Abstract
Crowdsourcing services like Amazon Mechanical Turk allow researchers to ask questions to crowds of workers and quickly receive high quality labeled responses. However, crowds drawn from the general public are not suitable for labeling sensitive and complex data sets, such as medical records, due to various concerns. Major challenges in building and deploying a crowdsourcing system for medical data include, but are not limited to: managing access rights to sensitive data and ensuring data privacy controls are enforced; identifying workers with the necessary expertise to analyze complex information; and efficiently retrieving relevant information in massive data sets. In this paper, we introduce a crowdsourcing framework to support the annotation of medical data sets. We further demonstrate a workflow for crowdsourcing clinical chart reviews including (1) the design and decomposition of research questions; (2) the architecture for storing and displaying sensitive data; and (3) the development of tools to support crowd workers in quickly analyzing information from complex data sets.
Introduction
Crowdsourcing has gained notoriety as services like Amazon Mechanical Turk (AMT)1–4 have enabled researchers to ask questions to crowds of workers and quickly receive labeled responses. These human labeled data sets are increasingly important for training supervised machine models, as labels do not exist for many important research questions and cannot be produced with automated methods. Unfortunately, crowds composed of individuals from the general public are inappropriate for numerous types of data sets that require crowdsourcing, such as clinical data, due to legislation (e.g., the Health Insurance Portability and Accountability Act of 1996) and organizational policies. In particular, privacy concerns prevent arbitrary users from accessing these data. Moreover, the subject matter being analyzed requires highly specialized training and expertise to accurately produce a label, which is often not available in a public crowd.
This paper outlines a crowdsourcing framework for medical data sets and one current deployment of the system. There are many components necessary for building such an environment to allow for scalable human computation on medical data sets. Broadly, the main components of the system include: (i) a crowdsourcing system that can be deployed within an organization that has the ability to specify workers’ attributes, roles and access controls, (ii) deidentification routines to perturb identifiers and meet ethical and legal requirements, (iii) graphical user interfaces to display sensitive data, (iv) and machine learning tools to assist workers to produce labels quickly. Moreover, beyond the technical components, this paper describes organizational processes that are needed to train researchers about crowdsourcing so they can construct well-defined questions for the crowd, and approaches to recruit skilled workers.
To demonstrate the challenges and complexities of developing and deploying a crowdsourcing system for sensitive data, this paper focuses on the important use case of clinical chart reviews5. Chart reviews are a common component of medical research in which medical students, staff or nurses comb through semi-structured electronic medical records (EMR) systems (which are designed for clinical treatment rather than research) for specific data. Unfortunately, scrolling through vast amounts of clinical text to produce labels is time-consuming and expensive. For example, at Vanderbilt University Medical Center, it currently costs $109 per hour for a service which pays a nurse to review patient charts and produce labels, where a large part of this fee goes to project management and other overhead. While some researchers have employed software scripts to infer labels from text data automatically6, the messiness and complexity of EMR systems’ semi-structured data7 make verifying the accuracy of the results difficult. More problematic is that the clinical text is filled with misspellings, medical acronyms, and abbreviations which make disambiguation difficult with natural language processing techniques8.
One major challenge for crowdsourcing workers is uncovering relevant information quickly from complex data sets. For example, in healthcare, patient charts are managed as a collection hundreds, if not thousands, of clinical documents, each of which may include tens of pages of information. Finding the specific paragraph related to a patient’s diabetes care history or cancer medication adherence is nontrivial. While keyword search can help find some content, variations in terminology and other clinical semantics make finding all relevant data challenging9. Moreover, identifying all relevant text in a single note related to, say, seizures remains time-consuming and requires extensive skimming. For these reasons, the crowdsourcing framework requires additional tools to assist workers in finding relevant content quickly, such as text highlighting and data visualization for summarization.
An effective crowdsourcing system for medical data sets can change how medical research is done and allow researchers to solve important problems. In our experience, the chart review process is often a key rate limiting step for modern studies; crowdsourcing has the ability to substantially lower the time to complete clinical studies. Additionally, the resulting labels are invaluable resources for supervised machine learning researchers that otherwise would be limited by smaller training data sets.
