Abstract
The automatic identification of relations between medical concepts in a large corpus of Electroencephalography (EEG) reports is an important step in the development of an EEG-specific patient cohort retrieval system as well as in the acquisition of EEG-specific knowledge from this corpus. EEG-specific relations involve medical concepts that are not typically mentioned in the same sentence or even the same section of a report, thus requiring extraction techniques that can handle such long-distance dependencies. To address this challenge, we present a novel frame work which combines the advantages of a deep learning framework employing Dynamic Relational Memory (DRM) with active learning. While DRM enables the prediction of long-distance relations, active learning provides a mechanism for accurately identifying relations with minimal training data, obtaining an 5-fold cross validationF1 score of 0.7475 on a set of 140 EEG reports selected with active learning. The results obtained with our novel framework show great promise.
Introduction
Clinical electroencephalography (EEG) is the most important investigation in the diagnosis and management of epilepsies and other types of brain disorders1. An EEG records signals measured along the scalp, which can be correlated with brain activity, enabling the diagnosis of brain-related illnesses. However, the complexity of EEG signals complicates its interpretation, usually documented in EEG reports. With the advent of big collections of clinical EEGs, the interpretation of EEG signals can be improved by providing neurologists with results of search for patients that exhibit similar EEG characteristics. Recently, Goodwin & Harabagiu(2016)2 have described the MERCuRY (Multi-modal EncephalogRam patient Cohort discoveRY) system that relies on deep learning to represent the EEG signal and operates on a multi-modal EEG index resulting from the automatic processing of both the EEG signal and the EEG reports. The MERCuRY system allows neurologist to search a vast data archive of clinical electroencephalography (EEG) signals and EEG reports, enabling them to discover patient populations relevant to queries like Q: Patients with shifting arrhythmic delta suspected of underlying cerebrovascular disease?
The identification of relevant patient cohorts satisfying the characteristics expressed in queries such as Q relies on the ability to automatically and accurately recognize both in the queries and throughout the EEG reports (a) various clinical concepts and their attributes as well as (b) relevant relations between them. An active deep learning paradigm capable of accurately identifying medical concepts such as (concept 1):“shifting arrhythmic delta”, an EEG activity, and (concept 2):“cerebrovascular disease”, a medical problem, mentioned in Q, was reported in Maldonado et al. (2017)3. The same system identifies EEG activities and events as well as medical problems and treatments in EEG with a past reports of patients from the desired cohort, e.g.:
CLINICAL HISTORY: 55 year old man admitted for [change in mental status]MEDICAL_PROBLEM, with a past medical history of [GI bleed]MEDICAL_PROBLEM, [anemia]MEDICAL_PROBLEM
[encephalopathy]MEDICAL_PROBLEM, and others.
MEDICATIONS: [Pantoprazole]TREATMENT, [Folic Acid]TREATMENT, [Carvedilol]TREATMENT
INTRODUCTION: Digital video EEG was performed at the bedside using standard 10-20 system of electrod eplacement with 1 channel EKG.
DESCRIPTION OF THE RECORD: The background EEG is characterized by [slowing]EEG_ACTIVITY and[disorganization]EEG_ACTIVITY. There is prominent shifting arrhythmic [delta activity]1EEG_ACTIVITY more prominent in the left mid to anterior temporal region. [Photic stimulation]EEG_EVENT generates scant[driving]EEG_ACTIVITY.
IMPRESSION: Abnormal EEG due to:
Marked background [slowing]EEG_ACTIVITY and [disorganization]EEG_ACTIVITY
Some arrhythmic [delta activity]1EEG_ACTIVITY
CLINICAL CORRELATION: These findings are supportive of a [bihemispheric disturbance of cerebral function]MEDICAL_PROBLEM. These are nonspecific findings which can be seen in a toxic and metabolic[encephalopathy] MEDICAL_PROBLEM and/or underlying [cerebrovascular disease]2MEDICAL_PROBLEM.
