Abstract
Cancer stage is one of the most important prognostic parameters in most cancer subtypes. The American Joint Com-mittee on Cancer (AJCC) specifies criteria for staging each cancer type based on tumor characteristics (T), lymph node involvement (N), and tumor metastasis (M) known as TNM staging system. Information related to cancer stage is typically recorded in clinical narrative text notes and other informal means of communication in the Electronic Health Record (EHR). As a result, human chart-abstractors (known as certified tumor registrars) have to search through volu-minous amounts of text to extract accurate stage information and resolve discordance between different data sources. This study proposes novel applications of natural language processing and machine learning to automatically extract and classify TNM stage mentions from records at the Utah Cancer Registry. Our results indicate that TNM stages can be extracted and classified automatically with high accuracy (extraction sensitivity: 95.5%–98.4% and classification sensitivity: 83.5%–87%).
Introduction
Cancer is the second leading cause of death in the United States and recently became the leading cause of death in 21 states, surpassing heart diseases. In the United States, about 595,690 cancer deaths are estimated to have occurred in 2016, which is about 1,630 people per day1. The burden of cancer on public health has mobilized national and international institutions to develop strategies to combat, prevent and control cancer2, 3.
One of the resources vital to the fight against cancer are cancer registries that collect critical information at the popu-lation level. Human abstractors, known as Certified Tumor Registrars (CTRs), are tasked with identifying reportable cancer cases and manually collecting data required for cancer registries. This is often a time-consuming and laborious sprocess that is prone to human error and affects quality, completeness and timeliness of cancer registry data. A study based on surveys conducted across European cancer registries as part of the EUROCOURSE project and covering a population of more than 280 million found that the median time to complete case ascertainment for the relevant year was 18 months, with an additional 3-6 months to publish data to national databases4,5.Though delay in completion is primarily related to clinical processes to evaluate the patients extent of disease, reduced time to ascertainment is very desirable. The Maryland Cancer Registry reported 13% of cases with missing staging information6. A similar study in the Ottawa Regional Cancer Centre found missing staging information in 10% of lymphoma cases and 38% of breast cancer cases7. In a prostate cancer study in Connecticut, about 23% of cases in the registry had incorrectly coded staging information8. A study conducted in Los Angeles County Cancer Surveillance Program (CSP) database found that 77% of cases with testicular cancer were coded with inaccurate stage group9. While there is a range of accuracy of stage determination, an automated or semi-automated process consisting of a systematic review of relevant text information could potentially improve accuracy.
The American Joint Committee on Cancer (AJCC) manual specifies criteria for staging each cancer site depending on primary tumor characteristics (T), number and location of lymph nodes involvement (N), and metastatic nature (M). Information about the cancer stage is critical for assessing prognosis and selection of treatment plans. Clinical guidelines require clinicians to assign TNM stages prior to initiating any treatment10. The clinical TNM stage is determined based on the results of physical exams, imaging (such as x-rays or CT scans), and tumor biopsies. The pathological TNM stage is determined based on surgery to remove a tumor or explore the extent of the cancer.
Natural Language Processing (NLP) coupled with Machine Learning (ML) are promising technologies to increase the efficiency of cancer registry data abstraction processes. NLP and statistical machine learning have been successfully applied in several medical domains to extract various types of information from clinical text. Their potential for increased efficiency and manual process automation have been demonstrated11. In the domain of cancer, several studies showed effectiveness of NLP and ML to mine the electronic health record for cancer-related information from a variety of report types12 and automatically discover reportable cancer cases based on analysis of pathology records13. In the study presented here, we use state-of-the-art Natural Language Processing (NLP) and Machine Learning (ML) to automatically extract TNM stage mentions from patient records collected at the Utah Cancer Registry. The TNM mentions are classified as either pathological or clinical depending on contextual information and will subsequently be used to automatically consolidate stage information and assign a stage group for each cancer case within the registry.
Related Work
Most previous studies have focused on extracting AJCC TNM stage information exclusively from pathology reports14–18. This would not support cancer registry efforts adequately because clinical staging is assigned prior to initiation of treatment and many patients do not immediately undertake resection of tumor and pathology examination. Newer editions of the AJCC TNM manual specifically include separate clinical stage (designated with cT, cN, and cM) and pathological stage (designated with pT, pN, pM) to reflect this time-sensitive staging mechanism. Cancer registries require the use of both clinical and pathological stage information to find the most accurate stage group. The former is primarily based on clinical examination tests and findings (e.g., imaging reports such as CT-scan) and cannot, by definition, be assigned based on information from pathology reports.
