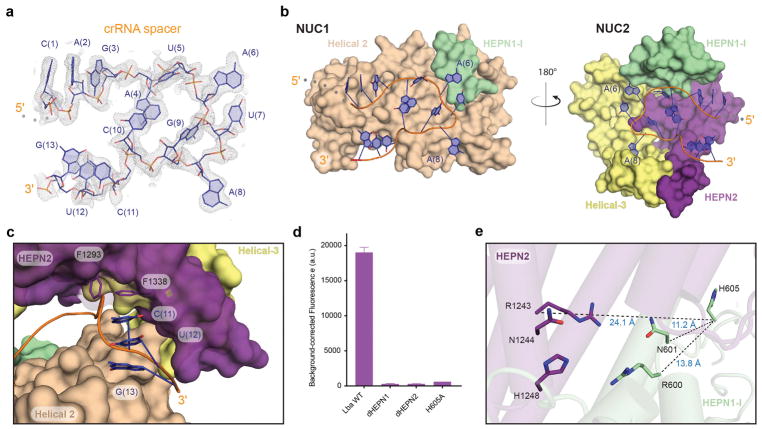
We observed unambiguous density for a contiguous stretch of 13 spacer nucleotides immediately proximal to the repeat (C(1)-G(13)) (Fig. 3a). The remaining 11 spacer nucleotides, while present in the crystal (Supplementary Fig. S1b), were disordered in the structure (Supplementary Fig. S1c). The repeat-proximal spacer adopts a distorted U-turn conformation in a pocket between NUC1 and NUC2 (Fig. 3b). Several sugar-phosphate interactions stabilize the highly-distorted RNA conformation within the NUC lobe (Supplementary Fig. S3a). While the discontinuous spacer nucleotides within LshCas13a appear to follow a similar trajectory, our structure provides a more holistic view of spacer orientation within the NUC lobe. The crystal structure of LshCas13a also revealed pre-ordered A-form spacer nucleotides bound at the surface of the NTD 16, which we did not observe in LbaCas13a.
Figure 3. Structure and recognition of the crRNA spacer.
a, Simulated annealing mFo-DFc omit electron density map of the crRNA spacer contoured at 2σ. b, Opposing perspectives of the two halves of the NUC lobe that distort the crRNA spacer into a stabilized U-turn conformation. The half of the NUC lobe projecting out of the page is omitted in each image for clarity. c, The 3′ spacer emerging from the NUC lobe where it interacts with HEPN2. The spacer is shown in cartoon representation (orange) with the A-form nucleobases C(11)-G(13) shown as sticks. The amino acids F1338 and F1293 are shown as sticks stacking with C(11) gating access to the upstream spacer. d, Endpoint total fluorescence values for ssRNA-targeting assay by LbaCas13a HEPN mutants (background-corrected mean ± st. dev, n=3). Fitted apparent rates for these reactions are presented in Supplementary Fig. S4e. e, View of the composite HEPN nuclease active-site with the catalytic residues (RX4H) shown in stick representation. Notably, H605 of HEPN1-I is removed from the surface exposed composite site with distances shown.
The conformation of the LbaCas13a spacer sequence led us to wonder if spacer length would dramatically impact the HEPN nuclease activity of LbaCas13a. Other homologs have demonstrated HEPN nuclease activation with a variety of spacer-lengths, ranging from 20–28 nucleotides 10,17. We did not observe a significant effect of spacer length on rates of LbaCas13a trans-ssRNA cleavage across this range (Supplementary Fig. S4b–c). We solved an additional structure of LbaCa13a complexed with a crRNA containing a 20-nt spacer (Table 1; Supplementary Fig. S1b). While the overall protein structure was identical to the 24-nt spacer containing complex, we observed disorder and partial occupancy of the crRNA spacer within the NUC lobe of the 20-nt spacer containing complex (Supplementary Fig. S1d). These observations suggest that LbaCas13a accommodates a range of spacer lengths, sequence identities, and conformations within the spacer cavity, consistent with its role as an adaptive RNA-guided immune system.