In order to identify a relevant patient, his or her EEG reports need to be relevant to the query. The identification of the medical concepts from the query in the EEG report is not sufficient, as many false positives can be produced. For example, not all patients having shifting arrhythmic delta activity(medical concept 1 from the Query) can be suspected of underlying cerebrovascular disease (concept 2 from the Query), unless a relation between these two medical concept scan be inferred from the EEG report. But, as shown in our EEG report example, mentions of the two concepts from the query Q in the EEG report, underlined and indexed with their respective concept numbers, do not appear in the same sentence, or even in the same section of the EEG report. Therefore, current state-of-the-art methods of identifying relations between medical problems4-6 cannot be used, as they expect the arguments of a relation to be present in the same sentence of a clinical document. Consequently, we needed to develop a novel method for identifying relations between medical concepts automatically recognized by the method reported in Maldonado et al. (2017)3. The relation identification approach reported in this paper operates on pairs of concepts from the same EEG report, that are not constrained to appear in the same sentence or section of the report. Our method identifies in the exemplified EEG report seven relations between medical concepts*: (R1) [delta activity]EEG_ACTIVITY-Evidences→[cerebrovascular disease]MEDICAL_PROBLEM; (R2) [slowing]EEG_ACTIVITY-Evidences→[bihemispheric disturbance of cerebral function]MEDICAL_PROBLEM; (R3) [disorganization]EEG_ACTIVITY-Evidences→[bihemispheric disturbance of cerebral function]MEDICAL_PROBLEM; (R4) [bihemispheric disturbance of cerebral function]MEDICAL_PROBLEM-Evidences→[encephalopathy]MEDICAL_PROBLEM; (R5) [Pantoprazole]TREATMENT-TREATMENT-FOR→[GI bleed]MEDICAL_PROBLEM; (R6) [Folic acid]TREATMENT-TREATMENT-FOR→[anemia]MEDICAL-PROBLEM and (R7) [photic stimulation]EEG_EVENT-Evokes→[driving]EEG_ACTIVITY. The relation R1 warrants the relevance of the exemplified EEG report to the query Q, identifying a patient from the desired cohort. When relations between medical concepts are recognized automatically in a large collection of EEG reports, they enable the generation of EEG-specific knowledge embeddings of high accuracy. High-quality embeddings have been shown recently7to be crucial in designing relevance models that rely on deep learning, and thus produce excellent results.
Background
In recent work8, we have proposed a novel paradigm for learning knowledge from a large corpus of EEG reports enabled by deep learning methods that observe the likelihood that medical concepts share certain relations, as evidenced by data. The resulting knowledge representation, called medical knowledge embeddings (MKE), resolves the semantic heterogeneity which arises when different terminology is used to refer to the same concepts or relations. For example, noted in Sahoo et al. (2014)9, a seizure with alteration of consciousness may be referred to as complex partial seizure, dialeptic seizure, or focal dyscognitive seizure by different epilepsy experts. An MKE representation should place all these expressions in a similar location of the multi-dimensional space, as it learns that they are involved in similar relations with other epilepsy-relevant concepts. When analyzing the results of the MKE produced from the EEG reports, it became clear that the quality of the identified relations needed to be improved8. Instead of considering all potential relations, only accurate relations between medical concepts should be used in the MKE.
Although it is now well established that automated extraction of knowledge from clinical notes involves accurately identifying not only the medical concepts, but also the various relationships in which they are involved10, the automatic identification of relations between medical concepts in EEG reports is hindered by two major obstacles. First, most of the successful techniques for automatically recognizing concepts in clinical texts considered only the target relations from the 2010 i2b2/VA challenge,11 which include relations from the following 3 categories: medical problem-treatment (TrP) relations, medical problem-test (TeP) relations, and medical problem-medical problem (PP)relations. As illustrated in the exemplified EEG report, TrP relations would be useful, but other types of relations relevant for the knowledge expressed in the report would be missed. The second hurdle is generated by the constraint that only relations between medical concepts observed in the same sentence can be identified with current methods, even those using deep learning methods capable of processing large corpora of clinical documents6. We found the solution of both these limitations by considering and extending RelNet12, a memory-augmented neural network in which medical concepts can be processed in abstract memory cells while relations between medical concepts are processed in separate relation memory cells. The memories implicitly model the current knowledge about medical concepts and the relations they share.
Data
In this work, relations between medical concepts were discovered in the EEG reports publicly available from the Temple University Hospital (TUH), comprising over 25,000 EEG reports from over 15,000 patients collected over 12 years. EEG reports are designed to convey a written impression of the visual analysis of the EEG along with an interpretation of its clinical significance. In accordance with the American Clinical Neurophysiology Society Guidelines for writing EEG reports, the reports from the TUH EEG Corpus start with a clinical history of the patient including information about the patient’s age, gender, current medical conditions (e.g. “change in mental status”), and relevant past medical conditions (e.g. “history of GI bleed”) followed by a list of medications the patient is currently taking (e.g. “Pantoprazole”). Together, these two initial sections depict the clinical picture of the patient, containing a “without heartrate”, “without heartrate”, wealth of medical concepts including medical problems (e.g. “encephalopathy”), symptoms (e.g. “without heartrate”, signs (e.g. “twitching” and treatments (e.g. “Depakote”, “pacemaker”). After the clinical picture of the patient is established, the introduction section of the EEG report describes the techniques used for the current EEG (e.g. “digitalvideo EEG using standard 10-20 system of electrode placement with one channel of EKG”), the patient’s condition at objective description the time of the record (e.g. fasting, asleep), and possible activating procedures carried out (e.g. “photic stimulation”). The description section is the mandatory part of the report, meant to provide a complete and objective description of the EEG, noting all observed EEG activities (e.g. “delta activity”), patterns (e.g. “slowing”), and EEG events (e.g. “photic stimulation”, “myoclonus”). The impression section indicates whether or not the EEG test is abnormal and, if so, lists the abnormalities in decreasing order of importance. These abnormalities are usually characteristic EEG activities (e.g. “arrhythmic delta activity”), but can also be EEG Events (e.g. “clinical seizures characterized by myoclonus”). Finally, the clinical correlation section explains what the EEG findings mean in terms of clinical interpretation, (e.g. “findings are supportive of bihemispheric disturbance of cerebral function”).
