Abstract
Tumor-specific CD4+ T helper 9 (TH9) cells, so-called because of their production of the cytokine interleukin-9 (IL-9), are a powerful effector T cell subset for cancer immunotherapy. We found that pretreatment of naïve CD4+ T cells with IL-7 further enhanced their differentiation into TH9 cells and augmented their antitumor activity. IL-7 markedly increased the abundance of the histone acetyltransferase p300 by activating the STAT5 and PI3K-AKT-mTOR signaling pathways and promoting the acetylation of histones at the Il9 promoter. As a result, the transcriptional regulator Foxo1 was dephosphorylated and translocated to the nucleus, bound to the Il9 promoter, and induced the production of IL-9 protein. In contrast, Foxp1, which bound to the Il9 promoter in naïve CD4+ T cells and inhibited Il9 expression, was outcompeted for binding to the Il9 promoter by Foxo1 and translocated to the cytoplasm. Furthermore, forced expression of Foxo1 or a deficiency in Foxp1 in CD4+ T cells markedly increased the production of IL-9, whereas a deficiency in Foxo1 inhibited the ability of IL-7 to enhance the differentiation and antitumor activity of TH9 cells. Thus, we identified the roles of Foxo1 as a positive regulator and Foxp1 as a negative regulator of TH9 cell differentiation and antitumor activity, which may provide potential targets for cancer immunotherapy.
INTRODUCTION
Naïve CD4+ T cells exit from quiescent status, become activated, and differentiate into different T helper (TH) cell subsets upon antigen stimulation in the presence of certain cytokines, including TH1, TH2, TH9, TH17, regulatory T (Treg) cells, and follicular helper T (TFH) cells, to mediate various immune responses (1). The cytokine interleukin-9 (IL-9) was previously considered to be a TH2-type cytokine and related to the TH2-type immune response (2). Although it was found that IL-4, together with transforming growth factor–β (TGF-β), stimulates activated T cells to produce IL-9 more than 20 years ago (3), this population of TH cells was only relatively recently identified as a specific subset called TH9 cells (4), and two transcriptional regulators that directly bind to the Il9 promoter [PU.1 and IRF4 (interferon regulatory factor 4)] were identified (5, 6). In addition to TH2 cell–like functions such as mediating allergic inflammation (7) and clearing intestinal parasites (8), the most prominent function of TH9 cells is their antitumor activity, especially against melanoma (9, 10). We and others showed that TH9 cells produce IL-9 and IL-21 to activate the antitumor function of CD8+ cytotoxic T cells (10, 11). Although transcriptional regulators [including PU.1, IRF4, GATA3, STAT5 (signal transducer and activator of transcription 5), and IRF1] are involved in regulating the differentiation of naïve CD4+ T cells into TH9 cells (4–6, 11, 12), TH9 cell–specific transcriptional factors are still unknown. It is unclear how the expression of Il9 is negatively regulated during TH9 cell differentiation.
IL-7 signaling is critical for thymocyte development, naïve T cell survival, and homeostasis in vivo (13). When normal mice are injected with IL-7, the number of T cells increases in peripheral lymph nodes. In addition, in mice that are irradiated in preparation for bone marrow transfer or rendered lymphopenic by cyclophosphamide, IL-7 accelerates T cell accumulation in peripheral lymph organs (14–16). Those features of IL-7 make it attractive for cancer therapy because many cancer patients experience cytotoxic chemotherapy–induced lymphopenia. When IL-7 is administered in vivo, it promotes TH1 cell differentiation and a T cell–dependent antitumor reactivity (17–20). In vitro, IL-7 prevents T cell death by increasing the abundance of the anti-apoptotic protein Bcl2 (21), which enables the increased numbers of T cells to be generated ex vivo for adoptive immunotherapy. The IL-7 receptor (IL-7R) consists of two subunits, the IL-7R α chain (IL-7Ra; also known as CD127) and the common γ chain (γC; also known as CD132). The main signaling pathways induced by IL-7 are mediated by STAT5, the serine and threonine kinases PI3K (phosphatidylinositol 3-kinase) and AKT, and the mitogen-activated protein kinase ERK (extracellular signal–regulated kinase). Both the STAT5 and PI3K-AKT pathways are indispensable for cell survival (22, 23).
Here, we found that IL-7–treated CD4+ T cells exhibit enhanced TH9 cell differentiation (hereafter referred to as IL-7–TH9 cells) and antitumor activity compared to TH9 cells that underwent differentiation in the absence of IL-7. Signaling of the IL-7R, including the STAT5 and PI3K-AKT–activated mTOR (mammalian target of rapamycin) pathways, was involved in histone acetylation at the Il9 promoter. The transcription factors Foxo1 (Forkhead box protein O1) and Foxp1 (Forkhead box protein P1) played reciprocal roles in regulating TH9 and IL-7–TH9 cell differentiation. We conclude that the enhanced antitumor function of IL-7– pretreated TH9 cells is Foxo1-dependent.
RESULTS
Pretreatment of naïve CD4+ T cells with IL-7 enhances their differentiation into TH9 cells
To evaluate the effect of IL-7 on TH9 cell differentiation, we cultured murine CD4+ T cells under TH9-polarizing conditions [that is, in the presence of the cytokines IL-4 and TGF-β, and a monoclonal antibody (mAb) against interferon-γ (IFN-γ)] with or without IL-7. We found that IL-7 did not enhance TH9 cell differentiation (fig. S1A), that is, the number of IL-9–expressing cells was not increased, which is consistent with a previous report (12). Considering the role of IL-7 in T cell homeostasis and survival, CD4+ T cells were pretreated with IL-7 for 48 hours and then cultured under TH9-polarizing conditions. The number of IL-9– and IL-21–producing cells (TH9 cells) was markedly increased in an IL-7 dose-dependent manner compared with cells that did not receive IL-7 pretreatment. Analysis by flow cytometry to measure the mean fluorescence intensity (MFI) of IL-9 in these cells confirmed these results (Fig. 1B). The enhanced generation of TH9 cells was also confirmed by IL-9–specific enzyme-linked immunosorbent assay (ELISA) and real-time polymerase chain reaction (PCR) analyses (Fig. 1, C and D). However, when CD4+ T cells were pretreated with other common γC receptor family cytokines, such as IL-2 or IL-15, TH9 cell differentiation was not enhanced (Fig. 1E). These data suggest that IL-7 exerts a specific effect on TH9 cell differentiation.
Fig. 1. Pretreatment of naïve CD4+ T cells with IL-7 enhances their differentiation into TH9 cells.
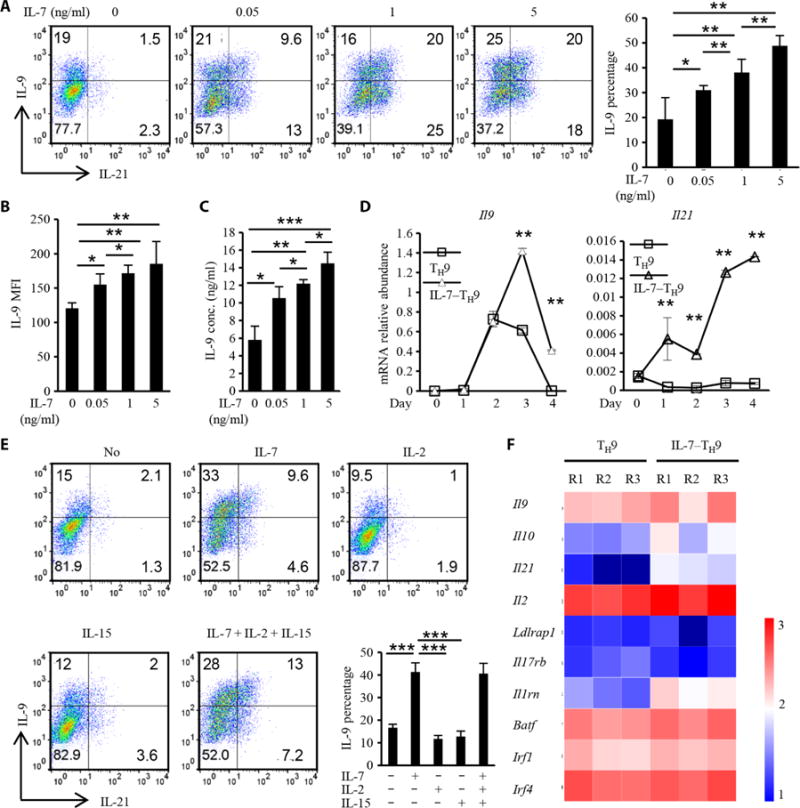
(Ato C) Naïve OT-II CD4+ T cells were cultured with the indicated concentrations of IL-7 for 2 days, washed twice with T cell medium, and then cultured for 4 days under the TH9-polarizing conditions. Freshly isolated naïve CD4+ T cells that were not pretreated with IL-7 were used as a control. The cells were then harvested and restimulated with PMA and ionomycin in the presence of brefeldin A for 4 hours. The cells were then analyzed by flow cytometry to detect IL-9 and IL-21 (A, dot plots), to determine the percentages of IL-9–expressing cells (A, bar graph), and to calculate the MFI of IL-9 (B). Alternatively, the cells were stimulated with anti-CD3 (1 μg/ml) for 12 hours, and the cell culture medium was analyzed by ELISA to determine the amount of secreted IL-9 (C). (D) TH9 cells (open squares) and IL-7–TH9 cells (open triangles) were harvested on the indicated days of TH9 cell differentiation, and RNA was extracted and used for real-time PCR analysis of the relative abundances of Il9 and Il21 mRNAs, which were normalized to that of mouse actb mRNA. (E) Naïve OT-II CD4+ T cells were pretreated with the indicated γC family cytokines for 2 days, washed twice, and then cultured under TH9-polarizing conditions for 4 days. The cells were then analyzed by flow cytometry to detect IL-9 and IL-21 (dot plots) and to determine the percentages of IL-9–expressing cells (bar graph). (F) TH9 and IL-7–TH9 cells were collected on day 3, and RNA was extracted for gene microarray analysis. The expression of TH9 cell–related genes, including those encoding cytokines and transcription factors, is shown. R1 to R3 indicate three replicates. Data in (A) to (E) are means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
To explore whether IL-7 also enhanced the differentiation of human naïve CD4+ T cells into TH9 cells, we conducted similar experiments and confirmed that IL-7 pretreatment also facilitated the generation of human TH9 cells (fig. S1, B and C), suggesting that it is possible to generate human polarized TH9 cells ex vivo for testing in clinical trials. The expression of Il9 was rapidly and markedly induced on day 2 of differentiation and suddenly decreased on day 4 in TH9 cells; however, in IL-7–TH9 cells, Il9 expression was substantially increased. A similar result was observed for Il21 expression (Fig. 1D). To understand the features of IL-7–TH9 cells, we performed a gene expression microarray to analyze TH9-related genes. Although no new potential TH9 cell– related transcription factors were identified (Fig. 1F), the abundances of TH9 cytokines, including IL-9, IL-21, IL-10, and IL-2, were increased, indicative of enhanced TH9 cell differentiation.