We examined the entirety of the bound spacer within LbaCa13a to identify interactions between the NUC lobe and crRNA spacer. The HEPN domains, Helical-2 and Helical-3 domains make a number of conserved contacts with the sugar-phosphate backbone of the spacer (Fig. 3c; Supplementary Fig. S3a–c). Notably, the 3′ end of the spacer (C(11)-G(13)) emerges from the NUC lobe, where it adopts an A-form conformation as C(11) stacks with F1293 and F1338 of HEPN2, effectively gating access to the proximal spacer sequence that is sandwiched within the NUC lobe (Fig. 3c). The nucleotides emerging from the NUC lobe adopt a similar conformation in LshCas13a and in our second structure (20nt-spacer) of LbaCas13a (Fig. 3c; Supplementary Fig. S1d–e). Additionally, there are two highly conserved pockets that extensively contact the sugar-phosphate backbone around spacer nucleotides A(6) and A(8), rendering the nucleobases solvent inaccessible (Supplementary Fig. S3b–c). These structural observations suggest that the Cas13a-crRNA complex orders the repeat-proximal spacer into a distorted U-turn while conformationally gating HEPN nuclease activation through an extensive network of interactions with the sugar-phosphate backbone. Further biochemical and structural studies will be required to determine how the spacer rearranges upon activator-binding and how this conformational change triggers Cas13a HEPN domain activation.
We also sought to understand how crRNA-bound LbaCas13a is competent for target search but not active for trans-ssRNA cleavage. Evidence from both biochemical and structural experiments has suggested that U-cleaving Cas13a enzymes are dependent on the formation of a composite active-site comprised of two RX4H motifs, one contributed by each HEPN domain 8,10,16,17. In agreement with the catalytic requirements of the U-cleaving subfamily, double mutation of the conserved arginine and histidine within either HEPN1 (R600, H605) or HEPN2 (R1243, H1248) abolished general RNase activity of LbaCas13a (Fig. 3d; Supplementary Fig. 4e, f). In the LbaCas13a structure, the HEPN1 motif adopts a conformation in which α2 of HEPN1-I is distorted, burying H605 of the RX4H motif distant from the putative active-site beneath a loop of HEPN2 (Fig. 3e). Given this remote and solvent inaccessible positioning, we questioned whether H605 participates in HEPN nuclease activity. Similar to double mutations in HEPN1 or HEPN2, the single H605A substitution was sufficient to render LbaCas13a catalytically inactive (Fig. 3d), suggesting that activation of the enzyme requires H605 to join the surface-exposed HEPN motif. Overall, these data demonstrate that the HEPN nuclease remains inactive prior to ssRNA-activator binding due to the crRNA-assisted separation of HEPN active site residues. These data extend the previous mechanistic understanding gained from the LshCas13a structure, where it was observed that the HEPN domains were surface exposed to form a composite active site 16. Activator binding must induce a synergistic conformational change involving the release of the spacer from the NUC lobe cavity and accommodation of the activator-spacer RNA duplex, similar to the target RNA binding mechanism observed in human Ago2 23.
Active site for Cas13a crRNA maturation
Pre-crRNA processing is broadly conserved within the Cas13a family, yet despite extensive mutagenesis studies, the precise active-site residues and chemical mechanism are unknown 10,14,16. Our binary LbaCas13a-crRNA structure represents the product-bound state of the pre-crRNA processing reaction, prompting us to examine the mature crRNA 5′-terminus for insights into the catalytic mechanism. The structure revealed that the 5′-flank (C(-24)-A(-28)) is directed away from the core of the enzyme and into a surface-exposed groove formed by the Helical-1 and HEPN2 domains (Fig. 4a). Both Helical-1 and HEPN2 extensively interact with the sugar-phosphate backbone of the 5′-flank, stabilizing the crRNA in the product-bound state (Supplementary Fig. S3a). The residues within LbuCas13a whose mutation results in processing defects are predicted by conservation to line this processing groove between Helical-1 and HEPN2 (Supplementary Fig. S5). The 5′ terminal nucleotide within the LbaCas13a structure, A(-28), has a well-resolved 5′-hydroxyl that is surrounded by H328, K435, and K1320, amino acids that are strictly conserved within the Cas13a A-cleaving subfamily (Fig. 4a; Supplementary Fig. S3a, Supplementary Fig. S5).