McCowan et. al.14 focused their work on extracting T and N stages from lung cancer pathology reports.The pathology reports were first preprocessed to standardize input followed by document-level bag-of-word classifiers to detect relevant reports that contain enough information for the T and N stage classification. They then used a series of rule-based and support vector machine (SVM-based) classifiers at the sentence level to detect phrases with relevant T and N stage information based on factors found in the TNM stage guidelines such as tumor dimension and lymph node involvement. The highest T and N stages detected by the sentence-level classifiers were assigned to each patient. Their approach achieved accuracies of 74.3% and 86.6% for T and N stage classification respectively when trained on a dataset of 710 cases, and evaluated on a held-out dataset of 179 cases. The authors used the manually-assigned TNM pathologic stages as gold standard to measure accuracy of their system. Since the approach to perform stage classification heavily relies on factors associated with lung cancer that were obtained using expert manual annotations, it would be difficult to generalize to other cancer sites without re-training the whole system on new annotations. Nguyen et al.15 used a similar lung cancer dataset and replaced the machine learning component of McCowan et. al. with a rule-based dictionary component. Their approach eliminated the need for expert manual annotations, and achieved comparable accuracies of 73% and 79% for T and N stages respectively.
Martinez et al.16 experimented with various machine learning approaches to identify TNM stages for colorectal cancer in reports obtained from the Royal Melbourne Hospital in Australia. A notable aspect of their experiment was assessment of the generalizability of their methods when using a colorectal cancer dataset from a different institution. Their results showed that accuracy dropped significantly from above 80% down to 50% to slightly above 60% when training and testing on the same corpus versus using corpora from different institutions (cross-corpora) for training and testing. They attribute this drop mainly to differences between the two corpora in expressing TNM labels (e.g., T1 for staging T). Based on feature selection analysis performed by the authors, these explicit TNM labels are among the top features for good performance within the same corpus and differences across corpora may lead to inconsistent predictions that could introduce many errors and hamper good performance.
Kim et al.17 focused on extracting TNM stage information from pathology notes of prostate cancer patients. Using a set of 100 radical prostatectomy specimen reports, they first created a gold standard using two blinded manual reviewers. They then used an NLP system developed to directly match TNM mentions like pT2 and achieved very high accuracies of 99%, 95% and 100% for T, N and M stages respectively. It is worth mentioning, however, that for the M stage, the dataset was highly skewed with all 100 cases from the randomly selected sample staged as MX.
Warner et al.18 implemented an NLP system that searched and directly found relevant phrases for summary stage information (i.e., stage I, stage II, early stage, etc.) from the entire EHR available at their institution. They successfully achieved high accuracy (Cohen’s kappa of 0.906) when comparing with stages manually determined at the cancer registry, and using a set of 2,323 cancer cases with about 751,880 documents.
Based on the prior scientific work cited above, we used a hybrid approach combining pattern-matching for extraction and supervised machine learning for classification of TNM stage mentions as either pathological or clinical. To the best of our knowledge, our study is the first to report about the extraction of TNM staging information from unstructured text found in records collected at the central Utah Cancer Registry (UCR).
Methods
Utah Cancer Registry Data
This study is based on data collected at the UCR which instructs and oversees more than 70 cancer facilities in the state of Utah. Each cancer treatment facility sends records abstracts containing the required data elements for a given cancer patient electronically to the Utah Cancer Registry for each newly diagnosed cancer case. Two types of reports were used for this study. The first type is the North American Association of Central Cancer Registries (NAACCR) abstract record19,20 which contains coded information required for reporting to national cancer databases such as the patient age, date of diagnosis, cancer tumor histology, and grade. NAACCR abstracts also contain unstructured text fields that include information such as the patient clinical history, clinical exam results, imaging study descriptions, and any potential staging information. The other type of record is the electronic surgical pathology report also known as E-path. Its content consist of mostly unstructured text fields about the tumor gross pathology, histology, and final diagnoses. Since a patient can be seen in multiple different facilities within a state or have multiple visits within the same facility, there are usually multiple NAACCR and E-path records available for each patient at the Utah Cancer Registry. We refer to these reports as unconsolidated records in this study. The role of registrars at the Utah Cancer Registry is to consolidate all information received by the registry for a given cancer case and to produce one final consolidated abstract that captures the most accurate information for final reporting to national authorities.