IL-7–TH9 cells have enhanced antitumor activity
To investigate whether IL-7–TH9 cells had enhanced antitumor function compared to normal TH9 cells, we used both prophylactic and therapeutic models. In the prophylactic model, C57BL/6 mice were challenged with B16-OVA (ovalbumin) melanoma cells by intravenous injection, which was followed by the adoptive transfer of TH cells on the same day. Lung metastasis was assessed on day 14 after tumor challenge (Fig. 2A). Consistent with our previous report (10), mice treated with TH9 cells had a decreased tumor burden compared to those that received TH1 cells (Fig. 2, B and C). Notably, IL-7–TH9 cells exhibited enhanced antitumor function; lung tumors were almost cleared in all of the mice by day 14 (Fig. 2, B and C). We found that more donor CD4+ T cells were in the lung tissue of mice that received IL-7–TH9 cells (Fig. 2D) than of those that received TH1 or TH9 cells, suggesting that IL-7–TH9 cells were more persistent in vivo. The numbers of CD8+ T cells were also increased in the lung tissue of mice that received IL-7–TH9 cells (Fig. 2E).
Fig. 2. IL-7–TH9 cells have enhanced antitumor activity.
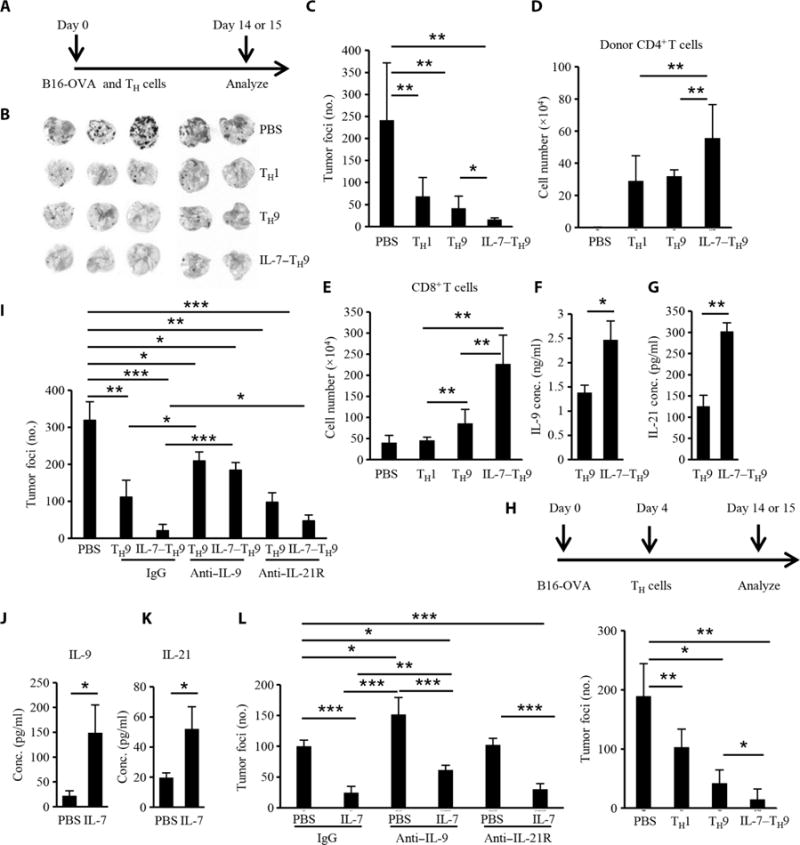
(A to G) C57BL/6 mice (five mice per group) were challenged with 2 × 105 B16-OVA cells, which was followed by the transfer of 2 × 106 TH1, TH9, or IL-7–TH9 cells from CD45.1 OT-II mice on the same day. On day 14, the mice were sacrificed and tissues were harvested for different assays. (A) Schematic showing the time course of the prophylactic model. (B) Images of lungs showing tumor development and foci in the different treatment groups. PBS, phosphate-buffered saline. (C) Lung tumor foci numbers according to the indicated treatment. (D) Numbers of adoptively transferred CD4+ T cells in the lungs of the recipient mice. (E) Numbers of CD8+ T cells that infiltrated the lungs for the indicated treatments. (F and G) Donor-derived CD4+ T cells from lung lymph nodes in the TH9 and IL-7–TH9 groups were sorted and stimulated with PMA and ionomycin. The amounts of IL-9 (F) and IL-21 (G) in the cell culture medium were determined by ELISA. (H) Therapeutic effect of TH cells in the B16-OVA lung model. Similarly to the prophylactic model, T cells were transferred to mice (four to five mice per group) on day 4 after tumor injection. (I) Numbers of tumor foci in the lungs according to the treatment group. In the therapeutic model (five mice per group), anti–IL-9 and anti–IL-21R antibodies (both at 200 μg per mice per injection) were injected intraperitoneally every other day, beginning 1 day before the TH9 cells were transferred, and tumor foci were analyzed on day 14. IgG, immunoglobulin G. (J to L) C57BL/6 mice (four to five mice per group) were injected with B16 tumor cells and received IL-7 (5 μg per injection) or PBS daily from the same day for 14 days. Anti–IL-9 or anti–IL-21R antibodies were injected every other day, and 1 day before IL-7 injection in the indicated groups. Then, mice were sacrificed on day 14, lung lymph nodes were collected from the PBS and IL-7 only groups, and total CD4+ T cells were isolated and stimulated with PMA and ionomycin before the amounts of IL-9 (J) and IL-21 (K) released into the cell culture medium were determined by ELISA. (L) The numbers of tumor foci were counted in all groups. Data in (B) to (L) were from three independent experiments and are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether these CD4+ T cells secreted IL-9 or other cytokines, donor-derived CD4+ T cells were sorted from the lung lymph nodes of the recipient mice and restimulated ex vivo with phorbol 12-myristate 13-acetate (PMA) and ionomycin, and the cell culture medium was collected for analysis by ELISA. We found that the IL-7–TH9 cells secreted more IL-9 and IL-21 than did the TH9 cells (Fig. 2, F and G). Moreover, in a therapeutic model, in which B16-OVA tumors were allowed to establish for 4 days before T cells were transferred into the mice (Fig. 2H), the IL-7–TH9 cells also exhibited a substantially greater antitumor function than did the TH9 cells. To investigate whether the enhanced antitumor function of the IL-7–TH9 cells was dependent on IL-9 or IL-21, neutralizing antibodies against either IL-9 or IL-21R were injected into the mice. Blocking IL-9 in vivo substantially inhibited the antitumor effect of both TH9 and IL-7–TH9 cells (Fig. 2I). Blocking IL-21R in vivo partially reduced the antitumor activity of the IL-7–TH9 cells but did not substantially affect the TH9 cells.
It has been reported that IL-7 alone or in combination with other drugs exhibits therapeutic effects in mouse tumor models, but it is not known whether IL-7 can stimulate IL-9 production in tumor models in vivo. To explore this question, we used the B16 tumor model. C57BL/6 mice were injected with B16 tumor cells and received IL-7 or PBS from that same day for a further 14 days. IL-7 exhibited a substantial antitumor effect (fig. S2A), and this effect was associated with the increased amounts of Il9 and Il21 mRNAs in the tumor bed (fig. S2, B and C), suggesting that IL-7 stimulates IL-9 and IL-21 production in vivo. To examine whether CD4+ T cells secreted these cytokines in response to IL-7, total CD4+ T cells were isolated from the lung lymph nodes of PBS- or IL-7–treated B16-bearing mice and restimulated ex vivo with PMA and ionomycin. IL-7 stimulated the CD4+ T cells to secrete IL-9 and IL-21 (Fig. 2, J and K). To determine whether the anti-tumor effect of IL-7 was dependent on IL-9 or IL-21, we used anti–IL-9 or anti–IL-21R antibodies in the presence or absence of IL-7 in the B16 tumor model. Blocking IL-9, but not IL-21R, in vivo not only rendered the mice more susceptible to developing lung melanoma but also compromised the antitumor effects of IL-7 (Fig. 2L).
IL-7 induces changes in the gene expression profile of CD4+ T cells
To understand the global effect of IL-7 on transcriptional regulation, we performed another gene microarray assay with naïve CD4+ T cells that were either untreated or treated with IL-7. IL-7 induced a global change in gene expression in these cells, with more than 4000 genes showing increased expression and 7000 genes showing decreased expression. Specifically, there was no difference between the two groups of cells in terms of the expression of genes involved in T cell–related transcriptional factors (Fig. 3A and fig. S3A), survival pathways, or metabolic pathways (fig. S3B). However, we found that the expression of more than two-thirds of the histone-encoding genes on the microarray was statistically significantly decreased in response to IL-7, which was suggestive of a marked change in chromatin structure to affect gene expression (fig. S4).