Figure 4. Structural basis for crRNA maturation by LbaCas13a.
a, The pre-crRNA processing groove formed between Helical-1 and HEPN2 domains of wild-type LbaCas13a (left). Close-up view of the pre-crRNA processing site in LbaCas13a (right). The 5′-hydroxyl and amino acids implicated in processing are shown as sticks with hydrogen-bonding interactions denoted by dashed lines. b, LbaCas13a-mediated pre-crRNA processing measured under single-turnover conditions for alanine substitutions within the pre-crRNA processing groove. For clarity, 60 min endpoints (mean ± st. dev, n=3) are graphed as the percentage of substrate remaining (uncleaved pre-crRNA). Time course data are plotted and rates calculated in Supplementary Fig. S4. c, Bar graph of LbaCas13a-mediated pre-crRNA processing under single-turnover conditions in the presence of EDTA or a modified pre-crRNA dG(-29). Percentage of uncleaved substrate was measured after 60 min (background correct mean ± st. dev, n=3). d, Close-up view of the pre-catalytic state of LbaCas13a (H328A) showing the pre-crRNA processing groove formed between Helical-1 and HEPN2 domains. The amino acids surrounding the scissile phosphate between A(-28) and G(-29) are shown as sticks with hydrogen-bonding interactions denoted by dashed lines. H328, marked with an asterisk, is shown in a ball-and-stick representation modeled in the conformation observed in the structure of wild-type LbaCas13a. e, Proposed model for acid-base-catalyzed maturation of pre-crRNA by LbaCas13a.
To test the involvement of H328, K435, and K1320 in crRNA maturation, we substituted alanine at these positions and assayed each point mutant’s capacity for pre-crRNA cleavage. All three single point mutants were completely inactive for pre-crRNA processing, confirming the role of H328, K435 and K1320 in catalysis of crRNA maturation (Fig. 4b; Supplementary Fig. S4d). Further biochemical testing of additional residues near A(-28) revealed that alanine substitutions at K432, D1268, or K1305 completely ablate activity, while mutations of W325 or N1232 modestly impact processing (Fig 4b; Supplementary Fig. S4d). To confirm that the observed mutant phenotypes are catalytic deficiencies and not substrate binding defects, we assayed mutant HEPN-nuclease activity and binding affinity of a pre-crRNA mimic substrate. With the exception of D1268A, the processing mutants maintained near wild-type levels of ssRNA trans-cleavage activity (Supplementary Fig. S4e–f). Furthermore, all of the catalytically implicated LbaCas13a mutants retained wild-type binding affinity for both the pre-crRNA mimic and the mature crRNA (Supplementary Fig. S4g–i).
These structural and biochemical observations are consistent with crRNA processing occurring via an acid-base mechanism, where the reaction proceeds through a cation metal-independent pathway in which the 2′-hydroxyl nucleophilically attacks the scissile phosphate in the 3′ position. To assess the role of divalent cations in pre-crRNA processing, we performed processing reactions in the presence of increasing concentrations of the metal chelator EDTA and observed minimal impacts on LbaCas13a processing activity (Fig. 4c; Supplementary Fig. S4j). To confirm the role of the 2′-hydroxyl of G(-29), we assayed the ability of LbaCas13a to process a pre-crRNA substrate in which the catalytic ribonucleotide is substituted for a 2′-deoxynucleotide (dG(-29) substrate). LbaCas13a was unable to cleave this pre-crRNA mimic substrate (Fig. 4f), providing biochemical evidence for the metal-independent acid-base mechanism of pre-crRNA processing.
To provide support for this model of crRNA processing, we crystallized the crRNA-processing deficient LbaCas13a H328A with a pre-crRNA substrate bearing two nucleotides 5′ to the scissile phosphate (Table1; Fig. 4d) Wild-type LbaCas13a can process this truncated pre-crRNA (Supplementary Fig. S4k), suggesting that this structure represents the pre-catalytic, substrate-bound state of crRNA processing. The structure revealed that K435 and K1320 are positioned around the scissile phosphate, while W325 and N1232 stabilize a distorted C2′-endo conformation of the G(-29) ribose. Furthermore, modeling of H328 into the mutant structure suggests that it would be positioned to contact the scissile phosphate. Interestingly, we did not observe any of the biochemically implicated residues within hydrogen-bonding distance of the ribose 2′-hydroxyl. The pre-crRNA-bound structure revealed that the 2′-hydroxyl of G(-29) contacts N1232 and a single water molecule that is coordinated by HEPN2 residues K1305 and K1320. These structural observations led us to hypothesize that the G(-29) 2′-hydroxyl is deprotonated by an activated water. Consistent with this hypothesis, K1305 is flanked by two carboxylate moieties (D1268 and E1235) that may enable it to act as a general base to activate the coordinated water.