Preprocessing. To identify medical concepts in each EEG report automatically, we relied on the active deep-learning system described in Maldonado et al. (2017)3, capable of accurately recognizing EEG Activities, EEG Events, medical activity problems, treatments, and tests. The International Federation of Clinical Neurophysiology defines an EEG activity as “any EEG wave or sequence of waves”, and an EEG event as “any stimulus that activates the record”13. For each of these medical concepts, a set of attributes is also recognized. For EEG activities we identified (a) general attributes of the waves, e.g. the MORPHOLOGY, the FREQUENCY BAND or the MAGNITUDE; (b) temporal attributes, e.g. RECURRENCE and (c) spatial attributes, e.g. DISPERSAL, HEMISPHERE and BRAIN LOCATION. As reported in Maldonado et al. (2017)3 we recognize 18 different attributes for each EEG Activity. Two of these attributes are the modality and the polarity of the EEG activity. When we considered the recognition of the modality, we took advantage of the definitions used in the 2012 i2b2 challenge on evaluating temporal relations in clinical text14. In that challenge, modality was used to capture whether a clinical event discerned from a medical record actually happens, is merely proposed, mentioned as conditional, or described as possible. We extended this definition such that the possible modality values of factual, possible, and proposed indicate that clinical concepts mentioned in the EEGs are actual findings, possible findings and findings that may be true at some point in the future, respectively. 2012 i2b2 For identifying polarity of clinical concepts in EEG reports, we relied on the same definition used in the 2012 i2b2challenge, considering that each concept can have either a positive or a negative polarity, depending on any absent or present negation of its finding. Through the identification of modality and polarity of the clinical concepts, we aimed the EEG to capture the neurologists beliefs about the clinical concepts mentioned in the EEG report. For example, the EEG activity “high amplitude spike and slow wave complexes seen anteriorly on the left” can be more completely described by noting its location (head region), hemisphere (side of the brain), magnitude (describes the activity’s amplitude), and morphology (the type or form of the EEG wave). The full attribute specification is described in Maldonado et al. (2017)3.
In addition, we also automatically identified EEG events, medical problems, treatments, and tests, which are characterized only by two attributes: modality and polarity. When preprocessing the corpus of EEG reports using the system developed in Maldonado et al. (2017)3, we were able to detect 365,218 medical concept mentions with 3,062,846 attributes in the TUH EEG corpus. For the remainder of this paper, the term concept will refer to a medical concept and its attributes. Moreover, because mentions of the same medical concept may be expressed with different words (e.g. “epileptic attack” and “epileptic seizure” are both mentions of the medical problem [epilepsy] MEDICAL_PROBLEM) we normalized each concept mention into a canonical form using the morphology attribute for EEG activities and the preferred name of each medical problem and treatment given by the Unified Medical Language System (UMLS)15. EEG events are normalized into a set of 10 events types using the same heuristic approach as in our previous work8. In this work, we use a subset of 140 EEG reports selected via active learning (explained in the Methods section) to train and evaluate our system for relation identification.
Methods
The automatic identification of relations between pairs of automatically identified medical concepts in EEG reports, regardless of their presence in the same sentence, section or across sentences and sections of the report, has been made possible by a novel deep learning system that we designed and implemented, namely the Memory-Augmented Active Deep Learning (MAADL) system. MAADL combines the strength of the Active Learning framework with the advantages of deep learning. While deep learning methods provide unprecedented performance in many tasks, active learning allows a deep learner to achieve this performance with less manually annotated training data, as it exposes the system to new examples on which its performance is still suffering. The operation of MAADL, which is illustrated in
Figure 1, relies on the availability of automatically recognized medical concepts in the corpus of EEG reports obtained with the deep-learning approach reported in Maldonado et al. (2017)3. The identification of relations between medical concepts in MAADL uses the following five steps:
- STEP 1:
The development of an annotation schema for relations between medical concepts in EEG reports;
- STEP 2:
Annotation of relations between medical concepts in the initial training data;
- STEP 3:
Design of a deep learning method for detecting relations between medical concepts in the EEG reports;
- STEP 4:
Development of sampling methods for the MAADL;
- STEP 5:
- Usage of the Active Learning system which involves:
- STEP 5.a:
- Accepting/Editing annotations of sampled examples of relations between medical concepts in EEG reports;
- STEP 5.b:
- Re-training the deep learning method and evaluating the re-trained system
Figure 1:
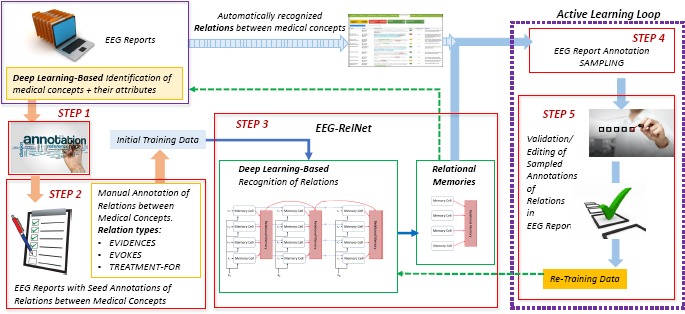
The Memory-Augmented Active Deep Learning (MAADL) system for automatically identifying relations between pairs of medical concepts in EEG reports.