Fig. 3. IL-7 promotes histone acetylation at the Il9 promoter.
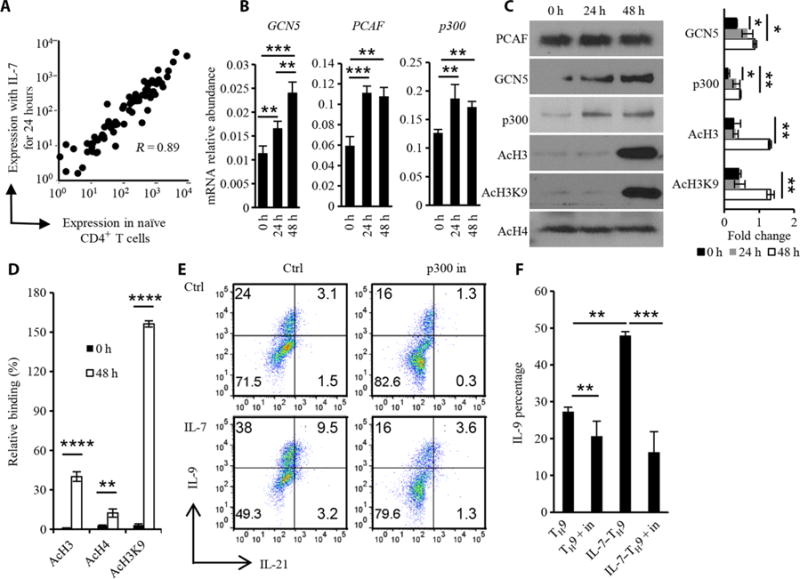
(A) Naïve OT-II CD4+ T cells were treated with IL-7 (1 ng/ml) for 0 or 24 hours, and RNA was extracted for gene microarray analysis. The expression of genes encoding selected T cell transcriptional factors with or without IL-7 treatment was determined. (B) Real-time PCR analysis was performed to determine the extent of expression of genes encoding the histone acetyltransferases GCN5, PCAF, and p300 in the T cells after IL-7 treatment for 24 or 48 hours. PCR results were normalized to mouse actb to show the mRNA relative abundance. (C) Naïve CD4+ T cells were treated with IL-7 for the indicated times before being analyzed by Western blotting with antibodies against the indicated proteins. (D) ChIP analysis of acetylated (Ac) H3 (AcH3), H4 (AcH4), and H3K9 (AcH3K9) at the Il9 promoter region in naïve CD4+ T cells that were untreated or were treated with IL-7 for 48 hours. Data were normalized to the control region. (E) Effect of a p300 inhibitor (p300 in) on TH9 and IL-7–TH9 cell differentiation. Naïve CD4+ T cells were pretreated with IL-7 for 2 days, washed twice, and polarized under TH9 conditions in the presence or absence of 5 μM C646 for 4 days. Naïve CD4+ T cells polarized under TH9 conditions without IL-7 pretreatment served as the TH9 control (Ctrl). Cells positive for IL-9 and IL-21 were determined by flow cytometry. (F) Percentages of IL-9+ T cells under the indicated conditions. Data in (C) to (F) are from three independent experiments and are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Because the expression of genes encoding the transcription factors required for T cell differentiation did not substantially change as a result of IL-7 exposure, we considered whether IL-7 might affect TH9 cell differentiation by modifying chromatin structure and gene locus accessibility, similar to a previous report on the TRG locus (24). Because histone modifications include methylation and acetylation (the latter is modified by histone acetyltransferases and acetylation facilitates gene expression), we analyzed whether there were changes in histone acetyltransferases in response to IL-7. The expression of some genes encoding histone acetyltransferases increased in IL-7–treated cells (fig. S5A). In addition, the abundances of GCN5 and p300 mRNA and proteins were also increased by IL-7, which resulted in an increase in total histone 3 (H3) acetylation (Fig. 3C), whereas total histone 4 (H4) acetylation did not change (Fig. 3, B and C). To understand whether histone acetylation occurred at the Il9 promoter locus, we performed chromatin immuno-precipitation (ChIP) assays and detected increased H3 acetylation and H4 acetylation at the Il9 (Fig. 3D) and Il21 promoter regions (fig. S5B). Studies showed that the acetylation of H3 Lys9 (H3K9) is enhanced in TH9 cells compared to that in other TH subset cells (5, 6), and we found that total acetylation of H3K9 (Fig. 3C) and acetylation of H3K9 at the Il9 promoter locus (Fig. 3D) were also increased in response to IL-7. To verify the importance of acetyltransferases in IL-7–induced TH9 cell differentiation, we performed experiments with a p300 inhibitor. We found that p300 inhibition slightly inhibited TH9 cell differentiation but substantially inhibited the increased TH9 cell differentiation induced by IL-7 (Fig. 3, E and F). Because of the global effect of p300 inhibition, other genes affected by p300 may also contribute to the decreased differentiation of TH9 cells.
The STAT5 and PI3K-AKT-mTOR signaling pathways contribute to histone acetylation on the Il9 promoter
Our earlier results suggest that IL-7 signaling modifies histone acetylation and may facilitate the access of transcription factors to the Il9 locus. How IL-7 controls histone acetylation in T cells is unclear. We found that signals downstream of IL-7, including PI3K-AKT, ERK, and STAT5, were activated, which is consistent with previous reports (23). A luciferase assay in 293T cells demonstrated that STAT5 bound directly to the p300 promoter and promoted its transcription (Fig. 4B), suggesting that STAT5 might contribute to the increased p300 abundance observed in IL-7–treated cells. To verify whether STAT5 directly bound to the p300 promoter in IL-7–treated CD4+ T cells, we performed a ChIP assay with an anti-STAT5 antibody. We found that IL-7 substantially increased the amount of STAT5 that bound to the p300 promoter in CD4+ T cells (Fig. 4C). In TH9 cells, increased amounts of STAT5 bound to the Il9 promoter were also detected after IL-7 treatment (fig. S6A). Western blotting analysis showed that STAT5 was substantially phosphorylated in CD4+ T cells on day 2 in the presence, but not absence, of IL-7, whereas the amounts of phosphorylated STAT5 (pSTAT5) were similar in TH9 cells with or without IL-7 pretreatment (fig. S6B). Moreover, the abundance of mTOR in CD4+ T cells increased after IL-7 treatment (Fig. 4A). Similarly to Foxo1, mTOR is a downstream target of the PI3K-AKT pathway.
Fig. 4. IL-7 promotes chromatin modification at the Il9 promoter and TH9 cell differentiation in a PI3K-AKT-mTOR–dependent manner.
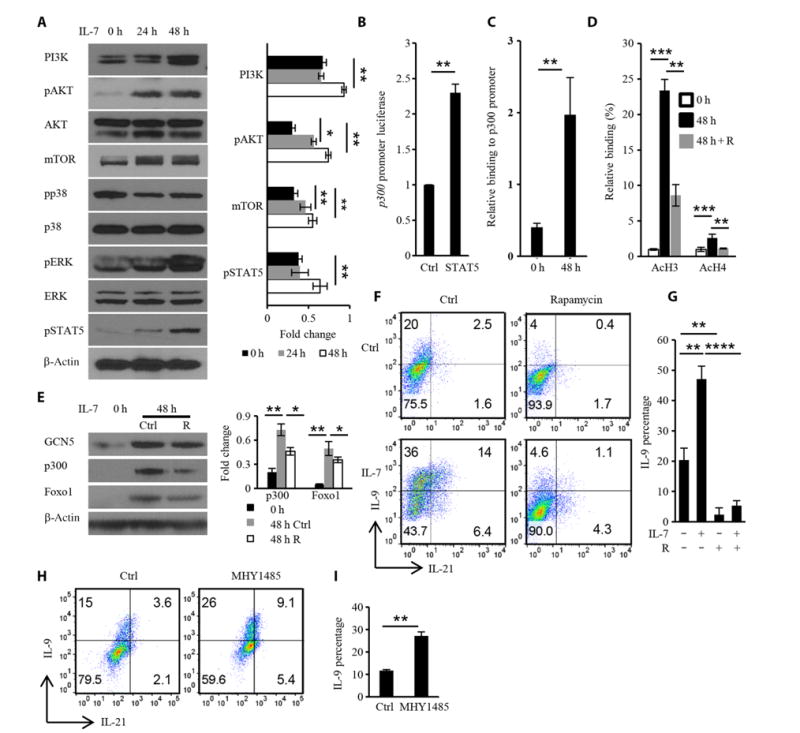
(A) Naïve OT-II CD4+ T cells were cultured for the indicated times with IL-7 (1 ng/ml), washed, and analyzed by Western blotting with antibodies against the indicated proteins to demonstrate signaling downstream of IL-7. Right: Densitometric analysis of the indicated band intensities. (B) EL4 cells were transfected with plasmids encoding the p300 promoter with or without the STAT5-expressing plasmid pcDNA3.1. Forty-eight hours later, the luciferase activity of the p300 promoter reporter was detected and normalized to that of Renilla luciferase. (C) ChIP analysis of the effect of IL-7 on the binding of STAT5 to the p300 promoter. (D) ChIP analysis of the inhibitory effect of rapamycin (R) on IL-7–induced histone acetylation after 48 hours of treatment. (E) Naïve OT-II CD4+ T cells were treated with IL-7 alone or in the presence of rapamycin for 48 hours before being analyzed by Western blotting with antibodies against the indicated proteins. (F to I) Naïve OT-II CD4+ T cells or IL-7–pretreated OT-II CD4+ T cells were cultured under TH9 conditions in the presence or absence of 30 nM rapamycin (F and G) or 1 μM MHY1485 (H and I) and then were analyzed by flow cytometry to detect IL-9 and IL-21. Data are from three independent experiments and are means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Considering the importance of mTOR in T cell differentiation (25), we wondered whether IL-7–induced histone acetylation and increased TH9 cell differentiation were also mTOR-dependent. Rapamycin, an mTOR complex 1 (mTORC1) inhibitor, inhibited histone acetylation at the Il9 promoter region, suggesting that mTOR plays a role in IL-7–induced histone acetylation (Fig. 4D). Rapamycin also inhibited the IL-7–induced increase in p300 abundance (Fig. 4E). Furthermore, rapamycin inhibited TH9 cell differentiation in both TH9 cell and IL-7–TH9 cell systems (Fig. 4, F and G), whereas the mTOR activator MHY1485 substantially promoted TH9 cell differentiation (Fig. 4, H and I). Together, these data suggest that the PI3K-AKT-mTOR pathway is important for the differentiation of naïve CD4+ T cells into TH9 cells and that both the STAT5 and PI3K-AKT-mTOR signaling pathways contribute to histone acetylation at the Il9 promoter region.