Consequently, as displayed in Figure 1, our goal is to build a lightweight, customizable pipeline that significantly reduces the cost and time to complete medical research while increasing reproducibility and accuracy, and maintaining privacy and security standards.
Figure 1:
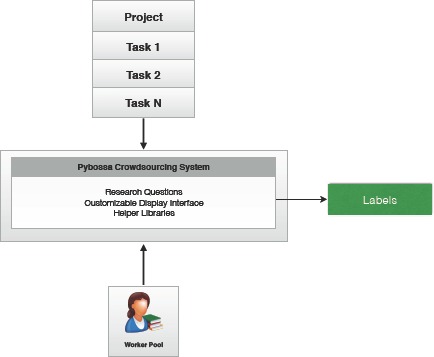
Overview of the crowdsourcing system.
2. Crowdsourcing Framework
This section introduces the main components of the crowdsourcing system, defines the workflow in which researchers develop crowdsourcing questions, and describes the process workers follow to produce labels.
-
1.
Questions Design: After recruiting and identifying a researcher with an amenable research project, we conducted a design workshop. The workshop included the researcher, medical personnel, computer science researchers and anthropologists. The workshop began by introducing the researcher to crowdsourcing preliminaries and non-healthcare crowdsourcing examples. Next, the team worked to clarify and decompose the research objective into atomic questions by refining the structure of the crowdsourcing project as in Figure 2. We discussed data needs (e.g., all notes or specific note types), question format (e.g., boolean, multiple choice or text snipping), scope of tasks (e.g., multiple questions per patient or a single one), and worker skills requirements for the tasks.
Crowdsourcing questions were constructed as narrowly as possible. For example:
-
(a)
Does a clinical note document patient conversations regarding diabetic diet alternatives? (Y/N)
-
(b)
Which of the following dietary alternatives were discussed with the patient? (Healthy oil choices, Sugar free sweets, Unsweetened tea, None)
-
(c)
Of the patient’s current diet choices listed in this note, rank them in terms of most problematic to their long-term health: (Soda, French fries, Dark chocolate, Broccoli)
In addition to true/false questions, multiple choice questions and ranking questions, researchers also asked that workers snip (or extract) text from notes that support the answer. We found these snippets were extremely helpful when experts were needed to adjudicate disagreements.
-
(a)
-
2.
Data Extraction and De-Identification: APIs are needed to extract data from the underlying data store and load them for analysis. In an academic setting, these APIs are configured to take an Institutional Review Board (IRB) number and return the set of medical record numbers in the study. For each medical record number (MRN), the associated charts are pulled and loaded into the system. Moreover, upon querying the charts, the APIs apply open-source, de-identification tools (e.g., the MITRE Identification Scrubber Toolkit13), to remove or scrub HIPAA-designated identifiers, such as patient name and residential addresses.
-
3.
Crowdsourcing System: At its core, a crowdsourcing system matches workers to questions and stores the answers (or labels) in a database. Instead of developing a crowdsourcing system from scratch, we leveraged the open-source Pybossa14 system. Pybossa provides many basic crowdsourcing features, such as: loading and styling questions (known as a presenter), registering workers, assigning workers to tasks, collecting answers, timing tasks and extracting aggregate statistics and labels.
Unfortunately, the default version of Pybossa lacks many of the privacy controls that are needed to manage sensitive data. Therefore, fine-grained access controls with two-factor authentication were added to limit access for each worker. Moreover, worker attributes (or properties) were added to the underlying worker data models so each worker could be categorized by his or her skill level and specialty. These attributes allow for fine-grained question assignment and weighting.
The resulting crowdsourcing system was deployed on an internal server within the Vanderbilt University Medical Center firewall. The site was not open to the public. All worker registration, task assignment, question answering and data extraction were managed through a web interface over HTTPS and the activity is logged.
The Pybossa system allows researchers to customize how questions are ‘presented’ to workers via basic HTML and JavaScript coding. These presenters are simple templated HTML forms that read from an API and populate question text and candidate answers.
-
4.
Customizable Display Interface: We specifically decided to separate the crowdsourcing system from the data display system. This abstraction separated the logic of presenting questions to workers from the task of effectively displaying data for the specific research project. Instead we used HTML IFrames as a means for Pybossa to point to data for analysis. These IFrames can load content from a given URL and provides the developer control over the data input, method of display and tools used to parse the data. The IFrame URL is another parameter specified when configuring a Pybossa project.