STEP 1: Annotation Schema for relations between pairs of medical concepts in EEG reports: The annotation of relations benefits from the definitions of relations developed in our previous work8. First, we decided to consider only binary relations between four types of medical concepts: (1) EEG events; (2) EEG activities; (3) medical problems and (4) treatments. Second, we decided to consider only three types of relations between medical concepts: EVIDENCES, EVOKES, and TREATMENT-FOR. The EVIDENCES relation considers (a) EEG events, EEG activities, treatments, and medical problems as providing evidence for (b) medical problems mentioned in the EEG report. The EVOKES relation represents the relationship where a medical concept evokes an EEG activity. EEG events, other EEG activities, medical problems and treatments can all evoke EEG activities. The TREATMENT-FOR relation links treatments to the medical problems for which they are prescribed. In addition, we made the decision to annotate relations between medical concepts, and not between their mentions in the EEG report. Because the same concept can be mentioned multiple times in the same EEG report, the representation of concepts achieved while pre-processing the EEG reports by (i) their normalized mention and (ii) their attributes made it possible to recognize co-referring mentions of the same concept by simply grouping concepts with the same normalized mention name and attribute values. Therefore, all co-referring mentions were considered a unique concept, and relations were annotated between unique concept pairs.
STEP 2: Initial Relation Annotations: A set of 40 EEG reports with 198 EVIDENCES relations, 146 EVOKES relations, and 72 TREATMENT-FOR relations were manually annotated and used as the initial training data for the relation detection system. This set of reports had previously been manually annotated with medical concepts and their attributes to ensure errors in concept/attribute detection did not effect relation detection.
STEP 3: Design of Deep Learning Architecture for the Memory-Augmented Active Deep Learning System: We designed a deep learning architecture, called EEG-RelNet, which provides an end-to-end detection of relations between medical concepts in each EEG report by using a neural network augmented with two types of memories: (i) a memory for each medical concept; and (ii) a memory for each relation between each pair of medical concepts. Moreover, the relational memory is dynamic as it changes to model the specific relations observed in each EEG report.
STEP 4: Active Learning Sampling Method: To improve the quality of the identified relations between medical concepts in EEG reports, as illustrated in Figure 1, an active learning loop is designed. In an active learning framework, the sampling method is used to automatically select examples of relations for human validation. Since this work is focused on relation detection between pairs of medical concepts, we chose a sampling method that only prioritizes relation detection performance, ignoring the quality of medical concepts and their attributes. Therefore, we do not use the rank combination protocol reported in our previous work3, opting for standard uncertainty sampling16 whereby EEG reports containing relations for which the model is most uncertain are selected for manual validation. The uncertainty of a report is measured at the report level by averaging the uncertainty of each relation classification decision in the report. The uncertainty of a relation classification decision is calculated using Shannon Entropy, H(R) = – Σt Rt log Rt, where R is a vector representing the probability distribution over possible relation types. These probability distributions are derived by EEG-RelNet from the learned dynamic relation memory, as shown in Figure 1.
STEP 5: Usage of the Memory-Augmented Active Deep Learning System: As shown in Figure 1, each iteration of active learning involves using the EEG-RelNet to make automatic relation annotations on the unlabeled EEG reports, selecting the most informative examples for manual validation, and re-training the EEG-RelNet using the new set of validated training examples.
EEG-RelNet: a Deep Learning Architecture for long-distance Relation Detection in EEG Reports
While medical concepts (EEG activities, EEG events, medical problems, treatments and tests) are available in each EEG report, due to the preprocessing that was applied to the entire TUH EEG corpus, inference of the EVIDENCES, EVOKES, and TREATMENT-FOR relations between pairs of such concepts was produced through dynamic memories based on neural networks, capable to capture the implicit participation of each medical concept in a relation of interest. Inspired by RelNet, a model reported in Bansal et al. (2017)12, we developed the EEG-RelNet, a deep neural network architecture that operates on the full text of an EEG report considering all medical concepts identified in the report to detect relations of the type EVIDENCES, EVOKES, and TREATMENT-FOR between any pair of concepts. More specifically, given the full text of an EEG report and the set of medical concepts identified in that report, EEG-RelNet can predict whether there is relation of type t, Rijt, between any pair of medical concepts ci and cj recognized in the report. To do so, EEG-RelNet processes the EEG report, one sentence at a time, reading its words, encoding the information from the sentence, processing the sentence information in the dynamic relational memory, and predicting each type of relation based on the dynamic memories after they have processed each sentence in the EEG report. The three modules of EEG-RelNet are:
the Input Encoding Module which encodes information from the report at concept- and sentence-level embedding vectors, which are used throughout the deep learning architecture;
the Dynamic Relational Memory Module which maintains and updates a set of hidden states called memories to capture accumulated information about each medical concept and potential relation in the EEG report;
the Output Module which uses the updated memories to determine the most likely relations (and their types) between medical concepts in the EEG report.