Foxo1 and Foxp1 play opposing roles in regulating IL-9 production
We next sought to determine which transcriptional factors directly regulated the differentiation of IL-7–TH9 cells in addition to the marked chromatin modification. With the chromatin modification, we hypothesized that the binding of positive transcriptional factors to the Il9 promoter would increase after IL-7 treatment, but IL-7 may decrease the binding of negative regulators to the Il9 promoter. Informatics analysis revealed two adjacent conserved Forkhead box transcription factor–binding sites on the Il9 promoter (Fig. 5A), and we detected both Foxo1 and Foxp1 during TH9 cell differentiation. Both Foxo1 and Foxp1 were found in naïve CD4+ T cells, and they increased in abundance during TH9 cell differentiation (Fig. 5B). A study showed that Foxp1 inhibits Il21 expression by directly binding to the Il21 promoter (26). Because we found that Il21 expression was increased in IL-7–TH9 cells, we wondered whether Foxp1 also regulated Il9 expression. Because Foxo1 is an important downstream target of PI3K-AKT-mTOR signaling and because Foxo1 and Foxp1 compete for binding to Il7r promoter to regulate IL-7 signaling in naïve T cells (27, 28), we hypothesized that Foxo1, Foxp1, or both might regulate Il9 expression. ChIP assays demonstrated that both Foxo1 and Foxp1 could bind to the Il9 promoter in naïve CD4+ T cells (Fig. 5, C and D). We overexpressed Foxo1 or knocked down Foxp1 in preactivated CD4+ T cells under TH9-polarizing conditions. Our results showed that Foxo1 overexpression substantially enhanced TH9 cell differentiation (Fig. 5, E and F), which also occurred as a result of Foxp1 knockdown (Fig. 5, G and H). These results were confirmed in mice in which Foxp1 or Foxo1 was conditionally knocked out in CD4+ T cells. TH9 cell differentiation was decreased as a result of Foxo1 deficiency (Fig. 5, I and J) but was substantially enhanced in Foxp1-deficient CD4+ T cells (Fig. 5, K and L).
Fig. 5. Reciprocal roles for Foxp1 and Foxo1 in TH9 cell differentiation.
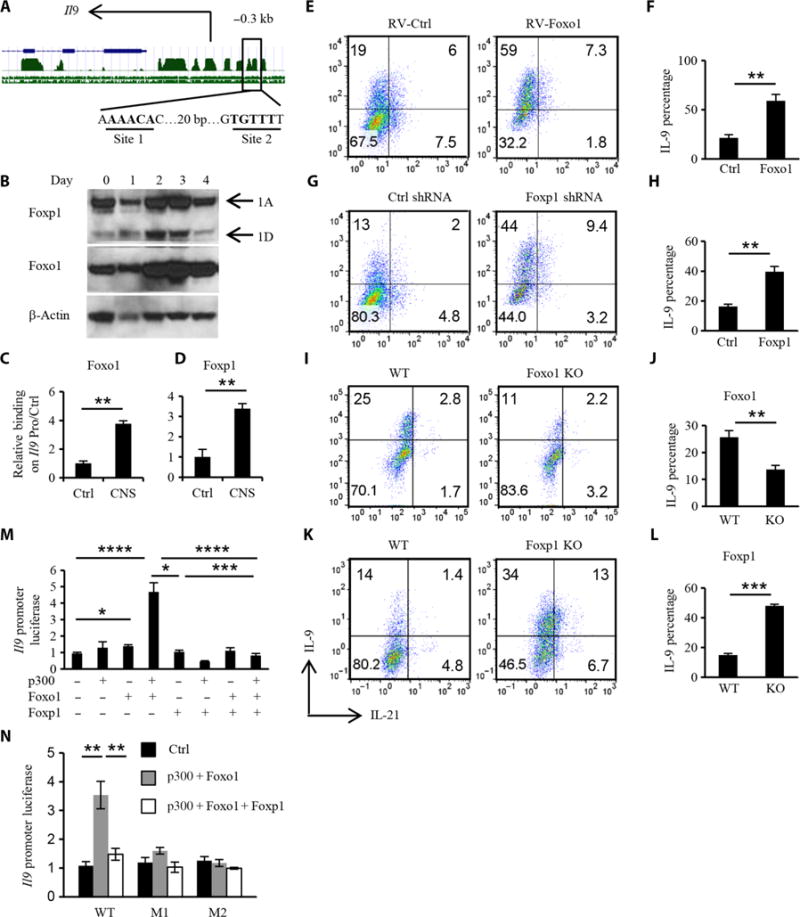
(A) Predicted Forkhead box transcription factor–binding sites on the Il9 promoter. Green peaks represent the conserved regions. The blue line represents the Il9 gene. Site 1 and site 2 are two predicted binding sites for Forkhead box transcription factors. bp, base pair. (B) Western blotting analysis of the abundances of Foxo1 and Foxp1 during TH9 cell differentiation over time. 1A, Foxp1A; 1D, Foxp1D. (C and D) ChIP analysis of the binding of Foxo1 and Foxp1 to the Il9 promoter in naïve OT-II CD4+ T cells. Data were normalized to the Il9 promoter control region. CNS, conserved noncoding sequence. (E and F) Naïve OT-II CD4+ T cells were cultured under the TH9 conditions for 24 hours, infected with control or mFoxo1 retrovirus (RV) in the presence of polybrene (10 μg/ml), and cultured for another 3 days under TH9 conditions. The cultured TH9 cells were then analyzed by flow cytometry to detect IL-9 and IL-21 in gated GFP+ (green fluorescent protein–positive) cells. (G and H) Preactivated CD4+ T cells were infected with control or Foxp1 shRNA (short hairpin RNA) lentivirus and differentiated for 4 days. Differentiated TH9 cells were analyzed by flow cytometry to detect IL-9 and IL-21 in gated GFP+ cells. (I and J) Naïve CD4+ T cells were isolated from tamoxifen-treated wild-type (WT) and FoxoF/F ERT2Cre mice and cultured under TH9 conditions. Total differentiated TH9 cells were analyzed by flow cytometry to detect IL-9 and IL-21. KO, knockout. (K and L) Effect of Foxp1 deficiency on TH9 cell differentiation. WT and Foxp1F/F ERT2Cre mice were treated with tamoxifen, and then isolated naïve CD4+ T cells were cultured under TH9 cell–differentiating conditions before total cells were analyzed by flow cytometry to detect IL-9 and IL-21. (M) Luciferase activity of the Il9 promoter combined with indicated factors to show the synergistic effect of Foxo1 and p300 and the inhibitory effect of Foxp1 on Foxo1-induced Il9 transcription. (N) Mutant Il9 promoters for site 1 (from AAACA to GAGTC, M1) and site 2 (from TGTTT to CCGGC, M2) were synthesized as indicated, and luciferase activity was analyzed to verify the specific binding locus of Foxo1 and Foxp1. Black column, control plasmids; gray column, p300 and Foxo1 plasmids; white column, p300, Foxo1, and Foxp1 plasmids. Data in (B) to (N) are from three independent experiments and are means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further test our hypothesis that Foxo1 and Foxp1 oppositely and competitively regulated Il9 expression in a direct manner, we performed an Il9 promoter luciferase assay. Foxo1 alone only slightly increased Il9 transcription (Fig. 5M); however, together with p300, Foxo1 substantially induced Il9 transcription, which was markedly inhibited by Foxp1 (Fig. 5M). To confirm whether Foxo1 and Foxp1 bound to the predicted sites on the Il9 promoter, promoters with point mutations were synthe-sized and used for luciferase assay. Both site 1 and site 2 were important binding sites for Foxo1 and Foxp1, because mutation of either site abolished the transcriptional activity of Foxo1 (Fig. 5N). Because Foxo family members in T cells include Foxo1, Foxo3, and Foxo4 and they have redundant functions in T cell regulation, we wondered whether Foxo3 and Foxo4 also played a role in TH9 cell differentiation. We found that Foxo4 directly activated Il9 transcription as determined with an Il9 promoter luciferase assay (fig. S7A). In addition, the abundance of Foxo4 increased during TH9 cell differentiation (fig. S7B). Ectopic over-expression of Foxo4 substantially increased TH9 cell differentiation (fig. S7, C and D), whereas Foxo4 deficiency in CD4+ T cells diminished TH9 cell differentiation (fig. S7, E and F). Furthermore, we found that Foxo4 bound to the Il9 promoter and became localized to the nucleus in response to IL-7 (fig. S7, G and H). Thus, these data suggest that Foxo1 and Foxp1 are transcriptional regulators that modulate TH9 cell differentiation in a reciprocal manner.