Together these data suggest a model for LbaCa13a-mediated crRNA processing in which conserved residues within the pre-crRNA processing groove between the Helical-1 and HEPN2 domains form an acid-base catalytic center (Fig. 4e). LbaCas13a binds to its cognate pre-crRNA, positioning the 5′-flank within the processing groove where W325 and N1232 stabilize the ribose of G(-29) in a distorted C2′-endo conformation. This coordination sets up the ribose 2′-hydroxyl for nucleophilic attack on the scissile phosphate, at which point K1305, an activated water, K1320, K435 and H328 participate in proton transfer to facilitate the formation of a 2′-3′-cyclic phosphate on G(-29), leaving behind a mature crRNA with a ribose 5′-hydroxyl on A(-28).
DISCUSSION
The signature proteins of Type VI-A CRISPR-Cas systems, Cas13a, are a remarkably diverse family of RNA-guided enzymes that degrade ssRNA non-specifically upon binding a spacer-complementary ssRNA-activator. In this work, we present high-resolution structures of the LbaCas13a:crRNA complex, revealing the active site for pre-crRNA processing and suggesting a structural gating mechanism of RNA-guided HEPN nuclease activity. We also uncovered the key conformational differences between two divergent Cas13a subfamilies, highlighting the molecular basis for orthogonal crRNA preferences. Finally, this study reveals that Cas13a ssRNA targeting activity is constrained during target search by both crRNA spacer conformation and HEPN domain arrangement.
Initial studies determined that residues important for pre-crRNA processing by two Cas13a homologs, LbuCas13a10 and LshCas13a16, are located in the groove between the Helical-1 and HEPN2 domains. Our study extends this work by establishing that the active site for pre-crRNA processing by all A-cleaving Cas13a homologs is in the Helical-1/HEPN2 interface based on structural, biochemical and phylogenetic data. Protein sequence divergence and differing sites of cleavage within pre-crRNA preclude direct comparison to all of the U-cleaving Cas13a subfamily members14. Nonetheless, biochemical evidence suggests that LbuCas13a pre-crRNA processing is also metal ion-independent and likely occurs in an active site located between the Helical-1 and HEPN2 domains10,14. These results are consistent with our identification of a conserved RNA processing groove that likely serves to stabilize the crRNA backbone.
After associating with its crRNA, the Cas13a:crRNA complex is competent for target surveillance. The high-resolution structures presented in this paper reveal conserved contacts between the spacer and the two halves of the NUC lobe, suggesting that these regions may function in conformational sensing of ssRNA-activator binding. The repeat-proximal portion of the spacer is highly distorted and sequestered within the core of the enzyme, making it unlikely that this pocket could accommodate a complementary activator RNA without significant conformational rearrangements. This contrasts with other RNA-guided nuclease complexes, including Cas9 and Argonaute, which pre-order their guides within an extended but accessible cavity 24,25. Due to the anchoring of the crRNA 5′ handle within the REC lobe and the U-turn conformation of the spacer, we expect that the activator-bound enzyme may contain distortions from a canonical double helix, similar to the spacer:target duplex observed in Type III Cmr RNA-targeting systems 26. We propose that both Cas13a and the distorted crRNA spacer must undergo a concerted conformational change upon ssRNA-activator association that drives HEPN domain dimerization, forming a composite active site and activating Cas13a for ssRNA cleavage.