Reference Standard
For development and evaluation of our system, a random subset of 100 cancer cases was selected from three different cancer primary sites (300 cases total): Colon, Lung and Prostate cancers. These three primary sites are among the most prevalent cancer types at the UCR and could therefore benefit the most from case review and consolidation automation. The text fields from NAACCR (see appendix for details) and e-path records for these 300 cases constituted the corpus for this study (see Table 1). Note that since each case may have multiple records (3 on average), the corpus contains far more documents than selected cancer cases. In our case, the corpus consisted of 810 NAACCR and e-path records as shown in Table 1. All text fields in these records were manually annotated for mentions of TNM staging information. Two human annotators who are certified tumor registrars conducted the annotation independently, and a third domain expert participated in the process for adjudication of differences between annotators when necessary. The annotation task was initiated by going through preliminary practice rounds in which annotators were given the same set of 25 documents to annotate followed by team meetings where agreement was discussed, and annotation guidelines revised to clarify ambiguous examples found during preceding practice sessions. Once an adequate level of agreement (K = 0.81) was observed and good understanding of the annotation task was achieved, we started the quality control phase in which a small subset (QCSET) of 20 cases from each cancer site (60 cases total) from the reference standard was selected for double-annotation. The inter-annotator agreement was calculated using Cohen’s kappa statistic and results are shown in Table 3. Disagreements in the QCSET were mostly due to either a missed TNM value mention by one annotator or discrepancies in the timing attribute. Given the excellent inter-annotator agreement observed, the remaining documents in our corpus (ABSTRACTION) were each annotated by one annotator only. The annotation project and reference standard development was managed using the WebAnno tool21.
Table 1:
Document types and counts for the corpus used in this study.
| Record Type | QCSET (n=60) | ABSTRACTION (n=240) | TOTAL (n=300) |
|---|---|---|---|
| NAACCR | 72 | 286 | 358 |
| E-path | 113 | 339 | 452 |
| TOTAL | 185 | 625 | 810 |
Table 3:
Inter-annotator agreement by document type in the QCSET. Method is Cohens Kappa for 2 raters.
| Document Type | Mentions annotated by both raters | Kappa | p-value |
|---|---|---|---|
| e-path | 60 | 0.658 | < 0:001 |
| NAACCR abstract | 125 | 0.9009 | < 0:001 |
| All | 185 | 0.8129 | < 0:001 |
The annotation schema included three categories of information to be annotated corresponding to T, N and M stage mentions. Each TNM stage mention was annotated with the following attributes:
Stage: The AJCC stage designation (e.g., T1, N1b).
Timing: This attribute is used to indicate if the staging is clinical or pathological as per the rules of the AJCC manual and according to the context of the mention.
Negation: The value to indicate if a TNM mention is within the scope of a negated context. The possible values are: affirmed (default, or most mentions), negated, and possible. Most mentions will have an affirmed value. The possible was selected when there was hedging involved within the context of mention.
Temporality: This attribute is used to capture historical or future mentions that do not necessarily represent current mentions valid at the point in time when the mention was stated at the patient record. The three possible values are: current (default, or most mentions), historical, and hypothetical (future mentions).
Subject: This is used to capture TNM mentions that are related to family relatives or others who are not the patient himself. Possible values include: patient (default), and other.
After completion of the reference standard development, we found that almost all TNM mentions were affirmed, recent and related to the patient (only 1 was negated, 3 were historical and 2 were related to someone other than the patient). Therefore, we decided to exclude negation, temporality and subject attributes from further training and analysis. Table 2 presents TNM annotations added by annotators in the reference standard. The reference standard was divided randomly into 3 subsets: Train (50%), Development (17%) and Test (33%).
Table 2:
TNM mentions extracted by annotators from corpus used as reference standard.
| Data Subset | Count of NAACCR and e-path Records | T | N | M | Total TNM annotations |
|---|---|---|---|---|---|
| Train (50%) | 405 | 235 | 192 | 86 | 513 |
| Development (17%) | 135 | 85 | 73 | 27 | 185 |
| Test (33%) | 270 | 139 | 119 | 52 | 310 |
| All | 810 | 459 | 384 | 165 | 1008 |
NLP and ML Systems
Figure 1 shows the complete NLP and ML system developed for this study. The pre-processing components were adapted from cTAKES22, a general clinical NLP application. Each text field is broken into smaller units (sentences and tokens such as words, numbers, and punctuation) and assigned parts-of-speech tags by pre-processing modules for further analysis. Both rule-based (pattern-matching) and ML-based (Conditional Random Fields) approaches were utilized for TNM mentions extraction and classification. The pattern matching component was developed using regular expressions based on human annotations from the training data to achieve high sensitivity. The Conditional Random Fields (CRF) component was developed using the CRFsuite package within ClearTK machine learning libraries. The Java-based Apache UIMA framework was used as the main development framework for this project.