Foxo1 and Foxp1 have reciprocal roles in the enhancement of IL-7–TH9 cell differentiation and antitumor activity
To verify whether Foxo1 and Foxp1 were also involved in enhanced differentiation of IL-7–TH9 cells, we first examined whether IL-7 affected the binding of Foxo1 or Foxp1 to the Il9 promoter. We found that IL-7 increased the amount of Foxo1 that bound to the Il9 promoter, whereas it increased the dissociation of bound Foxp1 from the Il9 promoter (Fig. 6A). IL-7 treatment did not substantially change the abundances of Foxo1 or Foxp1 mRNAs in TH9 cells (fig. S8A) but slightly increased the abundance of Foxo1 protein (Fig. 6B). Furthermore, the abundance of pFoxo1 was decreased in response to IL-7, suggesting that Foxo1 had increased nuclear localization and transcriptional activity. Confocal microscopy confirmed that IL-7 increased the amount of Foxo1 in the nucleus in CD4+ T cells (Fig. 6C). IL-7 signaling also did not affect the abundance of Foxp1 protein (Fig. 6B), but most of the nuclear Foxp1 protein was exported into the cytoplasm after IL-7 treatment (Fig. 6D). The increased abundance of Foxo1 and the decreased abundance of Foxp1 in the nucleus in response to IL-7 were confirmed by Western blotting analysis of fractioned nuclear and cytoplasmic compartments (Fig. 6E). These data suggest that IL-7 signaling increased the nuclear accumulation of Foxo1 and enhanced the translocation of Foxp1 from the nucleus into the cytoplasm, leading to increased Foxo1 binding to the Il9 promoter by competing with Foxp1.
Fig. 6. Reciprocal roles for Foxo1 and Foxp1 in regulating the differentiation of IL-7–TH9 cells and their antitumor activity.
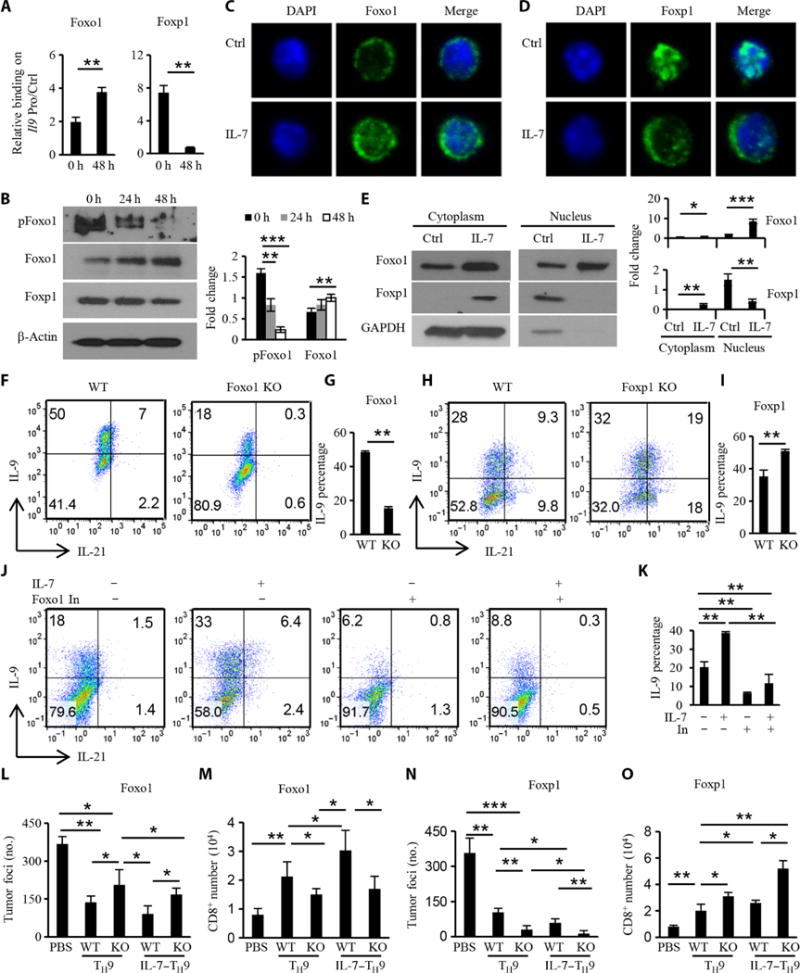
(A) ChIP analysis showing the effect of IL-7 on the binding of Foxp1 and Foxo1 to the Il9 promoter at 48 hours. Data were normalized to the Il9 promoter control region. (B) Western blotting analysis of the abundances of pFoxo1, Foxo1, and Foxp1 after treatment with IL-7 for the indicated times. (C and D) Effect of IL-7 on the localization of Foxo1 (C) or Foxp1 (D) in T cells after 48 hours of treatment. (E) Left: CD4+ T cells with or without IL-7 treatment were fractionated to generate nuclear and cytoplasmic compartments that were then analyzed by Western blotting to detect Foxo1 and Foxp1. Right: Densitometric analysis of the indicated bands. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F to I) Naïve CD4+ T cells that were untreated or were treated with IL-7 for 2 days were then stained with primary antibodies for Foxo1 and Foxp1, which was followed by incubation with FITC (fluorescein isothiocyanate)–labeled secondary antibody and DAPI (4′,6-diamidino-2-phenylindole). WT and Foxo1-knockout (F and G) or WT and Foxp1-knockout (H and I) naïve CD4+ T cells, pretreated with IL-7 for 2 days, were cultured under TH9 conditions for 4 days and then analyzed by flow cytometry. (J and K) Naïve CD4+ T cells or IL-7–pretreated CD4+ T cells were cultured under TH9 conditions with or without a Foxo inhibitor (100 nM) for 4 days and then were analyzed by flow cytometry. (L to O) Effect of Foxo1 (L) or Foxp1 (N) on the IL-7– enhanced antitumor function of TH9 cells. WT and Foxo1F/F OT-II ERT2Cre or WT and Foxp1F/F OT-II ERT2Cre mice were treated with tamoxifen before CD4+ T cells were isolated, left untreated or pretreated with IL-7, and then polarized under TH9 conditions. The antitumor activity of the different TH9 cells was detected in the therapeutic model as shown in Fig. 2H. Mice were sacrificed on day 14 to detect lung tumor foci. (M and O) Numbers of host CD8+ T cells in mouse lungs for the indicated treatments. Data in (A) to (O) are from three independent experiments and, where applicable, are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
To further confirm whether Foxo1 and Foxp1 were involved in the enhanced differentiation of IL-7–TH9 cells, we pretreated Foxo1- and Foxp1-knockout CD4+ T cells with IL-7 and then cultured them under TH9-polarizing conditions. Loss of Foxo1 almost completely abrogated the enhanced differentiation of IL-7–TH9 cells (Fig. 6, F and G), whereas loss of Foxp1 deficiency promoted their differentiation (Fig. 6, H and I). Because the Foxo family members are characterized by functional redundancy, we used a Foxo1 inhibitor, which can also inhibit Foxo3 and Foxo4 to some degree, to address the relative importance of the Foxo proteins in TH9 and IL-7–TH9 cell differentiation. Our results suggest that Foxo inhibition substantially decreased the differentiation of both TH9 and IL-7–TH9 cells (Fig. 6, J and K).
Because we showed that IL-7 increased the binding of Foxo1 but decreased the binding of Foxp1 to the Il9 promoter in naïve CD4+ T cells, we wondered whether Foxo1 and Foxp1 could still regulate Il9 expression during TH9 cell differentiation. We found that the extent of binding of Foxo1 to the Il9 promoter was substantially increased in IL-7–TH9 cells compared to that in TH9 cells (fig. S8B), which was consistent with the amount of IL-9 produced by these cells (Fig. 1A). TH9 and IL-7–TH9 cells did not differ in how much Foxp1 was bound to the Il9 promoter (fig. S8B). Foxo1 behaved similarly in its binding to the Il21 promoter as it did to the Il9 promoter, but Foxp1 did not inhibit Il21 transcription (Fig. 1D) in IL-7–TH9 cells, which was evident by the loss of binding of Foxp1 to the Il21 promoter in IL-7–TH9 cells (fig. S8C) and was consistent with the time course of Il21 expression (Fig. 1D). As expected, Foxo4 behaved similarly to Foxo1 (figs. S7, G and H, and S8).
Because IL-7–mediated enhancement of TH9 cell differentiation was tightly related with Foxo1 and Foxp1, we hypothesized that the enhanced antitumor activity of these T cells might also be Foxo1-dependent, whereas Foxp1 might inhibit the antitumor activity. Using the therapeutic mouse model, we found that Foxo1 knockout TH9 cells had compromised antitumor activity compared to WT TH9 cells (Fig. 6L). Furthermore, loss of Foxo1 also reduced the enhanced antitumor activity of IL-7–TH9 cells. Conversely, Foxp1 deficiency not only enhanced the antitumor activity of TH9 cells but also further increased the antitumor activity of the IL-7–TH9 cells (Fig. 6N). Furthermore, the antitumor activity of TH9 and IL-7–TH9 cells seemed to correlate with the number of tumor-infiltrating host CD8+ T cells in the mice (Fig. 6, M and O).
DISCUSSION
Although TH9 cells are considered to be a distinct TH subset, the transcriptional regulation of IL-9 production and TH9 cell differentiation remains poorly elucidated. Here, we demonstrate that IL-7–pretreated (programmed) CD4+ T cells exhibited enhanced TH9 cell differentiation potential. Mechanistic studies revealed that IL-7–stimulated STAT5 signaling increased the abundance of the acetyltransferase p300 in a direct transcriptional manner. In addition, IL-7–stimulated STAT5 and PI3K-AKT-mTOR signaling were necessary for the differentiation of both TH9 and IL-7–TH9 cells by promoting histone acetylation at the Il9 promoter. At the same time, p300 acted as a coactivator to enable Foxo1 to bind to the Il9 promoter to compete with Foxp1. Foxo1 played a positive role in Il9 transcription, whereas Foxp1 inhibited Il9 transcription, indicating that Foxp1 may serve as a negative regulator of Il9 expression in TH9 cells. The enhanced differentiation and antitumor activity of IL-7–TH9 cells were Foxo1-dependent, suggesting that Foxo1 is critical in TH9 cell differentiation and, consequently, in the antitumor activity of these cells.