Cas13a represents a novel paradigm within RNA-guided nucleases where the ordered region of the target-complementary sequence is sequestered within the enzyme, with disordered regions of the guide RNA exposed to solvent. Other RNA-guided nucleases, including the Argonaute family and Cas9, utilize a seed region within their guides for initial nucleic-acid recognition to direct target-binding into a pre-ordered A- or B-form guide 27,28. The pre-organization of the seed sequence accelerates target search and mismatch discrimination by decreasing the entropic cost of target binding 29. Cas13a is not following this model for target search, as the solvent-accessible crRNA regions are disordered and the ordered crRNA regions are incompatible with duplex formation. Due to the kinked and inaccessible spacer nucleotides, complementary ssRNA-activator binding can only occur once the system overcomes this additional energetic barrier, raising the question of what energetic parameter drives ternary complex formation.
The Cas13a enzyme family continues to be developed as a platform for RNA detection and programmable RNA binding 13,14. Bioengineering of Cas13a will require a complete understanding of how the enzyme carries out target search and is subsequently activated to become a general RNase. Our high-resolution target surveillance structure provides key details about crRNA recognition, processing and spacer ordering that will advance the development of LbaCas13a as valuable tool for RNA detection.
METHODS
LbaCas13a protein production and purification
The expression and purification of Lachnospiraceae bacterium NK4A179 Cas13a (LbaCas13a) was carried out as described elsewhere 14 with modifications, summarized here: His6-MBP-TEV-tagged Cas13a was prepared in E. coli Rosetta2 (DE3) grown in TB at 37°C. At an OD600 of 0.4–0.5, cultures were cooled on ice for 15 minutes prior to induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and expression overnight at 16°C. Soluble His6-MBP-TEV Cas13a was loaded onto a 5 mL HiTrap NiNTA column (GE Healthcare) and eluted over a linear imidazole (0.01–0.3 M) via FPLC (ÄKTA Pure). After overnight dialysis and TEV digestion, the eluted LbaCas13a was further purified by ion exchange (5 mL HiTrap SP, GE Healthcare) and size-exclusion chromatography (S200 16/60, GE Healthcare) with 2 mM TCEP supplemented into the gel filtration buffer.
In vitro transcription and purification of RNA
For biochemical experiments, RNA was transcribed and radiolabeled as described previously 14. For crystallization, pre-crRNA was transcribed in vitro from a PCR-amplified double-stranded DNA template encoding a hepatitis delta virus (HDV) ribozyme 3′ to the pre-crRNA sequence. Transcriptions were carried out as described previously 10 with the following modifications: pre-crRNAs were transcribed at 37°C overnight, followed by treatment with DNase I and 5 mM MgCl2 (37°C, 1 hr) to quench the reaction and drive ribozyme cleavage. Before complexing with LbaCas13a, pre-crRNA was annealed in 1X annealing buffer (10 mM Na-HEPES pH 7.4, 30 mM KCl, 1.5 mM MgCl2) by heating at 75°C for 5 min followed by rapid cooling on ice.
LbaCas13a:pre-crRNA complex formation
For crystallization, the LbaCas13a:crRNA or LbaCas13a (H328A) pre-crRNA complex was reconstituted in vitro by incubating purified LbaCas13a protein with pre-crRNA at a molar ratio of 1:1.3 at 37°C for 1 hr in complexing buffer (20 mM Na-HEPES pH 7.0, 200 mM KCl, 1 mM TCEP). The complex was purified by size-exclusion chromatography (S200 10/300 Increase, GE Healthcare) in complexing buffer and concentrated using a 10 kDa Centricon (Thermo Fisher) before immediate use in crystallization experiments. The purity and integrity of protein and RNA were assessed using SDS-PAGE (Coomassie blue staining) and 15% (v/v) TBE-urea-polyacrylamide gel electrophoresis (SYBR Gold straining), respectively (Supplementary Fig. S1b).