Figure 1:
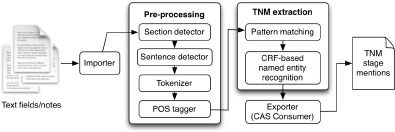
NLP and ML application high-level architecture.
The development of NLP included the following system variants:
- REGEX: based on direct pattern matching using regular expressions for TNM mentions, and rules for classifying timing attribute, as follows,
- If TNM mention has prefix letter ‘c’ then clinical else if it has prefix letter ‘p’ then pathological.
- If TNM mention has no ‘c’ or ‘p’ prefixes and TNM mention was extracted from pathology record then pathological, otherwise if extracted from NAACCR abstract then clinical.
CRF: based on a Conditional Random Fields (CRF) machine learning algorithm.
REGEX-CRF: hybrid system that combined regular expression output with CRF algorithm classification of timing attribute.
Results
Tables below detail the results of our evaluation of the NLP system variants. Reported metrics include: precision (equivalent to positive predictive value), recall (sensitivity) and the Fl-measure (harmonic mean of precision and recall). We report evaluations with both subsets: development and test. Results with the development subset were obtained by training with the train subset only while results with the test subset were obtained after training on both the train and development subsets. We used two evaluation approaches to compare the NLP system output with our reference standard: strict and partial matches. The former requires exact matching of the TNM mention sequence of characters predicted by the NLP system with the corresponding mention in the reference standard. The latter relaxes this restriction by allowing overlapping TNM mentions to be counted as true positives. For instance, if the NLP system extracted the mention “T1” and the reference standard had “cT1” then this would count as a true positive partial match. Table 4 shows the results of TNM mentions extraction. Table 5 shows the results of classifying the TNM mentions to pathological and clinical. Note the REGEX-CRF system uses REGEX to extract TNM mentions and CRF to classify them to pathological and clinical mentions.
Table 4:
TNM mentions extraction results.
| Evaluation Method | System | Development Set | Test Set | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-measure | Precision | Recall | F1-measure | ||
| Strict match | REGEX | 0.926 | 0.946 | 0.936 | 0.890 | 0.884 | 0.887 |
| CRF | 0.952 | 0.859 | 0.903 | 0.923 | 0.845 | 0.882 | |
| Partial match | REGEX | 0.958 | 0.984 | 0.971 | 0.961 | 0.955 | 0.958 |
| CRF | 0.988 | 0.897 | 0.940 | 0.989 | 0.906 | 0.946 | |
Table 5:
Pathological and clinical TNM classification results.
| Evaluation Set | System | Pathological | Clinical | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prec. | Recall | F1 | Prec. | Recall | F1 | Prec. | Recall | F1 | ||
| Development | REGEX | 0.952 | 0.688 | 0.798 | 1.000 | 0.025 | 0.049 | 0.529 | 0.543 | 0.536 |
| REGEX-CRF | 0.901 | 0.889 | 0.895 | 0.681 | 0.800 | 0.736 | 0.847 | 0.870 | 0.858 | |
| Test | REGEX | 0.934 | 0.536 | 0.681 | 1.000 | 0.051 | 0.097 | 0.386 | 0.384 | 0.385 |
| REGEX-CRF | 0.859 | 0.896 | 0.877 | 0.793 | 0.704 | 0.746 | 0.841 | 0.835 | 0.838 | |
Both the REGEX and CRF versions of the NLP system achieve comparable performance when detecting mentions of TNM staging (F1: 94.0%-97.1%), but the CRF version reached much higher accuracy when predicting the timing (clinical or pathological) attribute (F1: 83.8%-85.8%). In general, the REGEX (rule-based) version retrieved slightly more correct TNM mentions (i.e., had higher recall: 88.4%-98.4%) while the CRF version retained a higher precision (Precision: 92.3%-98.9%). The CRF version also performed better than the REGEX version when classifying the timing attribute. Our analysis of partial matching results indicate that the NLP system tends to miss words like “clinical” and “PATH” preceding TNM mentions as included by our annotators in the reference standard. This is despite the fact that our annotation guidelines did not in particular include specific instructions to the annotators on selecting contextual words preceding TNM mentions as in these cases. The NLP system was able to partially capture these longer multi-word TNM mentions found in the reference standard. The REGEX NLP system achieved very high sensitivity with partial matching and hence indicate that this approach is practical for extracting TNM mentions from NAACCR and E-path records. The hybrid REGEX-CRF NLP system combined the output of the REGEX version with the classification of timing by the CRF module, and achieved the highest F1-measure overall (F1: 83.8%-85.8%).