The IFrame design has proven to be extremely versatile as we have completed projects displaying different data types including clinical text and medical images.
-
5.
Helper Libraries: Perhaps the most important component of the system is a set of helper libraries that assist workers to produce labels. Example helper libraries include text highlighting tools, text search and document ranking.
-
6.
Worker Recruitment: When working with sensitive data, only certain individuals have the necessary credentials to access the data. For instance, in healthcare, only hospital employees (which includes faculty, staff, and trainees) can access medical records. While the pool of workers is limited (in contrast to the aforementioned public crowd on Amazon), there are often groups of highly motivated workers, such as medical students, who are willing to work given incentives12.
Our worker pool consists of mostly medical students and nursing students, with a small number of faculty. These workers were recruited through Grand Round presentations and IRB-approved email communications. For a worker to participate, he or she signed a data use agreement and, in some cases, was added as key personnel to the researcher’s IRB. We also recorded skill-level of each worker (e.g., medical student, intern, resident, fellow, attending, and nurse) and their specialization (if any), as these answers can impact which questions they are qualified to answer.
-
7.
Supervised Machine Learning: After the crowdsourcing task is complete, researchers can use the labels to train supervised machine learning models. In healthcare, popular prediction systems include clinical decision support15 and order recommendation16, among many others.
Figure 2:
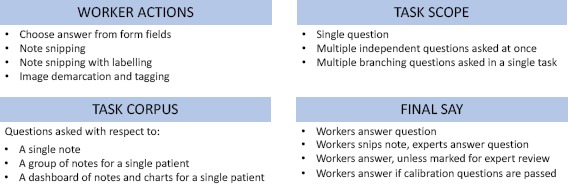
Agenda for crowdsourcing workshop with researchers.
3. Example Use Case
In this section, we present an example use case of the crowdsourcing framework.
Suppose a researcher needs to label notes from a diabetes cohort (e.g., patients with ICD code 250.*). For each note, a worker selects one of the following labels: not relevant, relevant, or partially relevant to diabetes care. Moreover, for a note with a relevant or partially relevant label, the researcher also wants to extract supportive snippets from the note.
After clarifying the research question, task scope, task corpus and worker action, a presenter is designed and loaded into Pybossa. Similarly, notes are extracted from the EMR system, de-identified and loaded into a chart review data system.
Workers are recruited and assigned to the project. A pre-test determines if each candidate worker has sufficient knowledge about diabetes to participate. Only candidates who pass the test are admitted into the worker pool and assigned tasks.
Admitted workers then begin reading notes assigned to them by Pybossa and producing labels. One note is shown to each worker at a time. The worker reads the content of the note, chooses a label, and selects relevant snippets from the note. This process continues until all notes are labeled. Depending on the coverage requirements, multiple workers might answer the same question.
To reduce the cognitive load of workers, the Pybossa presenter (as shown in Figure 3) only consists of the question, answer options and the input box of the search engine. Moreover, we only select necessary helper tools to assist workers find relevant information.
Figure 3:

Example of Pybossa presenter with a text search engine.
As shown in Figure 4, the interface highlights “diabetes” and semantically similar terms of “diabetes”, such as “hyperlipidemia” and “obesity.” On the left side of the note, a heat map displays the number of terms related to diabetes in each section of the note. When multiple notes are displayed to a worker in a single task, an EMR search engine automatically finds notes that contain diabetes or similar terms, and ranks the notes using an information retrieval metric.
Figure 4:
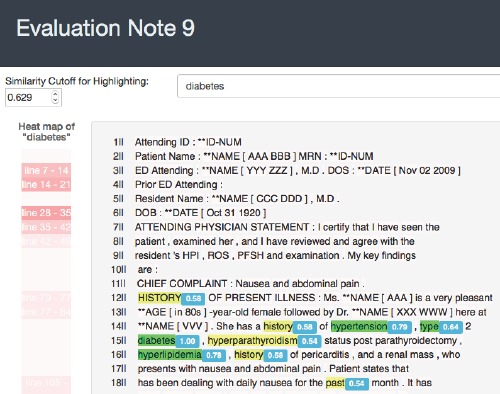
An example helper library: highlighting similar words in a note
After all tasks are completed, the researcher receives the labels and snippets. The researcher then utilizes the data in a supervised machine learning task, such as document classification.