LbaCas13a crystallization and data collection
Initial screening was carried out with JCSG Core Suites (Qiagen) in 96-well sitting drop vapor diffusion format (Intelli-plate, Hampton Research) at 293 ± 1 K. Diffraction-quality crystals of the LbaCas13a:crRNA complex (20-nt and 24-nt containing spacer) (A280=4.0) and LbaCas13a:pre-crRNA complex (24-nt containing spacer) (A280=5.7) grew in 0.2 M ammonium iodide and 20% (w/v) PEG 3,350. Crystals were cryoprotected in 10–15% (v/v) glycerol, and high-resolution diffraction data were collected at the Advanced Light Source (ALS) beam line 8.3.1 on a Pilatus3 S 6M (Dectris) detector under cryogenic conditions. To produce isomorphous crystals for phasing by native single-anomalous dispersion (SAD), sitting drop vapor diffusion experiments containing 2 μL purified LbaCas13a:crRNA complex (24-nt containing spacer) complex (A280=4.0) and 2 μL reservoir solution (0.2 M ammonium iodide, 20–25% (w/v) PEG 3,350) were prepared in 24-well Cryschem plates (Hampton Research). A total of 52 datasets were collected from 26 crystals (cryoprotected in 10% (v/v) glycerol) under cryogenic conditions at 6.0 keV (ALS beam line 8.3.1, Pilatus3 S 6M). Each dataset covered 360° of crystal rotation divided into 0.2° images collected in 45° inverse-beam wedges at 5 Hz. To minimize pixel readout error, the detector was moved by 10 mm before collecting a second 360°-pass from the same position of each crystal.
Structure determination by native SAD and model refinement
High-resolution data collected from LbaCas13a crystals at 11.1 keV were processed and scaled in XDS 30 before merging in AIMLESS 31,32. After processing with XDS, each SAD dataset was compared to every other dataset after applying each of the 27 possible re-indexing operations found by OTHERCELL 31, and then computing an isomorphous R-factor with a 4 Å resolution cut-off using SCALEIT 31. Pairs of datasets agreeing to better than 20% were grouped and merged using XSCALE 30 producing a final cluster containing 20 wedges in the space group C2 (data statistics summarized in Table 1). The merged anomalous data were prepared for substructure search in SHELXC 33, and input into SHELXD set to search for 65 sites using a high-resolution cutoff of 3.5 Å. Nine solutions were found after 25,000 trials and each solution was input separately into SHELXE. The correct substructure solution was found to contain a total of 115 sites, 32 of which were correct as determined using PHENIX.emma 34. A readily interpretable map (CC 25%) was obtained after 30 cycles of density modification and model building in SHELXE. PHENIX.AutoBuild 35 was used to extend the initial α-helical model, complemented with manual model-building in COOT 36 and RNA-modelling with RCrane 37. The atomic models were completed by iterative building and refinement in COOT and PHENIX.refine 38 against the high-resolution data using the 2mFo-DFc, mFo-DFc, anomalous Fourier difference and 2mFo-DFc composite omit 39 maps. The final models for LbaCas13a:crRNA (24-nt spacer), LbaCas13a:crRNA (20-nt spacer), and LbaCas13a(H328A):pre-crRNA (24-nt spacer) were validated in MOLPROBITY 40 and submitted to the Protein Data Bank (PDB) under the accession codes 5W1H, 5W1I, and 5WLH respectively. All figures were generated in PyMOL (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Radiolabeled crRNA maturation assays
Pre-crRNA processing assays were performed as previously described 14. Briefly, 100 nM LbaCas13a was incubated with <1 nM 5′-radiolabeled pre-crRNA substrates at 37°C in processing buffer (20 mM HEPES-Na pH 6.8, 50 mM KCl, 5 mM MgCl2, 10 μg/mL BSA, 100 μg/mL tRNA, 0.01% Igepal CA-630, and 5% glycerol). Reactions were quenched (100% formamide, 0.025% bromophenol blue, and 200 μg/mL heparin), denatured at 95°C for 5 min and resolved by 15% denaturing PAGE (0.5x TBE buffer). All bands were visualized by phosphorimaging (Typhoon, GE Healthcare) and quantified with ImageQuant (GE Healthcare). The percent cleavage was determined as the ratio of the intensity of the product band to the total intensity of both the product and uncleaved pre-crRNA bands, and normalized for background within each measured substrate. For clarity, in some figure panels the percentage of uncleaved substrates is presented (calculated as 100-%cleaved).