Discussion
Our effort outlined above showed that extraction of TNM mentions from unstructured text fields within records collected at the central Utah Cancer Registry can be automated with reasonable accuracy. One source of errors that was eliminated earlier during our development was matching of ‘TX’ mentions by the REGEX NLP system. Despite slight increase in sensitivity, there were numerous false positives and a decrease in precision when including this pattern since it could be confused with the commonly used ‘TX’ abbreviation for ‘treatment’ in this corpus (see Table 6, for examples). For this reason, we decided to exclude the pattern for ‘TX,’ especially since patient cases with no T stage mentions extracted from their records can be assigned ‘TX’ stage by default. Most strict matching errors were caused by missing contextual words preceding TNM mentions such as “CLINICAL:” and “PATHOLOGIC”. When considering partial matches, many errors were caused by spurious matching within alpha-numerical terms such as matching ‘T0’ or ‘N1’ in “T0012-9071” and “N13-129”. Other errors were due to confusion of text mentions related to MRI scans such as ‘T2’ inside the statement: “SUBTLE AREA OF FOCAL T2 SIGNAL LOSS”. Similarly, incorrectly matching ‘T1’ within a biomarker phrase as in the sentence:“weakly positive for WT1”. A third source of errors was the use of capital letter ‘O’ instead of the digit ‘0’ in some TNM mentions such as “NO” and “MO” instead of “N0” and “M0” stages, respectively. These errors could be addressed by including more contextual, lexical and character shape features to enable disambiguation and improve sensitivity while maintaining high precision.
Table 6:
Example statements containing the ‘TX’ abbreviation.
| Statements with TX abbreviations |
|---|
| DISCUSSED PALLIATIVE TX W/ CARBO/TAXL … |
| NEW LUNG CANCER F/U & TX … |
Although regular expressions were more robust for extracting TNM mentions, our range of features used with the CRF classifier were still limited and potential improvements may be observed if other more sophisticated feature patterns were used such as character N-grams. In addition, other machine learning algorithms could yield better performance than CRF and further investigation is required.
Results reported here were validated with patient records from various healthcare organizations (local and regional hospitals) in the state of Utah. We believe that despite potential differences in documentation style and use of linguistic patterns, the proposed NLP and ML systems were able to extract TNM mentions with high accuracy comparable to manual abstraction by humans. To investigate questions about the distribution of TNM mentions extracted across sites, the number of TNM mentions found for each patient case, and the percentage of patients without any TNM mentions in their records, we applied the REGEX NLP system to a selected set of 11,180 NAACCR and e-path records for a population of 4,117 patient cases available from the UCR database. Table 7 outlines the number of TNM mentions extracted across cancer sites. There were 14,560 mentions extracted in total from all cases records. In general, more patients had no M stage mentioned in their records, followed by N stage mentions, and finally T stage mentions. Across the three primary cancer sites, colon cases tended to have more TNM mentions in their records than prostate or lung cancer cases.
Table 7:
Count of TNM mentions extracted from a selected set of cases
| Site | TNM | count |
|---|---|---|
| Colon | T | 2814 |
| N | 2635 | |
| M | 1012 | |
| Lung | T | 1615 |
| N | 1407 | |
| M | 634 | |
| Prostate | T | 2341 |
| N | 1409 | |
| M | 693 | |
| Total | 14560 |
The number of TNM mentions extracted from patient records was distributed as shown in Figure 2. On average, there were about 5 mentions extracted per patient. The distribution is right skewed with a majority of patients having less than 10 mentions and then gradually fewer patients having more than 10. At the extreme right were patients with more than 30 TNM mentions. Note that this average excludes patients who have no TNM stage mentions in their records. Only patients with TNM stage mentions extracted from their records were considered.