In the remainder of this section, we provide a detailed description of each module of EEG-RelNet.
The Input Encoding Module. The role of this module is to learn (1) an embedding encoding each medical concept as well as each of its attributes and (2) an embedding encoding the information from each sentence in the EEG report. Formally, we represent an EEG report as a set of medical concepts, C = {c1,…,cd}, and a sequence of sentences, [si,…,sn]. Each medical concept, ci, is associated with several N-dimensional vectors called embeddings: (a) an embedding for the normalized concept name, and (b) separate embeddings for each of its A attributes values . Thus, the embedding for a medical concept is created by (1) concatenating the embedding for the name of the medical concept with the embedding for each of its attributes and (2) projecting this concatenated vector using a learned weight matrix , i.e. . In this way, each medical concept is represented by an embedding, , which is a vector in .
Participation of medical concepts in relations is informed by the context of each concept in the text of the EEG report. Contextual information is provided by the words of the sentence where the concept is mentioned, hence are presentation of words from each sentence as is also desirable. Therefore, we learn an embedding ei for each word wi in a sentence, enabling us to represent each sentence as a sequence of embeddings E = [e1, …, em] such that the elements of E occur in the same order as the words from the sentence †. While the traditional choice for combining and composing the embeddings in E into a single sentence embedding would be a Recurrent Neural Network (RNN), we instead adopt a more recent and significantly more efficient strategy, namely a positional mask12, 17 such that the k-th sentence from the EEG report is represented as: given that the sentence had m words, and the vectors [f1, …, fm], represent the learned positional mask and ⊙ is the element-wise product. It is important to note that the same vectors [f1, … , fm] are used when each new sentence is encoded and they are learned jointly with the other parameters of the deep learning model.
The Dynamic Relational Memory Module. Because EEG reports often contain long-distance relations between concepts we relied on a Dynamic Relational Memory12 (DRM) Module to keep track of the interactions between medical concepts in each report. The DRM accumulates information about medical concepts and the relations between them by processing each sentence encoded by the Input Module and updating a set of hidden states, called memories. Specifically, given a sentence embedding, and the corresponding set of concept embeddings , there are two scenarios for each : (scenario 1): the medical concept ci has not been mentioned in any previous sentence, thus its Concept Memory needs to be accounted for using a single, shared Concept Memory Cell; and (scenario 2): the concept ci has been previously mentioned, and thus its corresponding Concept Memory needs to be updated. Moreover, since each medical concept ci may participate in a relation, in (scenario 1), a unique Relation Memory needs to account for each relation in which the concept participates, whereas in (scenario 2) the corresponding Relation Memory needs to be updated. If an EEG report refers to d medical concepts, there will be d Concept Memory cells and d × (d – 1) Relation Memory cells. The Dynamic Relational Memory (DRM) consists of the entire set of Concept and Relation Memories in an EEG report.
The Concept Memories are organized as a Key-Value Memory Network18. Key-value paired memories are a generalization of the way context of concepts is stored in memory. In a Key-Value Memory Network, the lookup (addressing) stage is based on the key vector while the reading stage (giving the returned result) returns the value memory. Consequently, in EEG-RelNet, memory vectors are tied to so-called key vectors enabling the model to only update a memory vector when the input sentence has context that is relevant to the memory’s associated key vector. Henaff et al.19 have shown that when concept embeddings are used as key vectors, the associated memory vectors will accumulate information about those concepts. Consequently, in EEG-RelNet, concept embeddings are used as key vectors allowing the network to update each Concept Memory, hi, if an input sentence is relevant to the concept, ci. The Concept Memory Cell, illustrated in Figure 3, is used to update a Concept Memory, hi, given a medical concept embedding, ci, and a sentence encoding, , via the following equations:
| (1) |
| (2) |
| (3) |
Figure 3:
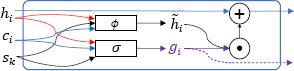
Concept Memory Cell
where Wu, Wv and Ws are trainable weight matrices in is the inner product, σ is the sigmoid function and ϕ is a Parametric Rectified Linear Unit (PReLU)20. Equation 1 is a gating function that determines how much the kth input sentence affects the ith Concept Memory such that values close to 1 indicate sentence sk is relevant to medical concept ci and values close to 0 indicate the opposite. Equation 2 defines the candidate Concept Memory that will be used to update the existing Concept Memory, hi, after it is scaled by gic as shown in equation 3.