TH9 cell differentiation is regulated by multiple transcriptional factors under different situations (29). IL-9 is the hallmark cytokine of TH9 cells, but Il9 transcription is temporary and short, so a negative regulatory mechanism that tightly controls Il9 transcription must exist. One of the factors that inhibit Il9 transcription is Bcl-6; Bcl6 expression can be induced by IL-21 but is suppressed by IL-2 (12, 30). However, there is no evidence that Bcl-6 inhibits Il21 expression, which IL-7–TH9 cells coexpressed with Il9. A study showed that IL-7 inhibits Bcl6 gene expression in activated CD4+ T cells (31), which is consistent with our finding that IL-7 inhibited Bcl6 transcription in naïve CD4+ T cells (fig. S4A). Foxp1 directly binds to the Il21 promoter during TFH cell differentiation (26), which we also confirmed here. Our results showed that Foxp1 directly repressed Il9 transcription and that the abundance of Foxp1 protein was increased in differentiating TH9 cells. Together, our findings suggest that Foxp1 may be a negative regulator of Il9 expression in TH9 cells.
IL-7 could temporarily cause bound Foxp1 to dissociate from the Il9 promoter and translocate from the nucleus into the cytoplasm. However, this cannot be the main mechanism for the enhanced TH9 cell differentiation that is induced by IL-7 pretreatment. We demonstrated the existence and role of Foxo1, a positive regulator of Il9 expression, during the differentiation of TH9 and IL-7–TH9 cells. Because Foxp1 and Foxo1 share the same conserved binding sequence on the Il9 promoter and play a reciprocal role in Il9 transcription, whichever one of them binds to the Il9 promoter site must be tightly regulated, probably through competition based on the relative amounts of these proteins or their modifications. IL-7 did not cause any change in Foxo1 mRNA abundance but increased the total amount of Foxo1 protein and decreased the amount of pFoxo1 in IL-7–treated CD4+ T cells, leading to the accumulation of more Foxo1 protein in the nucleus and its binding to the Il9 promoter, which further resulted in increased IL-9 production and enhanced TH9 cell differentiation. Thus, this study identifies Foxo1 as a positive regulator of Il9 transcription and IL-9 production in TH9 cells.
Histone acetylation is important for activating gene expression. For example, histone acetyltransferases, such as GCN5 and p300, positively regulate Il9 expression, whereas the histone deacetylase SIRT1 inhibits the differentiation of TH9 cells (32–34). A direct link between IL-7 signaling and increased histone modification has been reported for the TRG locus, and the increased histone acetylation favors the accessibility of Rag enzymes (24). Here, we found that IL-7 enhanced the production of p300 and histone acetylation in a STAT5- and PI3K-AKT-mTOR–dependent manner. It is well known that Foxo1 is activated by p300 through a direct interaction and that p300 also stabilizes Foxo1 protein and increases the amount of Foxo1 protein in the nucleus (35, 36). Although it was reported that IL-7 signaling activates the PI3K-AKT pathway, leading to Foxo1 phosphorylation and its translocation from the nucleus to cytoplasm (23), our studies showed that IL-7 reduced the phosphorylation of Foxo1 and increased its nuclear accumulation, which may be caused by the increased amount of p300. Thus, the increase in both the amount and activity of Foxo1 facilitated its binding to the Il9 promoter by competing with Foxp1, which was exported to the cytoplasm, to initiate Il9 transcription. Because T cells also express other Foxo family members, such as Foxo3 and Foxo4, and because the functions of these proteins in T cells are redundant (37), Foxo3 and Foxo4 may also have roles in TH9 cell differentiation. For this reason, we examined the role of Foxo4 and demonstrated that Foxo4 also enhanced TH9 cell differentiation. This finding may explain why TH9 cell differentiation could still be observed, albeit to a lesser extent, in Foxo1-deficient CD4+ T cells.
Forkhead box transcription factors are important in the differentiation of TH cells, including Treg cells, TH1 cells, and TFH cells, and the maintenance of T cell quiescence. Among the Forkhead box family members, Foxo1 and Foxp1 are the molecules most relevant to IL-7 signaling. The Foxo proteins are the major factors downstream of the IL-7-PI3K-AKT signaling pathway. In T cells, IL-7 signaling activates PI3K and AKT, which phosphorylate Foxo1, and pFoxo1 is exported from the nucleus to the cytoplasm to terminate its transcriptional activity. In B cells, however, the IL-7R–PI3K–AKT pathway regulates the activities of Foxo family proteins in an IL-7 concentration–dependent manner (38). These results may indicate that Foxo1 activity is tightly controlled by different mechanisms. Among those, p300 is one of the most important cofactors for Foxo1. Studies have shown that under different conditions, Foxo1 may exhibit totally opposing activities because of differences in its interaction with p300 (35). Foxo1 and Foxp1 are critical negative regulators of the differentiation of TFH cells (26, 39), and Foxo1 is also involved in the negative regulation of TH1 and TH17 cell differentiation and the positive regulation of Treg cell differentiation (40, 41). Our study demonstrates that Foxo1 and Foxp1 are important positive and negative regulators of TH9 cell differentiation and activity.
The production of IL-9 and IL-21 is important for the antitumor functions of TH9 cells (10, 11). We found that IL-7–TH9 cells produced more IL-21 than did TH9 cells. In mouse tumor models, IL-21 is important for the survival and proliferation of cytotoxic CD8+ T cells and natural killer cells (20, 42). Clinical trials using IL-21 or IL-21 combined with chemotherapy or biological agents are ongoing to treat different tumor types and exhibit great therapeutic potential (43). Compared with the systemic administration of cytokines, the advantage of cellular therapy includes the secretion of multiple cytokines and the potential infiltration of the tumor bed by T cells and their cytokines to exert anti-tumor effects. We suggest that IL-7–TH9 cells displayed enhanced anti-tumor activity by secreting large amounts of not only IL-9 but also IL-21 and that they could present an effective approach for the development of improved immunotherapeutics.
MATERIALS AND METHODS
Reagents
The Foxo1 inhibitor AS1842856 and the mTOR activator MHY1485 were purchased from Calbiochem; rapamycin and the p300 inhibitor C646 were purchased from Sigma.
Mice and cell lines
C57BL/6 mice were purchased from the National Cancer Institute, and OT-II (C57BL/6-Tg[TcrαTcrβ]425Cbn/J), Foxo1F/F, Foxo4F/F, CD4Cre, CD4CreERT2, and Il21r−/− mice were purchased from The Jackson Laboratory. Foxp1F/F mice were a gift from H. Hu (Department of Microbiology, University of Alabama at Birmingham). Six- to 8-week-old mice were used at the beginning of each experiment. B16-OVA melanoma cell lines were cultured in Iscove’s modified Dulbecco’s medium (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific), penicillin-streptomycin (100 U/ml), and 2 mM L-glutamine (both from Invitrogen). All experiments complied with protocols approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic.
In vitro differentiation of TH1 and TH9 cells
Naïve T cells purified from the spleens and lymph nodes of WT or Foxo1 and Foxp1 knockout OT-II mice by FACS (fluorescence-activated cell sorting) sorting or kit (STEMCELL Technologies) were differentiated into TH1 or TH9 cells with plate-bound anti-CD3 (2 μg/ml) and soluble anti-CD28 (1 μg/ml) antibodies together with different cytokine combinations: For TH1 cells, IL-2 (30 ng/ml; R&D Systems), IL-12 (4 ng/ml; R&D Systems), and anti–IL-4 mAb (10 μg/ml; eBioscience) were added; for TH9 cells, IL-4 (10 ng/ml; R&D Systems), TGF-β (1 ng/ml; R&D Systems), and anti–IFN-γ mAb (10 μg/ml; eBioscience) were added. After 3 days of culture, the differentiated TH cells were collected, washed, and allowed to continue to differentiate in the presence of the appropriate cytokines for 1 day. For IL-7 treatment, naïve T cells were cultured with IL-7 (PeproTech) for 2 days and then washed twice before being differentiated under TH9-polarizing conditions.
Human CD4+ T cell purification and differentiation
Naïve CD4+ T cells were isolated from peripheral blood mononuclear cells by negative selection with a kit (Miltenyi Biotec) and stimulated with human T-activator CD3/CD28 Dynabeads (Invitrogen) together with human IL-4 (10 ng/ml), human TGF-β1 (1 ng/ml), and anti– IFN-γ (10 μg/ml) for 5 days. Cells were restimulated with PMA and ionomycin for 4 hours with brefeldin A for intracellular staining of IL-9 with PE (phycoerythrin)–conjugated anti-human IL-9 antibody (MH9A4, BioLegend).
Induction of B16 lung melanoma
As was described previously (10), mice were injected intravenously with 2 × 105 B16-OVA cells. For adoptive transfer experiments, mice were injected intravenously with 2 × 106 OVA-specific TH1, TH9, or IL-7–TH9 cells on the same day in the prophylactic model or 4 days after tumor injection in the therapeutic model. At day 14 or 15 after tumor injection, mice were sacrificed for enumeration of metastatic lung foci. All lung lobes were evaluated under a tissue dissect microscope. For some experiments, anti–IL-9 and anti–IL-21R (both from Bio X Cell) and IL-7 antibodies were used as indicated.
Gene microarrays
Total RNA was extracted with TRIzol reagent from naïve CD4+ T cells that had been treated with or without IL-7 or from differentiated TH9 or IL-7–TH9 cells. The microarray was performed with the MouseRef-8 v2.0 Expression BeadChip Kit in the Cleveland Clinic Genomic Core.