Figure 2:
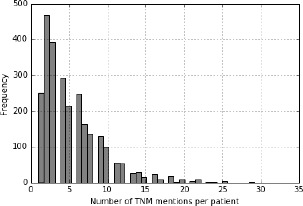
Frequency of TNM stage mentions extracted per patient.
There were about 6,485 records (out of a total of 11,180) with no TNM mentions, belonging to 1,443 patient cases (out of 4,117). When considering each T, N and M mention individually, about 37% of the patients had no T stage mentions (1558/4117), 44% had no N stage mentions and 63.5% had no M stage mentions.
Conclusion
The study presented here showed that automated extraction of TNM stage information using NLP and ML approaches could achieve high accuracy, at levels comparable with manual abstraction by humans. In a future study, we plan to use the NLP pipeline developed for TNM stage information extraction to then perform cancer stage consolidation at the patient level for cases from the three primary cancer sites included in this study. The automated stage consolidation will be compared with the consolidated stages assigned by human registrars manually in the central registry. Our aim is to eventually assess whether NLP and machine learning could be implemented with sufficient accuracy to automatically consolidate cancer stage and support the work of cancer registrars.
Acknowledgement
The authors would like to thank SuAnn McFadden, Kay McCandless and Jacque Clarken at the Utah Cancer Registry for spending the time to participate in the annotation effort and creation of the reference standard. This work has been supported by contract number HHSN261201300017I from the National Cancer Institute Surveillance, Epidemiology, and End Results (SEER) program.
A. NAACCR Text Fields
The table below lists the NAACCR column names and numbers for the free text fields used in the study.
Table 8.
| NAACCR Item Number # | Text Field Name |
|---|---|
| 2520 | Text–Dx Proc–PE |
| 2530 | Text–DX Proc–X-ray/scan |
| 2540 | Text–DX Proc–Scopes |
| 2550 | Text–DX Proc–Lab Tests |
| 2560 | Text–DX Proc–Op |
| 2570 | Text–DX Proc–Path |
| 2580 | Text–Primary Site Title |
| 2590 | Text- Histology Title |
| 2600 | Text–Staging |
| 2610 | RX Text–Surgery |
| 2620 | RX Text–Radiation (Beam) |
| 2630 | RX Text–Radiation Other |
| 2640 | RX Text–Chemo |
| 2650 | RX Text–Hormone |
| 2660 | RX Text–BRM |
| 2670 | RX Text–Other |
| 2680 | RX Text–Remarks |
| 2690 | Text–Place of Diagnosis |
References
- 1.Rebecca L Siegel, Kimberly D Miller, Ahmedin Jemal. Cancer statistics. CA: a cancer journal for clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.BWKP Stewart, Christopher P Wild, et al. World cancer report 2014. Health. 2017 [Google Scholar]
- 3.Donald M Parkin. The evolution of the population-based cancer registry. Nature reviews. Cancer. 2006;6(8):603. doi: 10.1038/nrc1948. [DOI] [PubMed] [Google Scholar]
- 4.Jan Willem Coebergh, Corina van den Hurk, Stefano Rosso, Harry Comber, Hans Storm, Roberto Zanetti, Lidia Sacchetto, Maryska Janssen-Heijnen, Melissa Thong, Sabine Siesling, et al. Eurocourse lessons learned from and for population-based cancer registries in europe and their programme owners: improving performance by research programming for public health and clinical evaluation. European Journal of Cancer. 2015;51(9):997–1017. doi: 10.1016/j.ejca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 5.R Zanetti, I Schmidtmann, L Sacchetto, F Binder-Foucard, A Bordoni, D Coza, S Ferretti, J Galceran, A Gavin, N Larranaga, et al. Completeness and timeliness: cancer registries could/should improve their performance. European journal of cancer. 2015;51(9):1091–1098. doi: 10.1016/j.ejca.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Ann C Klassen, Frank Curriero, Martin Kulldorff, Anthony J Alberg, Elizabeth A Platz, Stacey T Neloms. Missing stage and grade in maryland prostate cancer surveillance data, 1992-1997. American journal of preventive medicine. 2006;30(2):S77–S87. doi: 10.1016/j.amepre.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Jonathan C Yau, Arlene Chan, Tamina Eapen, Keith Oirourke, Libni Eapen. Accuracy of the oncology patients information system in a regional cancer centre. Oncology reports. 2002;9(1):167–169. [PubMed] [Google Scholar]