As illustrated in Figure 2, when each sentence si is processed, the DRM uses and updates not only concept memories, but also a much larger set of relation memories. This is explained by the fact that unfortunately, maintaining a single memory vector for each concept is not sufficient for modeling concepts that participate in multiple relations, especially when those relations involve concepts that are mentioned at significant distance in the EEG report. Thus, to model the interactions each concept has with each other concept in the same EEG report, we maintain a set of Relation Memories corresponding to each pair of concepts from the EEG report, , where C is the set of medical concepts in the EEG report. Each Relation Memory is updated using the Relation Memory Cell illustrated in Figure 4 via the following equations:
| (4) |
| (5) |
| (6) |
Figure 2:
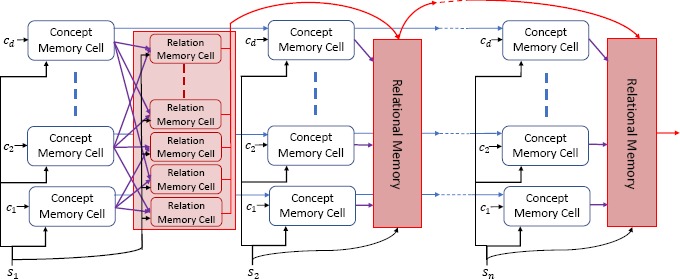
The Dynamic Relational Memory Module of EEG-RelNet. The Dynamic Relational Memory Module processes n sentences, updating a set of d Concept Memories and d (d – 1) Relation Memories for each sentence.
Figure 4.
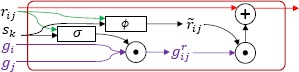
Relation Memory Cell
where WA and WB are trainable weight matrices in ℝN × N. As in the Concept Memory Cell, the Relation Memory Cell uses a gating function (equation 4) and a candidate memory (equation 5) to update the Relation Memory in a way that reflects how relevant the input sentence, sk, is to the concept pair, (ci, cj). To compute the gate value , the Relation Memory Cell uses the two concept gate values, from the Concept Memory Cells for concepts ci and cj, ensuring that input sentences that are relevant to either concept can be used to update the Relation Memory. By maintaining a memory vector for each pair of concepts and updating that memory vector as the model accumulates information across each sentence in an EEG report, EEG-RelNet can be interpreted as constructing a local latent knowledge graph12 for each EEG report, where each Relation Memory represents a possible relation in the graph.
The Output Module. The output module makes use of the Dynamic Relational Memory updated after processing the last sentence in the EEG report to identify relations (and their types) between any pair of medical concepts from the report. The relation prediction, between medical concepts ci and cj is produced by passing the Concept Memories associated with concepts ci and cj along with the Relational Memory rij to two fully connected PReLU layers followed by a soft max layer: where and are learned weight matrices, and ϕ is
| (7) |
a Parametric Rectified Linear Unit. Rij is a probability distribution over 4 possible relations: the 3 relation types described in the annotation schema and a 4th type indicating no relation. Consequently, the relation (if any) detected between concepts ci and cj is given by .
Results
To evaluate the Memory-Augmented Active Deep Learning (MAADL) system, we measured (1) the performance of EEG-RelNet and (2) the impact of Active Learning. To measure the impact of the EEG-RelNet architecture, we compare our system with two alternate configurations and one baseline:
EEG-RelNet_NRM is a deep neural network structured similarly to EEG-RelNet but without Relation Memories. Formally, we replace equation 7 with qij = ϕ (Wq [hi, hj]), and equations [4-6] are not used.
EEG-RelNet_NA is a deep neural network structured similarly to EEG-RelNet that ignores the attributes of each medical concept in the Input Module. Formally, EEG-RelNet_no-attr represents each concept embedding using only the embedding for the name of that concept, = .
Heuristic is a simple rule-based baseline from Maldonado et al. (2017)8 that uses medical concept type and section type to detect relations. EVIDENCES relations are created between any medical concept in an EEG report and medical problems in the clinical correlation section, EVOKES relations are created between any medical concept and an EEG activity, and TREATMENT-FOR relations are created between any treatment and medical problems in the history section of the EEG report.
We follow the evaluation procedure for relation classification reported in the 2012 Informatics for Integrating Biology at the Bedside (i2b2) shared task14. The Precision, Recall, and Fx measure for each relation type are calculated using 5-fold cross validation on the full set of 140 manually annotated EEG reports containing 1513 relations between 3691 medical concepts. Each EEG-RelNet configuration is trained for 10 epochs with the same random initialization, using N =100 as the embedding size.