4. Helper Libraries
Clinical notes contain vast amounts of unstructured information describing a patient’s past medical history and diagnoses. Ideally, crowd workers would be able to quickly parse through these data to answer their crowdsourcing questions. Unfortunately, searching for relevant data remains difficult. While workers deploy basic routines, such as starting with the most recent note or a specific note type (e.g., discharge summary), these basic approaches can miss relevant data.
The objective of the helper libraries is to make common crowdsourcing operations more efficient. In the case of chart reviews, this means more efficiently finding relevant clinical text. To that end, we have deployed a set of search tools to find and rank documents for review.
We have implemented and tested three types of search systems:
-
•
Keyword search: Return documents that contain the search term. Rank documents by the frequency of the search term in the document.
-
•
Expanded keyword search (Figure 5): Given a search term, expand the search to include terms that are semantically similar to the term (semantic similarity is defined by a word2vec model17, 18 trained on the medical records), and return documents with any of the similar terms. Rank documents by the number and extent of similar terms in the document.
-
•
Learning-to-rank keyword search19: A learning-to-rank system takes a few labeled examples, and then adjusts the similarity weights to prioritize certain terms over others. Documents are ranked like the expanded keyword search system, but with updated similarity weights. Clinicians with different specialties read notes with varying intentions. Learning-to-rank assists researchers identify content that is relevant to their specific problem.
Figure 5:
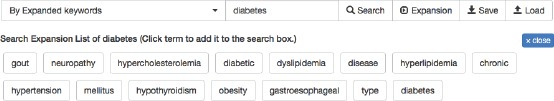
An example of expanded search terms for “diabetes”
In addition to search, we have found that highlighting relevant text within notes can help workers more quickly focus on important information. As Figure 4 shows, similar words to the search term are highlighted in dark green, and moderately similar terms are highlighted in yellow. The similarity value is determined by a word2vec model. Because clinical notes often contain many pages of information, highlighting allows workers to scroll to relevant text quickly.
5. Discussion and Next Steps
We have completed more than six crowdsourcing projects including projects that:
-
(a)
Analyze how well barriers to diabetes care are documented;
-
(b)
Analyze if a patient had dialysis two weeks prior to surgery;
-
(c)
Analyze if a note was relevant to a patient’s diabetes treatment.
For each project, we conducted workshops, and recruited medical students and nursing students to participate in the crowd (over a dozen have participated). We paid the workers a flat fee to complete each project, which was determined by multiplying an hourly rate times the expected number of hours of work.
For many projects, researchers have asked that workers snip the text used to make their decision. These snippets are then provided to an expert for validation. Even though this process requires an expert to review all answers, we find it is useful as the workers complete the time consuming task of scanning the entire document, while the expert simply reviews and approves snippets. If an expert’s time is limited and much more costly than workers, then this design can be effective.
6. Conclusion
In this paper, we presented a crowdsourcing framework for sensitive data sets, such as medical records. We developed a crowdsourcing platform that protects patient privacy and a set of helper libraries to assist workers complete tasks efficiently. Our aim is to help medical researchers attain high quality labels faster and more cheaply than previously possible. Future extensions of the framework include level-of-expertise weighted answers, quorum-detection, and machine learning prediction label assistance.
Acknowledgements:
Data was obtained from Vanderbilt University Medical Center’s Synthetic Derivative, which is supported by institutional funding and by the Vanderbilt CTSA grant ULTR000445 from NCATS/NIH.