Western blotting
Cells were washed with PBS twice and then lysed in 1× lysis buffer (Cell Signaling) with protease cocktail (Roche), which was followed by sonication. Cell lysates were resolved by SDS–polyacrylamide gel electrophoresis gel (Fisher) and then analyzed by Western blotting with the appropriate antibodies (all from Cell Signaling). For the fractionation of cells into nuclear and cytoplasmic fractions, the Cell Fractionation Kit (Cell Signaling) was used according to the manufacturer’s protocol.
Viral transduction
Complementary DNAs (cDNAs) encoding Foxo1 and Foxo4 were synthesized and subcloned into the MigR1 retroviral vector. Foxp1-specific shRNA (GCCCATTTCGTCAGCAGATAT) was synthesized by Sigma and subcloned into the pLKO.1-GFP lentiviral vector. Viruses were packaged in 293T cells transfected with Lipofectamine 2000 (Life Sciences). Virus-containing cell culture medium was harvested from the cells on days 1 to 3, filtered through a 0.45-μm filter, concentrated with PEG-it Virus Precipitation Solution, and stored at −80°C until use. Naïve or activated CD4+ T cells were mixed with virus and protamine sulfate (10 μg/ml; Sigma) in a 24-well plate, followed by centrifugation for 120 min at 500g at 32°C. After 2 hours of incubation, the activated CD4+ T cells were differentiated, as described earlier. Three days later, GFP+ cells were sorted by FACS for Western blotting analysis to check the efficiency of overexpression or knockdown efficiency (fig. S9). Total cells were harvested and analyzed by flow cytometry.
Flow cytometry
Polarized TH cells were stimulated with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (500 ng/ml; Sigma-Aldrich) and blocked by brefeldin A (BioLegend) for 4 hours. Cells were first stained with anti-CD4 antibody and then fixed and stained with the BD Cell Fixation/Permeabilization Kit (for cytokines) or the Foxp3/Transcription Factor Staining Kit (for Foxp3). Cells were analyzed with BD Calibur, BD Fortessa, or Miltenyi MACSQuant systems. Data were analyzed with FlowJo software (TreeStar). The mAbs used for flow cytometry were as follows: FITC-conjugated anti-CD4 (GK1.5, BD Biosciences) and PE-conjugated anti–IL-9 (D9302C12, BD Biosciences), IL-21R–Fc chimera (991-R2-100, R&D Systems), and Alexa Fluor 647–conjugated affinity-purified goat antibody [F(ab)′2 fragment] specific for the Fcγ fragment of human IgG (109-606-008, Jackson ImmunoResearch Laboratories).
Measurement of cytokines
Cells were harvested, counted, and restimulated with anti-CD3 or with PMA and ionomycin for 12 hours. Cell culture medium was collected and analyzed by ELISA to determine the concentrations of mouse IL-9 and IL-21 (all from R&D Systems) according to the manufacturer’s protocol.
ChIP assay
SimpleChIP Plus Enzymatic Chromatin IP Kits were used for ChIP assay according to the manufacturer’s protocol, and DNAs were measured by real-time quantitative PCR. Antibodies used for ChIP assay were against the following targets: IRF4, PU.1, Foxo1, Foxo4, and Foxp1 (all from Cell Signaling). Primers designed for the ChIP assay are as follows: Il9 promoter negative control (forward, 5′-GCCTGCAAGTTT-CTGGACAA-3′; reverse, 5′-GAATATGGGTGGGAGTGGGT-3′); CNS on the Il9 promoter (forward, 5′-AAAGATCTAGCCCCAACCCC-3′; reverse, 5′-TGACCCCTTCATTACCACCC-3′); Il21 promoter control region (forward, 5′-GCAGTAAGGGAAGAAGGTCAAG-3′; reverse, 5′-GGGCTGGATTTGTGGAAAGA-3′); and p300 promoter region (forward, 5′-AGGGATGGATAGAGTCCACAA-3′; reverse, 5′-GCTGCTTTACTCATTGCAGAAG-3′).
Luciferase transactivation assay
Mouse Il9 (from −895 to +5) and Il21 promoters were synthesized and subcloned into the pGL3 basic vector (Promega). 293T cells were transiently transfected for 24 hours with reporter plasmids, Renilla (for internal normalization), and the plasmid pcDNA3.1-Foxo1, pcDNA3.1-Foxp1, or pcDNA3.1-p300 (all from Addgene and subcloned into the pcDNA3.1 plasmid) with Lipofectamine 2000 (Invitrogen). EL4 cells were transfected with p300 reporter and pcDNA3.1-STAT5 plasmids by the Lonza Nucleofector 2b Device. Luciferase was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Real-time quantitative PCR
Total RNA from T cells was extracted with TRIzol, and cDNA was synthesized with M-MLV reverse transcriptase (Invitrogen) and analyzed by real-time quantitative PCR by SYBR Green (Applied Biosystems). The abundances of the mRNAs of interest were normalized to that of the mouse housekeeping gene actb. The primers used were as follows: Il9 (forward, 5′-CTGATGATTGTACCACACCGTGC-3′; reverse, 5′-GCCTTTGCATCTCTGTCTTCTGG-3′); Il21 (forward, 5′-AAGATT-CCTGAGGATCCGAGAAG-3′; reverse, 5′-GCATTCGTGAGCGTCTA-TAGTGTC-3′); Foxo1 (forward, 5′-CCGGAGTTTAACCAGTCCAA-3′; reverse, 5′-TGCTCATAAAGTCGGTGCTG-3′); Foxp1 (forward, 5′-CTGAATCTGGTATCAAGTGTCACCCTCT-3′; reverse, 5′-GATTCGA-GAATGGCCTGCCTGA-3′); Foxo4 (forward, 5′-TGACCAGTGCAGGT-TAGTGC-3′; reverse, 5′-CTGCTACACAGCCTTCCACA-3′); p300 (forward, 5′-TCACCTCCTAGCCGAGAAGA-3′; reverse, 5′-ACGGATCA-TAGACGGGTCAG-3′); GCN5 (forward, 5′-ACTGGTGCCTGAGAAGAG-GA-3′; reverse, 5′-CTCCCATGGAAGGACTGAAG-3′); PCAF (forward, 5′-AGTGCCATGGTTCCTTGTTC-3′; reverse, 5′-CCAAGTGAGAAACGT-GAGCA-3′); Bcl6 (forward, 5′-CACACCCGTCCATCATTGAA-3′; reverse, 5′-TGTCCTCACGGTGCCTTTTT-3′); IRF4 (forward, 5′-ACG-CTGCCCTCTTCAAGGCTT-3′; reverse, 5′-TGGCTCCTCTCGACC-AATTCC-3′); PU.1 (forward, 5′-GGAGAAGCTGATGGCTTGG-3′; reverse, 5′-CAGGCGAATCTTTTTCTTGC-3′); Tbx21 (forward, 5′-CAACA-ACCCCTTTGCCAAAG-3′; reverse, 5′-TCCCCCAAGCAGTTGACAGT-3′); Foxp3 (forward, 5′-CTCGTCTGAAGGCAGAGTCA-3′; reverse, 5′-TG-GCAGAGAGGTATTGAGGG-3′); Gata3 (forward, 5′-AGGGACA-TCCTGCGCGAACTGT-3′; reverse, 5′-CATCTTCCGGTTTCGGGTCTGG-3′); c-maf (forward, 5′-AGCAGTTGGTGACCATGTCG-3′; reverse, 5′-TGGA-GATCTCCTGCTTGAGG-3′); Batf (forward, 5′-GTTCTG-TTTCTCCAGGTCC-3′; reverse, 5′-GAAGAATCGCATCGCTGC-3′); RORgt (forward, 5′-TGAGGCCATTCAGTATGTGG-3′; reverse, 5′-CTTC-CATTGCTCCTGCTTTC-3′); Nfatc1 (forward, 5′-CTTCCAGC-CTGTCTTCTTGG-3′; reverse, 5′-TGCAAACACAAGCTCTGTCC-3′); and actb (forward, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′; reverse, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′).
Immunofluorescence microscopy
Naïve CD4+ T cells that were untreated or treated with IL-7 for 2 days were centrifuged onto glass coverslips, and the cells were then washed with PBS, fixed for 5 min in cold methanol, and permeabilized in 75% ethanol. After rehydration with PBS at 25°C, the coverslips were blocked and stained with primary antibodies for Foxo1 or Foxp1, followed by incubation with rhodamine-labeled secondary antibody and DAPI. Images were acquired with a Leica TCS-SP5II upright confocal microscope (Leica Microsystems).
Statistical analysis
A two-tailed Student’s t test was used for statistical analyses. P < 0.05 was considered to be statistically significant. Results are presented as means ± SD.
Supplementary Material
Fig. S1. Effect of IL-7 on TH9 cell differentiation from untreated mouse CD4+ T cells and pretreated human CD4+ T cells.
Fig. S2. Antitumor effect of IL-7 in the B16 tumor model.
Fig. S3. Effect of IL-7 on the expression of T cell–specific genes involved in transcription, apoptosis, and metabolism.
Fig. S4. Effect of IL-7 on the expression of histone-encoding genes.
Fig. S5. Effect of IL-7 on histone acetylation and binding to the Il21 promoter.
Fig. S6. Effect of IL-7 on the binding of STAT5 to the Il9 promoter in CD4+ T cells and on the activation of STAT5 in TH9 cells.
Fig. S7. Role of Foxo4 in TH9 cell differentiation.
Fig. S8. Effect of IL-7 on gene expression and the DNA binding of Foxo proteins and Foxp1 in TH9 cells.
Fig. S9. Analysis of Foxo1 and Foxo4 overexpression and Foxp1 knockdown.
Acknowledgments
We thank H. Hu (Department of Microbiology, University of Alabama at Birmingham) for providing the Foxp1F/F mice. We thank C. Talerico, a salaried employee of the Cleveland Clinic, for substantive editing and comments.