EEG-RelNet is able to successfully detect the three relation types, EVOKES, EVIDENCES, and TREATMENT-FOR, obtaining F1 scores of 0.8371, 0.6939, and 0.7116, respectively. Clearly, EEG-RelNet obtains the best performance on each relation type, demonstrating the importance of both the Dynamic Relational Memory and medical concept attributes when detecting relations. EEG-RelNet achieves significantly better performance when recognizing EVOKES relations compared to the other two relation types indicating that the network is able to correctly link medical problems with the EEG activities they evoke. The effect of the Dynamic Relational Memory is most obvious when considering the EVIDENCES relation type, increasing the F1 measure by nearly 20%. Interestingly, the removal of attribute information from the model drastically reduces performance when detecting the EVOKES relation type, but only slightly reduces performance on the other two types compared to the EEG-RelNet_NRM system. The Heuristic approach is able to achieve the highest recall on each relation type since it was specifically designed for high recall. However, due to the poor precision, the Heuristic baseline achieves by far the worst overall performance.
To evaluate the performance of Memory-Augmented Active Deep Learning (MAADL), we measured the change in performance after each additional round of active learning. Figure 5 presents these results, clearly showing a significant increase in performance from a macro-average F1 score of 0.6040 to 0.7475 (23.75%) with only 100 additional EEG reports annotated.
Figure 5.
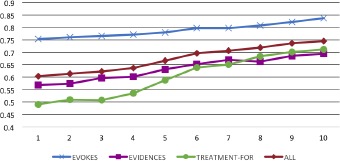
Learning curves for each relation type (and macro-averaged aggregate) shown over the first 100 EEG reports annotated, evaluated with Fi measure.
Discussion
In general, EEG-RelNet is able to correctly recognize relations between medical concepts as indicated by the macro-average Fi score of 0.7475. However, EEG-RelNet is clearly able to recognize EVOKES relations more accurately than TREATMENT-FOR and EVIDENCES relations, with F1 scores of 0.8371, 0.7116, and 0.6939, respectively. We believe the superior performance when detecting EVOKES relations may be explained by (1) the fact that EVOKES relations always involve an EEG activity and (2) the more sophisticated representation of EEG activities compared to the other medical concepts. Specifically, EEG activities have 18 semantic attributes that capture rich information, but medical problems, treatments, and EEG events only have two attributes: modality and polarity. This suggest that semantic attributes play an important role in detecting relations between medical concepts. We believe the performance of our model could be improved in the future by introducing more sophisticated representations of medical problems, treatments, and EEG events using neurological ontologies21 or other sources of medical knowledge, like the Unified Medical Language System (UMLS)15. For example, when determining if the concept [Lamictal]TREATMENT is a TREATMENT-FOR the concept [seizure]MEDICAL-PROBLEM, it would be beneficial to know that Lamictal is an anticonvulsant - knowledge contained in the UMLs. Another interesting phenomenon revealed by our experiments is that the recall when detecting TREATMENT-FOR relations is especially high (0.8953) while the precision is low (0.5905). A possible explanation for this phenomenon is that medications (the most common type of treatment in an EEG report) are listed contiguously in the list of medications section. Consequently, the model has difficulty determining which treatments from the same list are TREATMENT-FOR specific medical problems. This kind of error is especially prevalent at the beginning of active learning, but we can see from Figure 5 that performance sharply increases between annotation rounds 4-6, as the model is introduced to more TREATMENT-FOR relations. As described in the Background, in previous work8 we demonstrated that the types of relations detected by the MAADL system can be used to capture domain-specific medical knowledge in the form of Medical Knowledge Embeddings (MKE)8. However, one of the main limitations of the MKE was the poor quality of the automatically detected relations. Using MAADL to more accurately detect relations should enable higher quality MKE to be learned.
Conclusion
In this paper we describe a novel active deep learning framework for identifying relations between medical concepts discussed in the text of EEG reports by making use of the EEG-RelNet, a neural architecture capable of inferring relations between concepts through its dynamic relational memory. This deep learning architecture allowed us to identify relevant relations between medical concepts that were not mentioned in the same sentence or section of the EEG report, which is a key contribution of the framework presented in this paper. Most previous methods successfully employed for recognizing relations between medical concepts from clinical documents addressed only the case when the concepts were observed in the same sentence.