Funding:
Crowd Sourcing Labels from Electronic Medical Records to Enable Biomedical Research Award Number: 1 UH2 CA203708-01
References
- 1.Jenny J Chen, Natala J Menezes, Adam D Bradley. Opportunities for crowdsourcing research on amazon mechanical turk. Interfaces. 2011;5:3. [Google Scholar]
- 2.Gabriele Paolacci, Jesse Chandler, Pg Ipeirotis. Running experiments on amazon mechanical turk. Judgm. Decis. Mak. 2010;5(5):411–419. [Google Scholar]
- 3.M. Buhrmester, T. Kwang, S. D. Gosling. Amazon’s mechanical turk: a new source of inexpensive, yet high-quality, data? Perspect. Psychol. Sci. 2011;6(1):3–5. doi: 10.1177/1745691610393980. [DOI] [PubMed] [Google Scholar]
- 4.Winter Mason, Siddharth Suri. Conducting behavioral research on amazon’s mechanical turk. Behav. Res. Methods. 2012;44(1):1–23. doi: 10.3758/s13428-011-0124-6. [DOI] [PubMed] [Google Scholar]
- 5.Amy H Kaji, David Schriger, Steven Green. Looking through the retrospectoscope: reducing bias in emergency medicine chart review studies. Ann. Emerg. Med. 2014;64(3):292–298. doi: 10.1016/j.annemergmed.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira, Pedro L, Wei, Wei-Qi, Cronin, Robert M, Mo, Huan, VanHouten, Jacob P, et al. Evaluating electronic health record data sources and algorithmic approaches to identify hypertensive individuals. J. Am. Med. Inform. Assoc. 2017;24(1):162–171. doi: 10.1093/jamia/ocw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S Trent Rosenbloom, Joshua C Denny, Hua Xu, Nancy Lorenzi, William W Stead, Kevin B Johnson. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J. Am. Med. Inform. Assoc. 2011;18(2):181–6. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sujan Perera, Amit Sheth, Krishnaprasad Thirunarayan, Suhas Nair, Neil Shah. Challenges in understanding clinical notes. In Proc. 2013 Int. Work. Data Manag. Anal. Healthc. - DARE '13. 2013:21–26. [Google Scholar]
- 9.John B Smelcer, Hal Miller-Jacobs, Lyle Kantrovich. Usability of electronic medical records. J. Usability Stud. 2009;4(2):70–84. [Google Scholar]
- 10.Andre Esteva, Brett Kuprel, Roberto A Novoa, Justin Ko, Susan M Swetter, Helen M Blau, Sebastian Thrun. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yunzhu Li, Andre Esteva, Brett Kuprel, Rob Novoa, Justin Ko, Sebastian Thrun. Skin cancer detection and tracking using data synthesis and deep learning. arXivpreprint arXiv:1612.01074. 2016 [Google Scholar]
- 12.Fresne J, Youngclaus J, Shick M. Medical student education: debt, costs, and loan repayment fact card. Assoc. Am. Med. Coll. 2014 [Google Scholar]
- 13.John Aberdeen, Samuel Bayer, Reyyan Yeniterzi, Ben Wellner, Cheryl Clark, David Hanauer, Bradley Malin, Lynette Hirschman. The MITRE identification scrubber toolkit: design, training, and assessment. Int. J. Med. Inform. 2010;79(12):849–859. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Daniel Lombraa Gonzlez, alejandro dob, Marvin R., Martin Keegan, Rufus Pollock, Nigel Babu, et al. Scifabric/pybossa: v2.8.0. Zenodo. 2017 [Google Scholar]
- 15.Konstantina Kourou, Themis P. Exarchos, Konstantinos P. Exarchos, Michalis V. Karamouzis, Dimitrios I. Fotiadis. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin Wiesner, Daniel Pfeifer. Health recommender systems: concepts, requirements, technical basics and challenges. Int. J. Environ. Res. Public Health. 2014;11(3):2580–2607. doi: 10.3390/ijerph110302580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomas Mikolov, Kai Chen, Greg Corrado, Jeffrey Dean. Distributed representations of words and phrases and their compositionality. Adv. Neural Inf. Process. Syst. 2013:3111–3119. [Google Scholar]
- 18.Tomas Mikolov, Greg Corrado, Kai Chen, Jeffrey Dean. Efficient estimation of word representations in vector space. Proc. Int. Conf. Learn. Represent. (ICLR 2013) 2013:1–12. [Google Scholar]
- 19.Cao, Zhe, Qin, Tao, Liu, Tie-Yan, Tsai, Ming-Feng, Li, Hang. Learning to Rank: From Pairwise Approach to Listwise Approach. Proceedings of the 24th International Conference on Machine Learning. 2007 [Google Scholar]