Fluorescent ssRNA nuclease assays
Target cleavage assays were performed as previously described with minimal modification 14. Briefly, 100 nM LbaCas13a was complexed with 50 nM pre-crRNAs at 37°C in processing buffer for 30–60 mins. Alternatively, cleavage buffer (20 mM HEPES-Na pH 6.8, 50 mM KCl, 5 mM MgCl2) was used for reactions instead of processing buffer as noted in the figure legends, resulting in less fluorescent signal generated. Generally, 150 nM RNase Alert reporter (IDT) and 10 nM of ssRNA-activator were added to initiate the reaction. Reactions were incubated in a fluorescence plate reader (Tecan Infinite Pro F2000) for 60 min at 37°C, with fluorescence measurements taken every 5 min (λex: 485 nm; λem: 535 nm). Background corrected values were calculated by subtracting signal in the negative control containing no activating RNA. For endpoint analyzes, the background corrected florescence value from the 60 min time point is presented (mean ± st. dev., n= 3). Apparent rates were calculated according to a single-exponential decay using Prism7 (GraphPad) fitted to the entire curve, and calculated rates were plotted with their associated standard deviations (n=3).
Filter-binding assays
Filter-binding assays were carried out as previously described 10. Briefly, Cas13a and radiolabeled crRNA were incubated for 1 hr at 37°C in RNA-processing buffer (20 mM HEPES-Na pH 6.8, 50 mM KCl, 5 mM MgCl2, 10 μg/mL BSA, 100 μg/mL yeast tRNA, 0.01% Igepal CA-630 and 5% glycerol). Tufryn, Protran and Hybond-N+ membranes were assembled onto a dot-blot apparatus in the order listed. The membranes were washed twice with 50 μL cleavage buffer (20 mM HEPES-Na pH 6.8, 50 mM KCl, 5 mM MgCl2, and 5% glycerol) before the sample was applied to the membranes. Membranes were then washed with 50 μL cleavage buffer, dried, and visualized by phosphorimaging. Data were quantified with ImageQaunt TL Software (GE Healthcare) and fit to a binding isotherm using Prism7 (GraphPad). All experiments were carried out in triplicate.
Supplementary Material
Acknowledgments
We thank Nan Ma and Kaihong Zhou for technical assistance, Christine L. Gee for assistance with refinement, and members of the Doudna laboratory for helpful discussions. G.J.K. acknowledges support from the Howard Hughes Medical Institute. This work was supported in part by a Frontiers Science award from the Paul Allen Institute to J.A.D, and the National Science Foundation (MCB-1244557 to J.A.D.). J.A.D is an Investigator of the Howard Hughes Medical Institute. J.A.D. is a co-founder of Caribou Biosciences, Inc., Editas Medicine, and Intellia Therapeutics.
Footnotes
ONLINE CONTENT
The atomic coordinates and structure factors for the LbaCas13a-crRNA complexes have been deposited to the Protein Data Bank (PDB) under the accession codes 5W1H for LbaCas13a:crRNA complex (24-nt spacer), 5W1I for LbaCas13a:crRNA complex (20-nt spacer) and 5WLH for LbaCas13a(H328A):pre-crRNA complex (24-nt spacer).
AUTHOR CONTRIBUTIONS
G.J.K, A.E-S, M.R.O and J.A.D. designed the study. G.J.K., A.E-S., J.C.C. and E.C. expressed and purified proteins. G.J.K prepared and crystallized the Cas13a complexes with assistance from J.C.C, collected X-ray data, and determined the crystal structure with support from J.H. A.E.S carried out biochemical assays with assistance from G.J.K and J.C.C. G.J.K drafted the manuscript, and all authors reviewed and edited the manuscript.
References
- 1.Dy RL, Richter C, Salmond GP, Fineran PC. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu Rev Virol. 2014;1:307–31. doi: 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–44. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 7.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–6. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Molecular Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelles DA, et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–96. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.East-Seletsky A, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016 doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017 doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smargon AA, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017 doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gootenberg JS, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell. 2017;66:373–383 e3. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, et al. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell. 2017;168:121–134 e12. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:557. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–72. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Structural Principles of CRISPR RNA Processing. Structure. 2015;23:13–20. doi: 10.1016/j.str.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–41. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–13. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–11. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakanishi K. Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip Rev RNA. 2016;7:637–60. doi: 10.1002/wrna.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–30. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol Cell. 2009;33:204–14. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–81. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 29.Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr D Biol Crystallogr. 1999;55:1690–5. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–9. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keating KS, Pyle AM. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc Natl Acad Sci U S A. 2010;107:8177–82. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC, et al. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr D Biol Crystallogr. 2008;64:515–24. doi: 10.1107/S0907444908004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.