Table 1:
Performance of the MAADL system for detecting relations between medical concepts in EEG reports.
| Metric | EVOKES | EVIDENCES | ||||||
|---|---|---|---|---|---|---|---|---|
| EEG-RelNet | EEG-RelNet_NRM | EEG-RelNet_NA | Heuristic | EEG-RelNet | EEG-RelNet | NRM EEG-RelNet NA | Heuristic | |
| Precision | 0:8563 | 0:8037 | 0:6605 | 0:1960 | 0:7086 | 0:6325 | 0:6193 | 0:1750 |
| Recall | 0:8187 | 0:7500 | 0:6088 | 0:9771 | 0:6798 | 0:5365 | 0:5506 | 0:8624 |
| F1 | 0:8371 | 0:7759 | 0:6336 | 0:3265 | 0:6939 | 0:5805 | 0:5829 | 0:2910 |
| Metric | TREATMENT-FOR | All Relations (Macro Average) | ||||||
| EEG-RelNet | EEG-RelNet NRM | EEG-RelNet NA | Heuristic | EEG-RelNet | EEG-RelNet NRM | EEG-RelNet NA | Heuristic | |
| Precision | 0:5905 | 0:5932 | 0:5268 | 0:1715 | 0:7185 | 0:6764 | 0:6022 | 0:1808 |
| Recall | 0:8953 | 0:8845 | 0:8520 | 0:9856 | 0:7979 | 0:7237 | 0:6705 | 0:9417 |
| F1 | 0:7116 | 0:7101 | 0:6510 | 0:2921 | 0:7475 | 0:6888 | 0:6225 | 0:3032 |
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The definitions of the relations is provided in the annotation schema detailed in the Methods section.
Embeddings, ew, corresponding to words contained within a concept mention, ci are replaced with the embedding for that concept instead of the word, i.e. ew = . This is required to enable the Key-Value memory structures described in the next subsection
References
- 1.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin TR, Harabagiu SM. Multi-modal Patient Cohort Identification from EEG Report and Signal Data. In: Proceedings of the American Medical Informatics Association Annual Symposium (AMIA); November 2016; Chicago, IL, USA. pp. 1794–1803. [PMC free article] [PubMed] [Google Scholar]
- 3.Maldonado R, Goodwin TR, Harabagiu SM. Active Deep Learning-Based Annotation of Electroencephalography Reports for Cohort Identification. In: Proceedings of the American Medical Informatics Association Joint Summits on Clinical Research Informatics (AMIA-CRI); March 2017; San Francisco, CA, USA. pp. 229–238. [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y, Uzuner O, Szolovits P. Bridging semantics and syntax with graph algorithms state-of-the-art of extracting biomedical relations. Briefings in bioinformatics. 2016;18(1):160–178. doi: 10.1093/bib/bbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. Journal of the American Medical Informatics Association. 2011;18(5):594–600. doi: 10.1136/amiajnl-2011-000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo Y. Recurrent neural networks for classifying relations in clinical notes. Journal of Biomedical Informatics. 2017;72:85–95. doi: 10.1016/j.jbi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jameel S, Bouraoui Z, Schockaert S. MEmbER: Max-Margin Based Embeddings for Entity Retrieval. In: Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval; Shinjuku, Tokyo, Japan. 2017. pp. 783–792. [Google Scholar]
- 8.Maldonado R, Goodwin TR, Skinner MA, Harabagiu SM. Deep Learning Meets Biomedical Ontologies: Knowledge Embeddings for Epilepsy. In: Proceedings of the American Medical Informatics Association Annual Symposium (AMIA); November 2017; Washington DC, USA. pp. 1226–1235. [PMC free article] [PubMed] [Google Scholar]
- 9.Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, et al. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care. Journal of the American Medical Informatics Association. 2013;21(1):82–89. doi: 10.1136/amiajnl-2013-001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods of information in medicine. 1998;37(4-5):394. [PMC free article] [PubMed] [Google Scholar]
- 11.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bansal T, Neelakantan A, McCallum A. RelNet: End-to-end Modeling of Entities & Relations. arXiv preprint arXiv:170607179. 2017 [Google Scholar]
- 13.Noachtar S, Binnie C, Ebersole J, Mauguiere F, Sakamoto A, Westmoreland B. A glossary of terms most commonly used by clinical electroen-cephalographers and proposal for the report form for the EEG findings. The International Federation of Clinical Neurophysiology. Electroencephalography and clinical neurophysiology Supplement. 1999;52:21. [PubMed] [Google Scholar]
- 14.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. Journal of the American Medical Informatics Association. 2013;20(5):806–813. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Settles B. 55-66. Vol. 52. Madison: University of Wisconsin; 2010. Active learning literature survey; p. 11. [Google Scholar]
- 17.Sukhbaatar S, Weston J, Fergus R, et al. End-to-end memory networks. In: Advances in neural information processing systems. 2015:2440–2448. [Google Scholar]
- 18.Miller A, Fisch A, Dodge J, Karimi AH, Bordes A, Weston J. Key-value memory networks for directly reading documents. arXiv preprint arXiv:160603126. 2016 [Google Scholar]
- 19.Henaff M, Weston J, Szlam A, Bordes A, LeCun Y. Tracking the world state with recurrent entity networks. arXiv preprint arXiv:161203969. 2016 [Google Scholar]
- 20.He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision. 2015:1026–1034. [Google Scholar]
- 21.Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, et al. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care. Journal of the American Medical Informatics Association. 2014;21(1):82–89. doi: 10.1136/amiajnl-2013-001696. [DOI] [PMC free article] [PubMed] [Google Scholar]