- 8.Wen-Liang Liu, Stanislav Kasl, John T Flannery, Alric Lindo, Robert Dubrow. The accuracy of prostate cancer staging in a population-based tumor registry and its impact on the black-white stage difference (connecticut, united states) Cancer Causes and Control. 1995;6(5):425–430. doi: 10.1007/BF00052182. [DOI] [PubMed] [Google Scholar]
- 9.Kenneth D Faber, Victoria K Cortessis, Siamak Daneshmand. Urologic Oncology: Seminars and Original Investigations. Vol. 32. Elsevier; 2014. Validation of surveillance, epidemiology, and end results tnm staging for testicular germ cell tumor; pp. 1341–1346. [DOI] [PubMed] [Google Scholar]
- 10.Stephen B Edge, Carolyn C Compton. The american joint committee on cancer: the 7th edition of the ajcc cancer staging manual and the future of tnm. Annals of surgical oncology. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 11.Stephane M Meystre, Guergana K Savova, Karin C Kipper-Schuler, John F Hurdle, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;35(128):44. [PubMed] [Google Scholar]
- 12.Irena Spasic, Jacqueline Livsey, John A Keane, Goran Nenadic. Text mining of cancer-related information: review of current status and future directions. International journal of medical informatics. 2014;83(9):605–623. doi: 10.1016/j.ijmedinf.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 13.David A Hanauer, Gretchen Miela, Arul M Chinnaiyan, Alfred E Chang, Douglas W Blayney. The registry case finding engine: an automated tool to identify cancer cases from unstructured, free-text pathology reports and clinical notes. Journal of the American College of Surgeons. 2007;205(5):690–697. doi: 10.1016/j.jamcollsurg.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Iain A McCowan, Darren C Moore, Anthony N Nguyen, Rayleen V Bowman, Belinda E Clarke, Edwina E Duhig, Mary-Jane Fry. Collection of cancer stage data by classifying free-text medical reports. Journal of the American Medical Informatics Association. 2007;14(6):736–745. doi: 10.1197/jamia.M2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthony N Nguyen, Michael J Lawley, David P Hansen, Rayleen V Bowman, Belinda E Clarke, Edwina E Duhig, Shoni Colquist. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. Journal of the American Medical Informatics Association. 2010;17(4):440–445. doi: 10.1136/jamia.2010.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David Martinez, Lawrence Cavedon, Graham Pitson. Stability of text mining techniques for identifying cancer staging. Louhi; The 4th International Workshop on Health Document Text Mining and Information Analysis; NICTA, Canberra, Australia. 2013. [Google Scholar]
- 17.Brian J Kim, Madhur Merchant, Chengyi Zheng, Anil A Thomas, Richard Contreras, Steven J Jacobsen, Gary W Chien. Second prize: A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports. Journal of endourology. 2014;28(12):1474–1478. doi: 10.1089/end.2014.0221. [DOI] [PubMed] [Google Scholar]
- 18.Jeremy L Warner, Mia A Levy, Michael N Neuss, Jeremy L Warner, Mia A Levy, Michael N Neuss. Recap: Feasibility and accuracy of extracting cancer stage information from narrative electronic health record data. Journal of oncology practice. 2015;12(2):157–158. doi: 10.1200/JOP.2015.004622. [DOI] [PubMed] [Google Scholar]
- 19.A Stewart, A Hurlbut, LA Havener, F Michaud, S Capron, L Ries, et al. North american association of central cancer registries naaccr 2006 implementation guidelines and recommendations. D isponibleen. 2012. http://www.naaccr.org/SearchResults.aspx.
- 20.M Thornton, L OConnor. Springfield, IL: North American Association of Central Cancer Registries; 2012. Standards for cancer registries volume ii: Data standards and data dictionary, record layout version 12.2. [Google Scholar]
- 21.Seid Muhie Yimam, Iryna Gurevych, Richard Eckart de Castilho, Chris Biemann. Webanno: A flexible, web-based and visually supported system for distributed annotations. In ACL (Conference System Demonstrations) 2013:1–6. [Google Scholar]
- 22.Guergana K Savova, James J Masanz, Philip V Ogren, Jiaping Zheng, Sunghwan Sohn, Karin C Kipper-Schuler, Christopher G Chute. Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]