Funding: This work was supported by grants from the National Cancer Institute (R01 CA163881, R01 CA200539, R01 CA211073, and K99 CA190910), 4R00CA190910-03, the Leukemia and Lymphoma Society (6469-15), and the Multiple Myeloma Research Foundation.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/10/500/eaak9741/DC1)
Author contributions: Q.Y. and E.B. initiated the study, designed the experiments, and wrote the paper. E.B. performed most of the experiments and statistical analyses. X.M. did the Western blotting, confocal microscopy, and luciferase experiments. Y.L. helped with the animal experiments. Y.L. and X.M. read and edited the manuscript. M.Y. assisted in generating the transgenic mice. Q.W., G.X., S.W., and J.Q. provided critical suggestions.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Microarray data accession number is GSE86542.
REFERENCES AND NOTES
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gessner A, Blum H, Röllinghoff M. Differential regulation of IL-9-expression after infection with Leishmania major in susceptible and resistant mice. Immunobiology. 1993;189:419–435. doi: 10.1016/S0171-2985(11)80414-6. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kühn R, Müller W, Palm N, Rüde E. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 4.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho I-C, Khoury S, Oukka M, Kuchroo VK. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3− effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-β ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 7.Yao W, Zhang Y, Jabeen R, Nguyen ET, Wilkes DS, Tepper RS, Kaplan MH, Zhou B. Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity. 2013;38:360–372. doi: 10.1016/j.immuni.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faulkner H, Humphreys N, Renauld JC, van Snick J, Grencis R. Interleukin-9 is involved in host protective immunity to intestinal nematode infection. Eur J Immunol. 1997;27:2536–2540. doi: 10.1002/eji.1830271011. [DOI] [PubMed] [Google Scholar]
- 9.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, Clark RA, Kupper TS. Robust tumor immunity to melanoma mediated by interleukin-9–producing T cells. Nat Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Hong S, Li H, Park J, Hong B, Wang L, Zheng Y, Liu Z, Xu J, He J, Yang J, Qian J, Yi Q. Th9 cells promote antitumor immune responses in vivo. J Clin Invest. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Végran F, Berger H, Boidot R, Mignot G, Bruchard M, Dosset M, Chalmin F, Rébé C, Dérangère V, Ryffel B, Kato M, Prévost-Blondel A, Ghiringhelli F, Apetoh L. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat Immunol. 2014;15:758–766. doi: 10.1038/ni.2925. [DOI] [PubMed] [Google Scholar]
- 12.Liao W, Spolski R, Li P, Du N, West EE, Ren M, Mitra S, Leonard WJ. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc Natl Acad Sci U S A. 2014;111:3508–3513. doi: 10.1073/pnas.1301138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada K, Jameson SC. Naive T cell homeostasis: From awareness of space to a sense of place. Nat Rev Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 14.Boerman OC, Gregorio TA, Grzegorzewski KJ, Faltynek CR, Kenny JJ, Wiltrout RH, Komschlies KL. Recombinant human IL-7 administration in mice affects colony-forming units-spleen and lymphoid precursor cell localization and accelerates engraftment of bone marrow transplants. J Leukoc Biol. 1995;58:151–158. doi: 10.1002/jlb.58.2.151. [DOI] [PubMed] [Google Scholar]
- 15.Chung B, Dudl E, Toyama A, Barsky L, Weinberg KI. Importance of interleukin-7 in the development of experimental graft-versus-host disease. Biol Blood Marrow Transplant. 2008;14:16–27. doi: 10.1016/j.bbmt.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Morrissey PJ, Conlon P, Braddy S, Williams DE, Namen AE, Mochizuki DY. Administration of IL-7 to mice with cyclophosphamide-induced lymphopenia accelerates lymphocyte repopulation. J Immunol. 1991;146:1547–1552. [PubMed] [Google Scholar]
- 17.Tang J-C, Shen G-B, Wang S-M, Wan Y-S, Wei Y-Q. IL-7 inhibits tumor growth by promoting T cell-mediated antitumor immunity in Meth A model. Immunol Lett. 2014;158:159–166. doi: 10.1016/j.imlet.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Pellegrini M, Mak TW. Tumor immune therapy: Lessons from infection and implications for cancer—Can IL-7 help overcome immune inhibitory networks? Eur J Immunol. 2010;40:1852–1861. doi: 10.1002/eji.201040603. [DOI] [PubMed] [Google Scholar]
- 19.Colombetti S, Lévy F, Chapatte L. IL-7 adjuvant treatment enhances long-term tumor-antigen-specific CD8+ T-cell responses after immunization with recombinant lentivector. Blood. 2009;113:6629–6637. doi: 10.1182/blood-2008-05-155309. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Lizée G, Lou Y, Liu C, Overwijk WW, Wang G, Hwu P. IL-21 synergizes with IL-7 to augment expansion and anti-tumor function of cytotoxic T cells. Int Immunol. 2007;19:1213–1221. doi: 10.1093/intimm/dxm093. [DOI] [PubMed] [Google Scholar]
- 21.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 22.Seki Y-i, Yang J, Okamoto M, Tanaka S, Goitsuka R, Farrar MA, Kubo M. IL-7/STAT5 cytokine signaling pathway is essential but insufficient for maintenance of naive CD4 T cell survival in peripheral lymphoid organs. J Immunol. 2007;178:262–270. doi: 10.4049/jimmunol.178.1.262. [DOI] [PubMed] [Google Scholar]
- 23.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200:659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Durum SK, Muegge K. Cutting edge: Histone acetylation and recombination at the TCRg locus follows IL-7 induction. J Immunol. 2001;167:6073–6077. doi: 10.4049/jimmunol.167.11.6073. [DOI] [PubMed] [Google Scholar]
- 25.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, Chang LY, Sprout SL, Becker EK, Willen J, Tian L, Wang X, Xiao C, Jiang P, Crotty S, Victora GD, Showe LC, Tucker HO, Erikson J, Hu H. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15:667–675. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, Willen J, Bunte RM, Maika SD, Harriss JV, Caton AJ, Bhandoola A, Tucker PW, Hu H. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115:510–518. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan MH, Hufford MM, Olson MR. The development and in vivo function of T helper 9 cells. Nat Rev Immunol. 2015;15:295–307. doi: 10.1038/nri3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassil R, Orent W, Olah M, Kurdi AT, Frangieh M, Buttrick T, Khoury SJ, Elyaman W. BCL6 controls Th9 cell development by repressing Il9 transcription. J Immunol. 2014;193:198–207. doi: 10.4049/jimmunol.1303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald PW, Read KA, Baker CE, Anderson AE, Powell MD, Ballesteros-Tato A, Oestreich KJ. IL-7 signalling represses Bcl-6 and the TFH gene program. Nat Commun. 2016;7:10285. doi: 10.1038/ncomms10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goswami R, Kaplan MH. Gcn5 is required for PU.1-dependent IL-9 induction in Th9 cells. J Immunol. 2012;189:3026–3033. doi: 10.4049/jimmunol.1201496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao X, Shi X, Fan Y, Zhang X, Wu M, Lan P, Minze L, Fu YX, Ghobrial RM, Liu W, Li XC. GITR subverts Foxp3+ Tregs to boost Th9 immunity through regulation of histone acetylation. Nat Commun. 2015;6:8266. doi: 10.1038/ncomms9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Bi Y, Chen X, Li C, Li Y, Zhang Z, Wang J, Lu Y, Yu Q, Su H, Yang H, Liu G. Histone deacetylase SIRT1 negatively regulates the differentiation of interleukin-9-producing CD4+ T cells. Immunity. 2016;44:1337–1349. doi: 10.1016/j.immuni.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 35.van der Heide LP, Smidt MP. Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem Sci. 2005;30:81–86. doi: 10.1016/j.tibs.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol Endocrinol. 2005;19:2283–2298. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- 37.Hedrick SM, Michelini R Hess, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, Lin YC, Yang E, Goldrath AW, Li MO, Cantrell DA, Hedrick SM. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 2015;42:239–251. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lainé A, Martin B, Luka M, Mir L, Auffray C, Lucas B, Bismuth G, Charvet C. Foxo1 is a T cell–intrinsic inhibitor of the RORgt-Th17 program. J Immunol. 2015;195:1791–1803. doi: 10.4049/jimmunol.1500849. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, Peng M, Chan P, Ma Q, Mo Y, Meijer D, Zhao K, Rudensky AY, Atwal G, Zhang MQ, Li MO. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markley JC, Sadelain M. IL-7 and IL-21 are superior to IL-2 and IL-15 in promoting human T cell-mediated rejection of systemic lymphoma in immunodeficient mice. Blood. 2010;115:3508–3519. doi: 10.1182/blood-2009-09-241398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spolski R, Leonard WJ. Interleukin-21: A double-edged sword with therapeutic potential. Nat Rev Drug Discov. 2014;13:379–395. doi: 10.1038/nrd4296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Effect of IL-7 on TH9 cell differentiation from untreated mouse CD4+ T cells and pretreated human CD4+ T cells.
Fig. S2. Antitumor effect of IL-7 in the B16 tumor model.
Fig. S3. Effect of IL-7 on the expression of T cell–specific genes involved in transcription, apoptosis, and metabolism.
Fig. S4. Effect of IL-7 on the expression of histone-encoding genes.
Fig. S5. Effect of IL-7 on histone acetylation and binding to the Il21 promoter.
Fig. S6. Effect of IL-7 on the binding of STAT5 to the Il9 promoter in CD4+ T cells and on the activation of STAT5 in TH9 cells.
Fig. S7. Role of Foxo4 in TH9 cell differentiation.
Fig. S8. Effect of IL-7 on gene expression and the DNA binding of Foxo proteins and Foxp1 in TH9 cells.
Fig. S9. Analysis of Foxo1 and Foxo4 overexpression and Foxp1 knockdown.


